Abstract
Next Generation sequencing (NGS) applied to human papilloma viruses (HPV) can provide sensitive methods to investigate the molecular epidemiology of multiple type HPV infection. Currently a genotyping system with a comprehensive collection of updated HPV reference sequences and a capacity to handle NGS data sets is lacking. HPV-QUEST was developed as an automated and rapid HPV genotyping system. The web-based HPV-QUEST subtyping algorithm was developed using HTML, PHP, Perl scripting language, and MYSQL as the database backend. HPV-QUEST includes a database of annotated HPV reference sequences with updated nomenclature covering 5 genuses, 14 species and 150 mucosal and cutaneous types to genotype blasted query sequences. HPV-QUEST processes up to 10 megabases of sequences within 1 to 2 minutes. Results are reported in html, text and excel formats and display e-value, blast score, and local and coverage identities; provide genus, species, type, infection site and risk for the best matched reference HPV sequence; and produce results ready for additional analyses.
Availability
The database is available for free at http://www.ijbcb.org/HPV/
Keywords: Human papilloma virus, Genotyping, web-based, Blast search, Next Generation sequencing
Background
Human papilloma virus (HPV), the most common sexually transmitted infection, causes cervical cancer in women, contributes to anogenital cancers in men, and is associated with oropharyngeal cancers and genital warts in men and women [1]. Currently, PCR-based assays are applied to identify HPV prevalence, ranges of oncogenic and nononcogenic HPV types, and incidence of multiple type infection [2]. Next Generation sequencing (NGS) technology provides increased sensitivity for in depth analysis of HPV types, although large datasets of HPV sequences present considerable barriers for analyses. Available automated HPV genotyping tools, including Virus Sequence Database [3], REGA HPV Automated Subtyping Tool [4], and NCBI blastn are limited by either a restricted number of reference sequences with incomplete annotation or outdated nomenclature, an inability to classify short sequences, or an inadequate capacity to analyze efficiently large sequence data sets. To accelerate HPV genotyping of high-throughput NGS data, an automated system including a comprehensive collection of HPV mucosal and cutaneous reference sequences with updated nomenclature was developed.
Methodology
The web-based HPV-QUEST subtyping system uses PHP/HTML language, MYSQL, as the database management system for blast searches is available freely on http://www.ijbcb.org/HPV/. HPV-QUEST is able to processes up to 10 megabases (Mb) of sequences (around 6,500 sequences of 100 bp) per run, returns results within one to two minutes, and displays up to 150 hits with the top hit as default.
HPV genotyping is based on sequences from the L1 region comprised of 1500 nucleotides. HPV-QUEST includes a new HPV database with updated nomenclature for 150 annotated cutaneous and mucosal HPV L1 sequences, representing 5 genuses, 14 species, and 150 types, compiled from complete genomes, subgenomic regions containing the L1 region, or L1 region [5, 6] from NCBI Genebank [7], Virus Sequence Database [8], and Los Alamos HPV Sequence Database [9].
HPV-QUEST Input and Output
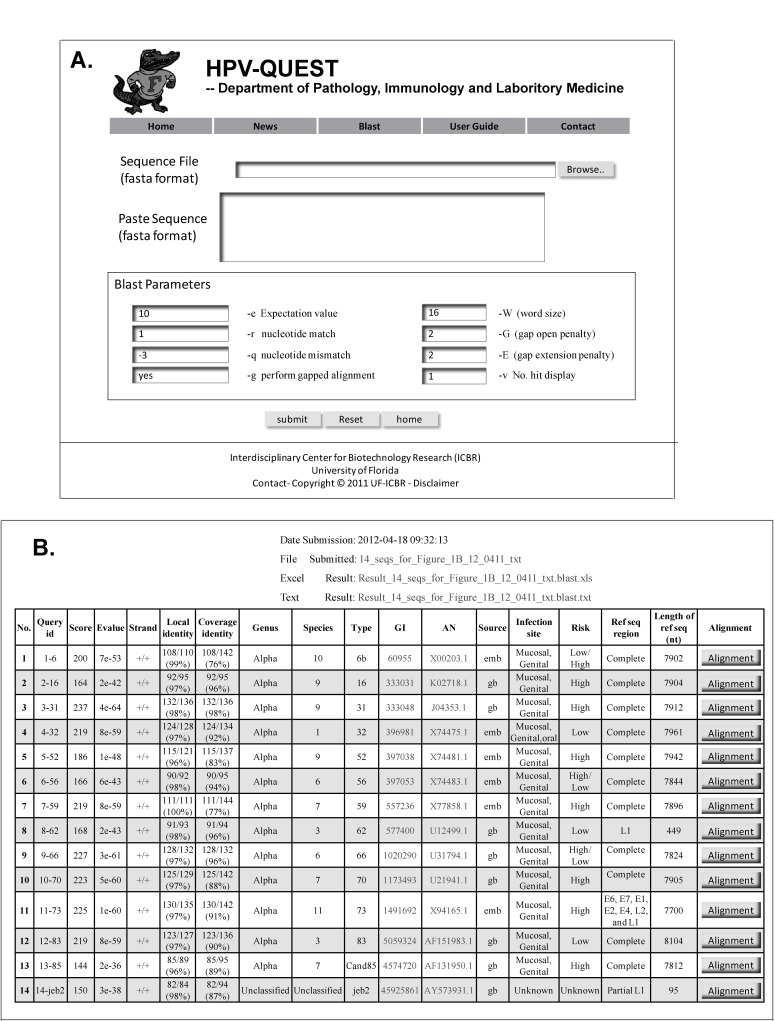
HPV-QUEST is password protected. User can obtain a log-in password for free access by visiting the website. Sequence files as large as 10 Mb containing forward or complementary reverse sequences are entered in fasta format by copy/pasting or uploading files. Pre-blast sequence cleaning to remove low quality reads is recommended. Blast parameters with suggested default values include: –e (expect value, default = 10), -r (nucleotide match, default = 1), -q (nucleotide mismatch, default = -3), -g (perform gapped alignment, default = yes), -W (word size, default = 16), -G (gap open penalty, default = 2), -E (gap extension penalty, default = 2), and -v (number of hits display, default = 1) (Figure 1A). A confirmation with submitted file name, date and time of submission and a link to view results and retrieve reports is generated and e-mailed to the user.
Figure 1.
Input and output files. (A) HPV-QUEST blast page. Users either paste or upload up to 10 Mb of sequences, chose desired parameters, click submit, and obtain the results in 1 to 2 minutes as html, excel or text files; (B) HPV-QUEST output. HPV-QUEST output includes a result page illustrating the No. (the query sequence serial number), Query id (fasta file header of the query sequence), Score (blast score), Evalue (expect value), Strand (+/+ or +/-), Local identity (percentage of matched nucleotides within alignment region), Coverage identity (percentage of nucleotides matched with reference sequence), Genus, Species, Type, GI (NCBI gene identification number), AN (NCBI accession number), Source (source of reference sequence), Infection site (mucosal or cutaneous or both), Risk (high or low or unknown), Ref seq region (reference sequence region in the genome), Length of ref seq (nt), and Alignment (alignment of query sequence with reference sequence). Date and time of submission is also displayed. Two result files in excel or text format are generated for download.
A set of Perl scripts is applied to parse the program output files and produce a result page in HTML format, and a report in both text- and excel-format containing: No. (the query sequence serial number), Query id (fasta file header of the query sequence), Score (blast score), Evalue (expect value), Strand (+/+ or +/-), Local identity (percentage of matched nucleotides within alignment region), Coverage identity (percentage of nucleotides matched with reference sequence), Genus, Species, Type, GI (NCBI gene identification number), AN (NCBI accession number), Source (source of reference sequence), Infection site (mucosal or cutaneous or both), Risk (high or low or unknown), Ref seq region (reference sequence region in the genome), Length of ref seq (nt), and Alignment (alignment of query sequence with reference sequence) (Figure 1B). The original query sequences are included in the report to eliminate the need to match query sequences with correspondent results. Query sequences failing to align with any known reference sequences in the HPV-QUEST are designated as “nd”. Any sequences that fail to blast, have low local identity, or with an e-value >1e-15 are considered as low quality, new recombination, or new genotype.
Testing and validation
HPV-QUEST version 1.0 was tested and validated in two ways. Firstly, reference sequences used to construct the database were blasted against themselves. The typing was 100% correct, and all e-values were 0 with local or coverage identities of 100%. Secondly, a test dataset of 18,000 quality HPV pyrosequences, generated by PGMY9/11 and GP5+/6+ primers using Titanium Amplicon Pyrosequencing technology from DNA extracted from genital swabs of 15 asymptomatic men recruited in an international study cohort, was processed by using HPV-QUEST and the results compared with typing by traditional NCBI blastn with an cutoff evalue of 1e-15 [10]. HPV genotypes and frequency distribution by using HPVQUEST coincided with results from NCBI blastn with significantly shorter processing time (less than 30 minutes versus more than 40 hours) to produce results ready for analysis.
Caveats and Future development
Although new HPV types are discovered continuously, HPV classification and nomenclature are updated periodically by the Reference Center for Human Papillomaviruses at the German Cancer Research Center in Heidelberg, which will be used to update HPV-QUEST. Version 2.0 will include HPV subgenomic regions other than L1, reference sequences for non-human papilloma viruses, and extensive data sets generated by next generation sequencing technology.
Acknowledgments
University of Florida Cancer Center /Moffitt Cancer Center Collaborative Initiative; NIH R01CA098803; Stephaney W. Holloway University Chair for AIDS Research.
Footnotes
Citation:Yin et al, Bioinformation 8(8): 388-390 (2012)
References
- 1.B Lu, et al. Cancer Epidemiol Biomarkers Prev. 2001;20:990. [Google Scholar]
- 2.M Plummer, et al. J Infect Dis. 2011;203:891. doi: 10.1093/infdis/jiq146. [DOI] [PubMed] [Google Scholar]
- 3. http://kcdc.labkm.net/vsd/main/index.jsp.
- 4. http://www.bioafrica.net/subtypetool/html/indexhpv.h tml.
- 5.EM de Villiers, et al. Virology. 2004;324:17. [Google Scholar]
- 6.HU Bernard, et al. Virology. 2010;401:70. [Google Scholar]
- 7. http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/ww wtax.cgi/?mode/=Tree/&id/=151340/&lvl/=3/&lin/=f/&keep/=1/&s rchmode/=1&unlock.
- 8. http://kcdc.labkm.net/vsd/database/gene/_search/_7.jsp/? orgId=7&reset=1.
- 9. http://hpv-web.lanl.gov/
- 10.AR Giuliano, et al. Cancer Epidemiol Biomarkers Prev. 2008;17:2036. doi: 10.1158/1055-9965.EPI-08-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]