Background: There is an ongoing debate on whether and how the ion pump NKA can be regulated by PKA.
Results: Phosphorylation of the PKA target Ser936 opens an intracellular ion pathway leading to the ion-binding sites
Conclusion: PKA phosphorylation has a drastic impact on NKA structure and dynamics.
Significance: The molecular mechanism of PKA regulation of NKA has been described for the first time.
Keywords: Molecular Dynamics; Na,K-ATPase; NAK-ATPase; Patch Clamp Electrophysiology; Protein kinase A (PKA); Ser936
Abstract
Phosphorylation is one of the major mechanisms for posttranscriptional modification of proteins. The addition of a compact, negatively charged moiety to a protein can significantly change its function and localization by affecting its structure and interaction network. We have used all-atom Molecular Dynamics simulations to investigate the structural consequences of phosphorylating the Na+/K+-ATPase (NKA) residue Ser936, which is the best characterized phosphorylation site in NKA, targeted in vivo by protein kinase A (PKA). The Molecular Dynamics simulations suggest that Ser936 phosphorylation opens a C-terminal hydrated pathway leading to Asp926, a transmembrane residue proposed to form part of the third sodium ion-binding site. Simulations of a S936E mutant form, for which only subtle effects are observed when expressed in Xenopus oocytes and studied with electrophysiology, does not mimic the effects of Ser936 phosphorylation. The results establish a structural association of Ser936 with the C terminus of NKA and indicate that phosphorylation of Ser936 can modulate pumping activity by changing the accessibility to the ion-binding site.
Introduction
Ion gradients across cellular membranes sustain electrical excitability, secondary transport, energy storage, and cell volume regulation. The plasma membrane contains numerous proteins, e.g. the ion channels that depend directly on the ion gradients created by ion pumps. Cation pumps such as the NKA2 use the energy of ATP hydrolysis to transport ions against their concentration gradients across the membrane. The NKA extrudes three Na+ ions in exchange of two K+ ions for each molecule of ATP hydrolyzed.
The NKA α-subunit has the conserved features of canonical P-type ion-ATPases, including cytoplasmic nucleotide, phosphorylation, and acutator domains and intramembrane ion-binding sites. X-ray crystallography has revealed the structure of NKA in the potassium-bound form (5, 6). In the sodium-bound pump, the two potassium-binding sites reorganize to achieve high sodium affinity, but the binding site for the third sodium ion remains a matter of debate (4, 5, 7).
PKA phosphorylation is one of several mechanisms for regulating NKA activity (1–3). Ser936 lies near the lipid-water interface, and in vitro phosphorylation depends on detergent, but antibodies raised against a peptide with the phosphorylated Ser936 show that the site is phosphorylated in vivo in response to PKA activation (for review, see Ref. 8).
In various cell types, induction of PKA and phosphorylation of NKA have been found to correlate inversely with NKA activity, whereas the amount of pumps at the plasma membrane remains unaffected, suggesting that phosphorylation of Ser936 directly lowers pump activity (2, 3, 9, 10, 12–14). Pump trafficking may, however, also be affected by PKA phosphorylation in some cell types, apparently via a cAMP-independent pathway responding to the intracellular sodium levels (15, 16).
Ser936 is only few angstroms from the C terminus, and we hypothesized that the phosphorylation may affect the C-terminal structure. In C-terminal mutants expressed in Xenopus oocytes, inwardly rectifying leak currents were observed in the presence of extracellular sodium (4, 17, 18), and similar though much smaller leak currents were observed with the phosphorylation-mimicking mutation S936E (8). However, the S936E mutation might underestimate the electrostatic and steric effects of Ser936 phosphorylation.
Here, the structural effects of Ser936 phosphorylation are investigated with Molecular Dynamics (MD) simulations. Ser936 lies on the cytoplasmic loop connecting transmembrane (TM)8 and TM9, which is close to the C-terminal interaction network that controls a pathway to the ion-binding sites. We examine the effects of Ser936 phosphorylation on the C-terminal network, on the ion-binding sites, and on transmembrane structure and dynamics. Upon phosphorylation, water molecules enter an intracellular pathway that leads to the ion binding sites. The pathway is similar to that described in C-terminal mutants (4). The finding is significant because it is the first example of a structural effect of phosphorylation on the NKA that may explain the marked functional effects of NKA phosphorylation by PKA.
EXPERIMENTAL PROCEDURES
MD Simulations
Simulations were implemented with the pig renal αβ dimer (Protein Data Bank (PDB) ID code 3B8E) (5) embedded in a fully hydrated 111 Å × 111 Å × 190 Å 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipid bilayer. POPC is a lipid well known model lipid used to model membranes both in simulations and in experiments in a large variety of systems, ranging from protein-membrane interactions such as ion-channel (19) and ion-pump simulations (4, 20–22) to drug-binding assays (23). A POPC membrane is in the fluid phase at room temperature. Important physical properties of the membrane, such as membrane thickness, area-per-lipid, and fluidity, all of which may impact protein-lipid interactions, are captured accurately by the POPC lipid model. Finally, lipids diffuse in membranes on time scales of 100 s of nanoseconds. Modeling a mixture of lipids will therefore bias the simulation toward their initial placement of lipids in the membrane.
Simulations were performed using GROMACS version 4.5.3 (24–26) using the modified Berger force field for lipids (27) and the OPLS-AA all-atom force field for proteins (28). The dihedral angles of the Berger force field were reoptimized to permit their combination with the OPLS-AA protein force field (29). The method reproduced accurate free energy of transfer of amino acids between aqueous and hydrophobic lipid phases and yielded reasonable protein-lipid interactions for a range of systems, including transmembrane peptides and proteins such as OmpF. Furthermore, the Berger force field retains some of the philosophy of the OPLS united-atom force field in its treatment of Lennard-Jones interactions (27). The two publicly available force fields to run protein-membrane simulations in the NPT ensemble are CHARMM and the OPLS-Berger combination. Simulations run more efficiently with the latter because it uses united atoms for lipids. Because of the original implementation of the OPLS-Berger force field combination, several other important studies have been carried out using the same force field combination, see for example (30–32), and reviewed in (33). Therefore, we use the OPLS-AA protein force field in combination with the Berger for field for lipids. It should be noted that depending upon the system of investigation, different protein force fields may perform differently, for example, in terms of predicting the transition states and the folding rates of proteins (34).
There are no major structural differences in the C-terminal and ion-binding region of the low resolution pig renal structure (3B8E) and the higher resolution shark enzyme (6) (PDB ID code 2ZXE), other than the presence of two extra water molecules near the ion-binding sites in the latter. The water molecules appeared spontaneously in the binding site during our simulations. All residue numbering follows the sequence of the pig renal protein. Asp926 and Glu954 were kept protonated, as argued earlier (4). All other aspartate and glutamate residues were deprotonated, and all arginine and lysine residues carried a charge of +1. The simulation cell was kept electrostatically neutral by using Na+ ions. The simulations were implemented in the isobaric-isothermal ensemble. Temperature coupling was performed using the Berendsen thermostat (35) separately for the protein, lipids, and the solvent with a reference temperature of 310 K and a time constant of 0.1 ps for all subgroups. A semi-isotropic pressure coupling was applied using Berendsen barostat (35). A coupling constant of 1.0 ps and a reference pressure of 1.0 bar were used in all directions. Because the compressibility, κ, for the systems was not known, the value for pure water, 4.5·10−5 bar−1 was used. For the production run, the leap-frog integrator (36) was used with a time step of 2 fs. All bonds containing hydrogen were constrained using the LINCS algorithm (37, 38), and water molecules were constrained with SETTLE (39). Periodic boundary conditions were applied in all directions. A neighbor list with a 10 Å cutoff was used for nonbonded interactions and was updated every 20 fs. The van der Waals interactions were truncated with a cutoff of 10.0 Å and the electrostatic interactions were treated with the Particle Mesh Ewald (40) method using default parameters. The center of mass translation was removed at every step of the simulation. Trajectories were sampled every 10 ps. Simulations were run for at least 60 ns. For calculation of ensemble-averaged properties, the first 20 ns of the production run of each simulation were discarded. The analysis was carried out using GROMACS or custom-made programs. Visualization and snapshots were rendered using VMD (41).
The wild-type system was equilibrated using harmonic constraints on lipids, water, and protein. The constraints were sequentially released, followed by constraint-free equilibration for 20 ns. The final conformation from this equilibrated system was used to start the production runs for the wild type (WT), the S936E mutant, and the protein with Ser936 phosphorylated (Ser(P)936). Two copies of the S936E and Ser(P)936 simulations were implemented to collect better statistics. Most of the results from the two copies were identical. Simulation snapshots will be presented only for one copy for brevity. Although the simulation time scales are indeed short by current standards, they still successfully capture the phenomena of interest (opening of water pathway), which takes place on time scales of 10 s of nanoseconds. For robust results, each system was simulated twice, and the results were reproducible. Furthermore, the simulations are in good agreement with electrophysiological data, which indicates that the risk of drawing invalid conclusions was minimized despite the short simulation time scales.
Electrophysiology
Pumps were expressed in oocytes from Xenopus laevis by co-injecting cRNAs encoding ouabain-insensitive hsNKAα2 and hsNKAβ1 (4). After 1–3 days at 19 °C, oocytes were loaded with sodium in 95 mm sodium, 90 mm sulfamic acid, 5 mm HEPES, 10 mm tetramethyl-ammonium chloride, 0.1 mm EGTA, pH 7.6. Two-electrode voltage clamping was performed in 115 mm sodium, 110 mm sulfamic acid, 1 mm MgCl2, 0.5 mm CaCl2, 5 mm BaCl2, 10 mm HEPES, 1 μm ouabain, pH 7.4. To determine pre-steady-state currents, a series of 200-ms voltage steps was run with and without 10 mm ouabain, and single exponentials were fitted to the difference traces to determine the amount of charges moved (Q) and the inverse time constants (R).
RESULTS
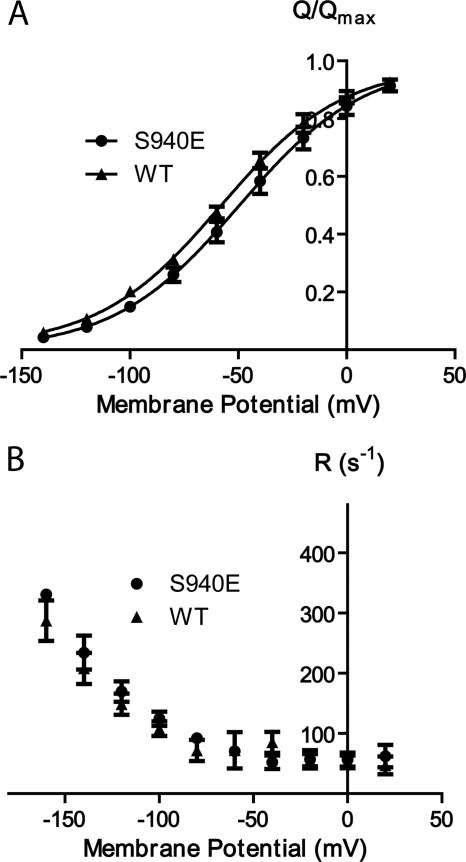
In oocytes, the mutation of the PKA-targeted serine (number 940 in the human α2 expressed) to glutamate led to increased leak currents in the presence of sodium at negative membrane potentials, an effect reminiscent of the effect of C-terminal mutations, but much milder (4, 17). The C-terminal mutants studied so far furthermore exhibit severely reduced sodium affinity and altered kinetics of extracellular sodium binding and release (4, 17, 42). These effects are not observed for the S940E mutant, which has extracellular sodium affinity and relaxation rate constants similar to the wild-type values as determined from pre-steady-state currents after expression in Xenopus oocytes (Fig. 1). α2-Expressing oocytes were treated with forskolin and isobutylmethylxanthine to activate PKA as described previously (43, 44). Phosphorylation was not observed under these conditions.
FIGURE 1.
Voltage dependence of transient currents of S940E mutated Na/K-ATPase. A, charge translocation (Q/Qmax) of mutated and wild-type pumps determined from the off pulses of a series of 20-mV voltage steps and fitted with Boltzmann curves. B, relaxation rate constants of mutated and wild-type pumps.
The limited effect of the S940E mutation in the electrical recordings correlates poorly with the inhibition of NKA observed upon PKA phosphorylation (9, 10). A likely reason for this discrepancy is that the introduction of glutamate does not fully replicate the larger steric and electrostatic effect of explicit phosphorylation, as reported previously for a cyclin-dependent kinase (45). Treatment of oocytes with the adenylyl cyclase activator forskolin did not alter the pump characteristics (data not shown). It was previously reported that NKA phosphorylation in oocyte homogenates, where PKA was activated by cAMP, depended on detergent treatment (46), suggesting that forskolin may be insufficient to initiate phosphorylation of NKA in the oocytes.
To study the structural effects of explicit phosphorylation of the PKA site in the sodium pump, we therefore implemented MD simulations of the pig renal α1-β dimer in a fully hydrated POPC membrane, examining the WT pump as well as pumps with the PKA target site Ser936 phosphorylated.
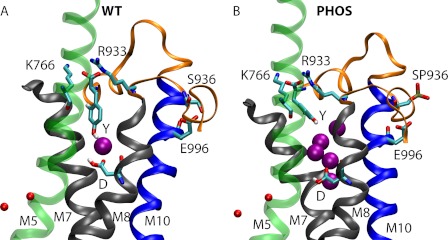
In the WT protein, the Ser936 side chain is hydrogen-bonded to the carboxylate of Glu996 (see Fig. 4A), which is 20 residues upstream of the C terminus on helix M10. Phosphorylation of Ser936 breaks the hydrogen bond and electrostatically repels Glu996. In the WT, Glu996 also interacts with Arg1003, but phophorylation leads to an electrostatic salt bridge between Ser(P)936 and Arg1003. The resultant movement of the M8-M9 loop displaces M8 slightly away from the third sodium-binding pocket and rearranges the interaction network in the ion-binding pockets.
FIGURE 4.
Simulations snapshots showing the entry of water between helices M5, M7, and M8 for the (A) WT and (B) PHOS simulation at 55 ns. Water molecules within 8 Å of Asp926 and within the M5-M7-M8 cavity are shown as purple spheres. K+ ions are shown as small red spheres. Y and D refer to residues Tyr1016 and Asp926, respectively. Helices M5, M7, M8, and M10 are shown in transparent green, black, black and blue, respectively. The loop connecting M8 and M9, and the last 13 C-terminal residues are shown as orange ribbons. A larger number of water molecules populate the cavity between M5, M7, and M8 in the PHOS simulation. Please refer to supplemental Movies 1, 2, and 3 for dynamics of entry of water molecules.
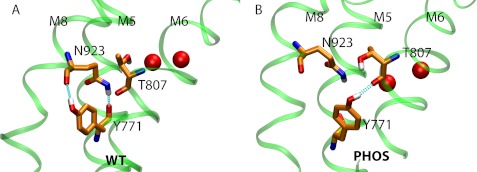
Two residues in the ion-binding region, Tyr771 and Asn923, are close together in all of the four available crystal structures of NKA (PDB ID codes 3A3Y, 3KDB, 3N23, and 2ZXE). In the MD simulations of the WT pump, the side chain of Tyr771 even hydrogen bonds with the backbone of Asn923 and vice versa (Fig. 2A). The movement caused by Ser936 phosphorylation in the MD simulations breaks the Tyr771-Asn923 interaction, and the Tyr771 side chain occasionally hydrogen bonds to the backbone of Thr807 that is part of the first Na+-binding site.
FIGURE 2.
Simulation snapshots showing hydrogen bonding between Asn923 on M8 and Tyr771 on M5 in (A) WT and (B) PHOS simulation at t = 55 ns. The side chain of each residue is hydrogen-bonded to the backbone of the other in the WT. Hydrogen bonds are shown as cyan springs. The hydrogen bond is broken as a result of M8 movement upon phosphorylation.
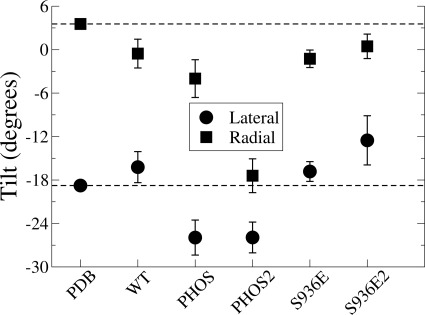
The reorientation of the TM helices in Ser(P)936 is captured in the changes in the angle between helices M7 and M8 in Fig. 3. The radial and axial tilt angles between M7 and M8 increase, resulting in the opening up of helices M5, M7, and M8 to a cavity that becomes hydrated as far as residue Asp926 (Fig. 4 and supplemental Movies 1, 2, and 3; coloring scheme same for movies and Fig. 4). In the WT, no water enters during the simulations.
FIGURE 3.
Relative axial and radial tilt angles between helices M7 and M8. Dashed lines correspond to the values in the starting PDB structure. Axial tilt simply represents the angle between the axes of the two helices. Radial tilt is the angle between the axis of M7 with the projection of the M7 axis on the plane through the M8 vector and the midpoint of the M7 axis. The two vectors describe the relative orientation of helices in the helix bundle. The magnitudes of both the radial and axial tilt angles increase in the PHOS simulations.
To examine whether the limited functional effect of the S936E (S940E) mutation (Fig. 1A) correlates with a limited structural effect of the mutation compared with the phosphorylation, we performed MD simulations of the S936E mutant. In S936E, the S936E-R1003 interaction and the S936E-E996 repulsion are much weaker compared with Ser(P)936 because of the lower size and charge (−1) of S936E compared with Ser(P)936 (−2). Therefore, the movement of the M8-M9 loop observed in Ser(P)936 is much smaller with S936E, and consequently, as observed with the WT, water does not enter between M5, M7, and M8.
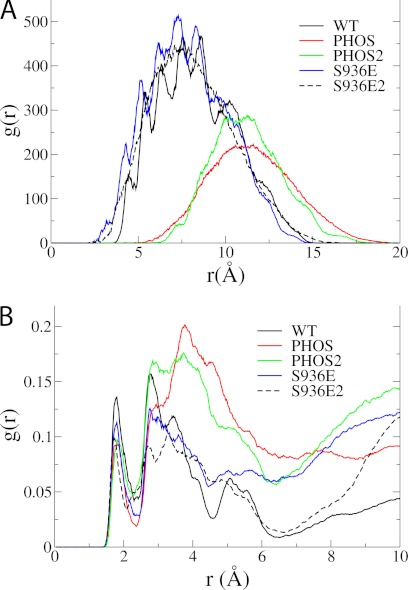
In the WT and S936E simulations, the Tyr1016 side chain is bridged to the protonated Asp926 residue by a single water molecule not observed in the crystal structures (Fig. 4A). Phosphorylation increases the distance between Tyr1016 and Asp926 significantly (Fig. 5A), and the space between the two residues is filled by a large number of water molecules (Fig. 4A). The radial distribution functions of Asp926 and water (Fig. 5B) illustrate that the phosphorylated enzyme has a peak at 1.8 Å and maximal values at 3–5 Å from Asp926, indicating substantial hydration, whereas the WT and S936E simulations only show peaks at 1.8 Å and 3 Å as characteristic of a single water molecule (Fig. 5B)
FIGURE 5.
Radial distribution functions of (A) Tyr1016 and (B) water around Asp926. The function calculates the local density of one group around the other in concentric spheres around the first group. It is thus a histogram made over the distance data between two groups over all trajectory frames. A 5-point running average is shown. The Asp-Tyr distance increases significantly in the PHOS mutations, and the resulting “gap” is filled with water molecules, which are manifest in the higher secondary peaks in B, showing greater hydration of Asp926.
It is interesting to note that the interactions of the C-terminal Tyr1016 residue with Lys766 and Arg933 seen in the crystal structure and in the WT and S936E simulation also remain largely unaffected by Ser936 phosphorylation.
DISCUSSION
MD simulations have been used to investigate the molecular effects of phosphorylation of Ser936 in NKA, a well documented PKA substrate. The simulations show that in the E2P state, phosphorylation resulted in movement of the M8-M9 loop away from the functionally important C terminus, causing an altered interaction network of loops close to the C terminus that allowed water to gain access to a cavity between helices M5, M7, and M8. Additionally, the simulations show that the phosphorylation caused rearrangements of residues suggested to be involved in Na+ binding, which may reflect a weakening of ion binding, so even though Ser936 is distant from the ion-binding sites, ion binding is likely to be affected by PKA phosphorylation.
The opening of the C-terminal pathway in the simulations upon Ser936 phosphorylation is similar to the opening seen with C-terminal mutations. Disruption of the C-terminal structure was previously shown to strongly affect both pre-steady-state and steady-state currents as measured with electrophysiology (4, 17, 18, 47). Based on the leak currents in C-terminal mutants and MD simulations that showed water entering a C-terminal pathway, an alternative mechanism of pumping was earlier proposed which involved proton entry through the C-terminal pathway. The same pathway opens in the current set of Ser(P)936 simulations, suggesting that leak currents, similar to those in C-terminal mutants, should be observed in the S936E mutant. The electrophysiological data presented here suggest that the S936E mutation has little or no effect on the pump properties; the mutant has affinity for extracellular sodium similar to the wild type and only leaks a little more in the presence of extracellular sodium, which is in contrast to the significant leak measured with C-terminal mutants under the same conditions. However, the electrophysiology data correlate well with the simulations of the same mutant showing that the C-terminal pathway does not open in S936E, unlike what was observed with the phosphorylated pump. Thus, the Ser to Glu mutation is unable to mimic the effects of a phosphorylation in the current case. The possible limitations of a glutamate mutation in mimicking phosphorylation have been documented earlier, e.g. that glutamate, unlike phosphorylation, did not act like phosphothreonine in the activation loop of the protein cyclin-dependent kinase 2 (45).
Of the proposed sodium pump phosphorylation sites, the PKA site is uniquely conserved; the consensus PKA recognition sequence, Arg-Arg-Xaa-Ser/Thr-Φ (where Φ is hydrophobic), is Arg-Arg-Asn-Ser-Val/Leu/Ile in all pump isoforms from Caenorhabditis elegans to man. From different experimental set-ups, phosphorylation of the PKA site has been found in either NKAα1 (48), NKAα2 (49), or NKAα3 (13). PKA phosphorylation even appears to be conserved in another ATPase, namely the closely related colonic H,K-ATPase (50). All three isoforms are phosphorylated by PKA (51). So although different isoforms are certainly regulated differently by PKA depending e.g. on their cellular localization and interaction partners, our studies address the molecular mechanistic consequences on ATPase function of the PKA phosphorylation, and they will not be subject to variation between the isoforms or even between the different ATPases. We argue that our finding of PKA phosphorylation affecting the C-terminal configuration will be valid for all X,K-ATPases because they share both the PKA site and the characteristic C terminus.
In the WT, the hydrophilic side chains of Asn923 and Tyr771 are stabilized by hydrogen bonding to the backbone atoms of each other. Although Asn923 and Tyr771 are proximal in structures, the internal hydrogen bonding arrangement is not apparent. Such a scheme of internal mutual hydrogen bonding could be generally applicable for stabilizing ion-binding hydrophilic residues in hydrophobic environments in absence of the bound ion.
The mutation Y1016F did not alter the sodium-binding affinity of the protein in membrane extracts (42), whereas Y1016A reduced the apparent affinity 5-fold (42), suggesting that the hydroxyl group is not a primary determinant of sodium affinity, whereas the aromatic ring is required. It has been suggested that Arg933 and Tyr1016 interact via a cation-π bond and not via the Tyr1016 hydroxyl group (11, 42). However, no configurations from the MD simulations support a direct cation-π interaction between Arg933 and Tyr1016 or Tyr1015, either stacked or “T” interaction, although the residues are proximal enough in the crystal structure to facilitate such interactions. Cation-π interactions in the E1 form or other intermediate states are also possible explanations to the importance of an aromatic side chain on the C-terminal residue.
Based on the evidence form MD simulations presented here, we suggest that PKA phosphorylation of Ser936 in NKA affects the structure of the C terminus and promotes hydration of a cavity in the transmembrane region that connects the cytoplasm and the ion-binding sites. The cavity has been previously identified to be similarly opened by mutations of residues in the C-terminal network that severely lower the sodium affinity of the pump. Our results can thus explain why PKA activation was reported to inhibit the NKA pumping activity (2, 3, 9, 10, 12–14), and we are currently pursuing a further structural and functional understanding of the role of NKA phosphorylation by PKA.
MEMPHYS-Center for Biomembrane Physics is supported by the Danish National Research Foundation. The computations were done at the SDU node of the Danish Center for Scientific Computing (DCSC). PUMPKIN is a Center of Excellence funded by the Danish National Research Foundation.

This article contains supplemental Movies 1–3.
- NKA
- Na+/K+-ATPase
- MD
- Molecular Dynamics
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- PDB
- Protein Data Bank
- TM
- transmembrane.
REFERENCES
- 1. Therien A. G., Blostein R. (2000) Mechanisms of sodium pump regulation. Am. J. Physiol. Cell Physiol. 279, C541–566 [DOI] [PubMed] [Google Scholar]
- 2. Bertorello A. M., Aperia A., Walaas S. I., Nairn A. C., Greengard P. (1991) Phosphorylation of the catalytic subunit of Na+,K+-ATPase inhibits the activity of the enzyme. Proc. Natl. Acad. Sci. U.S.A. 88, 11359–11362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cheng X. J., Fisone G., Aizman O., Aizman R., Levenson R., Greengard P., Aperia A. (1997) PKA-mediated phosphorylation and inhibition of Na+-K+-ATPase in response to β-adrenergic hormone. Am. J. Physiol. Cell Physiol. 273, C893–901 [DOI] [PubMed] [Google Scholar]
- 4. Poulsen H., Khandelia H., Morth J. P., Bublitz M., Mouritsen O. G., Egebjerg J., Nissen P. (2010) Neurological disease mutations compromise a C-terminal ion pathway in the Na+/K+-ATPase. Nature 467, 99–102 [DOI] [PubMed] [Google Scholar]
- 5. Morth J. P., Pedersen B. P., Toustrup-Jensen M. S., Sørensen T. L., Petersen J., Andersen J. P., Vilsen B., Nissen P. (2007) Crystal structure of the sodium-potassium pump. Nature 450, 1043–1049 [DOI] [PubMed] [Google Scholar]
- 6. Shinoda T., Ogawa H., Cornelius F., Toyoshima C. (2009) Crystal structure of the sodium-potassium pump at 2.4 angstrom resolution. Nature 459, 446–450 [DOI] [PubMed] [Google Scholar]
- 7. Morth J. P., Pedersen B. P., Buch-Pedersen M. J., Andersen J. P., Vilsen B., Palmgren M. G., Nissen P. (2011) A structural overview of the plasma membrane Na+,K+-ATPase and H+-ATPase ion pumps. Nat. Rev. Mol. Cell Biol. 12, 60–70 [DOI] [PubMed] [Google Scholar]
- 8. Poulsen H., Morth P., Egebjerg J., Nissen P. (2010) Phosphorylation of the Na+,K+-ATPase and the H+,K+-ATPase. FEBS Lett. 584, 2589–2595 [DOI] [PubMed] [Google Scholar]
- 9. Andersson R. M., Cheng S. X., Aperia A. (1998) Forskolin-induced down-regulation of Na+,K+-ATPase activity is not associated with internalization of the enzyme. Acta Physiol. Scand. 164, 39–46 [DOI] [PubMed] [Google Scholar]
- 10. Venosa R. A. (2005) Protein kinases A and C stimulate the Na+ active transport in frog skeletal muscle without an appreciable change in the number of sarcolemmal Na+ pumps. Acta Physiol. Scand. 185, 329–334 [DOI] [PubMed] [Google Scholar]
- 11. Morth J. P., Poulsen H., Toustrup-Jensen M. S., Schack V. R., Egebjerg J., Andersen J. P., Vilsen B., Nissen P. (2009) The structure of the Na+,K+-ATPase and mapping of isoform differences and disease-related mutations. Philos. Trans. R. Soc. B Biol. Sci. 364, 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng S. X., Aizman O., Nairn A. C., Greengard P., Aperia A. (1999) [Ca2+]i determines the effects of protein kinases A and C on activity of rat renal Na+,K+-ATPase. J. Physiol. 518, 37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu Z. Q., Chen J., Chi Z. Q., Liu J. G. (2007) Involvement of dopamine system in regulation of Na+,K+-ATPase in the striatum upon activation of opioid receptors by morphine. Mol. Pharmacol. 71, 519–530 [DOI] [PubMed] [Google Scholar]
- 14. Oliveira M. S., Furian A. F., Rambo L. M., Ribeiro L. R., Royes L. F., Ferreira J., Calixto J. B., Otalora L. F., Garrido-Sanabria E. R., Mello C. F. (2009) Prostaglandin E2 modulates Na+,K+-ATPase activity in rat hippocampus: implications for neurological diseases. J. Neurochem. 109, 416–426 [DOI] [PubMed] [Google Scholar]
- 15. Kristensen B., Birkelund S., Jorgensen P. L. (2003) Trafficking of Na,K-ATPase fused to enhanced green fluorescent protein is mediated by protein kinase A or C. J. Membr. Biol. 191, 25–36 [DOI] [PubMed] [Google Scholar]
- 16. Vinciguerra M., Deschênes G., Hasler U., Mordasini D., Rousselot M., Doucet A., Vandewalle A., Martin P. Y., Féraille E. (2003) Intracellular Na+ controls cell surface expression of Na,K-ATPase via a cAMP-independent PKA pathway in mammalian kidney collecting duct cells. Mol. Biol. Cell 14, 2677–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meier S., Tavraz N. N., Dürr K. L., Friedrich T. (2010) Hyperpolarization-activated inward leakage currents caused by deletion or mutation of carboxy-terminal tyrosines of the Na+/K+-ATPase α-subunit. J. Gen. Physiol. 135, 115–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yaragatupalli S., Olivera J. F., Gatto C., Artigas P. (2009) Altered Na+ transport after an intracellular α-subunit deletion reveals strict external sequential release of Na+ from the Na/K pump. Proc. Natl. Acad. Sci. U.S.A. 106, 15507–15512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang H. L., Cheng X., Taylor P., McCammon J. A., Sine S. M. (2008) Control of cation permeation through the nicotinic receptor channel. PLoS Comp. Biol. 4, e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Law R. J., Munson K., Sachs G., Lightstone F. C. (2008) An ion gating mechanism of gastric H,K-ATPase based on molecular dynamics simulations. Biophys. J. 95, 2739–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Espinoza-Fonseca L. M., Thomas D. D. (2011) Atomic-level characterization of the activation mechanism of SERCA by calcium. PLoS One 6, e26936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Musgaard M., Thøgersen L., Schiøtt B. (2011) Protonation states of important acidic residues in the central Ca2+ ion binding sites of the Ca2+-ATPase: a molecular modeling study. Biochemistry 50, 11109–11120 [DOI] [PubMed] [Google Scholar]
- 23. Witzke S., Duelund L., Petersen M., Kongsted J., Mouritsen O. G., Khandelia H. (2010) The inclusion of terpenoid plant extracts in lipid bilayers investigated by molecular dynamics simulations. J. Phys. Chem. B 114, 15825–15831 [DOI] [PubMed] [Google Scholar]
- 24. Berendsen H. J. C., van der Spoel D., van Drunen R. (1995) GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 91, 43–56 [Google Scholar]
- 25. Hess B., Kutzner C., van der Spoel D., Lindahl E. (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation J. Chem. Theoret. Comput. 4, 435–447 [DOI] [PubMed] [Google Scholar]
- 26. Spoel D. V. D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. C. (2005) GROMACS: fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 [DOI] [PubMed] [Google Scholar]
- 27. Berger O., Edholm O., Jähnig F. (1997) Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 72, 2002–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jorgensen W. L., Maxwell D. S., TiradoRives J. (1996) Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 118, 11225–11236 [Google Scholar]
- 29. Tieleman D. P., Maccallum J. L., Ash W. L., Kandt C., Xu Z., Monticelli L. (2006) Membrane protein simulations with a united-atom lipid and all-atom protein model: lipid-protein interactions, side chain transfer free energies, and model proteins. J. Phys. Condens. Matter 18, S1221–1234 [DOI] [PubMed] [Google Scholar]
- 30. Chakrabarti N., Neale C., Payandeh J., Pai E. F., Pomès R. (2010) An iris-like mechanism of pore dilation in the CorA magnesium transport system. Biophys. J. 98, 784–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Provasi D., Bortolato A., Filizola M. (2009) Exploring molecular mechanisms of ligand recognition by opioid receptors with metadynamics. Biochemistry 48, 10020–10029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ramachandran S., Serohijos A. W., Xu L., Meissner G., Dokholyan N. V. (2009) A structural model of the pore-forming region of the skeletal muscle ryanodine receptor (RyR1) PLoS Comp. Biol. 5, e1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stansfeld P. J., Sansom M. S. (2011) Molecular simulation approaches to membrane proteins. Structure 19, 1562–1572 [DOI] [PubMed] [Google Scholar]
- 34. Piana S., Lindorff-Larsen K., Shaw D. E. (2011) How robust are protein folding simulations with respect to force field parameterization? Biophys. J. 100, L47–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Berendsen H. J. C., Postma J. P. M., Vangunsteren W. F., Dinola A., Haak J. R. (1984) Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 [Google Scholar]
- 36. Leach A. R. (2001) Molecular Modelling: Principles and Applications, 2nd Ed., pp. 356–357, Pearson Prentice Hall, Harlow, UK [Google Scholar]
- 37. Hess B. (2008) P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theoret. Comput. 4, 116–122 [DOI] [PubMed] [Google Scholar]
- 38. Hess B., Bekker H., Berendsen H. J. C., Fraaije J. G. E. M. (1997) LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 [Google Scholar]
- 39. Miyamoto S., Kollman P. A. (1992) Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 13, 952–962 [Google Scholar]
- 40. Darden T., York D., Pedersen L. (1993) Particle mesh Ewald: An Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 41. Humphrey W., Dalke A., Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graphics 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 42. Toustrup-Jensen M. S., Holm R., Einholm A. P., Schack V. R., Morth J. P., Nissen P., Andersen J. P., Vilsen B. (2009) The C Terminus of Na+,K+-ATPase controls Na+ affinity on both sides of the membrane through Arg935 J. Biol. Chem. 284, 18715–18725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Awayda M. S., Ismailov I. I., Berdiev B. K., Fuller C. M., Benos D. J. (1996) Protein kinase regulation of a cloned epithelial Na+ channel. J. Gen. Physiol. 108, 49–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee Y. S., Lee J. A., Jung J., Oh U., Kaang B. K. (2000) The cAMP-dependent kinase pathway does not sensitize the cloned vanilloid receptor type 1 expressed in Xenopus oocytes or Aplysia neurons. Neurosci. Lett. 288, 57–60 [DOI] [PubMed] [Google Scholar]
- 45. Groban E. S., Narayanan A., Jacobson M. P. (2006) Conformational changes in protein loops and helices induced by post-translational phosphorylation. PLoS Comp. Biol. 2, e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chibalin A. V., Vasilets L. A., Hennekes H., Pralong D., Geering K. (1992) Phosphorylation of Na,K-ATPase α-subunits in microsomes and in homogenates of Xenopus oocytes resulting from the stimulation of protein kinase A and protein kinase C. J. Biol. Chem. 267, 22378–22384 [PubMed] [Google Scholar]
- 47. Tavraz N. N., Friedrich T., Dürr K. L., Koenderink J. B., Bamberg E., Freilinger T., Dichgans M. (2008) Diverse functional consequences of mutations in the Na+/K+-ATPase a2-subunit causing familial hemiplegic migraine type 2. J. Biol. Chem. 283, 31097–31106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Feschenko M. S., Sweadner K. J. (1994) Conformation-dependent phosphorylation of Na,K-ATPase by protein kinase A and protein kinase C. J. Biol. Chem. 269, 30436–30444 [PubMed] [Google Scholar]
- 49. Teixeira V. L., Katz A. I., Pedemonte C. H., Bertorello A. M. (2003) Isoform-specific regulation of Na+,K+-ATPase endocytosis and recruitment to the plasma membrane Ann. N.Y. Acad. Sci. 986, 587–594 [DOI] [PubMed] [Google Scholar]
- 50. Codina J., Liu J., Bleyer A. J., Penn R. B., DuBose T. D. (2006) Phosphorylation of S-955 at the protein kinase A consensus promotes maturation of the α- subunit of the colonic H+,K+-ATPase. J. Am. Soc. Nephrol. 17, 1833–1840 [DOI] [PubMed] [Google Scholar]
- 51. Blanco G., Mercer R. W. (1998) Isozymes of the Na-K-ATPase: heterogeneity in structure, diversity in function. Am. J. Physiol. 275, F633–650 [DOI] [PubMed] [Google Scholar]