Background: Dyn2 and Nup159 interact to induce oligomerization of the Nup82 cytoplasmic fibril complex.
Results: Dyn2 homodimers symmetrically bind Nup159 target sites through an antiparallel β-strand interaction.
Conclusion: Dyn2 homodimers bind diverse, arrayed Nup159 target sites, locally directing Nup159 homodimerization.
Significance: Learning how nucleoporins interact to form fibril architecture is crucial for understanding how the nuclear pore complex gates traffic.
Keywords: Dynein, Nuclear Pore, Nuclear Transport, Structural Biology, X-ray Crystallography, Dynein Light Chain, Isothermal Titration Calorimetry, Nucleoporin
Abstract
The nuclear pore complex gates nucleocytoplasmic transport through a massive, eight-fold symmetric channel capped by a nucleoplasmic basket and structurally unique, cytoplasmic fibrils whose tentacles bind and regulate asymmetric traffic. The conserved Nup82 complex, composed of Nsp1, Nup82, and Nup159, forms the unique cytoplasmic fibrils that regulate mRNA nuclear export. Although the nuclear pore complex plays a fundamental, conserved role in nuclear trafficking, structural information about the cytoplasmic fibrils is limited. Here, we investigate the structural and biochemical interactions between Saccharomyces cerevisiae Nup159 and the nucleoporin, Dyn2. We find that Dyn2 is predominantly a homodimer and binds arrayed sites on Nup159, promoting the Nup159 parallel homodimerization. We present the first structure of Dyn2, determined at 1.85 Å resolution, complexed with a Nup159 target peptide. Dyn2 resembles homologous metazoan dynein light chains, forming homodimeric composite substrate binding sites that engage two independent 10-residue target motifs, imparting a β-strand structure to each peptide via antiparallel extension of the Dyn2 core β-sandwich. Dyn2 recognizes a highly conserved QT motif while allowing sequence plasticity in the flanking residues of the peptide. Isothermal titration calorimetric analysis of the comparative binding of Dyn2 to two Nup159 target sites shows similar affinities (18 and 13 μm), but divergent thermal binding modes. Dyn2 homodimers are arrayed in the crystal lattice, likely mimicking the arrayed architecture of Dyn2 on the Nup159 multivalent binding sites. Crystallographic interdimer interactions potentially reflect a cooperative basis for Dyn2-Nup159 complex formation. Our data highlight the determinants that mediate oligomerization of the Nup82 complex and promote a directed, elongated cytoplasmic fibril architecture.
Introduction
The Saccharomyces cerevisiae nuclear pore complex (NPC)2 is a 66-MDa structure composed of ∼30 different proteins that embed in the nuclear envelope and facilitate transport across this barrier (1). NPC proteins are highly conserved in function and sequence across eukaryotes and carry out biologically conserved functions: mRNA export into the cytoplasm and gated transport of specific proteins into and out of the nucleus.
The proteins that make up this highly coordinated and complex structure form an eight-fold symmetrical pore from a limited number of structural folds (2). The types of domains in the nuclear pore proteins, or nucleoporins (Nups), are primarily α-solenoids, β-propellers, Phe-Gly rich repeats, coiled-coil domains, and transmembrane domains (3, 4). Transmembrane domains traverse the double nuclear envelope membrane and underlie the NPC core topology and biogenesis (5, 6). Phe-Gly repeats are primarily concentrated in the core interior where they function as a physical or entropic barrier to entering proteins while reversibly binding nuclear transport receptors and selectively allowing their passage (1, 7). On either side of the core, asymmetrically distributed elements are positioned to facilitate asymmetric, unidirectional transport. The nucleoplasmic side of the NPC contains proteins tethered into a basket-like structure that protrudes 95 nm into the nucleus, potentially serving as a molecular checkpoint for pre-mRNA before it exits the nucleus (8, 9). On the cytoplasmic surface, NPC fibrils stretch 50 nm into the cytoplasm (8). Cytoplasmic fibrils are primarily composed of nucleoporins from the Nup82 complex that bind translation initiation factors and mRNA export machinery (10). The Nup82 complex consists of Nup82, Nup159, and Nsp1 that work with Nup116, Nup42, Gle1, and Nup100 to mediate mRNA export in concert with the mRNA nuclear export receptor Mex67 and the DEAD box RNA helicase Dbp5 (4, 11–13).
Nup159 is a prime component of the Nup82 complex and plays a directed role in coordinating nucleoporins involved in mRNA export rather than protein trafficking between the cytoplasm and nucleus (14). Nup159 has an extended multicomponent architecture that facilitates its roles in mRNA export, as well as filament localization in the NPC structure (14–17). The Nup159 N-terminal domain constitutes a seven-bladed β-propeller that extends into the cytoplasm and mediates Dbp5 binding (18). Deletion of the Nup159 N-terminal domain results in a temperature-sensitive phenotype, lethal at 37 °C and hallmarked by Dbp5 mislocalization and constitutive mRNA export defects at 23 °C (14, 16). The Nup159 central 700 amino acids form a Phe-Gly-rich repeat domain. C-terminal to the Phe-Gly-rich repeats is a 100-amino acid region termed the dynein light chain-interacting domain (DID) that uses a pentameric array of dynein light chain binding motifs to bind the yeast dynein light chain Dyn2 (11). C-terminal to the DID, Nup159 contains a predicted helical region (19) that is essential for the stability and localization of Nup159 on the NPC and has recently been shown to form a heterotrimeric structure with Nup82 and Nup116 (14, 16, 20). Higher order oligomerization of the Nup82 complex requires both the Nup159 DID region as well as Dyn2 (11). The functional role of a dynein light chain at the nuclear pore is independent of its role in the cytoplasmic dynein microtubule motor complex (11).
The dynein light chain is a promiscuous protein, involved in a diversity of protein target interactions, only a subset of which involve binding to the cytoplasmic dynein microtubule motor complex. In S. cerevisiae, Dyn2 interacts with the dynein intermediate chain, Pac11, through tandem canonical 10–12-residue stretches, each containing a conserved QT motif (21). Similar Dyn2 binding motifs are arrayed in Nup159, where class averaged electron microscopy of the Nup159 DID region saturated with Dyn2 showed a stack of five densities like beads on a string, effectively mediating the Nup159 dimerization (11). As with Dyn2, the higher dynein light chain orthologs, dynein light chain 8 (LC8) and dynein light chain (DYNLL), bind partners outside of the dynein motor complex including the signaling molecules neuronal NOS and Pak1, the apoptosis regulator Bim/Bmf, and the mRNA localization protein Swallow (22–24). Given the diverse set of dynein light chain partners, it has been postulated that the dynein light chain functionally serves as a dimerization machine. This role correlates with the structures of higher Dyn2 orthologs that show dynein light chains complexed 2:2 with a variety of target peptides (23, 25–28).
Although studies to date have biophysically characterized dynein light chains from Drosophila, rat, and human, molecular details of the S. cerevisiae dynein light chain have remained outstanding. S. cerevisiae is a leading model system for biophysical, biochemical, and genetic investigations of the NPC and the cytoplasmic dynein motor complex. To further our molecular understanding of Dyn2 and its functional role in the NPC, we determined the x-ray crystal structure of Dyn2 in complex with a Nup159 target site. We couple structural data with gel filtration, multiangle light scattering, and isothermal titration calorimetry to derive a model for the Dyn2-Nup159 interaction and the role of Dyn2 as a dimerization machine.
EXPERIMENTAL PROCEDURES
Cloning and Expression of Full-length Dyn2 from S. cerevisiae
Full-length Dyn2 was cloned from S. cerevisiae S288c into the pGEX-6P2 expression vector (GE Healthcare) using the polymerase chain reaction and BamHI- and EcoRI-engineered flanking restriction sites. The Dyn2 insert was sequence-verified against GenBankTM accession NC_001136. pGEX-6P2-Dyn2 was transformed into Escherichia coli BL21 DE3 (pLysS) and grown under ampicillin selection in 6 liters of LB medium at 37 °C. At an optical density of 0.8 (600 nm), GST-Dyn2 expression was induced using 0.1 mm isopropyl-1-thio-β-d-galactopyranoside for 16 h at 18 °C. Cells were harvested by centrifugation at 2100 × g for 10 min at 4 °C, and the pellets were resuspended in buffer A (150 ml of 25 mm HEPES, pH 6.8, 300 mm NaCl, and 0.1% β-mercaptoethanol) and stored at −20 °C.
Protein Purification
Resuspended cell pellets were thawed and lysed by sonication at 4 °C. 0.1 mm phenylmethylsulfonyl fluoride was added to the lysate, and cell debris was pelleted by centrifugation at 23,000 × g for 45 min. Supernatant was loaded onto a 5-ml glutathione-Sepharose Fast Flow affinity column (GE Healthcare). GST-tagged Dyn2 was eluted from the glutathione column with 100 ml of 3 mm glutathione, pH 8.0, in buffer A. The GST-Dyn2 eluate was exchanged into buffer B (25 mm HEPES, pH 6.8, and 0.1% β-mercaptoethanol) using an Amicon Ultra 10-kDa spin concentrator (Millipore) and incubated for 16 h with PreScission protease (GE Healthcare). The cleaved protein was loaded onto an SP Sepharose Fast Flow column (GE Healthcare) and eluted over a linear 0–1 m NaCl gradient in buffer B. Dyn2 peak fractions were pooled and exchanged into 50 mm NaCl, 25 mm HEPES, pH 6.8, and 0.1% β-mercaptoethanol using an Amicon Ultra 3-kDa spin concentrator (Millipore) and concentrated to 5 mg/ml, snap-frozen in liquid nitrogen, and stored at −80 °C. All purification procedures were executed at 4 °C. The final, purified Dyn2 protein contained an N-terminal GPLGS cloning artifact.
Synthesis of Nup159 Peptides
Nup159 pep1 (YSADFDVQTSL, residues 1103–1113), pep2 (NYAESGIQTDL, residues 1116–1126), pep3 (YVKHNSTQTVK, residues 1141–1151), pep4 (YAVDNGLQTEP, residues 1153–1163), and pep5 (YTCNFSVQTFE, residues 1165–1175) (see Fig. 1C) were synthesized at the University of North Carolina Microprotein Sequencing and Peptide Synthesis Facility. Pep1, pep3, pep4, and pep5 were designed with an amino-terminal tyrosine to quantify the peptide concentration once solubilized. Lyophilized peptides were solubilized in 50 mm NaCl, 25 mm HEPES, pH 6.8, and 0.1% β-mercaptoethanol.
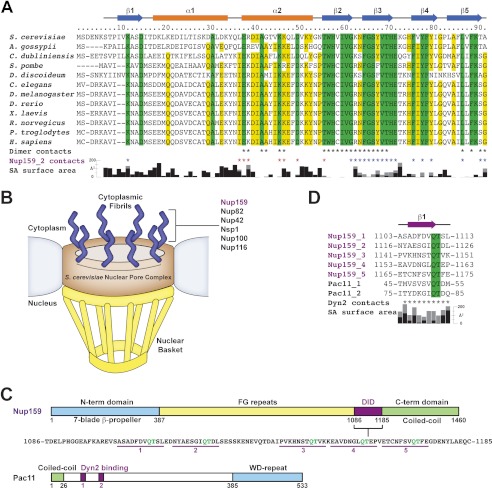
FIGURE 1.
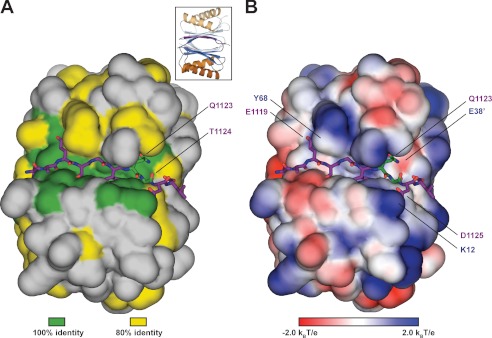
S. cerevisiae Dyn2 is a conserved dynein light chain involved in diverse macromolecular complexes including the nuclear pore complex and cytoplasmic dynein motor complex. A, sequence alignment of 12 dynein light chain family members ranging from S. cerevisiae to human. Residues aligned with 100 and 80% identity are colored green and yellow, respectively. Amino acid numbers and secondary structure elements, based on the S. cerevisiae Dyn2 structure, are shown above the alignment. Residues involved in Dyn2 dimerization and Dyn2-Nup159 pep2 binding are indicated below the alignment by asterisks based on European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI) PDBe Protein Interfaces, Surface and Assemblies (PISA). Black, Dyn2-Dyn2 chain A-C interactions; red, Dyn2-Nup159_2 chain A-D or chain C-B interactions; blue, Dyn2-Nup159_2 chain A-B or chain C-D interactions. Solvent-accessible (SA) surface area for respective Dyn2 chain C residues is indicated below the alignment, calculated in the presence (black) and absence of the Nup159_2 chain D (gray) using the Accessible Surface Area Analysis tool in CCP4 (41). A. gossypii, Ashbya gossypii; C. dubliniensis, Candida dubliniensis; S. pombe, Schizosaccharomyces pombe; D. discoideum, Dictyostelium discoideum; D. rerio, Danio rerio; X. laevis, Xenopus laevis; R. norvegicus, Rattus norvegicus; P. troglodytes, Pan troglodytes; H. sapiens, Homo sapiens. B, diagram of the nuclear pore complex illustrating the cytoplasmic localization of Nup159 and the Nup82 complex to cytoplasmic fibrils. C, domain architecture of known Dyn2-binding proteins: Nup159 and Pac11. Nup159 is composed of an N-terminal (N-term) β-propeller domain involved in Dbp5 binding, central Phe-Gly-rich repeats common to nucleoporins, a DID composed of five QT consensus motifs (residues 1103–1177) with Dyn2 binding activity, and a C-terminal (C-term) region involved in Nup159 anchoring to the nuclear pore complex (11, 14, 15). Pac11, the yeast dynein intermediate chain, shares architectural similarities with Nup159, composed of an N-terminal coiled-coil domain, tandem Dyn2 QT binding motifs, and a C-terminal WD-40 repeat domain, predicted to be a β-propeller. D, sequence alignment of the Dyn2 binding motifs from Nup159 and Pac11 highlighting the invariant QT motif. Nup159_2 secondary structure is indicated above the alignment. Nup159_2 residues involved in Dyn2 binding are indicated by asterisks below the alignment, as is solvent-accessible surface area, calculated in the presence (black) and absence of Dyn2 chains (gray).
Crystallization
1.0 mm Dyn2 was incubated with 1.5 mm Nup159 pep2 in 50 mm NaCl, 25 mm HEPES, pH 6.8, and 0.1% β-mercaptoethanol for 30 min on ice. Crystallization followed the hanging drop protocol using 1 μl of the Dyn2-Nup159 pep2 mixture and 1 μl of the 1-ml well solution: 0.3 m ammonium acetate, pH 5.5, 5% methyl pentanediol, and 35% polyethylene glycol 4000. Crystals grew at 20 °C to 200 × 200 × 600 μm over the course of a week. Crystals were transferred to Fomblin oil (Sigma) for cryoprotection and flash-frozen in liquid nitrogen.
Data Collection, Structure Determination, and Refinement
Dyn2-Nup159 pep2 crystals were maintained at 100 K under a cryo-cooled nitrogen stream, and diffraction data were collected using a Rigaku MicroMax 007HF x-ray generator (copper anode, 1.54 Å wavelength), Osmic mirrors, and a Rigaku Saturn 944+ charge-coupled device in the University of North Carolina Macromolecular X-Ray Crystallography Core Facility. 0.5° oscillations were collected over 160° from a single crystal. Data were indexed, integrated, and scaled using HKL2000 (29) (see Table 1). The structure was determined using the AutoMR molecular replacement program (PHENIX crystallographic suite (30)) and a modified 2PG1 (26) coordinate file in which a monomeric, apo Drosophila LC8 search model was used. The model was built using AutoBuild (PHENIX) (30) and refined iteratively through manual builds in Coot (31) followed by refinement runs using phenix.refine (PHENIX) (30). Refinement statistics were monitored using a Free R, calculated using 10% of the data, randomly excluded from refinement (32). The final model includes two Dyn2 molecules (chains A and C: residues 7–92), two Nup159 pep2 molecules (chain B: residues 1117–1126; chain D: residues 1116–1126 with Asn-1116 modeled as alanine), and 217 water molecules.
TABLE 1.
Data collection and refinement statistics
Parentheses list statistics for the high resolution shell.
| Data collection | |
| Wavelength (Å) | 1.54178 |
| Space group | P212121 |
| Cell dimensions (Å) | |
| a | 33.9 |
| b | 48.0 |
| c | 110.7 |
| Resolution (Å) | 50.0–1.85 (1.92–1.85) |
| Reflections | |
| Measured | 80,909 |
| Unique | 15,121 |
| Completeness (%) | 93.9 (62.7) |
| Mean redundancy | 5.4 (2.5) |
| I/σ | 24.8 (5.9) |
| Rsyma | 0.05 (0.15) |
| Refinement | |
| Resolution (Å) | 29.3–1.85 (1.92–1.85) |
| Rb/Rfree (%)c | 15.1 (18.0)/18.7 (24.7) |
| No. of reflections, R/Rfree | 13,573/1508 |
| Total atoms | 1758 |
| Protein/water | 1541/217 |
| Stereochemical ideality (r.m.s.d.) | |
| Bonds/angles (Å/°) | 0.006/1.02 |
| Mean B-factors (Å2) | |
| MC/SC/water | 13.8/19.9/28.5 |
| B-factor r.m.s.d. (Å2) | |
| MC/SC | 1.4/2.5 |
| Ramachandran analysis | |
| Favored/allowed (%) | 98.4/1.6 |
a Rsym = ΣhΣi|Ii(h) − 〈 l(h) 〉|/ΣhΣiIi(h) where Ii(h) is the integrated intensity of the ith reflection with the Miller Index h and 〈I(h)〉 is the average over Friedel and symmetry equivalents.
b R value = Σ(|Fobs| − k|Fcalc|)/Σ|Fobs|.
c Rfree is calculated using a 10% subset of the data that are removed randomly from the original data and excluded from refinement.
Size Exclusion Chromatography and Multiangle Light Scattering
100 μl of 200 μm Dyn2 was injected onto a Wyatt WTC030S5 silicone size exclusion column (for elution of 5,000–1,250,000-Da proteins) in 50 mm NaCl, 25 mm HEPES, pH 6.8, 0.1% β-mercaptoethanol, and 0.2 g/liter sodium azide and passed in tandem through a Wyatt DAWN HELEOS II light scattering instrument and a Wyatt Optilab rEX refractometer. The light scattering and refractive index data were used to calculate the weight-averaged molar mass and the mass fraction in each peak using the Wyatt Astra V software program (Wyatt Technology Corp.) (33).
Isothermal Titration Microcalorimetry
Isothermal titration calorimetry experiments were carried out at 15 °C in buffer C: 50 mm NaCl, 25 mm HEPES, pH 6.8, and 0.1% β-mercaptoethanol on a MicroCal AutoITC200 (GE Healthcare). Peptides were exchanged into buffer C using G-25 Sephadex quick spin columns (Roche Applied Science). 17 × 2-μl injections of 1 mm pep2 or pep4 were automatically injected into 200 μl of 100 μm Dyn2. The resulting binding isotherms (see Fig. 6, A and B) were analyzed using the Origin 7.0 software package (OriginLab) and were fit to a one-site binding model. Experiments were conducted in triplicate and averaged to determine respective mean KD values with standard deviations as shown.
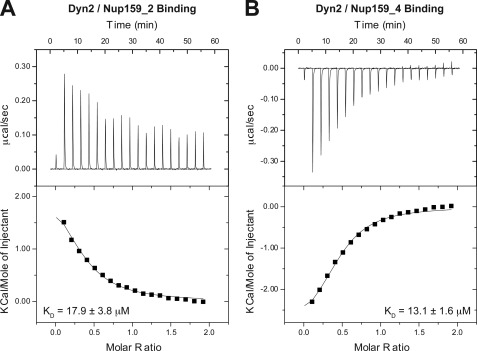
FIGURE 6.
The Dyn2 interactions with Nup159 pep2 and pep4 occur in a 1:1 stoichiometry and exhibit similar affinities but differ in their thermal binding modes. A, 17 × 2 μl of 1 mm Nup159 pep2 was injected into 200 μl of 100 μm Dyn2. The thermogram (upper panel) displays μcal/sec over the injection period (min). B, 17 × 2 μl of 1 mm Nup159 pep4 was injected into 200 μl of 100 μm Dyn2. Dyn2 binding to Nup159 pep2 (A) displayed an endothermic binding isotherm, whereas Dyn2 binding to Nup159 pep4 (B) showed exothermic binding. Thermograms (upper panels) were integrated, and the resulting isotherm was fit to a one-site binding model (lower panels) through iterative fitting. KD values presented (inset, lower panel) are the average of three independent experiments: Dyn2-Nup159 pep2, KD = 17.9 ± 3.8 μm, ΔH = 2500 cal/mol, ΔS = 31 cal/mol/deg, n = 0.33 sites; Dyn2-Nup159 pep4, KD = 13.1 ± 1.6 μm, ΔH = −4000 cal/mol, ΔS = 8.6 cal/mol/deg, n = 0.39 sites.
RESULTS
S. cerevisiae Dyn2 Is a member of the Conserved Dynein Light Chain Family
The dynein light chain, a component of the cytoplasmic dynein motor complex, is highly conserved from yeast to human (Fig. 1A). The dynein light chain is 90% identical across higher eukaryotes ranging from Caenorhabditis elegans to human, with significant identity extending to lower eukaryotes, as exemplified by the 50% identity between S. cerevisiae Dyn2 and Drosophila melanogaster LC8. Across organisms, evidence points to the role of the dynein light chain as a factor that promotes substrate dimerization. Although the dynein light chain is a component of the dynein microtubule motor complex, it is not exclusive to this complex. Recent work has shown that ∼25% of the S. cerevisiae dynein light chain member, Dyn2, is associated with the nuclear pore complex. Dyn2 associates with the Nup82 cytoplasmic fibril complex, binding to pentavalent motifs arrayed in the Nup159 DID (Fig. 1, B–D) (11). The Dyn2 binding motifs share a canonical QT motif with variable flanking components. Similar tandem binding sites have recently been mapped in the dynein intermediate chain, Pac11, and shown to mediate Dyn2 interaction (Fig. 1, C and D) (21). To understand the molecular basis of the Dyn2-Nup159 interaction, we cloned Dyn2 from S. cerevisiae (S288c) genomic DNA into the E. coli expression vector pGEX-6P2 and expressed and purified Dyn2 to homogeneity, removing the N-terminal GST tag. Nup159 peptides corresponding to the second and fourth Dyn2 DID binding sites (pep2 and pep4) were synthesized, purified by HPLC chromatography, and verified by mass spectrometry analysis. Pep4 incorporated an N-terminal tyrosine to facilitate concentration determination, whereas pep2 concentration was determined using its endogenous tyrosine.
The Dyn2 Homodimer Forms Two Composite Substrate Binding Sites Using a Central β-Sandwich and Flanking α2-Helices
To elucidate the structural determinants underlying the Dyn2-Nup159 interaction, we screened mixtures of Dyn2 and Nup159 pep2 and pep4 for co-crystallization. We obtained crystals of the Dyn2-Nup159 pep2 (residues 1116–1126) complex using a 1:1.5 molar ratio of Dyn2 and Nup159 pep2, respectively. The crystals diffracted to 1.85 Å resolution and belonged to the space group P212121. We solved the structure by the molecular replacement method using a peptide-free monomeric chain derived from the Drosophila dynein light chain (2PG1) that showed 50% sequence identity with Dyn2 (26). Two Dyn2 chains occupy the asymmetric unit, homodimerized around a noncrystallographic two-fold axis. Clear electron density was evident for two Nup159 pep2 chains, each bound to the Dyn2 homodimer. The R and Rfree values for the Dyn2-Nup159 pep2 structure are 15.1 and 18.0%, respectively. Crystallographic data and refinement statistics are presented in Table 1.
Dyn2 homodimerizes across a composite central β-sandwich (Fig. 2A). Each β-sheet is composed of five β-strands arranged in an antiparallel organization: β1-β4-β5-β2-β3′ in which the final β3′ strand is provided by the homodimeric mate. The prime interface between Dyn2 molecules is mediated by the antiparallel β2-β3′ strand interaction. Here, the β2-β3′ and β2′-β3 strand interactions encompass the noncrystallographic two-fold operator that relates each Dyn2 molecule. Flanking the central β-sandwich, each Dyn2 molecule contributes an α1 and α2 helix that bridge β1 and β2. The α1-α2 helix-turn-helix motifs symmetrically pack against the two β-sheets that form the central β-sandwich. The Dyn2 homodimer symmetrically binds two Nup159 peptides; the basis for the interaction is an extension of each β-sheet through an antiparallel strand that is stabilized through buttressing interactions with the neighboring α2-helix.
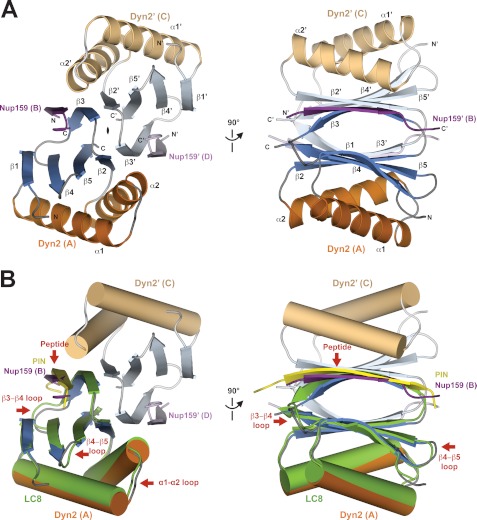
FIGURE 2.
Structure of Dyn2-Nup159 pep2 complex shows a quaternary complex composed of a Dyn2 dimer, bound to two Nup159 peptides through parallel, composite β-sheets. A, diagram of the Dyn2-Nup159 complex. Dyn2 chain A is shown in orange (α-helices) and dark blue (β-strands), and Dyn2 chain C is shown in beige (α-helices) and light blue (β-strands). The two-fold noncrystallographic symmetry operator that relates the Dyn2 and Nup159 chains in the asymmetric unit is indicated about the z axis. The image at right shows the complex after a 90° rotation about the y axis. B, the complex as shown in the two orientations in A, with Dyn2 chain A superimposed on the human dynein light chain, LC8 (light green) bound to a PIN peptide (yellow) (PDB 1CMI) after a least squares fit with an r.m.s.d. of 0.6 Å over 87 aligned Cα atoms (34). Helices are shown in cylindrical format. Structural differences between Dyn2 and human LC8 are indicated by red arrows and are dominated by loop regions as well as the bound peptides.
Dyn2 architecture is homologous to other dynein light chain structures determined to date, with the highest structural homology to the human LC8 complexed with a peptide from the protein inhibitor of neuronal nitric oxide synthase (PIN) (Protein Data Bank (PDB) 1CMI), 0.6 Å Cα r.m.s.d. over 87 residues (47% identity, Fig. 2B), and ranged among dynein light chain structures to 2.3 Å Cα r.m.s.d. over 81 residues when compared with the dynein light chain structure 1YO3 from Plasmodium falciparum (37% identity) (34, 35). The main elements that show structural diversity between Dyn2 and the Drosophila LC8 structure (1CMI) are restricted to loop regions, specifically the α1-α2 loop, the β3-β4 loop, and the β4-β5 loop. The core secondary structure elements of the domain show little plasticity. Diversity of allowable residues in the target peptide N-terminal to the canonical QT motif in turn shows structural diversity in the target β-strand backbone bound to the dynein light chain, as shown in the overlay of the Dyn2-Nup159 peptide structure with the human LC8-PIN peptide structure (Fig. 2B, supplemental Fig. 1).
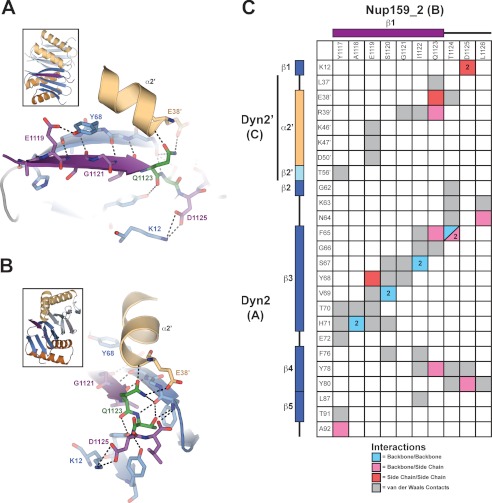
The Dyn2 homodimerization interface involves an extensive hydrophobic and electrostatic interface that buries ∼940 Å2 of solvent-accessible surface area on each Dyn2 molecule. Core β-strand-β-strand hydrogen-bonding networks extend the antiparallel sheets across homodimeric mates (β2-β3′and β2′-β3), augmented through additional backbone-side chain electrostatic interactions as well as van der Waals contacts between side chains (Fig. 3A). Helix α2′ packs against the β3 strand and buttresses the dimerization interface through the use of charged side chains, primarily Glu-38′ and Lys-46′, that afford van der Waals contacts as well as hydrogen bonding to β3 residues Asn-64 and Thr-70, respectively (Fig. 3, A and B). Overall, the homodimerization interface involves a pseudo-symmetric set of reciprocal interactions involving conserved residues (Figs. 1A and 3A).
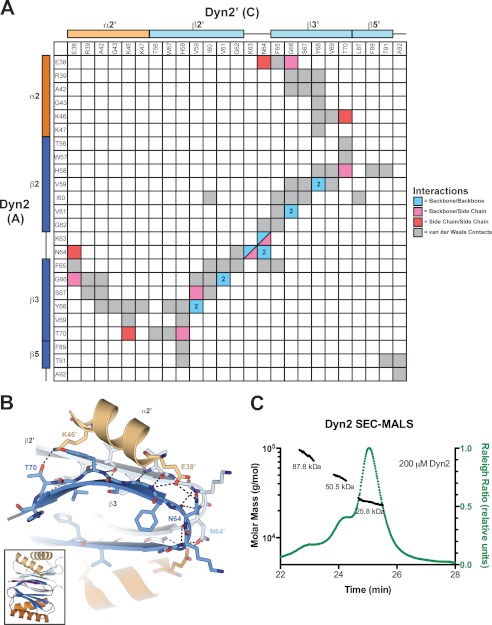
FIGURE 3.
Dyn2 homodimerizes via an extensive network of van der Waals contacts and hydrogen bonds. A, interaction matrix, showing the pseudo-symmetric bonding and contact networks formed between Dyn2 protomers A and C in the complex. Secondary structure elements corresponding to the residues of each protomer are indicated along the axes of the matrix. Backbone-backbone, backbone-side chain, side chain-side chain, and van der Waals interactions are indicated in blue, pink, red, and gray, respectively, and correlate with distances less than or equal to 3.5 (hydrogen bonds) and 4.5 Å (van der Waals contacts). Numbers in cells indicate the total number of hydrogen bonds (greater than one) between 2 residues. B, diagram of key residues and structural elements involved in the Dyn2-Dyn2 interface. The Dyn2 homodimer is shown as colored in Fig. 1. Specific Dyn2 residues mediating homodimerization are shown in stick format. Hydrogen bonds are indicated as dashed lines. The interface involves extensive antiparallel β-strand-β-strand interactions as well as contributions from the α2 helices that flank the central β-sandwich. The inset shows the relative orientation of the complex. C, size exclusion chromatography and multiangle light scattering (SEC-MALS) analysis of Dyn2, injected at an initial concentration of 200 μm (green) in 100 μl. The Raleigh Ratio elution profile was normalized. Dyn2 predominantly forms a dimer in solution at pH 6.8, with additional, higher order tetrameric and octameric species detected as well. The Dyn2 construct analyzed has a calculated monomeric molecular mass of 10,852 Da.
Dyn2 Exists as a Multimer in Solution
Although the crystal structure of the Dyn2-Nup159 pep2 complex showed Dyn2 in a homodimeric state, we wanted to determine whether this dimeric form existed in solution in the absence of bound peptide. To determine the oligomeric state of Dyn2, we utilized size exclusion chromatography coupled with multiangle light scattering. We analyzed the elution and mass profiles of purified Dyn2 injected at an initial concentration of 200 μm. The Dyn2 elution profile contained three main peaks with masses respectively calculated at 25.8, 50.5, and 87.8 kDa. On average, 87% of the eluted mass fraction was in the 25.8-kDa peak (Fig. 3C). The theoretical calculated molecular mass of our Dyn2 construct is 10,852 Da. Thus, under the conditions analyzed, peptide-free Dyn2 was found primarily as a homodimer with the remaining population in higher order oligomeric states.
The Dyn2 Homodimer Binds Parallel Nup159 Peptides Using a Conserved Composite Binding Site
Nup159 contains a pentameric array of Dyn2 binding sites (11). In the structure we present here, the Dyn2 homodimer is complexed with two Nup159 peptides corresponding to the second Dyn2 binding site in the Nup159 DID. The Nup159 peptide binds in a conserved pocket formed at the Dyn2 homodimer interface, consisting of both hydrophobic and charged residues (Fig. 4B). Nup159 pep2 (chain B) buries 911 Å2 of solvent-accessible surface area, whereas each of the Dyn2 molecules bury 512 and 129 Å2, for a collective 641 Å2 of solvent-accessible surface area buried at a single Nup159 pep2 binding site. 16 of the 25 Dyn2 residues involved in Nup159 peptide binding are 100% invariant across the 12 species shown in Fig. 1A, and 22 of the 25 are at least 80% invariant across these species (Fig. 4A). In Nup159, the glutamine (Gln-1123) and threonine (Thr-1124) that constitute signature dynein light chain binding determinants bind to the periphery of the peptide binding cleft in an area of high dynein light chain conservation. Analysis of the Dyn2 electrostatic surface shows that the peptide binding cleft is composed of mixed charges near the peptide N-terminal region, whereas positive charges dominate the electrostatic potential at the C-terminal region of the peptide. Key salt bridges in the complex include interactions between the invariant Dyn2 Lys-12 and Nup159 Asp-1125 as well as Dyn2 Glu-38′ and Nup159 Gln-1123. The Nup159 peptides form a β-strand interaction, extending the central β-sheets formed by Dyn2 homodimerization. The Nup159 β-strand runs antiparallel to the Dyn2 β3 strand and extends across 7 residues, terminating at the glutamine, Gln-1123, that composes the QT motif (Figs. 4 and 5). The Nup159 Gln-1123 side chain forms a network of hydrogen bonds with the start of the neighboring Dyn2 α2′ helix, capping the end through interactions with the Arg-39′ backbone amine as well as one of the Glu-38′ side chain carboxyl oxygens. In addition, the Gln-1123 side chain forms a hydrogen bond with the Phe-65 backbone amine on β3. The Gln-1123 backbone is stabilized through a hydrogen bond to the Dyn2 Tyr-78 hydroxyl group (Fig. 5, A–C). Thr-1124 from the Nup159 QT motif forms extensive contacts with Dyn2 Phe-65, engaging the Phe-65 backbone carbonyl and amine through hydrogen bonds from its own backbone amine and side chain hydroxyl group. The Thr-1124 side chain γC also forms van der Waals contacts with the Phe-65 benzene ring. Preceding the QT motif, the 6 N-terminal Nup159 residues primarily use an antiparallel β-strand-β-strand hydrogen bond network as well as van der Waals contacts to bind the conserved Dyn2 groove, indicative of the highly variable composition accepted in dynein light chain targets. Overall, the Nup159 pep2-Dyn2 interface is mediated by extensive hydrogen bonding and van der Waals contacts involving 10 residues in the Nup159 peptide. All Nup159 residues modeled contact one Dyn2 protomer, and 6 of these 10 residues make additional contacts with the Dyn2′ homodimeric mate, indicating that high affinity Dyn2-substrate recognition is mediated via Dyn2 dimerization. The Nup159 pep2-Dyn2 interface buries 1565 Å2 of solvent-accessible surface area at each binding site. The two Nup159 peptides bound to the Dyn2 homodimer run parallel to each other, related by a two-fold symmetry axis, with their midpoints separated by ∼20 Å.
FIGURE 4.
Dyn2 binds substrates through a highly conserved, positively charged composite groove formed by Dyn2 dimerization. A, conservation, as highlighted in Fig. 1A (green, 100% identity; yellow, 80% identity, as determined across 12 diverse species), mapped on the Dyn2 dimer shown in surface representation. Nup159 pep2 is shown in stick format in purple, inserted in the highly conserved interdimer groove. The highly conserved QT substrate motif (green sticks) is located C-terminal to the Nup159_2 β-strand. Conservation, however, is equally distributed across the Dyn2 substrate binding region. The inset shows the relative orientation of the complex in graphic format colored as in Fig. 2A. B, electrostatic surface calculated using APBS to generate solvent-accessible surface potentials that are shown in kBT/e, colored according to the key shown (42). Nup159 pep2 is shown in purple stick format with specific Dyn2 residues involved in hydrogen bond contacts labeled. The conserved QT motif is shown in green stick format. Dyn2-Nup159 pep2 interactions include Tyr-68–Glu-1119, Glu-38′–Gln-1123, and Lys-12–Asp-1125. The complex is oriented as in A.
FIGURE 5.
The Dyn2-Nup159 pep2 interaction is mediated by an extensive interaction network that recognizes 10 contiguous Nup159 residues, dually conferring specificity and substrate plasticity. A and B, close-up of residues involved in the Dyn2-Nup159 pep2 interaction. Secondary structure elements are shown as in Fig. 2A, with specific residues that mediate the Dyn2-Nup159 pep2 interaction shown in stick format and their corresponding hydrogen-bonding network shown with dashed lines. C, interaction matrix, showing the contact networks formed between Dyn2 protomers A and C with Nup159. Secondary structure elements and protomer designation are indicated along matrix axes. Backbone-backbone, backbone-side chain, side chain-side chain, and van der Waals interactions are indicated in blue, pink, red, and gray, respectively, and correlate with distances less than or equal to 3.5 (hydrogen bonds) and 4.5 Å (van der Waals contacts).
Nup159 DID Sites Two and Four, Respectively, Bind Dyn2 with 17.9 and 13.1 μm Affinity, Using Differential Thermal Binding Modes
To determine the affinities between Dyn2 and the five Nup159 Dyn2 binding sites in the DID, we synthesized the respective peptides and performed isothermal titration calorimetry, titrating peptides into the calorimeter cell containing Dyn2. Each Nup159 peptide binding experiment was performed in triplicate, and the fitted values were averaged. Each individual binding experiment was best fit to a one-site model (using the Dyn2 monomer concentration) (33). Pep2 showed an endothermic isotherm (Fig. 6A), whereas pep4 showed an exothermic isotherm (Fig. 6B). The experimentally determined affinities between Dyn2 and Nup159 pep2 and pep4 are shown in Fig. 6 and have KD values equal to 17.9 and 13.1 μm, respectively. Pep3 did not show sufficient signal to noise and was not soluble at the concentrations needed to determine binding accurately. Pep1 and Pep5 are highly hydrophobic, and once solubilized, failed to show binding to Dyn2 as determined using isothermal titration calorimetry (data not shown). This may be due to the weaker binding affinities for these peptides as was qualitatively shown in the aforementioned PepScan assay (11) or due to a folded/aggregated state that precluded Dyn2 from binding.
Translational Arrangement of Dyn2 Homodimer Facilitates Contiguous Binding to Arrayed QT Motifs
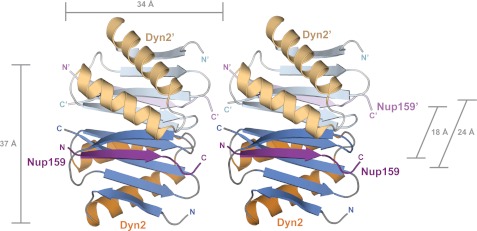
The arrangement of delineated QT motifs in Nup159 is nearly contiguous, separated by one or two amino acids except for a tentative QT region linking sites two and three that showed no Dyn2 binding activity in the previously mentioned PepScan assay (11). In the same investigation, electron microscopy of the Dyn2-Nup159 DID complex showed five densities arranged like beads on a string, leading the authors to propose a model in which five Dyn2 dimers bound parallel Nup159 DID arrays. Stelter et al. (11) modeled the bound Dyn2 dimers in a translational array. In the P212121 lattice presented here, we note a translational arrangement of Dyn2 dimers in the crystal lattice, which supports the Stelter et al. (11) Dyn2-Nup159 complex model. As shown in Fig. 7, Dyn2-Nup159 pep2 complexes are translationally arranged in the crystal, with a 34 Å translational component approximately collinear to the Nup159 peptide, effectively placing the C terminus of one Nup159 peptide proximal to the N terminus of the neighboring Nup159 peptide. Five Dyn2 dimers in this crystal lattice span 170 Å, on par with the 20-nm filaments observed by Stelter et al. (11) in electron micrographs of the Dyn2-Nup159 DID complex.
FIGURE 7.
Crystallographic contacts array Dyn2 dimers linearly in an arrangement that affords polarized binding to arrayed Dyn2 binding motifs. Dyn2-Nup159 crystallographic symmetry mates are shown in graphic representation, colored as in Fig. 2A. Dyn2 interdimer interactions coupled with parallel, arrayed binding motifs on Nup159 likely promote linear, cooperative binding activity between Dyn2 dimers and Nup159.
DISCUSSION
The dynein light chain, although a component of the cytoplasmic dynein motor complex, is promiscuous and has been identified as a component in numerous, diverse complexes. A universal role postulated for the dynein light chain is to serve as a dimerization machine. In S. cerevisiae, 25% of the Dyn2 cytoplasmic pool is found associated with the nuclear pore complex. Nup159, a component of the Nup82 complex of the cytoplasmic fibrils, was identified as a Dyn2 binding partner that promotes stable association of the Nup82 complex with the NPC (11). The Nup159 pentameric array of Dyn2 binding sites links the N-terminal Phe-Gly repeat region with the C-terminal NPC anchor region (20).
Our analysis of Dyn2 homodimer binding to individual Nup159 peptides showed similar affinities for pep2 and pep4, at 17.9 and 13.1 μm, respectively, whereas binding for pep1, pep3, and pep5 could not be experimentally determined based on properties of the individual peptides as synthesized. Binding curves fit best to a one-site binding model and are comparable with LC8 binding to peptides of similar size; DYNLL1 binds a 7-amino acid peptide from Bmf with a KD of 1.1 μm and similarly sized neuronal NOS peptide with a KD of 7.0 μm (36). The affinities determined between Dyn2 and the Nup159 peptides do not take into account potential cooperativity between arrayed Dyn2 homodimers based on interactions we observed in translational symmetry mates in the Dyn2-Nup159 crystal. The affinities and differential thermal binding modes determined for Nup159 pep2 and pep4 reflect plasticity in the Dyn2 binding pocket. The Dyn2 binding site does not have many steric occlusions and can thereby accommodate sequence diversity as observed with the LC8 family (25, 36, 37). The Dyn2-Nup159 crystal structure shows that extensive backbone-backbone interactions mediate the antiparallel β-sheet extension. This backbone-based interaction affords tight binding while simultaneously enabling diversity in the side chains that flank the core, conserved QT binding motif. The QT motif is present in most Dyn2/LC8 binding peptides characterized to date and constitutes the C-terminal flank of the β-strand of the target peptide. The QT motif contributes a network of hydrogen bonds and van der Waals contacts with the conserved groove of the dynein light chain, directly contacting residues from each subunit of the homodimer. The amino acid diversity flanking the QT motif likely underlies the differential affinities and thermal binding modes observed across dynein light chain targets. Pep4 exhibited exothermic binding, indicative of a strong enthalpic, electrostatically driven interaction, whereas pep2 exhibited endothermic binding, indicative of a hydrophobic, entropically driven interaction. Pep2 and pep4 each have electrostatic and hydrophobic residues. A key hydrophobic determinant that may underlie the endothermic binding observed with Nup159 pep2 is the tyrosine residue at position 1117, 6 residues upstream of the QT motif (i.e. Gln-6; Fig. 1D). The corresponding residue in Nup159 pep4 is an alanine. The Nup159 pep2 Gln-6 tyrosine makes numerous van der Waals contacts with the Dyn2 homodimer (Fig. 5C). Peptide-specific exothermic and endothermic binding has been observed with Dyn2 homologs from other species and highlights the sequence diversity within target sites that dynein light chains are capable of accommodating (27, 36–40).
Our work represents the first biophysical and structural characterization of the yeast dynein light chain, Dyn2. At physiological conditions, Dyn2 exists predominantly in the homodimeric state. As a homodimer, Dyn2 is positioned to interact with target sites and induce and stabilize parallel dimerization in these target proteins. Dimerization machines can cross-link targets, homo- or heterodimerize targets, serve to architecturally extend a target, as well as promote the avidity of a target for binding partners. The Dyn2-Nup159 structure creates a foundation for understanding the role of Dyn2 in the NPC as a dimerization machine that can scaffold Nup159 and extend the protein at least 170 Å (5 × 34 Å). Our structural and biophysical investigations of the Dyn2-Nup159 interaction have additional implications for the Dyn2 mode of interaction and function with the dynein intermediate chain, Pac11, and for its potential role in promoting Pac11 dimerization and aiding in the recruitment of the dynein activation complex, dynactin (21).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants 1R01GM094415 (to K. C. S.). This work was also supported by March of Dimes Grant 1-FY11-434 (to K. C. S.) and a research grant from the University Research Council, University of North Carolina at Chapel Hill (to K. C. S.).

This article contains supplemental Fig. 1.
The atomic coordinates and structure factors (code 4DS1) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- NPC
- nuclear pore complex
- Nup
- nucleoporin
- DID
- dynein light chain-interacting domain
- LC8
- dynein light chain 8
- PIN
- protein inhibitor of nitric oxide synthase
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., Chait B. T. (2000) The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148, 635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Maul G. G. (1971) On the octagonality of the nuclear pore complex. J. Cell Biol. 51, 558–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schwartz T. U. (2005) Modularity within the architecture of the nuclear pore complex. Curr. Opin. Struct. Biol. 15, 221–226 [DOI] [PubMed] [Google Scholar]
- 4. Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B. T., Sali A., Rout M. P. (2007) The molecular architecture of the nuclear pore complex. Nature 450, 695–701 [DOI] [PubMed] [Google Scholar]
- 5. Gerace L. (1988) Functional organization of the nuclear envelope. Ann. Rev. Cell Biol. 4, 335–374 [DOI] [PubMed] [Google Scholar]
- 6. Tcheperegine S. E., Marelli M., Wozniak R. W. (1999) Topology and functional domains of the yeast pore membrane protein Pom152p. J. Biol. Chem. 274, 5252–5258 [DOI] [PubMed] [Google Scholar]
- 7. Ribbeck K., Görlich D. (2001) Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20, 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fahrenkrog B., Hurt E. C., Aebi U., Panté N. (1998) Molecular architecture of the yeast nuclear pore complex: localization of Nsp1p subcomplexes. J. Cell Biol. 143, 577–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Galy V., Gadal O., Fromont-Racine M., Romano A., Jacquier A., Nehrbass U. (2004) Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116, 63–73 [DOI] [PubMed] [Google Scholar]
- 10. Allen N. P., Patel S. S., Huang L., Chalkley R. J., Burlingame A., Lutzmann M., Hurt E. C., Rexach M. (2002) Deciphering networks of protein interactions at the nuclear pore complex. Mol. Cell. Proteomics 1, 930–946 [DOI] [PubMed] [Google Scholar]
- 11. Stelter P., Kunze R., Flemming D., Höpfner D., Diepholz M., Philippsen P., Böttcher B., Hurt E. (2007) Molecular basis for the functional interaction of dynein light chain with the nuclear-pore complex. Nat. Cell Biol. 9, 788–796 [DOI] [PubMed] [Google Scholar]
- 12. Bailer S. M., Balduf C., Katahira J., Podtelejnikov A., Rollenhagen C., Mann M., Pante N., Hurt E. (2000) Nup116p associates with the Nup82p-Nsp1p-Nup159p nucleoporin complex. J. Biol. Chem. 275, 23540–23548 [DOI] [PubMed] [Google Scholar]
- 13. Hodge C. A., Tran E. J., Noble K. N., Alcazar-Roman A. R., Ben-Yishay R., Scarcelli J. J., Folkmann A. W., Shav-Tal Y., Wente S. R., Cole C. N. (2011) The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes Dev. 25, 1052–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Del Priore V., Heath C., Snay C., MacMillan A., Gorsch L., Dagher S., Cole C. (1997) A structure/function analysis of Rat7p/Nup159p, an essential nucleoporin of Saccharomyces cerevisiae. J. Cell Sci. 110, 2987–2999 [DOI] [PubMed] [Google Scholar]
- 15. Weirich C. S., Erzberger J. P., Berger J. M., Weis K. (2004) The N-terminal domain of Nup159 forms a β-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore. Mol. Cell 16, 749–760 [DOI] [PubMed] [Google Scholar]
- 16. Gorsch L. C., Dockendorff T. C., Cole C. N. (1995) A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J. Cell Biol. 129, 939–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schmitt C., von Kobbe C., Bachi A., Panté N., Rodrigues J. P., Boscheron C., Rigaut G., Wilm M., Séraphin B., Carmo-Fonseca M., Izaurralde E. (1999) Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 18, 4332–4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miller A. L., Suntharalingam M., Johnson S. L., Audhya A., Emr S. D., Wente S. R. (2004) Cytoplasmic inositol hexakisphosphate production is sufficient for mediating the Gle1-mRNA export pathway. J. Biol. Chem. 279, 51022–51032 [DOI] [PubMed] [Google Scholar]
- 19. Kraemer D. M., Strambio-de-Castillia C., Blobel G., Rout M. P. (1995) The essential yeast nucleoporin NUP159 is located on the cytoplasmic side of the nuclear pore complex and serves in karyopherin-mediated binding of transport substrate. J. Biol. Chem. 270, 19017–19021 [DOI] [PubMed] [Google Scholar]
- 20. Yoshida K., Seo H. S., Debler E. W., Blobel G., Hoelz A. (2011) Structural and functional analysis of an essential nucleoporin heterotrimer on the cytoplasmic face of the nuclear pore complex. Proc. Natl. Acad. Sci. U.S.A. 108, 16571–16576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stuchell-Brereton M. D., Siglin A., Li J., Moore J. K., Ahmed S., Williams J. C., Cooper J. A. (2011) Functional interaction between dynein light chain and intermediate chain is required for mitotic spindle positioning. Mol. Biol. Cell 22, 2690–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McCauley S. D., Gilchrist M., Befus A. D. (2007) Regulation and function of the protein inhibitor of nitric oxide synthase (PIN)/dynein light chain 8 (LC8) in a human mast cell line. Life Sci. 80, 959–964 [DOI] [PubMed] [Google Scholar]
- 23. Lightcap C. M., Sun S., Lear J. D., Rodeck U., Polenova T., Williams J. C. (2008) Biochemical and structural characterization of the Pak1-LC8 interaction. J. Biol. Chem. 283, 27314–27324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Benison G., Karplus P. A., Barbar E. (2007) Structure and dynamics of LC8 complexes with KXTQT-motif peptides: swallow and dynein intermediate chain compete for a common site. J. Mol. Biol. 371, 457–468 [DOI] [PubMed] [Google Scholar]
- 25. Fan J., Zhang Q., Tochio H., Li M., Zhang M. (2001) Structural basis of diverse sequence-dependent target recognition by the 8-kDa dynein light chain. J. Mol. Biol. 306, 97–108 [DOI] [PubMed] [Google Scholar]
- 26. Williams J. C., Roulhac P. L., Roy A. G., Vallee R. B., Fitzgerald M. C., Hendrickson W. A. (2007) Structural and thermodynamic characterization of a cytoplasmic dynein light chain-intermediate chain complex. Proc. Natl. Acad. Sci. 104, 10028–10033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benison G., Karplus P. A., Barbar E. (2008) The interplay of ligand binding and quaternary structure in the diverse interactions of dynein light chain LC8. J. Mol. Biol. 384, 954–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang W., Lo K. W., Kan H. M., Fan J. S., Zhang M. (2003) Structure of the monomeric 8-kDa dynein light chain and mechanism of the domain-swapped dimer assembly. J. Biol. Chem. 278, 41491–41499 [DOI] [PubMed] [Google Scholar]
- 29. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 30. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brünger A. T. (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 [DOI] [PubMed] [Google Scholar]
- 33. Wyatt P. J. (1993) Light scattering and the absolute characterization of macromolecules. Anal. Chim. Acta 272, 1–40 [Google Scholar]
- 34. Liang J., Jaffrey S. R., Guo W., Snyder S. H., Clardy J. (1999) Structure of the PIN/LC8 dimer with a bound peptide. Nat. Struct. Biol. 6, 735–740 [DOI] [PubMed] [Google Scholar]
- 35. Vedadi M., Lew J., Artz J., Amani M., Zhao Y., Dong A., Wasney G. A., Gao M., Hills T., Brokx S., Qiu W., Sharma S., Diassiti A., Alam Z., Melone M., Mulichak A., Wernimont A., Bray J., Loppnau P., Plotnikova O., Newberry K., Sundararajan E., Houston S., Walker J., Tempel W., Bochkarev A., Kozieradzki I., Edwards A., Arrowsmith C., Roos D., Kain K., Hui R. (2007) Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms. Mol. Biochem. Parasitol. 151, 100–110 [DOI] [PubMed] [Google Scholar]
- 36. Radnai L., Rapali P., Hódi Z., Süveges D., Molnár T., Kiss B., Bécsi B., Erdödi F., Buday L., Kardos J., Kovács M., Nyitray L. (2010) Affinity, avidity, and kinetics of target sequence binding to LC8 dynein light chain isoforms. J. Biol. Chem. 285, 38649–38657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rapali P., Radnai L., Süveges D., Harmat V., Tölgyesi F., Wahlgren W. Y., Katona G., Nyitray L., Pál G. (2011) Directed evolution reveals the binding motif preference of the LC8/DYNLL hub protein and predicts large numbers of novel binders in the human proteome. PLoS One 6, e18818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hódi Z., Németh A. L., Radnai L., Hetényi C., Schlett K., Bodor A., Perczel A., Nyitray L. (2006) Alternatively spliced exon B of myosin Va is essential for binding the tail-associated light chain shared by dynein. Biochemistry 45, 12582–12595 [DOI] [PubMed] [Google Scholar]
- 39. Wagner W., Fodor E., Ginsburg A., Hammer J. A., 3rd (2006) The binding of DYNLL2 to myosin Va requires alternatively spliced exon B and stabilizes a portion of the myosin's coiled-coil domain. Biochemistry 45, 11564–11577 [DOI] [PubMed] [Google Scholar]
- 40. Nyarko A., Hall J., Hall A., Hare M., Kremerskothen J., Barbar E. (2011) Conformational dynamics promote binding diversity of dynein light chain LC8. Biophys. Chem. 159, 41–47 [DOI] [PubMed] [Google Scholar]
- 41. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 42. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.