Background: asbABCDEF mediates petrobactin production and facilitates anthrax virulence.
Results: Purified AsbA-E proteins reconstituted petrobactin assembly in vitro. The crystal structure and enzymatic studies of AsbB highlight its function and role in the siderophore pathway.
Conclusion: AsbB characterization demonstrated reaction flexibility and substrate positions in the binding pocket.
Significance: Siderophore synthetases represent promising antimicrobial targets, and characterization of these versatile enzymes enables creation of novel compounds.
Keywords: Bacillus, Crystal Structure, Iron Metabolism, Secondary Metabolism, Siderophores, Bacillus anthracis, Iron Acquisition, Petrobactin, Synthetase Structure
Abstract
Petrobactin, a mixed catechol-carboxylate siderophore, is required for full virulence of Bacillus anthracis, the causative agent of anthrax. The asbABCDEF operon encodes the biosynthetic machinery for this secondary metabolite. Here, we show that the function of five gene products encoded by the asb operon is necessary and sufficient for conversion of endogenous precursors to petrobactin using an in vitro system. In this pathway, the siderophore synthetase AsbB catalyzes formation of amide bonds crucial for petrobactin assembly through use of biosynthetic intermediates, as opposed to primary metabolites, as carboxylate donors. In solving the crystal structure of the B. anthracis siderophore biosynthesis protein B (AsbB), we disclose a three-dimensional model of a nonribosomal peptide synthetase-independent siderophore (NIS) synthetase. Structural characteristics provide new insight into how this bifunctional condensing enzyme can bind and adenylate multiple citrate-containing substrates followed by incorporation of both natural and unnatural polyamine nucleophiles. This activity enables formation of multiple end-stage products leading to final assembly of petrobactin. Subsequent enzymatic assays with the nonribosomal peptide synthetase-like AsbC, AsbD, and AsbE polypeptides show that the alternative products of AsbB are further converted to petrobactin, verifying previously proposed convergent routes to formation of this siderophore. These studies identify potential therapeutic targets to halt deadly infections caused by B. anthracis and other pathogenic bacteria and suggest new avenues for the chemoenzymatic synthesis of novel compounds.
Introduction
Nearly all organisms on the earth have evolved to use iron as a cofactor in a variety of essential metabolic processes (1). Despite the ubiquity of this metal in biology, the acquisition of free iron from the environment and within the host is hampered by its inaccessibility in ferric complexes. Thus, to obtain necessary levels of iron, cells have developed diverse strategies, one of the most prominent among microbes being the biosynthesis of iron-specific, high-affinity chelators called siderophores (2).
Bacillus anthracis, the causative agent of anthrax and a known bioterrorism agent, is capable of synthesizing two siderophores, bacillibactin and petrobactin (3, 4). Bacillibactin, a catechol-containing trilactone, is generated by a non-ribosomal peptide synthetase pathway encoded by the dhb operon (5); however, this metabolite is dispensable during pathogenesis and likely not secreted during an anthrax infection (6–9). Conversely, a second siderophore, petrobactin (Fig. 1A), is essential for virulence within current infection models (6, 10) and has since become a focus of multiple microbiological, genetic, and biochemical studies (3, 7–18, 20). This mixed catechol-carboxylate siderophore was first isolated from the Gram-negative marine microbe Marinobacter hydrocarbonoclasticus (21). Transcriptional analysis of B. anthracis mutants deficient for growth in iron-depleted conditions enabled identification of a pathway responsible for siderophore biosynthesis with enzymatic machinery encoded by the B. anthracis siderophore biosynthesis (asb) operon and a metabolite identical to the marine-derived siderophore petrobactin (6, 7, 22).
FIGURE 1.
Structure and biosynthesis of petrobactin, the virulence-associated siderophore of Bacillus anthracis. A, petrobactin is a symmetrical, mixed catechol-hydroxy-carboxylate siderophore comprised of a central citric acid moiety, two spermidine arms, and two 3,4-DHBA moieties. The unique 3,4-position of the hydroxyl groups in the catechol confers the stealth characteristics of petrobactin during host infection. B, the asbABCDEF gene cluster, shown with B. anthracis strain Ames labels, encodes the enzymatic machinery required for biosynthesis of petrobactin. Products include NIS synthetases (gray), a nonribosomal peptide synthetase-like aryl transferase system (white), and a 3-dehydroshikimate dehydratase/3,4-DHBA synthase (black). C, shown is the proposed pathway for petrobactin biosynthesis. Left panel, AsbF converts 3-dehydroshikimate to 3,4-DHBA, which is in turn adenylated by AsbC and loaded onto the phosphopantetheine thiol of the aryl-carrier protein AsbD. In vitro, AsbE transfers the 3,4-dihydroxybenzoyl group from AsbD to the primary amino groups of spermidine. Right panel, through regioselective condensation of spermidine with citric acid, AsbA and AsbB form intermediates that subsequently react with 3,4-dihydroxybenzoyl units. The flexibility of AsbB in vitro suggests multiple possible routes for the biosynthesis of petrobactin.
Previous mutagenesis and biochemical studies have shown that products of the polycistronic operon consisting of the six genes asbABCDEF (Fig. 1B) contribute to assembly of petrobactin in bacteria. Furthermore, petrobactin arises from three simple metabolic precursors; they are the common primary metabolites citric acid and spermidine and the unique subunit 3,4-dihydroxybenzoic acid (3,4-DHBA) (Fig. 1C) (4, 10–14, 17). AsbA is a member of a growing family of gene products referred to as non-ribosomal peptide synthetase-independent siderophore (NIS) synthetases (12, 18, 23). Generally, in a multistep process, NIS synthetases function by binding and adenylating a substrate carboxylate group, thus activating it for condensation with a nucleophilic polyamine or amino alcohol resulting in formation of an amide or ester bond, respectively (24–26).
AsbA can be classified more specifically as a type A NIS synthetase due to its utilization of citrate as a substrate for adenylation. This is followed by condensation with spermidine leading to N8-citryl-spermidine (compound 1, Fig. 1C) as a first step in petrobactin biosynthesis (12). AsbB is similar in activity to AsbA but classified as a type C NIS synthetase due to its preference for advanced intermediates in siderophore biosynthesis as substrates including 1 (11, 17). Both AsbA and AsbB incorporate the asymmetric polyamine spermidine from only one (N8) of its two primary amino group termini (Fig. 1C) (12, 17, 18). Currently, the mechanism by which this regioselectivity is achieved is not well understood.
It is significant to note that the dependence of anthrax on petrobactin during host infection results from the iron chelator evading sequestration by the host innate immune protein siderocalin, which is largely capable of neutralizing most other catechol-containing siderophores. More specifically, petrobactin owes this “stealth siderophore” quality to steric hindrance created by its 3,4-DHBA subunit when interacting with siderocalin (27). Most other catecholate siderophores contain 2,3-dihydroxybenzoic subunits, which are effectively neutralized by this protein during the host response to infection (27–29). Previous work has shown that asbF, the final gene of the petrobactin biosynthetic operon, encodes a dehydratase responsible for conversion of 3-dehydroshikimate to 3,4-DHBA (10, 14) (Fig. 1C). Once available, linkage of the 3,4-DHBA moieties to an exposed primary amine is facilitated by interactions of three other asb products; AsbC catalyzes adenylation of 3,4-DHBA followed by transfer to the phosphopantetheinylated aryl carrier protein AsbD. Subsequently, AsbE functions to catalyze amide bond formation between 3,4-DHBA and molecules bearing a primary amine (13) (Fig. 1C). As there are several proposed petrobactin precursors that have an available amino group, we were motivated to ascertain which metabolic intermediates function as substrates for AsbE during petrobactin biosynthesis.
Studies on aspects of petrobactin assembly (10–14, 17, 18, 23–25), ligand binding (22, 30), and identification of the endogenous cellular receptor (20, 31) have enhanced our understanding of iron acquisition by B. anthracis during pathogenesis. The broad eubacterial distribution of NIS systems (10, 23, 25) indicate that new information about petrobactin biosynthesis and transport will provide insights into these iron acquisition strategies. In this work, we sought to resolve remaining questions about the petrobactin biosynthetic pathway. Individual enzymatic activities were combined to reconstitute siderophore assembly in vitro, thus demonstrating formation of a functional Asb biosynthetic enzyme system. Moreover, determining the crystal structure of AsbB provides the first complete enzymatic and structural characterization of a type C NIS synthetase, with mechanistic insights regarding substrate selection and catalytic activity. Furthermore, information derived from AsbB product formation supports a biosynthetic scheme in which multiple substrates of AsbB function as intermediates in the convergent biosynthesis of petrobactin. This information defines new targets that may facilitate development of effective antibiotics and expands our fundamental knowledge of NIS synthetases as widely distributed, multicomponent biochemical machines for iron acquisition.
EXPERIMENTAL PROCEDURES
Chemical and Reagent Origin
Unless otherwise noted, consumable materials were obtained from Sigma and were of >97% purity. Hydrochloride salts of all polyamines used were either purchased or prepared by titration of stock solutions with HCl to neutral pH.
B. anthracis Strains and Culture Conditions
Targeted mutants of B. anthracis Sterne 34F2 were prepared by the allelic exchange method described by Lee et al. in 2007 (11). ΔasbB spores were germinated overnight in brain-heart infusion broth at room temperature with shaking. After 16–18 h, vegetative bacilli were diluted 1:100 in brain-heart infusion broth and grown 1–2 h to allow cells to enter log phase. Actively growing bacilli were pelleted by centrifugation at 1600 × g for 20 min at 25 °C and then washed 5× with iron-depleted medium (6) to ensure removal of nutrients and potential iron sources carried over from brain-heart infusion. Washed bacteria were then used to inoculate 0.5-liter cultures of iron-depleted medium in 2-liter polyethylene flasks at an A600 nm = 0.05. These were grown overnight (16–18 h) at 37 °C with shaking. Vegetative bacilli were removed from the culture supernatant using a 0.22-μm vacuum filtration apparatus (Corning Glass). Catechol presence in the medium was determined using the Arnow assay (32) before metabolite extraction.
Gene Cloning, Expression, and Protein Purification for Enzymology
Supplemental information for expression strain, plasmid, and cloning information is provided in supplemental Table 1. Constructs for the expression of AsbB residue replacement mutant enzymes were produced using protocols provided with the QuikChange (Stratagene) site-directed mutagenesis kit. The addition of 2.5 μl of DMSO to 50 μl of PCR reactions was required in some instances. Cloning and mutagenesis was confirmed by sequencing (UM DNA Sequencing Core).
To isolate protein for enzymatic analysis, “Z-competent” (Zymo Labs) BLR (Novagen) Escherichia coli (DE3) cells were transformed individually with respective asb gene-containing plasmids. 5-ml overnight Miller modified lysogeny broth (LB) cultures were used to inoculate 2-liter baffled flasks containing 500 ml of terrific broth with 4% (v/v) glycerol and shaken at 37 °C. Upon reaching an A600 nm of ∼0.8, expression cultures were acclimated to 18 °C for 1.5 h before induction with isopropyl β-d-1-thiogalactopyranoside to a final concentration of 0.25 mm in the medium. In the instance of asbA, terrific broth cultures were also supplemented upon isopropyl β-d-1-thiogalactopyranoside induction with citric acid and spermidine to a final concentration of 1 mm. Overexpression was allowed to continue at 18 °C for ∼16 h before cells were pelleted and stored at −80 °C before lysis. All five overexpressed proteins for enzymatic analysis were purified using standard His6 Ni2+ affinity chromatography and are described in detail in supplemental Methods.”
Gene Cloning, Expression, and Protein Purification for Crystallization
The asbB gene was cloned into the pMCSG7 vector with an N-terminal His6 tag (33) as well as the pMCSG28 vector with a C-terminal His6 tag (34) and overexpressed in E. coli BL21 (DE3)-Gold (Stratagene) harboring an extra plasmid encoding three rare tRNAs (AGG and AGA for Arg, ATA for Ile). To obtain protein from both constructs, recombinant E. coli cells were grown using selenomethionine-containing enriched M9 medium (pink medium) under conditions known to inhibit methionine biosynthesis (35, 36). Cell cultures were grown at 37 °C to an A600 nm of ∼0.95 then cooled down to 18 °C before adding selenomethionine and 0.5 mm isopropyl β-d-1-thiogalactopyranoside to induce and maintained at 18 °C overnight. Protein was purified by two-step Ni2+ affinity chromatography following the standard protocol described previously (37) and in supplemental Methods.
The His6 tag was removed using recombinant tobacco etch virus protease in a 1:30 ratio by incubating at 4 °C for 6 h. The tobacco etch virus protease cleavage left six artificial residues (ENLYFQ) at the C terminus for the pMCSG28 construct protein; however, the His6 tag failed to be cleaved from the N terminus of the pMCSG7 construct protein. AsbB protein was then further purified by a 5-ml manually packed Ni2+-superflow affinity column (GE Healthcare). The protein eluted as a flow-through from the column in lysis buffer with 20 mm imidazole. Samples were dialyzed in crystallization buffer containing 20 mm HEPES, pH 8.0, 150 mm NaCl, 2 mm DTT, and concentrated to 60 mg/ml using an Amicon Ultra centrifugal filter device (Millipore). The protein was then further purified by size exclusion chromatography using a Superdex 200 16/60 HiLoad (GE Healthcare) column in the same crystallization buffer.
Protein Crystallization
Crystallization was set up for all AsbB proteins. They were screened for crystallization conditions using a Mosquito robot (TTP Labtech) on sitting drops in 96-well plates (Greiner) at 16 °C. Each 0.8-μl drop consisted of 0.4 μl of protein from the pMCSG7 or pMCSG28 vectors and 0.4 μl of each crystallization condition. A number of commercially available screens including Index (Hampton Research) and ANL-1 and ANL-2 (Qiagen) were used for crystallization. AsbB with an N-terminal His6 tag failed to yield a diffraction quality crystal. The best crystals of AsbB from the C-terminal construct appeared after 3 days from Index #61 containing 0.2 m l-proline, 0.1 m HEPES, pH 7.5, and 10% polyethylene glycol 3350. Before data collection, crystals were flash-frozen in liquid nitrogen in the presence of a number of different cryo-protectants. Chunky jewel shape crystals displaying 0.25 × 0.15 × 0.1-mm sides with the ethylene glycol cryoprotectant diffracted to about 2.3 Å and were used for data collection.
Data Collection, Structure Determination, Refinement, and Deposition
The single wavelength anomalous dispersion data near the selenium absorption edge at the peak wavelength 0.9793 Å up to 2.4 Å were collected from a single selenomethionine-labeled protein crystal at 100 K at the 19ID beam line of the Structural Biology Center at the Advanced Photon Source, Argonne National Laboratory. The crystal was exposed for 5 s per 1.0° rotation of ω circle with the crystal to detector distance of 340 mm. The data were recorded on a CCD detector Q315r from ADSC scanning a full 220° on ω using the SBC-Collect program for data collection and visualization of diffraction images. The space group was P212121 with cell dimensions of a = 64.26 Å, b = 155.92 Å, and c = 156.24 Å. All data were processed and scaled with HKL3000 (38) (Table 1).
TABLE 1.
Summary of the AsbB crystallographic data
| Data collection | |
|---|---|
| Space group | P212121 |
| Unit cell (Å) | a = 64.26 |
| b = 155.92 | |
| c = 156.24 | |
| Wavelength(Å) | 0.9793 |
| Highest resolution bin (Å) | 2.44–2.40 |
| Number of observed reflections | 62971 (3081) |
| Rsym(%)a | 9.0 (63.1)cb |
| Completeness (%) | 99.8 (100) |
| I/sI | 10.8 (3.2)b |
| Phasing | |
| Method | SAD |
| Phasing resolution range (Å) | 49.6-2.40 |
| Figure of merit/phasing power | 0.26/1.29 |
| Number of SeMet | 26 |
| Refinement | |
| Resolution range (Å) | 49.6-2.40 |
| Rcryst (%) | 17.4 |
| Rfree (%) | 22.7 |
| Number of protein residues | 1232 |
| Solvent molecules | 324 |
| Bond lengths (Å) | 0.012 |
| Bond angles (degree) | 1.49 |
| Dihedral angles (degree) | 17.2 |
| B-factors (Å2) | 59.8 |
| Protein main chain | 55.5 |
| Protein side chain | 62.2 |
| Others (water, ADP, Ethylene glycol) | 59.6 |
| Wilson B-factor (Å2) | 50.9 |
| Ramachandran Plot (%)b | |
| Preferred | 96.4 |
| Generously allowed | 3.1 |
| Disallowed | 0.5 |
| PDB ID | 3TO3 |
a Rmerge = (ΣhklΣi|Ii − 〈I〉|)/(ΣhklΣi|〈I〉|), where Ii is the intensity for the ith measurement of an equivalent reflection with indices h, k, and l.
b Numbers in parentheses are values for the highest resolution bin.
c Defined in Coot.
The structure was determined by single wavelength anomalous dispersion phasing utilizing the anomalous signal from selenium atoms with AUTOSHARP (SHARP 2.2.0 and Sushi 3.4.0) (39) using the peak data to 2.40 Å. ARP/wARP (40) was used to build an initial model that included 495 (of total 616 residues including the C-terminal artifact residues) and 470 residues for each of two protein chains. Significant manual model building and the subsequent refinement were performed iteratively by phenix.refine (41) and manual adjustment using Coot (42) until it converged to the R factor of 0.174 and the free R of 0.227 with root mean square bond distances of 0.012 Å and root mean square bond angles of 1.49°. The final model included residues 1–600 of chain A and 0–600 of chain B of AsbB, 1 ADP, 1 Mg2+, 2 chloride ions, 3 ethylene glycol molecules, and 324 ordered water molecules. Residues on the surface such as 138–143 of chain A and 139–144 of chain B are not ordered; neither are the C-terminal residues, 601–610 for both chain A and chain B, including those residues introduced during cloning (34). The stereochemistry of the structure was checked with PROCHECK (43) and the Ramachandran plot. Atomic coordinates and experimental structure factors of AsbB have been deposited in PDB under the code 3TO3.
Purification of N1-(3,4-DHB)-N8-citryl-spermidine (3)
All procedures were performed in plasticware thoroughly rinsed with double-deionized water (ddH2O) or glassware rinsed generously with 6 m HCl followed by ddH2O. Supernatants from previously described B. anthracis Sterne 34F2 ΔasbB cultures were stirred overnight with 37.5 g of Amberlite XAD-16 resin per liter of supernatant. Before mixing with culture supernatants, the resin was equilibrated for an hour in ddH2O and filtered through a milk filter that had also been rinsed with ddH2O. The batch binding slurry was poured though a glass chromatography column. Retained resin was washed with 1 liter of ddH2O that was collected followed by 500 ml of methanol (MeOH), which was also collected.
Liquids were evaporated to dryness and redissolved in 4 ml of ddH2O before purification on a Beckman-Coulter System Gold HPLC, which was conducted as follows. Solvents contained a 0.1% (v/v) mixture of formic acid brought to pH 4 by the addition of triethylamine. Separation was conducted on a Waters X-Bridge C18 (10 × 250 mm, 5 μm) column at a flow rate of 2.5 ml/min with an initial concentration of 5% MeOH in ddH2O for 15 min followed by a linear gradient increase to 50% MeOH over the course of 25 min. 100% MeOH was applied for an additional 12 min as a cleaning step. Peaks with UV absorption maxima matching that of 3,4-DHBA moieties (259 and 290 nm) were collected, and their identity was confirmed by mass spectrometry. To achieve the high degree of purity required for kinetics analysis, collected 3 was evaporated and applied to a second round of HPLC using a Phenomenex Synergi Hydro-RP (10 × 250 mm, 4 μm) semipreparative column eluted with solvents containing 0.05% (v/v) formic acid at a flow rate of 2.5 ml/min. 100% ddH2O was applied for 15 min after which MeOH concentration was increased to 50% in a linear gradient over 30 min followed by a 100% MeOH cleanup phase. 3 was collected, confirmed by mass spectrometry, and lyophilized. Before use, metabolites were dissolved in ddH2O to create 100 mm stock solutions that were stored at −20 °C.
Synthesis of Substrates
5 and 1 were prepared as previously described (12, 17). Although 1 was synthesized similarly to previously reported methods, purification of the final product was performed by HPLC on a Beckman-Coulter System Gold in line with a Phenomenex Synergi Hydro-RP (10 × 250 mm, 4 μm) semipreparative column with a 100% aqueous mobile phase containing 0.05% (v/v) formic acid at a flow rate of 2.5 ml/min.
In Vitro Reconstitution of Metabolite Biosynthesis
Small-scale reactions were conducted in 1.7-ml microcentrifuge tubes. Reaction buffer contained 50 mm HEPES, pH 8, 15 mm MgCl2, 0.5 mm tris(2-carboxyethyl) phosphine, 10 mm ATP, 2 mm carboxylate substrate (citric acid or 3), 4 mm nucleophilic substrate (spermidine or analogs thereof), and 4 mm 3,4-DHBA where applicable. All enzymes were used at a concentration of 1 μm with the exception of AsbD, which was used at 5 μm. All reactions were conducted at 37 °C unless otherwise noted. Reactions were quenched in two ways. The first was by adding 0.5 reaction volume of 5% (w/v) trichloroacetic acid (TCA) solution. Reactions were inverted to mix and centrifuged at 16,000 × g in a tabletop centrifuge for 20 min. Resultant supernatant was placed into a new tube containing an equal volume of 0.6 m sodium borate buffer, pH 8.0. The second way was by adding nine reaction volumes of HPLC grade MeOH spiked with 25 μm N-(3-aminobenzoyl)-l-β-alanine as an internal standard. Reactions were inverted to mix and centrifuged at 16,000 × g on a tabletop centrifuge for 20 min. The resultant supernatant was placed into a new tube and evaporated to dryness by vacuum centrifugation. Samples were redissolved in ddH2O before analysis. Enzymatically derived petrobactin obtained using this method was purified by the same HPLC method applied to 3 (described in a previous section) and retained for further analysis.
Fluorescamine Derivatization of Polar Compounds
10 μl of TCA-quenched sample was mixed with 10 μl of 200 mm fluorescamine in LC-MS grade acetonitrile (MeCN). Reactions were vortexed, briefly centrifuged, and placed in the dark for 2 h. After incubation, reactions were centrifuged in 0.65-ml microcentrifuge tubes at 16,000 × g for 10 min. 15 μl of supernatant was then diluted 4-fold in 50:50 MeCN:H2O for analysis by LC-MS.
LC-MS Analysis
2 μl of MeOH-quenched in vitro reaction samples were injected onto a Shimadzu 2010EV HPLC in line with an electrospray quadrupole mass spectrometer running the following chromatographic method; all solvents were LC-MS grade and contained 0.1% (v/v) formic acid. Separation was conducted on a Phenomenex Luna C18 (250 × 4.6 mm, 5 μm) column at a flow rate of 0.125 ml/min with an initial concentration of 5% MeCN in ddH2O for 8 min followed by a linear gradient of 5% to 95% MeCN over the course of 32 min. 95% MeCN was then applied for an additional 12 min. Spectra were scanned in both positive and negative mode over the range of 200–1000 m/z. Predetermined m/z values were also monitored in positive selective ion monitoring mode. Similar methods were used for TCA-quenched reactions and biological samples; however, 5 μl were injected, and the first 10 min of the HPLC protocol was diverted to waste to avoid introduction of salt to the mass spectrometer.
Metabolite Analysis by Tandem Mass Spectrometry (MS/MS)
Enzymatically prepared products and culture-derived N1-(3,4-DHB)-N8-citryl-spermidine (3) were collected from HPLC purification and stored at −20 °C before analysis. Using an Agilent Technologies 6500 quadrupole time of flight (Q-TOF) mass spectrometer, 1–3 μl of sample was applied by direct infusion with a 10% MeCN, 0.1% (v/v) formic acid solvent mixture at a flow rate of 0.4 ml/min. Electrospray ionization was used, scanning in positive mode between 100 and 1000 m/z. Target ions matching the predicted m/z values for analytes were fragmented by collision-induced dissociation at 20 V. An alternative LC-MS/MS method is described in supplemental Fig. 5.
2-Amino-6-mercapto-7-methylpurine Ribonucleoside (MESG) Continuous Pyrophosphate Detection Assay
Reactions were a modification of a previously described protocol (44, 45). For kinetics reactions, standard conditions were 50 mm Tris, pH 8.0, 15 mm MgCl2, 0.5 mm tris(2-carboxyethyl) phosphine, 6 mm ATP, 40 mm spermidine, 0.001 units/μl purine nucleoside phosphorylase, 0.0004 units/μl inorganic pyrophosphatase, 0.4 mm MESG (Berry and Associates, Dexter, MI), and 0.2 μm purified enzyme. Varying concentrations of carboxylate-containing substrate (citric acid or 3) were used to obtain enzymatic parameters. A standard curve was created under these reaction conditions, with the omission of enzyme and including varying concentrations of Na4P2O7. Change in A360 nm in response to pyrophosphate (PPi) content remained linear up to 0.05 mm, and this relation could be described by the equation 0.394 × ΔA360 nm = mm PPi. The relative polyamine acceptance assay utilized similar reaction conditions however, 20 mm analogs of spermidine and 3 mm carboxylate-containing substrate were used.
Assays were conducted in 96- or 384-well plate format prepared in 80 or 40 μl volumes, respectively. A360 nm readings were gathered every 15 s over the course of 10 min using a Molecular Devices SpectraMax M2e plate reader. Kinetics reactions were conducted in triplicate. The initial linear rate under each condition was calculated using the slope function in Microsoft Excel. Subsequent analyses were performed by plotting initial rate versus substrate concentration in Kaleidagraph (Synergy Software) and using nonlinear regression to fit to the Michaelis-Menten equation. Experiments to determine substrate preference were conducted in duplicate; initial rates were zeroed to that of no-enzyme controls and normalized based on the assumption that spermidine acts as a nucleophile to confer 100% activity.
RESULTS
In Vitro Reconstitution of Petrobactin Biosynthesis
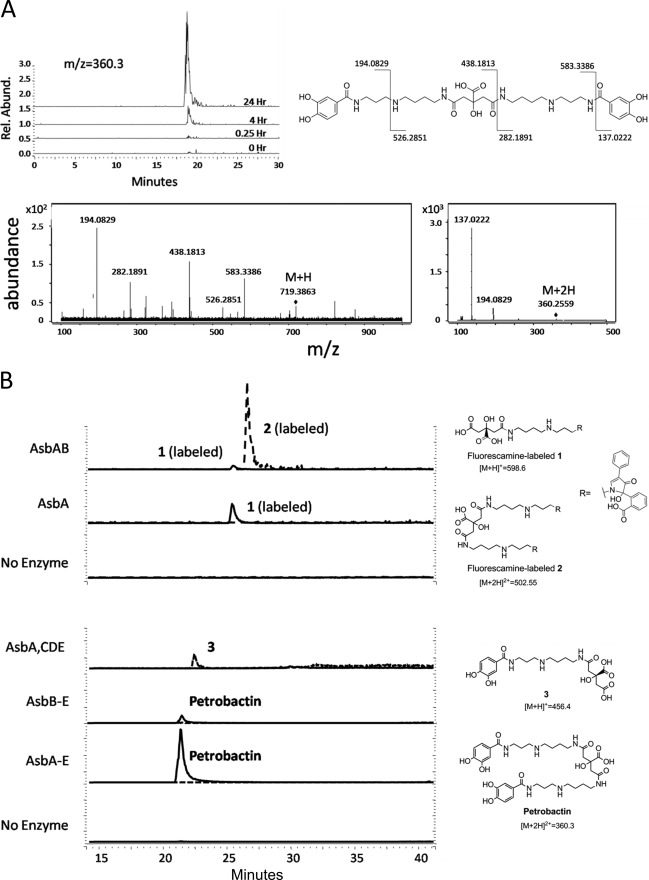
Previous studies on individual petrobactin biosynthetic enzymes have assigned a metabolic function to all products of the asb operon (Fig. 1B) (10–14, 17, 18, 23–25); however, whether these proteins are sufficient for petrobactin production in vitro has not been determined. asbA-asbE were individually amplified from B. anthracis Sterne strain 34F2 genomic DNA by PCR and cloned into individual expression vectors. All five genes were overexpressed, and the resulting proteins were purified by metal affinity chromatography for use in enzymatic assays. Recombinant His6-tagged AsbA, AsbB, AsbC, AsbD, and AsbE were incubated with ATP, MgCl2, and the three metabolic precursors to petrobactin: spermidine, citric acid, and 3,4-DHBA. This reaction was quenched at varying time points and subjected to LC-MS analysis, which revealed an accumulating product with m/z matching petrobactin (expected m/z for [M+2H]2+: 360.40; observed: 360.35) (Fig. 2A). This compound was not detected in reactions lacking ATP or all five recombinant proteins (Fig. 2B). The enzymatically derived product was confirmed to be petrobactin through tandem mass spectrometry, yielding a fragmentation pattern and spectra (Fig. 2A) that is identical to that of a previously characterized authentic standard (22). Thus, the asb gene products are sufficient for producing petrobactin from 3,4-DHBA, spermidine, and citrate and furthermore act in trans when individually purified to reconstitute siderophore biosynthesis in vitro.
FIGURE 2.
In vitro reconstitution of petrobactin biosynthesis. Heterologously expressed asb products were combined with metabolic precursors spermidine, citric acid, and 3,4-DHBA and the necessary substrate ATP and cofactor Mg2+. A, LC-MS data demonstrates accumulation of m/z = 360.3 over time, corresponding to the [M+2H]2+ of petrobactin. The compound associated with this peak was analyzed by MS/MS and shown to have a fragmentation pattern identical to that of authentic petrobactin. B, products from various combinations of asb enzymes were investigated by LC-MS. To detect zwitterionic intermediates generated by the NIS synthetases AsbA and AsbB, reactions were acid quenched, and products were derivatized with fluorescamine (top two traces). Other organic products were extracted with methanol for analysis (lower traces). Omission of AsbA still results in a modest accumulation of petrobactin, suggesting a compensatory role is filled by the type C NIS synthetase AsbB. Traces show relative intensity of selected m/z for each compound.
Isolation of Pathway Components in Petrobactin Biosynthesis
Additional pathway reconstitution experiments were motivated by previous characterization of B. anthracis mutant strains that demonstrated a detectable amount of petrobactin to be generated even in the absence of the asbA gene (11), the product of which has been shown to catalyze condensation of spermidine and citric acid (Fig. 1C) (12). In an effort to investigate this phenomenon in vitro, the biosynthetic reconstitution reaction was performed using AsbB, AsbC, AsbD, and AsbE. After incubation with the required substrates (spermidine, citrate, and ATP) and co-factor (Mg2+), a readily detectable, albeit small, amount of petrobactin was observed by LC-MS (Fig. 2B). In this instance, the remaining NIS synthetase AsbB is able to generate N8-citryl-spermidine (1), thus functioning as a partial substitute for AsbA.
Further analysis revealed that no other single-enzyme omissions yielded petrobactin. Therefore, AsbA does not play a compensatory role for the missing enzymatic activity of other asb gene products. Instead, removal of AsbB from the reaction conditions again resembles a phenotype of B. anthracis grown in iron-depleted medium (Fig. 2B), where the ΔasbB mutant accumulates the proposed intermediate N1-(3,4-dihydroxybenzoyl)-N8-citryl-spermidine (3) (11). Indeed accumulation of 3 occurs due to its confirmed role as a substrate for AsbB in vitro (17).
N8-Citryl-spermidine (1) and N8,N′8-citryl-bis(spermidine) (2) are products of AsbA and AsbB, respectively, and likely intermediates in petrobactin biosynthesis (4, 12, 17). Because they are zwitterionic molecules that are difficult to separate from other salts for analysis by mass spectrometry, a rapid derivatization method was developed to label the primary amines of 1 and 2 with fluorescamine. With the addition of this label to compound 2 or compound 1, m/z values corresponding to the fluorescaminylated derivatives were observed by LC-MS (di-fluorescaminylated 2, [M+2H]2+ = 502.55; fluorescaminylated 1, [M+H]+ = 598.6) (supplemental Fig. 1). Similarly, direct addition of fluorescamine to the in vitro pathway reconstitution mixture (AsbA or AsbA and AsbB, with substrates and co-factors) enabled ready confirmation of the presence of the derivatized forms of enzymatically produced 1 and 2 by LC-MS (Fig. 2B, top two traces).
Crystal Structure of AsbB
To gain greater insight into the structural basis for petrobactin formation, the crystal structure of AsbB was solved at 2.40 Å resolution (Fig. 3A). The monomer of the AsbB structure is similar to that of AlcC, alcaligin biosynthesis protein C (PDB IDs 2X0O, 2X0P, 2X0Q) from Bordetella bronchiseptica (46, 47) and that of AcsD (PDB IDs 2W02, 2W03, 2W04) from Pectobacterium chrysanthemi (48). AcsD was previously described as a type A NIS synthetase (47), which catalyzes one of the initial steps in the biosynthesis of achromobactin, a siderophore implicated in phytopathogenicity of P. chrysanthemi (49, 50).
FIGURE 3.
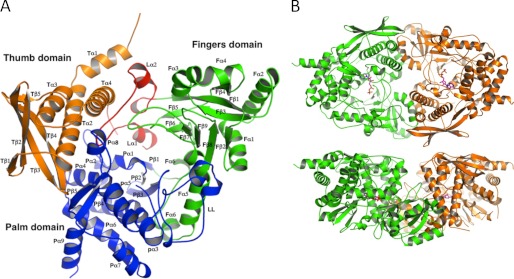
Structure of the type C NIS synthetase AsbB. A, overall monomer structure of AsbB shows the numbering scheme. The N-terminal thumb domain (orange, 1–136) is composed of four α-helices (a small N-terminal helix followed by a three-helix bundle) and a four-stranded anti-parallel β-sheet. A long loop (red, 137–183) consisting of three small α-helices and two short α-helices with connecting turns is a part of the substrate binding pocket and separates the thumb domain and the fingers domain. Sandwiched between these domains, the extended loop is stabilized by a number of hydrophilic and hydrophobic interactions involving several well conserved residues (supplemental Fig. 2). The fingers domain (green, 184–384) contains five α-helices and an 8-stranded antiparallel β-sheet that twists to form part of the cup-shape binding/active site. The C-terminal palm domain (blue, 380–601) of 10 α-helices and five β-strands (2 β-sheets; 3 stranded antiparallel and a β-ribbon) connects the other two domains and forms a major part of the substrate-binding pocket. B, shown is the dimeric form of AsbB as the asymmetric unit. The top model shows the large substrate binding pocket; the bottom model is rotated around the x axis by 90° relative to the top model and highlights the pocket formed by the two dimers.
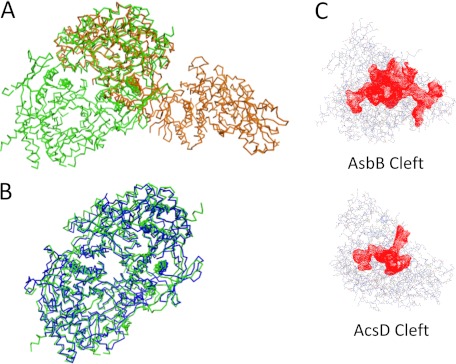
Two chains of the AsbB polypeptide occupy the asymmetric unit of the orthorhombic P212121 crystal as a dimer (Fig. 3B). A monomer of the AsbB structure superposes with AcsD (PDB ID 2W02) (47) closely (root mean square deviation 2.52 Å, 448 α-carbon atoms over 597 AsbB residues), although there is only a 20% sequence identity (Fig. 4A and supplemental Fig. 2). Briefly, AsbB has three domains, the thumb domain, the palm domain, and the fingers domain (after the AcsD notation), arranged in a structure resembling a cupped hand. Although the overall fold is similar, as expected from the low sequence identity, the details of each domain are different from AcsD. All three domains together make up a round-bottom, cup-shaped active site and are described in detail in Fig. 3A. Additionally, in the pocket, partial electron density corresponding to an ADP molecule (although not fully ordered) is found in chain B (supplemental Fig. 3) that is consistent in conformation with the ATP molecule of the AlcC structure (PDB ID 2X0Q) (48).
FIGURE 4.
Comparison of AsbB to other NIS synthetases. Calculations regarding structural similarity were made using PDBeFold (69). A, superposition of AsbB (green) and the type A NIS synthetase AcsD (orange) demonstrates similar monomeric structures with different dimers. Between the two proteins, the sequence identity is 20.5%, and the mean square deviation on α-carbon atom positions is 2.52 Å with 448 amino acids compared. B, superposition of AsbB (green) and the type C NIS synthetase AlcC (blue) indicates the same -fold with similar dimers. For these two proteins, the sequence identity is 24.6%, and the root mean square deviation on α-carbon atom positions is 1.79 Å with 516 amino acids compared. C, ligand binding pockets (red) of AsbB and AcsD are shown. The cleft sizes of AsbB and AcsD are 8614 and 3593 Å3, respectively, as calculated by Profunc (52, 70) and plotted by Jmol (19).
The positioning of the homodimer interface between AsbB (or AlcC) and AcsD varies considerably (Fig. 4, A and B). However, as in the case with AcsD, a dimeric AsbB found in the crystal structure is corroborated by size exclusion chromatography and computational analysis, suggesting that the dimeric form exists in vivo as well. In the AsbB structure, helices of Tα1, Tα2, and Tα4 in the thumb domain from one chain and a curved helix spanning Fα3-Fα4 in the fingers domain from the other chain bundle up to form a symmetric half of the dimer interface with a number of hydrophobic or hydrophilic residues exchanging interactions. In the AlcC structure, all the same matching helices maintain interactions to form a similar dimer (Fig. 4B). However, in the AcsD structure, one of the key helices (Tα1) is missing. Instead, a loop (42–50, 52–61) containing a turn of α-helix from each chain is inserted between Tα1 and Tα2 and contributes to a major part of the interface to make up a dimer, which is quite different from that of the AsbB structure (Fig. 4A).
Although AcsD is a type A NIS synthetase and employs citric acid as the substrate for adenylation, AsbB (and AlcC) is a type C NIS synthetase. The fact that AsbB must adenylate a substrate that is more complex than citrate is reflected in the structural differences between these two highly related proteins. AsbB has a significantly larger binding pocket with about 8500 Å3 encompassing 18.3 Å at its widest part compared with AcsD that bears a 13.9 Å binding pocket measured between identical amino acid residues with the cleft volume of ∼3500 Å3 (Fig. 4C). Furthermore, a solvent-exposed channel visibly connects the active site/binding pocket of one AsbB monomer to the corresponding pocket in the other monomer (Fig. 3B).
AsbB Substrate Binding Site
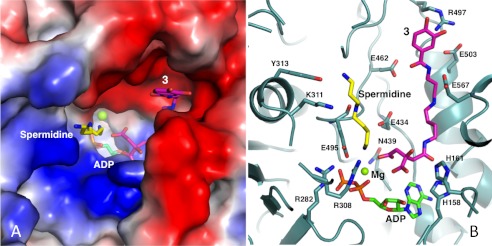
Our efforts to co-crystallize AsbB with citryl substrate, spermidine, ATP, and/or ADP were not successful. However, it is reasonable to suggest that these ligands assume positions in the binding pocket similar to those of AcsD or AlcC considering the presence of several highly conserved residues, including nearly all residues implicated in catalytic activity of NIS synthetases (48, 49) (supplemental Fig. 2). Like most other enzymes, the majority of the conserved residues within AsbB reside in the loops; in fact, the binding pocket is made of a number of loops likely arranged to provide flexibility as well as specificity in ligand binding. In this scenario, the substrate conformation is somewhat different from that of AcsD. The number of basic residues in AsbB (i.e. Arg-308, Arg-282, Lys-296) clustered in the loops of residues 296–308 and 280–284, provide a stable binding site for ATP/ADP at the bottom of the pocket as in the cases for AcsD and AlcC (48, 49) (Fig. 5, A and B, and supplemental Fig. 3).
FIGURE 5.
Modeling the substrate binding pocket of AsbB. A, the binding pocket of the AsbB monomer is shown in a charge potential surface drawing with ADP (green) as well as a manually modeled spermidine (yellow) and compound 3 (pink). For the ligands, oxygen, nitrogen, and phosphorous atoms are indicated by red, blue, and orange, respectively. B, hypothetical interactions of AsbB with substrates in the active site are shown. Compound 3 is positioned so that the carboxyl end of the citrate moiety is poised to attack the α-phosphate of ATP. The N8 of spermidine is near the same phosphate, poised to attack the ester bond of adenylated 3 for formation of compound 5. Subsequent molecular modeling allows the N1-(3,4-dihydroxybenzoyl)-spermidine tail of 3 and spermidine to stretch out within the pocket between the finger and palm domain in the approximate fashion pictured. Potential protein-substrate interactions include the N1 and N8 of spermidine with Tyr-313 and Glu-459, respectively, and N1 and N4 within the spermidinyl moiety of 3 with Glu-503 and Glu-567, respectively. Residues including His-158, His-161, Arg-282, Lys-308, and Asn-439 are highly conserved among NIS synthetases (supplemental Fig. 2) and implicated in coordinating adenylation of the citryl moiety as the first step of catalysis.
The preferred activity of AsbB, a type C NIS synthetase, requires binding of the relatively complex substrates compound 1 or its dihydroxybenzoylated counterpart 3. With the location of the triphosphate established, citrate or the citryl moiety of compound 3 most likely positions near the top of the α-phosphate as shown in the co-complex structure of AcsD with ATP and N-citryl-ethylenediamine from P. chrysanthemi (PDB ID 2X3J) (51). In this scenario the substrate, which is much bulkier in the case of AsbB, requires a larger space for the spermidinyl (in the case of 1) and/or 3,4-DHB-spermidinyl (in the case of 3) moiety to be stretched out within the active/binding site. One possible model is depicted in Fig. 5, A and B. ATP is positioned as suggested by partial electron density of ADP and comparison to the ATP-AlcC co-crystal structure. By adopting orientation and approximate location of the citryl moiety of the AcsD product N-citryl-ethylenediamine in the co-complex structure (PDB ID 2X3J) (48), the dihydroxybenzoyl-spermidine “tail” of 3 can be stretched out to the direction of the AsbB palm domain, following the groove formed between the β loop of Pβ4, Pα8, and Pβ5, and the long α-helix Pα3. Along the way, a number of protein residues can interact with the substrate; for example, acidic residues Glu-503 and Glu-567 are close to N1and N4, respectively, of the spermidinyl portion of 3, whereas Arg-497 is near the dihydroxybenzoyl moiety distal to the adenylation active site.
The nucleophilic substrate spermidine is incorporated as a final step, approaching the other side of the citryl moiety of 1 or 3 (near the α-phosphate of ATP where adenylation is likely to occur), and may extend out in the space between the fingers domain and palm domain, contacting Tyr-313 along the way. Acidic residues Glu-459 and/or Glu-434 are in position to recruit and stabilize spermidine during the reaction (Fig. 5B). Indeed, the structure reveals sufficient space to accommodate both substrates, a spermidine and an N8-citryl-spermidine (1) or N1-(3,4-DHB)-N8-citryl-spermidine (3) along with a co-substrate (ATP), without a major change in the main chain conformation of the binding pocket or the additional pocket formed between the two monomers in the AsbB dimer.
Enzymatic Characterization of NIS Synthetases AsbA and AsbB
The crystal structure of AsbB prompted us to further explore aspects of its enzymatic activity and that of its partner NIS synthetase AsbA. Previous work has shown these enzymes catalyze formation of the two amide bonds to the central citrate moiety of petrobactin (Fig. 1) (11, 12, 17). AsbA regioselectively catalyzes amide bond formation between citric acid and spermidine (12, 18). In the case of AsbB, a preference for 1 (the product of AsbA) and spermidine, respectively, fill this role (17). This was demonstrated previously by comparing relative reaction rates using different substrate sets and also revealed that AsbB readily accepts the 3,4-dihydroxybenzoylated form of 1 (compound 3), which had been previously proposed as a substrate (Fig. 1C) (11, 17).
Although relative rates of AsbA and AsbB have been demonstrated under different reaction conditions, kinetic parameters for these enzymes have not been determined. To accomplish this, ATP turnover from synthetase activity was monitored using an enzyme-coupled assay involving conversion of the reporter molecule MESG to its purine base (44, 45). This assay was used to empirically establish saturating levels of spermidine, ATP, and Mg2+ for AsbA to determine reaction rates dependent exclusively on citric acid.
Enzymatic parameters were approximated for AsbA by observing initial reaction rates under varying concentrations of citric acid. Initial experiments suggested that high levels of the substrate inhibited AsbA. Citric acid sequesters divalent cations (including Mg2+) at high concentrations, however, and titration of additional MgCl2 to 0.8 stoichiometric equivalents of citrate restored AsbA activity. The resultant enzymatic activity curve (supplemental Fig. 4) was sufficient to extrapolate an apparent kcat of 8.90 × 10−1± 0.074 s−1, a Km of 5.65 × 10−3 ± 0.0016 m, and an apparent kcat/Km = 1.58 × 102 m−1s−1 for purified AsbA-His6.
Because AsbA and AsbB exhibit such high sequence similarity, particularly between residues in the active sites (supplemental Fig. 2), and near-identical enzymatic activity (Fig. 1C), optimal spermidine, ATP, and Mg2+ concentration was predicted to be similar between the two enzymes. However, substrate-dependent supplementation of additional Mg2+ was not required, suggesting the citryl moieties of 1 and 3 do not sequester divalent cations as effectively as citric acid alone. We sought to determine kinetic parameters for AsbB-dependent on the citryl substrate 3 (supplemental Fig. 4), but UV interference associated with this substrate prevented observation of the reaction at starting concentrations higher than 12 mm. Thus, enzymatic parameters were extrapolated from initial rates occurring between 0–12 mm 3, which suggested a Vmax/Km = 2.40 × 102 m−1s−1.
Probing Acceptance of Unnatural Nucleophiles for Condensation by AsbA and AsbB
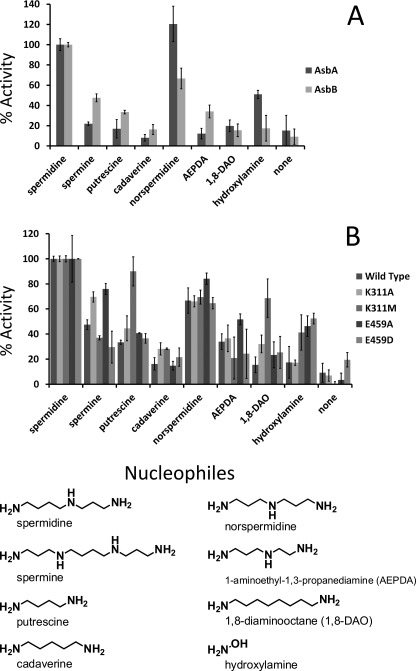
Previous mass spectrometric studies have demonstrated the capacity of AsbA to incorporate a variety of polyamines analogous to spermidine (18). Such innate substrate flexibility motivated us to investigate the ability of AsbB to incorporate unnatural ligands as well. Using the MESG assay described above, both AsbA and AsbB enzymes were incubated individually with ATP, MgCl2, and citric acid or 3 to compare activity with various nucleophiles related to the preferred substrate spermidine (Fig. 6A). Generally, for both AsbA and AsbB, spermidine remained the preferred nucleophile of the compounds tested. The one apparent exception was the relatively high activity of AsbA with norspermidine, which has one less carbon atom and thus bears symmetrical arms. Reaction rate comparison revealed a generally higher promiscuity of AsbB with unnatural substrates. Despite this, the general trend in substrate preference remained consistent between the two NIS synthetases.
FIGURE 6.
AsbA and AsbB display varying selectivity for nucleophilic substrates. A MESG-coupled pyrophosphate detection assay compared relative rate of incorporation of linear nucleophiles by AsbA and AsbB. Data are normalized to the preferred substrate spermidine. A, AsbB is more promiscuous by comparison; however, the general trend in substrate tolerance is conserved between the two NIS synthetases. B, residues in the AsbB substrate binding cleft hypothesized to interact with spermidine, Lys-311 and Glu-459, were subjected to site directed mutagenesis. AsbB mutants were compared for relative nucleophile acceptance.
Structural Basis for Polyamine Selectivity in AsbB
In addition to showing a similar profile for distinguishing polyamine nucleophiles as substrates, both AsbA and AsbB regioselectively incorporate spermidine at the N8 terminus in the biosynthesis of a symmetrical petrobactin molecule (12, 17, 18). To elucidate the basis for this selectivity, a model of the AsbB-substrate complex (Fig. 5B) was used to guide site-directed mutagenesis of residues hypothesized to confer an orientation-specific spermidine binding pocket in both AsbA and AsbB. Among these residues, Lys-311 and Glu-459 of AsbB align, respectively, with Lys-315 and Gln-468 of AsbA. This predictive modeling of the monomeric AsbA polypeptide also suggests that the side chains of Lys-315 and Gln-468 are solvent-exposed in the presumed substrate binding pocket (I-TASSER) (53, 54) (data not shown).
An AsbB mutant with alanine substituted for Lys-311 was expressed heterologously and purified for comparison of its nucleophile selectivity to that of wild type AsbB (Fig. 6B). Although preference for spermidine was not wholly abrogated, activity was elevated in conditions where unnatural nucleophiles were presented. In particular, polyamines of greater length (spermine) or lacking a secondary amine (l,8-diaminooctane and cadaverine), provided a slightly higher ATP turnover relative to spermidine with the AsbB K311A mutant than with wild type. Substitution at this position with methionine retains some of the space occupied by the original Lys-311 but presents a different charge to the proposed binding pocket. A K311M mutant displayed exclusion of the relatively large spermine molecule as observed with wild type AsbB, but increased acceptance of polyamines lacking a secondary amine still occurred.
Indeed, in the model presented, it appears that the side chain of Lys-311 “pushes” down on spermidine, with multiple positive charges in this space forcing contact between the aliphatic portion of lysine and an alkyl arm of the polyamine. Meanwhile, this conformation facilitates electrostatic interaction between the secondary amine of the substrate and the backbone carbonyl of Glu-459. It is possible that partial loss of this interaction would generate more favorable conditions for binding of substrates lacking a secondary amine.
In addition to the backbone oxygen of Glu-459, importance of this residue side chain in nucleophile selectivity was probed. Substituting this position with alanine demonstrated increased relative acceptance of spermine, norspermidine, and 1-aminoethyl-1,3-propanediamine. Conversely, a less drastically altered E459D mutant displayed selectivity closer to that of wild type. Considering the substrate conformation model presented (Fig. 5B), the side chain of Glu-459 is roughly the same length of the 4-carbon terminus of spermidine and is poised to be positioned parallel to the substrate, extending the N8 primary amine to the active site for condensation with adenylated 1 or 3 (Fig. 5B).
Multiple Petrobactin Biosynthesis Intermediates as Substrates for AsbCDE
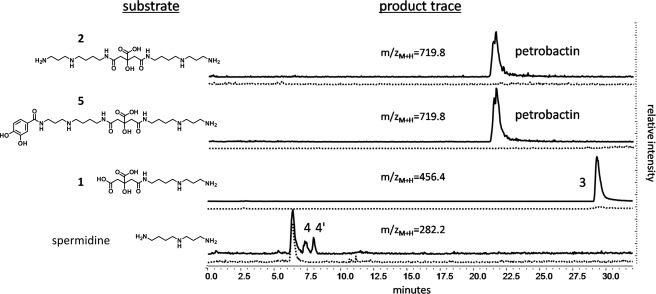
Spermidine is a nucleophilic substrate of the AsbC-AsbD-AsbE aryl transferase machinery, forming the 3,4-dihydroxybenzoylated product N1-(3,4-dihydroxybenzoyl)-spermidine (compound 4, Fig. 1C) and its regioisomer N8-(3,4-dihydroxybenzoyl)-spermidine (compound 4′) (11, 13) (Fig. 7, bottom trace). However, a moiety resembling 4′ is not observed within the petrobactin structure (21, 22), and more recent relative rate studies demonstrate 4 serves as a relatively poor substrate for AsbA or AsbB (12, 17, 18). Indeed, LC-MS analysis of petrobactin generated by in vitro pathway reconstitution supports this finding, revealing two peaks with m/z ([M+H]+ = 282.36) corresponding to accumulation of unincorporated 4 and 4′. Given an abundance of 3,4-DHBA, this reaction will also yield a product with an m/z and a fragmentation pattern corresponding to N1,N8-di-(3,4-dihydroxybenzoyl)-spermidine (compound 6, supplemental Fig. 5).
FIGURE 7.
3,4-Dihydroxybenzoylation by the aryl transferase components encoded by asbCDE. Multiple proposed intermediates of petrobactin biosynthesis contain primary amines and serve as substrates for AsbE. In vitro reactions of individually purified His6-AsbC, His6-AsbD, and His6-AsbE with ATP and 3,4-DHBA were extracted using methanol and analyzed by LC-MS. HPLC chromatograms show selected ion monitoring of predicted m/z of products. Dashed traces indicate no-enzyme controls.
Based on the finding that AsbA and AsbB are both capable of forming products with spermidine (12, 17, 18), we were motivated to investigate how these products could be utilized in subsequent biosynthetic steps. Therefore, we considered a more recent hypothesis that, like spermidine, the spermidinylated intermediates 1 and 2 serve as substrates for AsbE in later stages of biosynthesis (24). In both molecules, protection of the 4-carbon N terminus of spermidine (N8) through amide bond formation with the central citric acid moiety assures that only the primary N1 amine is available for AsbE-catalyzed condensation with 3,4-DHBA (Fig. 1C).
To test this hypothesis directly, 1, 2, and 5 were incubated with 3,4-DHBA, ATP, MgCl2, and recombinant His6-AsbC, His6-AsbD, and His6-AsbE. Products with m/z matching those predicted for 3,4-dihydroxybenzoylation of the substrates were observed by LC-MS (Fig. 7); compound 1, formed by AsbA, was converted to 3. As shown previously, 3 has been isolated from iron-depleted cultures of a B. anthracis ΔasbB mutant (11). The products of AsbB, 2 and 5, functioned as substrates for AsbE as well, participating in amide bond formation with 3,4-DHBA to form petrobactin (Fig. 7). In the case of 2, petrobactin formation likely occurs in two cycles, the first creating 5, which does not appear to accumulate but instead may be rapidly converted by a second cycle to form petrobactin as the final product. As expected, none of these compounds was observed when AsbC, AsbD, and AsbE were omitted from the reaction mixture.
DISCUSSION
All products of the asb operon implicated in assembly of petrobactin from spermidine, citric acid, and 3,4-DHBA have been heterologously expressed, purified, and reconstituted to generate a fully functional petrobactin biosynthetic pathway (Fig. 2A). The absence of AsbA from this reaction resulted in low, but detectable levels of petrobactin formation, suggesting that the similar sequence and enzymatic activity of the partner NIS synthetase AsbB can rescue its function to a modest extent (Fig. 2B). The enantioselective abilities of NIS synthetases have been highlighted previously (17, 18, 47); however, AsbB may tolerate both configurations of the chiral citryl moiety of 1 depending on whether this compound exists as a product of AsbA-like activity or as a substrate for condensation to a second spermidine.
The enzymatic activity of AsbA and AsbB during petrobactin assembly was confirmed by LC-MS analysis facilitated by fluorescamine derivatization of siderophore zwitterionic intermediates. Indeed, intermediates of other NIS pathways, including those for rhizoferrin (55), staphyloferrin B (56), and achromobactin (23), have similar charge issues that could be resolved by an analogous derivatization protocol.
By solving the crystal structure of AsbB, we disclose the second structural model of a type C NIS synthetase and the first to be characterized through concurrent biochemical analysis. Highlighted are residues conserved among other type C enzymes as well as the closely related AsbA. These structural similarities solidify a proposed adenylation mechanism in which attack of the terminal carboxylate within the citryl moiety of one substrate by the α-phosphate of ATP is made energetically favorable by the basic conditions established by surrounding histidines and an arginine (47) (Fig. 5, A and B, and supplemental Fig. 2). The abundance of conserved polar and charged residues indicates that proper coordination of the substrates is crucial for this first step of the reaction.
Comparison of AsbB to the structures of AlcC (a type C NIS synthetase) and AcsD (a type A NIS synthetase) (Fig. 4) highlights differences in substrate binding pocket size and exposure to solvent, which may determine interactions with relatively bulky pathway intermediates as opposed to primary metabolites. In particular, this includes the predominantly acidic face that may tightly accommodate the spermidinyl moiety of 1 or 3 (Fig. 5, A and B). Considering the structural similarity of the two substrates of AsbB (1 or 3 and spermidine), it is also possible that they exchange locations and retain the capacity to form an amide bond. This would require deviation from the citrate conformation of previous NIS synthetase-substrate co-crystal structures but is conceivable considering charge distribution in the AsbB pocket is subtly distinct from that of AcsD, in particular, where a citrate molecule can bind.
High nucleotide sequence identity between asbA and asbB (46.7%, compared with the 38.4–41.0% observed between asbA and type A NIS synthetases of other species) (57) likely results from duplication of an asbA-like predecessor gene during evolution of the petrobactin biosynthetic pathway. In support of this, relative catalytic activity values for AsbA and AsbB are now shown to be similar, lying within the same order of magnitude, an observation that is explained by the highly similar primary sequence, substrate structure, and functional activity shared by the two enzymes.
Comparing the relative rates of polyamine incorporation by AsbA and AsbB displayed a similar substrate preference between the two enzymes. Generally, the results demonstrate that both chain length and general charge are influential factors in nucleophile recruitment to the active site (Fig. 6A). Comparative analysis of the crystal structures for AsbB, AcsD, and AlcC suggests the region likely interacting with nucleophilic substrates extends on the surface of the large solvent-exposed binding cleft from the adenylation active site “upward” toward the solvent, loosely parallel to how 1 or 3 is thought to be binding (Fig. 5, A and B). Thus, the multiple charged residues along this surface can be envisioned to interact with the amines of spermidine and analogous linear compounds.
Of the residues suggested in binding and selection of nucleophiles in AsbB, Glu-434 and Glu-459 align closely with Glu-442 and Glu-466 (respectively) of the type A NIS synthetase AcsD (supplemental Fig. 2). Docking experiments in the AcsD active site implicate the utility of Glu442 in stabilizing the nucleophile l-serine, whereas Glu-466 resides between the nucleophilic alcohol of this substrate and the β/γ-phosphate of ATP (47). Indeed, conservation of charged residues at these positions is observed between AsbA, AsbB, AcsD, and AlcC, suggesting their importance in coordination of a nucleophilic substrate. Surface-exposed AsbB residues Lys-311 and Glu-459 reside in this patch (Fig. 5B) and also align with similar residues of AsbA (supplemental Fig. 2). Taking this into account, relative increase in substrate promiscuity with AsbB K311A, K311M, and E459A mutants (Fig. 6B) indicated that these residues, in coordination with Tyr-313 interacting with the distal amine of spermidine, might generate a conformation conducive to spermidine recruitment (Fig. 5B).
Activity studies with single amino acid substitutions can partially explain the shared regioselective preference of AsbA and AsbB for spermidine as a nucleophile; however, factors that cannot be extrapolated from primary sequence undoubtedly play a role, including conserved overall fold and charge distribution within the “fingers” domain. Direct confirmation of whether a loss of substrate specificity is coincident with a loss of regioselectivity will require further investigation and remains an important factor in the pursuit of chemoenzymatic creation of novel compounds using reconstituted biosynthetic systems.
Work highlighting the incorporation of spermidine by AsbA and AsbB has enabled development of a logical pathway proposal in which 3,4-DHBA is incorporated during late stages of petrobactin biosynthesis exclusively at the N1 terminus of spermidine moieties (Fig. 1C) (4, 12, 17, 24). In vitro results now support this pathway, demonstrating reactivity of AsbE with exposed amino groups of proposed intermediates (Fig. 7). 4 and 4′, the previously described products of AsbE aryl transferase activity, and the newly observed but related 6 accumulate during in vitro reconstitution of petrobactin biosynthesis (supplemental Fig. 5), suggesting they are side products that are not efficiently incorporated into the siderophore. Although 6 may be an artifact of the promiscuity AsbE displays in vitro, bis-(dihydroxybenzoyl)-spermidines have been isolated from natural sources including spider venom (58) and from bacterial cultures (59), and a synthetic compound matching the structure of 6 has shown potential as an antimalarial agent (60). Indeed, applications have only been superficially explored for the specific activity combined with substrate flexibility of transferases such as AsbE and siderophore synthetases like AsbB; thus their potential for expanding chemical diversity warrants additional efforts.
The asb operon encodes six separate peptides, the products of which clearly can operate in trans to form petrobactin. However, interactions between different petrobactin biosynthesis proteins within the cell may specify an exact order of steps in a pathway that appears to include accumulation of unincorporated side products (e.g. 4′) and multiple convergent routes in vitro (Fig. 1C) (4, 17, 25, 26). Indeed, recent pulldown experiments show specific interaction of the His6-labeled carrier protein AsbD with untagged AsbC and AsbE after co-expression of asbCDE in E. coli.5 Understanding how protein-protein interactions between products of asbABCDEF confer substrate specificity and reaction efficiency are key future objectives of our group.
The severe virulence of B. anthracis combined with its ability to form spores highly resistant to environmental stresses has made it a persisting global health concern and viable security threat (61, 62). Current methods for preventing or treating anthrax infection remain limited (63, 64). Because abrogation of petrobactin production or uptake has been shown to severely attenuate B. anthracis virulence (6, 10, 15), enzymes in the biosynthetic pathway remain attractive targets for identification of new antimicrobials. Gaining deeper insights into the details of NIS-associated enzyme function might provide access to mechanism-based inhibitors, as have been demonstrated for nonribosomal peptide synthetase-derived siderophore adenylation domains (13, 65, 66). Moreover, development of the chemoenzymatic approach described in this work toward creating new siderophore analogs could be applied to generate a class of “Trojan horse” antibiotics, bearing improved target effects and IC50 values compared with conventional chemotherapeutics (67, 68). These practical considerations further highlight the importance of expanding our understanding of iron acquisition mechanisms employed by a vast array of pathogenic microbes.
Supplementary Material
Acknowledgments
We thank all members of the Midwest Center for Structural Genomics and the Structural Biology Center at Argonne National Laboratory and members of the Life Sciences Institute at the University of Michigan for lending equipment and help in conducting experiments.
This work was supported, in whole or in part, by National Institutes of Health Grant GM074942 (to A. J.). This work was also supported by the United States Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357 (to A. J.), by the Biotechnology and Biological Sciences Research Council through Grant BB/F013760/1 (to G. L. C.), and by Great Lakes Regional Center for Excellence (GLRCE) in Biodefense and Emerging Infectious Disease Grant U54-AI-057153 and the H. W. Vahlteich Professorship (to D. H. S.).

This article contains supplemental Methods, Table 1, and Figs. 1–5.
The atomic coordinates and structure factors (code 3TO3) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
T. D. Nusca, Y. Kim, N. Maltseva, W. Eschenfeldt, L. Stols, M. M. Schofield, J. B. Scaglione, S. D. Dixon, D. Oves-Costales, G. L. Challis, P. C. Hanna, B. F. Pfleger, A. Joachimiak, and D. H. Sherman, unpublished data.
- 3,4-DHBA
- 3,4-dihydroxybenzoic acid
- NIS
- nonribosomal peptide synthetase-independent siderophore
- ddH2O
- double-deionized water
- MESG
- 2-amino-6-mercapto-7-methylpurine ribonucleoside.
REFERENCES
- 1. Lindley P. F. (1996) Iron in biology: a structural viewpoint. Rep. Prog. Phys. 59, 867–933 [Google Scholar]
- 2. Andrews S. C., Robinson A. K., Rodríguez-Quiñones F. (2003) Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237 [DOI] [PubMed] [Google Scholar]
- 3. Wilson M. K., Abergel R. J., Raymond K. N., Arceneaux J. E., Byers B. R. (2006) Siderophores of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. Biochem. Biophys. Res. Commun. 348, 320–325 [DOI] [PubMed] [Google Scholar]
- 4. Hotta K., Kim C. Y., Fox D. T., Koppisch A. T. (2010) Siderophore-mediated iron acquisition in Bacillus anthracis and related strains. Microbiology 156, 1918–1925 [DOI] [PubMed] [Google Scholar]
- 5. May J. J., Wendrich T. M., Marahiel M. A. (2001) The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J. Biol. Chem. 276, 7209–7217 [DOI] [PubMed] [Google Scholar]
- 6. Cendrowski S., MacArthur W., Hanna P. (2004) Bacillus anthracis requires siderophore biosynthesis for growth in macrophages and mouse virulence. Mol. Microbiol. 51, 407–417 [DOI] [PubMed] [Google Scholar]
- 7. Koppisch A. T., Browder C. C., Moe A. L., Shelley J. T., Kinkel B. A., Hersman L. E., Iyer S., Ruggiero C. E. (2005) Petrobactin is the primary siderophore synthesized by Bacillus anthracis strain Sterne under conditions of iron starvation. Biometals 18, 577–585 [DOI] [PubMed] [Google Scholar]
- 8. Wilson M. K., Abergel R. J., Arceneaux J. E., Raymond K. N., Byers B. R. (2010) Temporal production of the two Bacillus anthracis siderophores, petrobactin and bacillibactin. Biometals 23, 129–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee J. Y., Passalacqua K. D., Hanna P. C., Sherman D. H. (2011) Regulation of petrobactin and bacillibactin biosynthesis in Bacillus anthracis under iron and oxygen variation. PLoS One 6, e20777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pfleger B. F., Kim Y., Nusca T. D., Maltseva N., Lee J. Y., Rath C. M., Scaglione J. B., Janes B. K., Anderson E. C., Bergman N. H., Hanna P. C., Joachimiak A., Sherman D. H. (2008) Structural and functional analysis of AsbF. Origin of the stealth 3,4-dihydroxybenzoic acid subunit for petrobactin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 105, 17133–17138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee J. Y., Janes B. K., Passalacqua K. D., Pfleger B. F., Bergman N. H., Liu H., Håkansson K., Somu R. V., Aldrich C. C., Cendrowski S., Hanna P. C., Sherman D. H. (2007) Biosynthetic analysis of the petrobactin siderophore pathway from Bacillus anthracis. J. Bacteriol. 189, 1698–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oves-Costales D., Kadi N., Fogg M. J., Song L., Wilson K. S., Challis G. L. (2007) Enzymatic logic of anthrax stealth siderophore biosynthesis. AsbA catalyzes ATP-dependent condensation of citric acid and spermidine. J. Am. Chem. Soc. 129, 8416–8417 [DOI] [PubMed] [Google Scholar]
- 13. Pfleger B. F., Lee J. Y., Somu R. V., Aldrich C. C., Hanna P. C., Sherman D. H. (2007) Characterization and analysis of early enzymes for petrobactin biosynthesis in Bacillus anthracis. Biochemistry 46, 4147–4157 [DOI] [PubMed] [Google Scholar]
- 14. Fox D. T., Hotta K., Kim C. Y., Koppisch A. T. (2008) The missing link in petrobactin biosynthesis. asbF encodes a (−)-3-dehydroshikimate dehydratase. Biochemistry 47, 12251–12253 [DOI] [PubMed] [Google Scholar]
- 15. Koppisch A. T., Dhungana S., Hill K. K., Boukhalfa H., Heine H. S., Colip L. A., Romero R. B., Shou Y., Ticknor L. O., Marrone B. L., Hersman L. E., Iyer S., Ruggiero C. E. (2008) Petrobactin is produced by both pathogenic and non-pathogenic isolates of the Bacillus cereus group of bacteria. Biometals 21, 581–589 [DOI] [PubMed] [Google Scholar]
- 16. Koppisch A. T., Hotta K., Fox D. T., Ruggiero C. E., Kim C. Y., Sanchez T., Iyer S., Browder C. C., Unkefer P. J., Unkefer C. J. (2008) Biosynthesis of the 3,4-dihydroxybenzoate moieties of petrobactin by Bacillus anthracis. J. Org. Chem. 73, 5759–5765 [DOI] [PubMed] [Google Scholar]
- 17. Oves-Costales D., Kadi N., Fogg M. J., Song L., Wilson K. S., Challis G. L. (2008) Petrobactin biosynthesis: AsbB catalyzes condensation of spermidine with N8-citryl-spermidine and its N1-(3,4-dihydroxybenzoyl) derivative. Chem. Commun. (Camb), 4034–4036 [DOI] [PubMed] [Google Scholar]
- 18. Oves-Costales D., Song L., Challis G. L. (2009) Enantioselective desymmetrization of citric acid catalysed by the substrate-tolerant petrobactin biosynthetic enzyme AsbA. Chem. Commun. (Camb) 1389–1391 [DOI] [PubMed] [Google Scholar]
- 19. Hanson R. M. (2010) Jmol-a paradigm shift in crystallographic visualization. J. Appl. Crystallogr. 43, 1250–1260 [Google Scholar]
- 20. Zawadzka A. M., Abergel R. J., Nichiporuk R., Andersen U. N., Raymond K. N. (2009) Siderophore-mediated iron acquisition systems in Bacillus cereus. Identification of receptors for anthrax virulence-associated petrobactin. Biochemistry 48, 3645–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barbeau K., Zhang G., Live D. H., Butler A. (2002) Petrobactin, a photoreactive siderophore produced by the oil-degrading marine bacterium Marinobacter hydrocarbonoclasticus. J. Am. Chem. Soc. 124, 378–379 [DOI] [PubMed] [Google Scholar]
- 22. Liu H., Håkansson K., Lee J. Y., Sherman D. H. (2007) Collision-activated dissociation, infrared multiphoton dissociation, and electron capture dissociation of the Bacillus anthracis siderophore petrobactin and its metal ion complexes. J. Am. Soc. Mass. Spectrom. 18, 842–849 [DOI] [PubMed] [Google Scholar]
- 23. Challis G. L. (2005) A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochem 6, 601–611 [DOI] [PubMed] [Google Scholar]
- 24. Barry S. M., Challis G. L. (2009) Recent advances in siderophore biosynthesis. Curr. Opin. Chem. Biol. 13, 205–215 [DOI] [PubMed] [Google Scholar]
- 25. Oves-Costales D., Kadi N., Challis G. L. (2009) The long-overlooked enzymology of a nonribosomal peptide synthetase-independent pathway for virulence-conferring siderophore biosynthesis. Chem. Commun. (Camb), 6530–6541 [DOI] [PubMed] [Google Scholar]
- 26. Kadi N., Challis G. L. (2009) Chapter 17. Siderophore biosynthesis a substrate specificity assay for nonribosomal peptide synthetase-independent siderophore synthetases involving trapping of acyl-adenylate intermediates with hydroxylamine. Methods Enzymol. 458, 431–457 [DOI] [PubMed] [Google Scholar]
- 27. Abergel R. J., Wilson M. K., Arceneaux J. E., Hoette T. M., Strong R. K., Byers B. R., Raymond K. N. (2006) Anthrax pathogen evades the mammalian immune system through stealth siderophore production. Proc. Natl. Acad. Sci. U.S.A. 103, 18499–18503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fischbach M. A., Lin H., Liu D. R., Walsh C. T. (2006) How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat. Chem. Biol. 2, 132–138 [DOI] [PubMed] [Google Scholar]
- 29. Abergel R. J., Moore E. G., Strong R. K., Raymond K. N. (2006) Microbial evasion of the immune system. Structural modifications of enterobactin impair siderocalin recognition. J. Am. Chem. Soc. 128, 10998–10999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Abergel R. J., Zawadzka A. M., Raymond K. N. (2008) Petrobactin-mediated iron transport in pathogenic bacteria. Coordination chemistry of an unusual 3,4-catecholate/citrate siderophore. J. Am. Chem. Soc. 130, 2124–2125 [DOI] [PubMed] [Google Scholar]
- 31. Carlson P. E., Jr., Dixon S. D., Janes B. K., Carr K. A., Nusca T. D., Anderson E. C., Keene S. E., Sherman D. H., Hanna P. C. (2010) Genetic analysis of petrobactin transport in Bacillus anthracis. Mol. Microbiol. 75, 900–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arnow L. E. (1937) Colorimetric Determination of the Components of 3,4-dihydroxyphenylalaninetyrosine mixtures J. Biol. Chem. 118, 531–537 [Google Scholar]
- 33. Stols L., Gu M., Dieckman L., Raffen R., Collart F. R., Donnelly M. I. (2002) A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 25, 8–15 [DOI] [PubMed] [Google Scholar]
- 34. Eschenfeldt W. H., Maltseva N., Stols L., Donnelly M. I., Gu M., Nocek B., Tan K., Kim Y., Joachimiak A. (2010) Cleavable C-terminal His-tag vectors for structure determination. J. Struct. Funct. Genomics 11, 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. (1993) Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 [DOI] [PubMed] [Google Scholar]
- 36. Walsh M. A., Evans G., Sanishvili R., Dementieva I., Joachimiak A. (1999) MAD data collection. Current trends. Acta Crystallogr. D Biol. Crystallogr. 55, 1726–1732 [DOI] [PubMed] [Google Scholar]
- 37. Kim Y., Dementieva I., Zhou M., Wu R., Lezondra L., Quartey P., Joachimiak G., Korolev O., Li H., Joachimiak A. (2004) Automation of protein purification for structural genomics. J. Struct. Funct. Genomics. 5, 111–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. (2006) HKL-3000. The integration of data reduction and structure solution. From diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 39. Bricogne G., Vonrhein C., Flensburg C., Schiltz M., Paciorek W. (2003) Generation, representation and flow of phase information in structure determination. Recent developments in and around SHARP 2.0. Acta Crystallogr. D Biol. Crystallogr. 59, 2023–2030 [DOI] [PubMed] [Google Scholar]
- 40. Morris R. J., Perrakis A., Lamzin V. S. (2003) ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 374, 229–244 [DOI] [PubMed] [Google Scholar]
- 41. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 43. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK-a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 44. Wilson D. J., Aldrich C. C. (2010) A continuous kinetic assay for adenylation enzyme activity and inhibition. Anal. Biochem. 404, 56–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Upson R. H., Haugland R. P., Malekzadeh M. N., Haugland R. P. (1996) A spectrophotometric method to measure enzymatic activity in reactions that generate inorganic pyrophosphate. Anal. Biochem. 243, 41–45 [DOI] [PubMed] [Google Scholar]
- 46. McMahon S. A., Oke M., Liu H., Johnson K. A., Carter L., Kadi N., White M. F., Challis G. L., Naismith J. H. (2008) Purification, crystallization, and data collection of Pectobacterium chrysanthemi AcsD, a type A siderophore synthetase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64, 1052–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schmelz S., Kadi N., McMahon S. A., Song L., Oves-Costales D., Oke M., Liu H., Johnson K. A., Carter L. G., Botting C. H., White M. F., Challis G. L., Naismith J. H. (2009) AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis. Nat. Chem. Biol. 5, 174–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Oke M., Carter L. G., Johnson K. A., Liu H., McMahon S. A., Yan X., Kerou M., Weikart N. D., Kadi N., Sheikh M. A., Schmelz S., Dorward M., Zawadzki M., Cozens C., Falconer H., Powers H., Overton I. M., van Niekerk C. A., Peng X., Patel P., Garrett R. A., Prangishvili D., Botting C. H., Coote P. J., Dryden D. T., Barton G. J., Schwarz-Linek U., Challis G. L., Taylor G. L., White M. F., Naismith J. H. (2010) The Scottish Structural Proteomics Facility. Targets, methods, and outputs. J. Struct. Funct. Genomics 11, 167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Franza T., Mahé B., Expert D. (2005) Erwinia chrysanthemi requires a second iron transport route dependent of the siderophore achromobactin for extracellular growth and plant infection. Mol. Microbiol. 55, 261–275 [DOI] [PubMed] [Google Scholar]
- 50. Berti A. D., Thomas M. G. (2009) Analysis of achromobactin biosynthesis by Pseudomonas syringae pv. syringae B728a. J. Bacteriol. 191, 4594–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schmelz S., Botting C. H., Song L., Kadi N. F., Challis G. L., Naismith J. H. (2011) Structural basis for acyl acceptor specificity in the achromobactin biosynthetic enzyme AcsD. J. Mol. Biol. 412, 495–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Laskowski R. A., Watson J. D., Thornton J. M. (2005) Protein function prediction using local three-dimensional templates. J. Mol. Biol. 351, 614–626 [DOI] [PubMed] [Google Scholar]
- 53. Zhang Y. (2008) I-TASSER server for protein three-dimensional structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Roy A., Kucukural A., Zhang Y. (2010) I-TASSER. A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Holinsworth B., Martin J. (2009) Siderophore production by marine-derived fungi. Biometals 22, 625–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cheung J., Beasley F. C., Liu S., Lajoie G. A., Heinrichs D. E. (2009) Molecular characterization of staphyloferrin B biosynthesis in Staphylococcus aureus. Mol. Microbiol. 74, 594–608 [DOI] [PubMed] [Google Scholar]
- 57. Thompson J. D., Higgins D. G., Gibson T. J. (1994) ClustalW. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pereira L. S., Silva P. I., Jr., Miranda M. T., Almeida I. C., Naoki H., Konno K., Daffre S. (2007) Structural and biological characterization of one antibacterial acylpolyamine isolated from the hemocytes of the spider Acanthocurria gomesiana. Biochem. Biophys. Res. Commun. 352, 953–959 [DOI] [PubMed] [Google Scholar]
- 59. Tait G. H. (1975) The identification and biosynthesis of siderochromes formed by Micrococcus denitrificans. Biochem. J. 146, 191–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pradines B., Ramiandrasoa F., Basco L. K., Bricard L., Kunesch G., Le Bras J. (1996) In vitro activities of novel catecholate siderophores against Plasmodium falciparum. Antimicrob. Agents Chemother. 40, 2094–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Casadevall A., Relman D. A. (2010) Microbial threat lists. Obstacles in the quest for biosecurity? Nat. Rev. Microbiol. 8, 149–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bouzianas D. G. (2009) Medical countermeasures to protect humans from anthrax bioterrorism. Trends Microbiol. 17, 522–528 [DOI] [PubMed] [Google Scholar]
- 63. Navas E. (2002) Problems associated with potential massive use of antimicrobial agents as prophylaxis or therapy of a bioterrorist attack. Clin. Microbiol. Infect. 8, 534–539 [DOI] [PubMed] [Google Scholar]
- 64. Cegelski L., Marshall G. R., Eldridge G. R., Hultgren S. J. (2008) The biology and future prospects of antivirulence therapies. Nat. Rev. Microbiol. 6, 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Drake E. J., Duckworth B. P., Neres J., Aldrich C. C., Gulick A. M. (2010) Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis. Biochemistry 49, 9292–9305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gupte A., Boshoff H. I., Wilson D. J., Neres J., Labello N. P., Somu R. V., Xing C., Barry C. E., Aldrich C. C. (2008) Inhibition of siderophore biosynthesis by 2-triazole substituted analogues of 5′-O-[N-(salicyl)sulfamoyl]adenosine. Antibacterial nucleosides effective against Mycobacterium tuberculosis. J. Med. Chem. 51, 7495–7507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Miethke M., Marahiel M. A. (2007) Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71, 413–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Möllmann U., Heinisch L., Bauernfeind A., Köhler T., Ankel-Fuchs D. (2009) Siderophores as drug delivery agents. Application of the “Trojan Horse” strategy. Biometals 22, 615–624 [DOI] [PubMed] [Google Scholar]
- 69. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 70. Laskowski R. A., Watson J. D., Thornton J. M. (2005) ProFunc. A server for predicting protein function from three-dimensional structure. Nucleic Acids Res. 33, W89–W93 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.