Background: EKLF is a transcription factor that is critical for erythroid gene expression.
Results: The EKLF in vivo interaction with Ppm1b phosphatase has been identified after co-immunoprecipitation.
Conclusion: The effect of the interaction is to modulate EKLF stability and activity.
Significance: Our data uncover a novel interaction and reveal a multilayered and contextual difference in how Ppm1b alters EKLF regulation of its downstream targets.
Keywords: Erythropoiesis, Protein Phosphatase, Protein Stability, Transcription Elongation Factors, Transcription Factors
Abstract
Erythroid Krüppel-like factor (EKLF; KLF1) is an erythroid-specific transcription factor required for the transcription of genes that regulate erythropoiesis. In this paper, we describe the identification of a novel EKLF interactor, Ppm1b, a serine-threonine protein phosphatase that has been implicated in the attenuation of NFκB signaling and the regulation of Cdk9 phosphorylation status. We show that Ppm1b interacts with EKLF via its PEST1 sequence. However, its genetic regulatory role is complex. Using a promoter-reporter assay in an erythroid cell line, we show that Ppm1b superactivates EKLF at the β-globin and BKLF promoters, dependent on intact Ppm1b phosphatase activity. Conversely, depletion of Ppm1b in CD34+ cells leads to a higher level of endogenous β-globin gene activation after differentiation. We also observe that Ppm1b likely has an indirect role in regulating EKLF turnover via its zinc finger domain. Together, these studies show that Ppm1b plays a multilayered role in regulating the availability and optimal activity of the EKLF protein in erythroid cells.
Introduction
EKLF2 is an erythroid-specific transcription factor that plays an important role in the global regulation of erythroid transcription (1). ChIP-seq experiments have shown that EKLF, sometimes in concert with SCL/TAL 1 or GATA 1, is bound to promoter or intronic elements of almost all genes that are associated with processes regulating the survival, differentiation, or biology of erythroid cells (2, 3).
EKLF function is tightly controlled by protein/protein interactions and post-translational modifications. Much of the prior work that elucidated the role of EKLF in protein/protein interactions was motivated by attempts to study the role of EKLF in transcription regulation (4–11). These studies were driven by knowledge of the erythroid-specific transcription factors or co-activator proteins that compose the essential transcriptional machinery and were designed to find out if EKLF interacted with these known molecules. Although this approach shed much light on the molecular mechanisms of EKLF function, it also limited the extent to which one could elucidate unexpected modes of EKLF function or regulation.
Several lines of evidence hint at gaps in our knowledge of EKLF regulation and function. First, although EKLF has two nuclear localization signals and exhibits nuclear localization as expected for its function as a transcription factor (12, 13), a significant proportion of EKLF is localized to the cytoplasm (14, 15). Second, Quadrini and Bieker (16) showed that the stabilities of both nuclear and cytoplasmic EKLF could be regulated by the proteasome. To date, candidate proteins that regulate EKLF stability have not been reported in the literature.
Third, various studies have ascribed paradoxical roles of EKLF in cell cycle regulation. EKLF activates transcription of E2f2, through which it regulates the entry of erythroid progenitors into the cell divisions that precede terminal erythroid differentiation (17, 18). At the same time, EKLF has also been shown to be a transcriptional activator of cell cycle inhibitors like p18INK4c (19) and p21WAF1/CIP1 (20). Given the restricted tissue specificity and temporal regulation of EKLF expression in vivo, it is possible that post-translational regulation and/or protein interactions govern which of the opposing roles EKLF plays in the cell divisions that lead to terminal erythroid differentiation. In addition, EKLF is expressed over a period that encompasses several different stages in hematopoiesis (21, 22). Although Pilon et al. (2) have demonstrated that EKLF binding to various sites on the genome is dynamic and dependent on differentiation stage, it is not clear which signals trigger these changes. Exposing primary human erythroid progenitor cells to RB7, a short chain fatty acid derivative, can induce EKLF binding to the CACCC site of the γ-globin promoter, concomitant with induction of γ-globin expression (6). As EKLF does not activate γ-globin transcription in definitive erythroid cells (2), it is possible that changes in EKLF protein/protein interactions or post-translational modifications may modify its ability to activate transcription at such promoters.
Given the circumstantial evidence in the literature, most compellingly the cytoplasmic localization of EKLF, we hypothesized that EKLF might interact with, or be regulated by, proteins that are not typically part of the transcription machinery. To address this question, we immunoprecipitated FLAG-tagged EKLF that was expressed in MEL cells and identified associated proteins using mass spectrometry. In this paper, we show that EKLF interacts with Ppm1b, a protein phosphatase that belongs to the PP2C family of protein phosphatases (23), and we describe its ability to modulate EKLF function by various means, including regulation of EKLF protein stability and transcriptional function.
EXPERIMENTAL PROCEDURES
Cell Culture
The zinc-inducible FLAG-EKLF MEL stable cell line (7) was maintained in DMEM supplemented with 10% FBS in the presence of 200 μg/ml G418. FLAG-EKLF expression was induced by treating cell cultures with 100 μm ZnCl2 for 18 h. shPpm1b and scr stably transfected JK-1 cells were maintained in RPMI 1640 medium supplemented with 20% FBS in the presence of 1 μg/ml puromycin (Sigma).
Immunoprecipitation
To identify novel EKLF interactors, a stable MEL line containing a Zn2+-inducible FLAG-EKLF plasmid (7) was used to identify EKLF-associated complexes in MEL cells. 2.0 × 108 actively growing cells, seeded at a density of 1.0 × 106 cells/ml DMEM, were induced with 100 μm ZnCl2 for 18 h. Treatment with PBS was used as a negative control. Cells were washed with ice-cold PBS and lysed in a lysis buffer (1% Nonidet P-40, 300 mm NaCl, 50 mm Tris, pH 8.0, protease inhibitor mixture (Roche Applied Science), 0.5 mm iodoacetamide, 1 mm sodium butyrate, 0.5 mm sodium fluoride, and 1 mm sodium vanadate). Lysate, normalized to 42 mg of protein in 10 ml of lysis buffer, was precleared with rabbit IgG-agarose (Sigma) for 60 min at 4 °C. The resultant supernatants were immunoprecipitated with 250 μl of M2-agarose (Sigma) for 2 h at 4 °C (24). Following immunoprecipitation, the beads were washed three times with lysis buffer. FL-EKLF and associated proteins were eluted three times by incubating the beads in 1.25 ml of PBS containing 25 μg of FLAG peptide (Sigma) for 15 min. Eluates were concentrated to 50 μl with a centrifugal filter unit with a 10-kDa cutoff (Millipore) and diluted with 2× Laemmli sample buffer (Bio-Rad) with 10 mm DTT prior to loading on a 10% Tris-HCl acrylamide gel. The gel was stained with colloidal blue (Invitrogen) and destained in deionized water overnight. The protein bands were excised and analyzed by the mass spectrometry facility at the Mt. Sinai School of Medicine.
Co-immunoprecipitations of exogenous EKLF and Ppm1b utilized protein lysate obtained from a 10-cm dish of transfected 293T cells. Cells were washed in ice-cold PBS and lysed in buffer (1% Nonidet P-40, 300 mm NaCl, 50 mm Tris, pH 8.0) in the presence of protease inhibitor mixture (Roche Applied Science). 2 mg of lysate was precleared with rabbit IgG-agarose and immunoprecipitated with anti-FLAG M2-agarose. Following immunoprecipitation, the beads were washed three times with lysis buffer. Proteins were eluted with Laemmli sample buffer (Bio-Rad).
Transfection
Transfections were performed using DMRIE-C (Invitrogen) according to the manufacturer's protocol. For co-immunoprecipitation experiments, EKLF constructs or pSG5 empty vector were co-transfected with Ppm1b at a 1:1 ratio. Protein stability assays were carried out by co-transfecting EKLF constructs with Ppm1b, Ppm1b R/G (phosphatase-inactive), or pSG5 empty vector with GFP at a 2:2:1 ratio. Transfection of K562 and JK-1 cells was carried out with 4 μg of plasmid DNA and 8 μl of DMRIE-C per 200,000 cells.
Selection of Stably Transfected JK-1 Cell Lines
100,000 JK-1 cells were transfected with shPpm1b or control plasmids carrying a puromycin resistance gene (gifts of J. Yang). Puromycin was added to a final concentration of 1 μg/ml 48 h post-transfection. Culture media was changed 8 days post-transfection, and surviving cells were divided into smaller pools for further analysis.
Semi-quantitative RT-PCR
cDNA was synthesized using the either the Promega Reverse Transcription System (A3500) with a combination of random primers and dT (15), using 1 μg of RNA as starting material, or the Qiagen Sensiscript RT kit, using 40 ng of RNA as starting material. γ-Globin and β-globin were detected after 25 and 29 PCR cycles, respectively, using primers described by Blau et al. (25). EKLF was detected after 29 PCR cycles using primers described by Perrine et al. (6). GAPDH was detected after 25 PCR cycles using primers described by Narla et al. (26).
Antibodies
The following antibodies were used for Western blotting: M2-HRP conjugate (Sigma), anti-Ppm1b (A300–887A; Bethyl Laboratories), anti-Ppm1a antibody (ab76574; Abcam), anti-β-tubulin antibody (sc-58886; Santa Cruz Biotechnology), anti-GFP antibody (Living Colors A.v. monoclonal JL-8; Clontech), and anti-HSP90 antibody (53547; Anaspec Inc.). Anti-EKLF 7B2 antibody was generated in-house (21).
Primary Cell Culture and Nucleofection
Human CD34+ hematopoietic stem cells derived from peripheral blood were obtained from All Cells. CD34+ cells were plated at a density of 0.4 × 106 cells/ml in expansion media consisting of serum-free expansion medium (SFEM; StemCell Technologies) that was supplemented with 2% penicillin/streptomycin, recombinant human thrombopoietin (50 ng/ml), SCF (25 ng/ml), and Flt-3 ligand (50 ng/ml) (27). Media were changed when cell density reached 1 × 106 cells/ml. After 7 days of expansion, 1.5 × 106 cells per sample were nucleofected with 200 nm siPpm1b (supplemental Table S3) or untargeted siRNA (Qiagen) using the Amaxa Human CD34+ nucleofection kit (program U-008) (28). Erythroid differentiation was induced with Epo (4 units/ml) and SCF (100 ng/ml) (29) in SFEM media 24 h after nucleofection. Cells were collected 72 h after differentiation for analysis by RT-PCR.
RESULTS
EKLF Physically Interacts with Ppm1b
To elucidate novel interactors of EKLF, we employed an MEL cell line that was stably transfected with a pSVneoHMT-IIA plasmid expressing FLAG-EKLF under the control of a human metallothionein (hMT-IIa) promoter, containing sequence elements that rendered the promoter inducible by zinc (7, 30). EKLF was immunoprecipitated from whole cell extracts with anti-FLAG M2-agarose (Sigma). Bound complexes were eluted, resolved, and identified by mass spectrometry (supplemental Fig. S1 and Table 1). We excluded common contaminants seen in mass spectrometric analysis from further consideration. We also eliminated from consideration proteins that could have interacted with FL-EKLF as a response to cell stress. The induction of exogenous EKLF expression by ZnCl2 has the potential to evoke the expression of proteins that respond to cell stress or proteins involved in the unfolded protein response, which could be induced in response to increased protein synthesis. For this reason, we excluded Scyl2 (also known as coated vesicle-associated kinase of 104 kDa, CVAK104) (31) and Hspa8 (32).
We focused our study on Ppm1b, as 10 of the peptides, derived from three bands on the gel (supplemental Table 2) and providing the largest number of peptides in this dataset that corresponded with a single protein, were identified as derived from Ppm1b.
Many studies show a significant functional overlap between Ppm1b and the closely related Ppm1a. To confirm that the peptides detected by mass spectrometric analysis derive from Ppm1b, rather than Ppm1a, we performed a ClustalW sequence alignment analysis. Although several of the identified peptides are in regions where Ppm1a and -1b were identical, most are located in regions where there are several amino acid differences between the two proteins (Fig. 1). One identified peptide is located in the C-terminal region of Ppm1b, which has no analogous region in Ppm1a. The sequence alignment shows that the identified protein is indeed Ppm1b. The fact that the peptides were identified from three differently migrating bands isolated from an SDS gel raised the possibility that EKLF may interact with more than one Ppm1b isoform. A ClustalW sequence alignment revealed that 9 of the 10 peptides identified by the mass spectrometric analysis reside in Ppm1b regions that do not differ between isoforms (supplemental Fig. S2). One of the peptide sequences only exists in Ppm1b isoform 1. This result suggests that EKLF may interact with multiple Ppm1b isoforms.
FIGURE 1.
Identification of Ppm1b as an EKLF-interacting protein. Amino acid alignment of Ppm1b and Ppm1a are shown. Boxed regions correspond to Ppm1b peptides that were identified by mass spectrometry of the gel in supplemental Fig. S1 and tabulated in supplemental Table S2. Most are unique to Ppm1b and thus distinguish this protein from Ppm1a.
Ppm1b Depletion in Hematopoietic Stem Cells Alters β-Globin Transcription after Differentiation
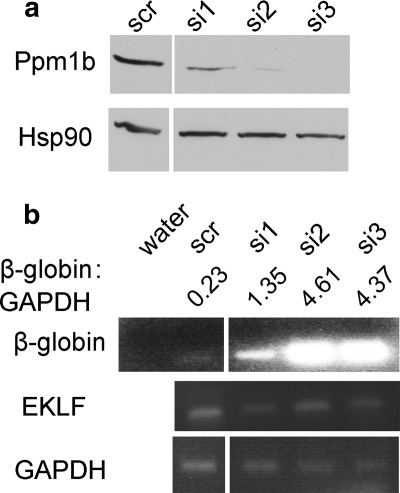
Evidence for a potential role of Ppm1b in gene regulation is complex. Ppm1b has been shown to dephosphorylate Cdk9 at Thr-186, acting either as a negative regulator of pTEF-b function (33) or as a positive effector that enables pTEF-b release from its inhibitory complex (34). As removal of Cdk9 (35) or inhibition of its activity (36) results in a defect in erythropoiesis, and because Cdk9 binds to the β-globin locus in vivo (37), we postulated that silencing of Ppm1b would alter erythropoietic gene expression. To test this hypothesis, we used three different siRNAs to silence Ppm1b in CD34+ hematopoietic stem cells (HSCs) (Fig. 2a), followed by differentiation of HSCs into erythroid progenitors (29). Consistent with data suggesting that Ppm1b is a negative regulator of Cdk9, whose activity is required for globin expression during both zebrafish and human erythropoiesis (36), we observed an increase in β-globin RNA concomitant with a decrease in Ppm1b protein levels (Fig. 2b). As depletion of Ppm1b did not affect induction of EKLF transcription, it seems likely that Ppm1b is functioning downstream of EKLF to regulate β-globin transcription upon erythroid induction.
FIGURE 2.
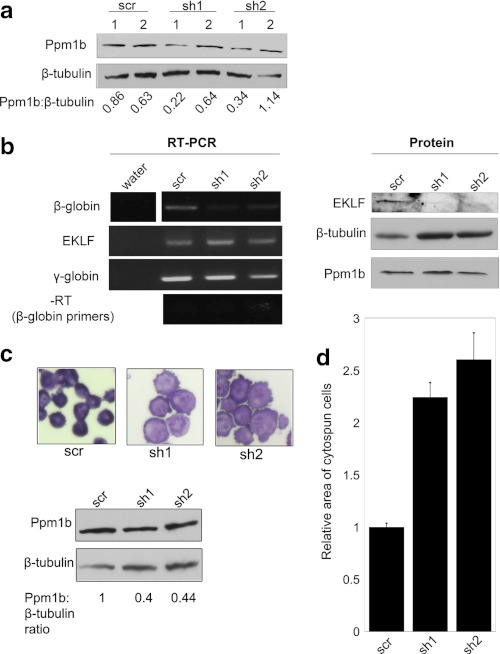
Knockdown of Ppm1b in human CD34+ HSCs. a, CD34+ cells were nucleofected with scrambled siRNA (scr) or siPpm1b (si1–3; supplemental Table S3), and Ppm1b protein expression was monitored by Western blot 24 h post-nucleofection (top). Hsp90 was used as a loading control (bottom panel). b, semi-quantitative RT-PCR was used to determine the effect of siPpm1b on β-globin (top panel) and EKLF (middle panel) RNA levels after 3 days of erythroid differentiation with Epo/SCF. GAPDH RNA levels were used as a normalization control (bottom panel). The normalized level of β-globin expression at each condition is calculated (β-globin:GAPDH).
Ppm1b Decreases the Rate of EKLF Turnover
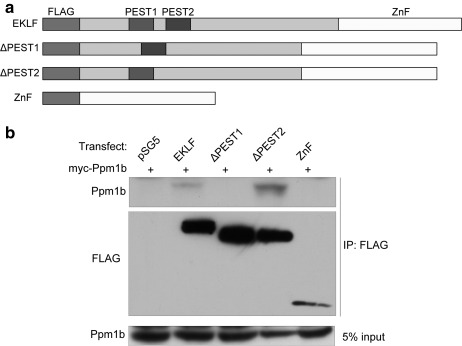
To further characterize the role of Ppm1b in erythropoiesis, and elucidate its structural or functional interaction with EKLF, we identified the region in EKLF that interacts with Ppm1b by co-expression of exogenous Myc-tagged Ppm1b with FLAG-tagged EKLF, or EKLF mutants in 293T cells, followed by immunoprecipitation with M2-agarose (Sigma). We included PEST sequence deletion mutants in this set of experiments because they have been shown to play a role in regulating EKLF stability (16). The PEST1 region of EKLF overlaps with the minimal activation domain of EKLF (16, 38) and the PEST2 region overlaps with the transactivation subdomain involved in binding Tfb1PH, a subunit of TFIIH (39). Fig. 3 shows that the PEST1 sequence of EKLF is important for mediating the interaction between EKLF and Ppm1b.
FIGURE 3.
Mapping of regions on EKLF that interact with Ppm1b. a, schematic of FLAG-EKLF constructs used in co-immunoprecipitation experiments. PEST sequences are as defined in Ref. 16. b, interaction of Ppm1b with FLAG-EKLF after co-transfection of the indicated constructs into 293T cells followed by immunoprecipitation (IP) with anti-FLAG antibody. pSG5 is the empty expression vector. The blot was probed with anti-Ppm1b (top panel) or anti-FLAG (middle panel). The bottom panel shows the input Ppm1b levels. Ppm1b interacts with full-length EKLF but not its zinc finger alone (ZnF) or EKLF ΔPEST1, suggesting that the PEST1 sequence on the N-terminal domain of EKLF is necessary for its interaction with Ppm1b.
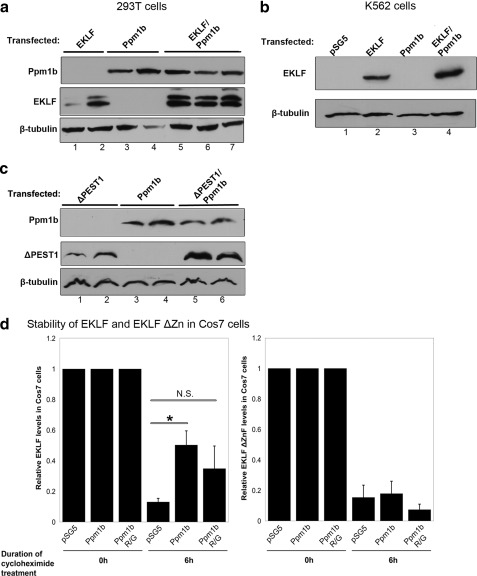
In the course of our studies, we noticed that EKLF levels increased when exogenous Ppm1b was co-expressed (compare lanes 1 and 2 with lanes 5–7 in Fig. 4a and compare lanes 2 and 4 in Fig. 4b). Because expression of each EKLF variant is under the control of the same promoter within each experiment, it seemed unlikely that the increase in EKLF levels is due to changes in transcription levels. Ppm1b also increases the EKLF protein levels even in the absence of the EKLF PEST1 sequence that is important for mediating the EKLF interaction with Ppm1b (compare lanes 1 and 2 with lanes 5 and 6 in Fig. 4c). This suggests that Ppm1b stabilizes EKLF via an indirect mechanism.
FIGURE 4.
Overexpression of Ppm1b stabilizes EKLF. For a–c, transfected material is as indicated at the top, and the antibody used for probe is on the left. β-Tubulin was used as a loading control. a, co-expression of Ppm1b increases the levels of exogenous EKLF in 293T cells. Lanes 1 and 2, 3 and 4, and 5–7 are derived from biological replicates of transfections. b, Ppm1b also increases exogenous EKLF levels in K562 cells (compare lanes 2 and 4). c, deletion of the PEST1 sequence that is important for the physical interaction of EKLF and Ppm1b does not abrogate this functional effect. Lanes 1 and 2, 3 and 4, and 5 and 6 are biological replicates of transfections. d, stability of protein expressed from the indicated constructs (as indicated below) after transfection into COS7 cells was monitored after cycloheximide treatment as indicated. Left panel shows the data with full-length EKLF and right panel shows the data from the EKLF ΔZnF variant. Ppm1b decreases the turnover of full-length EKLF (p < 0.05) but has no effect on the stability of EKLF ΔZnF. The data presented here are representative of three independent experiments. Error bars represent the mean ± S.E. NS, not significant.
To test the hypothesis that Ppm1b regulates EKLF stability, we co-transfected EKLF with empty vector, Ppm1b, or its phosphatase-inactive Ppm1b (R179G) variant (40) along with a plasmid encoding GFP as a transfection normalization control. 24 h post-transfection, cells were treated with cycloheximide and harvested 0 and 6 h after cycloheximide treatment (based on Ref. 16), and EKLF levels in cell lysates were monitored and quantitated by Western blotting. We found that transfection of exogenous Ppm1b reduces the degradation of EKLF (Fig. 4d). Co-transfection of EKLF and Ppm1b (R179G) trends toward an increase in EKLF stability, but the difference is not statistically significant (p = 0.218).
We then determined if there is a specific EKLF domain required for Ppm1b-mediated stabilization of EKLF. Having ruled out the EKLF PEST1 domain as being important in mediating this process, we tested the effect of Ppm1b in mediating the rate of EKLF turnover in the absence of the EKLF zinc finger domain. We found that Ppm1b is unable to stabilize EKLF in the absence of its zinc finger domain, despite the fact that it does not directly interact with this domain (Fig. 4d).
These data suggest that Ppm1b regulates EKLF protein expression by regulating its turnover. This effect appears to be indirect, perhaps requiring an intermediate, as it is independent of the presence of a region important for the interaction of EKLF and Ppm1b. The data also suggests that the stabilization of EKLF by Ppm1b is at least partially independent of Ppm1b's phosphatase activity, hinting that the role of Ppm1b in the stabilization of EKLF follows from elements that are inherent to the tertiary structure of Ppm1b, rather than from its enzymatic ability.
shPpm1b Decreases Transcription Activity of EKLF at the β-Globin and Ahsp Promoters
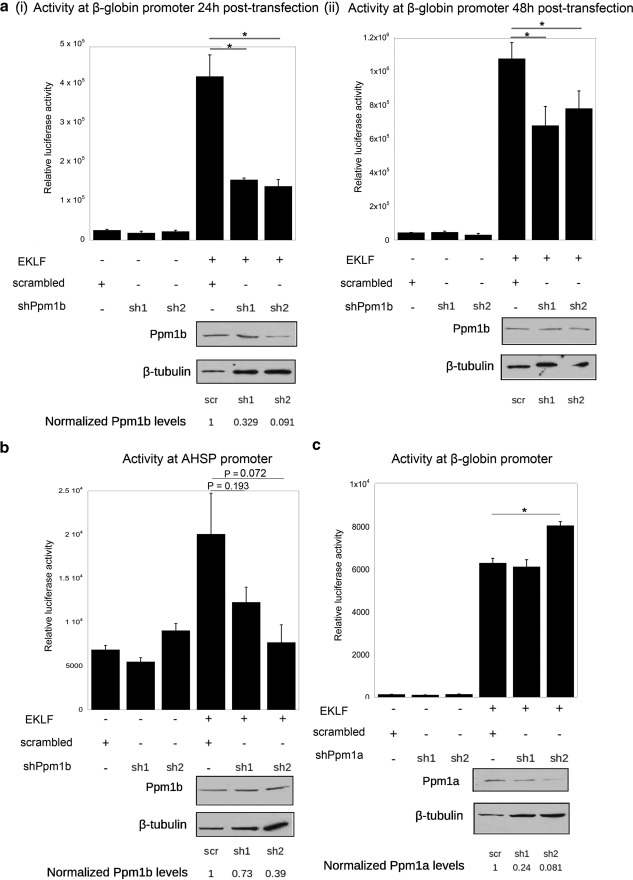
The previous set of experiments suggested that Ppm1b stabilizes the EKLF protein. We turned our attention to determining whether a decrease in Ppm1b leads to a decrease in EKLF transactivation. We co-transfected shPpm1b constructs with EKLF and examined their effects on EKLF-dependent promoters in K562 cells, an erythroleukemic cell line that does not express EKLF (41). Transfections included a plasmid encoding FLAG-EKLF, one of two different shPpm1b constructs (sh1 and sh2) (40) or a scr control, and firefly luciferase reporters under the control of either Ahsp or β-globin promoters. The efficacy of shRNA knockdown was monitored by Western blotting for Ppm1b and normalizing to β-tubulin levels.
Ppm1b knockdown is most apparent 24-h post-transfection rather than at 48 h post-transfection (Fig. 5a), and sh2 knocks down Ppm1b levels more efficiently than sh1. In either case, the efficacy of Ppm1b knockdown directly correlates with a decrease in β-globin promoter activity (Fig. 5a). Silencing Ppm1b also reduces the activity of EKLF at the Ahsp promoter (Fig. 5b); again, the extent to which Ahsp promoter activity decreased corresponds with the degree of Ppm1b knockdown, with sh2 causing a greater decrease than sh1. Neither sh1 nor sh2 had any effect on basal activity at the β-globin and Ahsp promoters, suggesting that decreasing the level of endogenous Ppm1b protein levels specifically affects EKLF-mediated transcription activation. The data suggest that EKLF-dependent reporter activity varies in direct proportion to Ppm1b levels.
FIGURE 5.
Effect of decreasing Ppm1b or Ppm1a levels on EKLF activity at target promoters. Co-transfection assays were performed in erythroleukemic K562 cells that do not express EKLF. Decrease in Ppm1b levels by specific shRNAs (sh1 or sh2) (40) compared with a scrambled control (scr) leads to a decrease in the following: a, β-globin promoter activity 24 h (panel i) or 48 h (panel ii) post-transfection; b, AHSP promoter activity 24 h post-transfection. Westerns blots of Ppm1b protein are shown below each panel, along with quantitation of Ppm1b levels from a comparison to β-tubulin. c, however, decreasing Ppm1a with specific shRNAs (sh1 or sh2) does not decrease EKLF activity, but rather it leads to a small but significant (p < 0.05) increase of β-globin promoter activity with sh2. Westerns blots of Ppm1a protein are shown below, along with quantitation of Ppm1a levels from a comparison with β-tubulin. Data are obtained from three independent experiments. Error bars are representative of S.E. *, p < 0.05.
We repeated the luciferase reporter assay in the presence of shPpm1a to determine whether it would also elicit a similar response in EKLF-dependent transcription. This was motivated by the fact that many of the documented functions of Ppm1b overlap significantly with Ppm1a and that Ppm1a and Ppm1b share a high degree of sequence similarity. We find that two shPpm1a constructs efficiently decrease the amount of endogenous shPpm1a, but neither construct leads to down-regulation of EKLF transcriptional activity at the β-globin locus (Fig. 5c). These assays show that Ppm1b plays a specific role in optimizing the activity of EKLF as a transcriptional activator.
Together, this set of experiments suggests that cellular Ppm1b levels are strongly correlated with EKLF transcriptional activity and possibly exert their effect by regulating EKLF stability. This may in turn affect the consequent availability of EKLF protein in the cell to activate transcription.
Reduction in Ppm1b Down-regulates EKLF Protein Levels in Vivo, Resulting in a Decrease of β-Globin Transcription
The experiments described above assayed the effect of Ppm1b on exogenous EKLF expressed in K562 or COS7 cells. We were therefore interested in elucidating the effect of shPpm1b on the expression of endogenous EKLF and the effect of decreased Ppm1b on the transcription of endogenous EKLF targets.
To carry out these experiments, we created cell lines that stably express shPpm1b and decrease levels of endogenous Ppm1b. Based on the erythroid expression profiles of selected human cell lines (42), we used the JK-1 erythroleukemia cell line, which was obtained from a patient diagnosed with chronic myelogenous leukemia at blast crisis (43). JK-1 expresses EKLF RNA and protein and expresses both β- and γ-globin (42), fitting the criteria necessary for carrying out the next stage of our study.
We transfected JK-1 cells with plasmids encoding scrambled RNA (scr), shPpm1b (sh1), and shPpm1b (sh2) and subjected transfected cells to puromycin selection. Small pools of puromycin-resistant cells were analyzed; we chose one pool from each for further analysis where Ppm1b was effectively knocked down (Fig. 6a).
FIGURE 6.
In vivo effect of decreasing Ppm1b. Pools of stably transfected human leukemic JK-1 cells that express EKLF were analyzed after shPpm1b (sh1 and sh2) or scrambled control (scr) transfection and selection with puromycin for 7 days. a, Ppm1b levels were monitored from two pools of cells from each selection (lanes 1 and 2). We selected pooled scr-1, sh1–1, and sh2–1 for further analysis. b, semi-quantitative RT-PCR was used to determine the effect of stable shPpm1b on EKLF, β-globin, and γ-globin RNA levels or on EKLF or Ppm1b protein levels; β-tubulin was used as a loading control. The decrease in Ppm1b correlates with a decrease in β-globin RNA, but there is no significant change in EKLF or γ-globin RNA expression. Expression of EKLF protein is significantly decreased in cells in which Ppm1b levels are decreased (relative to β-tubulin). c, stably transfected JK-1 cells expressing either scrambled (scr) or shPpm1b (sh1 or sh2) were cytospun and stained with May-Grunwald/Giemsa stain. Photographs were taken at the same magnification. Western blot and ratios are shown below. d, shPpm1b (sh1 and sh2)-transfected JK-1 cells were 2–2.5× the size of scr-transfected cells. Data are representative of the areas of 33 (sh1-transfected) or 34 (scr and sh2-transfected) cells; error bars are representative of S.E.
We analyzed expression of EKLF targets in these pools of cells using semi-quantitative RT-PCR. Consistent with the data in Fig. 5, β-globin expression is down-regulated in JK-1 cells expressing shPpm1b, suggesting that Ppm1b regulates the expression of EKLF target genes in vivo. EKLF and γ-globin RNA expression remain fairly constant in these cells, consistent with previous data suggesting that a decrease in Ppm1b specifically affects EKLF-dependent transcription at EKLF targets in erythroid cells (Fig. 6b). Western blotting of protein lysates from these cells indicates that EKLF protein levels are significantly decreased in cells that express decreased levels of Ppm1b, despite the fact that EKLF mRNA expression is not affected. The data strongly suggest that shPpm1b affects the transcription of EKLF targets by decreasing the stability, and thus the levels, of EKLF protein.
Because erythroid cell morphology is dysregulated in the absence of EKLF due to a loss in expression of erythroid membrane and cytoskeletal proteins (3, 44–46), we examined whether the decrease of EKLF protein in shPpm1b-expressing JK-1 cells results in a change of cell morphology. Fig. 6c shows that cells expressing shPpm1b are larger (quantified in Fig. 6d) and have ruffled cell membranes, consistent with the idea that the shPpm1b-mediated decrease in EKLF levels may alter the transcription of erythroid membrane and cytoskeletal protein targets, leading to phenotypic changes.
Ppm1b Superactivates EKLF Activity in a Phosphatase-dependent Manner
To this point, the data suggest that Ppm1b may modulate the expression of erythroid-specific genes by regulating the stability of EKLF protein or by regulating the phosphorylation status of Cdk9. Wang et al. (33) showed that Ppm1b was able to dephosphorylate p-Cdk9 in vitro when the inhibitory 7SK RNA was depleted from the P-TEFb complex. Their data raised the possibility that Ppm1b was a negative regulator of transcription, results consistent with our observations in differentiating CD34+ cells (Fig. 1). Yet surprisingly, decreasing Ppm1b levels led to a decrease in EKLF-dependent, but not basal, transcription activity at EKLF target promoters in K562 (Fig. 5) and JK-1 (Fig. 6) erythroleukemic cells, results more consistent with the idea that Ppm1b plays a role in Cdk9 activation (34) or that Ppm1b regulates transcription in committed erythroid cells by regulating the stability of EKLF (Figs. 4 and 6). In light of these data, we hypothesized that Ppm1b might behave as an activator of EKLF transcription when tested in a committed erythroid cell context.
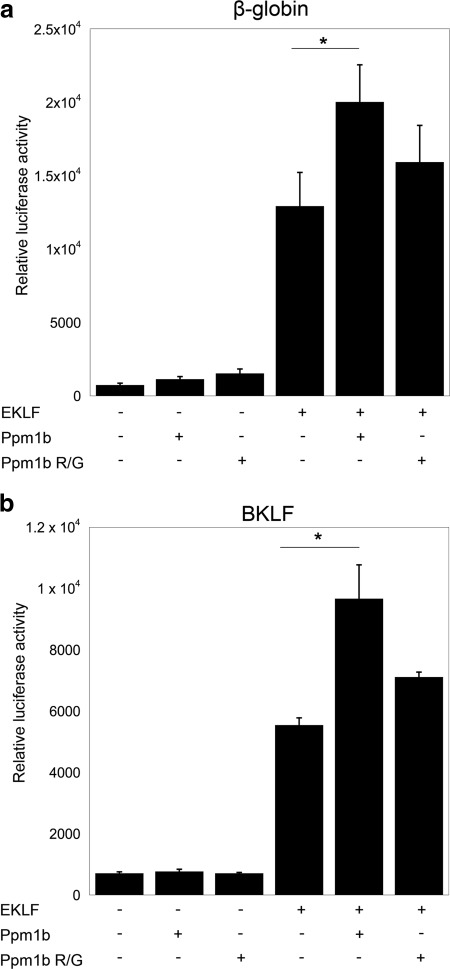
To test this idea, we examined the effect of exogenous Ppm1b on the ability of EKLF to activate transcription at the β-globin and Bklf promoters after co-transfection in K562 cells. Consistent with the data in Figs. 4c and 5, Ppm1b superactivates EKLF activity at the β-globin (Fig. 7a) and Bklf promoters (Fig. 7b). This effect is not observed when the phosphatase-inactive Ppm1b Arg/Gly mutant is co-expressed with EKLF. The data suggest that phosphatase activity of Ppm1b is required for it to regulate the transcriptional activity of EKLF, possibly via a mode that is distinct from the ability of Ppm1b to stabilize EKLF.
FIGURE 7.
Ppm1b superactivates EKLF. Activity of the β-globin (a) or BKLF (b) promoters was monitored after co-transfection of EKLF and Ppm1b or its phosphatase-inactive variant (R/G) into K562 cells as indicated. * indicates p < 0.05.
In summary, the data in Fig. 7 suggest that exogenous Ppm1b increases the transcriptional activity of EKLF at the β-globin and Bklf promoters, dependent on presence of the Ppm1b phosphatase activity. Additionally, we have found that overexpression of Ppm1b in K562 cells does not cause a global down-regulation of transcription (33); rather, we have demonstrated that the phosphatase activity of Ppm1b enhances the activity of a critical transcription factor in erythroid cells.
DISCUSSION
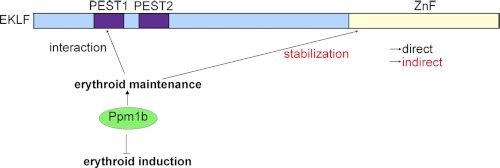
The experiments in this paper build on previous work that aimed to dissect modes of EKLF regulation via its interaction with other proteins. In this paper, we have described the identification of Ppm1b as a novel interactor that regulates the induction and maintenance of erythropoiesis via mechanisms that include regulation of EKLF stability and function (Fig. 8).
FIGURE 8.
Model for regulation of erythropoiesis by Ppm1b. Ppm1b functions as an inhibitor of gene transcription during the induction of the erythroid lineage, yet it plays a positive role in the maintenance of the erythroid lineage by functionally interacting with EKLF. Ppm1b interacts with the PEST1 sequence in EKLF (amino acids 50–70). Ppm1b also mediates EKLF stability via its zinc finger domain, thereby providing a second mechanism by which it can regulate EKLF transcription. It is likely that this mode of regulation is indirect, as Ppm1b does not interact with EKLF zinc finger domain.
Consistent with studies showing that Ppm1b negatively regulates Cdk9 function (33, 47), we have shown that depletion of Ppm1b augments the induction of β-globin expression when human hematopoietic stem cells are induced to differentiate to form erythroid progenitors. These data suggest that Ppm1b functions as a negative regulator of erythroid induction.
In contrast but consistent with other studies showing that Ppm1b activity is required for release of Cdk9 from its inhibitory complex (34), we have shown that Ppm1b can superactivate EKLF transcription activity at the BKLF and β-globin promoters in an erythroid cell line and that its phosphatase activity is required for this mode of EKLF regulation. It is unlikely that the ability of Ppm1b to superactivate EKLF activity is entirely due to its stabilizing effects on EKLF protein, because phosphatase-inactive Ppm1b, which still stabilizes EKLF, does not have a significant effect on EKLF activity.
Finally, we show that in nonerythroid cell lines and differentiated erythroid cells, Ppm1b indirectly regulates the stability of exogenous and endogenous EKLF via the EKLF zinc finger domain, thereby regulating the expression of EKLF target genes. Our work suggests that Ppm1b does not stabilize EKLF by masking its PEST sequences, which are implicated in regulation of protein stability (48), as both PEST sequences are intact in the EKLF ΔZn construct. On the contrary, Ppm1b stabilizes EKLFΔPEST1.
Regulation of β-Globin Induction by Ppm1b
Previous studies have demonstrated that depletion or inhibition of Cdk9 results in a specific defect in definitive hematopoiesis (35) and erythroid induction (36). This may be by virtue of the fact that Cdk9 associates in an erythroid-specific transcription complex with SCL, Ldb1, Runx1, and GATA1 (35, 36). In addition, Cdk9 localization to the murine β-globin promoter and coding region increases when MEL cells are induced to terminally differentiate (37). These data suggest that Cdk9 activation is involved in the induction and terminal differentiation of the erythroid lineage.
Ppm1b has been shown to dephosphorylate Cdk9 at Thr-186 (33). On the one hand, phosphorylation at this site is essential for the ability of the Cdk9-CycT complex to phosphorylate the C-terminal domain of RNA polymerase II (47). Ppm1b can therefore function as a negative regulator of transcriptional elongation. On the other hand, Cdk9 containing phosphorylated Thr-186 is mainly found in its inactive pTEFb complex, and this site is specifically dephosphorylated after treatment with hexamethylene bisacetamide, leading to dissociation from its inhibitory complex with HEXIM-7SK (49, 50). In this context, Ppm1b would play a positive role in releasing active Cdk9 (34).
We used siRNA to knock down Ppm1b in CD34+ HSCs, prior to erythroid differentiation with Epo/SCF. In the Ppm1b-depleted samples, we observed a marked increase in β-globin RNA levels upon erythroid induction. Our data provide the first evidence that Ppm1b selectively regulates the transcription of β-globin during erythroid induction, an example of a late erythroid gene, without affecting the transcription of EKLF, which is induced earlier in differentiation. Our data suggest that Ppm1b plays a negative role in the regulation of β-globin transcription during erythroid induction.
Regulation of EKLF Protein Stability by Ppm1b
In contrast to the role of Ppm1b in erythroid induction, Ppm1b plays a positive role in maintenance of the erythroid lineage by regulating the stability of EKLF protein.
Ppm1b, in concert with its regulatory subunit, GAS-41, has recently been shown to regulate p53 stability by specifically facilitating the dephosphorylation of p53 at Ser-366 (51). Our work introduces a second example of Ppm1b-mediated protein stability. In addition to showing that a drop in Ppm1b levels can decrease EKLF stability, we show that shPpm1b can decrease transcription levels of EKLF targets in a promoter-reporter system, as well as endogenous sites in vivo, potentially affecting erythroid differentiation. Furthermore, our work shows that down-regulation of EKLF-dependent transcription at exogenous promoters is specific to a decrease in Ppm1b and not in Ppm1a. The mechanism(s) by which Ppm1b regulates EKLF stability is yet unknown. Prior to the work of Park et al. (51), the PP2C family of protein phosphatases was thought to function as a monomer (23). The discovery that GAS-41 was a required cofactor in the dephosphorylation of p53 Ser-366 by Ppm1b proved to be the first instance in which a PP2C family member was regulated by an interacting protein. As Ppm1b stabilizes EKLF via a domain with which it does not directly interact, it is likely that additional cofactors are required for Ppm1b to stabilize EKLF via its zinc finger domain.
Using an shRNA approach, we show that Ppm1b regulates EKLF stability, thereby regulating the transcription of β-globin in vivo. Dysregulation of erythropoiesis caused by the knockdown of Ppm1b is also evident by the ruffled morphology and larger size of the shPpm1b-transfected JK-1 cells, reminiscent of circulating blood cells in E13.5 EKLF−/− murine embryos (44), as well as primitive erythroid cells from obtained from E11.5 (45) and E12.5 (46) EKLF−/− embryos. There is a large body of work that describes how EKLF is involved in various processes that govern the development and function of erythroid cells (1, 52), and we would expect that a decrease in EKLF stability would also lead to a decrease in the transcription of all or most EKLF targets. It will interesting to test if the stable shPpm1b-transfected JK-1 cells also recapitulate the increased transcription of megakaryocytic specific genes GpIX and Fli-1 that was observed in MEL cells expressing shEKLF (53).
A decrease in Ppm1b levels is associated with a decrease in EKLF activity, and the extent of Ppm1b knockdown correlates with the extent of decrease in EKLF activity as compared with EKLF activity in scrambled controls, where even knockdown of Ppm1b to 73% of its levels in scr-transfected cells is sufficient to cause a decrease in EKLF activity. This is consistent with the idea that Ppm1b regulates EKLF stability via the structural elements in the Ppm1b protein. Under physiological conditions, enzyme active sites are not typically saturated with substrate (54), and a relatively small drop in enzyme levels is not likely to cause a discernible effect in substrate processing. It is unlikely that a decrease of Ppm1b to 73% of its original levels would cause a detectable effect if Ppm1b regulated EKLF stability solely via its enzymatic activity. This analysis is highlighted by our experiments examining the effect of shPpm1a on EKLF activity. Unlike the effect of Ppm1b, knockdown of Ppm1a to 24% of scr levels did not cause any discernible effect in the activity of EKLF at the β-globin promoter.
Regulation of EKLF Transcriptional Activity by Ppm1b
Ouyang et al. (5) have shown that EKLF is phosphorylated on serines and threonines in MEL cells. In vitro kinase assays suggested that upon differentiation induction by HMBA, some kinases in MEL cells undergo a decrease in their ability to phosphorylate His-EKLF. In addition, the level of CKII/EKLF interaction decreases dramatically in lysates derived from HMBA-treated MEL cells (5). These data suggest that HMBA-induced differentiation of MEL cells is associated with decreased kinase activity or possibly increased phosphatase activity. The article in question focused on the structural and functional interaction between CKII and EKLF, but it was not clear if the decreased interaction between EKLF and CKII, and the decrease in kinase activity of CKII upon HMBA induction, was due to a decrease in CKII protein levels or other factors. Although the authors presented data showing that GAL/EKLF (amino acids 20–120) fusion, containing a CKII target site (Thr-41), could be superactivated by CKII, unpublished data3 suggest that phosphorylation of this site has an inhibitory effect on full-length unfused EKLF. An additional consideration is that the minimal activation domain of EKLF is proline-rich and has many serines and threonines that are potential candidates for phosphorylation, either individually or in combination. This suggests that EKLF may undergo phosphorylation reactions that selectively inhibit DNA binding at some promoters. It therefore follows that such inhibition could potentially be relieved by an increase in cellular phosphatase activity.
Ppm1b and Ppm1a have both been shown to be potential phosphatases of Cdk9 at Thr-186, which is part of the P-TEFb kinase complex that enhances the processivity of RNA polymerase II by phosphorylation of its C-terminal domain (33). Ppm1a and -1b may be functionally redundant in mediating the decrease in RNA polymerase II phosphorylation, potentially acting as global repressors of transcription.
Our paper, which examines the role of Ppm1b in erythroid cells, suggests that the role of Ppm1b in transcription regulation is more nuanced. In contrast to the predictions of the study by Wang et al. (33), overexpression of Ppm1b significantly increases EKLF activation at the EKLF and BKLF promoters, implying that Ppm1b functions as a transcriptional activator in committed erythroid cells.
Our work is one of the first studies to suggest that Ppm1a and -1b are not functionally redundant, at least with respect to regulation of the transcription ability of EKLF. Interestingly, shPpm1a (sh2), which knocks down Ppm1a levels to 9% of scr-transfected Ppm1a levels, causes a statistically significant increase in EKLF activation at the β-globin promoter. This is reminiscent of the mode of regulation suggested by Wang et al. (33) in which the silencing of Ppm1a increases the levels of phosphorylated Cdk9 2.5-fold, potentially increasing global levels of phosphorylated RNA polymerase II and increasing overall transcription. The specifics of how Ppm1a may regulate EKLF function is a fascinating question that has yet to be explored. It is likely that p-Cdk9, EKLF, and RNA polymerase II chromatin immunoprecipitations on erythroid promoters in cells that are stably transfected with scr or shPpm1a will shed light on this question.
Model of the Role of Ppm1b in Erythroid Development
In this study, we have demonstrated that Ppm1b plays different roles during the processes of erythroid induction and maintenance. After depleting Ppm1b in primary CD34+ cells prior to erythroid differentiation with Epo/SCF, there is an increase in β-globin RNA expression after differentiation. The data suggest that prior to differentiation, Ppm1b may play a role in altering the potential or kinetics of CD34+ cell differentiation along the erythroid lineage in response to cytokines. Although the induction of EKLF was not affected by Ppm1b depletion, the lack of Ppm1b in differentiating hematopoietic cells could have augmented the ability of EKLF to activate transcription of β-globin.
In contrast, Ppm1b acts as an activator of EKLF targets in committed erythroid cells. In committed erythroid cells, EKLF and Ppm1b interact physically and functionally at steady state. Depletion of Ppm1b from committed erythroid cells results in a decrease in EKLF stability, leading to a decrease in erythroid-specific transcription. In addition, its phosphatase activity is able to up-regulate EKLF transactivation function.
Although the existence of paradoxical roles in Ppm1b in erythroid development seems surprising, there is increasing evidence that EKLF itself plays various roles in erythroid populations at different stages of development (2, 4, 55). It is highly probable that these roles are due to different protein/protein interactions and post-translational modifications that take place over the course of development. In this study, we demonstrate that the functional interaction of Ppm1b with EKLF is dependent on the differentiation stage of hematopoietic cells. We also provide yet another example of a cellular factor that may modify EKLF function in a differentiation stage-specific manner.
Supplementary Material
Acknowledgments
We thank Georgia Dolios and Rong Wang at the MSSM mass spectrometry facility for assistance with obtaining mass spectrometry data; Jianhua Yang (Baylor College of Medicine) for the Ppm1b, Ppm1b R/G, scr, shPpm1a, and shPpm1b plasmids; Roxana Mesias and Deanna Benson for the use of their laboratory's nucleofector and technical advice on nucleofection; and Xiajun (John) Li and Scott Friedman for discussions that contributed to this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant DK46865 from USPHS. This work was also supported by NYSTEM Contract C026435 (to J. J. B.).

This article contains supplemental Figs. S1 and S2 and Tables S1–S3.
M. Siatecka and J. J. Bieker, unpublished observations.
- EKLF
- erythroid Krüppel-like factor
- HSC
- hematopoietic stem cell
- scr
- scrambled
- Epo
- erythropoietin
- SCF
- stem cell factor.
REFERENCES
- 1. Siatecka M., Bieker J. J. (2011) The multifunctional role of EKLF/KLF1 during erythropoiesis. Blood 118, 2044–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pilon A. M., Ajay S. S., Kumar S. A., Steiner L. A., Cherukuri P. F., Wincovitch S., Anderson S. M., NISC Comparative Sequencing Center, Mullikin J. C., Gallagher P. G., Hardison R. C., Margulies E. H., Bodine D. M. (2011) Genome-wide ChIP-Seq reveals a dramatic shift in the binding of the transcription factor erythroid Kruppel-like factor during erythrocyte differentiation. Blood 118, e139–e148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tallack M. R., Whitington T., Yuen W. S., Wainwright E. N., Keys J. R., Gardiner B. B., Nourbakhsh E., Cloonan N., Grimmond S. M., Bailey T. L., Perkins A. C. (2010) A global role for KLF1 in erythropoiesis revealed by ChIP-seq in primary erythroid cells. Genome Res. 20, 1052–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen X., Bieker J. J. (2004) Stage-specific repression by the EKLF transcriptional activator. Mol. Cell. Biol. 24, 10416–10424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ouyang L., Chen X., Bieker J. J. (1998) Regulation of erythroid Krüppel-like factor (EKLF) transcriptional activity by phosphorylation of a protein kinase casein kinase II site within its interaction domain. J. Biol. Chem. 273, 23019–23025 [DOI] [PubMed] [Google Scholar]
- 6. Perrine S. P., Mankidy R., Boosalis M. S., Bieker J. J., Faller D. V. (2009) Erythroid Kruppel-like factor (EKLF) is recruited to the γ-globin gene promoter as a co-activator and is required for γ-globin gene induction by short chain fatty acid derivatives. Eur. J. Haematol. 82, 466–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Siatecka M., Xue L., Bieker J. J. (2007) Sumoylation of EKLF promotes transcriptional repression and is involved in inhibition of megakaryopoiesis. Mol. Cell. Biol. 27, 8547–8560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sengupta T., Chen K., Milot E., Bieker J. J. (2008) Acetylation of EKLF is essential for epigenetic modification and transcriptional activation of the β-globin locus. Mol. Cell. Biol. 28, 6160–6170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sengupta T., Cohet N., Morlé F., Bieker J. J. (2009) Distinct modes of gene regulation by a cell-specific transcriptional activator. Proc. Natl. Acad. Sci. U.S.A. 106, 4213–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang W., Bieker J. J. (1998) Acetylation and modulation of erythroid Krüppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc. Natl. Acad. Sci. U.S.A. 95, 9855–9860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang W., Kadam S., Emerson B. M., Bieker J. J. (2001) Site-specific acetylation by p300 or CREB-binding protein regulates erythroid Krüppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol. Cell. Biol. 21, 2413–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Quadrini K. J., Bieker J. J. (2002) Krüppel-like zinc fingers bind to nuclear import proteins and are required for efficient nuclear localization of erythroid Krüppel-like factor. J. Biol. Chem. 277, 32243–32252 [DOI] [PubMed] [Google Scholar]
- 13. Pandya K., Townes T. M. (2002) Basic residues within the Kruppel zinc finger DNA binding domains are the critical nuclear localization determinants of EKLF/KLF-1. J. Biol. Chem. 277, 16304–16312 [DOI] [PubMed] [Google Scholar]
- 14. Shyu Y. C., Lee T. L., Wen S. C., Chen H., Hsiao W. Y., Chen X., Hwang J., Shen C. K. (2007) Subcellular transport of EKLF and switch-on of murine adult β maj globin gene transcription. Mol. Cell. Biol. 27, 2309–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Quadrini K. J., Gruzglin E., Bieker J. J. (2008) Nonrandom subcellular distribution of variant EKLF in erythroid cells. Exp. Cell Res. 314, 1595–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Quadrini K. J., Bieker J. J. (2006) EKLF/KLF1 is ubiquitinated in vivo, and its stability is regulated by activation domain sequences through the 26 S proteasome. FEBS Lett. 580, 2285–2293 [DOI] [PubMed] [Google Scholar]
- 17. Pilon A. M., Arcasoy M. O., Dressman H. K., Vayda S. E., Maksimova Y. D., Sangerman J. I., Gallagher P. G., Bodine D. M. (2008) Failure of terminal erythroid differentiation in EKLF-deficient mice is associated with cell cycle perturbation and reduced expression of E2F2. Mol. Cell. Biol. 28, 7394–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tallack M. R., Keys J. R., Humbert P. O., Perkins A. C. (2009) EKLF/KLF1 controls cell cycle entry via direct regulation of E2f2. J. Biol. Chem. 284, 20966–20974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tallack M. R., Keys J. R., Perkins A. C. (2007) Erythroid Kruppel-like factor regulates the G1 cyclin-dependent kinase inhibitor p18INK4c. J. Mol. Biol. 369, 313–321 [DOI] [PubMed] [Google Scholar]
- 20. Siatecka M., Lohmann F., Bao S., Bieker J. J. (2010) EKLF directly activates the p21WAF1/CIP1 gene by proximal promoter and novel intronic regulatory regions during erythroid differentiation. Mol. Cell. Biol. 30, 2811–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lohmann F., Bieker J. J. (2008) Activation of Eklf expression during hematopoiesis by Gata2 and Smad5 prior to erythroid commitment. Development 135, 2071–2082 [DOI] [PubMed] [Google Scholar]
- 22. Frontelo P., Manwani D., Galdass M., Karsunky H., Lohmann F., Gallagher P. G., Bieker J. J. (2007) Novel role for EKLF in megakaryocyte lineage commitment. Blood 110, 3871–3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shi Y. (2009) Serine/threonine phosphatases. Mechanism through structure. Cell 139, 468–484 [DOI] [PubMed] [Google Scholar]
- 24. Woo A. J., Moran T. B., Schindler Y. L., Choe S. K., Langer N. B., Sullivan M. R., Fujiwara Y., Paw B. H., Cantor A. B. (2008) Identification of ZBP-89 as a novel GATA1-associated transcription factor involved in megakaryocytic and erythroid development. Mol. Cell. Biol. 28, 2675–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blau C. A., Barbas C. F., 3rd, Bomhoff A. L., Neades R., Yan J., Navas P. A., Peterson K. R. (2005) γ-Globin gene expression in chemical inducer of dimerization (CID)-dependent multipotential cells established from human β-globin locus yeast artificial chromosome (β-YAC) transgenic mice. J. Biol. Chem. 280, 36642–36647 [DOI] [PubMed] [Google Scholar]
- 26. Narla G., Difeo A., Reeves H. L., Schaid D. J., Hirshfeld J., Hod E., Katz A., Isaacs W. B., Hebbring S., Komiya A., McDonnell S. K., Wiley K. E., Jacobsen S. J., Isaacs S. D., Walsh P. C., Zheng S. L., Chang B. L., Friedrichsen D. M., Stanford J. L., Ostrander E. A., Chinnaiyan A. M., Rubin M. A., Xu J., Thibodeau S. N., Friedman S. L., Martignetti J. A. (2005) A germ line DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res. 65, 1213–1222 [DOI] [PubMed] [Google Scholar]
- 27. Wu M. H., Smith S. L., Danet G. H., Lin A. M., Williams S. F., Liebowitz D. N., Dolan M. E. (2001) Optimization of culture conditions to enhance transfection of human CD34+ cells by electroporation. Bone Marrow Transplant. 27, 1201–1209 [DOI] [PubMed] [Google Scholar]
- 28. Bianchi E., Zini R., Salati S., Tenedini E., Norfo R., Tagliafico E., Manfredini R., Ferrari S. (2010) c-Myb supports erythropoiesis through the transactivation of KLF1 and LMO2 expression. Blood 116, e99–e110 [DOI] [PubMed] [Google Scholar]
- 29. Chen J., Peterson K. R., Iancu-Rubin C., Bieker J. J. (2010) Design of embedded chimeric peptide nucleic acids that efficiently enter and accurately reactivate gene expression in vivo. Proc. Natl. Acad. Sci. U.S.A. 107, 16846–16851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shoji W., Yamamoto T., Obinata M. (1994) The helix-loop-helix protein Id inhibits differentiation of murine erythroleukemia cells. J. Biol. Chem. 269, 5078–5084 [PubMed] [Google Scholar]
- 31. Düwel M., Ungewickell E. J. (2006) Clathrin-dependent association of CVAK104 with endosomes and the trans-Golgi network. Mol. Biol. Cell 17, 4513–4525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Newmyer S. L., Christensen A., Sever S. (2003) Auxilin-dynamin interactions link the uncoating ATPase chaperone machinery with vesicle formation. Dev. Cell 4, 929–940 [DOI] [PubMed] [Google Scholar]
- 33. Wang Y., Dow E. C., Liang Y. Y., Ramakrishnan R., Liu H., Sung T. L., Lin X., Rice A. P. (2008) Phosphatase PPM1A regulates phosphorylation of Thr-186 in the Cdk9 T-loop. J. Biol. Chem. 283, 33578–33584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cho S., Schroeder S., Ott M. (2010) CYCLINg through transcription. Post-translational modifications of P-TEFb regulate transcription elongation. Cell Cycle 9, 1697–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meier N., Krpic S., Rodriguez P., Strouboulis J., Monti M., Krijgsveld J., Gering M., Patient R., Hostert A., Grosveld F. (2006) Novel binding partners of Ldb1 are required for hematopoietic development. Development 133, 4913–4923 [DOI] [PubMed] [Google Scholar]
- 36. Bai X., Kim J., Yang Z., Jurynec M. J., Akie T. E., Lee J., LeBlanc J., Sessa A., Jiang H., DiBiase A., Zhou Y., Grunwald D. J., Lin S., Cantor A. B., Orkin S. H., Zon L. I. (2010) TIF1γ controls erythroid cell fate by regulating transcription elongation. Cell 142, 133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Song S. H., Kim A., Ragoczy T., Bender M. A., Groudine M., Dean A. (2010) Multiple functions of Ldb1 required for β-globin activation during erythroid differentiation. Blood 116, 2356–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen X., Bieker J. J. (1996) Erythroid Krüppel-like factor (EKLF) contains a multifunctional transcriptional activation domain important for inter- and intramolecular interactions. EMBO J. 15, 5888–5896 [PMC free article] [PubMed] [Google Scholar]
- 39. Mas C., Lussier-Price M., Soni S., Morse T., Arseneault G., Di Lello P., Lafrance-Vanasse J., Bieker J. J., Omichinski J. G. (2011) Structural and functional characterization of an atypical activation domain in erythroid Kruppel-like factor (EKLF). Proc. Natl. Acad. Sci. U.S.A. 108, 10484–10489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sun W., Yu Y., Dotti G., Shen T., Tan X., Savoldo B., Pass A. K., Chu M., Zhang D., Lu X., Fu S., Lin X., Yang J. (2009) PPM1A and PPM1B act as IKKβ phosphatases to terminate TNFα-induced IKKβ-NF-κB activation. Cell. Signal. 21, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Donze D., Townes T. M., Bieker J. J. (1995) Role of erythroid Kruppel-like factor in human γ- to β-globin gene switching. J. Biol. Chem. 270, 1955–1959 [DOI] [PubMed] [Google Scholar]
- 42. Bieker J. J. (1996) Isolation, genomic structure, and expression of human erythroid Krüppel-like factor (EKLF). DNA Cell Biol. 15, 347–352 [DOI] [PubMed] [Google Scholar]
- 43. Drexler H. G., Matsuo Y., MacLeod R. A. (2004) Malignant hematopoietic cell lines. In vitro models for the study of erythroleukemia. Leuk. Res. 28, 1243–1251 [DOI] [PubMed] [Google Scholar]
- 44. Nilson D. G., Sabatino D. E., Bodine D. M., Gallagher P. G. (2006) Major erythrocyte membrane protein genes in EKLF-deficient mice. Exp. Hematol. 34, 705–712 [DOI] [PubMed] [Google Scholar]
- 45. Hodge D., Coghill E., Keys J., Maguire T., Hartmann B., McDowall A., Weiss M., Grimmond S., Perkins A. (2006) A global role for EKLF in definitive and primitive erythropoiesis. Blood 107, 3359–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Drissen R., von Lindern M., Kolbus A., Driegen S., Steinlein P., Beug H., Grosveld F., Philipsen S. (2005) The erythroid phenotype of EKLF-null mice. Defects in hemoglobin metabolism and membrane stability. Mol. Cell. Biol. 25, 5205–5214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Baumli S., Lolli G., Lowe E. D., Troiani S., Rusconi L., Bullock A. N., Debreczeni J. E., Knapp S., Johnson L. N. (2008) The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J. 27, 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rechsteiner M., Rogers S. W. (1996) PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21, 267–271 [PubMed] [Google Scholar]
- 49. Chen R., Yang Z., Zhou Q. (2004) Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J. Biol. Chem. 279, 4153–4160 [DOI] [PubMed] [Google Scholar]
- 50. Chen R., Liu M., Li H., Xue Y., Ramey W. N., He N., Ai N., Luo H., Zhu Y., Zhou N., Zhou Q. (2008) PP2B and PP1α cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 22, 1356–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Park J. H., Smith R. J., Shieh S. Y., Roeder R. G. (2011) The GAS41-PP2Cβ complex dephosphorylates p53 at serine 366 and regulates its stability. J. Biol. Chem. 286, 10911–10917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tallack M. R., Perkins A. C. (2010) KLF1 directly coordinates almost all aspects of terminal erythroid differentiation. IUBMB Life 62, 886–890 [DOI] [PubMed] [Google Scholar]
- 53. Bouilloux F., Juban G., Cohet N., Buet D., Guyot B., Vainchenker W., Louache F., Morlé F. (2008) EKLF restricts megakaryocytic differentiation at the benefit of erythrocytic differentiation. Blood 112, 576–584 [DOI] [PubMed] [Google Scholar]
- 54. Berg J. M., Tymoczko J. L., Stryer L. (2002) Biochemistry, W. H. Freeman & Co., New York [Google Scholar]
- 55. Stadhouders R., Thongjuea S., Andrieu-Soler C., Palstra R. J., Bryne J. C., van den Heuvel A., Stevens M., de Boer E., Kockx C., van der Sloot A., van den Hout M., van Ijcken W., Eick D., Lenhard B., Grosveld F., Soler E. (2012) Dynamic long range chromatin interactions control Myb proto-oncogene transcription during erythroid development. EMBO J. 31, 986–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.