Background: Nitrogen-starvation and other stresses induce triacylglycerol (TAG) accumulation in algae, but the relevant enzymes and corresponding signal transduction pathways are unknown.
Results: RNA-Seq and genetic analysis revealed three acyltransferases that contribute to TAG accumulation.
Conclusion: TAG synthesis results from recycling of membrane lipids and also by acylation of DAG.
Significance: The genes are potential targets for manipulating TAG hyperaccumulation.
Keywords: Algae, Bioenergy, Biofuel, Glycerolipid, Lipid Metabolism, Plant Biochemistry, RNA-Seq, Triacylglycerol
Abstract
Algae have recently gained attention as a potential source for biodiesel; however, much is still unknown about the biological triggers that cause the production of triacylglycerols. We used RNA-Seq as a tool for discovering genes responsible for triacylglycerol (TAG) production in Chlamydomonas and for the regulatory components that activate the pathway. Three genes encoding acyltransferases, DGAT1, DGTT1, and PDAT1, are induced by nitrogen starvation and are likely to have a role in TAG accumulation based on their patterns of expression. DGAT1 and DGTT1 also show increased mRNA abundance in other TAG-accumulating conditions (minus sulfur, minus phosphorus, minus zinc, and minus iron). Insertional mutants, pdat1-1 and pdat1-2, accumulate 25% less TAG compared with the parent strain, CC-4425, which demonstrates the relevance of the trans-acylation pathway in Chlamydomonas. The biochemical functions of DGTT1 and PDAT1 were validated by rescue of oleic acid sensitivity and restoration of TAG accumulation in a yeast strain lacking all acyltransferase activity. Time course analyses suggest than a SQUAMOSA promoter-binding protein domain transcription factor, whose mRNA increases precede that of lipid biosynthesis genes like DGAT1, is a candidate regulator of the nitrogen deficiency responses. An insertional mutant, nrr1-1, accumulates only 50% of the TAG compared with the parental strain in nitrogen-starvation conditions and is unaffected by other nutrient stresses, suggesting the specificity of this regulator for nitrogen-deprivation conditions.
Introduction
Rising fossil fuel prices and dwindling reserves have led to renewed interest in developing alternative fuel sources (1–7). Although biomass from terrestrial plants can be processed and used as a feedstock for generating alcohols and other fuels (8) oil from photoautotrophically grown algae or seeds represents another fuel source, with algae offering distinct advantages over traditional oil crops (9, 10). The green alga Chlamydomonas reinhardtii accumulates increased amounts of triacylglycerols (TAGs)5 under conditions of growth-inhibiting macro- or micronutrient deficiency such as sulfur (11), nitrogen (12–20), phosphorus (15), zinc, or iron deficiency (21) or other growth-inhibiting stressors such as high salt (22) or high light (23). With a sequenced genome (24) and the potential for application of metabolic engineering (25), C. reinhardtii is a powerful reference organism for discovering green algal TAG biosynthetic pathway(s) and the ways in which they are regulated by nutrient deprivation.
The lipid composition of C. reinhardtii is representative of a typical photosynthetic eukaryote with the notable exception that the betaine-containing lipid diacylglycerol-trimethylhomoserine replaces phosphatidylcholine in extraplastidic membranes (26). The genes associated with membrane lipid and TAG synthesis have been annotated in Chlamydomonas, relying primarily on BLAST analysis of the genome (1, 27). Nevertheless, the gene models tend to be incomplete, and function has not been established, especially in situations where multiple isoforms are predicted or where multiple metabolic routes exist.
Research in other eukaryotic organisms, such as yeast, Arabidopsis, and mouse, has identified several key genes in TAG synthesis. One is a diacylglycerol acyltransferase (DGAT), which catalyzes the final step in de novo TAG synthesis. The protein was identified in the mouse and was found to have sequence homology to sterol:acyl-CoA acyltransferase (28); DGATs of this type are now known as type-1 DGATs. Two other DGAT enzymes with little homology to type-1 DGATs were subsequently found in the oleaginous fungus Morteriella ramanniana; this type is classified as type-2 DGATs (29). Both type-1 and type-2 DGAT proteins are localized to the endoplasmic reticulum (30–32), and a study in Vernicia fordii (Tung tree) indicated that the two types of enzymes localize to different subdomains of the endoplasmic reticulum, implying nonredundant functions (33). Type-1 DGATs in plants are expressed in various cells and tissues, and type-2 DGATs are expressed highly in developing oilseeds (33–35), suggesting the importance of type-2 DGATs in TAG-accumulating organs. Likewise, type-2 DGAT in mouse is important since mice lacking this enzyme are severely depleted in both tissue and plasma TAG, which leads to early death (36). Genes representing a third class (type-3) of DGAT, which is a soluble cytosolic enzyme, have been identified in Arachis hypogaea (peanut) (37) and Arabidopsis thaliana (38).
Another major contributor to TAG synthesis is phospholipid:diacylglycerol acyltransferase (PDAT), which is an acyl-CoA independent enzyme that transfers the acyl group from the sn-2 position of a phospholipid to the sn-3 position of a diacylglycerol. In Saccharomyces cerevisiae, the two major contributors to TAG production in stationary and exponential cultures are type-2 DGAT and PDAT, respectively (encoded by DGA1 and LRO1); two sterol acyltransferases (ARE1 and ARE2) also contribute a small amount to the production of TAG (39, 40).
In Chlamydomonas, homology searches identified 5 type-2 DGATs, encoded by DGTT1–DGTT5. DGTT1 is up-regulated in response to nitrogen starvation, but its function could not be validated (18, 41). Nevertheless, proteomic studies of membrane fractions from Chlamydomonas enriched for lipid droplets identified DGTT1, PDAT1, and a structural protein (named MLDP for major lipid droplet protein) (19, 42). To assess the role of these proteins in TAG accumulation in Chlamydomonas, we used RNA-Seq to describe the transcriptome during a time course of nitrogen starvation. The resulting coverage was used for annotation of DGTT1 and PDAT1 gene models, discovery, and annotation of a type-1 DGAT and a candidate transcription factor. The functions of DGTT1 and PDAT1 were validated by heterologous complementation in yeast, and the respective contributions of the enzymes encoded by PDAT1 and of the candidate regulator were assessed in loss of function mutants.
EXPERIMENTAL PROCEDURES
Chlamydomonas Culture Conditions
C. reinhardtii strains CC-3269/2137 (wild-type nit2 mt+), D66+ (CC-4425 nit2 cw15 mt+) (43, 44), nrr1 (nrr1 nit2 cw15 mt+), and pdat1 (pdat1 nit2 cw15 mt+) were cultured at 24 °C (agitated at 180 rpm at a photon flux density of 90 μmol/m2/s provided by cool white fluorescent bulbs at 4100 K and warm white fluorescent bulbs at 3000 K used in the ratio of 2:1) in tris acetate-phosphate (TAP) medium unless otherwise noted (45). Nutrient-deficient media were prepared as follows. NH4Cl was omitted in nitrogen-free TAP; MgSO4 was omitted in sulfur-free TAP; ZnSO4 was replaced with ZnCl2; potassium phosphate was replaced with 1 mm KCl in phosphorus-free medium (46); iron-free Hutner's trace element solution was supplemented with 0.25 μm FeCl3 for low iron medium (47) and zinc-free Kropat's trace elements for zinc-deficient TAP medium (21).
Nitrogen Starvation Time Course Experiments
Several independent time course experiments of nitrogen starvation were performed to identify changes in transcript abundance at different time scales (0–1 h, 0–8 h, and 0–24 h). For each experiment, cells were grown in replete medium to a density of ∼4 × 106 cells ml−1, collected by centrifugation (2170 × g, room temperature for 5 min), washed, and resuspended in nitrogen-free TAP medium to 2 × 106 cells ml−1. For the 8-h time course, samples were taken at 0, 0.5, 4, and 8 h after transfer to nitrogen-free medium for RNA, chlorophyll, starch, and lipid analyses. The experiment was performed in triplicate. One of the three sets of RNA samples was used for RNA-Seq, and all three RNA samples were used for validation by real time PCR of cDNA. The first set of experiments indicated that some responses to nitrogen starvation occurred very quickly (<1 h), and others were much slower (>8 h). Therefore, two more time courses were performed. For the 48-h time course, samples were taken at 0, 2, 8, 12, 24, and 48 h after transfer to nitrogen-free medium. Independent cultures were grown in triplicate and were processed as described for the 8-h time course. A 1-h time course was also performed to capture the fast dynamics of nitrogen starvation response. Samples were taken before washing (0′) and at 0, 2, 4, 8, 12, 18, 24, 30, 45, and 60 min after transfer to nitrogen-free medium. This experiment was performed in duplicate, with one set of RNA samples used for RNA-Seq and both for validation by real time PCR of cDNA.
Nitrogen Limitation and Cell Density Experiments
For growth in reduced nitrogen, nitrogen-free TAP medium was supplemented with NH4Cl to 7, 3.5, 1.75, 0.7, or 0.35 mm. Cultures of strain CC-3269 were inoculated to 1 × 105 cells ml−1 and collected for RNA at 1 × 106 cells ml−1, when the growth rates of all cultures were identical. For assessing the impact of cell density, cultures were inoculated at 1 × 104 cells ml−1 in replete medium and sampled at 5 × 105 cells ml−1 and at each doubling thereafter until the culture reached a final density of 8 × 106 cells ml−1.
Other Nutrient-deficient Experiments
Nitrogen-, sulfur-, or phosphorus-starved cells were grown to ∼4 × 106 cells ml−1, collected by centrifugation (2170 × g, room temperature, 5 min), washed, and resuspended in the same medium to 2 × 106 cells ml−1 (46, 48). For iron or zinc deficiency, cells were grown in the appropriate medium (1 μm Fe or 0 μm Zn) to generate an inoculum (2 × 104 cells ml−1) for the experimental culture (49, 50). Sulfur- and phosphorus-deficient cultures were sampled for lipid analysis at 0 and 48 h after the cells were transferred to medium devoid of these nutrients; iron- and zinc-deficient cultures were collected at 3 × 106 and 5 × 106 cells ml−1, respectively.
Saccharomyces Culture Conditions
S. cerevisiae strains BY4742 (wild type; MATa his3Δ1 leu2Δ0 lys Δ0 ura Δ0) and YPD1078 (MATahis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ycr048wΔ::KanMX4 ynr019wΔ::KanMX4 yor245cΔ::KanMX4 ynr008w Δ::KanMX4), gifts from Sepp Kohlwein, University of Graz, were grown in YPD medium. Transformants were selected on a minimal medium containing 0.17% (w/v) yeast nitrogen base without amino acids, carbohydrate, or ammonium sulfate and 0.2% drop-out synthetic mix minus uracil (both US Biological), 0.5% (w/v) ammonium sulfate, 2% (w/v) dextrose, plus 1.5% (w/v) agar for solid medium. Counter-selection of the ura3 marker was performed using 5′-fluoroorotic acid at a final concentration of 1 mg ml−1. Functional complementation assays were performed on YPD agar containing oleic acid (0.009% w/v).
RNA Preparation and Analysis
Cells (50 ml) were collected by centrifugation (3440 × g, 5 min, 22 °C) and resuspended in 2 ml of water. Two ml of 2× lysis buffer was added, and the suspension was rocked for 20 min before it was flash-frozen in liquid N2 and stored at −80 °C. Frozen samples were subsequently heated at 65 °C for 3 min, and RNA was extracted as described previously (51). The RNA concentration was measured on a NanoDrop 2000 (Thermo Scientific), and the quality of the RNA was assessed on an Agilent 2100 Bioanalyzer and by blot hybridization for CBLP (RACK1) mRNA (52). Other probes amplified from genomic DNA were for PDAT1, DGTT1, and NRR1 (see supplemental Table S2 for primer sequences). Samples from the 8-h time course (0, 0.5, 2, 4, 6, and 8 h) and the 48-h time course (0, 0.5, 2, 4, 6, 8, 12, and 48 h) were pooled for generation of 454 EST tags at Joint Genome Institute (JGI). The data are deposited under accession SRX038871. The sequences were aligned to the reference 4.0 assembly and were visualized on a local installation of the UCSC browser. RNAs were also sequenced at Illumina and Los Alamos National Laboratory on a GAIIx system for estimating transcript abundance in nitrogen deficiency time course experiments or in cells grown in various amounts of ammonium supplements. Sequence data from a previous publication (18) were re-analyzed on the same pipeline for the purpose of comparison with the results presented here. The reads were aligned using bowtie (53) in single-end mode and with a maximum tolerance of three mismatches to the Au10.2 transcripts sequences, corresponding to the version 4.0 assembly of the Chlamydomonas genome. Expression estimates were obtained for each individual run in units of reads/kb of mappable transcript length/million mapped reads (RPKMs) (54) after normalization by the number of aligned reads and transcript mappable length (49).
DNA Gel Blot Hybridization
Genomic DNA was analyzed by hybridization as described previously (55). The probe used to detect the AphVIII cassette was the AphVIII ORF (805 bp) amplified from pSL18 (56) using primers AphVIII-F plus AphVIII-R (see supplemental Table S2), and Pfx DNA polymerase (Invitrogen) as directed by the manufacturer. The PCR product was isolated and labeled with [α-32P]dCTP (57) to a specific activity of 2 × 109 dpm μg−1. Chlamydomonas DNA-specific probes were produced by Klenow extension of overlapping 60-mer oligonucleotides (SP52-F and SP52-R; SP218-F and SP218-R; SPNR-F and SPNR-R, see supplemental Table S3) in the presence of [α-32P]dCTP; probes had specific activities of 1 × 109 dpm μg−1. Blots were exposed to film (Blue-sensitive X-ray; Henry Schein) at −80 °C using two intensifying screens.
Estimation of Relative mRNA Abundance
Complementary DNA synthesis and real time PCR were performed as described previously (50). Primers are listed in supplemental Table S4. Data are presented as the relative mRNA abundance and are normalized to the endogenous reference gene CBLP using LinRegPCR analysis (58, 59). Values were generated from three experimental replicates (for the 8-h experiment) and two experimental replicates (for the 1-h experiment); each replicate was analyzed three times (technical replicates). Real time reverse transcriptase PCR procedures and analyses follow the MIQE guidelines (60).
Sequencing of the DGAT1-containing BAC on the Illumina Platform
BAC REI35-B03 containing ∼100 kb of C. reinhardtii DNA, including the DGAT1 locus (a gift from Hudson Alpha Institute for Biotechnology, Huntsville, AL), was prepared from Escherichia coli. The DNA concentration was determined by Qubit dsDNA BR assay kit (Invitrogen). 100 ng of purified BAC DNA was sheared by the S-220 Adaptive Focused Acoustics system (Covaris, Woburn, MA) with the following conditions: 5% duty cycle, 20-watt peak incident power, 200 cycles/burst, 35 s. The ends of the resulting DNA fragments were repaired, and adaptors were ligated using reagents in the Illumina DNA TruSeq kit version 1 (Illumina, Inc. San Diego). At the gel isolation step, four bands of different sizes were isolated and purified according to the standard protocol. Individually, the four libraries of various sizes were subjected to 20 rounds of PCR amplification using the Illumina buffers, primers, and conditions. To determine the sizes of the four libraries, each was run on a BioAnalyzer DNA High Sensitivity chip (Agilent Technologies, Waldbronn, Germany). The four libraries (minus 126 nt for the Illumina adaptors) were 155, 335, 556, and 1014 nt long. The concentration of each was determined by Qubit dsDNA BR assay kit as before, and a 2 pm solution of each library was sequenced in 50 + 50-nt paired-end runs using the Illumina HiSeq 2000 platform with a TruSeq cBot PE cluster generation kit version 3. We obtained 6 × 105 raw reads (300,237 for 155 nt, 146,563 for 335 nt, 89,590 for 556 nt, and 62,318 for 1014 nt) with the correct Illumina adaptor sequence for a total of 6 × 107 base calls.
Gap-filling DGAT1 Sequence
The DGAT1 gene model was built from the JGI version 3, 4, and 5 assemblies by re-assembly and filling of gaps in the vicinity with an iterative series of hand-directed analyses that relied on data from multiple sources, including dozens of Illumina RNA-Seq reads from this and other projects (GEO accession numbers GSE25124 and GSE35305), genomic DNA re-sequencing projects, and traces from the original JGI genome project (retrieved on March 15, 2011, by FTP to NCBI TraceDB) with manual re-calling of bases in selected chromatograms. Tools included the following: ABySS (61) for de novo assemblies (both of genome and transcriptome) at both a whole genome/transcriptome level as well as targeted on select reads; traditional low throughput assembly/alignment tools; GMAP (62) numerous ad hoc assembly tools/analyses; BLAST and NCBI nonredundant nucleotide and protein databases; UCSC (63) and IGV (64) genome browsers; and manual inspection. Considerable additional sequence was recovered relative to the JGI version 3 and 4 assemblies; the gene model was refined, and the uncertainty is now restricted to the 5′ end of the transcript and its corresponding genomic sequence (see “Results”). The stretches of high C + G content (causing orders of magnitude drop in sequence coverage), tandem repeats in the vicinity, and inclusion of low complexity sequence in the coding region make sequencing very difficult by all sequencing technologies tried. PSI-BLAST analysis supports the model presented in this work, because protein sequence to the right of the gap aligns well with several type-1 DGAT sequences from oil-accumulating plants.
Lipid and Fatty Acid Analysis
For quantitative total fatty acid analysis, 5 ml of cell culture was collected on Whatman GF/A 25-mm circular glass filters, frozen in liquid nitrogen, and lyophilized. The filters (with cells) were transferred to reaction tubes and subjected to fatty acid methyl ester reactions and gas chromatography as described below. For TAG content, 20 ml of cell culture was collected by centrifugation (2170 × g, 2 min), washed in 1 ml of water, collected by centrifugation (13,780 × g, 5 min) in a 1.5-ml screw cap microcentrifuge tube, and frozen in liquid nitrogen. Prior to analysis, frozen cells were resuspended in 1 ml of methanol/chloroform/formic acid (2:1:0.1 (v/v/v)). Extraction solution (1 m KCl, 0.2 m H3PO4; 0.5 ml) was added and mixed by vortexing. Cell debris was removed by centrifugation for 3 min at 13,780 × g.
Lipids were separated by thin layer chromatography using petroleum ether/diethyl ether/acetic acid (8:2:0.1 (v/v/v)). Samples were run on baked Si60 silica plates (EMD Chemicals) for 5 min. After separation, lipids were reversibly stained by exposure to iodine vapor. The TAG band was located below pigments but above free fatty acid and DAG bands. TAG bands were quantitatively recovered by scraping the plate with a razor blade and transferring the material to a fatty acid methyl ester reaction tube. HCl in methanol (1 n, 1 ml) was added to each tube, and 5 μg of an internal standard (fatty acids 15:0, working concentration 50 μg/ml in methanol) was added. The head space in the tube was purged with nitrogen and the cap tightly sealed. Samples were incubated at 80 °C for 25 min and allowed to cool to room temperature. Aqueous NaCl (0.9%, 1 ml) and n-hexane (1 ml) were added, and the samples were shaken vigorously. After centrifugation at 1690 × g for 3 min, the hexane layer was removed, transferred to a new tube, and dried under a N2 gas stream. The resulting fatty acid methyl esters were dissolved in 100 μl of hexane. Fatty acid content and composition of the extracts were determined by gas chromatography with flame ionization detection, as described previously (65). The capillary DB-23 column (Agilent) was operated as follows: initial temperature 140 °C, increased by 25 °C/min to 160 °C, then by 8 °C/min to 250 °C, and held at 250 °C for 4 min.
Gene Model Verification
Gene models were derived by adjusting FM4 and Au5 models based on 454 ESTs (SRA020135) and verified experimentally by amplification of cDNAs and sequencing (Beckman Coulter Genomics, Danvers, MA). Primers used for this can be found in the supplemental material. The latest release of Chlamydomonas gene models Au10.2 at Phytozome includes these adjustments.
Microscopy
Chlamydomonas and yeast (1 ml from the culture) were stained with Nile Red® (Sigma) by adding the dye to a final concentration of 1 mg/ml and agitating the suspension for 5 min. Cells were collected by centrifugation (1000 × g, 2 min), resuspended in 200 μl of the growth medium, and immobilized by 1:1 mixing with 1.5% low melting agarose (total volume of 16 μl). Images were acquired with a Leica TCS-SPE confocal microscope using an APO ×63.0 water objective with a numerical aperture of 1.15. The Nile Red® signal was captured using a laser excitation line at 488 nm, and emission was collected between 554 and 599 nm (gain, 700; offset, 0). Chlorophyll autofluorescence was excited at 635 nm and captured between 670 and 714 nm. Differential interference contrast images were acquired in the PM Trans channel (gain, 371; offset, 0). Images were colored using Leica confocal software.
Generation and Screening of Chlamydomonas Mutants
A library of 25,000 insertional mutants in CC-4425 (D66+), created by insertion of a 1.7-kb PCR fragment containing the selectable marker gene AphVIII (66), was prepared as described previously (67). Screening for mutants was performed according to González-Ballester (68). Briefly, transformants picked from selective agar were transferred to 96-well microtiter plates containing 100 μl of TAP medium. Plates were wrapped in parafilm to minimize evaporation and grown for 5 days. Aliquots from the wells of each plate were pooled so that each pool represented the 96 mutants on one plate. Genomic DNA was isolated using the standard phenol/chloroform method (69). Equal amounts of genomic DNA from each of 10 pools were combined to create a “superpool” of DNA at a final concentration of 100 ng/μl. To screen for mutants with inserts in genes of interest, the superpool DNA was used as the template for PCR. Each PCR was performed using a gene-specific primer (the gene for which a disruption is sought; see supplemental Table S5) and an AphVIII construct-specific primer (RB2, 5′-TACCGGCTGTTGGACGAGTTCTTCTG-3′). A number of gene-specific primers, annealing at different positions within the target gene, were employed (listed in supplemental Table S5).
PCRs (final volume 25 μl) contained 0.5 unit of TaqDNA polymerase (Qiagen), 1× PCR buffer, 1× “Q” solution, DMSO (5% v/v), dNTPs (200 μm each), primers (300 nm each), and pooled genomic DNAs (100 ng). Template DNA was denatured for 5 min at 95 °C followed by amplification (30 s at 95 °C, 45 s at 60 °C, and 2 min at 72 °C, 40 cycles). PCR products were separated by electrophoresis in 1% agarose gels. To identify DNA regions adjacent to the right border of the AphVIII construct, PCR products were isolated (QIAquick gel extraction kit, Qiagen) and sequenced. Once a positive clone in a superpool was identified, the PCR screen was used to determine the specific microtiter plate and then the row and column within that plate associated with the mutant strain of interest; this localizes the mutant to a single well.
PCR Analysis of AphVIII Cassette Insertion Structures in Mutant Lines
Amplification across the 5′ side junction between the AphVIII cassette and Chlamydomonas genomic DNA in mutant pdat1-1 was performed using primers PDAT1-1-F1 plus RIM5-2 (supplemental Table 5). Reactions contained 1× ThermalAce buffer (Invitrogen), dNTPs (160 μm each), primers (0.5 μm each), genomic DNA (100 ng), and Taq (1 unit). Template DNA was denatured for 5 min at 95 °C followed by amplification (30 s at 95 °C, 30 s at 55 °C, and 60 s at 72 °C, 36 cycles). The PCR product was isolated and sequenced using primers PDAT1-1-F1 and RIM5-2. Amplification across the AphVIII cassette in mutant pdat1-2 was performed using primers PDAT1-2-F1 plus PDAT-SR7, and Phusion DNA polymerase (Finnzymes) as directed by the manufacturer. Reactions contained 1× GC buffer, DMSO (3% v/v), dNTPs (300 μm each), primers (0.5 μm each), genomic DNA (100 ng), and polymerase (1 unit). Template DNA was denatured for 30 s at 98 °C followed by amplification (10 s at 98 °C and 155 s at 68 °C, 30 cycles). The PCR product was isolated and sequenced using primer PDAT1-2-F1. Location of the insert is shown in supplemental Table S6.
Plasmid Construction
Plasmids were introduced into and amplified in E. coli XL1-Blue (endA1 gyrA96(nalR) thi-1 recA1 relA1 lac glnV44 F′[::Tn10 proAB+ lacIq Δ(lacZ)M15] hsdR17(rK− mK+). Synthetic genes encoding DGTT1 and PDAT1 were codon-optimized for expression in S. cerevisiae by GenScript (Piscataway, NJ). The sequences are shown in supplemental Figs. S1 and S2 and a codon use table is provided in supplemental Table S7. Codon-optimized genes were cloned in pUC57 (GenScript, Piscataway, NJ) and subsequently subcloned into the SpeI and XhoI sites of pGPD417-LNK to generate pIKB506 and pIKB507. pIKB505 was generated by amplifying LRO1 from purified S. cerevisiae genomic DNA using primers 5′-ACGTAGAATTCATGGGCACACTGTTTCGAAGA-3′ and 5′-ACGTAGCATGCTTACATTGGGAAGGGCATCTGAG-3′ and inserted (after digestion) between the EcoRI and SphI sites of pCGCU. Plasmids were introduced into S. cerevisiae and selected on the basis of uracil prototrophy or kanamycin resistance as appropriate.
RESULTS
Time Course of TAG Accumulation
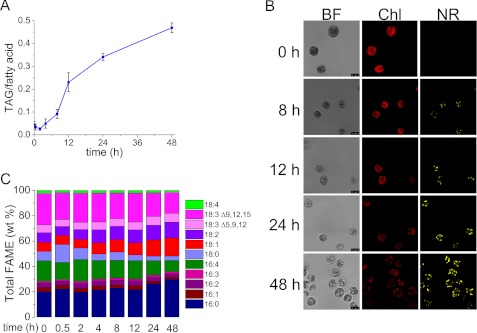
Several stress conditions promote the accumulation of TAG-containing lipid bodies in Chlamydomonas (11–15, 17–19, 21, 70). To identify changes in gene expression associated with TAG accumulation, we sought to analyze the transcriptome under nitrogen starvation conditions because it is the most effective inducer of TAG accumulation. To identify suitable time points for transcriptome analysis, we monitored TAG accumulation in a time course following transfer of wild-type cells from nitrogen-replete to nitrogen-deficient conditions (Fig. 1). Quantitative gas chromatography indicates that the fraction of fatty acids (out of total per cell) associated with TAG is noticeably increased at 8 h of starvation and continues to increase through 48 h (Fig. 1A). Student's t test indicates that 8-, 12-, 24-, and 48-h samples are statistically different from the 0-h sample with 98% confidence. This agrees well with confocal fluorescence microscopy, which shows increased abundance of Nile Red-stained lipid bodies with a concomitant decrease in chlorophyll fluorescence (Fig. 1B). In contrast, the replete cells did not accumulate Nile Red staining bodies (supplemental Fig. S3). Composition analysis indicates that there is a progressive increase in saturated and mono-unsaturated fatty acids (16:0 and 18:1) and a decrease in the proportion of polyunsaturated ones (16:4, 18:3) (Fig. 1C). After 48 h of starvation, the total amount of saturated fatty acids was 20% more (w/w) than at 0 h.
FIGURE 1.
Time course of increased TAG accumulation in nitrogen-starved C. reinhardtii wild type (CC-3269) cells. A, fraction of fatty acids in TAG in nitrogen-starved cells during a 48-h time course of nitrogen starvation on a mol/mol basis. Error bars represent standard deviation from three biological replicates. Two-tailed Student's t test on experimental triplicates for each time point indicate that 8-, 12-, 24-, and 48-h samples are statistically different from the 0-h sample at 98% confidence. B, confocal microscopy of nitrogen-starved cells to visualize TAG by Nile Red staining. BF, bright field; Chl, chlorophyll autofluorescence; NR, Nile Red fluorescence. C, fatty acid composition of lipids at each time point was measured by gas chromatography; the average of three experimental replicates is shown. Microscopy of cells in replete medium is given in supplemental Fig. S3.
Using the rationale that changes in mRNA abundance should precede changes in TAG accumulation, we isolated RNA for transcriptome analysis in three time course experiments as follows: 0–1 h, 0–8 h, and 0–48 h following transfer of cells from nitrogen-replete to nitrogen-deficient medium, referred to subsequently as short, medium, and long time courses. For the short time course, we sampled cells at two “reference” zero points: one labeled 0′, corresponding to the nitrogen-replete cells at a density of 4 × 106 cells ml−1, and the other labeled 0, corresponding to nitrogen-replete cells just immediately after centrifugation and transfer to fresh nitrogen-free medium at a density of 2 × 106 cells ml−1 (see under “Experimental Procedures”). This allowed us to distinguish changes related to the handling of cells from changes resulting from nitrogen starvation. RNAs were analyzed by sequencing (Illumina platform) and by real time PCR on cDNA generated by reverse transcriptase for validation of changes in abundance of select mRNAs (see under “Experimental Procedures”).
Impact on Fatty Acid and Lipid Synthesis
We augmented the existing annotations (27, 71, 72) of genes corresponding to enzymes involved in fatty acid and membrane and neutral lipid biosynthesis and queried the abundance of the corresponding mRNAs (select genes and time points given in Table 1, for all time points see supplemental Table S1). We noted that with the exception of a putative palmitoyl-protein thioesterase-encoding gene (Cre01.g067300), which may function to liberate fatty acids for lipid/TAG synthesis and whose expression is increased nearly 3-fold, most of the genes (ACX1, ACX2, BCC1, BCC2, ACP1, and ACP2) encoding enzymes (carboxyltransferases, acetyl-CoA biotin carboxyl carrier, acyl carrier protein, malonyl-CoA-acyl carrier protein transacylase) in de novo fatty acid synthesis, as well as genes (SQD1, SQD2, MGD1, DGD1, SAS1, BTA1, ECT1, PGP3, INO3, and PIS1) encoding functions in membrane lipid synthesis, are down-regulated. One of the most dramatic changes is in Cre14.g621650 mRNA (a malonyl-CoA-acyl carrier protein transferase), whose abundance decreased from 160 RPKM to 19.7 RPKM at 48 h of nitrogen starvation.
TABLE 1.
Nitrogen starvation impacts many genes associated with fatty acid metabolism
Many genes involved in glycerolipid metabolism were identified previously (18, 27, 72). Genes without functional annotation are indicated with Au10.5 and Au5 transcript IDs.
| Au10.2 | Au5 | Gene name | Description | RPKM |
||||
|---|---|---|---|---|---|---|---|---|
| 0 h | 2 h | 12 h | 24 h | 48 h | ||||
| Cre12.g519100 | 513248 | ACX1 | α-Carboxyltransferase | 140.5 | 68.2 | 81.3 | 71.6 | 70.2 |
| Cre12.g484000 | 512497 | BCX1 | β-Carboxyltransferase | 103.5 | 53.4 | 35.1 | 35.4 | 31.4 |
| Cre17.g715250 | 517403 | BCC1 | Acetyl-CoA biotin carboxyl carrier | 293.9 | 129.6 | 54.7 | 74.7 | 68.9 |
| Cre01.g037850 | 511392 | BCC2 | Acetyl-CoA biotin carboxyl carrier | 271.5 | 89.8 | 43.9 | 55.9 | 58.1 |
| Cre01.g063200 | 511929 | ACP1 | Acyl carrier protein | 211.0 | 220.6 | 171.6 | 157.3 | 113.7 |
| Cre13.g577100 | 514476 | ACP2 | Acyl carrier protein | 3513.1 | 1740.6 | 1110.7 | 1777.4 | 1399.1 |
| Cre16.g656400 | 516156 | SQD1 | UDP-sulfoquinovose synthase | 119.4 | 88.4 | 36.7 | 49.8 | 61.7 |
| Cre01.g038550 | 511409 | SQD2 | Sulfoquinovosyldiacylglycerol synthase | 10.3 | 7.6 | 3.4 | 6.7 | 2.1 |
| Cre16.g689150 | 516856 | SQD3 | Sulfoquinovosyldiacylglycerol synthase | 0.3 | 0.4 | 0.0 | 0.0 | 0.0 |
| Cre13.g585300 | 514645 | MGD1 | Monogalactosyldiacylglycerol synthase | 16.1 | 1.4 | 6.3 | 5.3 | 3.4 |
| Cre13.g583600 | 514612 | DGD1 | Digalactosyldiacylglycerol synthase | 29.0 | 31.2 | 52.4 | 42.5 | 36.4 |
| Cre06.g250200 | 522823 | SAS1 | S-Adenosylmethionine synthetase | 2334.8 | 1167.6 | 283.0 | 401.4 | 578.3 |
| Cre07.g324200 | 524437 | BTA1 | Diacylglyceryl-N,N,N-trimethylhomoserine synthesis protein | 120.1 | 129.7 | 82.5 | 79.1 | 68.6 |
| Cre12.g539000 | 513669 | ECT1 | CDP-ethanolamine synthase | 17.4 | 20.7 | 25.2 | 17.1 | 16.9 |
| Cre03.g162600 | 520698 | PGP3 | Phosphatidylglycerolphosphate synthase | 7.3 | 6.6 | 1.4 | 2.8 | 4.8 |
| Cre03.g180250 | 521070 | INO1 | Myoinositol-1-phosphate synthase | 194.9 | 158.7 | 305.3 | 282.2 | 188.5 |
| Cre10.g419800 | 509612 | PIS1 | Phosphatidylinositol synthase | 29.5 | 22.6 | 27.0 | 29.2 | 26.9 |
| Cre14.g621650 | 515418 | MCT1 | Malonyl-CoA:acyl carrier protein transacylase | 159.9 | 137.8 | 35.2 | 10.8 | 19.7 |
| Cre01.g045900 | 511566 | DGAT1 | Diacylglycerol acyltransferase, DAGAT type-1 | 3.3 | 11.7 | 15.0 | 9.1 | 20.8 |
| Cre12.g557750 | 514063 | DGTT1 | Diacylglycerol acyltransferase, DAGAT type-2 | 0.8 | 7.9 | 11.1 | 17.5 | 24.6 |
| Cre02.g121200 | 519435 | DGTT2 | Diacylglycerol acyltransferase, DAGAT type-2 | 22.2 | 21.1 | 24.5 | 26.2 | 26.8 |
| Cre06.g299050 | 523869 | DGTT3 | Diacylglycerol acyltransferase, DAGAT type-2 | 35.5 | 33.4 | 31.5 | 40.7 | 32.5 |
| Cre03.g205050 | 521604 | DGTT4 | Diacylglycerol acyltransferase, DAGAT type-2 | 4.6 | 7.5 | 7.5 | 2.9 | 3.3 |
| Cre02.g079050 | 518531 | DGTT5 | Diacylglycerol acyltransferase, DAGAT type-2 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 |
| Cre02.g106400 | 519119 | PDAT1 | Phospholipid diacylglycerol acyltransferase | 3.4 | 9.4 | 8.5 | 9.7 | 8.0 |
Surprisingly, genes LIPG2, Cre05.g234800, and Cre02.g127300, encoding candidate TAG lipases, are up-regulated, which has also been noted in previous transcriptome and proteome studies (18, 42). One possibility is that these enzymes are not TAG lipases but rather play a role in releasing fatty acids from membrane lipids for de novo TAG synthesis. Membrane recycling has been reported to supply ∼30% of TAG accumulation during nitrogen starvation (73). Several other candidate lipases are down-regulated, such as LIPG1, Cre10.g422850, and Cre05.g248200, and these may represent the authentic TAG lipases instead.
The expression of genes encoding various desaturases is either unchanged or decreased in nitrogen starvation conditions. DES6, one of the most abundant mRNAs in Chlamydomonas (rank 88 of 17,303 in nitrogen-replete cells) is decreased 7-fold at 48 h. FAD5a mRNA shows a similar 6.6-fold decrease, with a 1.6-fold decrease noted as early as 2 h after initiation of nitrogen starvation. These changes are consistent with the observation that the level of saturation of fatty acids is increased in response to nitrogen starvation (Fig. 1C).
Three Acyltransferases with Increased Expression
Three different DGAT enzymes are found in plants, two are membrane-associated and a third is soluble. The membrane-associated DGATs are of two types; the one corresponding to the DGAT1 gene family is related to the mammalian DGAT enzyme, and the others are similar to the steryl acyltransferases Are1p/Are2p from S. cerevisiae and a distinct type-2 family that encodes enzymes related to the S. cerevisiae Dga1p. In Chlamydomonas, these genes are named DGTT for diacylglycerol acyltransferase type-2. The function of the third soluble enzyme has not been investigated.
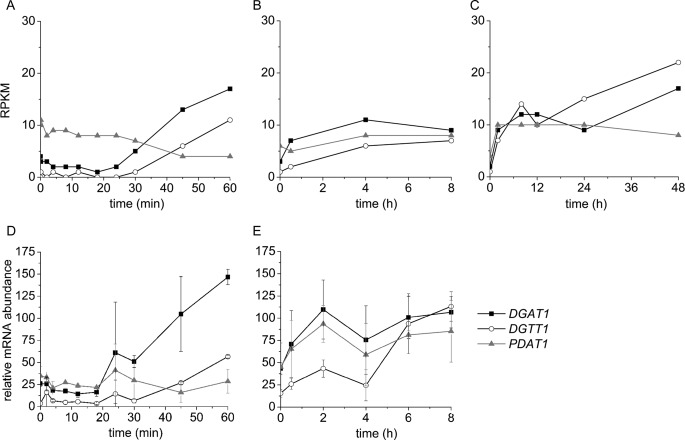
Of the five previously annotated DGTT genes, DGTT1 is the only one responsive to nitrogen starvation. DGTT2 and DGTT3 are expressed at moderate levels, and these levels remain approximately constant over the time courses of the experiments presented here. DGTT4 mRNA abundance is low (about 10–20% relative to DGTT2 and DGTT3), and DGTT5 expression has not been detected. These findings agree with a previous study of the response of Chlamydomonas to nitrogen starvation (18). The increase in DGTT1 mRNA is noted between 30 and 60 min of nitrogen starvation, and the level remains high for up to 48 h (Fig. 2, A–C). Patterns of expression noted in the mRNA-Seq analysis were validated in RNAs isolated up to 8 h after nitrogen starvation by real time PCR of cDNAs in triplicate experiments (Fig. 2, D and E, and supplemental Fig. S4). After 8 h of nitrogen starvation, nearly all transcripts were changed in abundance, and therefore we were unable to find a suitable “control” gene. Nevertheless, the magnitude of the increase in expression at 48 h is similar to the increase noted in previous work relative to the replete condition, time 0 (see Table 1) (18).
FIGURE 2.
Increased expression of DGAT1, DGTT1, and PDAT1 encoding three nitrogen deficiency-responsive acyltransferases. mRNA abundance was reported in RPKM in a 1-h time course (A), 8-h time course (B), and 48-h time course (C) after the onset of nitrogen starvation. Relative mRNA abundance in a 1-h time course (D) and 8-h time course (E) was measured by real time PCR. Error bars on D and E are standard deviations from two biological samples analyzed in technical triplicate.
In addition to DGTT1, we noted increased expression of a gene, which we named DGAT1, that encodes a type-1 DGAT (Fig. 2). This sequence was not annotated in previous assemblies of the Chlamydomonas genome because of a large sequence gap that obscured the 5′ half of the gene model. The pattern of expression of DGAT1 is very similar to that of DGTT1, which motivated us to improve the assembly (see “Experimental Procedures”). The sequence information now covers ∼90% of the locus (sequence is given in supplemental Fig. S5 and predicted protein structure is given in supplemental Fig. S6). Nevertheless, the region is refractory to amplification and cloning, and two small gaps remain in the predicted coding region (see supplemental Figs. 5 and 6). In terms of absolute abundance, DGTT1 and DGAT1 mRNAs are approximately the same, and they attain levels that are comparable with those observed for DGTT2 and DGTT3, whose levels are already high in nitrogen-replete cells and remain high throughout the time course (Table 1). In photoautotrophic cells, DGTT3 and DGTT4 mRNAs are also increased in nitrogen deficiency (74).
A third acyltransferase, which we named PDAT1 because of orthology to Arabidopsis PDAT1, also shows a small but reproducible increase in abundance in nitrogen-starved cells. In contrast to DGAT1 and DGTT1, whose mRNAs increase progressively throughout the 48-h time course, the increase in PDAT1 mRNA plateaus by 2 h. This may be related to the cell density-dependent decrease in PDAT1 mRNA abundance (supplemental Fig. S8). Increases in DGAT1 and PDAT1 mRNA were verified by real time PCR of cDNAs (Fig. 2, D and E) and also by blot hybridization (supplemental Fig. S7).
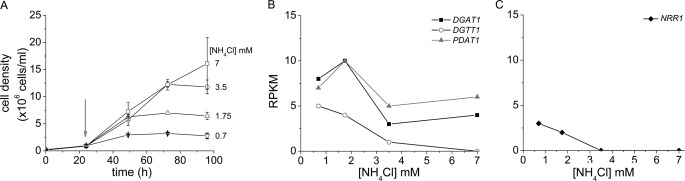
In a separate experiment, we monitored transcript abundance in cells acclimated to low nitrogen content. Chlamydomonas cells were washed with nitrogen-free medium and inoculated into medium supplemented with various amounts of NH4Cl (up to 7 mm, which is the concentration in TAP medium). Cells were grown to a density of 1 × 106 cells ml−1 prior to collecting the cells for RNA isolation. Because the cells grew at approximately the same rate up to this point, independent of NH4Cl concentration, this corresponded to about four divisions for each culture. When we monitored transcript abundance for the three acyltransferases, we noted that all three respond to low nitrogen content and that the abundances of PDAT1 and DGAT1 mRNAs were very similar, again suggesting that these three enzymes are responding to nitrogen deficiency (Fig. 3). In contrast, in these same experiments the amounts of DGTT2 and DGTT3 mRNAs appear to be independent of nitrogen content of the medium.
FIGURE 3.
Impact of nitrogen nutrition on cell growth (A) and mRNA abundance of DGAT1, DGTT1, and PDAT1 (B), and NRR1 (C). Chlamydomonas cells were grown with various amounts of ammonium chloride (see under “Experimental Procedures”), and RNA was sampled at a cell density of 1 × 106 cells ml−1 (indicated with an arrow) when cells were still growing at the same rate.
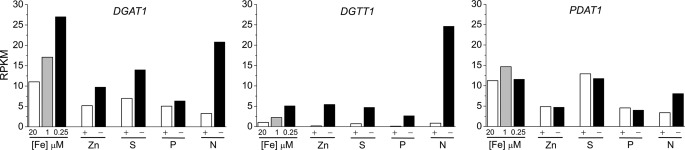
Because limitation for several different nutrients can promote TAG accumulation (11–15, 17–19, 21, 70), we tested whether low concentrations of a variety of different nutrients impact DGTT1, DGAT1, and PDAT1 expression (Fig. 4). DGTT1 mRNA increased under each nutrient limitation condition tested (minus sulfur, minus phosphorus, minus zinc, and low iron), although not to the same extent as during nitrogen starvation, which correlates with the relative amounts of TAG accumulated in each condition. Similar to DGTT1, DGAT1 is also up-regulated when the cells are limited for the nutrients tested in this experiment, with the exception of phosphorus deprivation, suggesting that at least some distinct signaling pathways may impact acyl-CoA-dependent TAG synthesis pathways and validating the relevance of both these enzymes in TAG biosynthesis. The PDAT1 transcript, however, only increases in nitrogen-starved cells. This may mean that fatty acid recycling from membrane lipids into TAG is more relevant in a subset of stress conditions or that PDAT activity can also be controlled by post-transcriptional processes in other stress situations.
FIGURE 4.
Response of DGAT1, DGTT1, and PDAT1 mRNA abundance to other TAG-accumulating conditions. Cells were grown in TAP medium under replete (20 μm iron, zinc, sulfur, phosphorus, and nitrogen), deficient (1 or 0.25 μm iron), or nutrient starvation (minus zinc, minus sulfur, minus phosphorus, and minus nitrogen) conditions as described under “Experimental Procedures.” mRNA abundance is expressed in RPKM.
Nitrogen Response Regulator
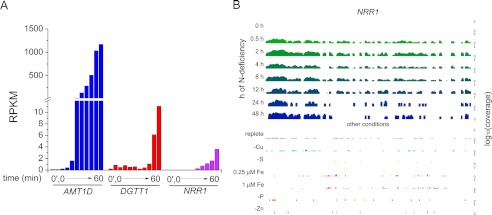
To identify candidate regulators of nitrogen deficiency-induced TAG accumulation, we searched among the differentially expressed RNAs for those encoding proteins with DNA binding domains. One transcript was of particular interest because of the magnitude of its regulation and co-expression with the DGTT1 mRNA (Fig. 5A). The gene model, which was named NRR1 for nitrogen response regulator, was manually curated based on RNA-Seq and 454 EST coverage (Fig. 6) and verified by sequencing (supplemental Fig. S9). A SQUAMOSA promoter-binding protein domain, named for its occurrence in the founding member (the SQUAMOSA promoter-binding protein), is predicted at its N terminus, and the SQUAMOSA promoter-binding protein domain is conserved in other algae and plants (supplemental Fig. S10). When we examined the abundance of the NRR1 transcript in other TAG-accumulating conditions, we found that the increase is specific for the nitrogen starvation response (Fig. 5B, for a larger version see supplemental Fig. S11). The gene is also expressed in cells acclimated to low nitrogen (Fig. 3C). Changes in mRNA abundance were validated by real time PCR of cDNA in triplicate experiments (supplemental Fig. S4).
FIGURE 5.
Pattern of expression of the nitrogen-responsive regulator, NRR1. A, mRNA abundance of an ammonium transporter AMT1D, an acyltransferase DGTT1, and NRR1. B, browser view of the Chlamydomonas NRR1 locus; nitrogen starvation mRNA abundance is shown at the top (0–48 h) and other conditions (replete, minus copper, and minus sulfur, 0.25 μm iron, 1 μm iron, minus phosphorus, and minus zinc sampled as described under “Experimental Procedures”) are below. Note that the y axis, representing absolute expression (RPKM) is presented on a log10 scale (0–3 in each case, covering 3 orders of magnitude); the differences in RNA abundance between minus nitrogen and the other deficiency conditions is substantial. A larger version of this figure is shown in supplemental Fig. S11, and the complete data set is available via the UCSC browser on line.
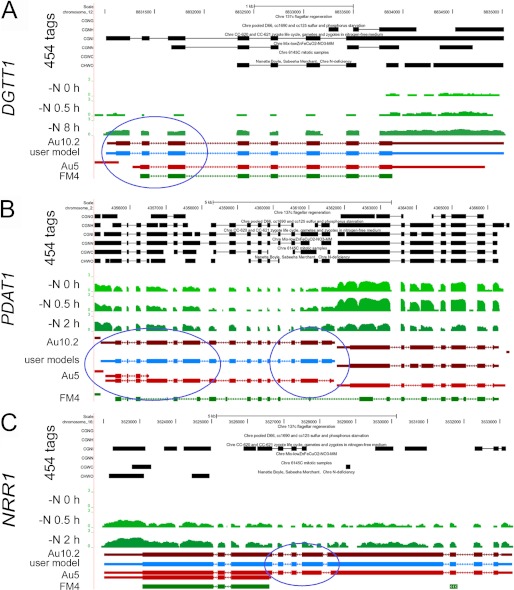
FIGURE 6.
Browser view of Chlamydomonas DGTT1 (A), PDAT1 (B), and NRR1 (C) loci. The UCSC browser was used to view RNA-Seq coverage. The genome coordinates are shown at the top. The next six tracks labeled 454 ESTs represent sequences, collapsed into a single track, from the UCLA/JGI EST project (accession SRX038871). The tracks shown in green represent RNA-Seq coverage, on a log scale, for the locus at three time points (0, 0.5, and 2 h) after Chlamydomonas cells were transferred to nitrogen-depleted medium. The JGI best gene model for the V4 assembly, FM4, and two different gene models predicted by the Augustus algorithm (Au5 and Au10.2), are shown in green and red, respectively. Thick blocks represent exons, and thin blocks represent UTRs, and arrowed lines represent introns. RNA-Seq, 454, and EST coverage were used to inform the manual construction of a single gene model at each locus, labeled user model and shown in blue. The PDAT1 gene is on the left, and the gene model on the right represents an expressed hypothetical protein whose expression is decreased in nitrogen starvation. The blue circles highlight new exons suggested by the patterns of RNA-Seq coverage and incorporated by manual curation into revised gene models. The complete data set is available via the UCSC browser on line.
Functional Validation
Because the gene models (FM4 or Au5 on the JGI browser) for the version 4 assembly of the Chlamydomonas genome were largely computationally derived and based only on a few hundred thousand Sanger-sequenced ESTs, we used EST data derived from a pooled sample of RNA from nitrogen-deficient cells (see under “Experimental Procedures”) and other recently obtained 454 ESTs to curate the gene models (Fig. 6). The manually curated models (Fig. 6, blue) are consistent with the RNA-Seq coverage graphs that show increased expression of each exon in nitrogen deficiency (see DGTT1 and NRR1 loci in particular). Each model was validated by sequencing the entire cDNA (e.g. for PDAT1) or by sequencing amplified regions of the cDNAs across exon-exon junctions (supplemental Fig. S7). For DGTT1, we noted the inclusion of an additional exon at the 5′ end, which is likely important for function (circled in Fig. 6). For PDAT1, the size of the transcript predicted for the revised model matches well with the size (∼3.6 × 102 nt) estimated from blot hybridization (supplemental Fig. S2). The new models are included in the latest set of Au10.2 gene models at Phytozome, which also relies on the 454 ESTs.
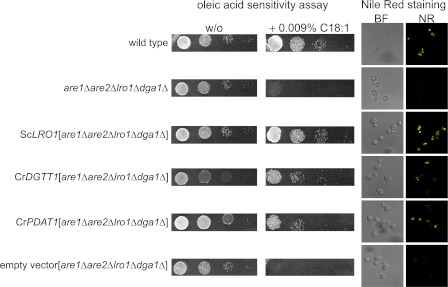
We tested the biochemical activities of DGTT1 and PDAT1 in a heterologous system by complementation of an S. cerevisiae mutant carrying a quadruple deletion in four acyltransferases (encoded by DGA1, LRO1, ARE1, and ARE2) that contribute to TAG synthesis (75). The resulting strain is sensitive to oleic acid, and introduction of either a DGAT- or PDAT-encoding sequence will rescue the strain (76). Codon-optimized versions of Chlamydomonas DGTT1 and PDAT1 were tested for function in this assay and indeed render the mutant resistant to oleic acid (Fig. 7). When we stained the yeast strains with Nile Red, only strains complemented with acyltransferases showed lipid body accumulation, validating the function of these enzymes. When the complementing plasmids were cured by 5′-fluoroorotic acid counter-selection, the strains regained the oleic acid sensitivity phenotype establishing a causal connection.
FIGURE 7.
Functional assessment of DGTT1 and PDAT1 in yeast. Left panel, oleic acid (C18:1) sensitivity assay of wild type, acyltransferase-deficient yeast, and complemented strains. Yeast strains were grown on agar supplemented with oleic acid (0.009% (w/v)) or not (w/o). Wild type, are1Δare2Δlro1Δdga1Δ (quadruple mutant lacking all diacylglycerol acyltransferase activity), and mutant strains complemented with ScLRO1 (encoding PDAT), codon-optimized CrDGTT1, codon-optimized PDAT1, or an empty vector were grown to an A600 = 1 diluted 10-fold and plated in serial 10-fold dilutions. Right, Nile Red staining of lipid bodies in each yeast strain described above.
Reverse Genetic Validation
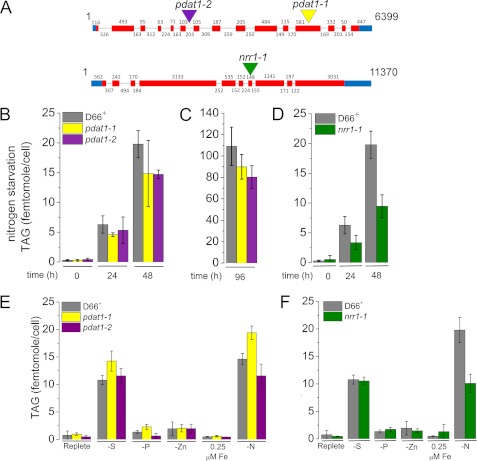
We took advantage of a collection of insertional mutants to identify candidate loss of function alleles in PDAT1 and NRR1 by PCR (67, 68). The position of the inserts was verified by sequencing PCR products from the 3′ side insertion junction; in pdat1-1 the AphVIII cassette is integrated in the 12th exon, in pdat1-2 in the 6th intron, and in nrr1-1 in the 7th exon (Fig. 8A). Sequencing across the 5′ side insertion junctions in the pdat mutants indicates that the integration events were accompanied by no or minimal deletion of Chlamydomonas DNA (0 bp in pdat1-2 and 4 bp in pdat1-1) (supplemental Fig. S1). Southern blotting verified that each mutant carries a single copy of the cassette per genome and also verified the position of the integrated DNA in each of the mutants (supplemental Fig. S4). For nrr1, we used multiple gene-specific probes to demonstrate that the insertion is not accompanied by a large deletion at the 5′ end of the locus. If there is a deletion, its size is <250 bp and does not extend into the neighboring gene(s) (supplemental Fig. S12).
FIGURE 8.
Phenotype of C. reinhardtii strains carrying inserts in the PDAT1 and NRR1 genes. A, site of insert of AphVIII into genomic DNA in the PDAT1 and NRR1 mutants is indicated with a colored triangle (see supplemental Table 6 for exact position of inset). B, TAG content of pdat1-1 (yellow), pdat1-2 (purple), and the parent strain CC-4425 (D66+) (gray) measured at 0, 24, and 48 h after nitrogen starvation by quantitative gas chromatography as described under “Experimental Procedures.” C, TAG content of pdat1-1 (yellow) and pdat1-2 (purple) after 96 h of nitrogen starvation. D, TAG content of nrr1 (green) and the parent strain CC-4425 (D66+) (gray) measured at 0, 24, and 48 h after nitrogen starvation. E, phenotype of pdat1-1 and pdat1-2 mutants in other TAG-accumulating conditions (−PO4 and −SO4, 0.25 μm iron, and −zinc). F, phenotype of nrr1-1 mutants in other TAG accumulating conditions (−PO4 and −SO4, 0.25 μm iron, and minus zinc). Error bars represent standard deviation from three biological replicates except for C, which is from six biological replicates. Two-tailed Student's t test on the data indicate that pdat1-1 and pdat1-2 are statistically different from the parent strain after 96 h of nitrogen starvation at 93 and 99% confidence levels, respectively; nrr1-1 is also significantly different at 48 h of nitrogen starvation at 99% confidence.
Each mutant was tested relative to the parental wild-type strain CC-4425 for TAG accumulation after transfer to nitrogen-free medium. Both pdat1-1 and pdat1-2 mutants show a 25% decrease in TAG content at 48 h. The difference is statistically significant at the 93 and 99% level, respectively (as determined by a two-tailed Student's t test), for the 96-h time point (Fig. 8B). The impact of the mutation is similar to that noted for the pdat1 mutant in Arabidopsis and the lro1 mutant in S. cerevisiae. For the nrr1 mutant, we saw a greater impact with a 52% decrease in TAG content at 48 h, which is statistically significant at the 99% confidence level. When we tested the impact of other nutrient deficiencies on TAG accumulation in the mutants, there was no effect (Fig. 8C), which confirms that PDAT1 and NRR1 are nitrogen starvation-responsive genes with a role in TAG accumulation.
DISCUSSION
Several studies have noted that many algae, when stressed to the point of growth inhibition, accumulate TAGs, a phenomenon of interest to researchers studying biofuels. Work with other organisms, including S. cerevisiae, Arabidopsis, and mammals, has identified key enzymes, which enable the identification of homologs and orthologs in algae now that genome sequences are available or are in the pipeline (1). Nevertheless, an assessment of the role of these homologs in individual or multiple stress situations requires functional studies. More importantly, if the signaling molecules that initiate the diversion of carbon and lipid metabolism toward TAG accumulation can be identified (referred to as the “lipid trigger”), it would enable the stress-free production of a biofuel precursor without accepting a productivity decline. With these goals, we used a transcriptome-based approach to identify three loci encoding acyltransferases that might be important for TAG accumulation in Chlamydomonas, which we are using as a reference organism.
DGAT
There are five genes encoding type-2 DGATs in Chlamydomonas; the occurrence of multigene families for these type-2 enzymes is common in the algae (for example, there are three in Volvox carteri, two in Chlorella sp. NC64A, and seven in Micromonas pusilla). Previous work has assigned function to DGTT2 through DGTT5 from Chlamydomonas via heterologous expression in yeast but not for DGTT1 (18, 41). Here, we have been able to take advantage of ESTs from nitrogen-deficient cells and RNA-Seq coverage (Fig. 6) to adjust previous gene models. This revealed an extra exon in DGTT1, which perhaps facilitated the expression of a functional protein (Fig. 7). DGTT1 is highly responsive at the level of mRNA abundance to both nitrogen concentration and time after removal of the nitrogen source (Figs. 2 and 6). Orthologs of Chlamydomonas DGTT1 in yeast and Arabidopsis, Dga1p and DGAT2, respectively, are established as significant contributors to oil accumulation. The yeast dga1 mutant produces 25% less TAG compared with the wild-type, and Arabidopsis DGAT2 is expressed highly in seed where oil accumulates. A phylogenetic tree of DGTT1 and related proteins is presented in supplemental Fig. S13. DGTT1 has also been reported to be highly induced after 1 day of nitrogen starvation in phototrophically grown Chlamydomonas cells (74).
DGAT1 also shows increased expression in nitrogen-deficient cells. The reason that DGAT1 was not characterized previously is attributed to a gap in the sequence coverage in the version 4 assembly of the genome sequence, which obscures about half the gene model. We reduced this gap substantially but were unable to close it completely even with targeted Sanger sequencing because it is difficult to amplify across the two remaining gaps either using genomic DNA or cDNA as the template. Msanne et al. (48) report that DGAT1 is not responsive to nitrogen starvation when Chlamydomonas is grown phototrophically, which may indicate this response is specific to growth on reduced carbon sources, as it is also more highly expressed in seeds of Arabidopsis (77). This difference could also be due to strain differences, which have already been noted to impact TAG accumulation (20). These acyltransferases were not identified in a study of the proteome of nitrogen-starved Thalassiosira pseudonana (78). In future studies it will be useful to understand the distinction between DGAT1 and DGTT1 with respect to kinetic parameters, substrate specificity, pattern of expression in the life cycle of Chlamydomonas, and subcellular location.
When we look at the absolute abundance of all DGAT-encoding mRNAs in Chlamydomonas, we note that the increase after 48 h of nitrogen starvation is only 62% because two other DGTT genes are constitutively expressed at relatively high levels (∼20–30 RPKM). This indicates that factors beyond mRNA abundance, such as compartmentalization of enzymes, post-transcriptional regulatory mechanisms that act on the protein(s), and metabolite flux, could contribute to TAG accumulation (1).
What is the source of substrate for the acylation reactions catalyzed by DGATs? Previous studies have provided evidence for fatty acid recycling, and this is consistent with increased expression of lipases in nitrogen-starved cells (18, 42, 73). Some of these are annotated as TAG lipases, which would be counter-intuitive, but they may well be membrane lipid lipases that release fatty acids that are subsequently incorporated into TAGs. Indeed, a recombinant tomato enzyme showed equal activity on TAG as on phosphatidylcholine, and a putative TAG lipase from Aspergillus oryzae showed activity on mono- and diacylglycerols as well (79, 80).
PDAT
The acyl-CoA independent pathway, dependent on PDAT1, is less responsive to nitrogen starvation at the level of RNA abundance, which prompted us to use a reverse genetic approach to assess its contribution to TAG accumulation (Fig. 8). The phenotype of Chlamydomonas pdat1 mutants is very similar to that noted for analogous mutants in yeast, specifically that TAG abundance is decreased but not eliminated. As for DGTT1, PDAT1 is also functional in yeast (Fig. 7); this is relevant because phosphatidylcholine, the preferred substrate of PDAT, is replaced by diacylglycerol trimethylhomoserine in Chlamydomonas membranes (26, 81, 82). Interestingly, a recent study showed that DGAT1, DGAT2, and PDAT1 genes were induced in nitrogen-deficient Arabidopsis seedlings as well, with a pattern (in terms of magnitude) similar to that noted in this study.
Regulators
NRR1, encoding a SQUAMOSA promoter-binding protein domain protein (and hence most likely a transcription factor), was identified based on its increased expression within half an hour of nitrogen starvation and its very tight regulation (essentially no RNA in nitrogen-replete cells). The timing of RNA increase is coincident with the increase in DGTT1 mRNAs and precedes substantially the appearance of Nile Red staining bodies (Fig. 1). The nrr1 mutant (likely loss of function because of the presence of a large insert in an exon) loses 50% of its TAG accumulation ability, which speaks to the importance of this regulator in the nitrogen-starvation response. A previous study identified Cre14.g624800 as a candidate regulator, although function was not tested by reverse genetics in that work (83). This gene is up-regulated in our study as well; however, it does not have any conserved domains so the function cannot be predicted. It will be interesting to identify the sought after lipid trigger and test whether its constitutive expression would be sufficient for stimulation of TAG accumulation and lipid body biogenesis.
Supplementary Material
Acknowledgments
We thank Jane Grimwood (Hudson Alpha) for access to the version 5 assembly prior to release and Sepp Kohlwein (University of Graz) for yeast strains and plasmids. Some sequencing (454 ESTs) was conducted by the United States Department of Energy Joint Genome Institute, which is supported by the Office of Science of the United States Department of Energy under Contract No. DE-AC02-05CH11231.
This work was supported, in whole or in part, by National Institutes of Health Grants T32 GM07185 and T32 ES015457 (to S. J. K. and I. K. B). This work was also supported by Air Force Office of Science Research Grants FA 9550-10-1-0095 (to S. M. and M. P.) and FA9550-08-0165 (to C. B.), Department of Energy Contract DE-EE0003046 (to the National Alliance for Advance Biofuels and Bioproducts Consortium, including S. M., M. P., and S. J.), and the German Academic Exchange Service Grant D/08/47579 (to A. H.).

This article contains supplemental Tables S1–S7, Figs. S1–S13, and additional references.
- TAG
- triacylglycerol
- DGAT
- diacylglycerol acyltransferase
- PDAT
- phospholipid:diacylglycerol acyltransferase
- nt
- nucleotide
- RPKM
- reads per kilobase per million reads
- TAP
- tris acetate-phosphate
- EST
- expressed sequence tag
- JGI
- Joint Genome Institute.
REFERENCES
- 1. Merchant S. S., Kropat J., Liu B., Shaw J., Warakanont J. (2012) TAG, You're it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr. Opin. Biotechnol. 23, 1–12 [DOI] [PubMed] [Google Scholar]
- 2. Chisti Y. (2008) Biodiesel from microalgae beats bioethanol. Trends Biotechnol. 26, 126–131 [DOI] [PubMed] [Google Scholar]
- 3. Chisti Y. (2007) Biodiesel from microalgae. Biotechnol. Adv. 25, 294–306 [DOI] [PubMed] [Google Scholar]
- 4. Scott S. A., Davey M. P., Dennis J. S., Horst I., Howe C. J., Lea-Smith D. J., Smith A. G. (2010) Biodiesel from algae. Challenges and prospects. Curr. Opin. Biotechnol. 21, 277–286 [DOI] [PubMed] [Google Scholar]
- 5. Schenk P., Thomas-Hall S., Stephens E., Marx U., Mussgnug J., Posten C., Kruse O., Hankamer B. (2008) Second generation biofuels. High efficiency microalgae for biodiesel production. BioEnergy Res. 1, 20–43 [Google Scholar]
- 6. Mata T. M., Martins A. A., Caetano N. S. (2010) Microalgae for biodiesel production and other applications. A review. Renewable Sustainable Energy Rev. 14, 217–232 [Google Scholar]
- 7. Griffiths M., Harrison S. (2009) Lipid productivity as a key characteristic for choosing algal species for biodiesel production. J. Appl. Phycol. 21, 493–507 [Google Scholar]
- 8. Steen E. J., Kang Y., Bokinsky G., Hu Z., Schirmer A., McClure A., Del Cardayre S. B., Keasling J. D. (2010) Microbial production of fatty acid-derived fuels and chemicals from plant biomass. Nature 463, 559–562 [DOI] [PubMed] [Google Scholar]
- 9. Waltz E. (2009) Biotech's green gold? Nat. Biotechnol. 27, 15–18 [DOI] [PubMed] [Google Scholar]
- 10. Rosenberg J. N., Oyler G. A., Wilkinson L., Betenbaugh M. J. (2008) A green light for engineered algae. Redirecting metabolism to fuel a biotechnology revolution. Curr. Opin. Biotechnol. 19, 430–436 [DOI] [PubMed] [Google Scholar]
- 11. Matthew T., Zhou W., Rupprecht J., Lim L., Thomas-Hall S. R., Doebbe A., Kruse O., Hankamer B., Marx U. C., Smith S. M., Schenk P. M. (2009) The metabolome of Chlamydomonas reinhardtii following induction of anaerobic H2 production by sulfur depletion. J. Biol. Chem. 284, 23415–23425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Z. T., Ullrich N., Joo S., Waffenschmidt S., Goodenough U. (2009) Algal lipid bodies. Stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot. Cell 8, 1856–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Y., Han D., Hu G., Dauvillee D., Sommerfeld M., Ball S., Hu Q. (2010) Chlamydomonas starchless mutant defective in ADP-glucose pyrophosphorylase hyper-accumulates triacylglycerol. Metab. Eng. 12, 387–391 [DOI] [PubMed] [Google Scholar]
- 14. Li Y., Han D., Hu G., Sommerfeld M., Hu Q. (2010) Inhibition of starch synthesis results in overproduction of lipids in Chlamydomonas reinhardtii. Biotechnol. Bioeng. 107, 258–268 [DOI] [PubMed] [Google Scholar]
- 15. Weers P. M., Gulati R. D. (1997) Growth and reproduction of Daphnia galeata in response to changes in fatty acids, phosphorus, and nitrogen in Chlamydomonas reinhardtii. Limnol. Oceanogr. 42, 1584–1589 [Google Scholar]
- 16. Hu Q., Sommerfeld M., Jarvis E., Ghirardi M., Posewitz M., Seibert M., Darzins A. (2008) Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54, 621–639 [DOI] [PubMed] [Google Scholar]
- 17. Work V. H., Radakovits R., Jinkerson R. E., Meuser J. E., Elliott L. G., Vinyard D. J., Laurens L. M., Dismukes G. C., Posewitz M. C. (2010) Increased lipid accumulation in the Chlamydomonas reinhardtii sta7-10 starchless isoamylase mutant and increased carbohydrate synthesis in complemented strains. Eukaryot. Cell 9, 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miller R., Wu G., Deshpande R. R., Vieler A., Gaertner K., Li X., Moellering E. R., Zauner S., Cornish A., Liu B., Bullard B., Sears B. B., Kuo M. H., Hegg E. L., Shachar-Hill Y., Shiu S. H., Benning C. (2010) Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 110.165159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moellering E. R., Benning C. (2010) RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot. Cell 9, 97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goodson C., Roth R., Wang Z. T., Goodenough U. (2011) Structural correlates of cytoplasmic and chloroplast lipid body synthesis in Chlamydomonas reinhardtii and stimulation of lipid body production with acetate boost. (2011) Eukaryot. Cell 10, 1592–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kropat J., Hong-Hermesdorf A., Casero D., Ent P., Castruita M., Pellegrini M., Merchant S. S., Malasarn D. (2011) A revised mineral supplement increase biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 66, 770–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Siaut M., Cuiné S., Cagnon C., Fessler B., Nguyen M., Carrier P., Beyly A., Beisson F., Triantaphylidès C., Li-Beisson Y., Peltier G. (2011) Oil accumulation in the model green alga Chlamydomonas reinhardtii. Characterization, variability between common laboratory strains, and relationship with starch reserves. BMC Biotechnol. 11, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhekisheva M., Boussiba S., Khozin-Goldberg I., Zarka A., Cohen Z. (2002) Accumulation of oleic acid in Haematococcus pluvialis (Chlorophyceae) under nitrogen starvation or high light is correlated with that of astaxanthin esters. J. Phycol. 38, 325–331 [Google Scholar]
- 24. Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., Witman G. B., Terry A., Salamov A., Fritz-Laylin L. K., Maréchal-Drouard L., Marshall W. F., Qu L. H., Nelson D. R., Sanderfoot A. A., Spalding M. H., Kapitonov V. V., Ren Q., Ferris P., Lindquist E., Shapiro H., Lucas S. M., Grimwood J., Schmutz J., Cardol P., Cerutti H., Chanfreau G., Chen C. L., Cognat V., Croft M. T., Dent R., Dutcher S., Fernández E., Fukuzawa H., González-Ballester D., González-Halphen D., Hallmann A., Hanikenne M., Hippler M., Inwood W., Jabbari K., Kalanon M., Kuras R., Lefebvre P. A., Lemaire S. D., Lobanov A. V., Lohr M., Manuell A., Meier I., Mets L., Mittag M., Mittelmeier T., Moroney J. V., Moseley J., Napoli C., Nedelcu A. M., Niyogi K., Novoselov S. V., Paulsen I. T., Pazour G., Purton S., Ral J. P., Riaño-Pachon D. M., Riekhof W., Rymarquis L., Schroda M., Stern D., Umen J., Willows R., Wilson N., Zimmer S. L., Allmer J., Balk J., Bisova K., Chen C. J., Elias M., Gendler K., Hauser C., Lamb M. R., Ledford H., Long J. C., Minagawa J., Page M. D., Pan J., Pootakham W., Roje S., Rose A., Stahlberg E., Terauchi A. M., Yang P., Ball S., Bowler C., Dieckmann C. L., Gladyshev V. N., Green P., Jorgensen R., Mayfield S., Mueller-Roeber B., Rajamani S., Sayre R. T., Brokstein P., Dubchak I., Goodstein D., Hornick L., Huang Y. W., Jhaveri J., Luo Y., Martinez D., Ngau W. C., Otillar B., Poliakov A., Porter A., Szajkowski L., Werner G., Zhou K., Grigoriev I. V., Rokhsar D. S., Grossman A. R. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wijffels R. H., Barbosa M. J. (2010) An outlook on microalgal biofuels. Science 329, 796–799 [DOI] [PubMed] [Google Scholar]
- 26. Giroud C., Gerber A., Eichenberger W. (1988) Lipids of Chlamydomonas reinhardtii. Analysis of molecular species and intracellular sites(s) of biosynthesis. Plant Cell Physiol. 29, 587–595 [Google Scholar]
- 27. Riekhof W. R., Sears B. B., Benning C. (2005) Annotation of genes involved in glycerolipid biosynthesis in Chlamydomonas reinhardtii. Discovery of the betaine lipid synthase BTA1Cr. Eukaryot. Cell 4, 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cases S., Smith S. J., Zheng Y. W., Myers H. M., Lear S. R., Sande E., Novak S., Collins C., Welch C. B., Lusis A. J., Erickson S. K., Farese R. V. (1998) Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. U.S.A. 95, 13018–13023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lardizabal K. D., Mai J. T., Wagner N. W., Wyrick A., Voelker T., Hawkins D. J. (2001) DGAT2 is a new diacylglycerol acyltransferase gene family. Purification, cloning, and expression in insect cells of two polypeptides from Mortierella ramanniana with diacylglycerol acyltransferase activity. J. Biol. Chem. 276, 38862–38869 [DOI] [PubMed] [Google Scholar]
- 30. Stymne S., Stobart A. K. (1987) in The Biochemistry of Plants (Stumpf P. K., ed) pp. 175–214, Academic Press, New York [Google Scholar]
- 31. Lacey D. J., Hills M. J. (1996) Heterogeneity of the endoplasmic reticulum with respect to lipid synthesis in developing seeds of the Brassica napus L. Planta 199, 545–551 [Google Scholar]
- 32. Stobart A. K., Stymne S., Höglund S. (1986) Safflower microsomes catalyze oil accumulation in vitro. A model system. Planta 169, 33–37 [DOI] [PubMed] [Google Scholar]
- 33. Shockey J. M., Gidda S. K., Chapital D. C., Kuan J. C., Dhanoa P. K., Bland J. M., Rothstein S. J., Mullen R. T., Dyer J. M. (2006) Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18, 2294–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li R., Yu K., Hildebrand D. (2010) DGAT1, DGAT2, and PDAT expression in seeds and other tissues of epoxy and hydroxy fatty acid accumulating plants. Lipids 45, 145–157 [DOI] [PubMed] [Google Scholar]
- 35. Banilas G., Karampelias M., Makariti I., Kourti A., Hatzopoulos P. (2011) The olive DGAT2 gene is developmentally regulated and shares overlapping but distinct expression patterns with DGAT1. J. Exp. Bot. 62, 521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stone S. J., Myers H. M., Watkins S. M., Brown B. E., Feingold K. R., Elias P. M., Farese R. V. (2004) Lipopenia and skin barrier abnormalities in DGAT2-deficient mice. J. Biol. Chem. 279, 11767–11776 [DOI] [PubMed] [Google Scholar]
- 37. Saha S., Enugutti B., Rajakumari S., Rajasekharan R. (2006) Cytosolic triacylglycerol biosynthetic pathway in oilseeds. Molecular cloning and expression of peanut cytosolic diacylglycerol acyltransferase. Plant Physiol. 141, 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peng F. Y., Weselake R. J. (2011) Gene coexpression clusters and putative regulatory elements underlying seed storage reserve accumulation in Arabidopsis. BMC Genomics 12, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sandager L., Gustavsson M. H., Ståhl U., Dahlqvist A., Wiberg E., Banas A., Lenman M., Ronne H., Stymne S. (2002) Storage lipid synthesis is nonessential in yeast. J. Biol. Chem. 277, 6478–6482 [DOI] [PubMed] [Google Scholar]
- 40. Oelkers P., Cromley D., Padamsee M., Billheimer J. T., Sturley S. L. (2002) The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J. Biol. Chem. 277, 8877–8881 [DOI] [PubMed] [Google Scholar]
- 41. Benning C., Miller R., Moellering E. R. (2010) Enzyme-directed oil biosynthesis in microalgae, Michigan State University Board of Trustees [Google Scholar]
- 42. Nguyen H. M., Baudet M., Cuiné S., Adriano J. M., Barthe D., Billon E., Bruley C., Beisson F., Peltier G., Ferro M., Li-Beisson Y. (2011) Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii, with focus on proteins involved in lipid metabolism. Proteomics 11, 4266–4273 [DOI] [PubMed] [Google Scholar]
- 43. Pollock S. V., Colombo S. L., Prout D. L., Jr., Godfrey A. C., Moroney J. V. (2003) Rubisco activase is required for optimal photosynthesis in the green alga Chlamydomonas reinhardtii in a low CO2 atmosphere. Plant Physiol. 133, 1854–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schnell R. A., Lefebvre P. A. (1993) Isolation of the Chlamydomonas regulatory gene NIT2 by transposon tagging. Genetics 134, 737–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Harris E. H. (1989) The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use, Academic Press, Inc., San Diego: [DOI] [PubMed] [Google Scholar]
- 46. Quisel J. D., Wykoff D. D., Grossman A. R. (1996) Biochemical characterization of the extracellular phosphatases produced by phosphorus-deprived Chlamydomonas reinhardtii. Plant Physiol. 111, 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moseley J., Quinn J., Eriksson M., Merchant S. (2000) The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 19, 2139–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. González-Ballester D., Casero D., Cokus S., Pellegrini M., Merchant S. S., Grossman A. R. (2010) RNA-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell 22, 2058–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Moseley J. L., Allinger T., Herzog S., Hoerth P., Wehinger E., Merchant S., Hippler M. (2002) Adaptation to Fe deficiency requires remodeling of the photosynthetic apparatus. EMBO J. 21, 6709–6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Allen M. D., del Campo J. A., Kropat J., Merchant S. S. (2007) FEA1, FEA2, and FRE1, encoding two homologous secreted proteins and a candidate ferrireductase, are expressed coordinately with FOX1 and FTR1 in iron-deficient Chlamydomonas reinhardtii. Eukaryot. Cell 6, 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hill K. L., Li H. H., Singer J., Merchant S. (1991) Isolation and structural characterization of the Chlamydomonas reinhardtii gene for cytochrome c6. Analysis of the kinetics and metal specificity of its copper-responsive expression. J. Biol. Chem. 266, 15060–15067 [PubMed] [Google Scholar]
- 52. Quinn J. M., Merchant S. (1998) in Methods in Enzymology (Lee M., ed) pp. 263–279, Academic Press, New York: [DOI] [PubMed] [Google Scholar]
- 53. Langmead B., Trapnell C., Pop M., Salzberg S. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Meth. 5, 621–628 [DOI] [PubMed] [Google Scholar]
- 55. Xie Z., Merchant S. (1996) The plastid-encoded ccsA gene is required for heme attachment to chloroplast c-type cytochromes. J. Biol. Chem. 271, 4632–4639 [DOI] [PubMed] [Google Scholar]
- 56. Depège N., Bellafiore S., Rochaix J. D. (2003) Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299, 1572–1575 [DOI] [PubMed] [Google Scholar]
- 57. Feinberg A. P., Vogelstein B. (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132, 6–13 [DOI] [PubMed] [Google Scholar]
- 58. Ramakers C., Ruijter J. M., Deprez R. H., Moorman A. F. (2003) Assumption-free analysis of quantitative real time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66 [DOI] [PubMed] [Google Scholar]
- 59. Ruijter J. M., Ramakers C., Hoogaars W. M., Karlen Y., Bakker O., van den Hoff M. J., Moorman A. F. (2009) Amplification efficiency. Linking base line and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M. W., Shipley G. L., Vandesompele J., Wittwer C. T. (2009) The MIQE guidelines: minimum information for publication of quantitative real time PCR experiments. Clin. Chem. 55, 611–622 [DOI] [PubMed] [Google Scholar]
- 61. Simpson J. T., Wong K., Jackman S. D., Schein J. E., Jones S. J., Birol I. (2009) ABySS. A parallel assembler for short read sequence data. Genome Res. 19, 1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wu T. D., Watanabe C. K. (2005) GMAP. A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21, 1859–1875 [DOI] [PubMed] [Google Scholar]
- 63. Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., Haussler D. (2002) The human genome browser at UCSC. Genome Res. 12, 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G., Mesirov J. P. (2011) Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rossak M., Schäfer A., Xu N., Gage D. A., Benning C. (1997) Accumulation of sulfoquinovosyl-1-O-dihydroxyacetone in a sulfolipid-deficient mutant of Rhodobacter sphaeroides inactivated in sqdC. Arch. Biochem. Biophys. 340, 219–230 [DOI] [PubMed] [Google Scholar]
- 66. Sizova I., Fuhrmann M., Hegemann P. (2001) A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277, 221–229 [DOI] [PubMed] [Google Scholar]
- 67. Pootakham W., Gonzalez-Ballester D., Grossman A. R. (2010) Identification and regulation of plasma membrane sulfate transporters in Chlamydomonas. Plant Physiol. 153, 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gonzalez-Ballester D., Pootakham W., Mus F., Yang W., Catalanotti C., Magneschi L., de Montaigu A., Higuera J. J., Prior M., Galván A., Fernandez E., Grossman A. R. (2011) Reverse genetics in Chlamydomonas. A platform for isolating insertional mutants. Plant Methods 7, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sambrook J., Fritsch E., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 70. James G. O., Hocart C. H., Hillier W., Chen H., Kordbacheh F., Price G. D., Djordjevic M. A. (2011) Fatty acid profiling of Chlamydomonas reinhardtii under nitrogen deprivation. Bioresour. Technol. 102, 3343–3351 [DOI] [PubMed] [Google Scholar]
- 71. Moellering E. R., Miller R., Benning C. (2009) in Lipids in Photosynthesis: Essential and Regulatory Functions (Wada H., Murata N., eds) pp. 139–155, Spring Science + Business, Berlin [Google Scholar]
- 72. Riekhof W. R., Benning C. (2009) in The Chlamydomonas Sourcebook (Harris E. H., ed) pp. 41–68, Elsevier Science Publishers B.V., Amsterdam [Google Scholar]
- 73. Fan J., Andre C., Xu C. (2011) A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett. 585, 1985–1991 [DOI] [PubMed] [Google Scholar]
- 74. Msanne J., Xu D., Konda A. R., Casas-Mollano J. A., Awada T., Cahoon E. B., Cerutti H. (2012) Metabolic and gene expression changes triggered by nitrogen deprivation in the photoautotrophically grown microalgae Chlamydomonas reinhardtii and Coccomyxa sp. C-169. 75, 50–59 [DOI] [PubMed] [Google Scholar]
- 75. Petschnigg J., Wolinski H., Kolb D., Zellnig G., Kurat C. F., Natter K., Kohlwein S. D. (2009) Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J. Biol. Chem. 284, 30981–30993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lockshon D., Surface L. E., Kerr E. O., Kaeberlein M., Kennedy B. K. (2007) The sensitivity of yeast mutants to oleic acid implicates the peroxisome and other processes in membrane function. Genetics 175, 77–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhang M., Fan J., Taylor D. C., Ohlrogge J. B. (2009) DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21, 3885–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hockin N. L., Mock T., Mulholland F., Kopriva S., Malin G. (2012) The response of diatom central carbon metabolism to nitrogen starvation is different from that of green algae and higher plants. Plant Physiol. 158, 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Matsui K., Fukutomi S., Ishii M., Kajiwara T. (2004) A tomato lipase homologous to DAD1 (LeLID1) is induced in post-germinative growing stage and encodes a triacylglycerol lipase. FEBS Lett. 569, 195–200 [DOI] [PubMed] [Google Scholar]
- 80. Toida J., Arikawa Y., Kondou K., Fukuzawa M., Sekiguchi J. (1998) Purification and characterization of triacylglycerol lipase from Aspergillus oryzae. Biosci. Biotechnol. Biochem. 62, 759–763 [DOI] [PubMed] [Google Scholar]
- 81. Vieler A., Wilhelm C., Goss R., Süss R., Schiller J. (2007) The lipid composition of the unicellular green alga Chlamydomonas reinhardtii and the diatom Cyclotella meneghiniana investigated by MALDI-TOF MS and TLC. Chem. Phys. Lipids 150, 143–155 [DOI] [PubMed] [Google Scholar]
- 82. Harris E. H. (2009) The Chlamydomonas Sourcebook, 2 Ed., Elsevier, Amsterdam [Google Scholar]
- 83. Yohn C., Mendez M., Behnke C., Brand A. (November 8, 2011) Stress-induced Lipid Trigger Organization, Sapphire Energy, U.S. Patent no. US 2011 023406
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.