Abstract
Screening for mutations in human disease-causing genes in a molecular diagnostic environment demands simplicity with a view to allowing high throughput approaches. In order to advance these requirements, we have developed and applied a primer design program, termed BatchPD, to achieve the PCR amplification of coding exons of all known human Refseq genes. Primer design, in silico PCR checks and formatted primer information for subsequent web-based interrogation are queried from existing online tools. BatchPD acts as an intermediate to automate queries and results processing and provides exon-specific information that is summarised in a spreadsheet format.
Keywords: Primers, Refseq genes, single nucleotide polymorphisms, BatchPD
Background
The molecular confirmation of a clinical diagnosis in the context of human heritable and somatic mutation events largely involves sequencing-based technology. The principal focus of any diagnostic sequencing approach is to interrogate the coding region of those genes that are known to carry mutations that cause a clinical phenotype. Critically, the process of designing primers to achieve the amplification of exons for subsequent sequencing has mostly been a tedious and labour intensive process, with no universally adopted methods. For the most part, designs are carried out manually or in a semi-automated manner followed by experimental validation of primers to establish specificity and efficiency.
Various primer design software are available ranging from Primer 3 [1], derivatives of Primer 3 [2, 3], bioinformatics suites that incorporate Primer 3 such as Geneious [4], or standalone programs/software suites such as Vector NTI [5]. In the case of ExonPrimer [3], which provides the ability to design primers against intronic regions flanking exons with a preference for regions excluding SNPs, either manual sequence entry or a UCSC accession ID are required. Critically, it would be desirable to incorporate other features in a primer design process: in-silico PCR evaluation; the identification of single nucleotide polymorphisms (SNPs) that lie in the human target sequence against which each primer anneals that might result in allele bias in genomic amplification; and optional primer tailing in order to allow for downstream sequencing applications.
None of the currently available free primer design software or packages addresses the specific task of designing primers against all exons of a given gene accession input with the ability to carry out subsequent quality checks such as SNP checks, while providing validation information in a complete package. In order to address these issues, and to move one step closer to a potential universally adopted procedure, we developed a primer design program termed BatchPD using the Java programming language. Our program automates and streamlines the primer design process by interfacing with various existing online primer-design related tools to integrate primer quality checks, in-silico PCR evaluation, primer tailing options, optional formatted outputs for SNP checking via an online tool SNPCheck, as well as a standardised spreadsheet summary output containing the most relevant information for the targeted exons.
As part of our program validation we used NCBI's Homo sapiens RefSeq gene accession list to produce an archive of predesigned primers for all of the RefSeq genes that produce a coding mRNA using BatchPD. Our pre-designed archive of primers for approximately 9000 human RefSeq genes has been made public allowing other research institutions and laboratories free access [6]. By preparing a list of pre-designed primers for the RefSeq genes we avoid the potential issues of BatchPD being invalidated due to future updates of web services, or even decommissioning these services. In support of the Open-source software development movement and in the interest of sharing our work, we have made both our program and source code available [7].
Methodology
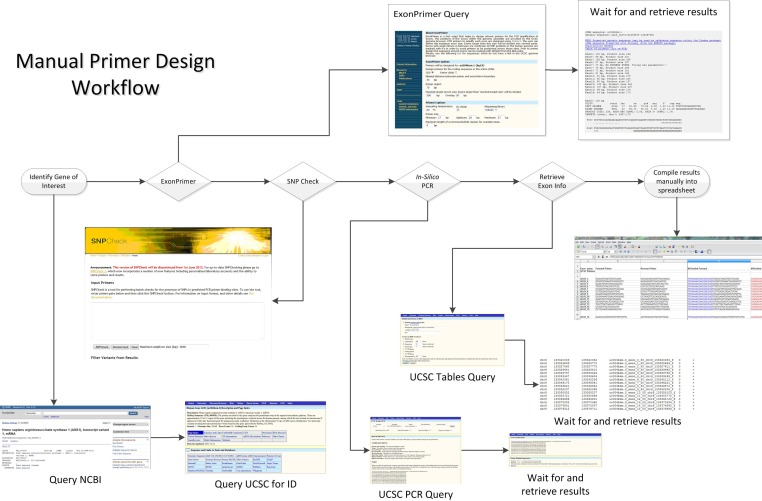
It was decided at the outset that the input for BatchPD should allow single, but preferably multiple, Refseq accession entries in a list format with an optional automatically-read starting input file (Figure 1).
Figure 1.
Manual primer design workflow shown with screen-captures of the relevant websites. The concept behind the design process is simple, but involves significant time waiting for results and processing them for subsequent steps.
In order to streamline downstream processing, primers are designed that allow annealing at 60°C. This removes the need for costly PCR optimisations saving on both technical time and reagent costs. The use of M13 tails allows for rapid sequencing of PCR products or user-defined custom tails for nextgeneration sequencing such as the Roche GSJunior or 454 platforms. We have validated the primers designed for approximately 80 genes using one set of PCR amplification conditions (35 cycles of 94°C for 45 seconds, 60°C for 30 seconds and 72°C for 30 seconds) using either of two buffers: a standard PCR buffer containing 2mM MgCl2, and another that is identical but optimised for GC-rich sequences [8, 9]. In our experience, the designed primers have a high probability of successful amplification with minimal need for redesigns.
The primer design process involves the use of RefSeq mRNA accession IDs, with a specific requirement for accessions starting with “NM_”. This can be achieved by querying the NCBI database [10] to identify the relevant RefSeq accession if present. In the case of batch queries using the full list of RefSeq accessions, we used the NCBI FTP archive [11].
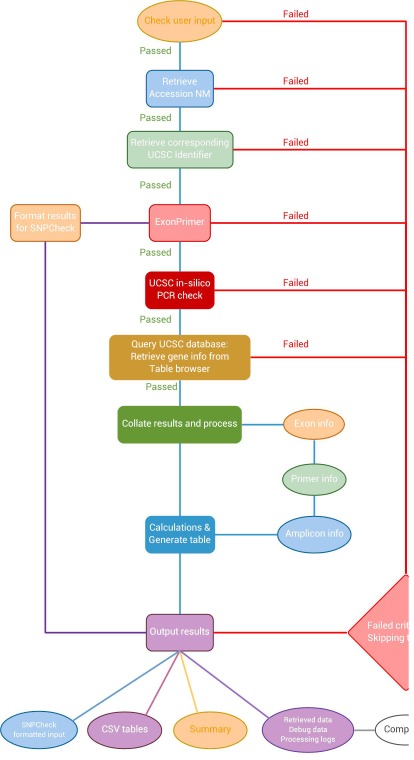
The RefSeq accession is used by BatchPD to retrieve the relevant GenBank file as well as to identify a corresponding UCSC accession number. Primers are designed via an online tool called ExonPrimer [3]. In order to simplify the design process the UCSC specific ExonPrimer script link is parsed from the University of California Santa Cruz (UCSC) genome browser's description and page index. The results from the ExonPrimer run are parsed for the relevant information and results. Should the run complete successfully the results are formatted for downstream work and optional SNP checking via SNPCheck [12]. The SNPCheck step requires user intervention to copy/paste the formatted inputs into the SNPCheck web service; this step was not integrated into the automated work flow (Figure 2).
Figure 2.
Flow diagram showing an overview of how BatchPD functions. Each individual accession query is fed through a series of steps with BatchPD automating the query, wait, results parsing and conversion as well as results retrieval procedure. Should any of the queries fail, the program will skip subsequent steps as appropriate and report the actions taken in the logs. Under some circumstances, BatchPD will attempt subsequent processing steps should a non-critical section fail internal checks.
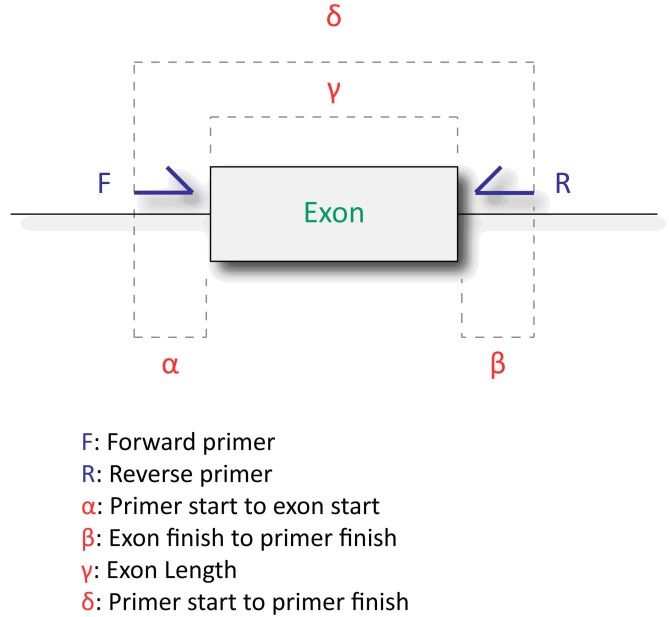
The UCSC in-silico PCR online tool was used in a simulated PCR of the human genome with the primers designed from ExonPrimer. This in-silico PCR evaluation determines whether a single amplicon would be amplified as opposed to multiple amplicons per primer pair. The results are then retrieved and parsed to identify relevant primer and PCR related information such as: number of amplicons, amplicon start position, amplicon end position, strand orientation, amplicon size(s) and chromosome location(s). (Figure 3)
Figure 3.
The distance values returned in the final output spread-sheet are illustrated here with distances being calculated relative to the primers and exon involved. α) Primer start to exon start is calculated from the 5' end of the forward primer to the 5' end of the exon. β) Exon finish to primer finish is calculated as the distance between the 3' end of the exon to the 5' end of the reverse primer. In the case of exons that include the start/stop of the coding sequence, these values are used to replace the exon 5'/3' ends, respectively.
At the end of the run, all relevant information is processed and formatted into a comma delimited file (.CSV) for importing into spreadsheet programs such as Microsoft Excel or LibreOffice Calc (formally OpenOffice Calc). For archival purposes, supporting data, webpage results and program logs are compressed into a zip-formatted file to conserve storage space. Relevant gene and exon information are retrieved from the UCSC Table Browser webpage to assist with the calculations in generating the .csv file.
Three result files are saved with every run in addition to a perbatch run log. The first file corresponds to results that are formatted for spreadsheet input (.csv); the second is a list of primary primer designs that are formatted for SNPCheck input; the third is a summary log of the particular gene run. Primer positions relative to relevant exon positions are calculated from the resulting data while taking into consideration codingsequence (CDS) start/stop positions. CDS start/stop positions are parsed from the GenBank data for each gene while exon coordinates are retrieved from the UCSC database. The positions returned from UCSC PCR are then used to calculate the primer start to exon start, as well as the exon finish to primer finish positions relative to exon start/stop. If the exon being queried contains the CDS start or stop positions then these are used in place of the exon start or stop for the appropriate calculations.
During the validation stage of BatchPD for both testing and debugging we took the Homo sapiens RefSeq gene database (retrieved March 8th, 2012) and processed them via BatchPD. The results from the run have been made available online [6] in the hope that this would allow access by other molecular diagnostic laboratories in order to save on technical hours spent attempting primer designs. The pre-compiled primer designs will be available until December 2014 at which time the RefSeq gene list should have changed enough to warrant a complete rerun to include all the new additions. With the pace at which new sequencing technologies are progressing, traditional Sanger-type sequencing methods and even amplicon sequencing via a “Next-Generation Sequencing” technology would be expected to be redundant or more costly than newer technologies that will be developed in the coming years.
Disscussion
The automated primer design program BatchPD has saved many technical hours for our laboratory staff, while using a standardised set of procedures that are both robust and stringent. In order to validate the program during development, we submitted the Homo sapiens RefSeq gene list of mRNA nucleotide identifiers into our program and produced a predesigned database of primers. This database and the corresponding BatchPD program have been made available online including the source code.
In hindsight, the program could have been developed to use offline copies of the databases and tools to lighten the burden on the free online services. The disadvantage of such an approach would be increased difficulty for the average laboratory technician, and the requirement of regular database updates. The primary advantage of the method adopted here in which BatchPD queries the online tools and parses the returned webpages is the ability to use the latest datasets without the need for complex offline mirrors that require periodic synchronization. The average laboratory technician can download the program, input a list of RefSeq mRNA identifiers, choose primer tailing options, click a button and carry on with other tasks while the program completes the task without user intervention. Alternatively a quick search of our pre-designed primer archive may suffice. Should the need arise, enterprising researchers can take the published BatchPD source code and modify it to accept a wider range of input variables, or redirect the program to use different web resources and tools.
Conclusion
BatchPD grew out the need to automate a primer design and evaluation process that we had developed manually over the past five years. Given the increasing need to sequence more genes based on clinical referrals, we decided to apply BatchPD to the design of primers for all exons of all archived Refseq genes with an NM accession ID. The added advantage of our approach is that it can equally be applied to next generation sequencing strategies that involve the pooling of targeted amplicons with defined tail-based sequences for amplicon capture.
Acknowledgments
We acknowledge the generosity of the developers of the UCSC genome browser and ExonPrimer in making their resources publicly available.
Footnotes
Citation:Lai & Love, Bioinformation 8(8): 365-368 (2012)
References
- 1.S Rozen, H Skaletsky. Methods Mol Biol. 2000;132:365. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 2.A Untergasser, et al. Nucleic Acids Res. 2007;35:71. [Google Scholar]
- 3. http://ihg2.helmholtzmuenchen.de/ihg/ExonPrimer.html.
- 4. http://www.geneious.com.
- 5. http://www.invitrogen.com/site/us/en/home/Productsand-Services/Applications/Cloning/vector-ntisoftware.html.
- 6. http://www.fos.auckland.ac.nz/~dlai008/
- 7. http://sourceforge.net/projects/batchpd/
- 8.R Marquis-Nicholson, et al. Gene. 2011;486:37. [Google Scholar]
- 9.R Marquis-Nicholson, et al. Genet Mol Res. 2010;9:1483. [Google Scholar]
- 10. http://www.ncbi.nlm.nih.gov/nuccore/
- 11. ftp://ftp.ncbi.nlm.nih.gov/refseq/H_sapiens/RefSeqGene/LRG_RefSeqGene.
- 12. https://ngrl.manchester.ac.uk/SNPCheckV2/snpcheck.htm.