Background: IL-9 is the signature cytokine of Th9 cells.
Results: NFAT1 deficiency or silencing of NF-κB (p65) impairs IL-9 production from Th9 cells.
Conclusion: NFAT1 creates an accessible chromatin platform for the recruitment of NF-κB (p65) onto the IL-9 promoter, resulting in increased IL-9 expression.
Significance: This is the first report elucidating the molecular mechanism of NFAT1- and NF-κB (p65)-mediated IL-9 expression in Th9 cells.
Keywords: Chromatin Remodeling, Cytokines/Interferon, Gene Regulation, T Cell, Transcription Factors, Transcription Regulation, IL-9, NF-kappaB, NFAT, p300
Abstract
IL-9 regulates diverse inflammatory immune responses. Although the functional importance of IL-9 has been investigated in various pathophysiological conditions, molecular mechanisms by which TCR stimulation induced IL-9 gene expression are still unclear. In this study, we investigated the functional importance of the NFAT1 and NF-κB (p65) in IL-9 gene transcription in CD4+ T cells. In vivo binding of NFAT1 and NF-κB (p65) to the IL-9 promoter was observed. NFAT1 binding induced a transcriptionally active chromatin configuration at the IL-9 promoter locus, whereas NF-κB (p65) binding transactivated the IL-9 promoter. Mouse deficient in NFAT1 shows a significant down-regulation of IL-9 expression that resulted from an inaccessible chromatin configuration at the IL-9 promoter. In parallel, knockdown of NF-κB (p65) also resulted in reduced IL-9 expression. In this process, NFAT1 plays a pivotal role as a core protein that creates an accessible platform for the assembly of transcription activators. The presence of NFAT1 correlates with recruitment of NF-κB (p65), p300, and active histone markers on the IL-9 promoter, resulting in a transcriptionally competent promoter. NFAT1 deficiency significantly reduced the recruitment of the above activation complex to the IL-9 promoter. In summary, our data suggest that functional cooperation of NFAT1 and NF-κB synergistically enhances IL-9 transcription in CD4+ T cells.
Introduction
Naive CD4+ T cells differentiate into diverse T helper subsets (Th1, Th2, Th9, Th17, and regulatory T cells) upon encountering antigens in a unique cytokine microenvironment. Whereas IL-12 and IL-4 induce the differentiation of IFN-γ-producing Th1 cells and IL-4, IL-5, and IL-13 secreting Th2 cells, respectively, TGF-β, IL-6, IL-1β, and IL-23 stimulate the development of IL-17-producing Th17 (1–4). Adding to this growing list of effector T helper cells are the Th9 cells. This newly discovered Th9 cell subset can be generated under a cytokine milieu enriched in IL-4 and TGF-β. Th9 cells are the major producer of IL-9 among the T helper cell subsets (5, 6). The biology and the function of IL-9 are complex and diverse (7). IL-9 functions through the IL-9 receptor, which shares the γ chain with IL-2, IL-4, IL-7, IL-15, and IL-21 cytokine receptors (8, 9). Initially, IL-9 was characterized as a growth factor for T cells and mast cells (10, 11). IL-9 promotes Th2-specific allergic responses, allergic inflammation, and asthmatic symptoms (12–15). IL-9 also promotes the differentiation of Th17 cells and has been implicated in the development and progression of experimental autoimmune encephalomyelitis (16–18). On the other hand, IL-9 increases the suppressive activity of regulatory T cells by yet undiscovered mechanisms (16, 19, 20). Thus, elucidating the underlying mechanism of IL-9 expression will further advance the understanding of the functional variety and diversity of IL-9-producing cells and IL-9 itself.
The activation and expression of specific transcription factors in the T helper cell subsets govern their signature cytokine expression. So far, Pu.1, IRF4, and STAT6 have been reported to play crucial roles in Th9 lineage commitment and regulation of IL-9 expression (21–23). However, these factors do not act alone and require the participation of other transcription factors that may play a pivotal role in inducing IL-9 following TCR stimulation (24). Ubiquitously expressed transcription factors are also known to induce subset-specific expression of cytokines. This includes the nuclear factor of activated T cells (NFAT)2 family members, which have diverse roles in T cell subsets (25). The NFAT proteins (NFAT1 to -4) are regulated by Ca2+ except for NFAT5 (TonEBP (tonicity element-binding protein) or OREBP (osmotic response element-binding protein)), which responds to osmotic stress (26, 27). TCR stimulation coupled with Ca2+ mobilization results in the activation of Ca2+-dependent NFATs via the calcium-calmodulin phosphatase pathway (28, 29). NFATs act in concert with other transcription factors to direct their target gene expression, often in a cell- and tissue-specific manner. For example, NFAT1/T-bet interaction directs the Th1-specific IFN-γ expression; NFAT1/IRF4 induces IL-4 and IL-10 expression in Th2 cells, whereas NFAT2/Smad3 interaction governs Foxp3 expression in regulatory T cells (30–33). NFATs also interact with active histone modifiers, CBP/p300, to direct changes in chromatin architecture (34, 35) and regulate the inducible gene expression of diverse cytokines in immune response (36). In the context of IL-9, previous reports suggested a potential link of NFAT with IL-9 expression. Treatment of cyclosporin A (CsA) to the mice infected with the parasite, Leishmania major, ablated antigen-specific IL-9 production (37). More specifically, umbilical cord blood T cells that express reduced NFAT1 compared with adult blood T cells produce a significantly reduced level of IL-9 (38, 39). However, no mechanistic investigation on the functional role of NFAT1 in IL-9 regulation has been conducted.
NF-κB family is another well known ubiquitous transcription factor family that regulates genes involved in diverse biological processes, such as growth and survival and also inflammatory immune responses. They bind to specific decameric sequences located within the target gene promoter or other regulatory regions (40–42). This family consists of five members (NF-κB1 (p50/p105), NF-κB2 (p52/p100), c-REL, RELA (p65), and RELB (I-REL)). Inhibitory proteins, inhibitory κB (IκB), regulate the activity of NF-κB family of proteins by sequestering them in the cytosol and preventing their nuclear translocation (43). This family requires the activity of transcription coactivators to execute their transactivity. CBP/p300 is one of the coactivators with intrinsic histone acetylase activity and is required for p65-mediated transactivation (44, 45). NF-κB has also been implicated in IL-9 expression in mouse macrophage cells upon LPS stimulation (46). Also, binding of NF-κB to the human IL-9 promoter in T cells has been demonstrated (24, 47). However, the role of NF-κB in regulating IL-9 expression in mouse Th9 cell is still unknown.
In this study, we identified the functional synergy of two transcription factors, NFAT1 and NF-κB (p65), in IL-9 expression by Th9 cells. NFAT1 is primarily involved in stabilizing and maintaining a transcriptionally competent chromatin structure, whereas NF-κB transactivates the IL-9 promoter.
EXPERIMENTAL PROCEDURES
Computational Analysis of IL-9 Locus
To identify potential transcription factor binding sites in the IL-9 promoter, comparative genomic analysis was performed. Genomic sequence of the IL-9 gene promoter was analyzed using the Web-based alignment software ECR Browser and VISTA Browser 2.0 (48).
Mice and Cell Lines
C57BL/6 mice 6–8 weeks of age were purchased from Orient Bio (Daejon, Korea). NFAT1-deficient mice (KO) were kindly provided by Dr. A. Rao (Harvard Medical School). Mice were housed in specific pathogen-free barrier facilities and used in accordance with protocols approved by the Animal Care and Ethics Committees of the Gwangju Institute of Science and Technology.
CD4+ T Cell Isolation, Differentiation, and Culture
CD4+ T cells were isolated, differentiated, and cultured as described previously (30). Briefly, CD4+ T cells were purified from the lymph nodes and spleen of 8–12-week-old mice with the use of magnetic beads (L3T4, Miltenyi). For T helper cell differentiation, CD4+ T cells (2–3 × 106/ml) were stimulated with 1 μg/ml plate-bound α-CD3 and 2 μg/ml soluble α-CD28 under Th1 skewing (10 ng/ml IL-12 plus 10 μg/ml α-IL-4), Th2 skewing (10 ng/ml IL-4, 10 μg/ml α-IFN-γ plus 10 μg/ml anti-IL-12), and Th9-skewing (10 ng/ml IL-4 and 1 ng/ml TGF-β) or left in unpolarized (CD4+ T cell blasts) (6) conditions in RPMI 1640 medium (Welgene) supplemented with 10% fetal bovine serum, l-glutamine, penicillin/streptomycin, non-essential amino acids, sodium pyruvate, vitamins, HEPES, and β-mercaptoethanol. 100 units/ml recombinant human IL-2 (rhIL-2) was added after 24 h. On day 3, cells were shifted to complete medium containing IL-2 and expanded. On day 5, they were restimulated with 50 ng/ml phorbol 12-myristate 13-acetate (PMA) plus 1 μm ionomycin or 1 μg/ml plate-bound α-CD3 and α-CD28 as mentioned in particular cases.
RNA Isolation, cDNA Synthesis, Quantitative RT-PCR (qRT-PCR), Conventional PCR, and ELISA
Total RNA was extracted from the stimulated or unstimulated cells using TRIzol reagent (Molecular Research Center, Cincinnati, OH) according to the manufacturer's protocol. For reverse transcription, 1 μg of total RNA was used, and cDNA was generated using oligo(dT) primer (Promega, Madison, WI) and Improm-II reverse transcriptase (Promega) in a total volume of 20 μl. The mRNA level was determined using 1 μl of cDNA by real-time PCR with SYBR green according to the manufacturer's protocol (Chromo4, MJ Research). Mouse hypoxanthine-guanine phosphoribosyltransferase primer was used for qRT-PCR to normalize the amount of cDNA used for each condition. The primer sequences used are as follows: hypoxanthine-guanine phosphoribosyltransferase (5′-TTA TGG ACA GGA CTG AAA GAC-3′ (forward) and 5′-GCT TTA ATG TAA TCC AGC AGG T-3′ (reverse)), IL-9 (5′-GTG ACA TAC ATC CTT GCC TC-3′ (forward) and 5′-GTG GTA CAA TCA TCA GTT GGG-3′ (reverse)). For ELISA, cell supernatants were collected from unstimulated and 24 h PMA/ionomycin- or α-CD3/CD28-stimulated cells, and cytokines were measured using IL-4, IL-9, and IFN-γ ELISA kits (eBiosciences, San Diego, CA).
Chromatin Immunoprecipitation (ChIP) Assay
ChIP analysis was carried out as described previously with minor modifications (30, 49–51). Chromatins prepared from Th9 cells after PMA/ionomycin stimulation were immunoprecipitated using antibodies against RNA Pol II (Santa Cruz Biotechnology, Inc.), acetyl histone H3 (AcH3), acetyl histone H4 (AcH4), dimethyl lysine histone H3 (H3K4me2) (Upstate, Lake Placid, NY), p300 (Millipore, Billerica, MA), NFAT1 (Santa Cruz Biotechnology, Inc.), p65 (Abcam, Cambridge, MA), and rabbit IgG (Sigma-Aldrich). After washing, samples were eluted in elution buffer (1% SDS and 0.1 m NaHCO3) for the first ChIP. For the second immunoprecipitation, complexes were eluted from the primary immunoprecipitation with 10 mm DTT at 37 °C for 30 min and diluted 1:50 in buffer (1% Triton X-100, 2 mm EDTA, 150 mm NaCl, 20 mm Tris-HCl, pH 8.1), followed by reimmunoprecipitation with second antibodies similarly to the first ChIP (52). Following reversal of cross-links, the presence of selected DNA sequences was assessed by qRT-PCR using SYBR Green PCR mix. As a loading control, PCR was done directly on input DNA purified from chromatin before immunoprecipitation. The primer pairs used for detecting IL-9 promoter are: (5′-CAT TAC CAC CCC TGT AAC TCA C-3′ (forward) and 5′-CTA CCA GCA TCT TCC AGT CTA GC-3′ (reverse)). A pair of primers corresponding to the non-relevant region (designated as NR; +1864/+2193), NR-F-5′ (ATG TAA CCA GGA ACA AGA TCA CTG CAG) and NR-R-5′ (GTG GAA GTA CTT ACC TAG ACC TTG GTG TC), was used as a control for the time-dependent ChIP experiment. The amount of chromatin precipitated by the indicated antibodies as detected by RT-PCR with primers against the target regions relative to that detected in the input (total chromatin) by the same primer pair is represented as a fraction of input or as negative images of EtBr-stained gels.
Chromatin Accessibility by Real-time PCR Assay
Th9 cells (5 × 106 cells/sample) differentiated from wild type (WT) and NFAT1−/− (KO) CD4+ T cells were left unstimulated or restimulated with PMA/ionomycin (PI) for 8 h and pelleted by centrifugation at 500 × g, washed in ice-cold PBS, and then resuspended in 100 μl of digestion buffer (10 mm Tris-HCl (pH 7.4), 15 mm NaCl, 60 mm KCl, 0.15 mm spermine, 0.5 mm spermidine, 1 mm CaCl2) with or without 5 units/ml micrococcal nuclease (MNase) (Roche Applied Science) and were incubated at 37 °C for 10 min. Reactions were terminated by adding 20 μl of Stop solution (100 mm EDTA, 10 mm EGTA, pH 8.1) and 10 ml of SDS 10% (w/v). DNA was isolated using the DNA blood genomic prep kit (Intron Biotechnology, Daejon, Korea) and eluted into 100 μl of TE (10 mm Tris-Cl, pH 7.5, 1 mm EDTA) buffer. DNAs recovered from MNase samples were checked for fragmentation (500–1000 bp) in 1% agarose gel. Primers used in the quantitative assays were validated by amplifying serially diluted genomic DNA as templates to create a standard curve for each primer set and analyzed using the quantification method. Untreated and MNase-treated samples were used in PCR assays to measure the relative abundance of target regions using the primer sets used in ChIP experiments. Actin promoter primers (5′-TTC CGA AAG TTG CCT TTT ATG GCT CGA-3′ (forward) and 5′-AAG GAG CTG CAA AGA AGC TGT G-3′ (reverse)) were used as controls. Chromatin accessibility values were calculated as the ratio of the undigested sample to the digested samples, and then the data were plotted as the ratio of accessibility observed in the digested DNA samples.
Plasmid Construction, Site-directed Mutagenesis, and Luciferase Reporter Assays
The promoter containing 5′ region of the murine IL-9 gene (36) encompassing nucleotides −366 to +48 (Fig. 1B) was amplified from T cell genomic DNA using primers 5′-CAT TAC CAC CCC TGT AAC TCA C (forward) and 5′-GTA TGT CAC CAA CAT GTT GAC (reverse) (indicated in Fig. 1B) and cloned into pXPG luciferase reporter vector. IL-9 promoter reporter construct that lacks the first NF-κB binding site (designated as ΔNF-κB1-Prom) was amplified using primer 5′-CCA GAA TTC CTG CTT TTA AAG (forward) and cloned into pXPG luciferase reporter vector. The sequences of cloned DNA fragments were confirmed by DNA sequencing. The two NF-κB binding sites at positions −315/−306 (named NT/κB1) and −48/−38 (named NT/κB2) were subjected to site-directed mutagenesis (NT/κB1 WT GGG GAA AAC ACA G to mutant GGG cAc AAC ACA G; NT/κB2 WT GTT TTT CCC GGT to mutant Gac aca CCC GGT). The sequences of cloned DNA fragments were confirmed by DNA sequencing. HEK-293 and EL-4 cells were transfected using GeneExpresso (Excellgen, Rockville, MD) according to the manufacturer's protocol, and primary Th9 cells were electroporated. Following PI stimulation for 8 h, cells were harvested, and luciferase activity was measured by the Dual-Luciferase assay system (Promega, Madison, WI). Wherever indicated, cyclosporin A (Calbiochem) was added at 0.1 mg/ml 20 min before PI stimulation. Data were normalized by the activity of Renilla luciferase, which was used as an internal control for transfection.
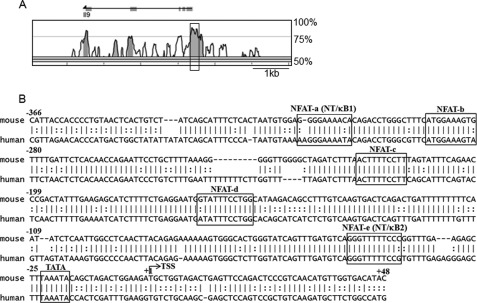
FIGURE 1.
Comparative bioinformatic analysis of the mouse and human IL-9 gene loci. A, rVISTA version 2.0 analysis depicting percentage conservation between mouse (as a base) and human IL-9 loci; the boxed region indicates the IL-9 promoter. B, nucleotide sequence comparison between the mouse (−366/+48) and human IL-9 promoter region. The predicted TATA site is boxed, and +1 denotes the transcription start site. The predicted NFAT sites with a matrix similarity of 0.8 are marked as follows: NFAT-a (−315/−306), NFAT-b (−290/−281), NFAT-c (−223/−214), NFAT-d (−165/−155), and NFAT-e (−48/−38), respectively. The composite sites for NFAT and NF-κB sites are marked as NT/κB1 (−315/−306) and NT/κB1 (−48/−38), respectively.
DNA Affinity Purification Assay
The DNA affinity purification assay was performed following protocols described previously with minor modifications (53). Briefly, biotinylated complementary oligonucleotides were annealed in TEN (10 mm Tris/HCl, pH 8.0, 1 mm EDTA, 100 mm NaCl) buffer. HEK-293 cells overexpressing NFAT1 and NF-κB (p65) were lysed by sonication in 200 μl of HKMG (10 mm Hepes, pH 7.9, 100 mm KCl, 5 mm MgCl2, 10% glycerol, 0.1% Nonidet P-40, and 1 mm DTT) buffer containing protease and phosphatase inhibitors (Roche Applied Science). The cellular debris was removed by centrifugation. The expression level of NFAT1 and NF-κB (p65) along with that of actin (control) was checked by blotting 30 μg of total cell lysate with α-NFAT1 (Santa Cruz Biotechnology, Inc.), α-p65, and α-actin (Abcam, Cambridge, MA) antibodies. The cell extracts (100–500 μg) were precleared with 10 μl of M-280 streptavidin beads (Invitrogen) for 1 h at 4 °C with gentle agitation. The cleared nuclear extracts were then incubated with 1 μg of biotinylated double-stranded probes and 10 μg of poly(dI-dC)·poly(dI-dC) overnight. 10–15 μl of M-280 streptavidin beads were used to pull down bound proteins for 1 h at 4 °C with gentle agitation. The beads were washed four times with cold HKMG buffer. SDS sample buffer was then added to the beads. The samples were boiled for 5 min and subjected to SDS-PAGE and Western blotting with α-NFAT1 (Santa Cruz Biotechnology, Inc.) and α-p65 (Abcam, Cambridge, MA) antibodies. The nucleotide sequences of oligonucleotides are as follows: NFAT-a and NT/κB-1 (encompassing region −315/−306; 5′-CTA ATG TGG AGG GGA AAA CAC AGA CCT GGG-3′ (forward) and 5′-CCC AGG TCT GTG TTT TCC CCT CCA CAT TAG-3′ (reverse)), NFAT-c (encompassing region −223/−214; 5′-GAT CTT TAA CTT TTC CTT TAG TAT TTC AG-3′ (forward) and 5′-CTG AAA TAC TAA AGG AAA AGT TAA AGA TC-3′ (reverse)), NFAT-e and NT/κB-2 (encompassing region −48/−38; 5′-GAT GTC AGG GTT TTT CCC GGT TTG-3′ (forward) and 5′-CAA ACC GGG AAA AAC CCT GAC ATC-3′ (reverse)), mtNT/κB-1 (5′-CTA ATG TGG AGG GcA cAA CAC AGA CCT GG-3′ (forward) and 5′-CCA GGT CTG TGT TgT gCC CTC CAC ATT AG-3′ (reverse)), mtNT/κB-2 (5′-GAT GTC AGG Gac aca CCC GGT TTG-3′ (forward) and 5′-CAA ACC GGG tgt gtC CCT GAC ATC-3′ (reverse)), Cons (mNF-AT1 + 2 (murine distal IL-2 NF-AT site); 5′-CCC AAA GAG GAA AAT TTG TTT CAT-3′ (forward) and 5′-ATG AAA CAA ATT TTC CTC TTT GGG-3′ (reverse) (54), and NR (5′-CCC AAA GAG GCC TTT GTT TCC AT-3′ (forward) and 5′-ATG GAA ACA AGG GCC TCT TTG GG-3′ (reverse).
NFAT1 Reconstitution in NFAT1−/− Th9 Cells and siRNA-mediated p65 Silencing in Wild Type Th9 Cells
For reconstitution assay, Th9 cells differentiated in vitro for 3 days from NFAT1−/− mice were transfected with 5 μg of NFAT1 expression construct or empty mock vector (control) into the NFAT1−/− Th9 cells. For the knockdown experiment, wild type Th9 cells were transfected with 100 pmol of p65 siRNA (Santa Cruz Biotechnology, Inc.) or scrambled siRNA (mock control) by using a T cell nucleofector kit (Lonza, Cologne, Germany) following the manufacturer's protocol. After 24 h of transfection, cells were restimulated with PI for 12 h for RT-PCR and immunoblot analysis and 24 h for ELISA, respectively.
Nuclear Extract Preparation and Immunoblotting
Nuclear extracts were prepared from PI-stimulated Th9 cells. Briefly, 2 × 107 cells were washed in ice-cold PBS and suspended in 1 ml of lysis buffer (10 mm Tris/HCl, 3 mm CaCl2, 2 mm MgCl2) containing protease inhibitor mixture (Roche Applied Science) for 10 min on ice. They were vortexed gently and incubated in 1 ml of Nonidet P-40 buffer (10 mm Tris/HCl, 3 mm CaCl2, 2 mm MgCl2, 1% Nonidet P-40) for 5 min at 4 °C and centrifuged at 3000 rpm for 10 min at 4 °C. Nuclei were washed with 1 ml of Buffer A (20 mm HEPES-KOH, 1.5 mm MgCl2, 10 mm KCl, 0.5 mm DTT, 0.5 mm PMSF), and 100 μl of Buffer C (20 mm HEPES-KOH, 25% glycerol, 420 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 5 mm DTT, 0.5 mm PMSF, 1% Triton X-100) was added to the pellets, and they were vortexed vigorously at 4 °C for 10 min. Thirty micrograms of the nuclear extracts were used for SDS-PAGE, and Western blot was carried out with α-NFAT1, α-Pu.1, α-IRF4, α-lamin B (Santa Cruz Biotechnology, Inc.), and α-tubulin (Abcam, Cambridge, MA) antibodies.
Statistical Analysis
Data are the mean ± S.E. of at least three independent experiments unless specified otherwise. Student's t test was used to calculate the statistical significance of the experimental data. The level of significance was set as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001. Significance was only indicated when appropriate.
RESULTS
Identification of Transcription Factor Binding Sites in IL-9 Promoter
To identify the potential transcription factors that are responsible for IL-9 gene expression, the genomic sequences of the human and mouse IL-9 promoter loci (−366/+48) were compared by bioinformatics analysis (Fig. 1A). Along with already identified Pu.1 and IRF4 (supplemental Fig. S1), five highly conserved (>85%) NFAT binding sites (marked as NFAT-a, -b, -c, -d, and -e, respectively) distributed in the IL-9 promoter were identified (Fig. 1B). Of the five NFAT binding sites, two of them encompassed the core NF-κB binding sites and are marked as NT/κB1 and NT/κB2, respectively (Fig. 1B). The IL-9 promoter showed highly conserved binding sites for other transcription factors such as AP1, STAT proteins, Smad2 and -3, ETS, Lef1, and Foxp3 (supplemental Fig. S1). Among the NFAT family members, we have mainly investigated the mechanism of NFAT1-mediated IL-9 gene expression because NFAT1 is the predominant NFAT protein in T cells and accounts for 90% of total NFAT DNA binding activity in wild type T cells (55).
NFAT1 Binds Directly to IL-9 Promoter in Th9 Cells
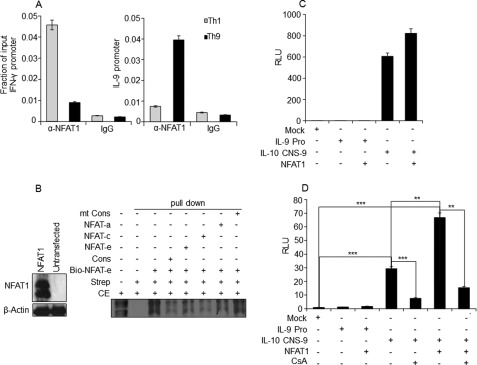
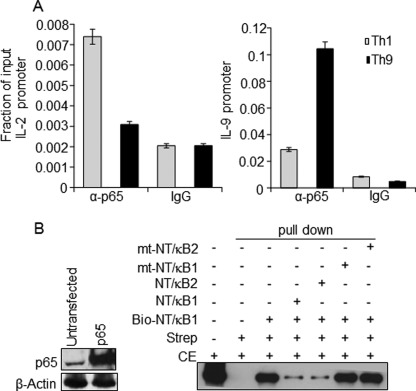
We utilized in vitro differentiated murine Th9 cells as our model, considering the fact that they are the major IL-9-producing T helper cell subset identified (5). To compare IL-9 expression among the T helper cell subsets, CD4+ T cells were cultured in vitro under Th1, Th2, and Th9 differentiation conditions by following the previously reported studies (6, 51). Expression level of IL-9 was measured among the various in vitro differentiated T helper subsets. Indeed, Th9 cells showed the highest IL-9 expression upon restimulation (supplemental Fig. S2, A–F). During in vitro Th9 cell differentiation, IL-9 expression was detectable on day 3 of culture and peaked at day 5 upon restimulation, and the highest expression of IL-9 was found between 8 and 12 h of stimulation and maintained until 24 h (supplemental Fig. S3, A and B) (56, 57). Next we performed a ChIP assay to detect a physical binding of NFAT1 to the predicted NFAT binding sites on the IL-9 promoter. NFAT1 enrichment was much higher in the IL-9 promoter of Th9 cells compared with Th1 cells, which correlates well with the IL-9 expression profiles between them (Fig. 2A). To further confirm the binding of NFAT1 to the IL-9 promoter, a DNA affinity purification assay was performed. Of the five NFAT binding sites, three probes were designed based on their highest matrix similarity (0.8): NFAT-a (−315/−306), NFAT-c (−223/−214), and NFAT-e (−48/−38). Biotinylated NFAT-e (−48/−38) probe efficiently pulled down NFAT1 protein from HEK-293 cell lysate overexpressing NFAT1 (Fig. 2B). In addition, the presence of nonbiotinylated competitor probes (NFAT-a (−315/−306), NFAT-c (−223/−214), and NFAT-e (−48/−38)) or consensus NFAT probe (Cons) from murine IL-2 promoter (described under “Experimental Procedures”) significantly reduced the pull-down efficiency (Fig. 2B). However, a mutated consensus competitor probe (mt-Cons) failed to inhibit the binding of biotinylated NFAT-e probe (lane 8 in Fig. 2B). This result confirmed that NFAT1 directly binds to the IL-9 promoter. Next we tested whether NFAT1 binding at the IL-9 promoter could enhance IL-9 transcription by performing IL-9 promoter reporter analysis. IL-9 promoter-driven luciferase activity was measured upon NFAT1 overexpression in HEK-293 and EL-4 T lymphoma cell lines. As a positive control for NFAT1-driven transactivation, CNS-9 containing IL-10 promoter was employed. Interestingly, overexpression of NFAT1 failed to transactivate the IL-9 promoter in both HEK-293 (Fig. 2C) and EL-4 T cells (Fig. 2D), whereas NFAT1 significantly increased the transactivity of CNS-9 containing IL-10 promoter in EL-4 cells. Reduction of CNS-9 containing IL-10 promoter activity in the presence of cyclosporin A further confirms the NFAT1-mediated transactivity of this CNS element (Fig. 2D) (30). These results suggest that although NFAT1 binds to the IL-9 promoter, it does not transactivate the IL-9 promoter.
FIGURE 2.
Physical association of NFAT1 with the IL-9 promoter. A, ChIP assay was performed with PMA/ionomycin-stimulated Th1 and Th9 cells using control IgG and NFAT1 antibody. The amounts of precipitated DNA were measured by quantitative PCR with primers specific for the IL-9 and IFN-γ promoter regions and are represented relative to their amount in total chromatin (input) as a fraction of input. B, left, antibodies against NFAT1 and actin (control) were used to perform Western blot for detecting NFAT1 expression in untransfected and NFAT1-transfected HEK cells. Right, biotin-conjugated probe corresponding to NFAT binding site (at −48/−38 in Fig. 1A; NFAT-e) were incubated with NFAT1-overexpressing HEK-293 cell lysate in the presence of the indicated non-biotinylated competitor probes. Cons and mt-Cons, conserved NFAT binding and NFAT nonbinding mutant probes, respectively. The protein DNA complexes were precipitated with streptavidin (Strep) and analyzed by immunoblotting with α-NFAT1 antibody. The first lane indicates input, which is 2.5% of the total cell extract (CE) used for pull-down. The data are representative of three independent experiments. C and D, HEK-293 (C) or EL-4 cells (D), respectively, were transfected with empty control (mock) or luciferase reporter constructs containing IL-9 promoter (IL-9 Pro; encompassing the −366/+48 region as shown in Fig. 1A) or enhancer containing IL-10 promoter (IL-10 CNS-9 (30)) in the presence of NFAT1 expression vector. Cells were stimulated with PMA/ionomycin for 8 h or with cyclosporin A (CsA) 20 min prior to PMA/ionomycin stimulation, and the luciferase assay was conducted. The luciferase activity was calculated relative to the activity of Renilla luciferase and represented in relative luciferase units (RLU) as a -fold difference relative to the control (mock/empty plasmid) value. The data shown are expressed as mean ± S.E. (error bars), n = 3; **, p < 0.01; ***, p < 0.001.
NFAT1-dependent IL-9 Expression in Th9 Cells
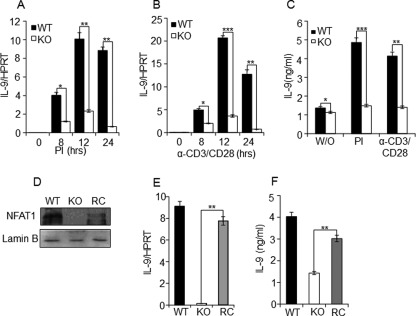
To elucidate the functional importance of NFAT1, we tested the effect of NFAT1 deficiency on IL-9 expression in Th9 cells. CD4+ T cells isolated from WT and NFAT1−/− (KO) mice were differentiated in vitro for 5 days into Th9 cells and were restimulated with PMA/ionomycin, or α-CD3/α-CD28. The expression levels of IL-9 transcript and protein between the groups were analyzed by qRT-PCR and ELISA, respectively. Compared with WT, the expression level of IL-9 in Th9 cells from NFAT1−/− mice was significantly decreased at all of the time points and under all of the stimulation conditions analyzed (Fig. 3, A and B). Down-regulated IL-9 mRNA level in NFAT1−/− Th9 cells is well correlated with the decreased level of IL-9 protein in the cell culture supernatants (Fig. 3C). To further validate the functional role of NFAT1 in IL-9 expression, we tested whether a reconstitution of NFAT1 into NFAT1−/− Th9 cells can restore IL-9 expression. NFAT1 expression plasmid was nucleofected in the Th9 cells derived from NFAT1−/− mice on day 3 of culture. After 48 h of nucleofection, cells were stimulated with PMA/ionomycin for 12 h. Reconstitution of NFAT1 into NFAT-deficient Th9 cells was confirmed by checking NFAT1 protein expression by Western blotting (Fig. 3D). Upon reconstitution (RC) of NFAT1, down-regulated IL-9 expression in NFAT1-deficient cells (KO) was successfully restored to the comparable levels of WT Th9 cells both in the mRNA (Fig. 3E) and protein levels (Fig. 3F). These results indicate the pivotal role of NFAT1 in IL-9 expression by Th9 cells.
FIGURE 3.
NFAT1 reconstitution restores IL-9 expression in the NFAT1-deficient Th9 cells. Th9 cells differentiated from WT and NFAT1−/− (KO) were stimulated by PI (A) or α-CD3/CD28 (B) for the indicated time, and IL-9 expression was measured by qRT-PCR by normalizing with the level of housekeeping gene hypoxanthine-guanine phosphoribosyltransferase. C, the IL-9 protein levels in culture supernatant that was either unstimulated (W/O) or restimulated for 24 h by PI or α-CD3/CD28 were quantified by ELISA. The data shown are expressed as mean ± S.E. (error bars) (n = 3); *, p < 0.05; **, p < 0.01; ***, p < 0.001. D, relative level of NFAT1 was analyzed in WT, KO, or NFAT1 reconstituted NFAT1−/− Th9 (RC) cells by Western blotting with α-NFAT1 antibody and α-lamin B (control). E and F, relative amount of IL-9 expression in the WT, NFAT1−/− (KO), and NFAT1 reconstituted NFAT1−/− Th9 (RC) cells was measured by qRT-PCR and ELISA, respectively. The data shown are expressed as mean ± S.E. (n = 3); **, p < 0.01.
Reduced Chromatin Accessibility of IL-9 Promoter in NFAT1−/− Th9 Cells
Previous studies showed that transcription factors IRF4, Pu.1, and STAT6 play decisive roles in Th9 cell lineage commitment. We tested whether a defect in IL-9 expression in NFAT1−/− Th9 cells was due to any changes in the expression levels of these transcription factors. In vitro differentiated Th9 cells from WT and NFAT1−/− (KO) mice were left unstimulated or restimulated with PMA/ionomycin for the indicated times, and nuclear extracts were used to check the nuclear level of NFAT1, Pu.1, IRF4, lamin B (nuclear control), and tubulin (cytosolic control) by Western blotting. However, no significant change was observed in the nuclear levels of IRF4 and Pu.1 between the WT and NFAT1−/− Th9 cells (Fig. 4A). We also confirmed that down-regulated IL-9 expression in NFAT1−/− condition is not mediated by alteration of Th9 development program because IL-9 expression was also significantly lower in the NFAT1−/− CD4+ T cell blasts compared with wild type counterparts (supplemental Fig. S4, A and B). Next, we questioned how the absence of NFAT1 could affect IL-9 gene expression without inducing any defect in Th9 differentiation or enhancing IL-9 promoter activation. We tested the possibility that NFAT1 is involved in the epigenetic modifications of the IL-9 promoter locus. One of the possible causes of reduced gene expression is the alteration of the chromatin architecture to a condensed and inaccessible form. This can be detected by analyzing accessibility of chromatin to an MNase-based PCR assay (58, 59). Thus, we analyzed the differential chromatin architecture of the IL-9 promoter region between the WT and NFAT1−/− Th9 cells by measuring relative MNase accessibility. Indeed, the IL-9 promoter region in NFAT1−/− Th9 cells was more resistant to MNase digestion compared with that of the WT cells (Fig. 4B). The accessibility of the IL-9 promoter from NFAT1−/− Th9 cells was restricted regardless of stimulation, whereas wild type Th9 cells showed a significant increase of accessibility upon stimulation (Fig. 4B). The actin promoter region was employed as a control for the constitutively active housekeeping gene and showed a similar accessibility regardless of NFAT1 deficiency (Fig. 4B).
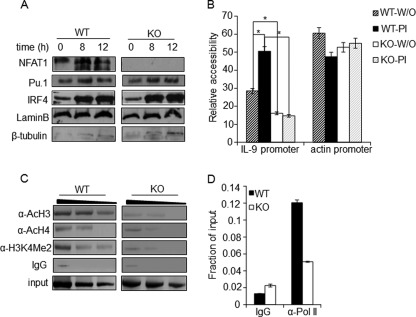
FIGURE 4.
NFAT1 regulates chromatin architecture at the IL-9 promoter. A, nuclear extracts were prepared from Th9 cells differentiated from WT and NFAT1−/− (KO) mice and stimulated with PMA/ionomycin for the indicated time. The nuclear levels of Pu.1, IRF4, lamin B (nuclear control), and β-tubulin (cytosolic control) were analyzed by immunoblotting with the respective antibodies. B, nuclei isolated from either unstimulated (W/O) or PI-stimulated WT and NFAT1−/− (KO) Th9 cells were left untreated or subjected to MNase digestion. Relative chromatin accessibility at the promoters of the IL-9 and actin promoter (as a control for the accessible region) was measured by qRT-PCR using specific primers. The results are represented as the ratio of PCR product obtained from digested samples normalized to the PCR products from undigested samples and mean ± S.E. (error bars) (n = 3); *, p < 0.05. C and D, PMA/ionomycin-stimulated Th9 cells from WT and NFAT1−/− (KO) mice were used for the ChIP assay with antibodies against acetylated histones (AcH3, AcH4, and H3K4Me2), RNA-Pol II, or control IgG. qRT-PCR with primer spanning the IL-9 promoter locus (−366/+48) was used to detect the precipitated DNA and represented as negative images of EtBR-stained gels (C) or relative to the amount in total chromatin (input) as fraction of input (D). All data are representative of at least three independent experiments.
In general, enrichment of acetyl histone H3 (AcH3), acetyl histone H4 (AcH4) and histone H3 lysine 4 dimethylation (H3K4Me2) to promoters correlates well with their transcriptionally active status (30, 50, 51). Thus, to further confirm the differential chromatin structure of the IL-9 promoter between WT and NFAT1−/− Th9 cells, relative amounts of recruited AcH3, AcH4, and H3K4Me2 levels were analyzed by a ChIP assay. Indeed, the enrichment levels of all of the tested active histone markers on the IL-9 promoter were significantly lowered in NFAT1−/− Th9 cells (Fig. 4C). We also measured RNA polymerase II (Pol II) binding to the IL-9 promoter because RNA Pol II enrichment marks actively transcribing promoter (34, 35). Correlating with the physiologically decreased binding of active histone markers, significantly lowered binding of Pol II was observed in NFAT1−/− Th9 cells (Fig. 4D). These results suggest that NFAT1 deficiency results in a transcriptionally inactive chromatin configuration at the IL-9 promoter.
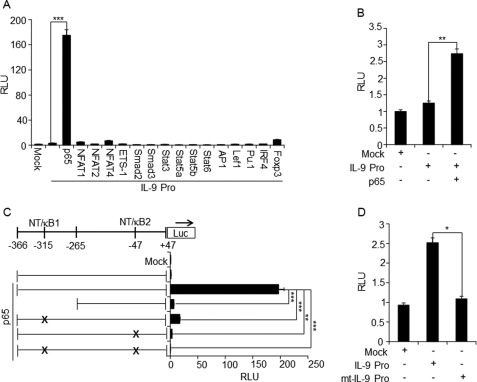
NF-κB (p65) Transactivates IL-9 Promoter
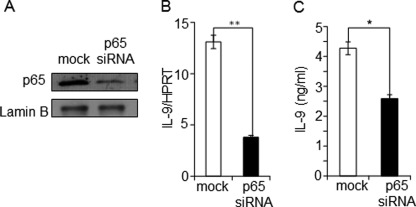
Generally, NFAT1 positively regulates transcription of a large number of inducible cytokine genes by directly binding to their promoters. ChIP and DNA affinity purification assay analysis showed the physiological binding of NFAT1 to the IL-9 promoter (Fig. 2, A and B). The findings that NFAT1 deficiency resulted in an inactive chromatin configuration (Fig. 4, B–D) and overexpression of NFAT1 failed to transactivate the IL-9 promoter (Fig. 2, C and D) suggest that NFAT1 may play a role in inducing or stabilizing the active chromatin status of the IL-9 promoter rather than activating it. Therefore, we asked if NFAT1 is not involved in the activation of IL-9 promoter, then which factor can drive IL-9 production? To identify candidate proteins that are subsequently recruited to the active IL-9 promoter and trigger the maximal expression of IL-9 upon stimulation, IL-9 promoter-driven luciferase reporter analysis was performed in the presence of the predicted transcription factors (supplemental Fig. S1) in HEK-293 cells. Among the tested transcription factors, only NF-κB (p65) significantly enhanced the IL-9 promoter activity (Fig. 5A). A significant increase of IL-9 promoter activity was also observed in EL4 T cells upon overexpression of NF-κB (p65) (Fig. 5B). To further confirm the NF-κB (p65)-dependent transactivation of IL-9 promoter, deletion or mutations were introduced in the NF-κB binding sites corresponding to the NT/κB1 (−315/−307) and NT/κB2 (−48/−38) (Fig. 1B and supplemental Fig. S1). A deletion or mutation of the predicted NF-κB binding sites (NT/κB1, NT/κB2, or both (NT/κB1,2)) completely abolished NF-κB-mediated transactivation of the IL-9 promoter in HEK-293 cells (Fig. 5C) and significantly reduced the activity of the IL-9 promoter in primary Th9 cells as well (Fig. 5D). Next we tested whether NF-κB (p65)-driven IL-9 enhancement is directly associated with physiological binding of NF-κB (p65) to the IL-9 promoter. Anti-NF-κB (p65) antibody was used to precipitate the chromatin prepared from in vitro differentiated and restimulated Th9 cells from WT and NFAT1−/− (KO) mice. Indeed, significant enrichment of NF-κB (p65) was observed at the IL-9 promoter in the Th9 cells compared with that of Th1 cells that produce negligible amounts of IL-9 (Fig. 6A). By performing a DNA affinity purification assay, we further confirmed the binding of NF-κB to the two predicted NF-κB (p65) binding sites at the IL-9 promoter. Efficient binding of NF-κB (p65) was detected when biotinylated NT/κB1 (−315/−306) probe was used to pull down NF-κB from HEK-293 cells that were transfected with NF-κB (p65) expression plasmid. Nonbiotinylated competitor probes significantly reduced NF-κB binding (lanes 4 and 5), whereas mutant competitors (lanes 6 and 7) failed to do so (Fig. 6B). To validate the functional involvement of NF-κB, the effect of p65 siRNA on IL-9 gene expression was tested. Th9 cells differentiated from WT mice were transfected with mock or p65 siRNA as described under “Experimental Procedures.” Transfection of p65 siRNA diminished the expression level of p65 compared with the cells transfected with control siRNA (mock), as confirmed by Western blotting (Fig. 7A). Indeed, knockdown of p65 expression significantly reduced IL-9 expression both at the mRNA and protein levels compared with scrambled siRNA-transfected cells (mock) (Fig. 7, B and C). These results suggest that the presence and physiological binding of NF-κB (p65) to the IL-9 promoter are essential for IL-9 expression in Th9 cells.
FIGURE 5.
NF-κB (p65) transactivates the IL-9 promoter. A, HEK-293 or B, EL-4 cells were transfected with the empty vector (Mock) or IL-9 promoter (−366/+48) (IL-9 Pro) containing luciferase reporter constructs in the presence of indicated expression vectors. C, HEK-293 cells were transfected with empty vector (Mock), IL-9 promoter, or mutant IL-9 promoter reporter constructs in the presence of NF-κB (p65) expression vector. D, Th9 cells were transfected with the empty vector (Mock), IL-9 promoter reporter construct, or mutated IL-9 promoter reporter construct that has a mutation in both NF-κB binding sites (mt-IL-9 Pro; NT/κB1 and NT/κB2). The luciferase activity was calculated relative to the activity of Renilla luciferase and represented in relative luciferase units (RLU) as a -fold difference relative to the control (mock/empty plasmid) value. The data are representative of at least three independent experiments. The data shown are expressed as mean ± S.E. (error bars) (n = 3); *, p < 0.05; **, p < 0.01; ***, p < 0.001.
FIGURE 6.
In vivo and in vitro binding of NF-κB (p65) to the IL-9 promoter. A, ChIP assay was performed with in vitro differentiated and PMA/ionomycin-stimulated Th1 and Th9 cells using control IgG and NF-κB (p65) antibodies. The amounts of precipitated DNA were measured by qRT-PCR with primers specific for the IL-9 and IL-2 promoter regions. Relative NF-κB (p65) enrichment in the precipitated samples compared with total chromatin (input) is shown as a fraction of input. B, left, antibodies against NF-κB (p65) and actin (control) were used to perform Western blot for detecting NF-κB (p65) expression in untransfected and NF-κB (p65)-transfected HEK cells. Right, biotin-conjugated probe corresponding to NF-κB (p65) binding site 1 (NT/κB-1; −315/−306 in Fig. 1B) were incubated with NF-κB (p65)-overexpressing HEK-293 cell lysate in the absence or presence of the indicated non-biotinylated competitor probes. The protein-DNA complexes were precipitated with streptavidin (Strep) and analyzed by immunoblotting with α-NF-κB (p65) antibody. The first lane indicates input, which is 2.5% of the total cell extract (CE) used for pull-down. The data are representative of three independent experiments.
FIGURE 7.
Knockdown of NF-κB (p65) reduces IL-9 expression. In vitro differentiated primary Th9 cells were transfected with siRNA against NF-κB (p65) or non-relevant siRNA (mock). A, the knockdown efficiency of p65 was confirmed by immunoblotting the respective cell lysates with antibodies against p65 and lamin B (control). B, relative level of IL-9 transcript in the mock- and p65 siRNA-transfected cells were analyzed by qRT-PCR and represented relative to the expression level of the housekeeping gene hypoxanthine-guanine phosphoribosyltransferase. C, p65 siRNA- or mock-transfected Th9 cells were stimulated for 24 h with PMA/ionomycin, and the IL-9 protein levels in the culture supernatant were measured by ELISA. The data shown are expressed as mean ± S.E. (error bars) (n = 3); *, p < 0.05; **, p < 0.01.
NFAT1-mediated Recruitment of Transcriptional Activation Complex Enhances IL-9 Expression
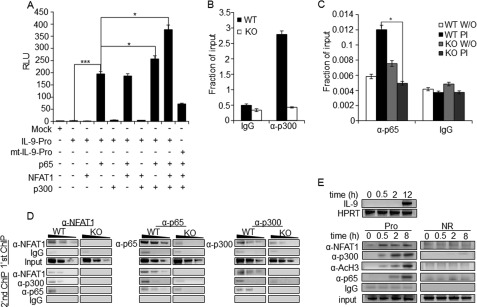
To further delineate the mechanism of functional synergy between the transcription factors in driving IL-9 expression, we tested the effect of overexpression of each transcription factors or their combinations on IL-9 promoter activity. Overexpression of NFAT1 alone or NFAT1 together with NF-κB failed to further enhance the NF-κB-induced IL-9 promoter activity (Fig. 8A). Interestingly, the histone acetyltransferase p300 has been reported to interact with both NFAT and NF-κB and is essential for NF-κB-mediated transactivation (34, 44, 45). Therefore, we tested the functional synergism between the factors p300, NF-κB (p65), and NFAT1 by performing IL-9 reporter assay. Indeed, co-expression of p300, NF-κB (p65), and NFAT1 significantly increased NF-κB (p65)-mediated IL-9 promoter activity (Fig. 8A, lane 9). However, the promoter construct with mutations in both of the NF-κB binding sites (mt-IL-9) abolished the enhancement of promoter activity in the presence of these three factors (Fig. 8A, lane 10). This result indicates that coactivator protein p300 acts as a bridge between the two transcription factors, leading to a functional synergy between them. This was further proved by the reduced binding of p300 to the IL-9 promoter in NFAT1−/− Th9 cells (Fig. 8B). Next, we tested the effect of NFAT1 deficiency on the recruitment of NF-κB (p65) to the IL-9 promoter. Upon stimulation, significant enrichment of NF-κB (p65) to the IL-9 promoter was observed in WT Th9 cells. However, NFAT1-deficient cells did not show such enrichment (Fig. 8C). These results suggest that NFAT1 is mainly involved in remodeling of the IL-9 promoter with transcriptional active status, where recruitment of p300 and NF-κB (p65) transactivates IL-9 promoter activity. We further tested the effect of NFAT1 deficiency on co-recruitment of p300 and NF-κB (p65) to the IL-9 promoter in Th9 cells by performing a ChIP-re-ChIP experiment. Chromatins were prepared from stimulated WT or NFAT1−/− Th9 cells, and a ChIP assay was performed with anti-NFAT1 (Fig. 8D, left), anti-p65 (Fig. 8D, middle), and anti-p300 (Fig. 8D, right) antibodies. The precipitated chromatin from each group was subjected to a second round of ChIP assay, using the indicated antibodies. Specific enrichment of the indicated factors was compared with control IgG. Next, to further characterize the functional role of NFAT1 as an active chromatin modifier and NF-κB (p65) as a transactivator, respectively, we tested the recruitment kinetics of activation complex (NFAT1, p300, and NF-κB (p65)) and its effect on IL-9 expression. Th9 cells were stimulated for the indicated time period, and the expression level of IL-9 (Fig. 8E, top) and relative recruitment of NFAT1 and other factors to the IL-9 promoter (Pro) or non-relevant region (NR) were analyzed (Fig. 8E, bottom). Compared with other proteins, earlier binding of NFAT1 was observed within 30 min of stimulation, and its level was maintained until a later period of stimulation (Fig. 8E, bottom). The binding levels of p300, AcH3, and NF-κB (p65) to the IL-9 promoter were increased with time, and the maximal binding of NF-κB p65 was detected at 8 h of stimulation (Fig. 8E, bottom). This result correlated well with the time-dependent increase of IL-9 expression that reaches a peak at later hours of stimulation (Fig. 8E, top, and supplemental Fig. S3B) (56, 57). A non-relevant control region (NR), however, failed to show enrichment with the tested proteins compared with control IgG antibody (Fig. 8E, bottom). Collectively, these results suggest that the activation complex assembled on the IL-9 promoter consists of NFAT1 (as a chromatin activator), p300 (as an active chromatin modifier), and NF-κB (p65) (as a transactivator). A functional synergy between these factors may potentiate high levels of IL-9 expression in Th9 cells.
FIGURE 8.
NFAT1 mediates the recruitment of activation complex to enhance IL-9 promoter activity. A, HEK-293 cells were transfected with empty control vector (Mock), IL-9 promoter (IL-9 Pro), or mutant IL-9 promoter (mt-IL-9 Pro (mutations in both NT/κB1 and NT/κB1 sites)) reporter constructs in the presence of the indicated expression vectors. Cells were stimulated with PMA/ionomycin for 8 h and harvested for the luciferase assay. The luciferase activity was calculated relative to the activity of Renilla luciferase and represented in relative luciferase units (RLU) as a -fold difference relative to the control (mock/empty plasmid) value. The data represent mean ± S.E. (error bars) (n = 3); *, p < 0.05; ***, p < 0.001. B and C, ChIP assay was performed with Th9 cells differentiated from WT and NFAT1−/− (KO) mice using control IgG and p300 (B) and p65 antibodies (C). The amounts of precipitated DNA were measured by qRT-PCR with primers specific for the IL-9 promoter region and are represented relative to the amount in total chromatin (input) as a fraction of input. W/O, untreated. The data represent at least three independent experiments. The data represent mean ± S.E. (n = 3); *, p < 0.05; **, p < 0.01. D, ChIP-re-ChIP experiment. Th9 cells from WT and NFAT1−/− (KO) mice were stimulated with PMA/ionomycin and subjected to ChIP with either α-NFAT1, α-p65, α-p300, or control IgG antibodies (first ChIP), and the immunoprecipitates were eluted either with elution buffer or 10 mm DTT and proceeded with second ChIP with the indicated antibodies. The amount of precipitated DNA was then detected by qRT-PCR with the primer spanning the IL-9 promoter locus and is presented as negative images of the EtBr-stained gel. Data are representative of at least three independent experiments. E, Th9 cells were restimulated with PMA/ionomycin for the indicated time periods, and IL-9 levels were measured by qRT-PCR (top). The ChIP assay was performed with the indicated antibodies, and the relative amount of precipitated IL-9 promoter DNA was then detected by qRT-PCR and is presented as negative images of EtBr-stained gel (bottom). Data are representative of at least three independent experiments.
DISCUSSION
The principle aim of our study is to elucidate the molecular mechanism of IL-9 gene transcription in Th9 cells. We demonstrated that functional cooperation of NFAT1 and NF-κB (p65) synergistically enhances IL-9 expression in Th9 cells. Binding of NFAT1 to the IL-9 promoter results in a transcriptionally competent IL-9 promoter by modifying it with active histone marks. This active chromatin configuration thereby favors the recruitment NF-κB (p65) to enhance IL-9 expression in Th9 cells. Consequently, NFAT1 deficiency or knockdown of NF-κB (p65) results in a significant down-regulation of IL-9 expression by affecting both the chromatin architecture and the formation of activation complex, respectively, at the IL-9 promoter.
Recent studies have identified the transcription factors involved in regulation of Th9 cell differentiation and IL-9 transcription. The ETS family and TGF-β-induced transcription factor, Pu.1, has been implicated in controlling IL-9 expression, and a high level of Pu.1 is essential for the Th9 cell differentiation program (21, 23, 24). T cell-specific deletion or knockdown of Pu.1 in mice or human T cells results in a significant reduction of IL-9 expression (21). The Pu.1-interacting and IL-4-up-regulated protein IRF4 is another transcription factor proven to be indispensable for Th9 cell development. The absence of IRF4 also prevented Th9 cell generation (22). STAT6 is yet another newly found transcription factor acting downstream of IL-4 signaling that affects Th9 lineage commitment by inducing IRF4 expression and repressing Tbet and FoxP3 expression in Th9 cells (23). However, the role of TCR-induced transcription factors responsible for acute IL-9 induction from Th9 cells is still unclear.
In this study, we have mainly identified two important TCR-induced transcription factors, NFAT1 and NF-κB, that functionally synergize to activate IL-9 gene transcription. NFAT1−/− Th9 cells display significantly reduced IL-9 expression (Fig. 3, A–C), whereas reconstitution of NFAT1 into NFAT1−/− Th9 cells restored IL-9 expression (Fig. 3, D–F). Based on these findings, we have investigated the underlying mechanism of NFAT1-mediated IL-9 expression in Th9 cells.
First, we tested whether NFAT1 deficiency could alter the Th9 differentiation program, thereby down-regulating IL-9 expression. However, we did not find any changes in the nuclear levels of reported Th9 lineage determinants, such as Pu.1 and IRF4, between the WT and NFAT1−/− Th9 cells (Fig. 4A). In addition, NFAT1 deficiency also decreased IL-9 expression in CD4+ T cell blasts that were not polarized into any specific T helper cell types (supplemental Fig. S4, A and B). This result thereby rules out the possibility that the absence of NFAT1 results in dysregulated Th9 differentiation, at least in the context of Pu.1 and IRF4 expression. Another possible outcome of NFAT1 deficiency-mediated IL-9 reduction could be the differential expression of the cytokines necessary for IL-9 production. IFN-γ has been shown to be a negative regulator of IL-9 expression in human Th9 cells (60). The absence of Th9 lineage factor IRF4 results in a huge increase in IFN-γ expression and hence could be another potential mechanism by which IL-9 expression is reduced in IRF4−/− mice (22). In this study, to rule out the possibility of IFN-γ involvement in dysregulated IL-9 expression in NFAT1−/− Th9 cells, Th9 cells were differentiated with IL-4 and TGF-β in the presence and absence of α-IFN-γ. Expression of IFN-γ was reduced upon neutralization of IFN-γ, and IL-9 expression was further increased both in the wild type and NFAT1−/− Th9 cells. Thus, IFN-γ signaling may not interfere with IL-9 expression in NFAT1−/− Th9 cells (supplemental Fig. S5, A and B). In addition to IFN-γ, IL-21 has been shown to be a positive regulator of IL-9 expression in human Th9 cells (60). IL-21 is itself regulated by NFAT1 (33), and thus this axis may also account for decreased IL-9 expression in NFAT1−/− cells.
Second, we tested the role of NFAT1 as a transcription factor that potentiates IL-9 promoter activity by directly binding to the IL-9 promoter. Indeed, NFAT1 binds to the NFAT1-responsive elements at the IL-9 promoter, but it failed to enhance its activity (Fig. 2). So we tested the possibility that NFAT1 could act as a chromatin remodeling factor. Previous studies have demonstrated that the N termini of NFATs interact physically with intrinsic histone acetylase CBP/p300, which enables NFAT to act as a chromatin-remodeling factor (34, 35). For example, in the case of GM-CSF enhancer, NFAT is responsible for the formation of a permissive chromatin architecture, thereby providing an access to AP1 that is obligatory for transactivating the GM-CSF enhancer (61). A recent report demonstrated that NFAT binding to the c-Myc promoter resulted in an increased acetylation of the c-Myc promoter via recruitment of the p300 coactivator. This acetylated c-Myc promoter subsequently binds to the ETS family transcription factor ELK-1 and triggers the maximal expression of c-Myc in the pancreatic cancer cell line (35). In our present study, we also found that NFAT1 deficiency reduced chromatin accessibility and recruitment of active histone markers and Pol II to the IL-9 promoter (Fig. 4, B–D).
The immediate follow-up question is as follows. What are the other factors that can potentiate IL-9 expression while acting in concert with NFAT1 in Th9 cells? In this context, we identified NF-κB (p65) as another TCR-induced transcription factor that works in functional synergy with NFAT1 and drives IL-9 expression. NF-κB serves as a transcriptional activator to enhance IL-9 gene expression in Th9 cells. Significant enrichment of NF-κB (p65) at the IL-9 promoter potentiated its activity, whereas siRNA-mediated knockdown of p65 in Th9 cells significantly decreased IL-9 expression (Figs. 5–7). In addition, we also elucidated the mechanism by which a functional synergy is achieved between NFAT1 and NF-κB in the context of IL-9 expression. NFATs often influence gene transcription, acting synergistically with several other transcription factors by assembling enhanceosomes together with coactivators CBP and p300 (25, 62–64). In addition to histone acetyltransferase activity, CBP/p300 has well known functions as scaffolding proteins, which leads to the formation of enhanceosomes onto gene promoter and regulatory elements (65). Incidentally, NF-κB (p65) also physically interacts with the p300/CBP, and this is essential and crucial for NF-κB (p65)-mediated transactivation (44, 45, 66, 67). Interestingly, in our study, we found that in the absence of NFAT1, there is a significant reduction of p300 recruitment to the IL-9 promoter (Fig. 8). This accounts for the creation of a hypoacetylated chromatin environment at the IL-9 locus and also manifests in the impaired and unstable binding of NF-κB (p65) to the IL-9 promoter (Fig. 8, B and C). However, the IL-9 promoter reporter plasmid showed similar activity in the NFAT1−/− Th9 cells as that in the wild type (supplemental Fig. S6). The activation of the IL-9 reporter construct with respect to the mock-transfected (empty vector) samples was ∼2.6- and 2.9-fold in the case of WT and KO cells, respectively (supplemental Fig. S6). The activity of the mutant promoter construct also decreased as in the case of transfection in WT cells (supplemental Fig. S6). This could be due the following facts. (i) An abundant amount of p65 protein was present both in the WT and NFAT1−/− Th9 cells (supplemental Fig. S6, inset). (ii) Because p65 and NFAT1 share the same binding sites (2 of the 5 NFAT binding sites) in the IL-9 promoter (Figs. 1A, 2B, and 6B), p65 can successfully maintain the promoter activity in the absence of NFAT1. Overlapping NFAT and NF-κB binding sites have been identified in HIV-1 LTR, IL-8, IL-13, GM-CSF, IFN-γ, TNF-α, etc, promoter and enhancer elements also (36, 68–71). This is due to the presence of a similar DNA binding domain (RHR domain) in the NFAT and NF-κB/Rel family of proteins (72). In the context of the IL-9 promoter, we have found that NFAT1-mediated chromatin activation is crucial for p65 binding to the IL-9 promoter, thereby indicating that the IL-9 promoter is responsive to both NFAT1 and NF-κB. The differential in vivo binding of p65 to the IL-9 promoter in the WT and NFAT1-deficient (KO) Th9 cells (Fig. 8, C and D) and the decrease in IL-9 expression upon p65 knockdown (Fig. 7) clearly establish the fact that both NFAT1 and p65 are indispensable for IL-9 expression. (iii) Additionally, the IL-9 reporter plasmid is an artificial in vitro designed construct used in the transient transfection assay. Thus, in this situation, its reporter activity is not susceptible to chromatin configuration changes unless it integrates into the genome as a transgene, which is possible only in a stable transfection system and not in a transient transfection assay (73, 74).
Our study also suggests a functional importance of cross-talk between the transcription factors and their co-activators. Physical association of NFAT1, NF-κB (p65), and p300 with the IL-9 promoter was confirmed by the ChIP-re-ChIP experiment (Fig. 8D). NFAT1 deficiency significantly reduced the recruitment of NFAT-interacting activation complexes, such as NF-κB and p300, to the IL-9 promoter, whereas co-overexpression of NFAT1, NF-κB (p65), and p300 significantly enhanced IL-9 promoter activity (Fig. 8, A and D). A recent paper suggested a two-step process in the assembly of a transcription factor complex on the IL-2 promoter. NFAT1, Jun, and Fos are the early and Oct2, Rel A (p65), c-Rel, and NFAT2 are the late binding factors, respectively (75). Thus, sequential binding of transcription factors and coactivators are probably essential and general series of events for the TCR-induced IL-9 expression also. Indeed, a time-dependent ChIP assay revealed that binding of NFAT1 to the IL-9 promoter is an initial event compared with other factors such as p300, AcH3, and NF-κB (p65) (Fig. 8E). NFAT1 binding to the IL-9 promoter region thus induces a net histone acetylation by co-recruitment of histone acetyltransferase p300, which results in the formation of a hyperacetylated, transcriptionally competent IL-9 promoter. This active promoter thereby recruits NF-κB (p65) and ensures maximal expression of IL-9. Therefore, we suggest that IL-9 transcription in Th9 cells is driven by a transcription activator complex consisting of NFAT1, p300, and NF-κB (p65), and in this process, p300 serves as a bridge that not only acetylates the promoter but also stabilizes the binding of NF-κB (p65). In this study, we have identified the functional importance of NFAT1 and NF-κB in enhancing IL-9 expression. However, we certainly cannot exclude the role of other transcription factors involved. The expression level of IL-9 is further enhanced under Th9 differentiation conditions compared with CD4+ T cell blasts, which suggests a possibility that NFAT1 may cooperate with Th9 lineage-determining transcription factors as well. IL-9 has also newly been described as a Th17 type cytokine, and subsequent roles of IL-9 in Th17-mediated inflammatory responses have also been implicated (18, 76). Interestingly, the HTLV-transformed cell line Hut-102 has an IL-9-producing Th17 phenotype. It is shown that in this cell, specific knockdown of NF-κB protein c-Rel results in reduced IL-9 production (77). We have also found that NFAT1-deficient Th17 cells have a defect in IL-9 expression (data not shown). Thus, the ubiquitous expression pattern of NFAT1 and NF-κB, yet a differential effect in the T helper cell subsets, can be exploited to elucidate their roles in regulating IL-9 expression in association with other transcription factor partners.
In summary, our study constitutes the first report showing that TCR-induced transcription factors NFAT1 and NF-κB (p65) play a crucial role in the regulation of IL-9 expression in Th9 cells. NFAT1 essentially creates an accessible platform for the assembly of transcriptional coactivators on the IL-9 promoter, whereas NF-κB (p65) potentiates maximal expression of IL-9. Thus, our study delineates an important role of NFAT1 and NF-κB (p65) in IL-9 expression by T lymphocytes.
Supplementary Material
Acknowledgments
We thank our colleagues in the laboratory for valuable comments on the work and Dr. Darren Williams (Gwangju Institute of Science and Technology) for critical review of the manuscript.
This work was supported by National Research Foundation Grant 2011-0028529, funded by the Korean government, and by a Systems Biology Infrastructure Establishment Grant provided by the Gwangju Institute of Science and Technology in 2012.

This article contains supplemental Figs. S1–S6.
- NFAT
- nuclear factor of activated T cells
- PMA
- phorbol 12-myristate 13-acetate
- qRT-PCR
- quantitative RT-PCR
- MNase
- micrococcal nuclease
- AcH3 and AcH4
- acetyl histone H3 and H4, respectively
- H3K4Me2
- dimethyl lysine histone H3
- Pol II
- polymerase II
- PI
- PMA/ionomycin.
REFERENCES
- 1. Korn T., Bettelli E., Gao W., Awasthi A., Jäger A, Strom T. B., Oukka M., Kuchroo V. K. (2007) IL-21 initiates an alternative pathway to induce proinflammatory TH17 cells. Nature 448, 484–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sutton C., Brereton C., Keogh B., Mills K. H., Lavelle E. C. (2006) A crucial role for interleukin (IL)-1 in the induction of IL-17-producing T cells that mediate autoimmune encephalomyelitis. J. Exp. Med. 203, 1685–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Komiyama Y., Nakae S., Matsuki T., Nambu A., Ishigame H., Kakuta S., Sudo K., Iwakura Y. (2006) IL-17 plays an important role in the development of experimental autoimmune encephalomyelitis. J. Immunol. 177, 566–573 [DOI] [PubMed] [Google Scholar]
- 4. Zhou L., Chong M. M., Littman D. R. (2009) Plasticity of CD4+ T cell lineage differentiation. Immunity 30, 646–655 [DOI] [PubMed] [Google Scholar]
- 5. Dardalhon V., Awasthi A., Kwon H., Galileos G., Gao W., Sobel R. A., Mitsdoerffer M., Strom T. B., Elyaman W., Ho I. C., Khoury S., Oukka M., Kuchroo V. K. (2008) IL-4 inhibits TGF-β-induced Foxp3+ T cells and, together with TGF-β, generates IL-9+ IL-10+ Foxp3− effector T cells. Nat. Immunol. 9, 1347–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Veldhoen M., Uyttenhove C., van Snick J., Helmby H., Westendorf A., Buer J., Martin B., Wilhelm C., Stockinger B. (2008) Transforming growth factor-β “reprograms” the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat. Immunol. 9, 1341–1346 [DOI] [PubMed] [Google Scholar]
- 7. Nowak E. C., Noelle R. J. (2009) Interleukin-9 and T cell subsets. Cell Cycle 8, 3798–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Renauld J. C., Druez C., Kermouni A., Houssiau F., Uyttenhove C., Van Roost E., Van Snick J. (1992) Expression cloning of the murine and human interleukin 9 receptor cDNAs. Proc. Natl. Acad. Sci. U.S.A. 89, 5690–5694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Renauld J. C., Kermouni A., Vink A., Louahed J., Van Snick J. (1995) Interleukin-9 and its receptor. Involvement in mast cell differentiation and T cell oncogenesis. J. Leukoc. Biol. 57, 353–360 [DOI] [PubMed] [Google Scholar]
- 10. Hültner L., Druez C., Moeller J., Uyttenhove C., Schmitt E., Rüde E., Dörmer P., Van Snick J. (1990) Mast cell growth-enhancing activity (MEA) is structurally related and functionally identical to the novel mouse T cell growth factor P40/TCGFIII (interleukin 9). Eur. J. Immunol. 20, 1413–1416 [DOI] [PubMed] [Google Scholar]
- 11. Uyttenhove C., Simpson R. J., Van Snick J. (1988) Functional and structural characterization of P40, a mouse glycoprotein with T-cell growth factor activity. Proc. Natl. Acad. Sci. U.S.A. 85, 6934–6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hauber H. P., Bergeron C., Hamid Q. (2004) IL-9 in allergic inflammation. Int. Arch. Allergy Immunol. 134, 79–87 [DOI] [PubMed] [Google Scholar]
- 13. Steenwinckel V., Louahed J., Lemaire M. M., Sommereyns C., Warnier G., McKenzie A., Brombacher F., Van Snick J., Renauld J. C. (2009) IL-9 promotes IL-13-dependent paneth cell hyperplasia and up-regulation of innate immunity mediators in intestinal mucosa. J. Immunol. 182, 4737–4743 [DOI] [PubMed] [Google Scholar]
- 14. Temann U. A., Laouar Y., Eynon E. E., Homer R., Flavell R. A. (2007) IL9 leads to airway inflammation by inducing IL13 expression in airway epithelial cells. Int. Immunol. 19, 1–10 [DOI] [PubMed] [Google Scholar]
- 15. Townsend J. M., Fallon G. P., Matthews J. D., Smith P., Jolin E. H., McKenzie N. A. (2000) IL-9-deficient mice establish fundamental roles for IL-9 in pulmonary mastocytosis and goblet cell hyperplasia but not T cell development. Immunity 13, 573–583 [DOI] [PubMed] [Google Scholar]
- 16. Elyaman W., Bradshaw E. M., Uyttenhove C., Dardalhon V., Awasthi A., Imitola J., Bettelli E., Oukka M., van Snick J., Renauld J. C., Kuchroo V. K., Khoury S. J. (2009) IL-9 induces differentiation of TH17 cells and enhances function of FoxP3+ natural regulatory T cells. Proc. Natl. Acad. Sci. U.S.A. 106, 12885–12890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li H., Nourbakhsh B., Ciric B., Zhang G. X., Rostami A. (2010) Neutralization of IL-9 ameliorates experimental autoimmune encephalomyelitis by decreasing the effector T cell population. J. Immunol. 185, 4095–4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nowak E. C., Weaver C. T., Turner H., Begum-Haque S., Becher B., Schreiner B., Coyle A. J., Kasper L. H., Noelle R. J. (2009) IL-9 as a mediator of Th17-driven inflammatory disease. J. Exp. Med. 206, 1653–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eller K., Wolf D., Huber J. M., Metz M., Mayer G., McKenzie A. N., Maurer M., Rosenkranz A. R., Wolf A. M. (2011) IL-9 production by regulatory T cells recruits mast cells that are essential for regulatory T cell-induced immune suppression. J. Immunol. 186, 83–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Noelle R. J., Nowak E. C. (2010) Cellular sources and immune functions of interleukin-9. Nat. Rev. Immunol. 10, 683–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chang H. C., Sehra S., Goswami R., Yao W., Yu Q., Stritesky G. L., Jabeen R., McKinley C., Ahyi A. N., Han L., Nguyen E. T., Robertson M. J., Perumal N. B., Tepper R. S., Nutt S. L., Kaplan M. H. (2010) The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat. Immunol. 11, 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Staudt V., Bothur E., Klein M., Lingnau K., Reuter S., Grebe N., Gerlitzki B., Hoffmann M., Ulges A., Taube C., Dehzad N., Becker M., Stassen M., Steinborn A., Lohoff M., Schild H., Schmitt E., Bopp T. (2010) Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells. Immunity 33, 192–202 [DOI] [PubMed] [Google Scholar]
- 23. Goswami R., Jabeen R., Yagi R., Pham D., Zhu J., Goenka S., Kaplan M. H. (2012) STAT6-dependent regulation of Th9 development. J. Immunol. 188, 968–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Perumal N. B., Kaplan M. H. (2011) Regulating Il9 transcription in T helper cells. Trends Immunol. 32, 146–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hermann-Kleiter N., Baier G. (2010) NFAT pulls the strings during CD4+ T helper cell effector functions. Blood 115, 2989–2997 [DOI] [PubMed] [Google Scholar]
- 26. Macian F. (2005) NFAT proteins. Key regulators of T-cell development and function. Nat. Rev. Immunol. 5, 472–484 [DOI] [PubMed] [Google Scholar]
- 27. Im S. H., Rao A. (2004) Activation and deactivation of gene expression by Ca2+/calcineurin-NFAT-mediated signaling. Mol. Cells 18, 1–9 [PubMed] [Google Scholar]
- 28. Cantrell D. (1996) T cell antigen receptor signal transduction pathways. Annu. Rev. Immunol. 14, 259–274 [DOI] [PubMed] [Google Scholar]
- 29. Weiss A., Littman D. R. (1994) Signal transduction by lymphocyte antigen receptors. Cell 76, 263–274 [DOI] [PubMed] [Google Scholar]
- 30. Lee C. G., Kang K. H., So J. S., Kwon H. K., Son J. S., Song M. K., Sahoo A., Yi H. J., Hwang K. C., Matsuyama T., Yui K., Im S. H. (2009) A distal cis-regulatory element, CNS-9, controls NFAT1 and IRF4-mediated IL-10 gene activation in T helper cells. Mol. Immunol. 46, 613–621 [DOI] [PubMed] [Google Scholar]
- 31. Rengarajan J., Mowen K. A., McBride K. D., Smith E. D., Singh H., Glimcher L. H. (2002) Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J. Exp. Med. 195, 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tone Y., Furuuchi K., Kojima Y., Tykocinski M. L., Greene M. I., Tone M. (2008) Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 9, 194–202 [DOI] [PubMed] [Google Scholar]
- 33. Mehta D. S., Wurster A. L., Weinmann A. S., Grusby M. J. (2005) NFATc2 and T-bet contribute to T-helper cell subset-specific regulation of IL-21 expression. Proc. Natl. Acad. Sci. U.S.A. 102, 2016–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. García-Rodríguez C., Rao A. (1998) Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/CREB-binding protein (CBP). J. Exp. Med. 187, 2031–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Köenig A., Linhart T., Schlengemann K., Reutlinger K., Wegele J., Adler G., Singh G., Hofmann L., Kunsch S., Büch T., Schäfer E., Gress T. M., Fernandez-Zapico M. E., Ellenrieder V. (2010) NFAT-induced histone acetylation relay switch promotes c-Myc-dependent growth in pancreatic cancer cells. Gastroenterology 138, 1189–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rao A., Luo C., Hogan P. G. (1997) Transcription factors of the NFAT family. Regulation and function. Annu. Rev. Immunol. 15, 707–747 [DOI] [PubMed] [Google Scholar]
- 37. Gessner A., Blum H., Röllinghoff M. (1993) Differential regulation of IL-9-expression after infection with Leishmania major in susceptible and resistant mice. Immunobiology 189, 419–435 [DOI] [PubMed] [Google Scholar]
- 38. Kadereit S., Mohammad S. F., Miller R. E., Woods K. D., Listrom C. D., McKinnon K., Alali A., Bos L. S., Iacobucci M. L., Sramkoski M. R. (1999) Reduced NFAT1 protein expression in human umbilical cord blood T lymphocytes. Blood 94, 3101–3107 [PubMed] [Google Scholar]
- 39. Kaminski B. A., Kadereit S., Miller R. E., Leahy P., Stein K. R., Topa D. A., Radivoyevitch T., Veigl M. L., Laughlin M. J. (2003) Reduced expression of NFAT-associated genes in UCB versus adult CD4+ T lymphocytes during primary stimulation. Blood 102, 4608–4617 [DOI] [PubMed] [Google Scholar]
- 40. Baldwin A. S., Jr. (1996) The NF-κB and IκB proteins. New discoveries and insights. Annu. Rev. Immunol. 14, 649–683 [DOI] [PubMed] [Google Scholar]
- 41. Baeuerle P. A., Henkel T. (1994) Function and activation of NF-κB in the immune system. Annu. Rev. Immunol. 12, 141–179 [DOI] [PubMed] [Google Scholar]
- 42. Grumont R. J., Gerondakis S. (2000) Rel induces interferon regulatory factor 4 (IRF-4) expression in lymphocytes. Modulation of interferon-regulated gene expression by Rel/nuclear factor κB. J. Exp. Med. 191, 1281–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sun S. C., Yamaoka S. (2005) Activation of NF-κB by HTLV-I and implications for cell transformation. Oncogene 24, 5952–5964 [DOI] [PubMed] [Google Scholar]
- 44. Gao Z., Chiao P., Zhang X., Zhang X., Lazar M. A., Seto E., Young H. A., Ye J. (2005) Coactivators and corepressors of NF-KB in IkBα gene promoter. J. Biol. Chem. 280, 21091–21098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Na S. Y., Lee S. K., Han S. J., Choi H. S., Im S. Y., Lee J. W. (1998) Steroid receptor coactivator-1 interacts with the p50 subunit and coactivates nuclear factor κB-mediated transactivations. J. Biol. Chem. 273, 10831–10834 [DOI] [PubMed] [Google Scholar]
- 46. Stassen M., Müller C., Arnold M., Hültner L., Klein-Hessling S., Neudörfl C., Reineke T., Serfling E., Schmitt E. (2001) IL-9 and IL-13 production by activated mast cells is strongly enhanced in the presence of lipopolysaccharide. NF-κB is decisively involved in the expression of IL-9. J. Immunol. 166, 4391–4398 [DOI] [PubMed] [Google Scholar]
- 47. Zhu Y. X., Kang L. Y., Luo W., Li C. C., Yang L., Yang Y. C. (1996) Multiple transcription factors are required for activation of human interleukin 9 gene in T cells. J. Biol. Chem. 271, 15815–15822 [DOI] [PubMed] [Google Scholar]
- 48. Frazer K. A., Pachter L., Poliakov A., Rubin E. M., Dubchak I. (2004) VISTA. Computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Demoulin J. B., Van Roost E., Stevens M., Groner B., Renauld J. C. (1999) Distinct roles for STAT1, STAT3, and STAT5 in differentiation gene induction and apoptosis inhibition by interleukin-9. J. Biol. Chem. 274, 25855–25861 [DOI] [PubMed] [Google Scholar]
- 50. Im S. H., Hueber A., Monticelli S., Kang K. H., Rao A. (2004) Chromatin-level regulation of the IL10 gene in T cells. J. Biol. Chem. 279, 46818–46825 [DOI] [PubMed] [Google Scholar]
- 51. Sahoo A., Lee C. G., Jash A., Son J. S., Kim G., Kwon H. K., So J. S., Im S. H. (2011) Stat6 and c-Jun Mediate Th2 Cell-specific IL-24 Gene Expression. J. Immunol. 186, 4098–4109 [DOI] [PubMed] [Google Scholar]
- 52. Lerner L., Henriksen M. A., Zhang X., Darnell J. E. (2003) STAT3-dependent enhanceosome assembly and disassembly. Synergy with GR for full transcriptional increase of the 2-macroglobulin gene. Genes Dev. 17, 2564–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xu L., Han C., Lim K., Wu T. (2006) Cross-talk between peroxisome proliferator-activated receptor and cytosolic phospholipase A2/cyclooxygenase-2/prostaglandin E2 signaling pathways in human hepatocellular carcinoma cells. Cancer Res. 66, 11859–11868 [DOI] [PubMed] [Google Scholar]
- 54. Timmerman L. A., Healy J. I., Ho S. N., Chen L., Goodnow C. C., Crabtree G. R. (1997) Redundant expression but selective utilization of nuclear factor of activated T cell family members. J. Immunol. 159, 2735–2740 [PubMed] [Google Scholar]
- 55. Macián F, García-Cózar F., Im S. H., Horton H. F., Byrne M. C., Rao A. (2002) Transcriptional mechanisms underlying lymphocyte tolerance. Cell 109, 719–731 [DOI] [PubMed] [Google Scholar]
- 56. Holz L. E., Jakobsen K. P., Van Snick J., Cormont F., Sewell W. A. (2005) Dexamethasone inhibits IL-9 production by human T cells. J. Inflamm. (Lond) 2, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Monteyne P., Renauld J. C., Van Broeck J., Dunne D. W., Brombacher F., Coutelier J. P. (1997) IL-4-independent regulation of in vivo IL-9 expression. J. Immunol. 159, 2616–2623 [PubMed] [Google Scholar]
- 58. Rao S., Gerondakis S., Woltring D., Shannon M. F. (2003) c-Rel is required for chromatin remodeling across the IL-2 gene promoter. J. Immunol. 170, 3724–3731 [DOI] [PubMed] [Google Scholar]
- 59. Workman J. L., Kingston R. E. (1998) Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67, 545–579 [DOI] [PubMed] [Google Scholar]
- 60. Wong M. T., Ye J. J., Alonso M. N., Landrigan A., Cheung R. K., Engleman E., Utz P. J. (2010) Regulation of human Th9 differentiation by type I interferons and IL-21. Immunol. Cell Biol. 88, 624–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Johnson B. V., Bert A. G., Ryan G. R., Condina A., Cockerill P. N. (2004) Granulocyte-macrophage colony-stimulating factor enhancer activation requires cooperation between NFAT and AP-1 elements and is associated with extensive nucleosome reorganization. Mol. Cell Biol. 24, 7914–7930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cron R. Q., Bartz S. R., Clausell A., Bort S. J., Klebanoff S. J., Lewis D. B. (2000) NFAT1 enhances HIV-1 gene expression in primary human CD4 T cells. Clin. Immunol. 94, 179–191 [DOI] [PubMed] [Google Scholar]
- 63. Farrow M. A., Kim E. Y., Wolinsky S. M., Sheehy A. M. (2011) NFAT and IRF proteins regulate transcription of the anti-HIV gene, APOBEC3G. J. Biol. Chem. 286, 2567–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gibson H. M., Hedgcock C. J., Aufiero B. M., Wilson A. J., Hafner M. S., Tsokos G. C., Wong H. K. (2007) Induction of the CTLA-4 gene in human lymphocytes is dependent on NFAT binding the proximal promoter. J. Immunol. 179, 3831–3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chan H. M., La Thangue N. B. (2001) p300/CBP proteins. HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114, 2363–2373 [DOI] [PubMed] [Google Scholar]
- 66. Gerritsen M. E., Williams A. J., Neish A. S., Moore S., Shi Y., Collins T. (1997) CREB-binding protein/p300 are transcriptional coactivators of p65. Proc. Natl. Acad. Sci. U.S.A. 94, 2927–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chen L., Mu Y., Greene W. C. (2002) Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. EMBO J. 21, 6539–6548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Macián F., López-Rodriguez C., Rao A. (2001) Partners in transcription. NFAT and AP-1. Oncogene 20, 2476–2489 [DOI] [PubMed] [Google Scholar]
- 69. Sica A., Dorman L., Viggiano V., Cippitelli M., Ghosh P., Rice N., Young H. A. (1997) Interaction of NF-kappaB and NFAT with the interferon-γ promoter. J. Biol. Chem. 272, 30412–30420 [DOI] [PubMed] [Google Scholar]
- 70. McCaffrey P. G., Jain J., Jamieson C., Sen R., Rao A. (1992) AT cell nuclear factor resembling NF-AT binds to an NF-κB site and to the conserved lymphokine promoter sequence cytokine-1. J. Biol. Chem. 267, 1864–1871 [PubMed] [Google Scholar]
- 71. Tsytsykova A. V., Rajsbaum R., Falvo J. V., Ligeiro F., Neely S. R., Goldfeld A. E. (2007) Activation-dependent intrachromosomal interactions formed by the TNF gene promoter and two distal enhancers. Proc. Natl. Acad. Sci. U.S.A. 104, 16850–16855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hogan P. G., Chen L., Nardone J., Rao A. (2003) Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 17, 2205–2232 [DOI] [PubMed] [Google Scholar]
- 73. Cakouros D., Cockerill P. N., Bert A. G., Mital R., Roberts D. C., Shannon M. F. (2001) An NF-κB/Sp1 region is essential for chromatin remodeling and correct transcription of a human granulocyte-macrophage colony-stimulating factor transgene. J. Immunol. 167, 302–310 [DOI] [PubMed] [Google Scholar]
- 74. Carey M., Smale S. T. (2002) Transcriptional Regulation in Eukaryotes: Concepts, Strategies, and Techniques, 1st Ed., pp. 144–145, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 75. Ishihara S., Schwartz R. H. (2011) Two-step binding of transcription factors causes sequential chromatin structural changes at the activated IL-2 promoter. J. Immunol. 187, 3292–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Stephens G. L., Swerdlow B., Benjamin E., Coyle A. J., Humbles A., Kolbeck R., Fung M. (2011) IL-9 is a Th17-derived cytokine that limits pathogenic activity in organ specific autoimmune disease. Eur. J. Immunol. 41, 952–962 [DOI] [PubMed] [Google Scholar]
- 77. Refaat A., Zhou Y., Suzuki S., Takasaki I., Koizumi K., Yamaoka S., Tabuchi Y., Saiki I., Sakurai H. (2011) Distinct roles of transforming growth factor-β-activated kinase 1 (TAK1)-c-Rel and interferon regulatory factor 4 (IRF4) pathways in human T cell lymphotropic virus 1-transformed T helper 17 cells producing interleukin-9. J. Biol. Chem. 286, 21092–21099 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.