Background: Gap junctions are intercellular plasma membrane domains enriched in channels that provide direct communication between adjacent cells.
Results: The gap junction channel protein connexin 43 is posttranslationally modified by SUMOylation.
Conclusion: SUMOylation of connexin 43 is a novel mechanism for regulating gap junctions.
Significance: The study has important implications for understanding how gap junctions are regulated under normal and pathological conditions.
Keywords: Cell Junctions, Connexin, Gap Junctions, Posttranslational Modification, SUMO, Ubiquitin
Abstract
SUMOylation is a posttranslational modification in which a member of the small ubiquitin-like modifier (SUMO) family of proteins is conjugated to lysine residues in specific target proteins. Most known SUMOylation target proteins are located in the nucleus, but there is increasing evidence that SUMO may also be a key determinant of many extranuclear processes. Gap junctions consist of arrays of intercellular channels that provide direct transfer of ions and small molecules between adjacent cells. Gap junction channels are formed by integral membrane proteins called connexins, of which the best-studied isoform is connexin 43 (Cx43). Here we show that Cx43 is posttranslationally modified by SUMOylation. The data suggest that the SUMO system regulates the Cx43 protein level and the level of functional Cx43 gap junctions at the plasma membrane. Cx43 was found to be modified by SUMO-1, -2, and -3. Evidence is provided that the membrane-proximal lysines at positions 144 and 237, located in the Cx43 intracellular loop and C-terminal tail, respectively, act as SUMO conjugation sites. Mutations of lysine 144 or lysine 237 resulted in reduced Cx43 SUMOylation and reduced Cx43 protein and gap junction levels. Altogether, these data identify Cx43 as a SUMOylation target protein and represent the first evidence that gap junctions are regulated by the SUMO system.
Introduction
Gap junctions are intercellular plasma membrane domains containing arrays of channels that provide exchange of ions and small molecules between neighboring cells (1). Gap junctions have fundamental roles in excitable tissues by allowing electrical transmission between adjacent cells (2, 3). Gap junction channel proteins are also expressed in nearly all cell types in non-excitable tissues and have critical roles in development, growth control, differentiation, and metabolic homeostasis (4, 5). Dysregulation of gap junctional intercellular communication has been linked to multiple human diseases, including heart failure, deafness, skin disorders, and neuropathologies (6). There is also substantial evidence that loss of cell-cell communication via gap junctions is involved in cancer development (7).
Gap junction channels are made of integral membrane proteins called connexins. The connexin gene family consists of 20 members in humans, of which the best-studied member is connexin 43 (Cx43)2 (8). Connexins are tetramembrane-spanning proteins having the N- and C termini located in the cytosol (9). Along their trafficking to the plasma membrane, individual connexins assemble into hexameric structures called connexons (10, 11). The connexons diffuse laterally in the plasma membrane until they dock with connexons in the adjacent cell membrane to form intact gap junction channels (12). The half-life of connexins is relatively short, ranging from 1.5 to 5 h in most tissue types (13, 14). During endocytosis of gap junctions, both membranes of the junction are internalized into one of the contacting cells, forming a double-membrane vesicle called an annular gap junction or connexosome (15–17). Connexins are then degraded in lysosomes (18–21).
Gap junction intercellular communication is regulated at multiple levels (22). The most rapid form of gap junction regulation involves changing the unitary conductance of single channels or altering their probability of opening. Slower forms of regulation is achieved by altering the number of channels present in gap junctions by changing the rate of transport of connexons to the plasma membrane, assembly of connexons into gap junctions, or connexin endocytosis and degradation. Posttranslational modifications of connexins play central roles in gap junction regulation (12, 23, 24). We and others have shown previously that Cx43 is posttranslationally modified by ubiquitin (25–29). There is increasing evidence that the ubiquitin system plays an important role in regulating Cx43 gap junction endocytosis as well as in the postendocytic trafficking of Cx43 to lysosomes (23).
The ubiquitin-like proteins have almost the same three-dimensional structure as ubiquitin and have similar enzymatic mechanisms for protein modification (30). The best-known ubiquitin-like protein is the small ubiquitin-related modifier (SUMO)2 (31). Yeast and invertebrates have a single SUMO-encoding gene, whereas vertebrates have four known SUMO isoforms, designated SUMO-1 to SUMO-4. Among these, only SUMO-1, -2, and -3 are expressed at the protein level. SUMO-2 and -3 share 97% sequence identity, whereas SUMO-1 shares ∼50% sequence identity with SUMO-2 and -3. The conjugation of SUMO to substrate proteins involves the sequential action of the E1 SUMO-activating enzymes and the E2 SUMO-conjugating enzyme 9 (Ubc9) (32–34). SUMOylation can affect the target proteins by multiple mechanisms; for instance, by altering the activity, localization, or turnover rate of the protein (31). The SUMO system has fundamental roles in numerous cellular processes, including DNA repair, transcription, cytoplasmic-nuclear transport, and chromosome separation (31). Most known SUMOylated proteins are located in the nucleus or perinucleus. However, important roles for SUMOylation in extranuclear processes, such as signal transduction and trafficking of membrane proteins, are emerging. Interestingly, SUMOylation has been found to play important roles in the regulation of certain integral membrane proteins, among which are the potassium channels K2P1 (35, 36) and Kv1.5 (37), the kainate receptor subunit GluR6 (38), the type I transforming growth factor β receptor (39), and insulin-like growth factor 1 receptor (40). There is increasing evidence that SUMOylation of integral membrane proteins might be important in cell growth control as well as in the control of the electrical properties of excitable cells (41).
Here, we show that Cx43 is posttranslationally modified by SUMOylation and demonstrate that the SUMO system regulates the Cx43 protein level and the level of Cx43 gap junctions at the plasma membrane. Evidence is provided that the membrane-proximal lysines at positions 144 and 237, located in the intracellular loop and in the C-terminal tail, respectively, act as SUMO conjugation sites. Collectively, these data identify Cx43 as a SUMOylation target protein and represent the first evidence that the SUMO system is involved in regulation of gap junctions.
EXPERIMENTAL PROCEDURES
Plasmids
The expression plasmid encoding rat Cx43 was a kind gift from Klaus Willecke (University of Bonn, Germany). The following plasmids were from Addgene: HA-SUMO-1 (plasmid 17359), HA-SUMO-2 (plasmid 17360), HA-SUMO-3 (plasmid 17361), HA-Ubc9 (plasmid 14438), FLAG-SENP1 (plasmid 17357), and FLAG-SENP2 (plasmid 18047). Mutations were generated by oligonucleotide-mediated site-directed mutagenesis on the basis of the conventional Kunkel's method (42). Each of the lysine residues (AAG) at positions 144 and 237 was mutated to arginine (AGG) by employing the oligonucleotides 5′-CCCTCATTTTCACCCTGCCGTGCTCTTC-3′ and 5′-CACGCGATCCCTAACGCCTTTG-3′, respectively. The introduced mutations were verified by DNA sequencing.
Antibodies
The anti-Cx43 antiserum was made in rabbits injected with a synthetic peptide consisting of the 20 C-terminal amino acids of Cx43 (43). The anti-HA antibody (6E2) was obtained from Cell Signaling Technology. Alexa Fluor 488-conjugated goat anti-rabbit IgG antibody was from Sigma. The goat anti-rabbit IgG antibody conjugated to horseradish peroxidase was obtained from Bio-Rad. The donkey anti-mouse IgG antibody conjugated to horseradish peroxidase was obtained from Immunolab.
Cell Culture and Transfection
HeLa-CCL2 cells were obtained from the ATCC. The HeLa cell clone used for quantitative dye transfer experiments was kindly provided by Dr. Jørgen Wesche (Oslo University Hospital). The cells were grown in DMEM supplemented with 10% (v/v) FBS (Invitrogen) and 2 mm l-glutamine (Sigma). Cells were plated onto 35-mm (4 × 105) or 100-mm (3,2 × 106) Petri dishes and transfected 24 h after seeding using Lipofectamine 2000 reagent (Invitrogen) according to the recommendations of the manufacturers. Prior to transfection, the medium containing FBS and l-glutamine was replaced with DMEM containing only l-glutamine. Five hours after transfection, DMEM containing only l-glutamine was replaced with DMEM containing both FBS and l-glutamine.
Cx43 SUMOylation Assay
Anti-Cx43 antibodies were covalently linked to protein A-Sepharose beads by incubating protein A-Sepharose beads in immunoprecipitation coupling buffer (dH2O, 100 mm NaHCO3, 50 mm NaCl, 1 mm PMSF (pH 8.3)) for 30 min and then mixed with anti-Cx43 antibodies and incubated for 1 h with continuous end-over-end gentle rotation. The beads were washed twice with 10 volumes of 0.2 m borate buffer (pH 9). Dimethyl pimelimidate dihydrochloride was added to a final concentration of 20 mm, and the mix was incubated for 30 min. The cross-linking reaction was terminated by washing the beads twice in 10 volumes of 0.2 m ethanolamine (pH 8). The beads were left in the last wash for 2 h with gentle mixing. The ethanolamine was discarded, and the beads were resuspended in the original volume of ice-cold PBS containing 1 mm PMSF. Forty-eight hours after transfection, HeLa cells were washed once with ice-cold PBS and added lysis buffer (PBS, 10% glycerol, 0.25% sodium deoxycholate, 0.45% sodium lauroyl sarcosine, 20 mm N-ethylmaleimide, protease and phosphatase inhibitor cocktails (Sigma), and 2 mm EDTA) for 5 min on ice. The lysates were precleaned by incubation with protein A-Sepharose beads (Invitrogen) at 4 °C for 30 min with shaking. Beads were pelleted by centrifugation at 7000 rpm for 5 min at 4 °C, and the supernatant was collected. To each sample, equal amounts of anti-Cx43 covalently linked to protein A-Sepharose beads were added. Preimmune serum from the same animal was used as a negative control. The reaction mixture was incubated at 4 °C for 2 h with shaking. The pellet was collected by centrifugation at 5000 rpm for 5 min at 4 °C and washed five times with ice-cold lysis buffer containing 20 mm N-ethylmaleimide. After the final wash, the pellet was resuspended in 15 μl Western sample buffer and heated to 95 °C for 5 min. The samples were centrifuged at 13000 rpm, and the supernatant was subjected to protein separation by 8% SDS-PAGE. Western blot analysis was performed as described below.
Western Blotting
Forty-eight hours after transfection, HeLa cells were washed with PBS and scraped in 300 μl of SDS electrophoresis sample buffer (10 mm Tris (pH 6.8), 15% w/v glycerol, 3% w/v SDS, 0.01% w/v bromphenol blue, and 5% v/v 2-mercaptoethanol). The cell lysates were sonicated and heated for 5 min at 95 °C. Samples were separated by 8% SDS-PAGE and transferred to nitrocellulose membranes as described (44). The membranes were developed with chemiluminescence (Lumiglo, Millipore) for Cx43 and β-actin and Super Signal West Femto maximum sensitivity substrate (Pierce) for anti-HA and imaged on a Bio-Rad image station. Bands were quantified using Bio-Rad Image Lab software.
Analysis of the Cx43 Detergent Solubility
The Triton X-100 solubility of Cx43 was evaluated on the basis of a method developed by VanSlyke and Musil (45), as described previously (46). Briefly, HeLa cells scraped in 1 ml of incubation buffer (136.8 mm NaCl, 5.4 mm KCl, 0.34 mm Na2HPO4, 0.35 mm KH2PO4, 0.8 mm MgSO4, 2.7 mm CaCl2, 20 mm Hepes (pH 7.4)) containing 10 mm N-ethylmaleimide and 200 μm phenylmethylsulfonyl fluoride and centrifuged at 3000 × g for 4 min at 4 °C. The cells were resuspended in 400 μl incubation buffer containing 10 mm N-ethylmaleimide, 200 μm phenylmethylsulfonyl fluoride, 22 μm leupeptin, and phosphatase mixture (Sigma Aldrich) and sonicated for 10 s. Triton X-100 was added to a final concentration of 1%, and the lysates were incubated for 30 min at 4 °C. 200 μl of the samples were centrifuged 100,000 × g for 50 min at 4 °C. The supernatant fraction was collected, and the pellet was resuspended in 200 μl of incubation buffer containing 10 mm N-ethylmaleimide and 200 μm phenylmethylsulfonyl fluoride. Immunoprecipitation of Cx43 from all the fractions was then performed, as described above.
Immunofluorescence Confocal Microscopy
HeLa cells grown in monolayer on coverslips were fixed and permeabilized with ice-cold methanol for 10 min. The cells were then rinsed twice in PBS and incubated with PBS containing 5% (w/v) dry milk and 0.1% Tween for 30 min. The cells were incubated for 1 h with anti-Cx43 (diluted 1:500), washed with PBS, and incubated with Alexa Fluor 488-conjugated goat anti-rabbit IgG antibodies (diluted 1:1000) for 1 h. The cells were then rinsed in PBS, and the nuclei were stained with Hoechst 33342 prior to mounting with Mowiol. The cells were analyzed with a LSM 710 confocal microscope (Carl Zeiss) equipped with a Plan Apochromat 63 × 1.4 NA oil immersion objective (Carl Zeiss). Images were acquired with the ZEN 2009 edition software and processed with Adobe Photoshop CS4.
Quantitative Scrape Loading Dye Transfer
Determination of gap junction intercellular communication by quantitative scrape loading was performed as described previously (47, 48). In these experiments we used a HeLa cell clone with high transfection efficiency that was well suited for functional dye transfer studies. Briefly, HeLa cells grown in 35-mm Petri dishes were washed twice with PBS and added 0.05% (w/v) Lucifer Yellow (Sigma) dissolved in PBS without Ca2+ and Mg2+. The monolayer was cut with a surgical scalpel, and after 3.5 min, the Lucifer Yellow solution was removed, and the dishes were rinsed four times with PBS. The cells were then fixed in 4% formaldehyde in PBS and mounted with a glass coverslip. Digital monochrome images were acquired by a COHU 4912 CCD camera (COHU, Inc., San Diego, CA) and a Scion LG-3 frame grabber card (Scion Corp. Frederick, MD). The levels of gap junctional intercellular communications were determined as the relative area of dye coupled cells, using National Institutes of Health image software. Exposing cells to 30 μm chlordane for 30 min results in a complete inhibition of gap junction intercellular communication. The fluorescent cells observed following such treatment have obtained the dye directly through the scrape process and was used to define zero communication. Values shown are the mean ± S.E. of four independent experiments in which 15 images were acquired for each experimental point.
RESULTS
Cx43 Is Covalently Modified by SUMO-1, -2, and -3
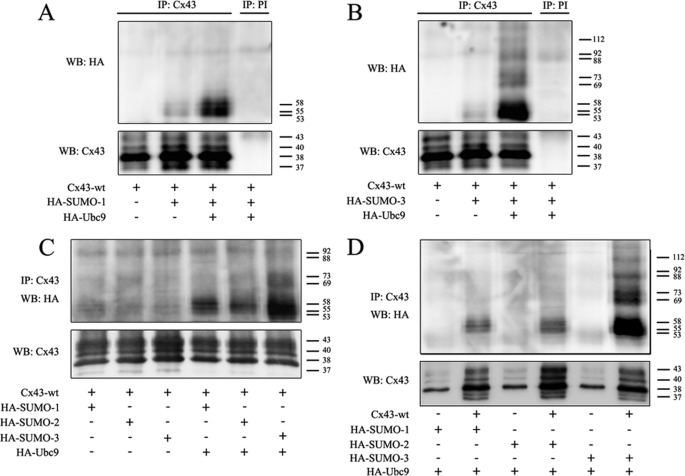
To determine whether Cx43 is SUMOylated, HeLa cells were cotransfected with plasmids encoding Cx43 and HA-tagged SUMO-1. Cell lysates were subjected to immunoprecipitation using antibodies against Cx43, followed by Western blotting using antibodies against Cx43 or HA. In accordance with previous studies, the immunoprecipitated Cx43 formed three major bands on SDS-PAGE (designated Cx43-P0, -P1, and -P2) with molecular masses of 38, 40, and 43 kDa, respectively, as well as a weaker band of 37 kDa (Fig. 1A) (28, 29). The anti-HA antibody detected three weak bands. These bands were not detected when cells were transfected with Cx43 alone or when the anti-Cx43 antibody was replaced with preimmune serum during immunoprecipitation, indicating that these bands represent SUMOylated Cx43 (Fig. 1A). Moreover, the intensity of the anti-HA bands was increased when cells were cotransfected with the SUMO E2 conjugating enzyme Ubc9, supporting the conclusion that these bands indeed represent SUMOylated Cx43 (Fig. 1A). The covalent attachment of SUMO to a target protein results in an increase of the molecular mass of the protein by 15–25 kDa (31). The approximate molecular mass of the bands detected by the HA antibody were 53, 55, and 58 kDa, suggesting that these bands represent SUMOylated forms of Cx43-P0, -P1, and -P2, respectively.
FIGURE 1.
Cx43 is post-translationally modified by SUMO-1, -2, and -3. A, HeLa cells were transfected with Cx43 alone or in combination with HA-SUMO-1 and HA-Ubc9. Cell lysates were subjected to immunoprecipitation (IP) using anti-Cx43 antibodies or preimmune serum (PI) as negative control. SUMOylated Cx43 was detected by Western blotting (WB) using anti-HA antibodies. The blot was stripped and reprobed using anti-Cx43 antibodies. B, as in A, but HA-SUMO-1 was replaced with HA-SUMO-3. C, HeLa cells were cotransfected with Cx43 and HA-SUMO-1, HA-SUMO-2, or HA-SUMO-3 with or without HA-Ubc9. Cell lysates were subjected to immunoprecipitation using anti-Cx43 antibodies. SUMOylated Cx43 was detected by Western blotting using anti-HA antibodies. The blot was stripped and reprobed using anti-Cx43 antibodies. D, HeLa cells were cotransfected with Cx43 or empty vector together with HA-SUMO-1, HA-SUMO-2, or HA-SUMO-3 and HA-Ubc9. Cell lysates were subjected to immunoprecipitation using an antibody against Cx43. SUMOylated Cx43 was detected by Western blotting using anti-HA antibodies. The blot was stripped and reprobed using anti-Cx43 antibodies. Molecular mass in kDa is indicated.
SUMO-3 has ∼50% sequence homology with SUMO-1. In contrast to SUMO-1, SUMO-3 is able to form polySUMOylation chains on the target protein (31). As shown in Fig. 1B, Cx43 was found to be covalently modified with SUMO-3. Cx43 conjugated to SUMO-3 was detected as three major bands with molecular mass of 53, 55 and 58 kDa, respectively, in accordance with the theoretical molecular mass of monoSUMOylated forms of Cx43-P0, -P1 and -P2. In addition, several slower-migrating HA-immunoreactive double bands were observed. These bands had a difference in relative molecular mass equivalent to the mass of one SUMO protein. These observations suggest that multiple SUMO-3 proteins are conjugated to Cx43, either in the form of multiple monoSUMOylation (single SUMO-3 proteins are conjugated to Cx43 at multiple sites), as polySUMOylation (a poly-SUMO-3 chain is conjugated to Cx43 on one site), or a combination of these two types of SUMO modifications.
To analyze the relative levels of the Cx43 protein pools modified with SUMO-1 and SUMO-3, immunoprecipitates containing Cx43 modified with HA-SUMO-1 and HA-SUMO-3 were applied on the same SDS-PAGE gel. The level of Cx43 modified with SUMO-3 was similar to the level of SUMO-1-conjugated Cx43 in cells not cotransfected with Ubc9 (Fig. 1C). However, in response to Ubc9 cotransfection, the level of the Cx43 pool modified by SUMO-3 was found to be considerably higher than the Cx43 pool modified by SUMO-1. We also examined whether Cx43 is modified with SUMO-2, which shares 97% sequence homology with SUMO-3. Interestingly, Cx43 was indeed found to be modified with SUMO-2, but the level of this pool of Cx43 was lower compared with the Cx43 pool modified with SUMO-1 or SUMO-3 (Fig. 1C). Moreover, we were unable to detect multiple SUMO bands with higher molecular weights, suggesting that SUMO-2 conjugates to Cx43 in a monoSUMOylated manner.
To verify that the bands detected by the anti-HA antibodies represent SUMOylation of Cx43 and not SUMOylation of proteins that are nonspecifically immunoprecipitated by the anti-Cx43 antibodies, plasmids expressing Cx43 were replaced by empty plasmids. Under these conditions, no anti-HA bands were detected (Fig. 1D). These results strongly indicate that the bands detected by the anti-HA antibodies represent SUMOylated forms of Cx43. In accordance with previous studies, HeLa-CCL2 cells expressed low levels of endogenous Cx43 (49).
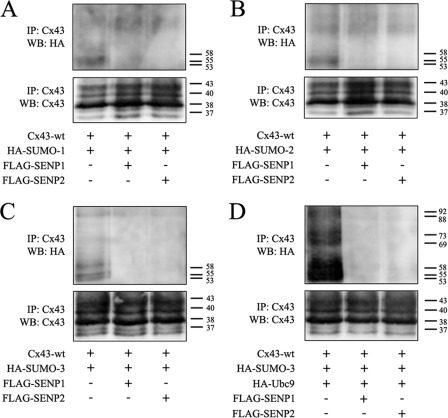
SUMOylation is a reversible process, and deconjugation of SUMO from the substrate protein is mediated by SUMO/sentrin-specific peptidase (SENP)1–3 and SENP5–7, of which SENP1 and -2 are the best-studied (50). SENP1 and -2 counteracted the conjugation of all three SUMO isoforms to Cx43 (Fig. 2, A–C). As shown in Fig. 2D, SENP1 and -2 also counteracted the conjugation of SUMO-3 to Cx43 under conditions where Ubc9 was overexpressed. Altogether, these results indicate that Cx43 is covalently modified by SUMO-1, SUMO-2, and SUMO-3. The data further indicate that Cx43 is modified by multiple SUMO-3 peptides but only single SUMO-1 and SUMO-2 peptides.
FIGURE 2.
Cx43 is deSUMOylated by SENP1/2. HeLa cells were cotransfected with Cx43 and HA-SUMO-1 (A), HA-SUMO-2 (B), or HA-SUMO-3 (C) with or without FLAG-SENP1 or FLAG-SENP2. Cell lysates were subjected to immunoprecipitation (IP) using an antibody against Cx43. SUMOylated Cx43 was detected by Western blotting (WB) using anti-HA antibodies. The blots were stripped and reprobed using anti-Cx43 antibodies. D, HeLa cells were cotransfected with Cx43, HA-SUMO3, and HA-Ubc9, with or without FLAG-SENP1 or FLAG-SENP2. Cell lysates were subjected to immunoprecipitation using anti-Cx43 antibodies. SUMOylated Cx43 was detected by Western blotting using anti-HA antibodies. The blot was stripped and reprobed using anti-Cx43 antibodies. Molecular mass in kDa is indicated.
The SUMO System Regulates Cx43 Gap Junctions
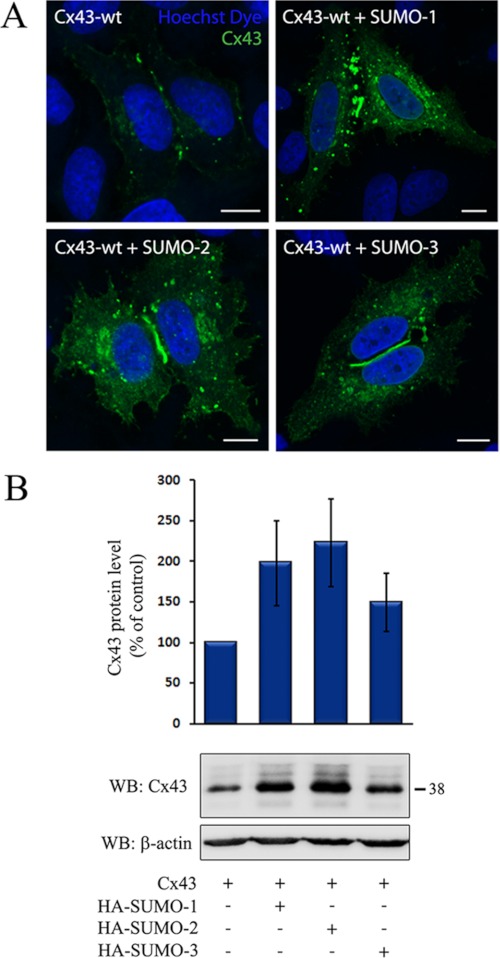
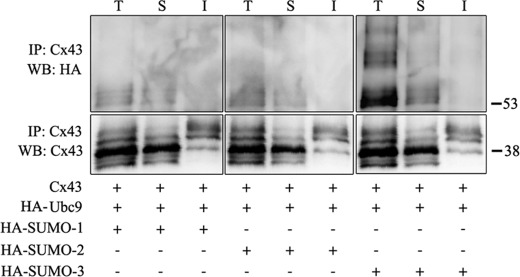
To investigate the role of SUMO in the regulation of Cx43 gap junctions, the effect of overexpression of SUMO-1, -2, or- 3 on Cx43 gap junction levels was determined. Importantly, overexpression of SUMO resulted in increased levels of Cx43 gap junctions at the plasma membrane, as determined by confocal microscopy (Fig. 3A). Increased Cx43 staining was also observed in intracellular vesicular structures. As determined by Western blotting, cotransfection of any of the three SUMO paralogs with Cx43 was associated with increased Cx43 protein levels (Fig. 3B). Among the three SUMO paralogs, SUMO-1 and SUMO-2 had the strongest effect on the Cx43 level, causing an approximate 2-fold increase, whereas cotransfection with SUMO-3 increased the Cx43 protein level by ∼50%. On the basis of these observations, we conclude that the SUMO system is involved in controlling the total cellular Cx43 protein level and Cx43 localized in gap junctions at the plasma membrane. To elucidate which subcellular pool of Cx43 is conjugated with SUMO, the detergent resistance of the SUMOylated Cx43 protein pool was determined. Cx43 organized in gap junction plaques is insoluble in Triton X-100 at 4 °C, whereas Cx43 not organized in gap junctions is Triton X-100-soluble (45). The majority of the SUMOylated form of Cx43 was found to be soluble in Triton X-100 (Fig. 4). These observations suggest that SUMOylated Cx43 is not organized in functional gap junctions.
FIGURE 3.
Cx43 is regulated by the SUMO system. HeLa cells were transfected with Cx43 alone or together with HA-SUMO-1, -2, or -3. A, cells were stained with anti-Cx43 antibodies and visualized using confocal microscopy. Scale bars = 10 μm. B, cell lysates were prepared, and equal amounts of total cell protein were subjected to SDS-PAGE. Cx43 was detected by Western blotting (WB) using Cx43 antibodies. The blot was stripped and reprobed with anti-β-actin antibodies. The intensities of the Cx43 bands were measured and normalized to the β-actin levels in the same samples. The total Cx43 protein level in cells cotransfected with Cx43 and HA-SUMO-1, -2, or -3 relative to cells transfected with only Cx43 is shown. Values shown are the mean ± S.D. of three independent experiments. Molecular mass in kDa is indicated.
FIGURE 4.
Analysis of the Triton X-100 solubility of SUMOylated Cx43. HeLa cells were transfected with Cx43; HA-Ubc9; and either HA-SUMO-1, -2, or -3. Cells were then subjected to a Triton X-100 solubility assay. The total cell lysate fractions (T), the Triton X-100-soluble fractions (S) and the Triton X-100-insoluble fractions (I) were subjected to immunoprecipitation using anti-Cx43 antibodies. Immunoprecipitates were subjected to SDS-PAGE and SUMOylated Cx43 was detected using anti-HA antibodies. The blots were stripped and reprobed with anti-Cx43 antibodies. Molecular mass in kDa is indicated.
Identification of Lys-144 and Lys-237 as Cx43 SUMOylation Sites
SUMOylation often occurs on a lysine residue within a consensus sequence ΨKXD/E, in which Ψ represents a large hydrophobic residue, K the acceptor lysine residue, and X any amino acid (31). The Cx43 C terminus contains nine lysine residues, whereas the intracellular loop contains 11 lysines, none of which reside within a SUMOylation consensus motif. To identify potential Cx43 SUMOylation sites, each of the lysines located in the intracellular loop and in the C-terminal tail of Cx43 was singly replaced with arginine, and the SUMOylation status of the various Cx43 mutants was then compared with the Cx43 wild type.
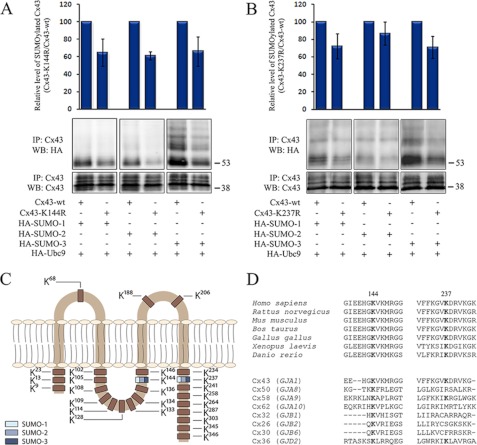
Importantly, mutation of Lys-144, located in the juxtamembrane region of the intracellular loop of Cx43, and mutation of Lys-237, located in the juxtamembrane region of the Cx43 C-terminal tail, resulted in reduced conjugation to all three SUMO paralogs (Fig. 5, A and B). Mutation of Lys-144 decreased the SUMOylation level by ∼40% for all three SUMO paralogs, whereas a reduction in SUMOylation of ∼20–25% was observed because of the Lys-237 mutation. On the basis of these experiments, we conclude that Lys-144 and Lys-237 act as Cx43 SUMO conjugation sites (Fig. 5C). Sequence comparison reveals that Lys-144 and Lys-237 are conserved through evolution (Fig. 5D). Interestingly, Lys-144 in Cx43 is also conserved in eight of the 20 known human connexin isoforms, including Cx26 and Cx32, two of the best-studied connexins beside Cx43. Moreover, Cx43 as well as six of the other connexin isoforms that harbor a lysine at position 144 contain a large hydrophobic amino acid directly downstream of the lysine residue and, thus, partly fulfill the requirement of being an inverted SUMOylation motif, as described previously for other proteins (51). Notably, among the 13 connexin isoforms that do not contain a lysine in this position, five contain an arginine residue instead of a lysine in the same position. Lys-237 is conserved in two other connexin isoforms, all of which contain a large hydrophobic amino acid directly upstream of the lysine residue (Fig. 5D). Altogether, these observations suggest that SUMOylation of connexins may be an evolutionary conserved mechanism for regulating gap junctions. Notably, our preliminary studies suggest that mutation of several other lysines in the Cx43 intracellular loop and C-terminal tail affects the Cx43 SUMOylation status, raising the possibility that multiple lysines in addition to Lys-144 and Lys-237 may act as SUMOylation sites3. The role of these lysines in the regulation of Cx43 was not investigated further in this study.
FIGURE 5.
Identification of Lys-144 and Lys-237 as Cx43 SUMOylation sites. HeLa cells were transfected with Cx43-WT (A, B), Cx43-K144R (A) or Cx43-K237R (B) in combination with HA-SUMO-1, -2, or -3 and HA-Ubc9, as indicated. Cell lysates were subjected to immunoprecipitation (IP) using anti-Cx43 antibodies. SUMOylated Cx43 was detected by Western blotting (WB) using anti-HA antibodies. The blots were stripped and reprobed using anti-Cx43 antibodies. The intensities of the HA signals were measured and normalized to the intensities of the Cx43 signals in the same samples. The SUMOylation level for Cx43-K144R and Cx43-K237R is expressed in percentage relative to Cx43-WT. Values shown are the mean ± S.D. of three independent experiments. Molecular mass in kDa is indicated. C, schematic overview of the location of Lys-144 and Lys-237 and other lysine residues in Cx43. Blue indicates that the lysine acts as a SUMO conjugation site. D, amino acid sequence of the human Cx43 regions that contain Lys-144 and Lys-237 and corresponding sequences in the indicated species homologs (upper panel) or in other connexin isoforms in humans (lower panel).
Cx43-K144R and Cx43-K237R Have a Reduced Ability to form Gap Junctions
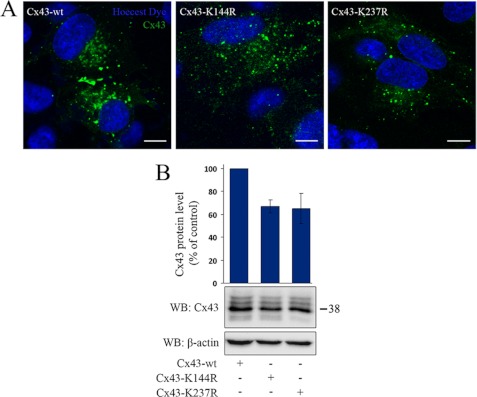
Importantly, replacement of Lys-144 or Lys-237 to arginines resulted in reduced Cx43 gap junction formation at the plasma membrane as determined by confocal microscopy (Fig. 6A). Mutation of Lys-144 or Lys-237 also resulted in reduced Cx43 protein levels, as determined by Western blotting (Fig. 6B). Taken together, these observations suggest that Lys-144 and Lys-237 play important roles in regulating the Cx43 protein level and the level of Cx43 gap junctions at the plasma membrane.
FIGURE 6.
Cx43-K144R and Cx43-K237R have a reduced ability to form gap junctions HeLa cells were transfected with Cx43-WT, Cx43-K144R or Cx43-K237R. A, cells were stained with anti-Cx43 antibodies and visualized using confocal microscopy. Scale bars = 10 μm. B, cell lysates were prepared, and equal amounts of total cell protein were subjected to SDS-PAGE. Cx43 was detected by Western blotting (WB) using anti-Cx43 antibodies. The blot was stripped and reprobed with anti-β-actin antibodies. The intensities of the Cx43 bands were measured and normalized to the β-actin levels in the same samples. The total Cx43 protein levels in cells transfected with Cx43-K144R and Cx43-K237R relative to cells transfected with Cx43-WT are shown. Values shown are the mean ± S.D. of three independent experiments. Molecular mass in kDa is indicated.
Effect of SUMOylation on Cx43 Gap Junction Channel Function
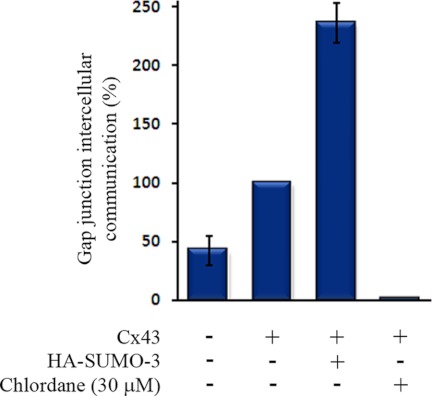
To elucidate the role of the SUMO system on Cx43 gap junction channel function, HeLa cells were transfected with Cx43 alone or in combination with HA-SUMO-3. The level of gap junction intercellular communication was determined by quantitative scrape loading dye transfer. Cotransfection of HA-SUMO-3 resulted in more than a doubling of the gap junctional communication compared with cells that were transfected with only Cx43 (Fig. 7).
FIGURE 7.
Effect of SUMO-3 on Cx43 gap junction channel function. HeLa cells were transfected with empty plasmids, Cx43 alone, or Cx43 together with HA-SUMO-3. The level of gap junctional communication was determined by quantitative scrape loading dye transfer. In accordance with previous studies, treatment of Cx43-transfected cells with chlordane (30 μm) for 30 min resulted in complete inhibition of gap junctional intercellular communication, and was used to define zero communication. Values shown are the mean ± S.E. of four independent experiments, relative to cells transfected with only Cx43. Note that cells transfected with empty plasmid were found to have low levels of gap junction intercellular communication, as they expressed low amounts of endogenous Cx43.
DISCUSSION
In this study, we demonstrate that Cx43 is posttranslationally modified and regulated by SUMOylation. The data indicate that the SUMO system has an important role in controlling the total cellular Cx43 protein level and the level of Cx43 gap junctions at the plasma membrane. Evidence is provided that the lysines at positions 144 and 237, located in the juxtamembrane regions of the Cx43 intracellular loop and C-terminal tail, respectively, act as SUMO conjugation sites. Altogether, these data represent the first evidence that SUMOylation is involved in regulation of gap junctions.
SUMO is known to play critical roles in the regulation of nuclear and perinuclear proteins (52, 53). There is also increasing evidence that SUMO is important in regulating extranuclear proteins, including proteins at the cell surface. However, only a few integral plasma membrane proteins have been shown to be modified by SUMO. Here we demonstrate that Cx43 is modified by SUMO-1, -2, and -3. The finding that all three SUMO paralogs conjugate to Cx43 raises the interesting possibility that they might affect Cx43 through distinct molecular mechanisms. Cx43 was found to be conjugated to single SUMO-1 and SUMO-2 proteins but multiple SUMO-3 proteins. Previous studies have shown that SUMO-3 is able to form polySUMOylation chains on substrate proteins in vivo (31). Thus, Cx43 may either be conjugated to single SUMO-3 peptides on multiple lysines, by a single polySUMO-3 chain, or a combination of these types of SUMO-3 modifications.
Our results indicate that overexpression of all of the three SUMO paralogs results in increased Cx43 protein levels and increased levels of gap junctions. The increase in Cx43 gap junction levels in response to overexpression of SUMO is likely to reflect an overall increase in Cx43 protein levels under these conditions, although it cannot be ruled out that SUMOylation of Cx43 might be involved in regulating the assembly of Cx43 into gap junctions or gap junction endocytosis. Replacement of the lysines in positions 144 or 237 with arginines resulted in reduced Cx43 SUMOylation, indicating that these two lysines act as Cx43 SUMO conjugation sites. As determined by confocal microscopy, Cx43-K144R and Cx43-K237R had a reduced ability to form gap junctions. At present, we do not know whether Cx43-K144R and Cx43-K237R have altered intracellular trafficking compared with Cx43-WT. It is also important to take into consideration that mutation of Lys-144 or Lys-237 could affect the Cx43 protein level and the ability of Cx43 to form gap junctions independently of the effect on the Cx43 SUMOylation status.
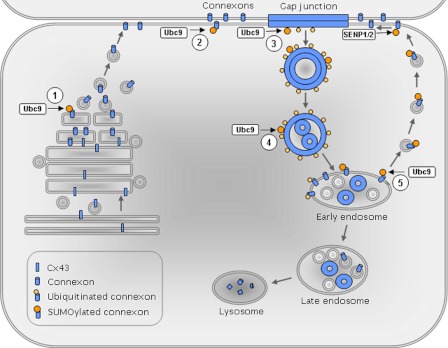
Further studies are required to determine in which subcellular compartments Cx43 SUMOylation occurs. The SUMOylated Cx43 pool was found to be soluble in Triton X-100, suggesting that the SUMOylated Cx43 is not organized in functional gap junctions. One possible scenario is that newly synthesized Cx43 undergoes SUMOylation along its trafficking from the Golgi/trans-Golgi network to the plasma membrane and then is subjected to deSUMOylation during or shortly after it has assembled into gap junction plaques (Fig. 8). We have shown previously that endocytosis of Cx43 gap junctions in response to activation of protein kinase C is associated with a loss of the Triton X-100 resistance of Cx43 organized in gap junctions (54, 55). The loss of the detergent resistance of Cx43 was found to be an early event in gap junction endocytosis. Thus, the finding that the SUMOylated Cx43 is Triton X-100-soluble might also indicate that SUMOylation of Cx43 occurs during gap junction endocytosis or along its postendocytic trafficking to lysosomes. The precise molecular mechanisms underlying the regulation of gap junctions by Cx43 SUMOylation remain to be determined. An increasing number of proteins have been shown to bind to SUMO via SUMO-interacting motifs (31). Proteins with SUMO-interacting motifs bind non-covalently to SUMOylated proteins by direct interaction with the SUMO moiety or the SUMO-substrate interface. Possibly, SUMOylation of Cx43 might result in non-covalent binding to one or more SUMO-interacting motif-containing proteins that promote the stability of Cx43. In an alternative scenario, SUMOylation of Cx43 at Lys-144 and Lys-237 might result in a conformation shift of the protein that promotes the stability of Cx43 gap junctions. A third possible scenario is that SUMOylation of Cx43 stabilizes the protein by competing with Cx43 ubiquitination.
FIGURE 8.
Working hypothesis for the regulation of Cx43 gap junctions by SUMOylation. Five possible subcellular locations for SUMOylation of Cx43 are indicated: 1) the Golgi/trans-Golgi network, 2) the plasma membrane outside gap junctions, 3) gap junctions, 4) connexosomes, and 5) early endosomes. At each of these locations, the SUMOylated Cx43 could possibly be recognized by yet unidentified SIM-containing proteins that control Cx43 trafficking and, hence, the level of Cx43 organized in gap junctions. Alternatively, SUMOylation of Cx43 might affect the level of Cx43 organized in gap junctions by competing with ubiquitination or other lysine modifications, such as acetylation. A third scenario is that SUMOylation of Cx43 affects gap junctions by altering the gap junction channel conformation.
Approximately 75% of known SUMOylation substrates are SUMOylated within a ΨKXD/E consensus motif (31). None of the lysines in the Cx43 C-terminal tail or intracellular loop reside within a SUMOylation consensus motif. Interestingly, however, Lys-237 is preceded by a valine residue, thus partly fulfilling the requirement for a SUMO consensus motif. Moreover, a valine residue is located directly upstream of Lys-144, which, hence, partly fulfills the requirement of residing within an inverted SUMOylation consensus motif (51). Importantly, Lys-144 is evolutionarily conserved in seven other connexin isoforms. Among these, six harbor a large hydrophobic amino acid directly upstream of the lysine residue. Lys-237 is conserved in two other connexin isoforms, both of which are preceded by a large hydrophobic amino acid. Taken together, these observations raise the possibility that SUMOylation could be a general mechanism for regulating connexins. Our preliminary studies suggest that also other lysines in the intracellular loop and C-terminal tail of Cx43 in addition to Lys-144 and Lys-237 might act as SUMO conjugation sites3. How SUMOylation of Cx43 at the various lysines might affect Cx43 gap junctions remains to be explored.
Our data indicate that both SENP1 and -2 induce deSUMOylation of Cx43. Notably, our preliminary studies suggest that overexpression of SENP-1 or -2 is not associated with reduced Cx43 protein levels or reduced levels of gap junctions at the plasma membrane3. Instead, overexpression of SENP1 or -2 appears to cause an increase in the Cx43 protein level. Because overexpression of SENP-1 and -2 is expected to induce global deSUMOylation, SENP1 and -2 could affect Cx43 indirectly, for instance by deSUMOylating Cx43-interacting proteins. Defining the role of SENP1 and -2 in the regulation of Cx43 gap junctions will be an important subject for future studies.
SUMOylation is tightly linked to other types of posttranslational modifications (31). In addition to being regulated by phosphorylation and ubiquitination, it was recently shown that Cx43 is also acetylated (56). An important subject for future work will be to elucidate the functional interplay between Cx43 phosphorylation, acetylation, ubiquitination, and SUMOylation in the regulation of gap junctions. To the best of our knowledge, this study represents the first evidence that the intracellular loop of Cx43 is subjected to posttranslational modification. Whether the intracellular loop of Cx43 undergoes other types of modifications in addition to SUMOylation, and how these modifications might interplay with Cx43 SUMOylation to control Cx43 gap junctions, remains to be determined.
In conclusion, this study identifies Cx43 as a SUMOylation substrate protein and demonstrates that the SUMO system is critically involved in regulating Cx43 gap junctions. SUMOylation of Cx43 is likely to have a significant impact on the control of electrical properties of excitable cells as well as on Cx43-mediated cell growth control. The finding that Cx43 is modified by SUMO adds another level to the complexity of gap junction regulation and has important implications for future studies aimed at uncovering the molecular basis underlying the dysregulation of gap junctions in human pathologies.
Acknowledgments
We thank Mette Eknæs and Merete Hektoen for excellent technical assistance. We also thank Stine A. Danielsen for assistance with DNA sequencing.
This work was supported by the Norwegian Cancer Society and the Norwegian Research Council.
A. Kjenseth, T. A. Fykerud, S. Sirnes, J. Bruun, Z. Yohannes, M. Kolberg, Y. Omori, E. Rivedal, and E. Leithe, unpublished data.
- Cx43
- connexin43
- SUMO
- small ubiquitin-related modifier
- SENP
- SUMO/sentrin-specific peptidase.
REFERENCES
- 1. Saez J. C., Berthoud V. M., Branes M. C., Martinez A. D., Beyer E. C. (2003) Plasma membrane channels formed by connexins. Their regulation and functions. Physiol. Rev. 83, 1359–1400 [DOI] [PubMed] [Google Scholar]
- 2. Severs N. J., Coppen S. R., Dupont E., Yeh H. I., Ko Y. S., Matsushita T. (2004) Gap junction alterations in human cardiac disease. Cardiovasc. Res. 62, 368–377 [DOI] [PubMed] [Google Scholar]
- 3. Connors B. W., Long M. A. (2004) Electrical synapses in the mammalian brain. Annu. Rev. Neurosci. 27, 393–418 [DOI] [PubMed] [Google Scholar]
- 4. Loewenstein W. R. (1979) Junctional intercellular communication and the control of growth. Biochim. Biophys. Acta 560, 1–65 [DOI] [PubMed] [Google Scholar]
- 5. Yamasaki H., Naus C. C. (1996) Role of connexin genes in growth control. Carcinogenesis 17, 1199–1213 [DOI] [PubMed] [Google Scholar]
- 6. Wei C. J., Xu X., Lo C. W. (2004) Connexins and cell signaling in development and disease. Annu. Rev. Cell Dev. Biol. 20, 811–838 [DOI] [PubMed] [Google Scholar]
- 7. Naus C. C., Laird D. W. (2010) Implications and challenges of connexin connections to cancer. Nat. Rev. Cancer 10, 435–441 [DOI] [PubMed] [Google Scholar]
- 8. Söhl G., Willecke K. (2003) An update on connexin genes and their nomenclature in mouse and man. Cell Commun. Adhes. 10, 173–180 [DOI] [PubMed] [Google Scholar]
- 9. Sosinsky G. E., Nicholson B. J. (2005) Structural organization of gap junction channels. Biochim. Biophys. Acta 1711, 99–125 [DOI] [PubMed] [Google Scholar]
- 10. Koval M. (2006) Pathways and control of connexin oligomerization. Trends Cell Biol. 16, 159–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Musil L. S., Goodenough D. A. (1993) Multisubunit assembly of an integral plasma membrane channel protein, gap junction connexin43, occurs after exit from the ER. Cell 74, 1065–1077 [DOI] [PubMed] [Google Scholar]
- 12. Laird D. W. (2006) Life cycle of connexins in health and disease. Biochem. J. 394, 527–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Berthoud V. M., Minogue P. J., Laing J. G., Beyer E. C. (2004) Pathways for degradation of connexins and gap junctions. Cardiovasc. Res. 62, 256–267 [DOI] [PubMed] [Google Scholar]
- 14. Hervé J. C., Derangeon M., Bahbouhi B., Mesnil M., Sarrouilhe D. (2007) The connexin turnover, an important modulating factor of the level of cell-to-cell junctional communication. Comparison with other integral membrane proteins. J. Membr. Biol. 217, 21–33 [DOI] [PubMed] [Google Scholar]
- 15. Jordan K., Chodock R., Hand A. R., Laird D. W. (2001) The origin of annular junctions. A mechanism of gap junction internalization. J. Cell Sci. 114, 763–773 [DOI] [PubMed] [Google Scholar]
- 16. Larsen W. J., Hai-Nan (1978) Origin and fate of cytoplasmic gap junctional vesicles in rabbit granulosa cells. Tissue Cell 10, 585–598 [DOI] [PubMed] [Google Scholar]
- 17. Piehl M., Lehmann C., Gumpert A., Denizot J. P., Segretain D., Falk M. M. (2007) Internalization of large double-membrane intercellular vesicles by a clathrin-dependent endocytic process. Mol. Biol. Cell 18, 337–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leithe E., Sirnes S., Fykerud T., Kjenseth A., Rivedal E. (2011) Biochim. Biophys. Acta. in press [DOI] [PubMed] [Google Scholar]
- 19. Lichtenstein A., Minogue P. J., Beyer E. C., Berthoud V. M. (2011) Autophagy. A pathway that contributes to connexin degradation. J. Cell Sci. 124, 910–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qin H., Shao Q., Igdoura S. A., Alaoui-Jamali M. A., Laird D. W. (2003) Lysosomal and proteasomal degradation play distinct roles in the life cycle of Cx43 in gap junctional intercellular communication-deficient and -competent breast tumor cells. J. Biol. Chem. 278, 30005–30014 [DOI] [PubMed] [Google Scholar]
- 21. VanSlyke J. K., Deschenes S. M., Musil L. S. (2000) Intracellular transport, assembly, and degradation of wild-type and disease-linked mutant gap junction proteins. Mol. Biol. Cell 11, 1933–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goodenough D. A., Paul D. L. (2009) Gap junctions. Cold Spring Harb. Perspect. Biol. 1, a002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kjenseth A., Fykerud T., Rivedal E., Leithe E. (2010) Regulation of gap junction intercellular communication by the ubiquitin system. Cell Signal. 22, 1267–1273 [DOI] [PubMed] [Google Scholar]
- 24. Lampe P. D., Lau A. F. (2000) Regulation of gap junctions by phosphorylation of connexins. Arch. Biochem. Biophys. 384, 205–215 [DOI] [PubMed] [Google Scholar]
- 25. Girão H., Catarino S., Pereira P. (2009) Eps15 interacts with ubiquitinated Cx43 and mediates its internalization. Exp. Cell Res. 315, 3587–3597 [DOI] [PubMed] [Google Scholar]
- 26. Kelly S. M., Vanslyke J. K., Musil L. S. (2007) Regulation of ubiquitin-proteasome system mediated degradation by cytosolic stress. Mol. Biol. Cell 18, 4279–4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Laing J. G., Beyer E. C. (1995) The gap junction protein connexin43 is degraded via the ubiquitin proteasome pathway. J. Biol. Chem. 270, 26399–26403 [DOI] [PubMed] [Google Scholar]
- 28. Leithe E., Rivedal E. (2004) Epidermal growth factor regulates ubiquitination, internalization and proteasome-dependent degradation of connexin43. J. Cell Sci. 117, 1211–1220 [DOI] [PubMed] [Google Scholar]
- 29. Leithe E., Rivedal E. (2004) Ubiquitination and down-regulation of gap junction protein connexin-43 in response to 12-O-tetradecanoylphorbol 13-acetate treatment. J. Biol. Chem. 279, 50089–50096 [DOI] [PubMed] [Google Scholar]
- 30. Hochstrasser M. (2009) Origin and function of ubiquitin-like proteins. Nature 458, 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gareau J. R., Lima C. D. (2010) The SUMO pathway. Emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Desterro J. M., Rodriguez M. S., Kemp G. D., Hay R. T. (1999) Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J. Biol. Chem. 274, 10618–10624 [DOI] [PubMed] [Google Scholar]
- 33. Desterro J. M., Thomson J., Hay R. T. (1997) Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 417, 297–300 [DOI] [PubMed] [Google Scholar]
- 34. Lee G. W., Melchior F., Matunis M. J., Mahajan R., Tian Q., Anderson P. (1998) Modification of Ran GTPase-activating protein by the small ubiquitin-related modifier SUMO-1 requires Ubc9, an E2-type ubiquitin-conjugating enzyme homologue. J. Biol. Chem. 273, 6503–6507 [DOI] [PubMed] [Google Scholar]
- 35. Plant L. D., Dementieva I. S., Kollewe A., Olikara S., Marks J. D., Goldstein S. A. (2010) One SUMO is sufficient to silence the dimeric potassium channel K2P1. Proc. Natl. Acad. Sci. U.S.A. 107, 10743–10748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rajan S., Plant L. D., Rabin M. L., Butler M. H., Goldstein S. A. (2005) Sumoylation silences the plasma membrane leak K+ channel K2P1. Cell 121, 37–47 [DOI] [PubMed] [Google Scholar]
- 37. Benson M. D., Li Q. J., Kieckhafer K., Dudek D., Whorton M. R., Sunahara R. K., Iñiguez-Lluhí J. A., Martens J. R. (2007) SUMO modification regulates inactivation of the voltage-gated potassium channel Kv1.5. Proc. Natl. Acad. Sci. U.S.A. 104, 1805–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martin S., Nishimune A., Mellor J. R., Henley J. M. (2007) SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature 447, 321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kang J. S., Saunier E. F., Akhurst R. J., Derynck R. (2008) The type I TGF-β receptor is covalently modified and regulated by sumoylation. Nat. Cell Biol. 10, 654–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sehat B., Tofigh A., Lin Y., Trocmé E., Liljedahl U., Lagergren J., Larsson O. (2010) SUMOylation mediates the nuclear translocation and signaling of the IGF-1 receptor. Sci. Signal. 3, ra10. [DOI] [PubMed] [Google Scholar]
- 41. Martin S., Wilkinson K. A., Nishimune A., Henley J. M. (2007) Emerging extranuclear roles of protein SUMOylation in neuronal function and dysfunction. Nat. Rev. Neurosci. 8, 948–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kunkel T. A., Roberts J. D., Zakour R. A. (1987) Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154, 367–382 [DOI] [PubMed] [Google Scholar]
- 43. Rivedal E., Mollerup S., Haugen A., Vikhamar G. (1996) Modulation of gap junctional intercellular communication by EGF in human kidney epithelial cells. Carcinogenesis 17, 2321–2328 [DOI] [PubMed] [Google Scholar]
- 44. Leithe E., Cruciani V., Sanner T., Mikalsen S. O., Rivedal E. (2003) Recovery of gap junctional intercellular communication after phorbol ester treatment requires proteasomal degradation of protein kinase C. Carcinogenesis 24, 1239–1245 [DOI] [PubMed] [Google Scholar]
- 45. VanSlyke J. K., Musil L. S. (2000) Analysis of connexin intracellular transport and assembly. Methods 20, 156–164 [DOI] [PubMed] [Google Scholar]
- 46. Sirnes S., Leithe E., Rivedal E. (2008) The detergent resistance of Connexin43 is lost upon TPA or EGF treatment and is an early step in gap junction endocytosis. Biochem. Biophys. Res. Commun. 373, 597–601 [DOI] [PubMed] [Google Scholar]
- 47. el-Fouly M. H., Trosko J. E., Chang C. C. (1987) Scrape-loading and dye transfer. A rapid and simple technique to study gap junctional intercellular communication. Exp. Cell Res. 168, 422–430 [DOI] [PubMed] [Google Scholar]
- 48. Opsahl H., Rivedal E. (2000) Quantitative determination of gap junction intercellular communication by scrape loading and image analysis. Cell Adhes. Commun. 7, 367–375 [DOI] [PubMed] [Google Scholar]
- 49. King T. J., Fukushima L. H., Donlon T. A., Hieber A. D., Shimabukuro K. A., Bertram J. S. (2000) Correlation between growth control, neoplastic potential and endogenous connexin43 expression in HeLa cell lines. Implications for tumor progression. Carcinogenesis 21, 311–315 [DOI] [PubMed] [Google Scholar]
- 50. Kim J. H., Baek S. H. (2009) Emerging roles of deSUMOylating enzymes. Biochim. Biophys. Acta 1792, 155–162 [DOI] [PubMed] [Google Scholar]
- 51. Matic I., Schimmel J., Hendriks I. A., van Santen M. A., van de Rijke F., van Dam H., Gnad F., Mann M., Vertegaal A. C. (2010) Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol. Cell 39, 641–652 [DOI] [PubMed] [Google Scholar]
- 52. Gill G. (2004) SUMO and ubiquitin in the nucleus. Different functions, similar mechanisms? Genes Dev. 18, 2046–2059 [DOI] [PubMed] [Google Scholar]
- 53. Schwartz D. C., Hochstrasser M. (2003) A superfamily of protein tags. Ubiquitin, SUMO and related modifiers. Trends Biochem. Sci. 28, 321–328 [DOI] [PubMed] [Google Scholar]
- 54. Leithe E., Brech A., Rivedal E. (2006) Endocytic processing of connexin43 gap junctions. A morphological study. Biochem. J. 393, 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Leithe E., Kjenseth A., Sirnes S., Stenmark H., Brech A., Rivedal E. (2009) Ubiquitylation of the gap junction protein connexin-43 signals its trafficking from early endosomes to lysosomes in a process mediated by Hrs and Tsg101. J. Cell Sci. 122, 3883–3893 [DOI] [PubMed] [Google Scholar]
- 56. Colussi C., Rosati J., Straino S., Spallotta F., Berni R., Stilli D., Rossi S., Musso E., Macchi E., Mai A., Sbardella G., Castellano S., Chimenti C., Frustaci A., Nebbioso A., Altucci L., Capogrossi M. C., Gaetano C. (2011) Nε-lysine acetylation determines dissociation from GAP junctions and lateralization of connexin 43 in normal and dystrophic heart. Proc. Natl. Acad. Sci. U.S.A. 108, 2795–2800 [DOI] [PMC free article] [PubMed] [Google Scholar]