Background: The scientific question that we address is how cytokine receptors organize their degradation.
Results: CHIP and Ubc13 are required for GH receptor endocytosis, implicating Lys63-specific ubiquitination.
Conclusion: This study shows how two ubiquitin ligases act in concert to allow receptor endocytosis.
Significance: Understanding this mechanism enables drug design to control GH signaling in fighting cancer and cachexia.
Keywords: E3 Ubiquitin Ligase, Growth Hormone, Receptor Desensitization, Receptor Endocytosis, Ubiquitin-conjugating Enzyme (Ubc), CHIP, Endocytosis, GHR, Lys63-linked Ubiquitin, Ubc13
Abstract
Growth hormone receptor (GHR) endocytosis is a highly regulated process that depends on the binding and activity of the multimeric ubiquitin ligase, SCFβTrCP (Skp Cullin F-box). Despite a specific interaction between β-transducin repeat-containing protein (βTrCP) and the GHR, and a strict requirement for ubiquitination activity, the receptor is not an obligatory target for SCFβTrCP-directed Lys48 polyubiquitination. We now show that also Lys63-linked ubiquitin chain formation is required for GHR endocytosis. We identified both the ubiquitin-conjugating enzyme Ubc13 and the ubiquitin ligase COOH terminus of Hsp70 interacting protein (CHIP) as being connected to this process. Ubc13 activity and its interaction with CHIP precede endocytosis of GHR. In addition to βTrCP, CHIP interacts specifically with the cytosolic tails of the dimeric GHR, identifying both Ubc13 and CHIP as novel factors in the regulation of cell surface availability of GHR.
Introduction
Growth hormone receptor (GHR)3 functions in longitudinal growth and metabolism. It is a prototypic member of the class I cytokine receptor family that, after binding to its natural ligand, growth hormone (GH), signals via Janus kinase 2 (Jak2). GH binding to the dimeric GHR induces a rotation in the GHR tail (1, 2) that subsequently leads to activation of Jak2 and, ultimately, to activation of signaling cascades via Stat5b, ERK, and MAP kinase pathways (reviewed in Ref. 3). Endocytosis and degradation of the GHR determine largely the sensitivity of cells for GH (4). GHR is endocytosed via clathrin-coated pits and transported via endosomes toward the lysosomes. To respond to different environmental signals, cells use specific tools to keep their responsiveness for growth factors and nutrients at the appropriate level. For many signaling receptors, entry into clathrin-coated pits depends on an active ubiquitination system. Although GHR ubiquitination is not required, GHR depends on the F-box protein β-transducing repeat-containing protein (βTrCP) for endocytosis (5). βTrCP binds to the ubiquitin-dependent endocytosis (UbE) motif of the GHR as part of the ubiquitin ligase complex SCFβTrCP (Skp1, Cullins, F-box proteins) (6). The same ligase is required to transport the GHR from endosomes to lysosomes (7).
There is emerging evidence for the involvement of Lys63-linked ubiquitination in the homeostasis of specific membrane proteins. In many instances, Ubc13 acts as the E2 enzyme that, together with pseudo E2s, UEV1A, or MMS2, assembles Lys63-linked ubiquitin chains on the substrates (8, 9). The dopamine transporter is provided with four-ubiquitin Lys63-linked chains, presumably via Nedd4-2 (10). UEV-1/Ubc13 is implicated in the regulation of an AMPA-type glutamate receptor in Caenorhabditis elegans (11). The yeast monocarboxylate transporter, Jen1, requires HECT-ubiquitin ligase Rsp5-dependent Lys63 ubiquitination for endocytosis (12). Short chain Lys63 ubiquitination mediates the regulated endocytosis of the aquaporin-2 water channel (13), and forced expression of ubiquitin mutants indicates that Lys63 ubiquitination of the prolactin receptor is important for its degradation (14). Chain assembly on substrates is an orchestrated interplay between an ubiquitin activating enzyme (E1), an conjugating enzyme (E2), and an ubiquitin ligase (E3). As exemplified, Ubc13/UEV1A utilizes several ubiquitin ligases to specifically ubiquitinate the substrates. In this study, we identified the COOH terminus of Hsp70 interacting protein (CHIP) as a specific E3 for GHR endocytosis. CHIP is a 35-kDa multi-domain protein containing an NH2-terminal tetratricopeptide repeat (TPR) and a COOH-terminal U-box domain. The U-box is related to the RING domain and acts passively as a scaffold for the E2, positioning it in proximity to the substrate. CHIP can act together with either UbcH5a or Ubc13/UEV1a to assemble either Lys48- or Lys63-linked chains, respectively. In both cases, the interaction is between the U-box and the conserved SPA motif of the E2 enzymes (15–17). CHIP binds with its TPR domain to the COOH-terminal EEVD sequence of the molecular chaperones Hsp70 and Hsp90 (18). Interaction of CHIP with the two E2s, UbcH5a and Ubc13, has distinct effects on the conformational dynamics of CHIP, suggesting different roles of the CHIP-E2 interaction in the ubiquitination of substrates (19). CHIP links the Hsp70/Hsp90 protein quality control/folding machinery with the ubiquitination/proteasomal degradation pathways, making it a fate-deciding point for proteins. In addition, functions independent of Hsp70 and Hsp90 have also been reported in glycoprotein quality control (with SCFFbx2), in the degradation of the Notch signaling factor Tal1 (with SCFSkp2), in controlling cellular levels of base excision repair enzymes, in the degradation of toxic forms of α-synuclein, and in the regulation of Smad1/5 proteins (20–25).
In this study, we describe a specific role of Lys63-linked ubiquitin chains and the E2/E3 pair Ubc13/CHIP in GHR endocytosis. Combining gene silencing and overexpressing approaches, the roles of the CHIP TPR domain, as well as the Ubc13 SPA motif in GHR endocytosis, were demonstrated. The GHR specificity is controlled by sequence information within and downstream of the UbE motif. We propose that the CHIP-Ubc13 activity occurs after the SCFβTrCP ubiquitin ligation activity and before GHR selection into clathrin-coated pits.
EXPERIMENTAL PROCEDURES
Materials, Antibodies, DNA Constructs, and Cell Lines
Antibody anti-GHR (T) was raised in rabbits against the cytoplasmic sequence between amino acids 271 and 381, as described previously (26). Anti-CHIP antibody was obtained from Calbiochem, monoclonal anti-HA antibody was from Babco (Richmond, CA), Cy3-GH was prepared as described previously (6), Alexa 488-transferrin was from Molecular Probes, and EGF-Alexa Fluor 488 streptavidin was from Invitrogen. DNA constructs CHIP and CHIPΔTPR were gifts from Dr. Douglas Cyr (University of North Carolina, Chapel Hill, NC), and Ubc13 and Ubc13 C78A in pEF-IRES-puro were gifts from Dr. James Chen (Southwestern University, Dallas, TX). The A98D mutation in Ubc13 was introduced using a QuikChange site-directed mutagenesis kit (Stratagene) according to the instructions of the manufacturer. The primers used for this reaction were 5′-TTGAAAGATAAGTGGTCCCCAGATCTCCAGATCCGCACAGTTCTG-3′ and 5′-CAGAACTGTGCGGATCTGGAGATCTGGGGACCACTTATCTTTCAA-3′. GST constructs were described previously (27). Dr. Matthias Mayer (Universität Heidelberg) kindly provided the CHIP overproducing strain (FI8202 transformed with pUHE21–2fdΔ12-hCHIP). U2OS cells expressing either GHR or both GHR and EGFR were described previously (7).
Cell Culture
U2OS cells were grown in DMEM (Invitrogen) supplemented with 10% FCS, 100 units/ml penicillin, and 0.1 mg/ml streptomycin. The U2OS GHR-expressing cells were grown in the same medium supplemented with hygromycin. The HEK293 cell line, expressing the tetracycline repressor (HEK293-TR) was a gift from Dr. Madelon Maurice (University Medical Center, Utrecht, The Netherlands). HEK293-TR cells were grown in DMEM supplemented with 10% FCS, 100 units/ml penicillin, 0.1 mg/ml streptomycin, and 12 μg/ml Blasticidin S (MP Biomedicals). All of the cells used were grown at 37 °C with 5% CO2. The cells were washed with PBS, detached from the flask with Trypsin-EDTA (Invitrogen), diluted in fresh growth medium, and split into new culture flasks twice a week.
Transfections
The cells were transfected with 40 nm validated small interfering RNAs specific for Ubc13 (5′-AATGGCAGCCCCTAAAGTACG-3′), CHIP (STUB,1; Ambion 289568) or Control#1 (Ambion AM4635) using Lipofectamine 2000 (Invitrogen) according to the instructions of the manufacturer. DNA transfections were performed using FuGENE 6 (Roche Applied Sciences) according to the instructions of the manufacturer.
Immunofluorescence Microscopy
GHR- or EGFR-expressing U2OS cells were grown on coverslips and allowed to take up Cy3-GH and/or Alexa 488-EGF at 37 °C for 30 min. The cells were washed and fixed with 3% paraformaldehyde. The cells were permeabilized with 0.2% Triton X-100 for 5 min and washed three times. The fixation was terminated in 0.5% BSA in PBS for 15 min. The coverslips were incubated with the primary antibody in 0.5% BSA in PBS for 30 min. After washing with 0.5% BSA in PBS, the cells were incubated with secondary antibody for 30 min or directly mounted using Prolong Gold DAPI and analyzed using a Zeiss LSM510 confocal microscope and Zen Software.
125I-GH Uptake
125I-Labeled human GH was prepared with the use of chloramine T (26), and iodinated GH was purified over a 500-μl Zeba spin column (Pierce Thermoscientific). For internalization studies, the cells were grown in 12-well plates, treated with siRNAs as described above; washed with DMEM; supplemented with 20 mm, Hepes, pH 7.4, and 0.1% BSA; and incubated in a water bath. 125I-GH (180 ng/ml) was bound on ice for 2 h. Subsequently, the cells were washed free of unbound ligand and incubated at 37 °C for 0–15 min. Membrane-associated ligand was removed by acid wash (0.15 m NaCl, 50 mm glycine, 0.1% BSA, pH 2.5) on ice. Internalized ligand was determined by measuring the radioactivity after solubilization of the acid-treated cells in 1 n NaOH in a LKB γ-counter.
Biotin-GH Pulldown and Immunoprecipitations
Cells were lysed with lysis buffer (1% Triton X-100, 1 mm EDTA, 1 mm PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1% BSA in PBS) on ice for 20 min. The lysates were clarified for 5 min, and the supernatants were incubated with biotin-GH or anti-GHR (T) for 1 and 2 h, respectively, followed by 1 h of incubation with streptavidin-agarose or protein A-Sepharose. Alternatively, for pulling the GHRs at the cell surface, HEK293-TR cells were placed on ice, and the culture medium was replaced with cold DMEM (20 mm Hepes, pH7.4), after which biotin-GH (180 ng/ml) was added. After 2 h of incubation, which results in saturating GHR binding to biotin-GH, the cells were washed three times with PBS, and lysed with lysis buffer. After 5 min of centrifugation, the supernatant was incubated for 1 h with streptavidin-agarose beads. The beads were washed three times with lysis buffer and three times with PBS. The samples were subjected to reducing SDS-PAGE and transferred to Immobilon-FL polyvinylidenedifluoride membrane (Millipore). Detection and quantification of the signals were performed with an Odyssey system (LI-COR Biosciences).
For ubiquitination experiments, the cells were lysed in 1% Triton X-100 with inhibitors (1 mm EDTA, 1 mm PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 10 mm N-ethylmaleimide, 1 mm Na3VO4 and 100 mm NaF). The cell lysates were centrifuged at maximum speed in an Eppendorf centrifuge for 5 min at 4 °C, and the supernatants were used for GHR isolation with anti-GHR antibody (anti-T) and protein A beads in 1% Triton X-100, 0.5% SDS, 0.25% sodium deoxycholate, 0.5% BSA, and inhibitors. Immunoprecipitates were subjected to reducing SDS-PAGE and Western blotting.
Recombinant CHIP and βTrCP Purification
Induction of CHIP expression was performed in 1.5-liter cultures of the bacterial host FI8202, at A680 = 0.8 with 250 μm isopropyl-1-thio-β-d-galactopyranoside, at 30 °C for 3 h. After spinning down the culture, the pellet was resuspended in 50 ml of buffer A (50 mm Hepes, pH 7.0, 50 mm KCl, 5 mm DTT, 10% glycerol, 1 mm PMSF), supplemented with protease inhibitors (10 μg/ml leupeptin, 10 μg/ml aprotinin) and 0.4 mg/ml lysozyme. The suspension was left on ice for 30 min, after which the cells were lysed by sonication. After removing the cell debris by centrifugation at 20,000 × g for 20 min, the supernatant was subjected to ultracentrifugation at 100,000 × g for 2 h. The supernatant was recovered and precipitated with 40% ammonium sulfate. After centrifugation at 10,000 × g for 30 min, the pellets were resuspended in 10 ml of buffer A and dialyzed against the same buffer overnight. The samples were subjected to ultracentrifugation at 100,000 × g for 1 h, and the filtered supernatant was loaded onto a anionic exchange column (DEAE FF16/10 1× 20 ml). The column was washed with 1 column volume of buffer A, and bound proteins were eluted with a linear gradient of 0 to 100% buffer B (50 mm Hepes, pH 7.0, 1 m KCl, 5 mm DTT, 10% glycerol, 1 mm PMSF) over 5 column volumes. CHIP-containing fractions were pooled, concentrated to 500 μl, and loaded on a S200 gel filtration column pre-equilibrated with 2 column volumes of buffer A. The gel filtration step resulted in three distinct peaks, corresponding to different molecular weight CHIP forms. A mixture of three forms was used in the experiments. βTrCP purification from insect cells is described elsewhere.4
GST Fusion Protein Production and Pulldown Competition Experiments
GST fusion proteins were produced and purified according to the GST fusion protocol (Qiagen). The plasmids were transfected in a BL21 Rossetta strain and grown at 37 °C. At A600 = 0.5, the expression was induced with 1 mm isopropyl-1-thio-β-d-galactopyranoside for 4 h at 30 °C. GST fusion proteins were purified from bacterial lysates with glutathione-Sepharose beads for 2 h. The beads were washed six times with lysis buffer and then added to either cell lysates containing CHIP, purified CHIP, or βTrCP in lysis buffer and incubated for 2 h. The beads were washed three times with lysis buffer and three times with PBS. For the competition experiments, the beads were additionally incubated for 2 h with the indicated concentration of competing protein and washed again. The beads were boiled in SDS sample buffer and analyzed by Western blotting. The proteins were separated using SDS-PAGE and transferred to blot by Western blotting. The blots were detected using indicated antibodies with an Odyssey system.
RESULTS
Lys63-linked Ubiquitin Chains Are Involved GHR Endocytosis
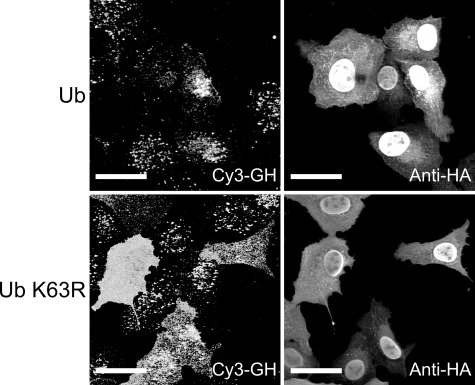
To investigate the role of Lys63-linked ubiquitin chains in GHR endocytosis, we expressed wild type and mutant K63R ubiquitin, unable to form Lys63-linked chains, in GHR-expressing U2OS cells. To monitor endocytosis, we incubated cells with Cy3-GH for 30 min. In cells overexpressing HA-ubiquitin K63R, we observed a clear increased cell surface labeling of Cy3-GH, compared with wild type ubiquitin, indicative of inhibition of GH degradation (Fig. 1). Because not all cells synthesize equal amounts of GHR, some HA-positive cells show a moderate response because of very low GH-binding sites. This finding indicates that Lys63-linked ubiquitin chains are involved in GHR degradation.
FIGURE 1.
Lys63 linkages are involved in GHR endocytosis/degradation. GHR-expressing U2OS cells were transfected with DNA constructs containing HA-tagged ubiquitin or ubiquitin K63R and incubated with Cy3-GH at 37 °C for 30 min. The cells were immune stained with anti-HA and Alexa 488-labeled anti-mouse IgG. Localization of Cy3-GH in HA-positive cells was determined with confocal microscopy. Bar, 5 μm. Ub, ubiquitin.
GHR Endocytosis Requires Both Ubc13 and CHIP
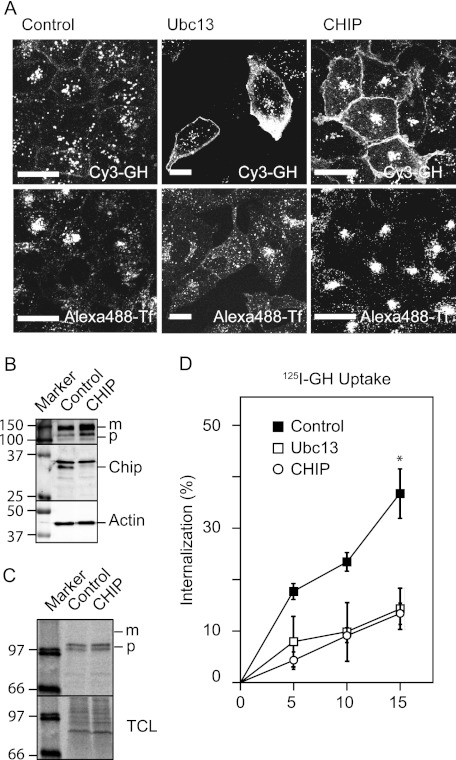
Because GHR endocytosis was Lys63-dependent GHR degradation, we asked whether Ubc13 is required for this process. In complex with the pseudo E2 enzymes UEV1 or MMS2, Ubc13 can assemble Lys63-linked ubiquitin chains (8). We assessed the uptake of Cy3-GH by confocal fluorescence microscopy in cells silenced for Ubc13. Fig. 2A shows a strong accumulation of Cy3-GH signal on the cell surface of cells that were treated with Ubc13-specific siRNA that did not occur in cells treated with control siRNA.
FIGURE 2.
GHR endocytosis requires Ubc13 and CHIP. A, U2OS cells expressing GHR were treated with Ubc13, CHIP, or control siRNA for 3 days; incubated with Cy3-GH and Alexa 488-Tf for 30 min; and fixed. Localization of Cy3-GH and transferrin was determined with confocal microscopy. Bars, 20 μm. B, the cells were treated with CHIP or control siRNA for 2 days. The cell lysates were boiled in sample buffer and analyzed using Western blot and immunostaining with anti-GHR(B), anti-CHIP, and anti-actin. C, cells were labeled for 15 min in the presence of [35S]methionine and then chased for 15 min. After lysis, GHR was immunoprecipitated using anti-GHR(B) and analyzed on a Storm imaging system after electrophoresis. TCL, total cell lysate. D, after silencing for 3 days, initial endocytosis rates were measured 5, 10, or 15 min after a 2-h incubation with 180 ng/ml 125I-GH on ice. The amount of intracellular radioactivity was measured and expressed as a percentage of total radio activity. All of the data in this figure are representative of four independent experiments. *, p < 0,001 compared with control.
To identify the E3 that is involved in this process, we transfected the cells with five siRNAs to silence the expression of Triad1, CHIP, CHFR, Parkin, and RNF85, known to interact with Ubc13. As seen in Fig. 2A (right panels), transfection of siRNA, specific for the ubiquitin ligase CHIP, revealed similar accumulation of Cy3-GH as in Ubc13-depleted cells, indicating that Ubc13 might function together with CHIP in the GHR endocytosis system. In the lower panels, we monitored transferrin uptake in control and Ubc-13- and CHIP-silenced cells; as expected, neither Ubc13 nor CHIP silencing affected transferrin uptake. In most cells at steady state, ∼80% of the transferrin receptors are inside. Because the total amount of Alexa 488-transferrin in the cells did not change upon silencing, we conclude that CHIP is neither involved in transferrin uptake, nor in the regulation of the outside/inside distribution of the transferrin receptor and recycling kinetics. However, CHIP silencing affected its intracellular distribution, very likely by redirecting transferrin to the so-called recycling compartment, which contains tubulovesicular structures and recycles EGF and transferrin with slow kinetics (28, 29). U2OS cells that did not express GHR showed no Cy3-GH labeling (not shown), indicating that the entire fluorescent label originated from GHR activity.
GHR is a rapidly degraded membrane protein with a half-life of ∼50 min (30–33). Inhibition of endocytosis leads to retention of GHR on the cell surface and prevents GHR from being transported to the lysosome for degradation.
Given the role of CHIP in ERAD, it is important to investigate a possible role in GHR synthesis and maturation (22, 34, 35). Previously, we showed that the GHR is efficiently folded in the endoplasmic reticulum (36). In Fig. 2B, we determined the effect of CHIP depletion on the steady state of GHR, and we asked whether endoplasmic reticulum maturation was impeded. The CHIP-depleted cells contained both more precursor and mature GHR compared with control cells, indicating that CHIP might be involved in endoplasmic reticulum maturation of the GHR. The increase of mature GHR was in agreement with the observation that CHIP depletion caused GHR accumulation at the cell surface (Fig. 2A). To exclude that the increased amount of precursor GH was due to increased biosynthesis, we labeled the cells for 15 min with [35S]methionine. CHIP depletion did not affect the amount of radioactivity incorporated in the GHR precursor molecules. Together, the results of Fig. 2 (B and C) implicate that CHIP might be involved in folding of the GHR during biosynthesis. Despite a delayed maturation, CHIP depletion caused accumulation of mature GHR.
To establish whether the accumulation of mature GHR is due to a change in endocytosis rate, we measured initial endocytosis rates of 125I-GH in U2OS cells, silenced for either of the two factors. After incubation with 125I-GH on ice, uptake kinetics, measured at 37 °C, revealed a 2.5-fold inhibition even at 5 min from the start, if either Ubc13 or CHIP were depleted (Fig. 2D). In general, the gene silencing of both Ubc13 and CHIP varied between 50 and 80%, estimated on Western blots (Fig. 2B).
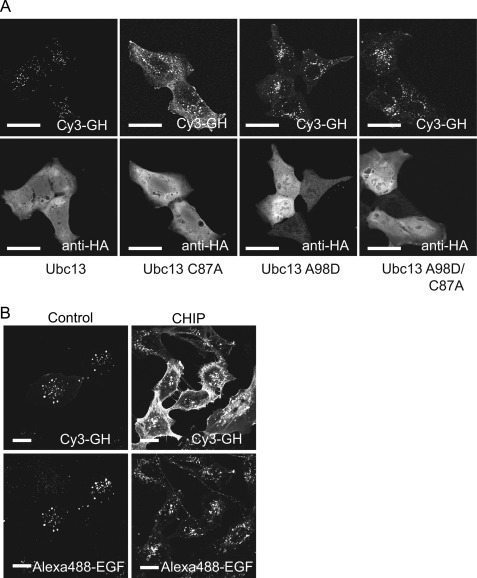
The results presented above show that depletion of either Ubc13 or CHIP inhibits GH endocytosis, as well as GHR degradation. To confirm that this is due to defective ubiquitin conjugation, we investigated the effect of mutations in critical residues of Ubc13 on GHR endocytosis. Cells contain sufficient Ubc13 activity to serve ongoing Lys63-linked ubiquitination: (over)expression of exogenous wild type Ubc13 does not induce phenotypic effects. However, if the active cysteine residue of Ubc13 is mutated to alanine (C87A), the conjugation activity is no longer functional, and expression of such a mutant acts as dominant negative for Ubc13-driven ubiquitination. As seen in Fig. 3A, exogenous expression of Ubc13(C87A) resulted in a clear inhibition of Cy3-GH uptake, whereas exogenous wild type Ubc13 expression did not interfere. This shows that enzyme activity of Ubc13 is essential for proper GHR endocytosis. The dominant negative effect indicates that Ubc13 C87A competes with endogenous Ubc13 for the binding to a ubiquitin ligase. Ubc13 interacts with CHIP through its SPA motif (15). To test whether the SPA motif on Ubc13 is essential for the dominant negative effect on GHR endocytosis, we introduced the A98D mutation into the Ubc13(C87A) mutant, disrupting Ubc13-CHIP binding. As expected, the dominant negative effect was abolished, and the cells endocytose Cy3-GH comparably with cells transfected with either wild type or Ubc13(A98D). Based on our screen results, showing that no other the Lys63-specific E3 showed a GHR-specific effect on Cy3-GH endocytosis, we conclude that both Ubc13 activity and the CHIP-Ubc13 interaction are needed for proper GHR endocytosis.
FIGURE 3.
Ubc13 activity is essential for GHR endocytosis. A, GHR-expressing U2OS cells were transfected with HA-tagged constructs of Ubc13 or Ubc13 mutants (C87A, A98D, C87A, and A98D) and incubated with Cy3-GH for 30 min at 37 °C. The cells were fixed and immune stained with anti-HA and a secondary antibody conjugated with Alexa 488. Localization of Cy3-GH and HA was determined by confocal microscopy. B, CHIP acts specifically on GHR endocytosis. U2OS cells expression both GHR and EGFR were transfected with siRNA specific for CHIP or control siRNA and incubated with Cy3-GH and Alexa 488 EGF for 30 min. The cells were fixed, and the labels were visualized on a LSM510 confocal microscope using Zen software. Bar, 20 μm. The data in this figure are representative of three independent experiments.
CHIP Silencing Does Not Inhibit General Clathrin-mediated Endocytosis
Because ubiquitination by CHIP has important general functions in protein homeostasis and chaperone activities in connection with Hsp70/90, it is important to ascertain that its action in GHR endocytosis is specific, rather than part of the general clathrin-mediated endocytosis machinery. In Fig. 2, we showed that transferrin receptor endocytosis requires neither CHIP nor Ubc13. To investigate whether CHIP is also involved in the endocytosis of the EGF receptor, prototypic for many receptor tyrosine kinases (37), we used U2OS cells expressing both GHR and EGF receptor. Fig. 3B shows that Alexa 488-EGF uptake is not affected in cells depleted for CHIP. Together with the observations for transferrin (Fig. 2), this implies that CHIP affects neither endocytosis of a prototypic receptor tyrosine kinase (EGF receptor) nor the endocytosis of transferrin.
CHIP Interacts with GHR
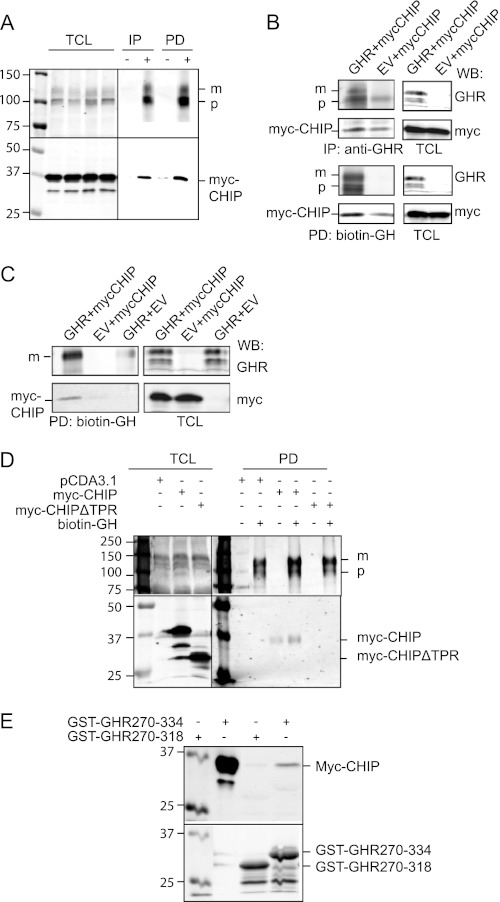
To unravel the role of CHIP in GHR endocytosis further, we analyzed the CHIP-GHR binding. We transiently expressed Myc-tagged CHIP in GHR-expressing U2OS cells and addressed the GHR-CHIP interaction using both biotin-GH and anti-GHR immunoprecipitation (Fig. 4A). Alternatively, we co-expressed GHR and Myc-CHIP in HEK293-TR cells and isolated the complexes in the same way (Fig. 4B). In both cases, specific interaction of Myc-labeled CHIP with GHR was detected, despite some background binding of CHIP to the beads in the absence of expressed GHR. We conclude that CHIP specifically binds to GHR. An important question is whether CHIP indeed interacts with GHR at the cell surface. To address this, we co-expressed GHR and Myc-CHIP in HEK293-TR cells and incubated the cells with biotin-GH on ice. Fig. 4C shows that CHIP specifically interacts with the GHR at the cell surface.
FIGURE 4.
CHIP interacts with GHR. A–C, Myc-CHIP was transiently transfected into GHR-expressing U2OS cells (A) or transfected together with wild type GHR or empty vector (EV) in HEK293-TR cells (B and C). A and B, the cell lysates were incubated with biotin-GH or anti-GHR(T), and the protein complexes were isolated on immobilized streptavidin (PD) and protein A beads (IP), respectively, and eluted with SDS sample buffer. C, cells were incubated for 2 h with biotin-GH on ice, and protein complexes from the cell surface were isolated using immobilized streptavidin, eluted with SDS sample buffer. D, cell lysates from Myc-CHIP, Myc-CHIPΔTPR, or empty vector transfected U2OS cells were incubated with biotin-GH or PBS (control), and protein complexes were isolated, washed on streptavidin beads, and analyzed for CHIP on Western blots. E, Myc-CHIP was transiently expressed in U2OS cells. The cells were lysed and incubated with glutathione beads preloaded with GST-GHR270–318 or GST-GHR270–334 fusion proteins. The beads were washed and analyzed for CHIP on a Western blot (WB). TCL, total cell lysate; m, mature GHR; p, precursor GHR. The data in this figure are representative of three independent experiments.
Previously, we reported the small glutamine-rich TPR-containing protein (SGT) to bind via its first TPR to the UbE motif of the GHR (27), but gene silencing of SGT did not result in an endocytosis phenotype.5 To examine whether the CHIP-GHR interaction is via the TPR domain, we used CHIP lacking the TPR domain. Using biotin-GH, we were able to pull down Myc-CHIP, but not Myc-CHIPΔTPR, together with full-length GHR, indicating that indeed the protein-protein interacting TPR domain is involved in binding CHIP to GHR (Fig. 4D). Attempts to detect binding between Ubc13 and GHR failed, probably because of the transient character of the interaction. Next, we investigated whether CHIP, present in total cell lysates, was able to bind purified GHR cytoplasmic tails. Using identical GST fusion proteins as in the SGT study, Fig. 4E shows specific interaction that depends on the presence of amino acid residues between 318 and 334 in the GST fusion protein, confirming that the binding is specific (27).
CHIP Binds GHR Independently of βTrCP
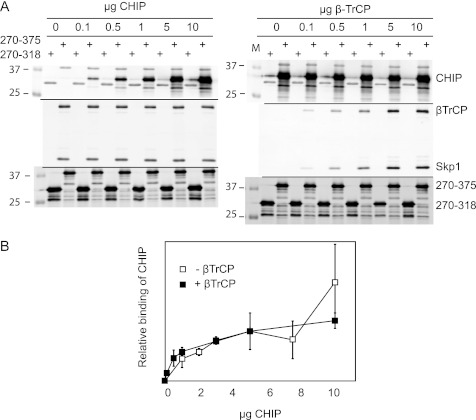
CHIP recognizes the acidic (M/I)EEVD consensus sequence in Hsp90 and Hsp70 (18). Additionally, CHIP can bind to exposed hydrophobic parts of unfolded proteins (38). The results from Fig. 4E suggest that the sequence between residues 318 and 334 is important for CHIP-GHR binding. We noticed that the GHR tail, between amino acids 320 and 350, contains an acidic-rich region that clearly extends beyond amino acid residue 334, the COOH-terminal amino acid residue of our GST fusion protein used in the Fig. 4. To ascertain that we test the GHR-CHIP interactions to the full extent, we used a longer GST-GHR fusion protein, ranging from amino acids 270 to 375, and untagged CHIP, produced in Escherichia coli. Using this assay, we addressed two questions: 1) Is the CHIP-GHR interaction indeed direct and saturable? 2) How does CHIP interact with GHR in the presence of SCFβTrCP, the other E3, involved in GHR endocytosis? As seen in Fig. 5A, both recombinant βTrCP and CHIP bound the GST-GHR270–375 in a concentration-dependent and saturable manner. The previously described UbE motif (321DDSWVEFIELDIDE334) was found to be involved in βTrCP binding (6). To address the second question and to investigate whether βTrCP and CHIP bind independently or cooperatively to the same region of GHR, we performed competition experiments. Beads coupled to GST-GHR270–375 were saturated with either CHIP or βTrCP (purified from SF9 cells) and incubated with increasing amounts of the “competing” interactor. We observed that CHIP binding to GHR could not be competed off by βTrCP, and vice versa. Additionally, we assessed the binding of CHIP to GST-GHR270-375 in the presence and absence of βTrCP. Fig. 5B shows no difference in CHIP binding. We conclude from these data that CHIP and βTrCP do not compete for binding on the GHR tails used and that no cooperativity exists between βTrCP and CHIP for binding to the (monomeric) GHR tails. This implies the existence of two different, independent binding sites for βTrCP and CHIP.
FIGURE 5.
CHIP binds GHR independently of βTrCP. A, glutathione beads were maximally loaded with recombinant GST-GHR270–375 or GST-GHR270–318 (illustrated in the lower panels). The beads were washed and incubated with saturating concentrations (10 μg/ml) of either purified βTrCP (left panels) or CHIP (right panels). The beads were extensively washed and incubated with indicated concentrations of competing ligand (CHIP, left panels; βTrCP, right panels) for 2 h at 4 °C. In both cases, maximal binding was achieved at 5–10 μg/ml similar to conditions without competing ligands. The antibodies used for detection are indicated in the figure. Binding was monitored by Western blotting. GST-GHR270–318 served as background binding. B, the effect of βTrCP on the binding of CHIP to GST-GHR270–375 was quantified. Glutathione beads saturated with GST-GHR270–375 were incubated with indicated concentrations of purified CHIP, with or without 10 μg/ml purified βTrCP, for 2 h at 4 °C. After extensive washing, the beads were boiled in sample buffer, and the binding was analyzed on Western blot. Signals for CHIP and GST were quantified with Odyssey software. The amount of bound CHIP was normalized against the GST signal; the amount of CHIP bound at 5 μg/ml was set at 100%. The data are representative of two independent experiments.
Ubc13 and CHIP Act after βTrCP during GHR Endocytosis
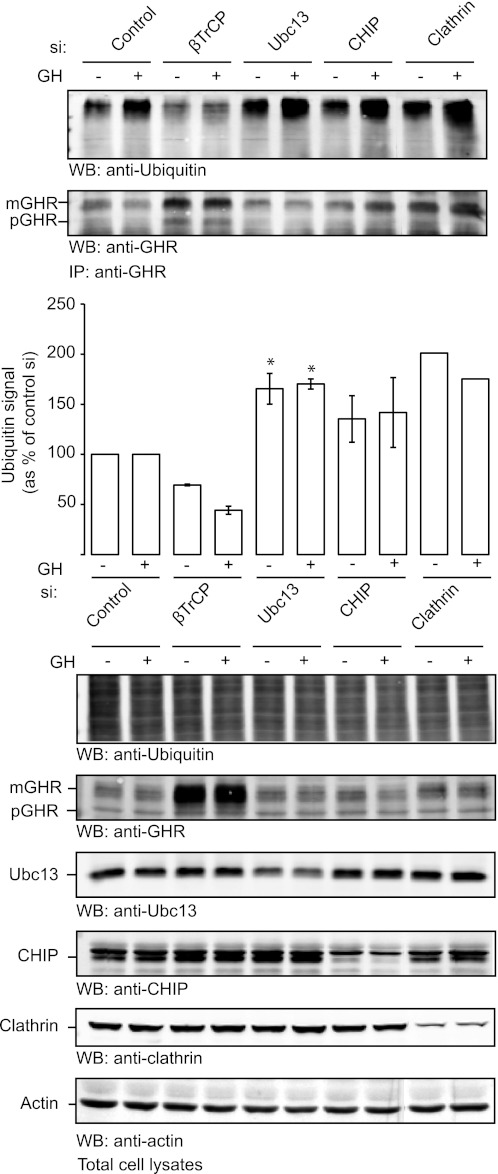
Once it is established that both βTrCP and CHIP are required for GHR endocytosis and that both bind to the cytosolic tail segment 318–375, it is important to determine their respective roles. Most probably both are required at a stage before GHR recruitment into clathrin-coated pits. Because both factors act in GH-dependent as well as independent (constitutive) endocytosis, there is no easy parameter to measure the progression toward recruitment into coated pits. Therefore, we used the ubiquitination state of GHR as a tool to assess the order of events in GHR endocytosis. As previously shown, βTrCP depletion abolishes GHR (Lys48) ubiquitination, indicating that GHR is a substrate for βTrCP. We also showed that depletion of clathrin not only inhibits GHR endocytosis but also causes a strong accumulation of ubiquitinated GHR at the cell surface (39). Taking GHR ubiquitination as a measure of the activity of SCFβTrCP, we tested the effect of both Ubc13 and CHIP depletion on the GHR ubiquitination. As seen in Fig. 6, depletion of Ubc13 caused a significant accumulation of ubiquitinated GHR, which is comparable with the accumulation seen upon clathrin silencing. CHIP silencing resulted in a modest increase in the amount of ubiquitinated GHR, but this accumulation was not statistically significant when compared with the control situation. Overall, these results suggest that the pair Ubc13/CHIP acts downstream of SCFβTrCP. These results are in line with our previous observation that K63R overexpression has little effect on the ubiquitination state of GHR (39). The results with anti-GHR (Fig. 6) show that gene silencing of neither Ubc13 nor CHIP increased the (130 kDa) GHR protein levels in contrast to the effects of silencing clathrin and βTrCP. This is possibly due to a shift of GHRs to higher molecular weight (ubiquitinated) species that are very difficult to quantify. Together, we conclude that, although CHIP and βTrCP can bind GHR independently, they act consecutively.
FIGURE 6.
CHIP acts after βTrCP. GHR-expressing U2OS cells were transfected with siRNA targeting βTrCP, Ubc13, CHIP, clathrin, or a control sequence. The cells were lysed and incubated with anti-GHR(T) in denaturing buffer to remove interacting proteins. Bound receptors were pulled from the lysates using immobilized protein A. The beads were extensively washed and boiled in SDS sample buffer. The ubiquitin signal of the receptor was analyzed by Western blotting, quantified, and expressed as arbitrary units relative to the amount of ubiquitination in control cells (expressed as 100%), representative of three experiments. For clathrin, quantitation of ubiquitination was highly variable. Statistical analysis revealed that Ubc13 silencing resulted in a significantly increased ubiquitination signal when compared with the control condition (p = 0.029) and with the βTrCP silencing condition (p = 0.008) (marked with asterisk). Although consistently higher, CHIP silencing did not result in a significant change in the ubiquitination status of GHR compared with the control or Ubc13 silencing conditions because of the variability of the measurement. The lower panels show the effect of gene silencing on the levels of ubiquitin conjugates, GHR, Ubc13, CHIP, clathrin, and actin in the total cell lysates. IP, immunoprecipitation; WB, Western blot.
DISCUSSION
We have shown that both βTrCP and Ubc13/CHIP are required for GHR endocytosis. Although their binding sites partly overlap, they bind independently. Although the binding of βTrCP is strictly confined to the UbE motif and is defined as unconventional compared with the conventional DSGXXS motif present in β-catenin and IκB,4 the CHIP-GHR interaction extends beyond the UbE motif. Our results show that GHR accommodates two completely different E3 enzymes, both required for its endocytosis, involved in different ubiquitination activities. Because GHR with all lysine residues replaced by arginines is endocytosed as wild type GHR (5), we assume that both E3s target other substrates than GHR itself.
The GHR-CHIP interaction depends on the presence of a TPR domain in CHIP. This domain consists of three copies of the helix-turn-helix TPR motif that, together with a seventh COOH-terminal helix, functions in many signaling protein complexes. NMR and CD spectroscopic studies have revealed that this domain is largely unfolded until it interacts with a MEEVD pentapeptide derived from Hsp90 (38) that results in a stably folded structure. Because of the high content of acidic amino acid residues in the presumed binding region of the GHR between amino acids 320 and 350 (three sets of di-acidic conserved sequences), it is tempting to assume that they are part of a CHIP-binding site, analogous to the MEEVD sequence in Hsp90. Binding experiments with an array of alanine point-mutated GST-GHR peptides did not reveal a CHIP-specific binding motif between amino acid residues 320 and 350 (not shown). This might be because mutating single amino acids is not sufficient to considerably weaken the interaction or because of a temperature-sensitive complex (38, 40). Because both βTrCP and CHIP can bind to the same extent without mutual interference, a possible binding site for CHIP would be the amino acid sequence IDDPDE (rabbit) or IDEPDE (human GHR), located immediately after the UbE motif. Our experiments show that CHIP binds both to (dimeric) GHR isolated from cells as to (monomeric) GST fusion proteins. As we performed the competition studies in the latter system, the results suggest that both E3s can bind to the same GHR polypeptide.
There are indications for a role of ubiquitination in the endocytosis machinery. This is best illustrated by the necessity of ubiquitin-binding domains (UBDs) in the endocytosis adaptors Epsin and Eps15 that can also act as scaffold proteins (41). Because both adaptors can also be monoubiquitinated, it has been proposed that they can be in a closed, inactive conformation if the UBD binds to the attached monoubiquitin moiety (42, 43). In the open configuration, they can bind to ubiquitinated cargo via the UBD (44). The role of ubiquitin in endocytosis remains unclear: on the one hand, cargo-specific ubiquitination may be required to enable binding to the UBD of an adaptor; on the other hand, monoubiquitination of the adaptor (Eps15, Epsin) may be inhibitory because of its closed conformation. Because CHIP ubiquitinates Epsin, it is probably a negative factor in the endocytosis of cargo that needs Epsin for endocytosis such as in the case of Notch and the EGF receptor (45–47). In this study, CHIP is clearly acting as a positive factor for GHR endocytosis.
The role of ubiquitination in endocytosis of specific membrane proteins is slowly emerging as recently reviewed in Ref. 48. For some membrane proteins, cargo-specific ubiquitination has been reported. The epithelial sodium channel requires the ubiquitination machinery directed via Nedd4-2 (49). The IGF-I receptor is ubiquitinated by both Mdm2 (Lys63) and c-Cbl (Lys48) (50). The prolactin receptor requires Lys63 polyubiquitination for endocytosis (14), as well as aquaporin-2 (13). Endocytosis of the EGF receptor is accommodated by c-Cbl, although discussion about the necessity of covalent ubiquitin attachment remains (51). In fact, except for the epithelial sodium channel, strict proof of the necessity of covalent attachment to the cytosolic tail of mammalian cargo molecules has not been established. For GHR endocytosis, ubiquitination is a major requirement, although the ubiquitin chains do not need to be covalently attached to the receptor. Remarkably, this is the case for both E3s described in this study.
In mammalian cells and in yeast, Lys63-linked ubiquitin chains are required for sorting in endosomes via the ESCRT complex, but there are few indications for their involvement in endocytosis (48, 52). In yeast, the monocarboxylate transporter, Jen1, requires Rsp5-dependent Lys63 ubiquitination for endocytosis (12). In mammalian cells, the Kaposi's sarcoma-associated herpesvirus-encoded ubiquitin E3 ligase, K3, ubiquitinates cell surface MHC class I molecules, causing the internalization and degradation of MHC I. In this process, K3 recruits the cellular Ubc13 to generate Lys63-linked polyubiquitin chains on MHC I, leading to its clathrin-mediated endocytosis. Our study is the first that reveals a cellular machinery that requires Lys63-linked ubiquitination in cargo selection at the cell surface. Previously, we demonstrated that SCFβTrCP is indispensable for GHR to pass the endosomes (7). Because CHIP and SCFβTrCP might act together at the cell surface, they might also do so in endosomes. Actually, Fig. 1 shows Cy3-GH accumulation, not only at the cell surface, but also intracellularly, if Ub K63R is overexpressed.
The likelihood that CHIP and Ubc13 act together in GHR endocytosis is strengthened by the experiments with overexpression of Ubc13 mutants, together with the observation that overexpression of K63R ubiquitin blocks GHR endocytosis. Insight in the conformational dynamics of CHIP with E2 enzymes and substrates other than chaperones indicate that Lys63 ubiquitination via CHIP regulate processes in membrane trafficking (53), and DNA repair (54). In contrast to CHIP in complex with UbcH5a, CHIP in complex with Ubc13 can bind to chaperones via the TPR domain, indicating that binding to a substrate like GHR through the TPR domain does not interfere with ability of CHIP to synthesize Lys63-linked ubiquitin chains. Another question in this regard is our previous observation that GHR, truncated at amino acid 334, requires SCFβTrCP for endocytosis (6). Because the binding site of CHIP extends downstream of amino acid 334, it might mean that removing this binding site also would eliminate the necessity of CHIP. This is reminiscent of the involvement of the proteasome: its action is required for the endocytosis of the full-length receptor but is dispensable for a receptor truncated after amino acid amino acid 366 (55). However, the GHR truncated at 334 might still have sufficient binding affinity for CHIP to act as a necessary factor for endocytosis of the GHR334.
On the cooperativity of CHIP with other E3s, two action modes have been proposed: 1) in neurons, CHIP acts with Fbx2 in glycoprotein quality control, where an NH2-terminal PEST sequence interacts with the TPR domain of CHIP to facilitate the F-box associated domain (FBA)-high mannose glycoprotein interaction (22), and 2) in the Notch-induced degradation of Tal1/SCL, where both CHIP and Skp2 can bind Tal1 (23). The latter case bears some similarity with the current study in that the substrate (GHR) binds two E3s, independently. However, a major difference is that in both models CHIP is involved in ubiquitin/proteasome dependent degradation, whereas in the current study CHIP and SCFβTrCP function in cargo selection for endocytosis. Indeed, we cannot exclude the possibility that in the course of this process, an as yet unknown (Lys48-ubiquitinated) substrate must be degraded to allow progression of the process, as we hypothesized previously (55). CHIP-Ubc13-Uev1a may work sequentially with SCFβTrCP, whereby the SCF ligase initiates ubiquitination, and the CHIP enzyme complex carries out Lys63-linked polyubiquitination on an as yet unidentified substrate (56). It was recently shown that CHIP/Ubc13 can extend a monoubiquitinated protein and synthesize Lys63-linked chains on these ubiquitin moieties (17). As in the absence of Ubc13/CHIP, where GHR is ubiquitinated to the same extent as in clathrin-depleted cells, it might indicate that Ubc13/CHIP might edit pre-existing, βTrCP-derived, ubiquitin chains on GHR. The opposite could be envisioned if we consider that the WD40 propeller domain of βTrCP was identified as a ubiquitin-binding domain, indicating that βTrCP might also recognize a ubiquitin moiety as its substrate (57). However, if we deplete CHIP, GHR is still ubiquitinated, presumably by βTrCP. Thus, CHIP activity is not required for GHR ubiquitination by βTrCP, suggesting that CHIP does not initiate this ubiquitination. Together, these observations lead to a model in which SCFβTrCP binding to the GHR initiates the endocytosis followed by the CHIP activity that results in clathrin-mediated endocytosis.
CHIP is known to function in the molecular chaperone system together with Hsp90 and Hsp70 (58, 59). For the mineralocorticoid receptor, it was reported that inhibition of Hsp90 results in increased CHIP-mediated ubiquitination and subsequent proteasomal degradation of the receptor (60). In another study, by Wang et al. (20), it was shown that Hsp70 competes with SMAD1/5 for the TPR-mediated binding of CHIP and subsequent ubiquitination of SMAD1/5. These examples show that under certain stress conditions affecting the activity of Hsp70/90, the activity of CHIP toward other substrates might increase. In such a model, higher cellular concentration of “free” CHIP might accelerate GHR endocytosis and negatively regulate the GH sensitivity of cells.
Acknowledgments
We thank Matthias Mayer for help with the CHIP production, Douglas Cyr for the Myc-CHIP constructs, and James Chen for the Ubc13 constructs. We thank all other members of the GHR group for the critical discussions.
This work was supported by the Netherlands Proteomics Centre NPC3.1, European Network of Excellence Grant LSHG-CT-2005-018683, and Marie Curie Network Grant MRTN-CT-2006-034555.
Ana C. da Silva Almeida, Henry G. Hocking, Rolf Boelens, Ger J. Strous, and Agnes G. S. H. van Rossum, manuscript in preparation.
P. van Kerkhof, personal communication.
- GHR
- growth hormone receptor
- GH
- growth hormone
- UbE
- ubiquitin-dependent endocytosis
- CHIP
- COOH terminus of Hsp70-interacting protein
- E3
- ubiquitin-protein isopeptide ligase
- E2
- ubiquitin carrier protein
- βTrCP
- β-transducing repeat-containing protein
- TPR
- tetratricopeptide repeat
- UBD
- ubiquitin-binding domain.
REFERENCES
- 1. Brown R. J., Adams J. J., Pelekanos R. A., Wan Y., McKinstry W. J., Palethorpe K., Seeber R. M., Monks T. A., Eidne K. A., Parker M. W., Waters M. J. (2005) Model for growth hormone receptor activation based on subunit rotation within a receptor dimer. Nat. Struct. Mol. Biol. 12, 814–821 [DOI] [PubMed] [Google Scholar]
- 2. Gent J., van Kerkhof P., Roza M., Bu G., Strous G. J. (2002) Ligand-independent growth hormone receptor dimerization occurs in the endoplasmic reticulum and is required for ubiquitin system-dependent endocytosis. Proc. Natl. Acad. Sci. U.S.A. 99, 9858–9863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhu T., Goh E. L., Graichen R., Ling L., Lobie P. E. (2001) Signal transduction via the growth hormone receptor. Cell Signal. 13, 599–616 [DOI] [PubMed] [Google Scholar]
- 4.Deleted in proof
- 5. Govers R., ten Broeke T., van Kerkhof P., Schwartz A. L., Strous G. J. (1999) Identification of a novel ubiquitin conjugation motif, required for ligand-induced internalization of the growth hormone receptor. EMBO J. 18, 28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van Kerkhof P., Putters J., Strous G. J. (2007) The ubiquitin ligase SCF(βTrCP) regulates the degradation of the growth hormone receptor. J. Biol. Chem. 282, 20475–20483 [DOI] [PubMed] [Google Scholar]
- 7. van Kerkhof P., Westgeest M., Hassink G., Strous G. J. (2011) SCF(TrCP) acts in endosomal sorting of the GH receptor. Exp. Cell Res. 317, 1071–1082 [DOI] [PubMed] [Google Scholar]
- 8. Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., Chen Z. J. (2000) Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103, 351–361 [DOI] [PubMed] [Google Scholar]
- 9. Brun J., Chiu R., Lockhart K., Xiao W., Wouters B. G., Gray D. A. (2008) hMMS2 serves a redundant role in human PCNA polyubiquitination. BMC Mol. Biol. 9, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vina-Vilaseca A., Sorkin A. (2010) Lysine 63-linked polyubiquitination of the dopamine transporter requires WW3 and WW4 domains of Nedd4-2 and UBE2D ubiquitin-conjugating enzymes. J. Biol. Chem. 285, 7645–7656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kramer L. B., Shim J., Previtera M. L., Isack N. R., Lee M. C., Firestein B. L., Rongo C. (2010) UEV-1 is an ubiquitin-conjugating enzyme variant that regulates glutamate receptor trafficking in C. elegans neurons. PLoS ONE 5, e14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Paiva S., Vieira N., Nondier I., Haguenauer-Tsapis R., Casal M., Urban-Grimal D. (2009) Glucose-induced ubiquitylation and endocytosis of the yeast Jen1 transporter. Role of lysine 63-linked ubiquitin chains. J. Biol. Chem. 284, 19228–19236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kamsteeg E. J., Hendriks G., Boone M., Konings I. B., Oorschot V., van der Sluijs P., Klumperman J., Deen P. M. (2006) Short-chain ubiquitination mediates the regulated endocytosis of the aquaporin-2 water channel. Proc. Natl. Acad. Sci. U.S.A. 103, 18344–18349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Varghese B., Barriere H., Carbone C. J., Banerjee A., Swaminathan G., Plotnikov A., Xu P., Peng J., Goffin V., Lukacs G. L., Fuchs S. Y. (2008) Polyubiquitination of prolactin receptor stimulates its internalization, postinternalization sorting, and degradation via the lysosomal pathway. Mol. Cell. Biol. 28, 5275–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu Z., Kohli E., Devlin K. I., Bold M., Nix J. C., Misra S. (2008) Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes. BMC Struct. Biol. 8, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang M., Windheim M., Roe S. M., Peggie M., Cohen P., Prodromou C., Pearl L. H. (2005) Chaperoned ubiquitylation. Crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell 20, 525–538 [DOI] [PubMed] [Google Scholar]
- 17. Soss S. E., Yue Y., Dhe-Paganon S., Chazin W. J. (2011) E2 conjugating enzyme selectivity and requirements for function of the E3 ubiquitin ligase CHIP. J. Biol. Chem. 286, 21277–21286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ballinger C. A., Connell P., Wu Y., Hu Z., Thompson L. J., Yin L. Y., Patterson C. (1999) Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 19, 4535–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Graf C., Stankiewicz M., Nikolay R., Mayer M. P. (2010) Insights into the conformational dynamics of the E3 ubiquitin ligase CHIP in complex with chaperones and E2 enzymes. Biochemistry 49, 2121–2129 [DOI] [PubMed] [Google Scholar]
- 20. Wang L., Liu Y. T., Hao R., Chen L., Chang Z., Wang H. R., Wang Z. X., Wu J. W. (2011) Molecular mechanism of the negative regulation of Smad1/5 protein by carboxyl terminus of Hsc70-interacting protein (CHIP). J. Biol. Chem. 286, 15883–15894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McDonough H., Charles P. C., Hilliard E. G., Qian S. B., Min J. N., Portbury A., Cyr D. M., Patterson C. (2009) Stress-dependent Daxx-CHIP interaction suppresses the p53 apoptotic program. J. Biol. Chem. 284, 20649–20659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nelson R. F., Glenn K. A., Miller V. M., Wen H., Paulson H. L. (2006) A novel route for F-box protein-mediated ubiquitination links CHIP to glycoprotein quality control. J. Biol. Chem. 281, 20242–20251 [DOI] [PubMed] [Google Scholar]
- 23. Nie L., Wu H., Sun X. H. (2008) Ubiquitination and degradation of Tal1/SCL are induced by notch signaling and depend on Skp2 and CHIP. J. Biol. Chem. 283, 684–692 [DOI] [PubMed] [Google Scholar]
- 24. Parsons J. L., Tait P. S., Finch D., Dianova I. I., Allinson S. L., Dianov G. L. (2008) CHIP-mediated degradation and DNA damage-dependent stabilization regulate base excision repair proteins. Mol. Cell 29, 477–487 [DOI] [PubMed] [Google Scholar]
- 25. Tetzlaff J. E., Putcha P., Outeiro T. F., Ivanov A., Berezovska O., Hyman B. T., McLean P. J. (2008) CHIP targets toxic α-synuclein oligomers for degradation. J. Biol. Chem. 283, 17962–17968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Strous G. J., van Kerkhof P., Govers R., Ciechanover A., Schwartz A. L. (1996) The ubiquitin conjugation system is required for ligand-induced endocytosis and degradation of the growth hormone receptor. EMBO J. 15, 3806–3812 [PMC free article] [PubMed] [Google Scholar]
- 27. Schantl J. A., Roza M., De Jong A. P., Strous G. J. (2003) Small glutamine-rich tetratricopeptide repeat-containing protein (SGT) interacts with the ubiquitin-dependent endocytosis (UbE) motif of the growth hormone receptor. Biochem. J. 373, 855–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nickerson D. P., Brett C. L., Merz A. J. (2009) Vps-C complexes. Gatekeepers of endolysosomal traffic. Curr Opin Cell Biol. 21, 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Dam E. M., Ten Broeke T., Jansen K., Spijkers P., Stoorvogel W. (2002) Endocytosed transferrin receptors recycle via distinct dynamin and phosphatidylinositol 3-kinase-dependent pathways. J. Biol. Chem. 277, 48876–48883 [DOI] [PubMed] [Google Scholar]
- 30. Baxter R. C. (1985) Measurement of growth hormone and prolactin receptor turnover in rat liver. Endocrinology 117, 650–655 [DOI] [PubMed] [Google Scholar]
- 31. Murphy L. J., Lazarus L. (1984) The mouse fibroblast growth hormone receptor. Ligand processing and receptor modulation and turnover. Endocrinology 115, 1625–1632 [DOI] [PubMed] [Google Scholar]
- 32. Roupas P., Herington A. C. (1988) Intracellular processing of growth hormone receptors by adipocytes in primary culture. Mol. Cell. Endocrinol. 57, 93–99 [DOI] [PubMed] [Google Scholar]
- 33. van Kerkhof P., Smeets M., Strous G. J. (2002) The ubiquitin-proteasome pathway regulates the availability of the GH receptor. Endocrinology 143, 1243–1252 [DOI] [PubMed] [Google Scholar]
- 34. Grove D. E., Rosser M. F., Ren H. Y., Naren A. P., Cyr D. M. (2009) Mechanisms for rescue of correctable folding defects in CFTRΔ F508. Mol. Biol. Cell 20, 4059–4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meacham G. C., Patterson C., Zhang W., Younger J. M., Cyr D. M. (2001) The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 3, 100–105 [DOI] [PubMed] [Google Scholar]
- 36. van den Eijnden M. J., Lahaye L. L., Strous G. J. (2006) Disulfide bonds determine growth hormone receptor folding, dimerisation and ligand binding. J. Cell Sci. 119, 3078–3086 [DOI] [PubMed] [Google Scholar]
- 37. Waterman H., Levkowitz G., Alroy I., Yarden Y. (1999) The RING finger of c-Cbl mediates desensitization of the epidermal growth factor receptor. J. Biol. Chem. 274, 22151–22154 [DOI] [PubMed] [Google Scholar]
- 38. Cliff M. J., Williams M. A., Brooke-Smith J., Barford D., Ladbury J. E. (2005) Molecular recognition via coupled folding and binding in a TPR domain. J. Mol. Biol. 346, 717–732 [DOI] [PubMed] [Google Scholar]
- 39. Putters J., da Silva Almeida A. C., van Kerkhof P., van Rossum A. G., Gracanin A., Strous G. J. (2011) Jak2 is a negative regulator of ubiquitin-dependent endocytosis of the growth hormone receptor. PLoS ONE 6, e14676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rosser M. F., Washburn E., Muchowski P. J., Patterson C., Cyr D. M. (2007) Chaperone functions of the E3 ubiquitin ligase CHIP. J. Biol. Chem. 282, 22267–22277 [DOI] [PubMed] [Google Scholar]
- 41. Maldonado-Báez L., Wendland B. (2006) Endocytic adaptors. Recruiters, coordinators and regulators. Trends Cell Biol. 16, 505–513 [DOI] [PubMed] [Google Scholar]
- 42. Polo S., Sigismund S., Faretta M., Guidi M., Capua M. R., Bossi G., Chen H., De Camilli P., Di Fiore P. P. (2002) A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416, 451–455 [DOI] [PubMed] [Google Scholar]
- 43. van Delft S., Govers R., Strous G. J., Verkleij A. J., van Bergen en Henegouwen P. M. (1997) Epidermal growth factor induces ubiquitination of Eps15. J. Biol. Chem. 272, 14013–14016 [DOI] [PubMed] [Google Scholar]
- 44. Hoeller D., Crosetto N., Blagoev B., Raiborg C., Tikkanen R., Wagner S., Kowanetz K., Breitling R., Mann M., Stenmark H., Dikic I. (2006) Regulation of ubiquitin-binding proteins by monoubiquitination. Nat. Cell Biol. 8, 163–169 [DOI] [PubMed] [Google Scholar]
- 45. Le Bras S., Loyer N., Le Borgne R. (2011) The multiple facets of ubiquitination in the regulation of notch signaling pathway. Traffic 12, 149–161 [DOI] [PubMed] [Google Scholar]
- 46. Timsit Y. E., Miller S. L., Mohney R. P., O'Bryan J. P. (2005) The U-box ligase carboxyl-terminus of Hsc 70-interacting protein ubiquitylates Epsin. Biochem. Biophys. Res. Commun. 328, 550–559 [DOI] [PubMed] [Google Scholar]
- 47. Kazazic M., Bertelsen V., Pedersen K. W., Vuong T. T., Grandal M. V., Rødland M. S., Traub L. M., Stang E., Madshus I. H. (2009) Epsin 1 is involved in recruitment of ubiquitinated EGF receptors into clathrin-coated pits. Traffic 10, 235–245 [DOI] [PubMed] [Google Scholar]
- 48. Hurley J. H., Stenmark H. (2011) Molecular mechanisms of ubiquitin-dependent membrane traffic. Annu Rev. Biophys. 40, 119–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Staub O., Gautschi I., Ishikawa T., Breitschopf K., Ciechanover A., Schild L., Rotin D. (1997) Regulation of stability and function of the epithelial Na+ channel (ENaC) by ubiquitination. EMBO J. 16, 6325–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sehat B., Andersson S., Girnita L., Larsson O. (2008) Identification of c-Cbl as a new ligase for insulin-like growth factor-I receptor with distinct roles from Mdm2 in receptor ubiquitination and endocytosis. Cancer Res. 68, 5669–5677 [DOI] [PubMed] [Google Scholar]
- 51. Huang F., Kirkpatrick D., Jiang X., Gygi S., Sorkin A. (2006) Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell 21, 737–748 [DOI] [PubMed] [Google Scholar]
- 52. Lauwers E., Erpapazoglou Z., Haguenauer-Tsapis R., André B. (2010) The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 20, 196–204 [DOI] [PubMed] [Google Scholar]
- 53. Galan J. M., Haguenauer-Tsapis R. (1997) Ubiquitin Lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 16, 5847–5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 [DOI] [PubMed] [Google Scholar]
- 55. van Kerkhof P., Alves dos Santos C. M., Sachse M., Klumperman J., Bu G., Strous G. J. (2001) Proteasome inhibitors block a late step in lysosomal transport of selected membrane but not soluble proteins. Mol. Biol. Cell 12, 2556–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Windheim M., Peggie M., Cohen P. (2008) Two different classes of E2 ubiquitin-conjugating enzymes are required for the mono-ubiquitination of proteins and elongation by polyubiquitin chains with a specific topology. Biochem. J. 409, 723–729 [DOI] [PubMed] [Google Scholar]
- 57. Pashkova N., Gakhar L., Winistorfer S. C., Yu L., Ramaswamy S., Piper R. C. (2010) WD40 repeat propellers define a ubiquitin-binding domain that regulates turnover of F box proteins. Mol. Cell 40, 433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cyr D. M., Höhfeld J., Patterson C. (2002) Protein quality control. U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem. Sci. 27, 368–375 [DOI] [PubMed] [Google Scholar]
- 59. Murata S., Chiba T., Tanaka K. (2003) CHIP. A quality-control E3 ligase collaborating with molecular chaperones. Int. J. Biochem. Cell Biol. 35, 572–578 [DOI] [PubMed] [Google Scholar]
- 60. Faresse N., Ruffieux-Daidie D., Salamin M., Gomez-Sanchez C. E., Staub O. (2010) Mineralocorticoid receptor degradation is promoted by Hsp90 inhibition and the ubiquitin-protein ligase CHIP. Am. J. Physiol. Renal Physiol. 299, F1462–F1472 [DOI] [PubMed] [Google Scholar]