Background: Information on the role of N-glycosylation is limited.
Results: The site and structure of N-glycosylation have evident effects on the activity and stability of CBH.
Conclusion: N-Glycosylation affects the characteristics of CBHI and also brings a new function to CBHI as a nonenzymatic synergism factor.
Significance: Understanding the effects of N-glycosylation is important for improvement of enzyme technology.
Keywords: Glycosylation, Post-translational Modification, Protein Engineering, Protein Expression, Protein Purification, CBH I, N-glycosylation, Penicillium decumbens, Post-translation Modification, Synergism
Abstract
Four cellobiohydrolase I (CBHI) glycoforms, namely, CBHI-A, CBHI-B, CBHI-C, and CBHI-D, were purified from the cultured broth of Penicillium decumbens JU-A10. All glycoforms had the same amino acid sequence but displayed different characteristics and biological functions. The effects of the N-glycans of the glycoforms on CBH activity were analyzed using mass spectrum data. Longer N-glycan chains at the Asn-137 of CBHI increased CBH activity. After the N-glycans were removed using site-directed mutagenesis and homologous expression in P. decumbens, the specific CBH activity of the recombinant CBHI without N-glycosylation increased by 65% compared with the wild-type CBHI with the highest specific activity. However, the activity was not stable. Only the N-glycosylation at Asn-137 can improve CBH activity by 40%. rCBHI with N-glycosylation only at Asn-470 exhibited no enzymatic activity. CBH activity was affected whether or not the protein was glycosylated, together with the N-glycosylation site and N-glycan structure. N-Glycosylation not only affects CBH activity but may also bring a new feature to a nonhydrolytic CBHI glycoform (CBHI-A). By supplementing CBHI-A to different commercial cellulase preparations, the glucose yield of lignocellulose hydrolysis increased by >20%. After treatment with a low dose (5 mg/g substrate) of CBHI-A at 50 °C for 7 days, the hydrogen-bond intensity and crystalline degree of cotton fibers decreased by 17 and 34%, respectively. These results may provide new guidelines for cellulase engineering.
Introduction
Lignocellulose is the most abundant renewable natural biological resource (1) whose enzymatic hydrolysis could be performed by cellulase. A complete cellulase system consists of three enzyme classes: cellobiohydrolase (CBH,2 EC 3.2.1.91), endoglucanase (EC 3.2.1.4), and β-glucosidase (EC 3.2.1.21). An enzymatic process may be accomplished through the synergism of these various enzymes. Cellulases are relatively costly enzymes. Thus, significant cost reduction is important for their commercial use in biorefinery. Fungal CBHs, which are the key enzymes in the total hydrolysis of biomass-based processes, belong to glycosidase hydrolase families 6 and 7 according to the carbohydrate-active enzymes database (2). In the culture media of typical cellulolytic fungi, CBHs are the most abundant among the secreted proteins, often accounting ∼60% w/w (3, 4). The cbh1 gene can represent different glycoforms, which may be a result of posttranslational modifications, through proteolytic cleavage by an endogenous enzyme or glycosylation (5). Protein glycosylation is the most complex posttranslational modification process, which includes N- and O-glycosylations (6). Both types of glycosylations are prevalent in CBHs. O-Linked glycosylation occurs at the linker abundant in the hydroxyl groups of serine (Ser) and threonine (Thr) residues, whereas N-linked glycosylation occurs at the asparagines (Asn) in the NX(S/T) consensus sequence motif in the catalytic domain (CD).
The isoforms observed in Trichoderma reesei Cel7A are partly due to the random substitution of glycans on N-glycosylation sites (7). N-Glycosylation patterns and sites are studied extensively in T. reesei cellulases. However, knowledge on the role of N-glycosylation is still limited. A large amount of information on the glycosylation of yeast proteins and heterologous expression in filamentous fungi has been published. However, the nature and effect of glycosylation on proteins in filamentous fungi are understood poorly (8). Recently, protein glycosylation has become widely accepted as an important factor affecting enzyme activity. CBHIs expressed in Saccharomyces cerevisiae and Pichia pastoris are hyperglycosylated, and neither exhibits any activity (9). By removing the Asn-384 glycosylation site and expression in Aspergillus niger, T. reesei CBHI shows 70% greater activity compared with the wild-type T. reesei enzyme (7). The role of glycosylation was studied previously using heterologous expression and rarely by homologous expression, which could not provide the most accurate information because the glycosylation of cellulase varies with the expression host and culture conditions (7). Accordingly, the current work studied the effect of glycosylation on CBH using homologous expression to produce the most accurate information regarding the effect of N-glycosylation on cellulase. In this study, the effects of N-glycosylation, together with its site and chain lengths, on the enzymatic activity of CBHI were investigated thoroughly.
EXPERIMENTAL PROCEDURES
Microorganism and Culture Condition
A fast-growing cellulolytic fungus, P. decumbens 114-2, was isolated from the soil (10). Its catabolite repression-resistant and cellulase-overproducing mutant, JU-A10, was obtained via physical and chemical mutagenesis from the wild strain 114-2 in our laboratory (11). The medium used for cellulase production was according to Gao et al. (12). PyrG-deficient P. decumbens 114-2 was used as the homologous expression host. Subsequently, 0.05% uridine was added to the aforementioned medium for cellulase production. Spore suspension was inoculated into 50 ml of liquid medium in a 300-ml conical flask and grown at 30 °C with rotatory shaking at 180 rpm.
Purification of Proteins
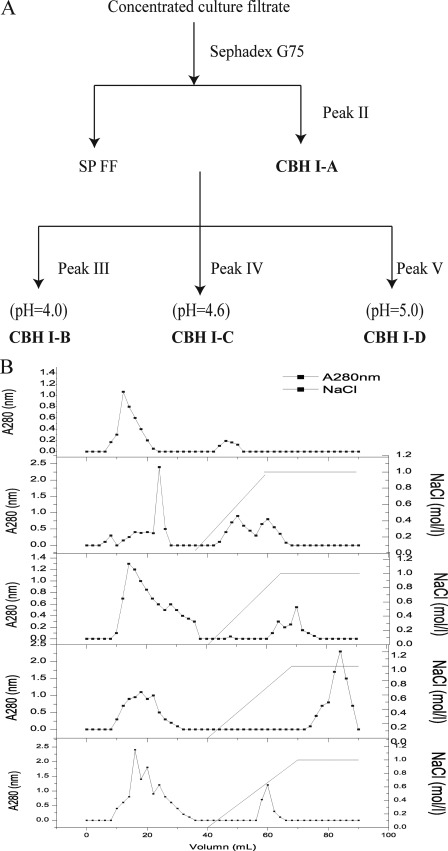
Concentrated culture filtrate was loaded onto the gel filtration chromatography column (Sephadex G-75, GE Healthcare). The column was equilibrated and washed with 20 mm sodium acetate buffer (pH = 4.8) at a 0.15 ml/min flow rate. Protein concentration was detected using a spectrometer at 280 nm absorbance.
The main fractions eluted from the gel filtration chromatography column were collected and concentrated using a Mr 10,000 cut-off Millipore membrane (Millipore). The concentrated fractions were separated using an anion exchange SP Fast Flow column (GE Healthcare), which resulted in three peaks. The collected fractions from the three peaks were used for further separation. The combined fractions from the peaks were loaded into a DEAE Fast Flow column (1.6 cm × 20 cm) (GE Healthcare) equilibrated with 20 mm sodium acetate buffer (pH 4.0, 4.6, and 5.0). The column was eluted with a linear NaCl gradient from 0 to 1 m in the same buffer at 2 ml/min flow rate, and fractions of 2 ml each were collected.
Determination of Protein Concentration
Protein concentrations were determined using a trace protein concentration determination kit via the bicinchoninic acid method (TransGen Biotech).
Internal Amino Acid of Purified Proteins via MALDI-TOF MS
Purified proteins were identified via tandem MS (MS/MS) by the Beijing Proteome Research Center and Tianjin Biochip Corp. in Nankai University.
Enzyme Assay of Purified Proteins
Culture supernatants were collected via centrifugation. Aliquots of the supernatants were diluted for the CBH activity assay following a method described previously (12).
Endoglucanase and β-glucosidase activities were determined using different cellulosic substrates at 1% concentration (w/v), namely, carboxymethyl cellulose and salicin. The reactions were performed in 50 mm sodium acetate buffer (pH 4.8) at 50 °C for 30 min. The total amount of reducing sugars in the supernatants was determined via the dinitrosalicylic acid method (13).
Effects of Temperature and pH on CBH Activity
The optimum temperature for purified proteins on CBH activity was measured from 30 to 80 °C in 50 mm sodium acetate buffer (pH 4.8). The optimum pH for purified proteins on CBH activity was measured at the pH range of 3 to 7. Thermostability was investigated by incubating the enzyme in a water bath at 50 °C in the same buffer without a substrate. Samples were collected at different time intervals, and the residual activity was assayed.
Synergism between Purified Proteins and Different Enzyme Samples
Supplementation experiments were performed in duplicate in a 10-ml centrifuge tube. The reaction mixtures contained 5% (w/v) pretreated corn stover (water washed, provided by DSM, Den Haag, Holland) and 20 FPU (Filter Paper Unit) of different commercial enzyme preparations per gram substrate in a total volume of 5 ml (NS50012 and NS50013 from Novozymes; Jan-A from DSM, Holland; Gen from Genencor). Supplementation dosage of 0.36 mg/g substrate of the four CBHI glycoforms was from P. decumbens JU-A10. Saccharification was performed at 50 °C for 15 h, and glucose concentration was determined using an SBA-4 Biological Sensor Analyzer (Biological Institute of Shandong Academy of Science, Jinan, China).
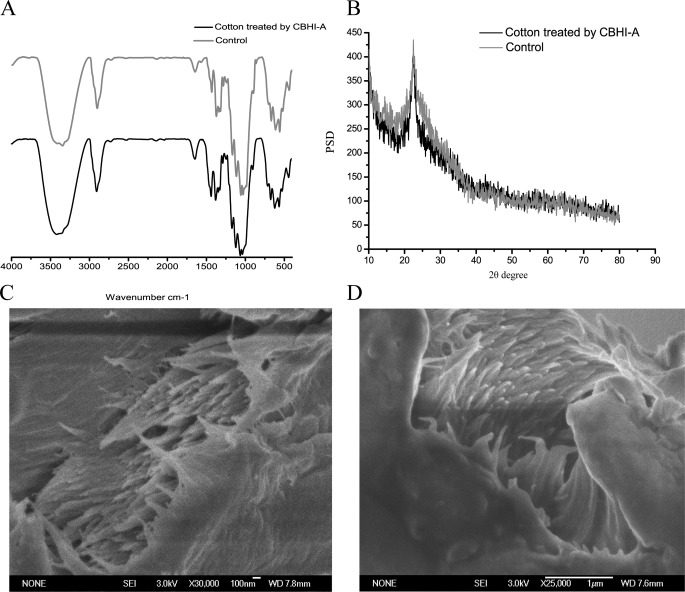
Analysis of Cotton Fibers Treated with Purified Proteins via FT-IR Spectroscopy, X-ray Diffraction (XRD), and SEM
Easily observable cotton fibers with high crystalline degrees were used as substrates. For 7 days of treatment, 100 μg of purified P. decumbens JU-A10 proteins was added separately to 20 mg of cotton fibers at 50 °C. The precipitate was acquired via centrifugation at 10,000 × g for 10 min and completely dried via progressive ethanol dehydration. The precipitate was dried at 50 °C for 10 h using a vacuum dryer (DZF-6020, Shanghai). The cotton fibers were analyzed via FT-IR and XRD according to methods described previously (14, 15). The hydrogen bond intensity and crystallinity of the cotton were calculated according to Liu et al. (14).
The cotton fibers treated with purified P. decumbens JU-A10 proteins were scanned via SEM to compare the changes in diameter. The cotton fibers were coated with platinum before and after treatment and then studied. Images were taken using a JEOL JSM-6700 SEM (JEOL, Tokyo, Japan), and the cotton fiber diameters were estimated using Sigma Pro software (version 5.0). The cotton fiber diameters were equal to the average of hundreds of cotton fibers.
Sugars released from the cotton fibers were determined using HPLC (Shimadzu, Kyoto, Japan) with a refractive index detector (Shimadzu, Kyoto, Japan). All samples were diluted and passed through a 0.45-μm filter before HPLC analysis. The saccharides were separated in an Aminex HPX-87P column (Bio-Rad) running at a flow rate of 0.6 ml/min at 78 °C, with water as eluent.
Analysis of N-Glycans Using Mass Spectra of Proteins
The mass spectra of the peptides were obtained after the digestion of proteins via SDS-PAGE with trypsin. The peptide masses of the purified P. decumbens JU-A10 proteins were analyzed using a peptide mass tool from ExPASy. Some peptide masses from the mass spectrum were more than the theoretical unmodified tryptic peptides. The peptide masses were taken into account and compared with the peptide masses with potential N-glycosylation sites (16). The N-glycan structure was speculated based on the previously reported molecular weight of glycans.
Modification of N-Linked Glycosylation Sites of cbh1 Gene
Genomic DNA was extracted from P. decumbens JU-A10 cultivated in Mandel's medium for 48 h. The cbh1 gene was amplified from JU-A10 via PCR using mutagenic primers. The two amplified gene fragments were purified and fused together via fusion PCR. The whole fragment containing the mutation site was amplified with ∼2,000-bp fragments upward and downward, which were used as homologous arms for transformation.
Fungal Transformation and Confirmation of cbh1 Gene Containing Mutation Site
The cbh1 fragment with homologous arms and ANIp-8 was cotransformed into the PyrG-deficient P. decumbens 114-2. The transformants were selected on plates without uridine via single spore culture for further analysis. The PyrG-deficient P. decumbens 114-2 was transformed according to the method described by Gomi et al. (17). Positive clones were verified though DNA sequencing. The DNA and RNA of transformants were acquired. The mutated cbh1 genes were amplified using DNA and cDNA from successful transformants. The cbh1 genes with mutation site were sequenced and compared with the wild-type cbh1 gene from P. decumbens using the Basic Local Alignment Search Tool at the National Center of Biotechnology Information.
Modeling of cbh1 Catalysis Domain with Different Types of Glycosylation
Using the amino acid sequence acquired through MALDI-TOF, the cbh1 gene was found in the genomic library of P. decumbens. The cbh1 gene structure was predicted based on a previously published T. reesei CBHI structure. The three-dimensional structure of the cbh1 CD was modeled using the automated mode of the SWISS-MODEL. The glycoproteins with four different glycans were built, solvated, and energy-minimized using Glycam (18). The Glycam output structures were visualized and analyzed using PyMOL (19).
RESULTS
Purification of P. decumbens JU-A10 Proteins
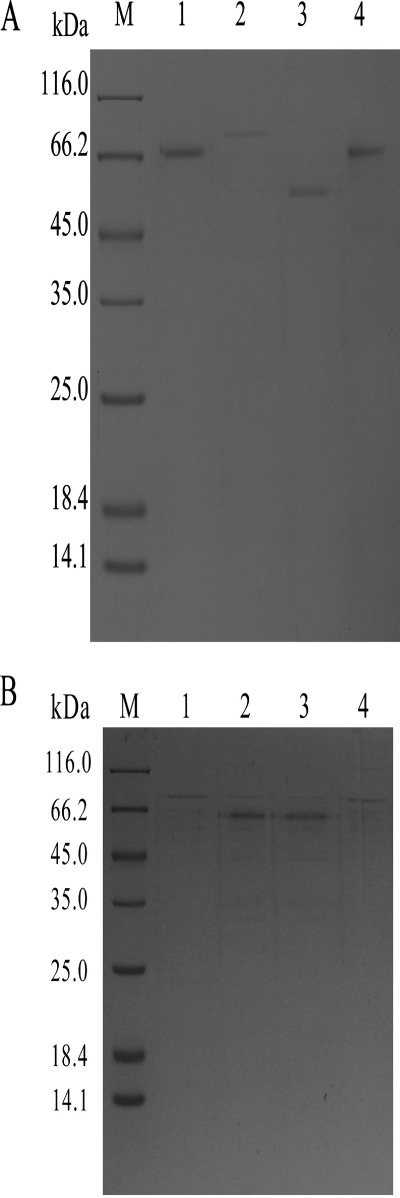
The whole purification scheme for P. decumbens JU-A10 proteins is shown in Fig. 1. The concentrated culture filtrate was loaded into the gel filtration chromatography column (Sephadex G-75; GE Healthcare). The main proteins (peak I) with CBH activity were first eluted. A small amount of protein (peak II) was absorbed on the gel filtration chromatography and finally eluted. Peak II protein was detected via SDS-PAGE as a single band with a molecular mass of 66 kDa (Fig. 2). The main fractions (peak I) were collected and filtrated using Millipore. The concentrated fractions were separated by an anion exchange SP Fast Flow column (GE Healthcare), which resulted in three peaks marked as peaks III, IV, and V. The collected fractions from the three peaks were used for further separation (Fig. 1). The combined fractions of each peak were separated using a DEAE Fast Flow column (GE Healthcare) at different pH values. The three proteins marked as peaks VI, VII, and VIII were all detected via SDS-PAGE as a single band. The molecular masses of the three P. decumbens JU-A10 proteins were 74, 57, and 68 kDa, respectively (Fig. 2A).
FIGURE 1.
Fractionation and purification scheme (A) and chromatograms (B) of CBHI-A, CBHI-B, CBHI-C, and CBHI-D produced by Penicillium decumbens JU-A10. CBHI-A, CBHI-B, CBHI-C, and CBHI-D were purified according to peaks II, VI, VII, and VIII, respectively.
FIGURE 2.
A, different CBHI glycoforms separated via SDS-PAGE. Lanes 1, 2, and 3 show CBHI-B, CBHI-C, and CBHI-D purified from Penicillium decumbens JU-A10, respectively. Lane 4 shows the nonhydrolytic protein CBHI-A corresponding to peak II via gel filtration chromatography (Sephadex G-75) from P. decumbens JU-A10. Lane M indicates the protein molecular weight marker. B, different rCBHIs separated via SDS-PAGE. Lanes 1, 2, 3, and 4 show purified rCBHI-N137D-470, rCBHI-N137D, rCBHI-N470D-137, and rCBHI-N470D expressed in P. decumbens.
Identification of Four Purified P. decumbens Proteins via MALDI-TOF (MS/MS)
The proteins purified from the extracellular P. decumbens JU-A10 proteins were identified through MS/MS at the Beijing Proteome Research Center and Tianjin Biochip Corp. in Nankai University. The internal amino acid sequences of the four proteins were analyzed via in-gel digestion of the protein. The four proteins were digested with trypsin, sequenced via MALDI-TOF, and identified as exoglucanase (CBH) 1 from Penicillium. Using the amino acid sequence acquired via MALDI-TOF, the cbh1 gene was found in the genomic library of P. decumbens 114-2 (unpublished data).3 Surprisingly, the four proteins shared identical gene and amino acid sequences of CBHI from P. decumbens 114-2. In agreement, peptide mass fingerprinting of the four CBHI glycoforms strongly suggested that the four CBHI glycoforms shared a common polypeptide core, which endured differential glycosylation. The four proteins were designated as CBHI-A (peak II), CBHI-B (peak VI), CBHI-C (peak VII), and CBHI-D (peak VIII).
Characteristic Assay of Four CBHI Glycoforms
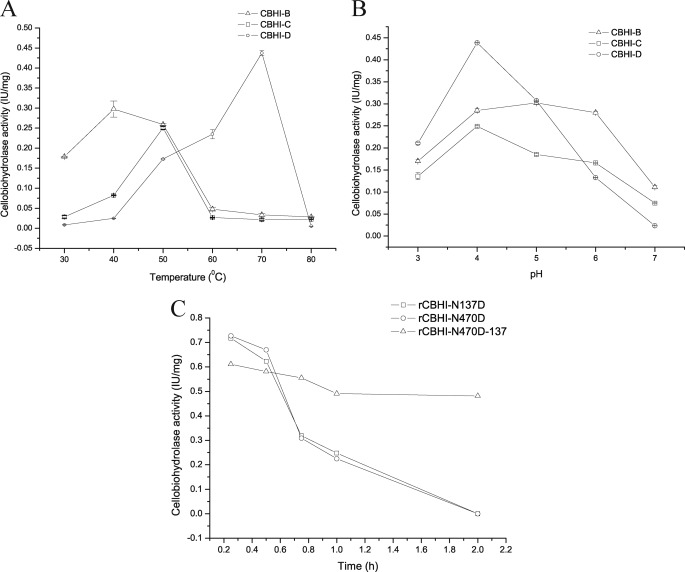
The enzyme activity against the p-nitrophenyl-β-d-cellobioside (pNPC) of the four CBHI glycoforms was tested. Interestingly, CBHI-A exhibited no detectable activity against pNPC, whereas CBHI-B, CBHI-C, and CBHI-D demonstrated 0.3, 0.25, and 0.44 IU/mg, respectively. The optimum temperatures for the purified CBHI-B, CBHI-C, and CBHI-D were 40, 50, and 70 °C, respectively (Fig. 3A). Optimum pH levels for CBHI-B, CBHI-C, and CBHI-D were 5, 4, and 4, respectively, as obtained by measuring their activities at the pH range of 3 to 7 (Fig. 3B). The purified CBHI-A exhibited no enzymatic activity against carboxymethyl cellulose, pNPC, and salicin. The four CBHI glycoforms, with identical amino acid sequences, showed different enzyme characteristics. The differential glycosylation is hypothesized to be responsible for the different biochemical and physical properties of the CBHI glycoforms.
FIGURE 3.
Shown are optimum temperatures (A) and pH levels (B) of the activities of the different purified CBHI glycoforms. C, thermal inactivation kinetics studied by incubating rCBHIs in the buffer (pH 4.8) at 50 °C. rCBHI-N470D-137 showed higher thermostability compared with rCBHI-N137D and rCBHI-N470D. RCBHI-N137D and rCBHI-N470D exhibited identical thermal inactivation profiles, and their half-lives decreased to ∼40 min.
Analysis of N-Glycans of Four CBHI Glycoforms Using Mass Spectrum Data
The MALDI-TOF mass spectrum of the peptides obtained after a tryptic digestion of the SDS-PAGE band is shown in supplemental Fig. 1. Using the peptide mass from the ExPASy proteomic site, specific tryptic peptides matching the amino acid sequences of CBHI were found. By comparing the experimental masses of glycopeptides with the theoretical ones, more masses were found. The molecular weights of the glycans detected using MALDI-TOF (20) were acquired from a previous paper and used to analyze the N-glycan structures of the different CBHI glycoforms.
The peak with m/z 1500.8 (supplemental Fig. 1) corresponded to the N-glycosylated peptide (LYLLENDTTYQK) in the amino acid sequences of CBHI. By comparing the experimental masses of CBHI with the theoretical ones, the additional peak with m/z 2639.2 cumulatively differed from the peak with m/z 1500.8 by 1138 Da. According to the molecular weight of the N-glycan structure detected using MALDI-TOF (20), it was found that (Man)3(GlcNAc)2 + GlcNAc bound to the Asn-137 of CBHI-A. Similar analyses of the MS data were performed for the other CBHI glycoforms. The glycans (Man)3(GlcNAc)2 + (Man)3GlcNAc, (Man)3(GlcNAc)2 + (Man)2(GlcNAc)2, and (Man)3(GlcNAc)2 + (Man)5GlcNAc were found bound to Asn-137 in CBHI-B, CBHI-C, and CBHI-D, respectively. The oligosaccharides seemed to represent the well known conserved core structure of (Man)3(GlcNAc)2 by forming high mannose and hybrid/complex glycans in the glycoproteins from different organisms. Furthermore, N-glycosylated proteins share a common structure, (Hex)x + (HexNAc)y(Man)3(GlcNAc)2, in Chrysosporium lucknowense (16). Comparing the glycans at Asn-137 with the CBH activities of the CBHI glycoforms, longer glycan chains at the Asn-137 of CBHI were found to have higher CBH activities.
Synergism between CBHI-A and Different Enzyme Samples
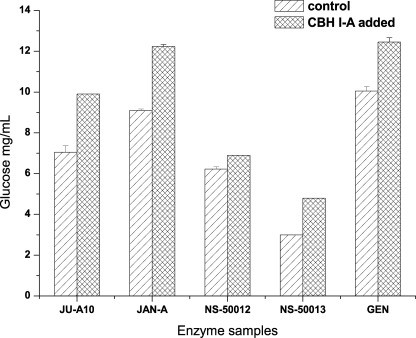
The four CBHI glycoforms were supplemented, respectively, to different commercial cellulases with a small dosage of 0.36 mg protein/g substrate in a total of 5 ml volume. A total of 5% pretreated corn stover (water-washed) was used as the substrate. Saccharification was performed at 50 °C for 15 h. Surprisingly, CBHI-B, CBHI-C, and CBHI-D exhibited no apparent synergism with the commercial cellulases (data not shown), whereas CBHI-A showed a strong synergism on cellulose degradation with the different enzyme samples. In the supplementation experiment, although the dosage of CBHI-A was small, the glucose yield realized from the pretreated corn stover (water-washed) increased from 20 to 40%. Surprisingly, the addition of CBHI-A to the NS50013 sample (incomplete cellulase system) from Novozymes increased glucose deliberation by ∼60%, with pretreated corn stover as the substrate (Fig. 4). This result implies that nonhydrolytic CBHI glycoforms have a new function and feature.
FIGURE 4.
Synergism between CBHI-A and different commercial cellulases (20 FPU/g substrate for each), including JU-A10, JAN-A, NS-50012, NS-50013, and GEN, in the hydrolysis of 5% (w/v) pretreated corn stover with a supplementation dosage of 0.36 mg CBHI-A from P. decumbens JU-A10.
Analysis of Cotton Fibers Treated with CBHI-A via FT-IR Spectroscopy and XRD
The rationale behind the synergism between CBHI-A and the different commercial cellulases, as well as the effect of CBHI-A on the substrate, still needs to be discussed. Structural changes in the cotton fibers treated with CBHI-A were analyzed via FT-IR spectroscopy, XRD, and SEM (Fig. 5). The absorbance of the hydrogen-bonded OH stretch was at 4000 cm−1 to 2995 cm−1 wave number range. FT-IR spectroscopy showed that at 4000 cm−1 to 2995 cm−1 wave number range, the transmittance of the cotton fibers treated with CBHI-A evidently increased, implying a decrease in absorbance. The hydrogen bond intensity and crystallinity of the cotton fibers treated with CBHI-A decreased by 19 and 34%. This result implies that the hydrogen bonds and crystalline of the cotton fibers were destroyed. The sugars released in the supernatant were determined through centrifugation and analyzed by HPLC. Cellobiose was the dominant product of hydrolysis. The hydrolysis of the cotton fibers by CBHI-A produced no cellobiose and reducing sugar, whereas the hydrolysis by CBHI-B, CBHI-C, and CBHI-D produced 14.2, 13.8, and 28.2 mg cellobiose/g cotton fibers, respectively. The fracture surfaces of the cotton fibers were observed using SEM, and the cotton fiber diameters were determined using the Sigma Pro software (version 5.0). The diameter of the cotton fibers treated with CBHI-A for 7 days was 96 nm, which was ∼7% larger than that of the cotton fibers before treatment. According to the AFM results (21, 22), enzymatic hydrolysis would perform in the outer surface of the cellulose and then in the next layer. According to the model, a single glycan chain should be removed from the microfiber to be effectively hydrolyzed by cellulase. CBHI-A may play an important role in destroying the hydrogen bonds of cellulose to spin off microfibers. N-Glycosylation may provide nonhydrolytic CBHI glycoform a new function and synergism.
FIGURE 5.
Structural changes in cotton treated with CBHI-A purified from Penicillium decumbens JU-A10. Changes were analyzed using FT-IR (A), XRD (B), and SEM before (C) and after (D) treatment.
Removal of N-Glycosylation Sites from CBHI and Expression of Mutants
The N-linked glycosylation sites in P. decumbens CBHI were indicated at Asn-137 and Asn-470 (supplemental Fig. 2). The mutated cbh1 gene was expressed in P. decumbens. The DNA and RNA of transformants were acquired. The mutated cbh1 genes were amplified using DNA and cDNA from successful transformants. The cbh1 genes with a mutation site were sequenced and compared with the wild-type cbh1 gene from P. decumbens. The results showed that the sequence of mutated cbh1 amplified from DNA was in agreement with that amplified from cDNA. The cbh1-containing mutation site was only expressed in the mutation strains at the gene and transcript level, whereas non-mutated cbh1 was not found at all. These results confirmed that the asparagine (Asn) has been successfully replaced by aspartate (Asp) in the mutant strains. The enzyme of the recombinant CBHI (rCBHI) was purified using SP and DEAE Fast Flow columns, and detected via SDS-PAGE (Fig. 2B). The removal of N-linked glycosylation at Asn-137 or Asn-470 sites all resulted in two different rCBHIs with different molecular weights. Both rCBHIs with glycosylation at Asn-470 or Asn-137 (rCBHI-N137D-470 or rCBHI-N470D-137) had the same molecular mass of 76 kDa, whereas others (rCBHI-N137D or rCBHI-N470D) had no glycosylation at Asn-470 or Asn-137 with a molecular mass of 57 kDa (Fig. 2B). The activities of the rCBHIs with no glycosylation were 0.72 IU/mg (Table 1), which was 65% higher than that of CBHI-D, the wild-type CBHI with the highest specific activity. However, the deglycosylated protein lost its activity in a half-month, even at 4 °C. Thermal inactivation kinetics was studied by incubating the rCBHIs in the buffer (pH 4.8) at 50 °C. The thermostability of rCBHI-N470D-137 was higher than that of rCBHI-N137D and rCBHI-N470D (Fig. 3C). The profiles of the thermal inactivations of rCBHI-N137D and rCBHI-N470D were identical, and their half-lives decreased to ∼40 min. Glycoproteins are known to be more resistant to denaturation than their deglycosylated equivalent (23).
TABLE 1.
Comparison of N-glycan structures at Asn-137 and CBHI activities between CBHIs with mutation expressed in P. decumbens and wild-type CBHIs from P. decumbens
N137D-N470D showed that Asn-137 and Asn-470 of CBHI from P. decumbens were both replaced with aspartate. N137D-470 and N470D-137 showed that Asn-137 or Asn-470 of CBHI from P. decumbens was replaced with aspartate. However, another putative N-glycosylation site (Asn-470 or Asn-137) was glycosylated. ND, not determined.
| Relative molecular mass | Cellobiohydrolase | Glycan structure at Asn-137 | |
|---|---|---|---|
| kDa | IU/mg | ||
| CBHI-A | 66 | 0 | (Man)3(GlcNAc)2 + GlcNAc |
| CBHI-B | 74 | 0.30 | (Man)3(GlcNAc)2 + (Man)3GlcNAc |
| CBHI-C | 58 | 0.25 | (Man)3(GlcNAc)2 + (Man)2(GlcNAc)2 |
| CBHI-D | 68 | 0.44 | (Man)3(GlcNAc)2 + (Man)5GlcNAc |
| rCBHI-N137D/rCBHI-N470D | 57 | 0.72 | |
| rCBHI-N470D-137 | 76 | 0.61 | ND |
| rCBHI-N137D-470 | 76 | 0 | ND |
The effect of N-glycosylation on CBH activity was determined by the N-glycosylation site. RCBHI-N470D-137 exhibited enzymatic activity at 0.61 IU/mg, which was 40% higher than that of CBHI-D. This result implies that glycosylation at Asn-137 could greatly improve CBH, which is in agreement with the results from the MS data. That is, longer glycan chains result in higher CBH activities.
RCBHI-N137D-470 has no enzymatic activity, which implies that the glycan at Asn-470 negatively affects CBH activity. The molecular weights of the rCBHIs with glycosylation at Asn-137 or Asn-470 were larger than any wild-type CBHIs.
Modeling of cbh1 Catalysis Domain with Different Types of Glycosylation
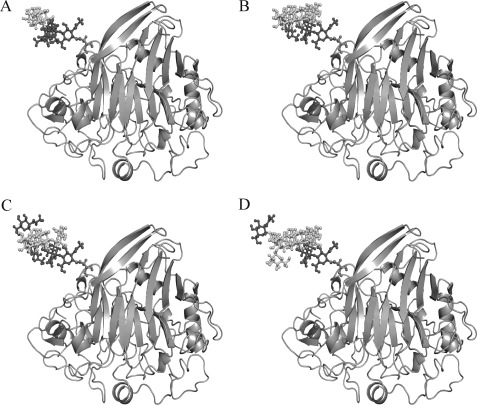
The structure of the cbh1 gene product was predicted based on the previously published structure of T. reesei CBHI (Protein Data Bank code 1Q9H). P. decumbens JU-A10 CBHI showed a high degree of sequence identity (73.7%) with the template 1Q9H via SWISS-MODEL. As shown in Fig. 6, different forms of glycans at the Asn-137 of CBHI were marked in the modeled three-dimensional structure of CBHI CD. Asn-137 was located on a loop at the back of the CD and near the entrance of the active-tunnel of CBHI. The modeled three-dimensional structure of CBHI CD showed that the glycan chain at Asn-137 extended between carbohydrate binding module and CD in the space position. The glycan chain may affect the conformational states and the collaboration between the two domains, thereby enhancing the processing activity and favorably improving the CBH activity (7). These data suggest that not only the active site but also the coordination and cooperation of the two domains of the enzyme affect the enzymatic activity. The effect of the glycan chain on the collaboration between the two domains and the enhancement of the enzymatic activity requires further investigation.
FIGURE 6.
Three-dimensional structure of CBHI-A (A), CBHI-B (B), CBHI-C (C), and CBHI-D (D) modeled using the automated mode of SWISS-MODEL. Glycoprotein models with different glycans at the Asn-137 site were built and marked in the modeled three-dimensional structure.
Asn-470 is located at the linker region of P. decumbens CBHI. Hence, the presence of the glycan at this position could limit the freedom of the linker. The absence of N-glycans could increase the flexibility of the linker, which may improve the binding of CBHI to the substrate and the threading of a cellulose chain into the CBHI tunnel.
DISCUSSION
The CBHs produced by the fungal species are similar in primary sequence (60% overall identity) but significantly different in their rates and extents of cellulose hydrolysis (7, 24). The reason behind the similar sequences among CBHIs but largely different enzymatic activities still needs to be discussed. Glycans could carry much more finely tuned information than other biomolecules because of their diversity (25). The roles of glycosylation in enzyme function have not been fully elucidated (26). For understanding why different forms of CBHI displayed different characteristics and biological functions, we analyzed the N-glycosylation structures CBHI from P. decumbens. We found that all forms of CBHI have the same amino acid sequence but have different N-glycosylation structures. Therefore, we presumed that N-glycosylation site and glycan structure may be the one of main reasons for their functional difference. The present work presents a systematic analysis of the effects of N-glycosylation on the enzymatic characteristics and functions of CBHI. In addition to the effects of glycosylation and N-glycan length on the enzymatic activity, the glycosylation site showed a significant role in CBH activity. By analyzing the effects of deleting N-glycosylation sites in the gene, the effects of N-glycsylation on enzyme activity were confirmed. Glycosylation at Asn-137 improved cellobiohydrolase, whereas glycosylation at Asn-470 completely inhibited cellobiohydrolase. Identical glycosylation sites of T. reesei CBHII have different degrees of glycosylation, and different glycosylation sites may have a competitive relationship (27). The degree of glycosylation at each site was not fixed; that is, it may be fully or partially glycosylated. The molecular weights of the rCBHIs with glycosylation at Asn-137 or Asn-470 were larger than any wild-type CBHIs, implying that the Asn-137 or Asn-470 of rCBHI was fully glycosylated. Many different wild-type CBHI glycoforms are products of different glycosylation at Asn-137 and Asn-470. Therefore, the specific CBH activity of the wild-type CBHI glycoform ranged between 0 and 0.71 IU/ml. The identity gene expressed different CBHI glycoforms by selecting different ways of post-translational modifications.
Glycosylation may provide the nonhydrolytic CBHI glycoform a new function and synergism with other cellulases. The current paper has reported a CBHI glycoform (CBHI-A) with no detectable enzymatic activity but with strong synergism with different commercial cellulases. These results demonstrate that CBHI-A could disrupt the hydrogen bonds of cellulose and make the high crystalline cellulose more amorphous. More than 60 years ago, Reese et al. (28) first proposed that the C1 factor destroying the crystalline of cellulose is beneficial for the hydrolysis of cellulose by cellulase. In the course of exploring the mechanism of cellulose degradation, many studies discovered several materials with a similar function to C1, including swollenin from T. reesei (29), CDH from Phanerichaete chrysoporiu (30), HO• from T. reesei (31), and the short fiber-generating factor from T. pseudokoningi (32). However, the above materials are subject to much debate because of their unapparent synergism with commercial cellulases. The present study found a real cause of the cellulose hydrogen bond destruction. During N-glycosylation as post-translational modification, part of the CBHI was modified with a high mannose glycan to generate CBHI-A. N-Glycosylation provided CBHI-A a new function of damaging the hydrogen bonds of the crystalline cellulose. We speculated that different N-glycosylation may affect the conformational states and change the pace of processing activity of CBHI. However, the real mechanism of N-glycosylation effect on CBHI function diversity is still not very clear and needs to be investigated farther in future.
Many scholars have been looking for synergism factors from cellulases but have neglected the original evolution of cellulase. The metabolic evolution process is economical and needs to be simplified (33). Therefore, a protein family always contains proteins with enzyme functions, whereas others may not function as enzymes at all (24). Expansin is a well known example. Expansin does not have a hydrolytic activity but has amino acid sequences similar to GH45 endoglucanase. Expansin could weaken the mechanical strength of the cellulosic filter paper by disrupting the hydrogen bonds between the cellulose chains and cell wall polysaccharides. Proteins of the same family, with similar or the same amino acid sequences, may have different biological functions. This phenomenon may be common in cellulase evolution. Evolution may select N-glycosylation as a modification to providing different functions from identical proteins.
Compared with transforming the cellulase and changing the ratio of different components of the cellulase system, glycosylation could significantly improve cellulase activity. Understanding the effect of glycosylation on cellulase is important for further improvement of the enzyme technology for biomass conversion. This conclusion could provide guidelines for the reconstruction of the cellulase system, including the selection of the type and component of cellulase with a higher activity.
Supplementary Material
Acknowledgments
We thank Dr. Manoj Kumar and Dr. Hein Stem in DSM White Biotechnology B.V., Netherlands, for support and advice on the screening of new features and synergism of the enzyme components.
This work was supported by National Basic Research Program of China Grant 2011CB707403 and National Natural Sciences Foundation of China Grants 31030001 and 31170107.

This article contains supplemental Figs. 1 and 2.
L. Gao, F. Gao, L. Wang, C. Geng, L. Chi, J. Zhao, and Y. Qu, unpublished data.
- CBH
- cellobiohydrolase
- rCBHI
- recombinant CBHI
- XRD
- x-ray diffraction.
REFERENCES
- 1. King A. J., Cragg S. M., Li Y., Dymond J., Guille M. J., Bowles D. J., Bruce N. C., Graham I. A., McQueen-Mason S. J. (2010) Molecular insight into lignocellulose digestion by a marine isopod in the absence of gut microbes. Proc. Natl. Acad. Sci. U.S.A. 107, 5345–5350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The carbohydrate-active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 37, 233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou J., Wang Y. H., Chu J., Zhuang Y. P., Zhang S. L., Yin P. (2008) Identification and purification of the main components of cellulases from a mutant strain of Trichoderma viride T 100-14. Bioresour. Technol. 99, 6826–6833 [DOI] [PubMed] [Google Scholar]
- 4. Kim K. H., Brown K. M., Harris P. V., Langston J. A., Cherry J. R. (2007) A proteomics strategy to discover β-glucosidases from Aspergillus fumigatus with two-dimensional page in-gel activity assay and tandem mass spectrometry. J. Proteome. Res. 6, 4749–4757 [DOI] [PubMed] [Google Scholar]
- 5. García R., Cremata J. A., Quintero O., Montesino R., Benkestock K., Ståhlberg J. (2001) Characterization of protein glycoforms with N-linked neutral and phosphorylated oligosaccharides: Studies on the glycosylation of endoglucanase 1 (Cel7B) from Trichoderma reesei. Biotechnol. Appl. Biochem. 33, 141–152 [DOI] [PubMed] [Google Scholar]
- 6. Wong C. H. (2005) Protein glycosylation: New challenges and opportunities. JOC Perspective 70, 4219–4225 [DOI] [PubMed] [Google Scholar]
- 7. Adney W. S., Jeoh S., Beckham G. T., Chou Y. C., Baker J. O., Michener W., Brunecky R., Himmel M. E. (2009) Probing the role of N-linked glycans in the stability and activity of fungal cellobiohydrolases by mutational analysis. Cellulose 16, 699–709 [Google Scholar]
- 8. Goto M. (2007) Protein O-glycosylation in fungi: Diverse structures and multiple functions. Biosci. Biotechnol. Biochem. 71, 1415–1427 [DOI] [PubMed] [Google Scholar]
- 9. Wu G. C., Wei L. G., Liu W. F., Lin J. Q., Wang l. S., Qu Y. B., Zhuang G. Q. (2010) Asn-64 glycosylation affects Hypocrea jecorina (syn. Trichoderma reesei) cellobiohydrolase Cel7a activity expressed in Pichia pastoris. World J. Microbiol. Biotechnol. 26, 323–328 [Google Scholar]
- 10. Qu Y. B., Gao P. J., Wang Z. N. (1984) Screening of catabolite repression-resistant mutants of cellulase producing Penicillium spp. Acta Mycol. Sinica. 3, 238–243 [Google Scholar]
- 11. Sun X. Y., Liu Z. Y., Zheng K., Song X., Qu Y. B. (2008) The composition of basal and induced cellulase systems in Penicillium decumbens under induction or repression conditions. Enzyme Microb. Technol. 42, 560–567 [Google Scholar]
- 12. Gao L., Wang F., Gao F., Wang L., Zhao J., Qu Y. (2011) Purification and characterization of a novel cellobiohydrolase (PdCel6A) from Penicillium decumbens JU-A10 for bioethanol production. Bioresour. Technol. 102, 8339–8342 [DOI] [PubMed] [Google Scholar]
- 13. Chahal D. (1985) Solid-state fermentation with Trichoderma reesei for cellulase production. Appl. Environ. Microbiol. 49, 205–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu K., Lin X., Yue J., Li X., Fang X., Zhu M., Lin J., Qu Y., Xiao L. (2010) High concentration ethanol production from corncob residues by fed-batch strategy. Bioresour. Technol. 101, 4952–4958 [DOI] [PubMed] [Google Scholar]
- 15. Yu C. T., Chen W. H., Men L. C., Hwang W. S. (2009) Microscopic structure features changes of rice straw treated by boiled acid solution. Ind. Crop. Prod. 29, 308–315 [Google Scholar]
- 16. Gusakov A. V., Antonov A. I., Ustinov B. B. (2008) N-Glycosylation in Chrysosporium lucknowense enzymes. Carbohydr. Res. 343, 48–55 [DOI] [PubMed] [Google Scholar]
- 17. Gomi K., Iimura Y., Hara S. (1987) Integrative transformation of Aspergillus oryzae with a plasmid containing the Aspergillus nidulans argB gene. Agric. Biol. Chem. 51, 2549–2555 [Google Scholar]
- 18. Wang J., Cieplak P., Kollman P. A. (2000) How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 21, 1049–1074 [Google Scholar]
- 19. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace: A web-based environment for protein structure homology modeling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 20. Kainz E., Gallmetzer A., Hatzl C., Nett J. H., Li H., Schinko T., Pachlinger R., Berger H., Reyes-Dominguez Y., Bernreiter A., Gerngross T., Wildt S., Strauss J. (2008) N-glycan modification in Aspergillus species. Appl. Environ. Microbiol. 74, 1076–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee I., Evans B. R., Woodward J. (2000) The mechanism of cellulase action on cotton fibers: Evidence from atomic force microscopy. Ultramicroscopy 82, 213–221 [DOI] [PubMed] [Google Scholar]
- 22. Igarashi K., Koivula A., Wada M., Kimura S., Penttilä M., Samejima M. (2009) High speed atomic force microscopy visualizes processive movement of Trichoderma reesei cellobiohydrolase I on crystalline cellulose. J. Biol. Chem. 284, 36186–36190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Benoit I., Asther M., Sulzenbacher G., Record E., Marmuse L., Parsiegla G., Gimbert I., Asther M., Bignon C. (2006) Respective importance of protein folding and glycosylation in the thermal stability of recombinant feruloyl esterase A. FEBS Lett. 580, 5815–5821 [DOI] [PubMed] [Google Scholar]
- 24. McQueen-Mason S., Cosgrove D. J. (1994) Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc. Natl. Acad. Sci. U.S.A. 91, 6574–6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mariño K., Bones J., Kattla J. J., Rudd P. M. (2010) A systematic approach to protein glycosylation analysis: A path through the maze. Nat. Chem. Biol. 6, 713–723 [DOI] [PubMed] [Google Scholar]
- 26. Beckham G. T., Dai Z., Matthews J. F., Momany M., Payne C. M., Adney W. S., Baker S. E., Himmel M. E. (2011) Harnessing glycosylation to improve cellulase activity. Curr. Opin. Biotechnol. 23, 1–8 [DOI] [PubMed] [Google Scholar]
- 27. Harrison M. J., Nouwens A. S., Jardine D. R., Zachara N. E., Gooley A. A., Nevalainen H., Packer N. H. (1998) Modified glycosylation of cellobiohydrolase I from a high cellulase-producing mutant strain of Trichoderma reesei. Eur. J. Biochem. 256, 119–127 [DOI] [PubMed] [Google Scholar]
- 28. Reese E. T. (1976) Polysaccharase and the hydrolysis of insoluble substrates. Proc. Sess. 6, 9–12 [Google Scholar]
- 29. Saloheimo M., Paloheimo M., Hakola S., Pere J., Swanson B., Nyyssönen E., Bhatia A., Ward M., Penttilä M. (2002) Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur. J. Biochem. 269, 4202–4211 [DOI] [PubMed] [Google Scholar]
- 30. Tanaka H., Yoshida G., Baba Y., Matsumura K., Wasada H., Murata J., Agawa M., Itakura S., Enoki A., (2007) Characterization of a hydroxyl radical-producing glycoprotein and its presumptive genes from the white-rot basidiomycete Phanerochaete chrysosporium. J. Biotechnol. 128, 500–511 [DOI] [PubMed] [Google Scholar]
- 31. Chandhoke V., Goodell B., Jellison J., Fekete F. A. (1992) Oxidation of 2-keto-4-thiomethylbutyric acid (KTBA) by iron-binding compounds produced by the wood decaying fungus Gloeophyllum irabeum. FEMS Microbiol. Lett. 90, 263–266 [Google Scholar]
- 32. Wang W., Liu J., Chen G., Zhang Y., Gao P. (2003) Function of a low molecular weight peptide from Trichoderma pseudokoningii S38 during cellulose biodegradation. Curr. Microbiol. 46, 371–379 [DOI] [PubMed] [Google Scholar]
- 33. Todd A. E., Orengo C. A., Thornton J. M. (1999) Evolution of protein function, from a structural perspective. Curr. Opin. Chem. Biol. 3, 548–556 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.