Background: Caspase-6 is a drug target against neurodegenerative diseases and is suppressed by phosphorylation at Ser257.
Results: S257E mutation inhibits caspase-6 activation by locking the protein in the “inhibited state” and inhibits caspase-6 activity by steric hindrance.
Conclusion: Phosphorylation inhibits caspase-6 through the same mechanism.
Significance: The study revealed the inhibition mechanism of caspase-6 phosphorylation and provided new strategies for drug discovery.
Keywords: Caspase, Crystal Structure, Molecular Dynamics, Neurodegenerative Diseases, Protein Phosphorylation, ARK5
Abstract
The apoptotic effector caspase-6 (CASP6) has been clearly identified as a drug target due to its strong association with neurodegeneration and axonal pruning events as well as its crucial roles in Huntington disease and Alzheimer disease. CASP6 activity is suppressed by ARK5-mediated phosphorylation at Ser257 with an unclear mechanism. In this work, we solved crystal structures of ΔproCASP6S257E and p20/p10S257E, which mimicked the phosphorylated CASP6 zymogen and activated CASP6, respectively. The structural investigation combined with extensive biochemical assay and molecular dynamics simulation studies revealed that phosphorylation on Ser257 inhibited self-activation of CASP6 zymogen by “locking” the enzyme in the TEVD193-bound “inhibited state.” The structural and biochemical results also showed that phosphorylation on Ser257 inhibited the CASP6 activity by steric hindrance. These results disclosed the inhibition mechanism of CASP6 phosphorylation and laid the foundation for a new strategy of rational CASP6 drug design.
Introduction
Caspases, a family of cysteine proteases that cleave substrates after an aspartate residue, are the major executioners of apoptosis and inflammation and are classified into inflammatory caspases, apoptotic initiators, and effectors (1, 2). CASP63 is classified to be an apoptotic effector by sequence similarity, and it is often activated by CASP3 during apoptosis (3). However, CASP6 also undergoes in vitro and in vivo self-activation (4, 5). CASP6 is expressed as the zymogen of a short pro-domain, a large subunit (p20), an intersubunit linker (L), and a small subunit (p10), and is activated by proteolytic processing at either or both intersubunit cleavage sites (4).
Increasing evidence has demonstrated the critical roles of CASP6 in neurodegenerative diseases, including Huntington disease (HD) and Alzheimer disease (AD) (for review, see Ref. 6). In vivo proteolytic cleavage of murine htt at the CASP6 cleavage site is a crucial and rate-limiting event in the pathogenesis of HD (7). Activation of CASP6 is observed before the onset of motor abnormalities in human and murine HD brain, and active CASP6 levels correlate directly with the CAG size and inversely with the age of onset (8). Active CASP6 is abundant in the neuropathological lesions of AD (9, 10). Furthermore, CASP6 mediates axon pruning through the death receptor 6 pathway during neural development (11), and many cytoskeletal proteins are targets of CASP6 (12), suggesting the importance of CASP6 in neurodegenerative processes.
Phosphorylation is critically important to caspase regulation (13). CASP6 is negatively regulated by AMP-activated protein kinase-related kinase 5 (ARK5, also known as NUAK1)-mediated phosphorylation at Ser257 (14), but the mechanism of suppression remains unclear. CASP6 is not inhibited by IAPs (inhibitors of apoptosis) (1, 2), and the alternatively spliced transcript product CASP6β only inhibits the activation of proCASP6, but not the activated CASP6 (15). So far, phosphorylation is the most efficient known inhibition mechanism for CASP6 because it inhibits both the activation and activity of CASP6 (14).
Several crystal structures of zymogen, free active and inhibitor-bound CASP6 have been published (5, 16–19). Our previous work has shown that the intersubunit cleavage site TEVD193 binds in the active site in CASP6 zymogen and revealed a unique self-activation mechanism for CASP6 through intramolecular cleavage at Asp193 (5). However, none of the above studies could explain the inhibitory effect of CASP6 phosphorylation. In this work, we used a CASP6 mutant to mimic the phosphorylated CASP6 and revealed the inhibition mechanism of CASP6 phosphorylation. We also used molecular dynamics (MD) simulations to test the effects of Ser257 mutation and phosphorylation on the structure of CASP6 zymogen, and the results showed that the mutant used in this study is a good mimic of CASP6 phosphorylation.
EXPERIMENTAL PROCEDURES
Mutagenesis of CASP6
The ΔproCASP6S257E and other mutants were generated by overlapping PCR, using WT CASP6 or ΔproCASP6S257E as template. The DVVD179NQ sites of D179Th mutants were replaced by the thrombin cleavage site, LVPR179GS.
Protein Preparation
All CASP6 mutants were cloned in pET21b vector with a C-terminal (His)6 tag and expressed in Escherichia coli Rosetta (DE3) strain at 18 °C for 20 h. Bacterial cells harboring CASP6 mutants were cultured in LB medium supplemented with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol at 37 °C to an A600 of 0.6–0.8. Protein expression was induced with 0.5 mm isopropyl β-d-1-thiogalactopyranoside at 18 °C for 20 h. Cells were harvested and resuspended in 20 mm Tris-HCl, pH 7.5, 500 mm NaCl, and 50 mm imidazole (equilibration buffer). Cells were sonicated and centrifuged at 50,000 × g for 60 min at 4 °C, and soluble fractions were loaded onto a 5-ml HisTrap HP column (GE Healthcare) equilibrated with equilibration buffer. The columns were washed with equilibration buffer containing 100 mm imidazole, and the target proteins were eluted with equilibration buffer containing 500 mm imidazole. ΔproCASP6S257E proteins used for crystallization were further purified by gel filtration (120-ml Superdex-75; GE Healthcare) in buffer containing 20 mm Tris-HCl, pH 7.5, and 150 mm NaCl, and 10 mm dithiothreitol (DTT, final concentration) was added after gel filtration. To obtain fully processed ΔproCASP6S257E, 5 μg/μl purified ΔproCASP6S257E protein was incubated with 5 ng/μl active CASP3 at 4 °C for 2 days. Other CASP6 mutants used for biochemical analysis were purified by nickel-chelating column as described above and transferred to the same final buffer as ΔproCASP6S257E by desalting column (5-ml HiTrap desalting column; GE Healthcare).
Crystallization and Data Collection
Both crystals of ΔproCASP6S257E and p20/p10S257E were grown using the sitting-drop diffusion method. Crystals of ΔproCASP6S257E were obtained by incubating 10 mg/ml protein (in 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 10 mm DTT) in 0.1 m MES, pH 6.6, 0.2 m sodium chloride, and 10% m/v polyethylene glycol (PEG) 3350 at 20 °C. Crystallization solution containing 10% dimethyl sulfoxide was used for cryoprotection. The crystal was flash-frozen and maintained at 100 K by nitrogen gas during data collection. Diffraction data were collected at a wavelength of 0.98 Å on the Beamline BL5A at the KEK, Photon Factory, Tsukuba, Japan. The data were processed by the program XDS (20). The crystals belong to space group P6522.
Crystals of p20/p10S257E were obtained by incubating 2 mg/ml protein (in 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 10 mm DTT) in 20 mm HEPES, pH 7.5, 2.0 m NaCl at 20 °C. Crystallization solution containing 10% glycerol was used for cryoprotection. The crystal was flash-frozen and maintained at 100 K by nitrogen gas during data collection. Diffraction data were collected at a wavelength of 0.98 Å on the Beamline BL17U at the SSRF (Shanghai Synchrotron Radiation facility), Shanghai, China. The data were processed by HKL2000 (21). The crystals belong to space group P21.
Structure Determination and Refinement
Both structures were determined by molecular replacement calculations with PHENIX AutoMR (22) using ΔproCASP6C163A dimer (Protein Data Bank ID code 3NR2) as the search model. The models were completed with the graphics program Coot (23) and refined using PHENIX (22). The refinement statistics are summarized in Table 1. The PDB access code for ΔproCASP6S257E is 3V6L and for p20/p10S257E is 3V6M. The data processing and refinement statistics are summarized in Table 1.
TABLE 1.
Data collection and statistics from crystallographic analysis
Values in parentheses are for the highest resolution shell.
| Parameter | ΔproCASP6S257E | p20/p10S257E |
|---|---|---|
| Crystal parameters | ||
| Wavelength (Å) | 0.98 | 0.98 |
| Space group | P6522 | P21 |
| Cell dimension | ||
| a, b, c (Å) | 126.4, 126.4, 165.8 | 81.1, 162.6 89.2 |
| α, β, γ (°) | 90, 90, 120 | 90, 95, 90 |
| Data collection | ||
| Resolution range (Å) | 50–2.2 (2.3–2.2) | 50–2.7 (2.75–2.7) |
| Rsyma (%) | 9.3 (66.1) | 8.3 (64.4) |
| Mean I/σI | 39.7 (7.2) | 38.1 (7.4) |
| Completeness (%) | 99.1 (93.7) | 93.7 (100) |
| Redundancy | 40.9 | 6.1 |
| Refinement statistics | ||
| Resolution range (Å) | 20–2.2 | 20–2.7 |
| Number of reflections | 39831 | 59021 |
| Rworkb/Rfreeb,c (%) | 16.9/20.2 | 21.5/25.9 |
| Average B-factors | 41.3 | 52.40 |
| Root mean square deviationd | ||
| Bond lengths (Å) | 0.008 | 0.015 |
| Bond angles (°) | 1.052 | 1.385 |
| Ramachandran plots | ||
| Most favored (%) | 96.6 | 95.5 |
| Allowed (%) | 3.2 | 4.5 |
| Disallowed (%) | 0.2 | 0 |
a Rsym = Σ|Iobs−Iavg|/ΣIobs.
b Rwork, free = Σ‖Fobs|−|Fcalc‖/Σ|Fobs|.
c Rfree values are calculated for a randomly selected 5% of the data that was excluded from the refinement.
d Root mean square deviation from ideal/target geometries.
MD Simulation of CASP6
The SANDER and PMEMD modules of AMBER 10.0 package (24) were utilized in simulations in conjunction with the AMBER ff03 all-atom force field (25) in an NVT ensemble. All MD simulations were performed as explicit solvation with approximately 3500 SPC/E waters (26). All MD simulations employed an integral time step of 2 fs, restraint of all hydrogen-containing bonds through the SHAKE algorithm (27), a cutoff of 10 Å for nonbonded interactions, and particle mesh Ewald treatment of long range electrostatic interactions (28). Because in the experimental structure, ΔproCASP6S257E, parts of the L2 and L4 loops are flexible and their electron density is missing, we first used the SWISS-MODEL server (29) to build the missing parts. To obtain a relaxed loop structure, a 2-ns MD simulation at 600 K with a constrained protein structure except for the L2 loop was performed. Equilibration simulation at 300 K was then performed starting from the resulting structure. The simulation temperature was maintained at 300 K using Langevin dynamics with a friction coefficient of 5 ps−1.
Self-activation Analysis of CASP6 Variants
Purified ΔproCASP6S257E and proCAPS6S257E were diluted to 1 μg/μl in the assay buffer (20 mm HEPES, pH 7.4, 50 mm NaCl, 2 mm EDTA, 0.1% CHAPS, and 5 mm DTT) and incubated at 25 °C for 15 days. The samples were analyzed by 15% SDS-PAGE.
Purified ΔproCASP6S257E and other mutants were diluted to 0.2 μg/μl in the assay buffer containing 2 μg/μl bovine serum albumin (BSA) and incubated at 37 °C for 14 h. Samples were separated by 15% SDS-PAGE, transferred to Immobilon-P polyvinylidene fluoride (PVDF) membranes (Millipore), and probed with 1:40,000 dilution of rabbit anti-CASP6 serum, 1:5,000 dilution of secondary anti-rabbit IgG-HRP (MBL), and detected with Metal Enhanced DAB Substrate kit (Thermo Scientific).
Proteolytic Processing of ΔproCASP6D179Th Variants by Thrombin
Purified ΔproCASP6D179Th variants were diluted to 0.5 μg/μl and incubated with 5 nanounits/μl thrombin (GE Healthcare) in the assay buffer at 25 °C for 24 h. Samples were analyzed by 15% SDS-PAGE.
Enzyme Activity Analysis of Processed CASP6 Variants
To obtain processed CASP6 variants, 0.5 μg/μl purified proteins were incubated with 0.5 ng/μl active CASP3 in the assay buffer at 25 °C for 14 h. Protein solutions were sampled and analyzed by 15% SDS-PAGE. The activities of processed proteins were assessed by fluorogenic assay using Ac-Val-Glu-Ile-Asp-7-amino-4-trifluoro-methylcoumarin (Ac-VEID-AFC) as substrate. The assay was performed with 50 μm Ac-VEID-AFC in a 96-well plate with a reaction volume of 100 μl, and each well contained 50 ng of processed proteins and 500 ng of BSA in the assay buffer. The activities were measured by infinite M200 multimode microplate reader (TECAN) with wavelengths of 400 nm for excitation and 505 nm for emission. The results were read at intervals of 1 min for up to 60 min. Fluorescence units were converted to the amount of moles of AFC released based on a standard curve of 0–20 μm free AFC. Cleavage rates were calculated from the linear phase of the assay.
RESULTS
To investigate the phosphorylation regulation mechanism of CASP6, Ser257 was mutated to glutamate to mimic the phosphorylated serine residue. The resulting mutant, proCASP6S257E, was only self-cleaved at Asp23, not at Asp193 or Asp179 (supplemental Fig. S1A), suggesting that Ser257 phosphorylation inhibits CASP6 self-activation. The pro-domain deleted mutant ΔproCASP6S257E did not undergo self-activation even after 15 days of incubation at 25 °C (supplemental Fig. S1B) and was used for crystallization. The structure was determined at 2.2 Å and refined to R factors Rwork/Rfree of 16.9%/20.2%. The data collection and statistics from crystallographic analysis are summarized in Table 1.
Structure Analyses of ΔproCASP6S257E
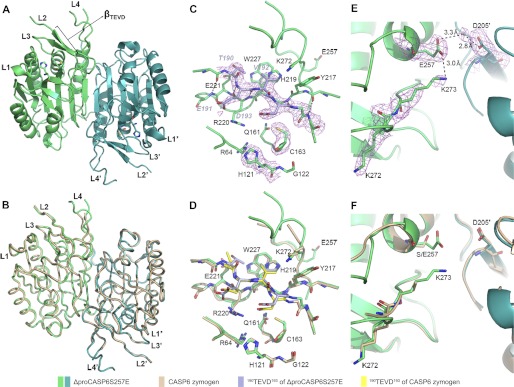
ΔproCASP6S257E exists as homodimer with each monomer assembled into a central six-stranded β-sheet flanked by five α-helices and two small β-strands, with four loops (L1–L4) protruding from the central β-sheet, forming the active site (Fig. 1A). The structure of ΔproCASP6S257E is almost identical to CASP6 zymogen (5), with root mean square deviations of 0.41 Å for all aligned Cα atoms (Fig. 1B). The intersubunit cleavage site TEVD193 also binds in the active site as a β-strand (βTEVD) in the ΔproCASP6S257E. The βTEVD containing ITEVDAA195 forms an anti-parallel β-sheet with YYSHRET222 of the L3 loop through six main chain hydrogen bonds (Fig. 1C). Superimposing the active sites of ΔproCASP6S257E and the zymogen, we found that residues forming the substrate binding pockets and TEVD193 both overlapped very well (Fig. 1D). The mutation site, S257E, is at the end of the helix connecting to the L4 loop. The Glu257 forms a “salt bridge” with Lys273 (Fig. 1E). By contrast, in CASP6 zymogen, Ser257 cannot stabilize Lys273, and the side chain of Lys273 is flexible without intact electron density (Fig. 1F and supplemental Fig. S2). These observations indicate that the structures of ΔproCASP6S257E and the zymogen are in the same conformation. The fact that ΔproCASP6S257E has the intact catalytic dyad Cys163/His121 but cannot undergo self-cleavage suggests that both the two structures are in the “inhibited state.” However, in the zymogen, certain conformational changes can trigger the intramolecular self-cleavage, whereas in ΔproCASP6S257E, the single mutation “locks” the protein in the inhibited state, and the salt bridge between Glu257 and Lys273 may play a key role for the locking mechanism.
FIGURE 1.
Structure of ΔproCASP6S257E. A, overall structure of ΔproCASP6S257E. B, structure overlay of ΔproCASP6S257E and CASP6 zymogen. C, active sites of ΔproCASP6S257E. D, active sites overlay of ΔproCASP6S257E and CASP6 zymogen. E and F, residues surrounding 257 site in ΔproCASP6S257E (E) and in structure overlay of ΔproCASP6S257E and CASP6 zymogen (F). The electron density map (2Fo − Fc maps) was shown at 1.0 σ, calculated by PHENIX.refine. The salt bridge and hydrogen bonds are represented by black dashed lines.
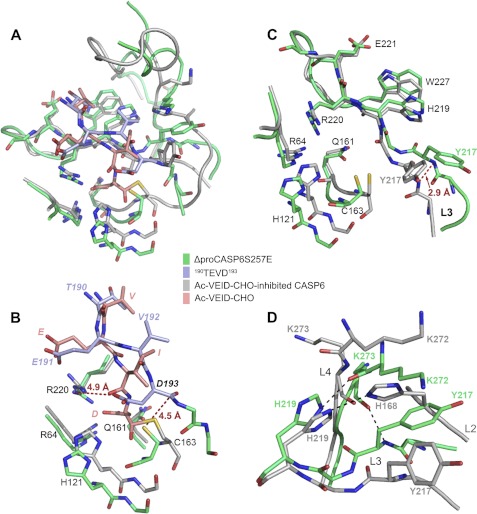
Compared with the Ac-VEID-CHO bound CASP6 (representing fully active enzyme, in the “active state”), in ΔproCASP6S257E, Asp193 does not fully bind in the S1 pocket (constructed by Arg64, Asn161, and Arg220; Fig. 2A). In the Ac-VEID-CHO-bound CASP6, the Arg64 and Arg220 are engaged in strong salt bridges with the P1 aspartate, which is further hydrogen-bonded to Asn161 (Fig. 2B). The carbonyl group of the P1 aspartate covalently bonds to sulfur of Cys163, and the carbonyl oxygen is stabilized in the “oxyanion hole” formed by the main chain amino nitrogen of Cys163 and imidazolyl nitrogen of His121. In contrast, in ΔproCASP6S257E, the side chain carboxyl oxygen of Asp193 is approximately 5 Å from the guanidinium group of Arg220 and even farther away from Arg64 and Asn161. The distance between the sulfur of Cys163 and the main chain carbonyl of Asp193 is approximately 4.5 Å, and the main chain carbonyl oxygen of Asp193 points to the opposite direction of the oxyanion hole. At this conformation, it is hard for Cys163 to nucleophilically attack the main chain carbonyl of Asp193 and to initiate the self-cleavage. Furthermore, the L3 loops of the two structures deviate at His219, and the main chain amino nitrogen of Tyr217 split about 2.9 Å (Fig. 2C). In ΔproCASP6S257E, the six hydrogen bonds between TEVD193 and the L3 loop fasten this cleavage site on the L3 loop, and therefore the conformation of TEVD193 varies with the conformation of the L3 loop. The phenolic group of Try217 hydrophobically interacts with the side chain of Lys272, and the imidazolyl nitrogen of His219 and the main chain amino nitrogen of Tyr217 form two hydrogen bonds with the main chain ketonic oxygens of Lys272 and Lys273, respectively (Fig. 2D). These interactions lead to the coupling between the L3 and L4 loops. More importantly, Glu257 forms a salt bridge with Lys273, which stabilizes the side chain of Lys273 and further locks TEVD193 in the inhibited state conformation. In Ac-VEID-CHO-bound CASP6, His168 of the L2 loop forms a hydrogen bond with His219, and with the insertion of His168 between the L3 and L4 loops, these two loops do not directly interact. These observations indicate that after cleavage at Asp179, the conformational changes of the L2 loop could break the interaction network between the L3 and L4 loops and release the CASP6 from the inhibited state.
FIGURE 2.
Active sites overlay of ΔproCASP6S257E and Ac-VEID-CHO bound CASP6. A–D, structure superimposition of whole active sites (A), substrates and S1 pockets (B), L3 loops, the S1 pockets, and the Cys/His dyads (C), and L3–L4 loop interaction (D). The black dashed lines represent the hydrogen bonds, and red dashed lines show the distance between two atoms.
MD Simulations Show Good Resemblance between S257E Mutation and Phosphorylation
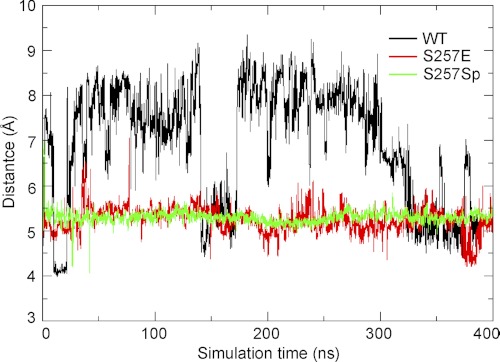
To test how well the S257E mutation resembles phosphorylation and to gain molecular detailed understanding of the effects of phosphorylation on the activation of CASP6, we performed MD simulations using the ΔproCASP6S257E crystal structure, the structure with the mutation of Glu257 to phosphorylated serine (representing the phosphorylated CASP6 zymogen), and the structure with Glu257 mutated back to serine (representing the WT CASP6 zymogen), respectively. The simulation results showed that the fluctuation of the distance between the sulfur of Cys163 and the main chain carbonyl of Asp193 in ΔproCASP6S257E or phosphorylated CASP6 zymogen is smaller than that in WT CASP6 zymogen (Fig. 3). The results indicated that both S257E mutation and phosphorylation have similar effects on freezing the motions of the active site, and the active site of the WT zymogen is much more flexible. The results suggested that S257E mutation is a good mimic of Ser257 phosphorylation and further confirmed that the self-activation of CASP6 zymogen is inhibited through the locking mechanism.
FIGURE 3.
MD simulation of ΔproCASP6S257E, phosphorylated CASP6 zymogen, and WT CASP6 zymogen. The distance between the sulfur of Cys163 and main chain carbonyl of Asp193 in three different mutant/state of CASP6 zymogen were calculated by MD simulation and are shown on the histogram as a function of time. S257Sp, Ser257 mutated to phosphorylated serine.
Biochemical Assays Agreed with Structure Analyses
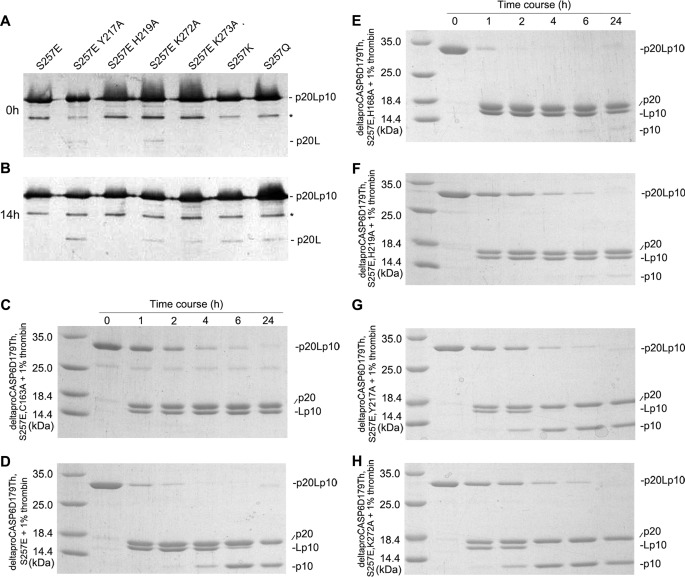
To determine further the specific contribution of each residue, the key residues involved in the interaction network around Glu257 were mutated to alanine separately. Purified ΔproCASP6S257E and ΔproCASP6(S257E,H219A) proteins did not self-activate (Fig. 4, A and B). By contrast, ΔproCASP6(S257E,Y217A) and ΔproCASP6(S257E,K272A) already contained little self-cleaved enzyme before incubation, indicating that these two mutants underwent self-activation during purification. These results demonstrated that CASP6S257E regained self-activation ability by breaking the hydrophobic interaction between Tyr217 and Lys272 and indicated that this hydrophobic interaction was essential in constructing the network. Furthermore, to disrupt the salt bridge between Glu257 and Lys273, Lys273 was mutated to alanine, or Glu257 was mutated to lysine or glutamine. The resulting ΔproCASP6(S257E,K273A), ΔproCASP6S257K, and ΔproCASP6S257Q also regained self-activation ability, which confirmed the key role of salt bridge between Glu257 and Lys273 in the inhibition mechanism.
FIGURE 4.
A and B, self-activation analysis of CASP6 variants from 0 (A) to 14 h (B) by Western blotting. The asterisk-labeled bands were a bacterial contamination protein. C–H, Coomassie Blue-stained SDS-polyacrylamide gels of ΔproCasp6S257E,D179Th,C163A (C), ΔproCasp6S257E,D179Th (D), ΔproCasp6S257E,D179Th,H168A (E), ΔproCasp6S257E,D179Th,H219A (F), ΔproCasp6S257E,D179TH,Y217A (G), and ΔproCasp6S257E,D179Th,K272A (H) incubated with thrombin for 24 h.
To investigate the influence of cleavage at DVVD179 on the phosphorylation-inhibited CASP6, DVVD179NQ was replaced by the thrombin cleavage site, LVPR179GS, and the resulting mutants will be referred to as D179Th. These mutants were cleaved at their thrombin sites and transferred to their active form, p20/Lp10, by incubating with thrombin. Thrombin could not cleave the TEVD193 site, which was evidence that thrombin did not cleave ΔproCASP6(S257E,D179Th,C163A) at TEVD193 (Fig. 4C). Notably, the TEVD193 site of ΔproCASP6(S257E,D179Th) was cleaved, as the p10 band appearing after the D179Th site was cleaved (Fig. 4D), indicating that ΔproCASP6S257E could self-cleave at TEVD193 after the 179 site was cleaved.
Structure analyses suggest that after cleavage at Asp179, the rotation of the L2 loop results in the insertion of His168 between the L3 and L4 loops and breaking their interaction network. Indeed, after cleavage at the D179Th site, ΔproCASP6(S257E,D179Th,H168A) and ΔproCASP6(S257E,D179Th,H219A) (Fig. 4, E and F) self-cleaved at Asp193 much slower than ΔproCASP6(S257E,D179Th), indicating that the formation of the hydrogen bond between His168 and His219 was crucial to the network disruption. By contrast, ΔproCASP6(S257E,D179Th,Y217A) and ΔproCASP6(S257E,D179Th,K272A) self-cleaved at Asp193 much faster (Fig. 4, G and H), further confirming the dominant role of the hydrophobic interaction between Tyr217 and Lys272 in constructing the network. The quantitative data of thrombin proteolytic processing assay (Fig. 4, C–H) are summarized in supplemental Table S1.
These results all agree with the conclusions drawn from the structural analyses which showed that CASP6 is locked in the inhibited state through the interaction network among TEVD193, the L3 and the L4 loops, as well as the salt bridge between Glu257 and Lys273. Meanwhile, the cleavage at Asp179 followed by insertion of His168 between the L3 and L4 loops breaks the interaction network, and further releases CASP6 from the inhibited state.
S257E Inhibits Activity of Cleaved CASP6 by Steric Hindrance
To investigate whether phosphorylation inhibits the activity of activated CASP6, ΔproCASP6S257E was processed by 0.1% active CASP3 for 14 h (supplemental Fig. S3A). The activity of the processed protein (referred as p20/p10S257E in the following discussion) was approximately 0.4% of that of WT CASP6 and was one third that of proCASP6D(23,179,193)A (representing the activity of CASP6 zymogen, supplemental Fig. S3B). The low activity of p20/p10S257E suggests that phosphorylation at Ser257 also inhibits the activity of activated CASP6. Furthermore, the activities of CASP3-processed ΔproCASP6(S257E,Y217A), ΔproCASP6(S257E,K272A), and ΔproCASP6(S257E,K273A) were as low as ΔproCASP6S257E (supplemental Fig. S3B), indicating the mutations that regained CASP6 self-activation ability cannot increase the activity of CASP6S257E and suggesting that phosphorylation inhibited the activity of processed CASP6 through a different mechanism.
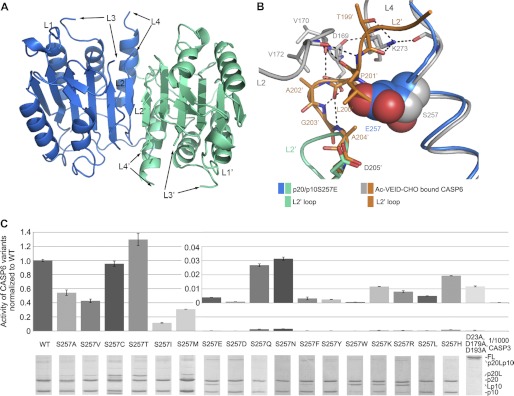
The p20/p10S257E was crystallized for structure determination. The crystals belong to space group P21, with four (p20/p10)2 dimers in one asymmetric unit. The four CASP6 dimers in the same asymmetric unit are almost identical, with root mean square deviations of between 0.11 and 0.22 Å for all aligned Cα atoms. For the following discussion, we will only refer to the A/B dimer. The structure of p20/p10S257E is very similar to that of free active CASP6 (18), with typical caspase α/β-fold, but most parts of the loops forming the active sites are flexible without electron density, namely residues 164–179, 194–197 of L2, residues 213–221 of L3, and residues 260–274 of L4 (Fig. 5A). In the free active CASP6, the N terminus of p10 is oriented toward the dimer interface (18), and substrate binding induces the N terminus of p10 to rotate 180° and form a loop bundle with the L2 and L4 loops as observed in the Ac-VEID-CHO-bound CASP6. Superimposing the structures of p20/p10S257E and Ac-VEID-CHO bound CASP6, we found that the loop bundle is well formed in the inhibitor-bound CASP6; however, the mutation site Glu257 of p20/p10S257E overlapping with the L2′ loop of inhibitor bound CASP6 (Fig. 5B). These observations indicate that with the S257E mutation, the loop bundle cannot be formed because of steric hindrance, which agrees well with the low activity of p20/p10S257E.
FIGURE 5.
S257E inhibits the activity of cleaved CASP6 by steric hindrance. A, overall structure of p20/p10S257E. B, superimposition of loop bundle region in Ac-VEID-CHO-bound CASP6 and p20/p10S257E. The hydrogen bonds are represented by black dashed lines, and Glu/Ser257 are shown as spheres. C, VEIDase activity of CASP6 variants. The activities of processed CASP6 variants were normalized to WT CASP6. The activities with low values are also shown in a histogram with appropriate maximum scale of vertical axis. Assays were done in triplicate, and error bars represent S.D. The activity of 1/1000 CASP3 is shown as a negative control. The processed CASP6 variants were analyzed by SDS-PAGE shown below the histogram.
To prove this hypothesis, Ser257 was mutated to 17 other amino acids except for proline and glycine, and the mutants were processed by CASP3. All of the mutants with small side chains, including S257A, S257V, S257C,S257T, and S257I, had a relatively high activity, comparable with WT CASP6; whereas all of the mutants with big side chains, including S257E, S257D, S257Q, S257N, S257F, S257Y, S257W, S257K, S257R, S257L, and S257H had a relatively low activity, <3% activity of the WT CASP6, and comparable with proCASP6D(23,179,193)A (Fig. 5C). The mutant S257M was the only exception; it has a long side chain, but its activity was relatively high, which may be due to the side chain flexibility of methionine. These results are consistent with the notion that the activities of CASP6 mutants depended on the side chain size of the 257 residue, confirming that phosphorylation inhibited CASP6 activity by steric hindrance.
DISCUSSION
In this work, we used the S257E mutation to mimic the Ser257-phosphorylated CASP6. Our structural and biochemical results revealed the S257E mutation inhibited self-activation of CASP6 zymogen and the activity of processed CASP6 through two different mechanisms. It inhibited self-activation of CASP6 zymogen by locking the CASP6 in the TEVD193-bound inhibited state. The salt bridge between Glu257 and Lys273 fixed the position of Lys273 and further locked the zymogen in the inhibited state through the interaction network among TEVD193, the L3 loop, and the L4 loop. It inhibited the activity of processed CASP6 by steric hindrance because the side chain of Glu257 hindered the formation of loop bundle, which is crucial for the activities of caspases in general. Based on the fact that phosphorylated serine has the same charge and a similar size with a glutamate, we propose that Ser257 phosphorylation inhibits CASP6 through the same mechanism as S257E mutation. This proposal was further validated by MD simulations, which showed that both the S257E mutant and the WT CASP6 with the phosphorylated Ser257 show similar structural and dynamical properties. Compared with the unphosphorylated WT protein, the active site in both protein forms became less flexible and prevented the close approach between the Cys163 and the Asp193, which is essential for the self-activation of the enzyme. ARK5 inhibits CASP6 activation by phosphorylation at Ser257 in vitro and in vivo, and this phosphorylation also suppresses the CASP3-mediated activation of CASP6 (14). The inhibitory mechanism of CASP6 phosphorylation revealed by our structural, biochemical, and MD simulation results agreed very well with those early studies.
This study has extended our understanding of the molecular mechanism of CASP6 activation and regulation. Our previous study revealed that CASP6 could undergo self-activation through intramolecular cleavage at Asp193, and we have proposed that the ordered TEVD193 conformation has both activation and inhibition function in regulating CASP6 activity (5). The presence of TEVD193 in the active site of zymogen inhibits the activation and activity of CASP6 by excluding other substrate binding and preventing this site from intermolecular cleavage; meanwhile, because TEVD193 is located very close to the catalytic site, a small conformational change would be enough to induce the intramolecular self-cleavage. The phosphorylated and unphosphorylated CASP6 zymogens are in the same TEVD193-bound inhibited state, but they have different fates. The unphosphorylated zymogen can initiate self-activation through certain conformational change, whereas the phosphorylated zymogen is locked in the inhibited state. These results further suggest that dephosphorylation by a specific phosphatase may be a trigger to the intramolecular self-cleavage.
Crucial roles for CASP6 in HD and AD have been well documented (6), and regulating the activation and activity of CASP6 could be an efficient method to develop pharmaceutical therapy against these neurodegenerative disorders. Most of reported caspase inhibitors are peptide-based and mimic the cleavage site in caspase substrates, but these inhibitors often lack selectivity and may act on more than one caspase target (30). Moreover, peptides have poor cell penetration properties and are unlikely to fulfill criteria for bioavailability after oral uptake (31). Therefore, specific and effective inhibitors are still in demand. Nonpeptide inhibitors have only recently been described for CASP6 (32, 33), but these inhibitors also target the active site of the enzyme. ARK5 inhibits both activation and activity of CASP6 by phosphorylation, and it regulates the expression level of CASP6 indirectly because it phosphorylates and suppresses p53 (34), and CASP6 is directly transactivated by p53 (35). These functions make AKR5 a potential therapeutic target. Meanwhile, because phosphorylation inhibits both self-activation of CASP6 and the activity of processed CASP6, and this phosphorylation site is unique for CASP6 (13, 14), phosphorylating CASP6 or mimicking the phosphorylation state may lead to a novel strategy for drug discovery rather than directly targeting the active site. Therefore, our results not only revealed the molecular mechanism of this inhibition but also provided the essential structural basis for rational drug design outside the active site.
Supplementary Material
Acknowledgments
We thank Dr. Xiang Liu and Dr. Thomas Earnest and Jian-Shi Jin for valuable discussion and proofreading and Wensheng Wei for providing the microplate reader.
This work was supported by grants from the National Basic Research Program of China 973 Grant 2011CB911103 (to X.-D. S.), National High Technology 863 Program 2006AA02A317 (to X.-D. S.), and National Natural Science Foundation of China (Canada–China Joint Health Initiative 30711120581 (to X.-D. S.) and 21125311, 91027044 (to Y.-Q. G.).

This article contains supplemental Figs. S1–S3 and Table S1.
The atomic coordinates and structure factors (codes 3V6L and 3V6M) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- CASP6
- caspase-6
- Ac-VEID-AFC
- Ac-Val-Glu-Ile-Asp-7-amino-4-trifluoromethylcoumarin
- AD
- Alzheimer disease
- ARK5
- AMP-activated protein kinase-related kinase 5
- HD
- Huntington disease
- L
- intersubunit linker
- MD
- molecular dynamics
- p10
- small subunit
- p20
- large subunit.
REFERENCES
- 1. Yan N., Shi Y. (2005) Mechanisms of apoptosis through structural biology. Annu. Rev. Cell Dev. Biol. 21, 35–56 [DOI] [PubMed] [Google Scholar]
- 2. Pop C., Salvesen G. S. (2009) Human caspases: activation, specificity, and regulation. J. Biol. Chem. 284, 21777–21781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Slee E. A., Harte M. T., Kluck R. M., Wolf B. B., Casiano C. A., Newmeyer D. D., Wang H. G., Reed J. C., Nicholson D. W., Alnemri E. S., Green D. R., Martin S. J. (1999) Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J. Cell Biol. 144, 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Klaiman G., Champagne N., LeBlanc A. C. (2009) Self-activation of caspase-6 in vitro and in vivo: caspase-6 activation does not induce cell death in HEK293T cells. Biochim. Biophys. Acta 1793, 592–601 [DOI] [PubMed] [Google Scholar]
- 5. Wang X. J., Cao Q., Liu X., Wang K. T., Mi W., Zhang Y., Li L. F., LeBlanc A. C., Su X. D. (2010) Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation. EMBO Rep. 11, 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Graham R. K., Ehrnhoefer D. E., Hayden M. R. (2011) Caspase-6 and neurodegeneration. Trends Neurosci. 34, 646–656 [DOI] [PubMed] [Google Scholar]
- 7. Graham R. K., Deng Y., Slow E. J., Haigh B., Bissada N., Lu G., Pearson J., Shehadeh J., Bertram L., Murphy Z., Warby S. C., Doty C. N., Roy S., Wellington C. L., Leavitt B. R., Raymond L. A., Nicholson D. W., Hayden M. R. (2006) Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell 125, 1179–1191 [DOI] [PubMed] [Google Scholar]
- 8. Graham R. K., Deng Y., Carroll J., Vaid K., Cowan C., Pouladi M. A., Metzler M., Bissada N., Wang L., Faull R. L., Gray M., Yang X. W., Raymond L. A., Hayden M. R. (2010) Cleavage at the 586-amino acid caspase-6 site in mutant huntingtin influences caspase-6 activation in vivo. J. Neurosci. 30, 15019–15029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Albrecht S., Bourdeau M., Bennett D., Mufson E. J., Bhattacharjee M., LeBlanc A. C. (2007) Activation of caspase-6 in aging and mild cognitive impairment. Am. J. Pathol. 170, 1200–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guo H., Albrecht S., Bourdeau M., Petzke T., Bergeron C., LeBlanc A. C. (2004) Active caspase-6 and caspase-6-cleaved tau in neurophil threads, neuritic plaques, and neurofibrillary tangles of Alzheimer's disease. Am. J. Pathol. 165, 523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nikolaev A., McLaughlin T., O'Leary D. D., Tessier-Lavigne M. (2009) APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature 457, 981–989 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12. Klaiman G., Petzke T. L., Hammond J., Leblanc A. C. (2008) Targets of caspase-6 activity in human neurons and Alzheimer disease. Mol. Cell. Proteomics 7, 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kurokawa M., Kornbluth S. (2009) Caspases and kinases in a death grip. Cell 138, 838–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Suzuki A., Kusakai G., Kishimoto A., Shimojo Y., Miyamoto S., Ogura T., Ochiai A., Esumi H. (2004) Regulation of caspase-6 and FLIP by the AMPK family member ARK5. Oncogene 23, 7067–7075 [DOI] [PubMed] [Google Scholar]
- 15. Lee A. W., Champagne N., Wang X., Su X. D., Goodyer C., Leblanc A. C. (2010) Alternatively spliced caspase-6B isoform inhibits the activation of caspase-6A. J. Biol. Chem. 285, 31974–31984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baumgartner R., Meder G., Briand C., Decock A., D'arcy A., Hassiepen U., Morse R., Renatus M. (2009) The crystal structure of caspase-6, a selective effector of axonal degeneration. Biochem. J. 423, 429–439 [DOI] [PubMed] [Google Scholar]
- 17. Vaidya S., Velázquez-Delgado E. M., Abbruzzese G., Hardy J. A. (2011) Substrate-induced conformational changes occur in all cleaved forms of caspase-6. J. Mol. Biol. 406, 75–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Müller I., Lamers M. B., Ritchie A. J., Park H., Dominguez C., Munoz-Sanjuan I., Maillard M., Kiselyov A. (2011) A new apo-caspase-6 crystal form reveals the active conformation of the apoenzyme. J. Mol. Biol. 410, 307–315 [DOI] [PubMed] [Google Scholar]
- 19. Müller I., Lamers M. B., Ritchie A. J., Dominguez C., Munoz-Sanjuan I., Kiselyov A. (2011) Structure of human caspase-6 in complex with Z-VAD-FMK: new peptide binding mode observed for the non-canonical caspase conformation. Bioorg. Med. Chem. Lett. 21, 5244–5247 [DOI] [PubMed] [Google Scholar]
- 20. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Macromol. Crystallography A 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 22. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24. Case D. A., Darden T. A., Cheatham T. E., 3rd, Simmerling C. L., Wang J., Duke R. E., Luo R., Crowley M., Walker R. C., Zhang W., Merz K. M., Wang B., Hayik S., Roitberg A., Seabra G., Kolossváry I., Wong K. F., Paesani F., Vanicek J., Wu X., Brozell S. R., Steinbrecher T., Gohlke H., Yang L., Tan C., Mongan J., Hornak V., Cui G., Mathews D. H., Seetin M. G., Sagui C., Babin V., Kollman P. A. (2008) AMBER 10, University of California, San Francisco [Google Scholar]
- 25. Yang L., Tan C. H., Hsieh M. J., Wang J., Duan Y., Cieplak P., Caldwell J., Kollman P. A., Luo R. (2006) New-generation amber united-atom force field. J. Phys. Chem. B 110, 13166–13176 [DOI] [PubMed] [Google Scholar]
- 26. Berendsen H. J. C., Grigera J. R., Straatsma T. P. (1987) The missing term in effective pair potentials. J. Phys. Chem. 91, 6269–6271 [Google Scholar]
- 27. Ryckaert J. P., Ciccotti G., Berendsen H. J. (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 [Google Scholar]
- 28. Darden T., York D., Pedersen L. G. (1993) Particle mesh Ewald: an Nlog(N)method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 29. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 30. McStay G. P., Salvesen G. S., Green D. R. (2008) Overlapping cleavage motif selectivity of caspases: implications for analysis of apoptotic pathways. Cell Death Differ. 15, 322–331 [DOI] [PubMed] [Google Scholar]
- 31. Lipinski C. A., Lombardo F., Dominy B. W., Feeney P. J. (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46, 3–26 [DOI] [PubMed] [Google Scholar]
- 32. Chu W., Rothfuss J., Chu Y., Zhou D., Mach R. H. (2009) Synthesis and in vitro evaluation of sulfonamide isatin Michael acceptors as small molecule inhibitors of caspase-6. J. Med. Chem. 52, 2188–2191 [DOI] [PubMed] [Google Scholar]
- 33. Ekici O. D., Li Z. Z., Campbell A. J., James K. E., Asgian J. L., Mikolajczyk J., Salvesen G. S., Ganesan R., Jelakovic S., Grütter M. G., Powers J. C. (2006) Design, synthesis, and evaluation of aza-peptide Michael acceptors as selective and potent inhibitors of caspases-2, -3, -6, -7, -8, -9, and -10. J. Med. Chem. 49, 5728–5749 [DOI] [PubMed] [Google Scholar]
- 34. Hou X., Liu J. E., Liu W., Liu C. Y., Liu Z. Y., Sun Z. Y. (2011) A new role of NUAK1: directly phosphorylating p53 and regulating cell proliferation. Oncogene 30, 2933–2942 [DOI] [PubMed] [Google Scholar]
- 35. MacLachlan T. K., El-Deiry W. S. (2002) Apoptotic threshold is lowered by p53 transactivation of caspase-6. Proc. Natl. Acad. Sci. U.S.A. 99, 9492–9497 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.