Background: Vinculin plays profound roles in cell adhesion and migration.
Results: Trapping or knockdown of the vinculin gene in chondrocytes leads to impaired differentiation by affecting multiple signaling pathways.
Conclusion: Vinculin has pleiotropic roles in chondrocytic differentiation.
Significance: This study reveals a novel, tissue-specific function of vinculin in chondrocytes.
Keywords: Cell Biology, Chondrogenesis, Extracellular Matrix, Insulin-like Growth Factor (IGF), Signal Transduction, Gene Trap, Vinculin
Abstract
To identify the genes involved in chondrocytic differentiation, we applied gene trap mutagenesis to a murine mesenchymal chondrogenic cell line ATDC5 and isolated a clone in which the gene encoding vinculin was trapped. The trapped allele was assumed to express a fusion protein containing a truncated vinculin lacking the tail domain and the geo product derived from the trap vector. The truncated vinculin was suggested to exert a dominant negative effect. Impaired functioning of vinculin caused by gene trapping in ATDC5 cells or knockdown in primary chondrocytes resulted in the reduced expression of chondrocyte-specific genes, including Col2a1, aggrecan, and Col10a1. The expression of Runx2 also was suppressed by the dysfunctional vinculin. On the other hand, the expression of Sox9, encoding a key transcription factor for chondrogenesis, was retained. Knockdown of vinculin in metatarsal organ cultures impaired the growth of the explants and reduced the expression of Col2a1 and aggrecan. Gene trapping or knockdown of vinculin decreased the phosphorylation of ERK1/2 but increased that of Src homology 2 domain-containing tyrosine phosphatase 2 (SHP2) and Akt during chondrocytic differentiation, suggesting a disturbance of signaling by insulin-like growth factor I (IGF-I). Knockdown of vinculin in the metatarsal organ culture abrogated the IGF-I-induced growth and inhibited the up-regulation of Col2a1 and aggrecan expression by IGF-I. Loss of vinculin function in differentiating chondrocytes impaired the activation of the p38 MAPK pathway also, suggesting its involvement in the regulation of chondrogenesis by vinculin. Our results indicate a tissue-specific function of vinculin in cartilage whereby it controls chondrocytic differentiation.
Introduction
Endochondral bone formation consists of the mesenchymal condensation of undifferentiated cells, the proliferation of chondrocytes, and differentiation into hypertrophic chondrocytes, followed by mineralization. Proliferating chondrocytes express type II, IX, and XI collagen and proteoglycans, such as aggrecan, whereas hypertrophic chondrocytes are characterized by a high level of alkaline phosphatase and the production of type X collagen. Various transcription factors and signaling molecules, including Sox9/5/6, Runx2, Indian hedgehog (Ihh), parathyroid hormone-related protein, insulin-like growth factors (IGFs), and fibroblast growth factors (FGFs), have been revealed to regulate the maturation of chondrocytes (1, 2). However, the molecular mechanisms underlying chondrogenesis have not been fully elucidated.
Gene trapping is a genome-wide approach used to clarify the roles of genes by the random insertion of a trapping vector that might disturb gene function (3). A gene trap vector consists of a promoterless reporter gene, a selectable marker, and a splice acceptor site immediately upstream of the reporter gene. When the vector is appropriately inserted in an endogenous gene, a fusion transcript, including the upstream coding sequence of the gene and the reporter gene, is generated by the promoter and enhancer of the trapped gene. This event is associated with the conversion of the trapped gene into a mutated gene and allows one to examine its expression via the reporter activity.
To identify the factors involved in chondrogenesis, we used a gene trap approach in the murine mesenchymal cell line ATDC5, an established cell model of endochondral bone formation (4). ATDC5 cells differentiate into proliferating chondrocytes and undergo cellular hypertrophy and mineralization. By applying gene trapping in ATDC5 cells, we have previously identified several factors involved in chondrocytic differentiation, including the p85a subunit of phosphatidylinositide 3-kinase (PI3K) and nuclear factor I transcription/replication factor (5, 6).
Vinculin is a 116-kDa cytoskeletal protein associated with focal adhesion and adherens junctions. It exists in multimolecular complexes, which function in adhesion and/or signaling between the extracellular milieu and the cell, via integrins and cadherins. The amino-terminal head domain of vinculin binds to talin (7), which in turn binds to β integrins, whereas the carboxyl-terminal tail domain binds to actin (8), phospholipids (9), and paxillin (10). This arrangement allows vinculin to function as a regulatory bridge between the extracellular matrix (ECM)2 and the actin cytoskeleton. Disruption of vinculin expression in mice results in embryonic death with severe defects in cardiac and brain development (11, 12). Cells depleted of vinculin have a reduced ability to adhere to a variety of ECM proteins, increased migration rates, and fewer and smaller adhesion sites compared with wild-type cells (12–14). In addition to the profound roles of vinculin in cell adhesion and motility, there might be some tissue-specific functions, which remain to be elucidated.
By gene trapping in chondrocytic ATDC5 cells, we isolated a clone in which the gene encoding vinculin was trapped. The clone expressed a truncated vinculin lacking the tail domain, which exerted a dominant effect. The clone's ability to differentiate into mature chondrocytes was impaired, leading us to hypothesize that vinculin plays a role in chondrocyte differentiation. Here we provide evidence for chondrocyte-specific roles of vinculin and describe possible molecular mechanisms by which vinculin regulates chondrocytic differentiation.
EXPERIMENTAL PROCEDURES
Cell Culture
ATDC5 cells and the trap clones were maintained in a 1:1 mixture of Dulbecco's modified Eagle's and Ham's F-12 (DMEM/F-12) medium (Sigma-Aldrich) supplemented with 5% fetal bovine serum (FBS) (Invitrogen) and 1% insulin-transferrin-selenium G supplement (ITS) (Invitrogen) at 37 °C in a 5% CO2 atmosphere. For chondrogenic induction, cells were inoculated into 6-well culture plates (5 × 104 cells/well). Three days later, the medium was changed to α-minimal essential medium supplemented with 5% FBS and ITS, and the culture plates were sealed to facilitate mineralization, as described previously (5, 6). The medium was replaced every 3 days.
COS7 cells were cultured in DMEM supplemented with 10% FBS. For transient transfections, FuGene6 reagent (Roche Applied Science) was utilized.
Isolation of Primary Chondrocytes
Animal protocols were approved by the Institutional Animal Care and Use Committee. Primary chondrocytes were isolated from the ribcages of 5-day-old ICR mice following a previous report (15). In brief, the cartilage dissected from ribcages was incubated with actinase E (2 mg/ml in PBS; Kaken Pharmaceutical Co. Ltd., Tokyo, Japan) for 30 min at 37 °C, rinsed three times with PBS, treated with collagenase (3 mg/ml, Wako, Osaka, Japan) in 10 ml of DMEM for 90 min at 37 °C, and then transferred to a 50-ml tube. After pipetting, the tube was stood for 5 min, and the supernatant containing soft tissue was discarded. The remaining cell clumps were washed and passed through a 100-μm cell strainer. The collected chondrocytes were cultured in DMEM supplemented with 10% FBS and 50 μg/ml ascorbic acid (Sigma-Aldrich).
Organ Culture of Mouse Metatarsals
The organ culture of mouse metatarsals was performed following a previous report (16). The central metatarsal rudiments were dissected from each hind limb of E15.5 ICR mouse embryos, placed into 48-well plates (designated day 0), and cultured in α-minimal essential medium containing 50 μg/ml ascorbic acid, 1 mm β-glycerophosphate, 0.2% bovine serum albumin, and 0.1% penicillin/streptomycin at 37 °C under a 5% CO2 atmosphere. The medium was changed on day 2 and day 4. When viral transfection was performed, the viruses were added to the medium on day 0 and when the medium was changed.
Gene Trapping
As the trap vector, we used pPT1-geo, which lacks its own promoter and enhancer but contains a lacZ gene as a reporter fused to a neomycin resistance gene as a selection marker, which was designated geo (17). After pPT1-geo was introduced into ATDC5 cells using the Gene Pulser II electroporation system (Bio-Rad), neomycin-resistant clones were selected and screened for β-galactosidase activity. Clones with a 10-fold higher level of β-galactosidase activity than the parental ATDC5 cells were then subjected to chondrogenic induction, followed by Alcian blue and Alizarin red staining to evaluate the production and mineralization of extracellular matrices, respectively.
Cell Staining
The cells were fixed with 95% ethanol and stained with 1% Alizarin red S (Sigma-Aldrich), Alcian blue stain solution, pH 2.5 (Nacalai Tesque, Kyoto, Japan), or 0.1% crystal violet solution (Kanto Chemical, Tokyo, Japan). Staining for β-galactosidase activity was performed using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) (Wako) as a substrate.
Southern Blot Analysis
Genomic DNA was extracted from parental ATDC5 cells and the trap clone and digested with the restriction enzyme SphI or PstI. The digested DNA was then electrophoresed, transferred to a Hybond-N+ membrane (Amersham Biosciences), and probed with a radiolabeled fragment of lacZ cDNA prepared by digestion of pPT1-geo with EcoRI/SacI. The restriction enzymes were purchased from New England Biolabs (Beverly, MA).
Identification of Trapped Genes by 5′-Rapid Amplification of cDNA Ends (RACE)
Total RNA was extracted from the trap clone with the RNeasy kit (Qiagen Inc., Valencia, CA), and messenger RNA was purified with oligo(dT) latex (OligotexTM-dT30 Super mRNA Purification Kit; Takara Biomedicals, Shiga, Japan). To identify the trapped gene, 5′-RACE was performed utilizing the 5′-RACE System for Rapid Amplification of cDNA Ends (Invitrogen), according to the manufacturer's instructions with some modifications. Briefly, first-strand cDNA was synthesized from mRNA (1 μg) using SuperScript II reverse transcriptase (Invitrogen) with a primer specific to lacZ cDNA in pPT1-geo: LacZ-GSP1, 5′-TGGCGAAAGGGGGATGTG-3′. After the first strand of cDNA was synthesized, the original mRNA template was removed by treatment with RNase, and the unincorporated dNTPs and the primer were separated from the cDNA using a GlassMAX Spin Cartridge (Invitrogen). Then a homopolymeric tail was added to the 3′-end of the cDNA using TdT and dCTP. This was followed by PCR amplification using Taq polymerase (Takara) and the following set of primers: 5′-RACE Abridged Anchor Primer, 5′-GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG-3′ (where I represents inosine); LacZ-GSP2, 5′-ATGTGCTGCAAGGCGATTAAGTTG-3′. The product served as the template for a second round of PCR using the primers LacZ-GSP3 (5′-CCAGGGTTTTCCCAGTC-3′) and 5′RACE-AUAP (5′-GGCCACGCGTCGACTAGTAC-3′). The product of the second PCR was then cloned into the vector pT7-Blue (Novagen, Madison, WI) and sequenced using an automated sequencer (model 377A; PerkinElmer Life Sciences).
Assay for Proliferation
The cells were plated onto 96-well culture plates at a density of 1 × 103 cells/well (designated as day 0) and cultured in DMEM/F-12 medium supplemented with 5% FBS and ITS. Then the cell number in each well was evaluated by a 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt assay performed using a CellTiter 96® Aqueous One solution cell proliferation assay kit (Promega, Madison, WI) according to the manufacturer's instructions. Proliferation of the cells was also assayed using a Calbiochem BrdU cell proliferation assay kit (Merck).
Reverse Transcription-Polymerase Chain Reaction (RT-PCR) and Real-time PCR
Total RNA (2.5 μg) treated with DNase (Qiagen) was reverse transcribed using random hexamer (Promega) and SuperScript II (Invitrogen). PCR was performed using Taq polymerase (Takara) and the specific primer sets summarized in supplemental Table S1. Amplification of the expected fragments was confirmed by sequencing of the products. For real-time PCR, we utilized TaqMan® gene expression assays with the 7300 real-time PCR system (Applied Biosystems). To generate a standard curve for real-time PCR, the amplicons of interest were first cloned into the pT7-Blue vector (Novagen), and serial 10-fold dilutions of the constructed plasmid were included in the assay. Samples were analyzed in triplicate. The copy number of the target cDNA in each sample was estimated by referring to the standard curve, which was standardized by that of Gapdh in each sample.
Plasmids and Recombinant Adenoviruses
The plasmid carrying a full-length cDNA for mouse vinculin (accession number AK077850) was obtained as the RIKEN® PHANTOMTM clone pFLC1-vinculin (clone ID 5930419L09). The fragment excised from pFLC1-vinculin by digestion with SfiI was inserted into pcDNA3.1-Zeo (Invitrogen), yielding pcDNA-vinculin(WT). We then constructed pcDNA-vinculin(WT)-V5 and pcDNA-vinculin(WT)-Myc encoding the carboxyl-terminal V5- and Myc-tagged forms of wild-type vinculin, respectively, by mutating the stop codon to introduce a new ClaI site using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), where annealed oligomers corresponding to the tags were inserted. The plasmid encoding the V5-tagged truncated mutant of vinculin (aa 1–624) was constructed by cloning of the corresponding fragment to pcDNA4.0/V5-His in frame (pcDNA4.0-vinculin(1–624)-V5).
To generate adenoviral vectors, the cDNAs corresponding to the wild-type vinculin and the mutant proteins were subcloned into pENTR11 (Invitrogen), and the inserts were transferred to the pAd/CMV/V5-DEST vector (Invitrogen). Recombinant adenoviruses were prepared with the ViraPowerTM Adenovirus Expression System (Invitrogen). For comparison, we constructed vectors encoding the carboxyl region of vinculin corresponding to aa 625–1066 also, by removing the fragment for aa 2–624 from pENTR-vinculin(WT).
To construct the expression plasmid encoding paxillin, we first performed PCR-based cloning of cDNA for mouse paxillin using RNA derived from ATDC5 cells. Then the cDNA was cloned in frame into a pcDNA4/Myc-His vector (Invitrogen) after the stop codon was mutated to a EcoRI recognition site, resulting in a plasmid encoding paxillin fused to a Myc tag at the carboxyl terminus (designated pcDNA-paxillin-Myc-His).
Gene Silencing
Gene silencing in ATDC5 cells was performed using the siPORT Amine transfection agent (Applied Biosystems) and Silencer® Select siRNAs (Applied Biosystems) by the reversal transfection method. As the vinculin-specific siRNA, Silencer® Select siRNAs s75918 and s75919 were utilized. A negative control siRNA with a scrambled sequence was also included in the experiments.
The knocking down of vinculin expression in primary chondrocytes and the organ culture of metatarsals was performed by adenovirus-mediated expression of microRNA (miRNA) using the BLOCK-iTTM Pol II miR RNAi Expression Vector Kit with EmGFP and BLOCK-iTTM Pol II miR RNAi Select (Mmi526082 and Mmi526083) (Invitrogen).
Western Blotting
Whole cell extracts were harvested in radioimmune precipitation buffer (1% Triton, 1% sodium deoxycholate, 0.1% SDS, 150 mm NaCl, 10 mm Tris-Cl (pH 7.4), 5 mm EDTA, 1 mm orthovanadate and protease inhibitor mixture (CompleteTM, Roche Applied Science)). The cell lysates containing 10 μg of each protein were then subjected to SDS-PAGE and transferred to PVDF membranes (Bio-Rad). After blocking with BlockAce reagent (Dainippon Pharmaceuticals, Osaka, Japan) or Blocking-one reagent (Nacalai Tesque), the membranes were incubated with the following primary antibodies: polyclonal antibody against the amino or carboxyl terminus of vinculin (N-19 and C-20, respectively; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), anti-β-galactosidase polyclonal antibody (Rockland, Gilbertsville, PA), anti-phospho-FAK antibody, anti-FAK antibody, anti-paxillin antibody (BD Transduction Laboratories, Franklin Lakes, NJ), anti-V5 antibody (Invitrogen), anti-cMyc antibody, anti-Sox9 antibody (Santa Cruz Biotechnology, Inc.), anti-phospho-Sox9 (Ser(P)-181) antibody (AnaSpec Inc., San Jose, CA), anti-phospho ERK1/2 antibody, anti-ERK1/2 antibody, anti-phospho-Akt antibody, anti-Akt antibody, anti-phospho-SHP2 (Src homology 2 domain-containing tyrosine phosphatase 2) antibody, anti-SHP2 antibody, anti-phospho-p38 MAPK antibody, anti-p38 MAPK antibody, anti-phospho-Smad antibodies, and anti-Smad antibodies (Cell Signaling, Beverly, MA). After incubation with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibody, the proteins were visualized using the enhanced chemiluminescence (ECL) detection system (Amersham Biosciences).
Immunoprecipitation
Whole cell lysates were harvested in a lysis buffer (5 mm EDTA, 150 mm NaCl, 0.5% Nonidet P-40, 10% glycerol, and 10 mm Tris-HCl, pH 8.0) containing a protease inhibitor mixture (CompleteTM, EDTA-free, Roche Applied Science). The lysates were incubated with an antibody, and the immunocomplex was immobilized on protein A/G-agarose conjugate at 4 °C overnight. The agarose beads were extensively washed, and the immunoprecipitates were subjected to Western blot analyses.
Immunohistochemistry
Tibiae obtained from E18.5 mouse embryos or the cultured metatarsal bones were fixed in 10% buffered formalin and embedded in paraffin. Sections were deparaffinized, rehydrated, and subjected to antigen retrieval. The quenching of endogenous peroxidase activity and immunohistochemical staining were performed using ImmunoCruz staining systems (Santa Cruz Biotechnology, Inc.). After blocking, the sections were incubated with anti-vinculin antibody (Santa Cruz Biotechnology, Inc.), anti-Col II mouse monoclonal antibody (Clone 2B1.5; Thermo Fisher Scientific, Waltham, MA), or anti-aggrecan antibody (Santa Cruz Biotechnology, Inc.). Normal IgG was used as a negative control. Thereafter, the sections were incubated with biotinylated secondary antibodies and with HRP-conjugated streptavidin. Development of the peroxidase staining was done with 3′,3-diaminobenzidine. Antigen retrieval was performed by incubating the sections in 10 mm citrate buffer (pH 6.0) at 95 °C for 15 min, in pepsin solution (Thermo Fisher Scientific) at 37 °C for 60 min, or in 0.02% hyaluronidase (Sigma-Aldrich) in 0.1 m sodium acetate (pH 5.0) at 37 °C for 60 min, for staining with the anti-vinculin antibody, anti-Col II antibody, or anti-aggrecan antibody, respectively. The sections were counterstained with hematoxylin.
Cell Attachment Assay
Cells were trypsinized, and the reaction was neutralized with a trypsin inhibitor (Invitrogen). Then the cells suspended in serum-free medium (5 × 105 cells) were plated on fibronectin-coated dishes. After 30, 60, or 90 min of incubation, the non-attached cells were removed by washing, and the attached cells were harvested by trypsinization and counted.
Cell Migration Assay
To evaluate the ability of the cells to migrate, we performed a wound healing assay using culture inserts intended for this purpose (Ibidi GmbH, München, Germany), according to a previous report (18). Each insert has two wells with a 500-μm gap. The inserts were placed on culture plates, and cells were plated in both wells (7 × 103 cells/well, whose growth area was 2 × 0.22 cm2). When the cells reached confluence, the medium was changed to serum-free medium. Ten hours later, the inserts were removed, and cells were incubated at 37 °C in a CO2 incubator. The cells were subjected to microscopy to evaluate the narrowing of the cell-free gap.
Statistical Analysis
Data were analyzed using one-way analysis of variance. The method of Tukey was used for post hoc tests. All statistical analyses were conducted using Dr. SPSS II software.
RESULTS
Isolation of Clone in Which Gene Encoding Vinculin Was Trapped
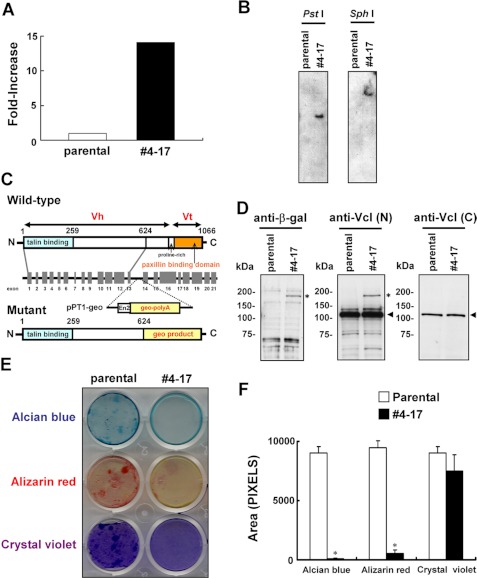
Because the trap vector pPT1-geo possesses the lacZ-neo fusion gene geo, clones can be isolated on selection with neomycin only when the trapped gene is expressed in ATDC5 cells. We obtained 815 neomycin-resistant clones and subjected the clones with 10-fold more β-galactosidase activity than the parental cells to chondrogenic induction. We then further analyzed clone 4-17, which exhibited fewer mineralized nodules. The β-galactosidase activity in clone 4-17 was ∼13-fold that in parental cells (Fig. 1A). The trap vector was inserted at only one location (Fig. 1B). A 5′-RACE analysis demonstrated that the vector was inserted into intron 13 of the vinculin gene (accession number NC_000080.5). The trapped allele was assumed to express a fusion protein containing the talin-binding head domain of vinculin and the product of geo but lacking the paxillin-binding tail domain of vinculin (Fig. 1C). Western blotting confirmed the production of the fusion protein (Fig. 1D). Interestingly, Western blotting using the antibody against the carboxyl terminus of vinculin, which was absent from the mutant protein, demonstrated comparable amounts of wild-type vinculin between the parental cells and clone 4-17 (Fig. 1D). Staining of clone 4-17 for β-galactosidase suggested the cytoplasmic distribution of the fusion product (data not shown). Clone 4-17 accumulated less cartilaginous matrix than the parental cells, as shown by Alizarin red and Alcian blue staining (Fig. 1, E and F). Staining with crystal violet verified the adhesion of the cells to the culture plates (Fig. 1, E and F). In contrast to clone 4-17, most of the other clones exhibited no obvious change in Alizarin red and Alcian blue staining (supplemental Fig. S1).
FIGURE 1.
Isolation of the clone with the trapped vinculin gene. A, the β-galactosidase activity in clone 4-17 and the parental ATDC5 cells. B, Southern blotting. Genomic DNA was digested with the restriction enzyme PstI or SphI, and subjected to hybridization with a radiolabeled fragment of lacZ cDNA prepared by EcoRI/SacI digestion of pPT1-geo. C, genomic organization of the murine vinculin gene and the insertional mutation resulting from the gene trapping. The locations of exons (gray boxes) and introns (horizontal lines between exons) are indicated. pPT1-geo was inserted into intron 13, resulting in a mutant protein in which vinculin lacking the tail domain was fused to the geo product. D, detection of the β-galactosidase fusion protein. Cell lysates were subjected to immunoblotting using antibodies against β-galactosidase (left) or amino-terminal (center) or carboxyl-terminal vinculin. The asterisk and the arrow indicate the signals corresponding to the fusion protein comprising part of vinculin and the geo product and that for wild-type vinculin, respectively. E, impaired chondrogenic nodule formation in clone 4-17. The cells were cultured for 8 weeks in chondrogenic medium and subjected to Alcian blue and Alizarin red staining. Clone 4-17 accumulated less cartilaginous matrix than parental cells. The cells were also stained with crystal violet to confirm the adhesion to culture plates. F, the stained area in E was quantitated using NIH Image 1.63 software. Data are shown as the mean ± S.D. (error bars) (n = 3). *, p < 0.001 versus parental cells.
Expression of Vinculin Increased during Chondrocyte Differentiation
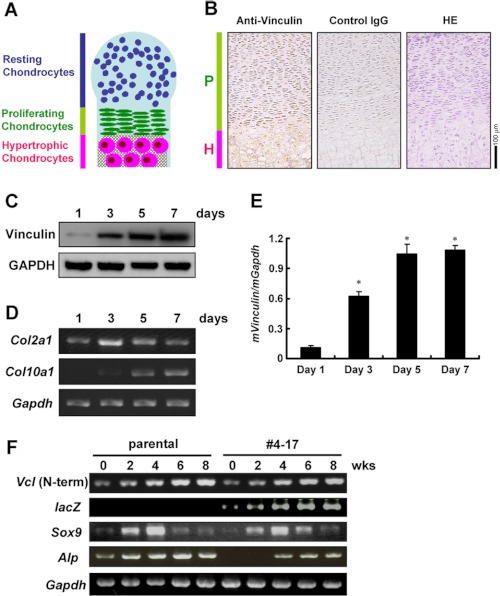
Immunostaining with the anti-vinculin antibody was performed using sections of tibiae harvested from E18.5 wild-type mouse embryos. Chondrocytes were positively stained, and the immunoreactivity was intense in hypertrophic chondrocytes (Fig. 2B).
FIGURE 2.
Increasing expression of vinculin during the differentiation of chondrocytes. A, illustration of endochondral bone formation displayed as a layered structure. B, immunostaining of mouse tibiae from an E18.5 embryo with anti-vinculin antibody. The immunoreactivity was intense in hypertrophic chondrocytes. P and H, proliferating and hypertrophic chondrocytes, respectively. HE, hematoxylin/eosin-stained section. C–E, increasing expression of vinculin during the maturation of primary chondrocytes. Chondrocytes isolated from mouse ribcages were cultured for the period indicated before being harvested for Western blotting (C) and real-time PCR (E). In E, data are shown as the mean ± S.D. (error bars). *, p < 0.05 versus day 1. The temporal expression of Col2a1 and Col10a1 analyzed by RT-PCR is shown in D. F, RT-PCR analyses for the temporal expression of the amino-terminal fragment of vinculin, Sox9, and Alp (alkaline phosphatase) during the chondrocytic differentiation of parental ATDC5 cells and clone 4-17. The primer set for amino-terminal vinculin amplifies the fragment derived from both the wild-type and trapped alleles.
We also examined the temporal expression pattern of vinculin in primary chondrocytes isolated from ribcages of wild-type newborns. The increasing expression of Col10a1, the gene for type X collagen and a marker for hypertrophic chondrocytes, indicated the progression of the differentiation during the culture (Fig. 2D). The expression of vinculin also was increased as the chondrocytes matured (Fig. 2, C and E).
To examine the temporal expression of vinculin in the parental ATDC5 cells and clone 4-17, these cells were cultured in the chondrogenic medium. The expression of Sox9, the key transcription factor for chondrocytic differentiation, reached a maximum at 4 weeks of culture in both cells. The expression of Alp (alkaline phosphatase) was lower in clone 4-17. To examine the expression of vinculin by RT-PCR, we used a pair of primers that amplified the fragment corresponding to the amino terminus of vinculin shared by the transcript from the wild-type allele and that from the trapped allele and found a temporal increase in both cells. In clone 4-17, the expression of lacZ was also elevated during the culture (Fig. 2F). To examine whether or not the trapping of vinculin altered the fate of the cells, we analyzed the expression of MyoD1 and Pparg, markers for myocytes and adipocytes, respectively, and found no expression (data not shown).
Alteration in Signaling Induced by Adhesion to ECM in Clone with Trapped Vinculin Gene
The increase in the expression of vinculin during chondrocyte differentiation suggested some role in chondrogenesis. To investigate this role, we further characterized clone 4-17.
To characterize the truncated vinculin derived from the trapped allele, we constructed plasmids encoding a V5-tagged wild-type vinculin (pcDNA-vinculin(WT)-V5) or a truncated protein corresponding to the mutant derived from the trapped allele (pcDNA-vinculin(1–624)-V5). For comparison, we constructed another plasmid encoding the carboxyl fragment of vinculin (pcDNA-vinculin(625–1066)-V5), which lacked the region retained in the trapped allele, also. Co-immunoprecipitation experiments confirmed that the truncated vinculin(1–624) exhibited impaired binding to paxillin (Fig. 3A). The binding to talin was markedly increased in the truncated vinculin(1–624) compared with in vinculin(WT) (Fig. 3B).
FIGURE 3.
Alteration in the signaling induced by adhesion to ECM in the clone 4–17 and the cells with the vinculin gene knocked down. A, the truncated vinculin derived from the trapped allele was unable to bind to paxillin. COS7 cells were transfected with the expression plasmid encoding Myc-tagged paxillin (paxillin-Myc) together with the plasmid encoding V5-tagged wild type (vinculin(WT)-V5), the truncated vinculin corresponding to the mutant derived from the trapped allele in clone 4-17 (vinculin(1–624)-V5), or the mutant encoding the carboxyl fragment of vinculin (vinculin(625–1066)-V5), and lysates were harvested for immunoprecipitation (IP) using anti-V5 antibody. The immunoprecipitates and the input protein (10 μg) were subjected to Western blotting using the indicated antibodies. B, the binding to talin was enhanced in the truncated vinculin derived from the trapped allele. COS7 cells were transfected with the plasmid encoding vinculin(WT)-V5, vinculin(1–624)-V5, or vinculin(625–1066)-V5, and lysates were used for immunoprecipitation with anti-talin antibody. The immunoprecipitates and the input protein (10 μg) were subjected to Western blotting with the indicated antibodies. C, increase in phosphorylation of FAK at Tyr-397 and ERK1/2 induced by adhesion to FN in clone 4-17. Parental ATDC5 cells or clone 4-17 were plated onto FN. Thirty minutes later, lysates were harvested for Western blot analyses. D and E, increase in phosphorylation of FAK at Tyr-397 and ERK1/2 induced by adhesion to Col II in clone 4-17. Cells were plated onto Col II, and lysates were harvested at the indicated time points. The densitometric ratio of the intensity of the signal corresponding to phosphorylated FAK to that of total FAK is shown in E. The data are shown as the mean ± S.D. (error bars) (n = 3). *, p < 0.05 versus 0 h; #, p < 0.05 versus parental cells. F, knockdown of vinculin in parental ATDC5 cells increased the adhesion-induced phosphorylation of FAK and ERK1/2, which was cancelled by simultaneous introduction of the expression plasmid encoding vinculin(WT). Lysates were harvested for Western blotting from the parental ATDC5 cells transfected with control siRNA, vinculin-specific siRNA with or without the expression plasmids encoding vinculin(WT) or lacZ, the non-transfection control, and clone 4-17 with the vinculin gene trapped 30 min after the plating onto FN. G, exogenous expression of the truncated vinculin(1–624) in parental ATDC5 cells increased the adhesion-induced phosphorylation of FAK, which was not abrogated by the simultaneous expression of exogenous wild-type vinculin. Lysates were harvested from the parental ATDC5 cells infected with the adenoviral vector for vinculin(1–624)-V5 together with that for lacZ or vinculin(WT)-V5 and the cells expressing lacZ alone 30 min after the plating onto FN. H, exogenous expression of the truncated vinculin(625–1066) in parental ATDC5 cells had no effects on the adhesion-induced phosphorylation of FAK. ATDC5 cells were infected with the vector for vinculin(625–1066)-V5 together with that for lacZ or vinculin(WT)-V5 and the cells expressing lacZ alone, and lysates were harvested 30 min after the plating onto FN. I, the truncated vinculin(1–624) interfered with the interaction between the wild-type vinculin and talin. COS7 cells were transfected with Myc-tagged wild-type vinculin (vinculin(WT)-Myc) together with vinculin(WT)-V5 or the mutant vinculin(1–624)-V5, and the lysates were subjected to immunoprecipitation using anti-talin antibody. The immunoprecipitates and the input protein (10 μg) were subjected to Western blotting using the indicated antibodies. In the immunoblot using anti-Myc antibody, the co-transfection of vinculin(1–624)-V5 reduced the signal for the co-immunoprecipitation of vinculin(WT)-Myc with talin. p-FAK, phosphorylated FAK; t-FAK, total FAK.
Mouse embryo fibroblasts isolated from vinculin-knock-out mouse embryos demonstrated increased phosphorylation of focal adhesion kinase (FAK) and extracellular signal-regulated kinase (ERK) in response to adhesion to ECMs (12). Therefore, we examined the phosphorylation of FAK at Tyr-397 and ERK1/2 induced by adhesion to ECMs in clone 4-17 and the parental ATDC5 cells. The phosphorylation of FAK and ERK1/2 induced by adhesion to both fibronectin (FN) and type II collagen (Col II) was increased in clone 4-17 (Fig. 3, C–E). The knockdown of vinculin expression in parental cells also resulted in an increase in the adhesion-induced phosphorylation of FAK and ERK1/2 (Fig. 3F). Because transfection of siRNA may have toxic effects, we confirmed that the expression of vinculin was completely reversed at 9 days after the transfection of gene-specific siRNA (supplemental Fig. S2). The data indicate that vinculin functions as a negative regulator of the signaling induced by adhesion to ECM. Considering the similarity in the increase in the adhesion-induced phosphorylation of FAK and ERK1/2 between the vinculin-silenced cells and clone 4-17, we assume that the truncated vinculin derived from the trapped allele has some dominant effect that interferes with the functions of wild-type vinculin. Consistent with this notion, exogenous expression of vinculin(1–624)-V5 into the parental cells also increased the adhesion-induced phosphorylation of FAK, and the simultaneous overexpression of wild-type vinculin failed to abolish the increase (Fig. 3G). On the other hand, exogenous expression of the carboxyl fragment vinculin(625–1066)-V5 into the parental cells had no obvious influence on the adhesion-induced phosphorylation of FAK (Fig. 3H).
To elucidate the mechanism by which the truncated vinculin(1–624)-V5 exerted a dominant negative effect on signaling, we examined whether the mutant affected the interaction between the wild-type vinculin and talin. COS7 cells were transfected with Myc-tagged wild-type vinculin (vinculin(WT)-Myc) together with vinculin(WT)-V5 or the mutant vinculin(1–624)-V5, and the lysates were subjected to immunoprecipitation using anti-talin antibody. Aliquots of the immunoprecipitates were subjected to immunoblotting with anti-Myc antibody, and the proteins were visualized using ECL PlusTM reagent (Amersham Biosciences) with high sensitivity. The co-transfection of vinculin(1–624)-V5 reduced the co-immunoprecipitation of vinculin(WT)-Myc with talin (Fig. 3I). These results suggest that vinculin(1–624) interferes with the interaction between the wild-type vinculin and talin, which might be the mechanism for the dominant negative effect of the mutant.
Effects of Trapping of Vinculin Gene on Cell Proliferation, Attachment, and Migration in Vitro
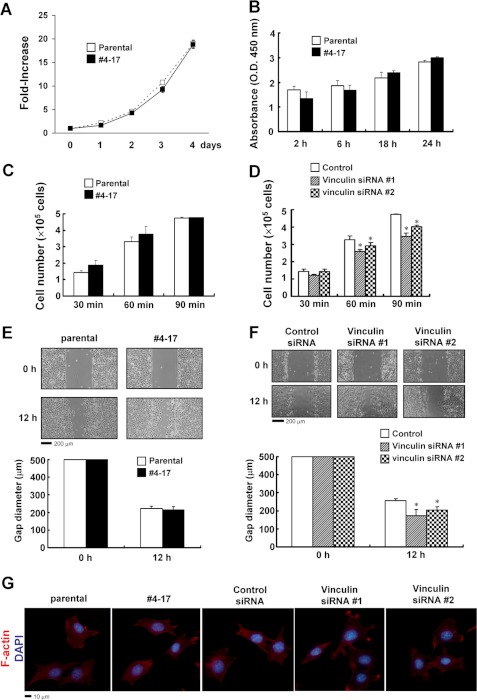
We next examined the effects of the trapping of the vinculin gene on cell behavior in vitro. The proliferation was comparable between the parental cells and the clone 4-17 with the trapped vinculin gene (Fig. 4, A and B). As for the attachment to the ECM and motility, there was no obvious difference between the parental cells and clone 4-17 (Fig. 4, C and E) either, which was probably due to the activity of the remaining wild-type allele in clone 4-17. We also examined the effects of the knockdown of vinculin in parental cells, which slightly reduced the attachment and increased the cell migration (Fig. 4, D and F). Trapping of the vinculin gene in clone 4-17 had no apparent effect on focal adhesion. Knockdown of vinculin in parental cells had no obvious influence on focal adhesion either. These observations suggest the residual activity of vinculin to be enough for focal adhesion in vitro (Fig. 4G).
FIGURE 4.
Effects of the trapping or knockdown of the vinculin gene on the increase in cell number, attachment and migration. A and B, no obvious difference in cell proliferation between parental ATDC5 cells and clone 4-17 with the trapped vinculin gene. In A, cells were plated in 96-well culture plates at a density of 1 × 103 cells/well (designated day 0) and cultured for the period indicated. The cell number in each well was evaluated by a 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt assay, and the -fold increase from the value on day 0 was calculated. The data are shown as the mean ± S.D. (error bars) (n = 4). In B, a BrdU cell proliferation assay was performed. C, trapping of the vinculin gene had no obvious effect on cell attachment. The parental ATDC5 cells and clone 4-17 were plated onto FN-coated dishes, and the attached cells were counted after the period indicated. The data are shown as the mean ± S.D. (n = 3). D, attachment assay in the ATDC5 cells transfected with control siRNA or vinculin-specific siRNA. Cells were plated onto FN-coated dishes, and the attached cells were counted after the indicated period. The data are shown as the mean ± S.D. (n = 3). *, p < 0.05 versus control siRNA. E, trapping of the vinculin gene had no obvious effect on cell migration. Microscopic images of the parental ATDC5 cells and clone 4-17 assayed for their ability to close a defined 500-μm gap in a cell migration assay. We used defined culture inserts, the removal of which generated a 500-μm gap between cells. The gap diameters are shown as the mean ± S.D. (n = 3). F, a cell migration assay was performed in the parental ATDC5 cells transfected with control siRNA or vinculin-specific siRNA. The gap diameters are shown as the mean ± S.D. (n = 3). *, p < 0.05 versus control siRNA. G, trapping or knockdown of the vinculin gene resulted in no obvious change in focal adhesion. Parental ATDC5 cells, clone 4-17, and the cells transfected with vinculin-specific siRNAs or control siRNA were plated onto FN-coated coverslips and, 5 h later, fixed and stained with Alexa FluorTM 555 phalloidin to examine the subcellular distribution of F-actin. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI).
Defects in Chondrocytic Differentiation in Clone with Trapped Vinculin Gene
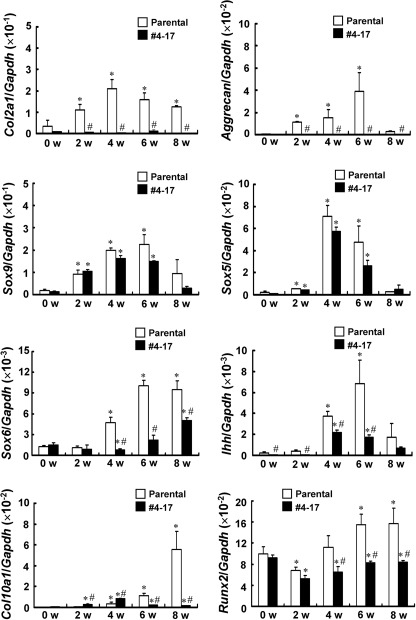
To clarify whether vinculin plays a role in chondrogenesis, we further analyzed the chondrocytic phenotype of clone 4-17. Parental ATDC5 cells and clone 4-17 were cultured in the chondrogenic medium, and the temporal expression of the chondrocytic marker genes was evaluated (Fig. 5). The expression of Sox9 was retained in clone 4-17. However, the expression of Col2a1 encoding the a1 chain of Col II as well as that of aggrecan was markedly reduced in clone 4-17. Sox5 and Sox6 are transcription factors that cooperate with Sox9 in chondrocytic differentiation. Although the expression of Sox5 was retained in clone 4-17, that of Sox6 was decreased (Fig. 5). The expression of Ihh, a marker for prehypertrophic chondrocytes, peaked at 6 weeks of culture in the parental cells. In clone 4-17, it reached a maximum at 4 weeks of culture and declined thereafter. The expression of Ihh in clone 4-17 suggested that at least some cells differentiated into prehypertrophic chondrocytes. The expression of Col10a1 gradually elevated during the differentiation of parental ATDC5 cells, whereas it markedly decreased in clone 4-17. Runx2 is a transcription factor that plays a critical role in chondrocyte maturation and directly controls Col10a1 expression (19). The expression of Runx2 increased during the later differentiation in the parental cells but not in clone 4-17 (Fig. 5).
FIGURE 5.
Altered gene expression in clone 4-17 with the trapped vinculin gene. Parental ATDC5 cells and clone 4-17 were cultured in the chondrogenic medium for the period indicated, before RNA was extracted for real-time PCR analyses. The copy number of the target cDNA in each sample was estimated by referring to a standard curve, which was standardized by that of Gapdh. The data are shown as the mean ± S.D. (error bars) (n = 3). *, p < 0.05 versus 0 weeks; #, p < 0.05 versus parental cells; w, weeks.
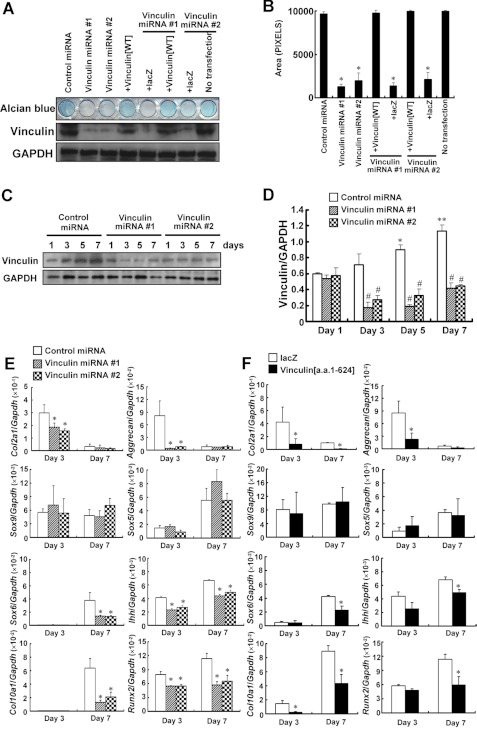
Knockdown of Vinculin or Expression of Mutant Vinculin(1–624) Caused Impaired Maturation in Primary Chondrocytes
We also examined the effects of knocking down the expression of vinculin expression using primary chondrocytes. Primary chondrocytes were plated on day 0 and, 24 h later, infected with an adenovirus encoding vinculin-specific miRNA or control miRNA. Alcian blue staining demonstrated less accumulation of cartilaginous matrix in the cells where the expression of vinculin was knocked down, a situation that was rescued by the simultaneous introduction of the adenovirus encoding wild-type vinculin (Fig. 6, A and B). The expression of neither Sox9 nor Sox5 was influenced, whereas that of Sox6 on day 7 was suppressed by knocking down the expression of vinculin. The expression of Col2a1 and aggrecan on day 3 was also markedly decreased, and that of Ihh was reduced on both day 3 and day 7 by the knockdown. As for the expression of Col10a1, which was detected on day 7 in the control cells, the knockdown of vinculin resulted in its reduction. The expression of Runx2 also was decreased by the knockdown (Fig. 6E). These alterations in gene expression were consistent with those in clone 4-17. Adenoviral transfer of the truncated vinculin(1–624)-V5 in primary chondrocytes also resulted in the reduced expression of Col2a1, aggrecan, Sox6, Ihh, Col10a1, and Runx2 and less accumulation of cartilaginous matrix (Fig. 6F and supplemental Fig. S3). On the other hand, adenoviral transfer of the carboxyl fragment vinculin(625–1066)-V5 in primary chondrocytes had no obvious influence on the accumulation of cartilaginous matrix (data not shown).
FIGURE 6.
Impaired maturation of the primary chondrocytes with the vinculin gene knocked down or the exogenous expression of the truncated mutant vinculin(1–624). A, knockdown of vinculin expression in primary chondrocytes reduced the accumulation of cartilaginous matrices, which was rescued by simultaneous transfer of the adenovirus encoding vinculin(WT). Primary chondrocytes were plated onto 48-well culture plates (2 × 104 cells/well). The next day (day 1), the cells were infected with adenovirus encoding control miRNA or vinculin-specific miRNA with or without the virus for vinculin(WT)-V5 or lacZ. On day 7, the cells were subjected to Alcian blue staining or Western blotting. B, the Alcian blue-stained area in A was quantitated. The data are shown as the mean ± S.D. (error bars) (n = 3). *, p < 0.05 versus control miRNA. C and D, efficient knockdown of vinculin expression by gene-specific miRNA in primary chondrocytes. Cells were infected with an adenovirus encoding vinculin-specific miRNA or control miRNA on day 1, and lysates were harvested for Western blotting at the indicated time points. The densitometric ratio of the intensity of the signals corresponding to vinculin and to GAPDH is shown in D. The data are shown as the mean ± S.D. (n = 3). *, p < 0.05; **, p < 0.01 versus day 1; #, p < 0.05 versus control miRNA. E, the knockdown of vinculin expression in primary chondrocytes reduced the expression of Col2a1, aggrecan, Sox6, Ihh, Runx2, and Col10a1. Cells were infected with the adenovirus encoding vinculin-specific miRNA or control miRNA on day 1. RNA was extracted for real-time PCR on day 3 and day 7. The data are shown as the mean ± S.D. (n = 3). *, p < 0.05 versus control miRNA. F, expression of the truncated vinculin(1–624) in primary chondrocytes also reduced the expression of Col2a1, aggrecan, Sox6, Ihh, Runx2, and Col10a1. The data are shown as the mean ± S.D. (n = 3). *, p < 0.05 versus lacZ.
Knockdown of Wild-type Vinculin as Well as Expression of Vinculin(1–624) Impaired Growth of Metatarsal Explants
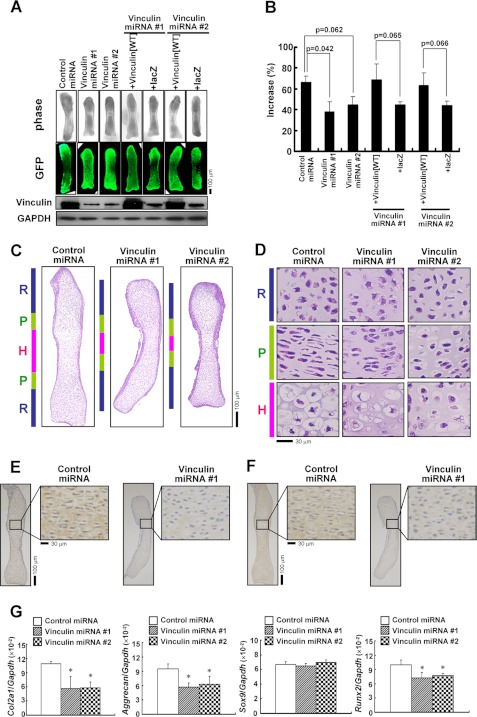
The role of vinculin in chondrogenesis was further investigated using organ cultures of metatarsal rudiments ex vivo. Metatarsals from E15.5 mouse embryos were infected with an adenovirus for vinculin-specific miRNA or control miRNA and cultured for 7 days. In this system, the rudiments grew during the culture. In the sections cut from the rudiments, the direct fluorescence of EmGFP derived from the vectors encoding miRNAs was detected in chondrocytes as well as in the perichondrium, demonstrating the infection in these cells (supplemental Fig. S4). We found that the extended length was shorter in the rudiments infected with vinculin-specific miRNA. Moreover, simultaneous infection with the virus encoding wild-type vinculin, but not the control virus encoding lacZ, restored the blunted growth due to the knockdown of vinculin (Fig. 7, A and B). Hematoxylin/eosin (H&E) staining of sections cut from the rudiments demonstrated that the columnar proliferation of the chondrocytes was disturbed, and the subsequent hypertrophic change was insufficient in the rudiments with the knockdown of vinculin (Fig. 7, C and D). In immunohistochemistry, immunoreactivity with the antibodies against Col II and aggrecan was decreased in the rudiments infected with vinculin-specific miRNA (Fig. 7, E and F). Real-time PCR analyses using RNA extracted from the rudiments demonstrated that the knockdown of vinculin caused the reduced expression of Col2a1 and aggrecan. The expression of Runx2 also was decreased by the knockdown of vinculin, whereas that of Sox9 was retained (Fig. 7G). Adenoviral transfer of the truncated vinculin(1–624)-V5 also impaired the growth of the metatarsal rudiments (supplemental Fig. S5).
FIGURE 7.
Knockdown of vinculin expression in metatarsal organ cultures resulted in impaired linear growth. A and B, knockdown of vinculin inhibited the linear growth of metatarsal rudiments. Metatarsals from E15.5 mouse embryos were infected with the adenovirus encoding control miRNA or vinculin-specific miRNA with or without the adenovirus for vinculin(WT) or lacZ after the measurement of initial length using a microscope and Image-Pro PLUS 6.3 software. On day 7, the length of the rudiments was measured again, and the protein was extracted for Western blotting. In A, representative images of the phase-contrast view and the direct fluorescence of EmGFP derived from the vector encoding miRNAs are depicted, together with the results of Western blotting using the indicated antibodies. In B, the increase in the length of the rudiments is shown as a percentage of the extended length during the culture to the initial length of each bone. The data are shown as the mean ± S.D. (error bars) (n = 3). C and D, HE-stained sections of metatarsal rudiments infected with adenovirus encoding control miRNA or vinculin-specific miRNA. R, P, and H, resting, proliferating, and hypertrophic zones, respectively. Higher power images are shown in D. E and F, immunostaining demonstrated the reduced expression of Col II (E) and aggrecan (F) in the metatarsal explants where the expression of vinculin was knocked down. G, real-time PCR for the genes indicated using RNA extracted from the metatarsal explants infected with vinculin-specific miRNA or control miRNA. Knockdown of vinculin suppressed the expression of Col2a1, aggrecan, and Runx2 in the metatarsals.
Altered Signaling during Chondrocytic Differentiation in Cells with Knocked Down or Trapped Vinculin Gene
Thus, vinculin appears to play an important role in chondrogenesis. Therefore, we next investigated the mechanism by which vinculin regulates chondrocyte differentiation.
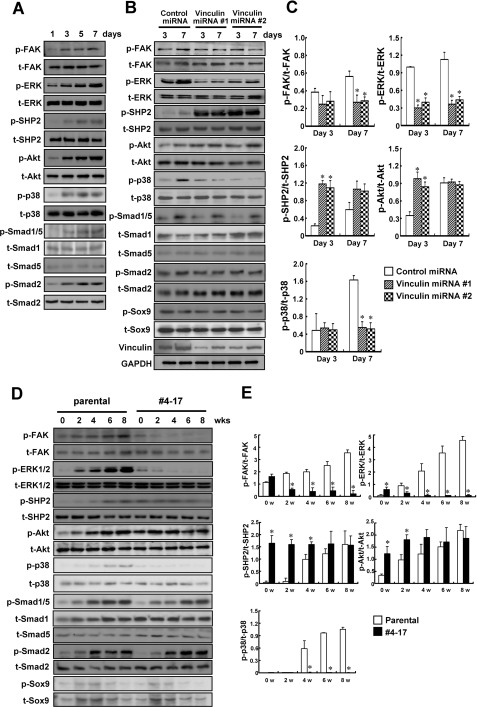
Several signaling pathways have been implicated in chondrogenesis. The phosphorylation of ERK1/2, SHP2, and Akt as well as that of FAK increased during the maturation of primary chondrocytes. In addition, the phosphorylation of p38 MAPK and some Smads also increased as the chondrocytes matured (Fig. 8A). Therefore, we examined whether the impaired functioning of vinculin affected the phosphorylation of these signaling molecules. Knockdown of vinculin in primary chondrocytes resulted in the decreased phosphorylation of ERK1/2 on both day 3 and day 7 of culture and reduced phosphorylation of FAK on day 7. On the other hand, the phosphorylation of SHP2 on day 3 and day 7 as well as that of Akt on day 3 increased with the knockdown of vinculin expression (Fig. 8, B and C). We also analyzed the phosphorylation of these proteins during the differentiation of parental ATDC5 cells and clone 4-17 with the trapped vinculin gene and obtained consistent results. Phosphorylation of FAK and ERK1/2 was decreased in clone 4-17 as compared with in parental cells after 2 weeks of culture. On the other hand, phosphorylation of SHP2 and Akt was stronger in clone 4-17 than in parental cells during weeks 0–4 (Fig. 8, D and E).
FIGURE 8.
Altered signaling during the chondrocytic differentiation in the cells with a knocked down or trapped vinculin gene. A, increasing phosphorylation of FAK, ERK1/2, SHP2, Akt, p38 MAPK, and Smads during the maturation of primary chondrocytes. B and C, knockdown of vinculin expression altered the phosphorylation of FAK, ERK1/2, SHP2, Akt, and p38 MAPK during the maturation of primary chondrocytes. Cells were infected with the adenovirus encoding control miRNA or vinculin-specific miRNA on day 1, and lysates were harvested on day 3 and day 7 for Western blotting with the indicated antibodies. The densitometric ratio of the intensity of the signals is demonstrated in C. The data are shown as the mean ± S.D. (error bars) (n = 3). *, p < 0.05 versus control miRNA. D and E, phosphorylation status of the signaling molecules during the differentiation of parental ATDC5 cells and clone 4-17 with the trapped vinculin gene. Parental ATDC5 cells and clone 4-17 were cultured in the differentiation medium for the period indicated (0–8 weeks) before lysates were harvested for Western blotting. The densitometric ratio of the intensity of the signals is demonstrated in E. The data are shown as the mean ± S.D. (n = 3). p, phosphorylated; t, total; w, weeks.
Interestingly, we found that the phosphorylation of p38 MAPK was markedly reduced in chondrocytes with vinculin knocked down as well as in clone 4-17, suggesting that dysfunctional vinculin impaired the activation of the p38 MAPK pathway in differentiating chondrocytes. We also examined the phosphorylation of some Smads, which was not affected by the dysfunctional vinculin (Fig. 8, B–E).
The reduced phosphorylation of FAK and ERK1/2 during the chondrocytic differentiation shown in Fig. 8 makes a striking contrast to the increase in adhesion-induced phosphorylation of FAK and ERK1/2 in the cells in which vinculin was knocked down or trapped (Fig. 3). Therefore, we performed another series of experiments to examine the phosphorylation of FAK, ERK1/2, and SHP2 at 0, 5, 10, 24, 48, 72, and 120 h after the cells were plated (supplemental Fig. S6). At the time points of 0–48 h, the phosphorylation of FAK and ERK was increased in the clone 4-17 compared with in parental cells, which was consistent with the results shown in Fig. 3, where the phosphorylation was examined 0.5–5 h after the cells were plated. On the other hand, at the time points of 72 and 120 h, FAK and ERK phosphorylation was stronger in the parental cells than in clone 4-17, which was consistent with the results in Fig. 8, where the phosphorylation was examined at 0–8 weeks. Thus, these results demonstrate that the phosphorylation of FAK and ERK was initially increased and then suppressed in the cells with a trapped vinculin gene compared with the parental cells after they adhered. The phosphorylation of SHP2 was increased in clone 4-17 at 72 and 120 h, which might be involved in the reduced phosphorylation of ERK and FAK (supplemental Fig. S6).
As shown in Fig. 5 and 6, trapping or knockdown of vinculin did not alter the mRNA expression of Sox9. Therefore, we examined whether the impaired functioning of vinculin affected the phosphorylation of the Sox9 protein. Neither trapping nor knockdown of the vinculin gene affected the phosphorylation of Sox9, suggesting that the function of Sox9 was retained in the cells with a dysfunctional vinculin (Fig. 8, B and D).
Impaired Functioning of Vinculin Blunted Responsiveness of Chondrocytes to IGF-I
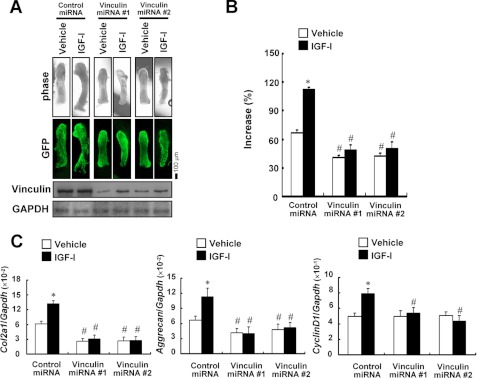
Changes in the phosphorylation of signaling molecules during chondrocytic differentiation suggested that the knockdown or trapping of vinculin has an influence not only on signaling from the ECM but also on other signaling pathways, including those of growth factors. IGF-I exerts its signals via PI3K/Akt and MEK/ERK pathways. In addition, IGF-I is known to regulate Col2a1 and aggrecan expression, both of which were down-regulated in the cells in which vinculin was trapped or knocked down. Therefore, we examined whether responsiveness to IGF-I was altered by the dysfunctional vinculin using the organ culture of metatarsal rudiments. The knockdown of vinculin expression impaired the IGF-I-induced growth in the rudiments (Fig. 9, A and B). Real-time PCR analyses revealed that treatment with IGF-I increased the expression of Col2a1, aggrecan, and a cell cycle-related gene, cyclin D1, in the control rudiments but not in those with the vinculin gene knocked down (Fig. 9C), suggesting that the dysfunctional vinculin blunted the responsiveness to IGF-I. The expression of IGF-I receptor was not altered by the knockdown of vinculin (data not shown).
FIGURE 9.
Dysfunctional vinculin caused a disturbance of IGF-I signaling. A and B, dysfunctional vinculin impaired the IGF-I-induced growth of metatarsal organ cultures. Metatarsals were infected with the adenovirus encoding control miRNA or vinculin-specific miRNA and treated with 100 ng/ml IGF-I or vehicle for 7 days to evaluate linear growth. Representative images of the phase contrast view and the direct fluorescence of EmGFP derived from the vector-encoding miRNAs are depicted, together with the results of Western blotting. In B, the increase in the length of the rudiments is shown as a percentage of the extended length during the culture to the initial length of each bone. The data are shown as the mean ± S.D. (error bars) (n = 3). *, p < 0.01 versus vehicle. #, p < 0.05 versus control miRNA. C, dysfunctional vinculin abrogated the IGF-I-induced up-regulation of Col2a1, aggrecan, and cyclin D1 expression in the metatarsal organ cultures. RNA was extracted from the metatarsals treated as in A and subjected to real-time PCR analyses for the genes indicated.
DISCUSSION
The ECM provides a cell type-specific microenvironment and is recognized and bound by integrins and cell surface transmembrane receptors. Vinculin is a major component of the focal adhesion complex that acts as a mechano-coupler and an actin-binding protein. Several functions, mostly related to cell adhesion and motility processes, have been ascribed to vinculin. Cells depleted of vinculin show reduced adhesion to various ECM proteins and increased migration rates compared with wild-type cells (12–14). Despite profound roles in cell adhesion and motility, little is known about the tissue-specific functions of vinculin.
Because vinculin-knock-out mice die in the embryonic stage before skeletogenesis is initiated (12), the roles of vinculin in skeletal development have remained unclear. Results shown here clearly indicate that vinculin plays a critical role in chondrogenesis. Impaired functioning of vinculin in chondrocytes caused the decreased expression of Col2a1 and aggrecan, which are markers for early chondrocytes (Figs. 5 and 6, E and F). It was interesting that the expression of Sox9 was retained both in the vinculin-trapped clone and in cells with vinculin knocked down because Sox9 is a central transcription factor in chondrogenesis and regulates the expression of several chondrocyte-specific genes, including Col2a1 and aggrecan (20, 21). Although the expression of Sox6 was decreased in the cells with vinculin trapped or knocked down, this was unlikely to be the main cause for the reduced expression of Col2a1 and aggrecan, because Sox6 expression was detected after that of Col2a1 and aggrecan in primary chondrocytes (Fig. 6E). In addition, Sox5 and Sox6 are functionally redundant, and the skeletal phenotype of Sox6-knock-out mice is mild (22). Our results suggest the complexity of the regulation of Col2a1 and aggrecan expression.
In addition to the markers for early chondrocytes, the expression of those for later stages, such as Col10a1 and Runx2, was also markedly reduced by a dysfunctional vinculin, and the mineralization was impaired. Because Runx2 is known to directly control the expression of Col10a1 in hypertrophic chondrocytes, the reduced expression of Runx2 is likely to be a cause for the decreased level of Col10a1. The activity of the Runx2 protein also might be impaired, because the suppression of Col10a1 was so remarkable.
The impaired differentiation of chondrocytes with a dysfunctional vinculin indicates the critical roles of the signaling from the ECM in chondrogenesis. The importance of the signaling from the ECM in chondrogenesis was also suggested by the chondrodysplasia-like phenotype of chondrocyte-specific β1 integrin-knock-out mice and integrin-linked kinase-knock-out mice (23, 24). However, reduced proliferation of the chondrocytes was the main cause for the skeletal phenotype in these mice, and the expression of chondrocyte-specific genes, such as Col2a1, was comparable with that in wild-type mice. Thus, the effects of the knockdown of vinculin on chondrogenesis seem to be different from those of β1 integrin or integrin-linked kinase.
Here we applied gene trapping in chondrocytic ATDC5 cells to identify vinculin as involved in chondrocytic differentiation. ATDC5 cells differentiate into mature chondrocytes in several weeks, which enables one to screen the trap clones by phenotype in a relatively short time. On the other hand, gene trapping in ATDC5 cells also has limitations; because the wild-type alleles remain, clones in which some recessive genes are trapped will be missed in the screening by phenotype. However, when the trapped genes have a dosage effect or the trapping results in a fusion protein with a dominant effect, one can expect the clones to exhibit a unique phenotype in terms of chondrocytic differentiation. In clone 4-17, Western blotting using anti-vinculin antibodies demonstrated that the phenotypical change of the clone was unlikely to be caused by a gene dosage effect (Fig. 1D). Instead, we assume that the truncated vinculin derived from the trapped allele has acquired a dominant effect (Fig. 3). The truncated mutant possesses the head domain (Vh) but lacks the tail domain of vinculin (Vt). Wild-type vinculin binds to the integrin-binding protein talin and α-actinin at Vh as well as to actin filaments and paxillin at Vt (25). The conformation of vinculin switches between an inactive “closed” state and an activated “open” state (26–28). In the “closed” state, Vh and Vt bind to each other, masking the binding sites for actin and talin. In the “open” state, Vh can bind to talin and α-actinin, and Vt can bind to actin filaments, thus supporting the linkage between the actin cytoskeleton and integrin adhesion junctions. It was recently reported that the expression of a recombinant Vh domain peptide in smooth muscle tissue inhibited the acetylcholine-induced recruitment of endogenous vinculin to the membrane and the interaction of vinculin with talin and also inhibited vinculin phosphorylation (29). These results indicated that the recombinant Vh peptide inhibited the function of endogenous vinculin. In our study as well, the truncated mutant in clone 4-17 exhibited stronger affinity for talin than did wild-type vinculin, and the mutant appeared to inhibit the interaction between the wild-type vinculin and talin (Fig. 3, B and I). This inhibition might be a mechanism for the dominant negative effect of the truncated mutant.
In our knockdown experiments, we used gene-specific siRNAs and miRNAs for vinculin. To verify that the effects of vinculin-specific siRNAs and miRNAs were not caused by the suppressed expression of so-called “off-targets,” we confirmed that the effects could be cancelled by co-transfection of the vectors encoding the cDNA for wild-type vinculin (Figs. 3F, 6 (A and B), and 7 (A and B)). Therefore, we prefer to think that the observed effects of the siRNAs and miRNAs were exerted by the decreased expression of vinculin, although we cannot completely exclude the possibility of off-target effects.
Adenoviral transfer of the vinculin-specific miRNA to the metatarsal rudiments markedly reduced the growth and suppressed the expression of Col2a1 and aggrecan. Because the direct fluorescence of EmGFP in the sections cut from the rudiments demonstrated the viral infection in chondrocytes as well as in perichondrium (supplemental Fig. S4), the suppressed expression of vinculin in chondrocytes was likely to result in the impaired differentiation and the blunted growth of the rudiments. It is possible that the knockdown of vinculin in perichondrium also affected the growth and chondrocyte differentiation by influencing the production of some humoral factors there.
Our results demonstrated that the impaired functioning of vinculin affected multiple signaling pathways involved in chondrocyte differentiation. When primary chondrocytes matured, the phosphorylation of FAK, ERK1/2, SHP2, Akt, p38 MAPK, and Smads was increased, suggesting the involvement of the signaling mediated by these molecules in chondrogenesis (Fig. 8A). Activation of ERK1/2 through expression of a constitutively active MEK1 resulted in achondroplasia-like dwarfism in mice (30), indicating the critical role of the MEK/ERK pathway in chondrogenesis. SHP2 encoded by the PTPN11 gene has been implicated in signal transduction initiated by multiple growth factor and cytokine receptors (31, 32). In humans, mutations in PTPN11 are responsible for Noonan syndrome and LEOPARD syndrome, characterized by clinical features including skeletal abnormalities (33). As for the PI3K/Akt pathway, studies suggested that it is required for normal hypertrophic cell differentiation and endochondral bone growth (34, 35). In our study, we found that a dysfunctional vinculin affected the MEK/ERK pathway and PI3K/Akt pathway (Fig. 8, B–E). Previous studies suggested SHP2 to be a positive regulator in mitogenic signal transduction upstream of ERK1/2 (36, 37). However, the phosphorylation of SHP2 at Tyr-542, which is responsible for the activation of the molecule, was unexpectedly increased by the knockdown of vinculin (Fig. 8, B and C). It was previously demonstrated that SHP2 was co-immunoprecipitated with vinculin and talin, suggesting that SHP2 associates with the focal adhesion complex (38). Thus, we speculate that the disruption of vinculin interferes with the function of SHP2 in the activation of ERK1/2. Phosphorylation of Akt also has been reported to be regulated by SHP2 (39).
IGF-I is known to regulate the expression of Col2a1 and aggrecan in chondrocytes via the PI3K/Akt and MEK/ERK pathways (40). Given that the trapping or knockdown of vinculin resulted in the reduced expression of Col2a1 and aggrecan and affected PI3K/Akt and MEK/ERK signaling, we speculated that the loss of vinculin function reduced the responsiveness of chondrocytes to IGF-I. Consistent with this notion, the IGF-I-induced growth and the up-regulation of Col2a1 and aggrecan expression were blunted in metatarsal rudiments with the vinculin gene knocked down (Fig. 9), suggesting that vinculin regulates chondrocyte differentiation at least partly through an impact on IGF-I signaling.
Of note, activation of the p38 MAPK pathway in differentiating chondrocytes also was impaired by the loss of vinculin function (Fig. 8, B–E). The importance of the p38 MAPK pathway in chondrogenesis was previously suggested by experiments with a transgenic mouse in which a dominant negative form of p38 MAPK was overexpressed in cartilage, resulting in shortened limbs (41). In addition, it was reported that hypoxia induced the expression of Col2a1 and the production of glycosaminoglycan in a mesenchymal cell line, C3H10T1/2, via activation of the p38 MAPK pathway independently of Sox9 (42). It has also been suggested that p38 MAPK signaling is required for hypertrophic chondrocyte differentiation (43). Considering these observations, the reduced activation of the p38 MAPK pathway also is likely to be responsible for the impaired chondrocytic differentiation caused by the dysfunctional vinculin.
In conclusion, we have demonstrated the tissue-specific function of vinculin in cartilage, by which vinculin controls chondrocytic differentiation. Our results indicate that vinculin has pleiotropic roles in chondrogenesis and regulates the expression of chondrocyte-specific genes via the integration of various signaling pathways. The orchestration of the signaling of humoral factors and the ECM by vinculin appears to be critical to chondrogenesis and should be considered a factor in the regeneration of cartilage.
Supplementary Material
Acknowledgments
We thank Drs. Noriyuki Tsumaki and Yoshinao Wada for discussions and Dr. Yukinao Shibukawa for technical advice.
This work was supported in part by Grants-in-aid for Scientific Research from the Ministry of Education, Science, and Culture of Japan (to T. M.) and a grant from the Foundation of Growth Science (to T. M.).

This article contains supplemental Table S1 and Figs. S1–S6.
- ECM
- extracellular matrix
- IGF
- insulin-like growth factor
- FN
- fibronectin
- Col II
- type II collagen
- miRNA
- microRNA
- FAK
- focal adhesion kinase
- Vh
- vinculin head domain
- Vt
- vinculin tail domain
- aa
- amino acids
- E15.5 and E18.5
- embryonic day 15.5 and 18.5, respectively.
REFERENCES
- 1. Zuscik M. J., Hilton M. J., Zhang X., Chen D., O'Keefe R. J. (2008) Regulation of chondrogenesis and chondrocyte differentiation by stress. J. Clin. Invest. 118, 429–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kronenberg H. M. (2003) Developmental regulation of the growth plate. Nature 423, 332–336 [DOI] [PubMed] [Google Scholar]
- 3. Stanford W. L., Cohn J. B., Cordes S. P. (2001) Gene trap mutagenesis. Past, present, and beyond. Nat. Rev. Genet. 2, 756–768 [DOI] [PubMed] [Google Scholar]
- 4. Shukunami C., Shigeno C., Atsumi T., Ishizeki K., Suzuki F., Hiraki Y. (1996) Chondrogenic differentiation of clonal mouse embryonic cell line ATDC5 in vitro. Differentiation-dependent gene expression of parathyroid hormone (PTH)/PTH-related peptide receptor. J. Cell Biol. 133, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ihara-Watanabe M., Uchihashi T., Miyauchi Y., Sakai N., Yamagata M., Ozono K., Michigami T. (2004) Involvement of phosphoinositide 3-kinase signaling pathway in chondrocytic differentiation of ATDC5 cells. Application of a gene trap mutagenesis. J. Cell. Biochem. 93, 418–426 [DOI] [PubMed] [Google Scholar]
- 6. Uchihashi T., Kimata M., Tachikawa K., Koshimizu T., Okada T., Ihara-Watanabe M., Sakai N., Kogo M., Ozono K., Michigami T. (2007) Involvement of nuclear factor I transcription/replication factor in the early stage of chondrocytic differentiation. Bone 41, 1025–1035 [DOI] [PubMed] [Google Scholar]
- 7. Jones P., Jackson P., Price G. J., Patel B., Ohanion V., Lear A. L., Critchley D. R. (1989) Identification of a talin binding site in the cytoskeletal protein vinculin. J. Cell Biol. 109, 2917–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Johnson R. P., Craig S. W. (1995) F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature 373, 261–264 [DOI] [PubMed] [Google Scholar]
- 9. Johnson R. P., Craig S. W. (1995) The carboxyl-terminal tail domain of vinculin contains a cryptic binding site for acidic phospholipids. Biochem. Biophys. Res. Commun. 210, 159–164 [DOI] [PubMed] [Google Scholar]
- 10. Turner C. E., Glenney J. R., Jr., Burridge K. (1990) Paxillin. A new vinculin-binding protein present in focal adhesions. J. Cell Biol. 111, 1059–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu W., Coll J. L., Adamson E. D. (1998) Rescue of the mutant phenotype by reexpression of full-length vinculin in null F9 cells. Effects on cell locomotion by domain-deleted vinculin. J. Cell Sci. 111, 1535–1544 [DOI] [PubMed] [Google Scholar]
- 12. Xu W., Baribault H., Adamson E. D. (1998) Vinculin knockout results in heart and brain defects during embryonic development. Development 125, 327–337 [DOI] [PubMed] [Google Scholar]
- 13. Coll J. L., Ben-Ze'ev A., Ezzell R. M., Rodríguez Fernández J. L., Baribault H., Oshima R. G., Adamson E. D. (1995) Targeted disruption of vinculin genes in F9 and embryonic stem cells changes cell morphology, adhesion, and locomotion. Proc. Natl. Acad. Sci. U.S.A. 92, 9161–9165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Volberg T., Geiger B., Kam Z., Pankov R., Simcha I., Sabanay H., Coll J. L., Adamson E., Ben-Ze'ev A. (1995) Focal adhesion formation by F9 embryonal carcinoma cells after vinculin gene disruption. J. Cell Sci. 108, 2253–2260 [DOI] [PubMed] [Google Scholar]
- 15. Lefebvre V., Garofalo S., Zhou G., Metsäranta M., Vuorio E., De Crombrugghe B. (1994) Characterization of primary cultures of chondrocytes from type II collagen/β-galactosidase transgenic mice. Matrix Biol. 14, 329–335 [DOI] [PubMed] [Google Scholar]
- 16. Haaijman A., D'Souza R. N., Bronckers A. L., Goei S. W., Burger E. H. (1997) OP-1 (BMP-7) affects mRNA expression of type I, II, X collagen, and matrix Gla protein in ossifying long bones in vitro. J. Bone Miner. Res. 12, 1815–1823 [DOI] [PubMed] [Google Scholar]
- 17. Mainguy G., Montesinos M. L., Lesaffre B., Zevnik B., Karasawa M., Kothary R., Wurst W., Prochiantz A., Volovitch M. (2000) An induction gene trap for identifying a homeoprotein-regulated locus. Nat. Biotechnol. 18, 746–749 [DOI] [PubMed] [Google Scholar]
- 18. Lopez J., John S. W., Tenev T., Rautureau G. J., Hinds M. G., Francalanci F., Wilson R., Broemer M., Santoro M. M., Day C. L., Meier P. (2011) CARD-mediated autoinhibition of cIAP1's E3 ligase activity suppresses cell proliferation and migration. Mol. Cell 42, 569–583 [DOI] [PubMed] [Google Scholar]
- 19. Li F., Lu Y., Ding M., Napierala D., Abbassi S., Chen Y., Duan X., Wang S., Lee B., Zheng Q. (2011) Runx2 contributes to murine Col10a1 gene regulation through direct interaction with its cis-enhancer. J. Bone Miner. Res. 26, 2899–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lefebvre V., Huang W., Harley V. R., Goodfellow P. N., de Crombrugghe B. (1997) SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro-α1(II) collagen gene. Mol. Cell. Biol. 17, 2336–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sekiya I., Tsuji K., Koopman P., Watanabe H., Yamada Y., Shinomiya K., Nifuji A., Noda M. (2000) SOX9 enhances aggrecan gene promoter/enhancer activity and is up-regulated by retinoic acid in a cartilage-derived cell line, TC6. J. Biol. Chem. 275, 10738–10744 [DOI] [PubMed] [Google Scholar]
- 22. Smits P., Li P., Mandel J., Zhang Z., Deng J. M., Behringer R. R., de Crombrugghe B., Lefebvre V. (2001) The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev. Cell 1, 277–290 [DOI] [PubMed] [Google Scholar]
- 23. Aszodi A., Hunziker E. B., Brakebusch C., Fässler R. (2003) β1 integrins regulate chondrocyte rotation, G1 progression, and cytokinesis. Genes Dev. 17, 2465–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Terpstra L., Prud'homme J., Arabian A., Takeda S., Karsenty G., Dedhar S., St-Arnaud R. (2003) Reduced chondrocyte proliferation and chondrodysplasia in mice lacking the integrin-linked kinase in chondrocytes. J. Cell Biol. 162, 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cohen D. M., Chen H., Johnson R. P., Choudhury B., Craig S. W. (2005) Two distinct head-tail interfaces cooperate to suppress activation of vinculin by talin. J. Biol. Chem. 280, 17109–17117 [DOI] [PubMed] [Google Scholar]
- 26. Ziegler W. H., Liddington R. C., Critchley D. R. (2006) The structure and regulation of vinculin. Trends Cell Biol. 16, 453–460 [DOI] [PubMed] [Google Scholar]
- 27. Chen H., Choudhury D. M., Craig S. W. (2006) Coincidence of actin filaments and talin is required to activate vinculin. J. Biol. Chem. 281, 40389–40398 [DOI] [PubMed] [Google Scholar]
- 28. Bakolitsa C., Cohen D. M., Bankston L. A., Bobkov A. A., Cadwell G. W., Jennings L., Critchley D. R., Craig S. W., Liddington R. C. (2004) Structural basis for vinculin activation at sites of cell adhesion. Nature 430, 583–586 [DOI] [PubMed] [Google Scholar]
- 29. Greenberg R. S., Bernstein A. M., Benezra M., Gelman I. H., Taliana L., Masur S. K. (2006) FAK-dependent regulation of myofibroblast differentiation. FASEB J. 20, 1006–1008 [DOI] [PubMed] [Google Scholar]
- 30. Murakami S., Balmes G., McKinney S., Zhang Z., Givol D., de Crombrugghe B. (2004) Constitutive activation of MEK1 in chondrocytes causes Stat1-independent achondroplasia-like dwarfism and rescues the Fgfr3-deficient mouse phenotype. Genes Dev. 18, 290–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Matozaki T., Murata Y., Saito Y., Okazawa H., Ohnishi H. (2009) Protein tyrosine phosphatase SHP-2. A proto-oncogene product that promotes Ras activation. Cancer Sci. 100, 1786–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chong Z. Z., Maiese K. (2007) The Src homology 2 domain tyrosine phosphatases SHP-1 and SHP-2. Diversified control of cell growth, inflammation, and injury. Histol. Histopathol. 22, 1251–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gelb B. D., Tartaglia M. (2006) Noonan syndrome and related disorders. Dysregulated RAS-mitogen activated protein kinase signal transduction. Hum. Mol. Genet. 15, R220–R226 [DOI] [PubMed] [Google Scholar]
- 34. Ulici V., Hoenselaar K. D., Gillespie J. R., Beier F. (2008) The PI3K pathway regulates endochondral bone growth through control of hypertrophic chondrocyte differentiation. BMC Dev. Biol. 8, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fujita T., Azuma Y., Fukuyama R., Hattori Y., Yoshida C., Koida M., Ogita K., Komori T. (2004) Runx2 induces osteoblast and chondrocyte differentiation and enhances their migration by coupling with PI3K-Akt signaling. J. Cell Biol. 166, 85–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tang T. L., Freeman R. M., Jr., O'Reilly A. M., Neel B. G., Sokol S. Y. (1995) The SH2-containing protein-tyrosine phosphatase SH-PTP2 is required upstream of MAP kinase for early Xenopus development. Cell 80, 473–483 [DOI] [PubMed] [Google Scholar]
- 37. Milarski K. L., Saltiel A. R. (1994) Expression of catalytically inactive Syp phosphatase in 3T3 cells blocks stimulation of mitogen-activated protein kinase by insulin. J. Biol. Chem. 269, 21239–21243 [PubMed] [Google Scholar]
- 38. MacGillivray M., Herrera-Abreu M. T., Chow C. W., Shek C., Wang Q., Vachon E., Feng G. S., Siminovitch K. A., McCulloch C. A., Downey G. P. (2003) The protein-tyrosine phosphatase SHP-2 regulates interleukin-1-induced ERK activation in fibroblasts. J. Biol. Chem. 278, 27190–27198 [DOI] [PubMed] [Google Scholar]
- 39. Ivins Zito C., Kontaridis M. I., Fornaro M., Feng G. S., Bennett A. M. (2004) SHP-2 regulates the phosphatidylinositide 3-kinase/Akt pathway and suppresses caspase 3-mediated apoptosis. J. Cell. Physiol. 199, 227–236 [DOI] [PubMed] [Google Scholar]
- 40. Yin W., Park J. I., Loeser R. F. (2009) Oxidative stress inhibits insulin-like growth factor-I induction of chondrocyte proteoglycan synthesis through differential regulation of phosphatidylinositol 3-kinase-Akt and MEK-ERK MAPK signaling pathways. J. Biol. Chem. 284, 31972–31981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Namdari S., Wei L., Moore D., Chen Q. (2008) Reduced limb length and worsened osteoarthritis in adult mice after genetic inhibition of p38 MAP kinase activity in cartilage. Arthritis Rheum. 58, 3520–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hirao M., Tamai N., Tsumaki N., Yoshikawa H., Myoui A. (2006) Oxygen tension regulates chondrocyte differentiation and function during endochondral ossification. J. Biol. Chem. 281, 31079–31092 [DOI] [PubMed] [Google Scholar]
- 43. Stanton L. A., Sabari S., Sampaio A. V., Underhill T. M., Beier F. (2004) p38 MAP kinase signaling is required for hypertrophic chondrocyte differentiation. Biochem. J. 378, 53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.