Background: Carbon skeleton rearrangements of acyl-CoA esters are catalyzed by coenzyme B12-dependent mutases.
Results: A bacterial mutase specifically catalyzes the isomerization of 2-hydroxyisobutyryl- and (S)-3-hydroxybutyryl-CoA.
Conclusion: Substrate affinity and enzyme activity depend strongly on the active site amino acid Ile90.
Significance: This is the first characterization of an enzyme isomerizing hydroxylated short chain carboxylic acids.
Keywords: Adenosylcobalamin, Bacterial Metabolism, Enzyme Catalysis, Enzyme Kinetics, Enzyme Mechanisms, Metalloenzymes, 2-Hydroxyisobutyric Acid, 3-Hydroxybutyric Acid, Acyl-CoA Mutases, Substrate Specificity
Abstract
Coenzyme B12-dependent acyl-CoA mutases are radical enzymes catalyzing reversible carbon skeleton rearrangements in carboxylic acids. Here, we describe 2-hydroxyisobutyryl-CoA mutase (HCM) found in the bacterium Aquincola tertiaricarbonis as a novel member of the mutase family. HCM specifically catalyzes the interconversion of 2-hydroxyisobutyryl- and (S)-3-hydroxybutyryl-CoA. Like isobutyryl-CoA mutase, HCM consists of a large substrate- and a small B12-binding subunit, HcmA and HcmB, respectively. However, it is thus far the only acyl-CoA mutase showing substrate specificity for hydroxylated carboxylic acids. Complete loss of 2-hydroxyisobutyric acid degradation capacity in hcmA and hcmB knock-out mutants established the central role of HCM in A. tertiaricarbonis for degrading substrates bearing a tert-butyl moiety, such as the fuel oxygenate methyl tert-butyl ether (MTBE) and its metabolites. Sequence analysis revealed several HCM-like enzymes in other bacterial strains not related to MTBE degradation, indicating that HCM may also be involved in other pathways. In all strains, hcmA and hcmB are associated with genes encoding for a putative acyl-CoA synthetase and a MeaB-like chaperone. Activity and substrate specificity of wild-type enzyme and active site mutants HcmA I90V, I90F, and I90Y clearly demonstrated that HCM belongs to a new subfamily of B12-dependent acyl-CoA mutases.
Introduction
The tertiary carbon-bearing 2-hydroxyisobutyric acid (2-HIBA)2 is rarely found in Nature and is not an intermediate of the main metabolic pathways. However, it is one of the urinary organic acids found in humans with lactic acidosis (1), and several metabolic sequences can be proposed leading to this unusual short chain carboxylic acid (Fig. 1). A major natural source of 2-HIBA might be the plant cyanoglycoside linamarin (2), as the nitrile corresponding to 2-HIBA is an intermediate of linamarin biosynthesis and catabolism. In addition, 2-HIBA could be produced during degradation of isobutane and isobutene via oxidation of the corresponding alcoholic and diolic metabolites. Besides, anthropogenic sources of 2-HIBA exist, because it is a pharmaceutical intermediate and by-product of industrial processes, e.g. the production of poly(methyl methacrylate) (PMMA) (3). It has also been identified as a metabolite in the bacterial degradation of the gasoline additive methyl tert-butyl ether (MTBE) (4), which is used at large scale since the 1990s as a fuel oxygenate for reducing carbon monoxide emissions and now threatens drinking water resources due to its persistence in contaminated aquifers (5). Although biodegradation of 2-HIBA and its methyl ester has already been observed in one early study in 1984 when investigating the bacterial degradation of wastewater compounds of a PMMA plant (6), convincing enzymatic steps for its conversion to common metabolites have not been proposed for >20 years.
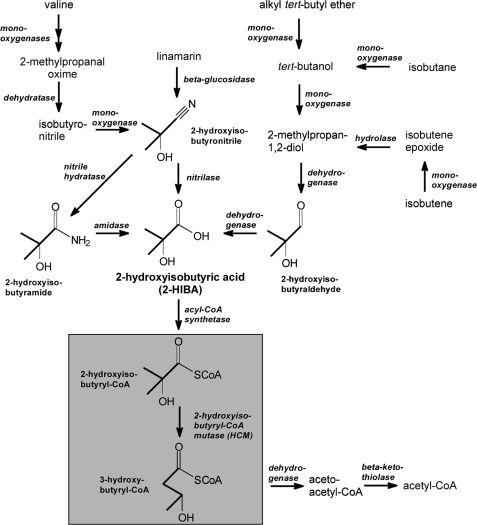
FIGURE 1.
Possible metabolic routes leading to 2-HIBA. Degradation of valine, linamarin, alkyl tert-butyl ethers, and branched C4 hydrocarbons could result in 2-HIBA formation. Further degradation proceeds via isomerization to the common metabolite 3-hydroxybutyric acid by the B12-dependent acyl-CoA mutase HCM discovered in the MTBE-degrading bacterial strain A. tertiaricarbonis L108 (7).
Recently, we found a coenzyme B12-dependent rearrangement reaction in the bacterial degradation pathway of MTBE via 2-HIBA, likely catalyzed by a novel acyl-CoA mutase (7). In this enzymatic step, the CoA ester of 2-HIBA is converted to 3-hydroxybutyryl-CoA (Fig. 1). Thus, in a single reaction the branched-chain α-hydroxy carboxylic acid is rearranged into an easily metabolizable linear isomer, now possessing an oxidizable secondary hydroxyl group at the β position. Consequently, the proposed 2-hydroxyisobutyryl-CoA mutase (HCM) would play a central role in MTBE metabolism enabling its complete mineralization. Similar rearrangements in carboxylic acids have already been observed with several acyl-CoA mutases (8, 9, 10), namely methylmalonyl-CoA mutase (MCM), isobutyryl-CoA mutase (ICM) and ethylmalonyl-CoA mutase (ECM). Recently, a variant of ICM (IcmF) has been characterized as a fusion of the ICM sequence with the G protein chaperone MeaI (11), a paralog of the MCM-associated MeaB protein. However, HCM would be distinct from all other known acyl-CoA mutases because it would catalyze the conversion of hydroxylated carboxylic acids.
This study aimed at elucidating the biochemistry of the acyl-CoA mutase activity from the MTBE-degrading bacterium Aquincola tertiaricarbonis L108. It was found that HCM consists of a large acyl-CoA-binding and a small B12-binding subunit, HcmA and HcmB, respectively. Destroying the corresponding genes hcmA and hcmB by insertional mutation resulted in complete loss of 2-HIBA-degrading capability. The wild-type mutase genes were cloned in Escherichia coli strains, and enzyme activity of heterologously expressed subunits was characterized. Catalysis by purified recombinant HCM was specific for 2-hydroxyisobutyryl- and (S)-3-hydroxybutyryl-CoA. Sequence comparison with known acyl-CoA mutases identified a single active site amino acid residue present in HcmA likely being important for determining substrate specificity. To understand the function of this residue, we analyzed activities of three mutant HcmA subunits having relevant amino acid substitutions.
EXPERIMENTAL PROCEDURES
Materials
Tert-butyl alcohol (TBA) (≥99%), 2-HIBA (>98%), tert-amyl alcohol (>99%), and anhydrides of butyric (98%) and isobutyric (98%) acids were purchased from Merck Schuchardt. 2-Methylpropan-1,2-diol was from Taros Chemicals at the highest purity available. Coenzyme B12 (≥97%), CoA (≥93%), methylmalonyl-CoA (≥96%, racemic mixture), and succinyl-CoA (≥94%) were purchased from Sigma. Enantiopure (R)- and (S)-3-hydroxybutyryl-CoA were provided by Evonik Industries (53–54% acyl-CoA enantiomer, >99.9 and 98.9% enantiomeric excess, respectively; 40% NaCl; 6–7% N-hydroxysuccinimide).
Syntheses
Synthesis of Acyl-CoA Esters
Acyl-CoA esters not commercially available were synthesized according to two methods. Isobutyryl-CoA and butyryl-CoA were prepared from their anhydrides (12). 2-Hydroxyisobutyryl-CoA was synthesized from its free acid via thiophenyl ester by the method of Padmakumar et al. (13).
Identification of Synthesis Products
Identity of synthesized acyl-CoA esters was verified by electrospray ionization-tandem mass spectrometry (ESI-MS/MS) using a 4000 QTRAP LC/MS/MS system consisting of a liquid chromatograph and a high performance triple quadrupole/linear ion trap mass spectrometer (Applied Biosystems). Acyl-CoA esters were detected directly by ESI-MS/MS. Isocratic elution was achieved using a mobile phase containing 17.5 mm ammonium acetate, 0.5 volume % acetic acid, and 30 volume % acetonitrile (14). Characteristic mass spectra for the respective acyl-CoA compounds were obtained (supplemental Fig. S1).
Bacterial Strains and Growth Conditions
Strain A. tertiaricarbonis L108 isolated from an MTBE-contaminated aquifer at Leuna, Germany (7, 15) was cultivated in liquid mineral salt medium (MSM) containing MTBE at concentrations of 0.3 g liter−1 as described previously (16). For growth and resting-cell studies, also tert-amyl alcohol, TBA, 2-methylpropan-1,2-diol, and 2-HIBA were supplied as sole source of carbon and energy at 0.5 g liter−1. E. coli TOP10 and ArcticExpress (DE3) were grown in Luria-Bertani broth. Growth was monitored by measuring optical density (OD) of cultures at 700 or 550 nm as indicated.
Sequencing of Genomic DNA from A. tertiaricarbonis
For sequencing a larger fragment of the hcm gene region of wild-type strain L108, genomic DNA was extracted using MasterPure DNA Purification kit (Epicenter) and sequenced by Illumina HiSeq 2000 technology (GATC Biotech). The obtained DNA sequences were analyzed for open reading frames using Rast (Rapid Annotation using Subsystem Technology). A 4.5 kb sequence including both hcm genes was obtained.
Cloning and Heterologous Expression of hcmA and hcmB from A. tertiaricarbonis in E. coli
The gene encoding for the large subunit of HCM (hcmA) was amplified from strain L108 genomic DNA by applying the forward primer 5′-AATG ACC TGG CTT GAG CCG CAG A-3′ and reverse primer 5′-TCCC GAA GAC CGG GTC TCG CGG-3′. PCR was accomplished with Pfu DNA polymerase (Promega) for 30 cycles, including denaturation at 94 °C for 1 min, annealing at 57 °C for 1 min, and extension at 72 °C for 2 min. The PCR product was cloned into expression vector pASG-IBA43 (IBA Goettingen) and transformed into E. coli TOP10 according to the protocol of IBA Goettingen. Induction was performed at OD550 of 0.5 with 200 μg liter−1 anhydrotetracycline for 3 h at 30 °C. Cells were centrifuged and suspended in Tris buffer (100 mm Tris, 150 mm NaCl, pH 8.0) for further analysis. The cloning vector pPR-IBA1::hcmB for the small subunit of HCM was purchased from DNA2.0 and transformed into 100 μl of electrocompetent E. coli ArcticExpress (DE3) (Novagen) at 300 mV for 5 ms in chilled 0.1-cm cuvettes in a MicroPulser (Bio-Rad). Induction was performed with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 20 h at 12 °C, after growth at 30 °C in Luria-Bertani medium containing 20 mg liter−1 gentamycin and 50 mg liter−1 kanamycin to an OD550 of 0.4 and further incubation at 12 °C to an OD550 of 0.5. Then, cells were centrifuged and suspended in Tris buffer.
Purification of Recombinant Proteins
For the purification of recombinant mutase subunits, crude extracts of induced E. coli cells were prepared by disruption using a mixer mill (MM 400, Retsch GmbH, Germany) with glass beads (212–300 μm, Sigma) at 30 s−1 for 30 min. The recombinant HcmA and HcmB subunits were purified with the help of their His and Strep tags, respectively. All purification steps were performed at 12 °C.
HcmA Subunit
Crude extracts of E. coli TOP10 pASG-IBA43::hcmA were loaded on a nickel-nitrilotriacetic acid Superflow 10-ml column (IBA Goettingen). After washing with 20 column volumes of imidazole buffer (50 mm sodium phosphate, 300 mm NaCl, 20 mm imidazole, pH 8.0), HcmA was eluted with the same buffer containing 250 mm imidazole. Fractions containing HcmA were concentrated via viva spin columns (30 kDa; GE Healthcare) and diluted with conservation buffer (50 mm potassium phosphate, 10% glycerol, pH 7.4).
HcmB Subunit
Crude extracts of E. coli ArcticExpress (DE3) pPR-IBA1::hcmB were loaded on a Strep-Tactin Superflow high capacity 10-ml column (IBA Goettingen). After washing with 20 column volumes of Tris buffer, HcmB was eluted with elution buffer (Tris buffer containing 2.5 mm desthiobiotin). Fractions containing HcmB were concentrated using viva spin columns (10 kDa) and diluted with conservation buffer.
Expression and Purification of Site-specific HcmA Mutants
The hcmA mutant genes cloned into pASG-IBA43 were purchased from GeneCust Europe. Site-directed mutagenesis resulted in HcmA I90Y, I90F, and I90V mutants (with the point mutations a268t plus t269a, a268t, and a268g, respectively). The vectors were transformed into E. coli TOP10. Expression and purification were performed as described for the wild-type recombinant HcmA.
Quantitative Enzymatic Measurements of Recombinant Enzyme
HCM Activity
Enzyme activity was routinely measured in 1–2 ml of 50 mm potassium phosphate buffer, pH 6.6, containing 10% glycerol, 833 μm coenzyme B12, and 10 mm MgCl2, at 30 °C in the dark. This reaction mixture was incubated in 10-ml headspace glass vials sealed with rubber stoppers. As variation of HcmA and HcmB ratios (up to 5:1 and 1:5, respectively) did not result in increased activities, the recombinant subunits were added at equimolar ratios (3 μm) throughout the study. After a 5-min preincubation in the presence of coenzyme B12, the reaction was started by adding acyl-CoA substrates. Throughout the experiments, oxygen concentrations were minimized by permanently flushing the incubation vials with nitrogen. For determination of pH optimum, pH values of the phosphate buffer and a phosphate/acetate buffer (phosphate buffer plus 50 mm sodium acetate buffer) were adjusted to values between 5.0 and 7.8. The temperature optimum was determined by incubating at temperatures between 20 and 55 °C.
HPLC Analysis of Acyl-CoA Esters
Concentrations of acyl-CoA ester substrates and products were quantified by ion-pair chromatography using an HPLC system (Shimadzu) with a Nucleosil 100–5 C18 column (250 mm × 3 mm, 5 μm; Macherey-Nagel) and a mobile phase of 14.5 vol% acetonitrile, 10 mm tetrabutylammonium hydrogen sulfate and 100 mm sodium phosphate at pH 4.5 for the separation of 2-hydroxyisobutyryl-, 3-hydroxybutyryl-, methylmalonyl-, and succinyl-CoA, with retention times of about 35, 24, 30, and 40 min, respectively. For separation of isobutyryl- and butyryl-CoA, acetonitrile content of the eluent was 21.6 vol%, resulting in retention times of 35 and 40 min, respectively (17). Eluent flow was adjusted to 0.6 ml min−1, and column oven temperature was 30 °C. Absorbance at 260 nm was used for quantifying acyl-CoA esters. In addition, spectra from 190 to 280 nm were recorded for distinguishing peaks of CoA esters from unspecific signals (18). Detection limit at 260 nm was 0.5 μm CoA ester. For stopping enzymatic catalysis, samples were mixed with an equal volume of stop buffer (100 mm acetate, pH 3.5) and incubated at 60 °C for 5 min prior to HPLC analysis.
Knock-out Mutant Construction and Strain Isolation
To establish the enzymatic function of HcmAB in 2-HIBA degradation, we generated knock-out mutants of strain A. tertiaricarbonis L108 by electroporation of 1 μl of EZ-Tn5<KAN-2>Tnp transposome (Epicenter Biotechnologies) to 70 μl of electrocompetent L108 cells in chilled cuvettes at 1.8 kV for about 5 ms (MicroPulser; Bio-Rad). Transformed cells were rescued in 5 ml of MSM amended with 10 mm fructose for 6 h at 30 °C and 150 rpm. Dilutions were plated on MSM fructose agar containing 50 μg ml−1 kanamycin and incubated for 2 days at 30 °C. As the transposome integrates randomly into the DNA, all colonies obtained had to be analyzed for loss of their capability to grow on MSM agar containing 0.5 g liter−1 TBA. In addition, copies of the colonies were maintained on MSM fructose agar for further analysis. Colonies with restricted or even lost TBA degradation potential were transferred on MSM 2-HIBA agar plates. The mutants that failed to grow were further analyzed.
The exact integration site of the kanamycin cassette into the genomic DNA was determined by direct DNA sequencing using flanking KAN-2 primers (Epicenter Biotechnologies) of 1 mg liter−1 high concentrated genomic DNA (MasterPure DNA Purification kit). Conditions were 4 min initial denaturation at 95 °C followed by 60 cycles of 30 s at 95 °C and 4 min 60 °C, finally cooled down to 8 °C. The products were cleaned with Centri-Sep columns (Applied Biosystems) and sequenced using an ABI PRISM 3100 Genetic Analyzer with the BigDye Terminator v1.1 Cycle sequencing kit (Applied Biosystems). The resulting sequences were analyzed with BLAST (19) and aligned via Sequencher 5.0 software (Gene Codes Corporation).
RESULTS
Insertional Inactivation of hcmA and hcmB, the Putative Genes Encoding for HCM Large and Small Subunits, and Characterization of Mutant Strains
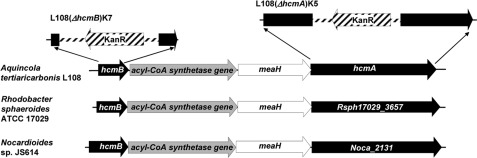
Sequencing of genomic DNA of wild-type A. tertiaricarbonis L108 revealed that a 4.5-kb fragment comprised both hcmA and hcmB separated by two genes encoding for a putative acyl-CoA synthetase and a MeaB-like G protein chaperone, respectively (Fig. 2). A highly similar sequence (>97% identity) having the same gene organization has already been found in the genome of the MTBE-degrading strain Methylibium petroleiphilum PM1 (20), supporting the hypothesis that HCM is involved in degradation of the MTBE metabolite 2-HIBA (7). However, the organization of hcm-like genes interrupted by genes likely encoding for a acyl-CoA synthetase and MeaB-like chaperone is also present in other bacteria, such as Rhodobacter sphaeroides ATCC 17029 and Nocardioides sp. JS614 (Fig. 2), thus far not related to fuel oxygenate ether degradation. As has already been suggested on the basis of transcriptome analysis of MTBE-grown cells of strain PM1 (21), the acyl-CoA synthetase could be involved in CoA activation of 2-HIBA (Fig. 1). The G protein chaperone may play a role in HCM assembly and stabilization, as has already found for MeaB and MeaI associated with MCM and ICM/IcmF, respectively (11, 22). Therefore, we propose for the MeaB/MeaI paralog associated with HCM the name MeaH (Fig. 2).
FIGURE 2.
HCM genetic structure. Comparison of the 4.5-kb hcm gene environment in A. tertiaricarbonis L108 with the closely related sequences of R. sphaeroides ATCC 17029 (NCBI locus tag Rsph17029_3654 to Rsph17029_3657) and Nocardioides sp. JS614 (Noca_2129 to Noca_2131). Genes of the HCM subunits hcmB and hcmA are drawn in black; the acyl-CoA synthetase gene is gray; and meaH, encoding for a MeaB-like chaperone, is white. The Ez-Tnp5<Kan-2>Tnp inserts from the two knock-out mutants L108(ΔhcmB)K7 and L108(ΔhcmA)K5 are shown fasciated, whereas the disrupted genes stay black. The different gene length and overlapping intensity between meaH and hcmA are based on the real data from NCBI. An identical organization of the hcm operon was also detected in the genomes of strains M. petroleiphilum PM1, R. sphaeroides KD131, S. novella DSM 506, Marinobacter algicola DG893, Mesorhizobium alhagi CCNWXJ12-2 and X. autotrophicus Py2.
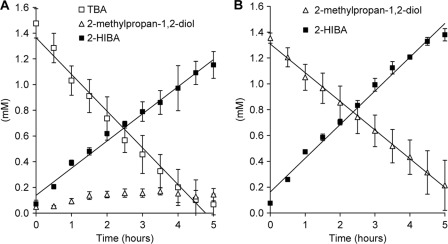
For precisely studying the role of HCM in bacterial 2-HIBA degradation, we tried to create hcmA and hcmB knock-out mutants of strain L108 by a site-specific mutagenesis approach via homologous recombination. However, even after several trials corresponding knock-out mutants were not obtained. On the other hand, unspecific transposon-mediated mutagenesis and screening for loss of capability to grow on 2-HIBA resulted in two hcm mutants, L108(ΔhcmA)K5 and L108(ΔhcmB)K7, bearing stable insertions of a functionally active kanamycin resistance gene in the hcmA and hcmB genes, respectively (Fig. 2). In contrast to the wild-type L108 strain, both mutant strains were not able to grow on TBA and 2-methylpropan-1,2-diol, precursors of 2-HIBA in the tert-butyl alkyl ether degradation pathway (Fig. 1), establishing the postulated central role of HCM in the degradation of organic compounds possessing the tert-butyl moiety (7). Accordingly, TBA and 2-methylpropan-1,2-diol were stoichiometrically converted to 2-HIBA by both knock-out mutants in resting-cell experiments (Fig. 3), indicating that only the 2-HIBA-metabolizing enzymatic activity was affected by the hcm knockouts. Contrarily, the mutants still grew well on tert-amyl alcohol, suggesting an alternative degradation pathway for this C5 homolog of TBA (23).
FIGURE 3.
Phenotypic characterization of hcm knock-out mutants. Nearly stoichiometric conversion of TBA (A) and 2-methylpropan-1,2-diol (B) to 2-HIBA by resting cells of the knock-out mutant strains L108(ΔhcmB)K7 and L108(ΔhcmA)K5 pre-grown on tert-amyl alcohol (mean values of experiments performed with both mutants) is shown. In the case of incubation with TBA, also small amounts of 2-methylpropan-1,2-diol accumulated.
Expression of hcm Genes in E. coli and Purification of HCM Subunits
For biochemical characterization, wild-type hcmA and hcmB as well as hcmA mutant genes were heterologously expressed in E. coli. N-terminal His-tagged and C-terminal Strep-tagged proteins, respectively, were purified from crude extracts by one-step affinity chromatography. Denaturing gel electrophoresis established the presence of the mutase subunits in crude extracts of induced E. coli cells and successful purification of the tagged proteins (supplemental Fig. S2). Yields of purified HcmA (HcmA Ile90 and HcmA mutants) and HcmB were about 100 and 80 mg liter−1 of E. coli culture, respectively.
Characterization of Reconstituted Hcm Enzyme Activity
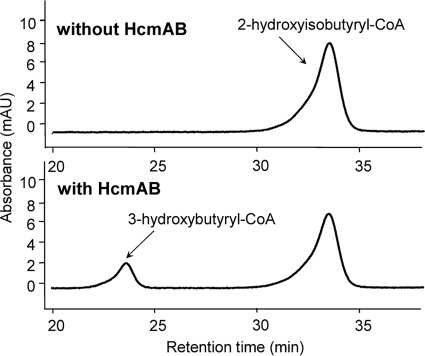
As has already been shown for the small and large ICM subunits (9), neither recombinant HcmA nor HcmB alone was able to catalyze an acyl-CoA mutase reaction. However, a combination of both purified subunits resulted in an enzymatic rearrangement of 2-hydroxyisobutyryl- into 3-hydroxybutyryl-CoA (Fig. 4). Under the experimental conditions applied, a linear increase in 3-hydroxybutyryl-CoA for about 10 min could be achieved, thus, enabling determination of turnover rates by tracing changes in substrate and product concentrations with HPLC (supplemental Fig. S3). Within a pH range of 5–8, significant enzymatic conversion of 2-hydroxyisobutyryl-CoA was observed, having a maximal activity at pH 6.6 (supplemental Fig. S4). The temperature optimum of the mutase activity was in the mesophilic range at about 30 °C. At 20 °C, however, still 50% of the maximal activity was achieved, whereas a complete loss of catalysis was shown at 45 °C (supplemental Fig. S5), which corresponds well to the temperature spectrum described for growth of wild-type strain L108 on TBA (24). Interestingly, as demonstrated by analyzing the 3-hydroxybutyryl-CoA esters produced, after derivatization to propyl esters and enantioselective separation on a chiral GC column (supplemental Fig. S6), 2-hydroxyisobutyryl-CoA was predominantly converted to (S)-3-hydroxybutyryl-CoA, and only minor amounts of about 20% of the (R)-enantiomer were obtained, indicating stereospecific catalysis.
FIGURE 4.
HPLC assay for the quantification of enzymatic transformation of 2-hydroxyisobutyryl-CoA. HPLC chromatograms of samples after 10 min of incubation show formation of 3-hydroxybutyryl-CoA only in the presence of reconstituted wild-type mutase subunits HcmA and HcmB.
Kinetic Parameters of HCM Catalysis
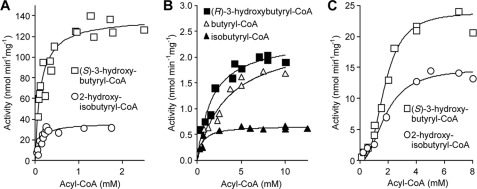
Possible enzymatic rearrangement of substrates other than 2-hydroxyisobutyryl-CoA was tested at optimal pH and temperature conditions (Table 1 and Fig. 5). As expected, purified wild-type HCM subunits were able to convert (S)-3-hydroxybutyryl-CoA at high rates, demonstrating the reversibility of the rearrangement reaction. Compared with Vmax obtained with 2-hydroxyisobutyryl-CoA, rates with (S)-3-hydroxybutyryl-CoA were nearly four times higher, indicating that at least in vitro not the direction toward degradation of tert-butyl group molecules (Fig. 1) but the reverse reaction is favored. As reconstituted HcmAB remained active for extended times, not only steady-state rates were measured but also equilibrium between 2-hydroxyisobutyryl- and 3-hydroxybutyryl-CoA could be reached (Fig. 6). Accordingly, with the higher specific activities obtained for (S)-3-hydroxybutyryl-CoA (Table 1), the equilibrium lies on side of 2-hydroxyisobutyryl-CoA, resulting in an equilibrium constant Keq of approximately 1.5. In line with the finding that conversion of 2-hydroxyisobutyryl-CoA was stereospecific for the (S)-isomer of 3-hydroxybutyryl-CoA (>80% enantiomeric excess), only very low activity was observed with pure (R)-3-hydroxybutyryl-CoA, resulting in approximately 2% of the Vmax value obtained with the (S)-enantiomer. As expected, reconstituted HcmAB did not show detectable conversion of the MCM substrates methylmalonyl- and succinyl-CoA. However, with the ICM substrates isobutyryl- and butyryl-CoA low rearrangement activities were observed, corresponding to approximately 0.5 and 2% of the (S)-3-hydroxybutyryl-CoA conversion rate. With the conservative HcmA I90V mutant, significant activities were only obtained with (S)-3-hydroxybutyryl- and 2-hydroxyisobutyryl-CoA, showing <20% of the wild-type rates (Table 1 and Fig. 5C). The nonconservative substitutions I90Y and I90F resulted in complete loss of enzyme activity.
TABLE 1.
Kinetic parameters of reconstituted HCM subunits HcmA (wild-type Ile90 and mutant I90V) and wild-type HcmB incubated with various acyl-CoA substrates at pH 6.6 and 30 °C (Fig. 5).
| Subunit and substrate | Vmaxa | Km | kcat | kcat/Km | Vmax/Km |
|---|---|---|---|---|---|
| nmol min−1mg−1 | μm | min−1 | mm−1 min−1 | liters min−1mg−1 | |
| HcmA Ile90 | |||||
| (S)-3-Hydroxybutyryl-CoA | 140 ± 5.6 (100)b | 128 ± 22 | 12 ± 0.5 | 90 ± 16 | 1,080 ± 190 |
| 2-Hydroxyisobutyryl-CoA | 36 ± 2.8 (26) | 104 ± 25 | 3.0 ± 0.2 | 29 ± 7.3 | 350 ± 87 |
| (R)-3-Hydroxybutyryl-CoA | 2.4 ± 0.15 (2) | 1,660 ± 367 | 0.20 ± 0.01 | 0.12 ± 0.03 | 1.4 ± 0.3 |
| Butyryl-CoA | 2.4 ± 0.26 (2) | 3,340 ± 880 | 0.20 ± 0.02 | 0.06 ± 0.02 | 0.7 ± 0.2 |
| Isobutyryl-CoA | 0.67 ± 0.03 (0.5) | 550 ± 140 | 0.06 ± 0.003 | 0.10 ± 0.03 | 1.2 ± 0.3 |
| HcmA I90V | |||||
| (S)-3-Hydroxybutyryl-CoA | 24 ± 1.1 (17) | 1,760 ± 240 | 2.0 ± 0.1 | 1.1 ± 0.16 | 14 ± 2.0 |
| 2-Hydroxyisobutyryl-CoA | 15 ± 1.1 (11) | 1,840 ± 370 | 1.2 ± 0.1 | 0.7 ± 0.14 | 8.1 ± 1.7 |
a Activities obtained with the wild-type HcmAB for methylmalonyl- and succinyl-CoA and with the HcmA I90V mutant reconstituted with wild-type HcmB for all acyl-CoA esters tested, except for (S)-3-hydroxybutyryl- and 2-hydroxyisobutyryl-CoA, were below detection limit (<0.01 nmol min−1 mg−1).
b For calculating relative activities, Vmax obtained with wild-type HcmAB for (S)-3-hydroxybutyryl-CoA was set to 100%.
FIGURE 5.
Kinetic plots of acyl-CoA rearrangement activities catalyzed by reconstituted HcmA and HcmB. A and B, conversion of 2-hydroxyisobutyryl- and (S)-3-hydroxybutyryl-CoA (A) and (R)-3-hydroxybutyryl-, butyryl-, and isobutyryl-CoA by wild-type HcmAB (B). C, conversion of 2-hydroxyisobutyryl- and (S)-3-hydroxybutyryl-CoA by HcmA I90V mutant reconstituted with the wild-type HcmB. All enzyme activities were measured at pH 6.6 and 30 °C. By using nonlinear regression analysis (Graph Pad Prism 5.0 software), Km and Vmax values were revealed (Table 1), applying either the Michaelis-Menten equation (A and B) or an allosteric sigmoidal model (C).
FIGURE 6.
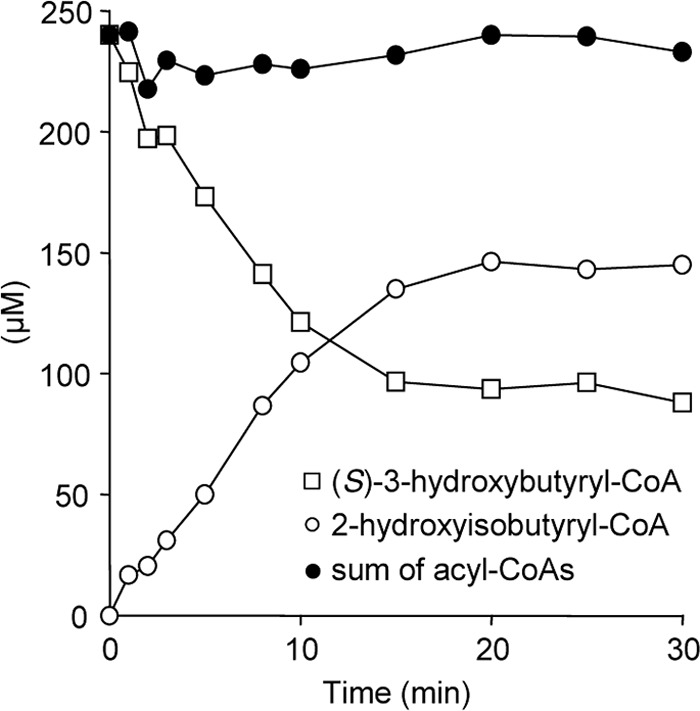
Equilibrium of HCM-catalyzed hydroxyacyl-CoA rearrangements. Appearance of equilibrium between (S)-3-hydroxybutyryl- and 2-hydroxyisobutyryl-CoA conversions by wild-type HcmAB after prolonged incubation is shown.
With the wild-type HcmA, the Km values for 2-hydroxyisobutyryl- and (S)-3-hydroxybutyryl CoA were both determined to be in the 100 μm range, whereas affinities to (R)-3-hydroxybutyryl-CoA and the ICM substrates were approximately 5–30 times lower (Table 1). Consequently, with these latter substrates the catalytic efficiency (kcat/Km) was approximately 1000-fold lower than with (S)-3-hydroxybutyryl-CoA. The substitution I90V resulted in Km values close to 2 mm and a nearly 100-fold diminution of the catalytic efficiency with (S)-3-hydroxybuytryl-CoA.
HCM Activity Defines New Subfamily of B12-dependent Acyl-CoA Mutases
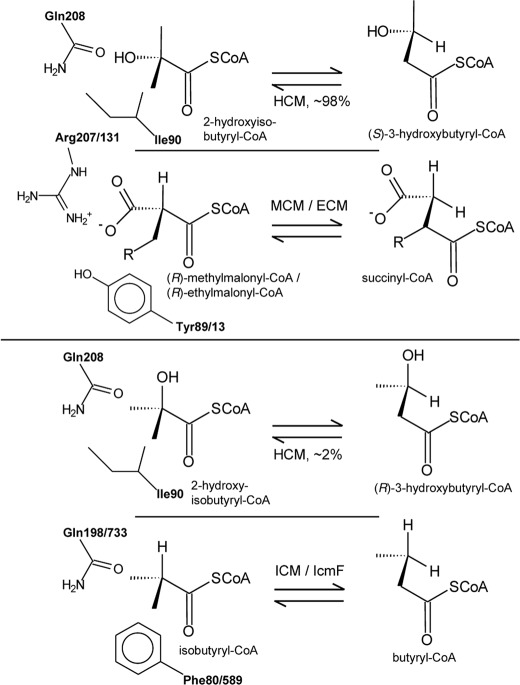
HcmAB showed only significant activity with 2-hydroxyisobutyryl- and (S)-3-hydroxybutyryl-CoA (Table 1). Likewise, narrow substrate specificity has also been demonstrated for the thus far known subfamilies of B12-dependent acyl-CoA mutases (8, 9, 10). Hence, the distinct specificity for hydroxylated short chain acyl residues found for HCM catalysis suggests that this enzyme represents a new mutase subfamily. This is also supported by a recent study on different IcmF enzymes demonstrating that 3-hydroxybutyryl-CoA is not converted by this type of mutase (25). BLAST analysis of L108 hcmA as query against the NCBI data base (March 2012) resulted in only a few HcmA-like proteins as closest matches with >60% identity to the L108 sequence. N-terminal regions of all matches were aligned with the corresponding sequences of representatives of the other mutase subfamilies ICM, IcmF, MCM, and ECM (supplemental Fig. S7). Particularly, comparison of two sequence sections containing amino acid residues known to be responsible for binding the acyl-CoA substrates (9) revealed a single Ile residue that clearly distinguishes HcmA-like sequences from other mutase proteins (Ile90 in the L108 sequence, Fig. 7). In addition, HCM, ICM, and IcmF sequences possess a Gln at position 208 (numbering as in HcmA of strain L108) corresponding to an Arg residue in MCM and ECM.
FIGURE 7.
ClustalW2 multiple sequence alignment of acyl-CoA binding domains of B12-dependent mutases. Comparison of HCM from A. tertiaricarbonis L108 with orthologous sequences from M. petroleiphilum PM1 (Mpe_B0541), R. sphaeroides KD131 (RSKD131_3116), R. sphaeroides ATCC 17029 (Rsph17029_3657), S. novella DSM 506 (Snov_2770), X. autotrophicus Py2 (Xaut_5021), M. alhagi CCNWXJ12–2 (ZP_09295256), M. algicola DG893 (MDG893_09606), and Nocardioides sp. JS614 (Noca_2131) is shown. In addition, the paralogous domains of ICM from S. cinnamonensis A3823.5 (icm, AAC08713), IcmF from Geobacillus kaustophilus HTA426 (GK3391), MCM from Propionibacterium freudenreichii subsp. shermanii CIRM-BIA1 (YP_003687736), and ECM from R. sphaeroides ATCC 17029 (Rsph17029_2621) were aligned. Bold amino acids show conserved residues directly involved in substrate binding (9). Residues highlighted in gray background are known to interact specifically with the CoA moiety. White-typed amino acids on black highlight two residues thus far identified to interact specifically with the acyl group of the substrates. Asterisks indicate identical residues in all sequences. A more extended alignment of these mutase sequences is presented in supplemental Fig. S7.
DISCUSSION
Role of HCM in 2-HIBA Catabolism
Comparison of the wild-type A. tertiaricarbonis L108 and the mutant strains L108(ΔhcmA)K5 and L108(ΔhcmB)K7 clearly shows that integrity of both HCM subunits is required for degradation of 2-HIBA. In line with the assigned role in the degradation pathway proposed for compounds bearing a tert-butyl group (7) (Fig. 1), reconstituted HcmAB specifically catalyzes the reversible rearrangement of 2-hydroxyisobutyryl- and 3-hydroxybutyryl-CoA. Conversion of substrates of other B12-dependent acyl-CoA mutases, such as methylmalonyl- and succinyl-CoA, is negligible, thus preventing interference of HCM activity with central carbon metabolism. However, Keq and kcat values determined for the purified recombinant enzyme reveal that not the catalysis of 2-hydroxyisobutyryl-CoA conversion but the reverse reaction is favored, suggesting that HCM is not only employed for channeling tert-butyl moieties toward central metabolism. A role in synthesis of 2-HIBA and related molecules, on the other hand, would be surprising, as these compounds are rarely found in Nature. Further investigations are required for testing whether catalysis of 3-hydroxybutyryl-CoA conversion is also favored under in vivo conditions. In this connection, interplay with other proteins has to be considered, e.g. it has previously been reported that interaction of MCM with the P-loop GTPase MeaB facilitates GTP-dependent assembly of the holo-MCM and protects against the radical intermediates formed during mutase catalysis (22). Accordingly, a strong operonic association of meaB and mcm genes has been observed (26). In case of HCM, a quite similar association is found (Fig. 2), indicating that MeaH also functions as chaperone with this mutase. Possibly, interaction with MeaH or other proteins could also modulate enzyme kinetics in favor of 2-HIBA degradation. On the other hand, an efficient processing of (S)-3-hydroxybutyryl-CoA by a suitable dehydrogenase, and further conversion into acetyl-CoA (Fig. 1) might be sufficient to shift the equilibrium of the mutase catalysis toward the 3-hydroxy carboxylic acid. In line with this, growth rate of the wild-type L108 strain on 2-HIBA is in the same range as found with other efficient substrates, such as fructose and lactic acid (24).
Retention of Configuration during HCM Catalysis
As already found with MCM and ECM, only converting (R)-methylmalonyl- and (R)-ethylmalonyl-CoA (10, 27), respectively, HCM catalysis is stereospecific, favoring the rearrangement of (S)-3-hydroxybutyryl-CoA versus the (R)-enantiomer. This specificity of catalysis may also shed light on the orientation of the hydroxyacyl substrate at the catalytic site of HCM. The rearrangement by MCM and ECM proceeds strictly with retention of configuration (Fig. 8). In the case of ICM, although its natural substrates isobutyryl- and butyryl-CoA do not possess chirality, catalysis is also predominantly stereospecific with retention of configuration, as has been revealed by testing labeled substrates in vivo (28) and with partially purified enzyme (29). Now, due to the structural similarities between the 2-hydroxyisobutyryl and isobutyryl substrate residues, in HCM an ICM-like orientation of the two methyl groups of the branched-chain carboxylic acid could be expected (30) which would result in catalysis predominantly toward (R)-3-hydroxybutyryl-CoA (Fig. 8). However, considering the observed stereospecificity of HCM catalysis favoring the (S)-enantiomer of 3-hydroxybutyryl-CoA, it can be concluded that in HCM the hydroxyacyl residue of its substrates is mainly oriented in the same way as the carboxyacyl substrate moiety in MCM and ECM. In HCM, the polar but uncharged hydroxyl group would specifically interact with the amido function of Gln208 (numbering as in HcmA of strain L108), whereas in MCM/ECM the negatively charged carboxyl group is close to the guanidinium ion of Arg207/Arg131 (numbering as in Figs. 7 and 8). Interestingly, a second polar amino acid residue corresponding to the Tyr89/Tyr13 in MCM/ECM is missing in HCM. Instead, the hydrophobic Ile90 likely interacts with the hydroxyl and methyl groups of the HCM acyl substrates. However, the ICM-like orientation of the methyl groups of the 2-hydroxyisobutyryl residue is not completely excluded, but takes place in about 2% of substrate binding events, as can be deduced from the transformation rates of (R)-3-hydroxybutyryl-CoA. This deviation from complete stereospecificity may also explain the low but significant conversion of the ICM substrates isobutyryl- and butyryl-CoA (Table 1), as it can be assumed that these substrates and (R)-3-hydroxybutyryl-CoA have a similar orientation in the substrate binding site of HCM (Fig. 8). Both HCM and ICM/IcmF share the uncharged Gln residue (Gln208 and Gln198/Gln733 in Fig. 8), suggesting that this amino acid is not determining substrate orientation and stereospecificity. However, the ICM-like substrate orientation is likely discriminated in HCM due to the replacement of the aromatic Phe80/Phe589 postulated to be specifically interacting with the substrate in ICM/IcmF (9, 11) with the smaller aliphatic Ile90 (Fig. 8). Accordingly, the conservative substitution I90V still possess activity toward (S)-3-hydroxybutyryl- and 2-hydroxyisobutyryl-CoA, whereas the nonconservative mutations I90F and I90Y completely lost rearrangement activity.
FIGURE 8.
Orientation of substrates and retention of configuration in the B12-dependent rearrangement of the carbon skeleton of acyl-CoA esters. The observed isomerization of the 3-hydroxybutyryl-CoA enantiomers catalyzed by HCM is compared with transformations catalyzed by MCM/ECM and ICM/IcmF. Active site amino acid residues proposed to interact specifically with acyl group of the substrates are indicated. Numbering of residues is as in Fig. 7.
In contrast to the observed deviation in the predominant substrate orientation at the catalytic sites, HCM is phylogenetically close to bacterial ICM, together forming a cluster within archaeal MCM sequences (10). In addition, HCM and ICM share the same subunit organization consisting of a small B12-binding and a large protein encoded by two distinct genes, whereas in bacterial MCM and ECM the B12- and acyl-CoA binding domains are both located on the same subunit (8, 10). The high sequence and structural similarity between HCM and ICM may allow studying substrate binding and catalysis of specific mutant enzymes. Comparable single amino acid mutations in ICM and MCM resulted mainly in loss of enzymatic activity (31), possibly due to the larger sequence and structural deviations between these mutase subfamilies. However, the activities of the amino acid mutants analyzed in this study clearly show that even in HCM single active site residue substitutions are not sufficient to change substrate specificity, e.g. achieving ICM activity with an HcmA I90F mutant.
Occurrence and Evolution of HCM
As already outlined, the HCM enzyme is, besides ICM, the second representative within the B12-dependent acyl-CoA mutase subfamilies thus far identified to be organized as small and large subunits, binding the coenzyme B12 and the acyl-CoA ester substrate, respectively. In addition, HCM activity is different, as this enzyme specifically catalyzes the rearrangement of hydroxylated carboxylic acids which has not been observed with other mutases. However, the origin of HCM is enigmatic. BLAST analysis revealed a quite heterogeneous group of bacteria, including phylogenetically distant proteobacteria and a Gram-positive strain, which all possess the four genes encoding for the HCM subunits plus a putative acyl-CoA synthetase and the chaperone MeaH organized in an operon-like structure (Figs. 2 and 7). The high sequence similarity and the identical gene grouping suggest an interchange of the complete hcm operon by horizontal gene transfer. Unlike strains A. tertiaricarbonis L108 and M. petroleiphilum PM1 (15, 32), the other bacterial strains with an hcm operon have not been associated with the degradation of fuel oxygenates MTBE and TBA. An alternative source for compounds bearing the tert-butyl group is the industrial production of PMMA (3, 6), as the corresponding wastewater contains 2-HIBA and its methyl ester. Accordingly, it has already been demonstrated that strains R. sphaeroides ATCC 17029, Xanthobacter autotrophicus Py2 and Nocardioides sp. JS614 are able to grow on 2-HIBA (33). However, worldwide production of PMMA at large scale started not earlier than in the late 1930s (34), whereas the hcm operon bearing strain Starkeya novella DSM 506 has already been isolated from uncontaminated agricultural soil in 1934 (35), indicating that other drivers than this kind of anthropogenic contamination for the evolution of HCM may exist. The other known B12-dependent acyl-CoA mutases are widely distributed among bacteria playing central roles in primary and secondary carbon metabolism, e.g. in branched-chain amino acid catabolism (8) and in the recently discovered ethylmalonyl-CoA pathway for acetate assimilation (10) as well as in the synthesis of macrolide and polyether antibiotics (9). In contrast, HCM would be the first mutase exclusively employed for the dissimilatory degradation of a single substrate, i.e. 2-HIBA. In this connection, several not yet identified mutases have been postulated to be involved in bacterial degradation pathways for the mineralization of alkanes, ethylbenzene, and the quaternary carbon-bearing pivalic acid (36, 37). Hence, it could be speculated that all of these mutases employed in dissimilatory pathways may have the same origin being adapted to their specific substrates by a moderate variation of the amino acid residues in the substrate binding site. The previously described close phylogenetic relationship of bacterial HCM and ICM sequences with archaeal mutases (10) is also interesting, as it suggests an origin of HCM outside the domain Bacteria.
Supplementary Material
Acknowledgments
We thank C. Schumann (Helmholtz Centre for Environmental Research, UFZ), M. Neytschev (UFZ) and S. Kluge (UFZ) for technical assistance and B. Würz (UFZ) for excellent analytical advice.
This work was supported by the Helmholtz Centre for Environmental Research within the Chemicals in the Environment program and by Evonik Industries.

This article contains supplemental Figs. S1–S7.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) JQ708092.
- 2-HIBA
- 2-hydroxyisobutyric acid
- ECM
- ethylmalonyl-CoA mutase
- HCM
- 2-hydroxyisobutyryl-CoA mutase
- ICM
- isobutyryl-CoA mutase
- IcmF
- isobutyryl-CoA mutase-fused
- MCM
- methylmalonyl-CoA mutase
- MSM
- mineral salt medium
- MTBE
- methyl tert-butyl ether
- PMMA
- poly(methyl methacrylate)
- TBA
- tert-butyl alcohol.
REFERENCES
- 1. Kumps A., Duez P., Mardens Y. (2002) Metabolic, nutritional, iatrogenic, and artifactual sources of urinary organic acids: a comprehensive table. Clin. Chem. 48, 708–717 [PubMed] [Google Scholar]
- 2. Forslund K., Morant M., Jørgensen B., Olsen C. E., Asamizu E., Sato S., Tabata S., Bak S. (2004) Biosynthesis of the nitrile glucosides rhodiocyanoside A and D and the cyanogenic glucosides lotaustralin and linamarin in Lotus japonicus. Plant Physiol. 135, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chisholm M. S. (2000) Artificial glass: the versatility of poly(methyl methacrylate) from its early exploitation to the new millenium. J. Chem. Edu. 77, 841–845 [Google Scholar]
- 4. Steffan R. J., McClay K., Vainberg S., Condee C. W., Zhang D. (1997) Biodegradation of the gasoline oxygenates methyl tert-butyl ether, ethyl tert-butyl ether, and tert-amyl methyl ether by propane-oxidizing bacteria. Appl. Environ. Microbiol. 63, 4216–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moran M. J., Zogorski J. S., Squillace P. J. (2005) MTBE and gasoline hydrocarbons in ground water of the United States. Ground Water 43, 615–627 [DOI] [PubMed] [Google Scholar]
- 6. Holowach L. P., Swift G. W., Wolk S. W., Klawiter L. (1994) in Polymers from Agricultural Coproducts (Fishman M. L., Friedman R. B., Huang S. J., eds) pp. 202–211, ASC, Washington, DC [Google Scholar]
- 7. Rohwerder T., Breuer U., Benndorf D., Lechner U., Müller R. H. (2006) The alkyl tert-butyl ether intermediate 2-hydroxyisobutyrate is degraded via a novel cobalamin-dependent mutase pathway. Appl. Environ. Microbiol. 72, 4128–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Birch A., Leiser A., Robinson J. A. (1993) Cloning, sequencing, and expression of the gene encoding methylmalonyl-coenzyme A mutase from Streptomyces cinnamonensis. J. Bacteriol. 175, 3511–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ratnatilleke A., Vrijbloed J. W., Robinson J. A. (1999) Cloning and sequencing of the coenzyme B12-binding domain of isobutyryl-CoA mutase from Streptomyces cinnamonensis, reconstitution of mutase activity, and characterization of the recombinant enzyme produced in Escherichia coli. J. Biol. Chem. 274, 31679–31685 [DOI] [PubMed] [Google Scholar]
- 10. Erb T. J., Rétey J., Fuchs G., Alber B. E. (2008) Ethylmalonyl-CoA mutase from Rhodobacter sphaeroides defines a new subclade of coenzyme B12-dependent acyl-CoA mutases. J. Biol. Chem. 283, 32283–32293 [DOI] [PubMed] [Google Scholar]
- 11. Cracan V., Padovani D., Banerjee R. (2010) IcmF is a fusion between the radical B12 enzyme isobutyryl-CoA mutase and its G-protein chaperone. J. Biol. Chem. 285, 655–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Simon E. J., Shemin D. (1953) The preparation of S-succinyl coenzyme A. J. Am. Chem. Soc. 75, 2520 [Google Scholar]
- 13. Padmakumar R., Gantla S., Banerjee R. (1993) A rapid method for the synthesis of methylmalonyl-coenzyme A and other CoA-esters. Anal. Biochem. 214, 318–320 [DOI] [PubMed] [Google Scholar]
- 14. Dalluge J. J., Gort S., Hobson R. (2002) Separation and identification of organic acid-coenzyme A thioesters using liquid chromatography/electrospray ionization-mass spectrometry. Anal. Bioanal. Chem. 374, 835–840 [DOI] [PubMed] [Google Scholar]
- 15. Lechner U., Brodkorb D., Geyer R., Hause G., Härtig C., Auling G., Fayolle-Guichard F., Piveteau P., Müller R. H., Rohwerder T. (2007) Aquincola tertiaricarbonis gen. nov., sp. nov., a tertiary butyl moiety-degrading bacterium. Int. J. Syst. Evol. Microbiol. 57, 1295–1303 [DOI] [PubMed] [Google Scholar]
- 16. Schäfer F., Muzica L., Schuster J., Treuter N., Rosell M., Harms H., Müller R. H., Rohwerder T. (2011) Formation of alkenes via degradation of tert-alkyl ethers and alcohols by Aquincola tertiaricarbonis L108 and Methylibium spp. Appl. Environ. Microbiol. 77, 5981–5987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Valentin H. E., Steinbüchel A. (1993) Expression of an α-galactosidase gene under control of the homologous inulinase promoter in Kluyveromyces marxianus. Appl. Microbiol. Biotechnol. 39, 309–317 [DOI] [PubMed] [Google Scholar]
- 18. Hermans-Lokkerbol A., van der Heijden R., Verpoorte R. (1996) Isocratic high-performance liquid chromatography of coenzyme A esters involved in the metabolism of 3S-hydroxy-3-methylglutaryl coenzyme A. Detection of related enzyme activities in Catharanthus roseus plant cell cultures. J. Chromatogr. A 752, 123–130 [Google Scholar]
- 19. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kane S. R., Chakicherla A. Y., Chain P. S., Schmidt R., Shin M. W., Legler T. C., Scow K. M., Larimer F. W., Lucas S. M., Richardson P. M., Hristova K. R. (2007) Whole-genome analysis of the methyl tert-butyl ether-degrading β-proteobacterium Methylibium petroleiphilum PM1. J. Bacteriol. 189, 1931–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hristova K. R., Schmidt R., Chakicherla A. Y., Legler T. C., Wu J., Chain P. S., Scow K. M., Kane S. R. (2007) Comparative transcriptome analysis of Methylibium petroleiphilum PM1 exposed to the fuel oxygenates methyl tert-butyl ether and ethanol. Appl. Environ. Microbiol. 73, 7347–7357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Padovani D., Banerjee R. (2006) Assembly and protection of the radical enzyme, methylmalonyl-CoA mutase, by its chaperone. Biochemistry 45, 9300–9306 [DOI] [PubMed] [Google Scholar]
- 23. Schuster J., Schäfer F., Hübler N., Brandt A., Rosell M., Härtig C., Harms H., Müller R. H., Rohwerder T. (2012) Bacterial degradation of tert-amyl alcohol proceeds via hemiterpene 2-methyl-3-buten-2-ol by employing the tertiary alcohol desaturase function of the Rieske nonheme mononuclear iron oxygenase MdpJ. J. Bacteriol. 194, 972–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Müller R. H., Rohwerder T., Harms H. (2008) Degradation of fuel oxygenates and their main intermediates by Aquincola tertiaricarbonis L108. Microbiology 154, 1414–1421 [DOI] [PubMed] [Google Scholar]
- 25. Cracan V., Banerjee R. (2012) Novel coenzyme B12-dependent interconversion of isovaleryl-CoA and pivalyl-CoA. J. Biol. Chem. 287, 3723–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Korotkova N., Lidstrom M. E. (2004) MeaB is a component of the methylmalonyl-CoA mutase complex required for protection of the enzyme from inactivation. J. Biol. Chem. 279, 13652–13658 [DOI] [PubMed] [Google Scholar]
- 27. Sprecher M., Clark M. J., Sprinson D. B. (1966) The absolute configuration of methylmalonyl coenzyme A and stereochemistry of the methymalonyl coenzyme A mutase reaction. J. Biol. Chem. 241, 872–877 [PubMed] [Google Scholar]
- 28. Reynolds K. A., O'Hagan D., Gani D., Robinson J. A. (1988) Butyrate metabolism in Streptomyces: characterization of an intramolecular vicinal interchange rearrangement linking isobutyrate and butyrate in Streptomyces cinnamonensis. J. Chem. Soc. Perkin Trans. 1, 3195–3207 [Google Scholar]
- 29. Moore B. S., Eisenberg R., Weber. C., Bridges A., Nanz D., Robinson J. A. (1995) On the stereospecificity of the coenzyme B12-dependent isobutyryl-CoA mutase reaction. J. Am. Chem. Soc. 117, 11285–11291 [Google Scholar]
- 30. Rohwerder T., Müller R. H. (2010) Biosynthesis of 2-hydroxyisobutyric acid (2-HIBA) from renewable carbon. Microbial Cell Fact. 9, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vlasie M. D., Banerjee R. (2004) When a spectator turns killer: suicidal electron transfer from cobalamin in methylmalonyl-CoA mutase. Biochemistry 43, 8410–8417 [DOI] [PubMed] [Google Scholar]
- 32. Nakatsu C. H., Hristova K., Hanada S., Meng X. Y., Hanson J. R., Scow K. M., Kamagata Y. (2006) Methylibium petroleiphilum gen. nov., sp. nov., a novel methyl tert-butyl ether-degrading methylotroph of the Betaproteobacteria. Int. J. Syst. Evol. Microbiol. 56, 983–989 [DOI] [PubMed] [Google Scholar]
- 33. Rohwerder T., Breuer U., Benndorf D., Lechner U., Müller R. H. (2007) in Proceedings of the 3rd European Conference on MTBE and Other Fuel Oxygenates (Bastiaens L., ed) pp. 11–14, VITO, Mol [Google Scholar]
- 34. Bauer W. (2002) in Ullmann's Encyclopedia of Industrial Chemistry, Vol. 21, pp. 585–597, Wiley-VCH, Weinheim [Google Scholar]
- 35. Starkey R. L. (1934) Cultivation of organisms concerned in the oxidation of thiosulfate. J. Bacteriol. 28, 365–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rohwerder T., Müller R. H. (2007) in Vitamin B: New Research (Elliot C. M., ed) pp. 81–98, Nova Science Publisher, Hauppauge [Google Scholar]
- 37. Probian C., Wülfing A., Harder J. (2003) Anaerobic mineralization of quaternary carbon atoms: isolation of denitrifying bacteria on pivalic acid (2,2-dimethylpropionic acid). Appl. Environ. Microbiol. 69, 1866–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.