Background: Ribonucleotide reductase M2 (RRM2) is a rate-limiting step for DNA synthesis. It is still unknown how RRM2 is involved in decidualization.
Results: RRM2 is highly expressed in the decidua and up-regulated by progesterone and DNA damage. Decidualization is significantly inhibited by specific RRM2 inhibitors.
Conclusion: RRM2 is essential for mouse decidualization.
Significance: This study will shed light on understanding the mechanism underlying decidualization.
Keywords: DNA Damage, MYC, Pregnancy, Progesterone, Uterus, Embryo Implantation, Decidualization, Ribonucleotide Reductase
Abstract
Embryo implantation into the maternal uterus is a crucial step for the successful establishment of mammalian pregnancy. Following the attachment of embryo to the uterine luminal epithelium, uterine stromal cells undergo steroid hormone-dependent decidualization, which is characterized by stromal cell proliferation and differentiation. The mechanisms underlying steroid hormone-induced stromal cell proliferation and differentiation during decidualization are still poorly understood. Ribonucleotide reductase, consisting of two subunits (RRM1 and RRM2), is a rate-limiting enzyme in deoxynucleotide production for DNA synthesis and plays an important role in cell proliferation and tumorgenicity. Based on our microarray analysis, Rrm2 expression was significantly higher at implantation sites compared with interimplantation sites in mouse uterus. However, the expression, regulation, and function of RRM2 in mouse uterus during embryo implantation and decidualization are still unknown. Here we show that although both RRM1 and RRM2 expression are markedly induced in mouse uterine stromal cells undergoing decidualization, only RRM2 is regulated by progesterone, a key regulator of decidualization. Further studies showed that the induction of progesterone on RRM2 expression in stromal cells is mediated by the AKT/c-MYC pathway. RRM2 can also be induced by replication stress and DNA damage during decidualization through the ATR/ATM-CHK1-E2F1 pathway. The weight of implantation sites and deciduoma was effectively reduced by specific inhibitors for RRM2. The expression of decidual/trophoblast prolactin-related protein (Dtprp), a reliable marker for decidualization in mice, was significantly reduced in deciduoma and steroid-induced decidual cells after HU treatment. Therefore, RRM2 may be an important effector of progesterone signaling to induce cell proliferation and decidualization in mouse uterus.
Introduction
Ribonucleotide reductase (RR)2 is the only enzyme responsible for the reduction of ribonucleotides to their corresponding deoxyribonucleotides and a rate-limiting enzyme for the production of 2′-deoxyribonucleoside 5′-diphosphates (dNTP) required for DNA synthesis and repair (1–3). RR consists of RR1 (encoded by the Rrm1 gene) and RR2 (encoded by the Rrm2 and Rrm2b genes) subunits, and both proteins are required for enzymatic activity. The cellular RRM1 protein level remains relatively constant throughout the cell cycle, whereas RRM2 is only expressed during the late G1/early S phase and degraded in late S phase (4). RRM2B is a newly identified small subunit of RR. However, human RRM2 has a 4.76-fold higher binding affinity for human RRM1 than human RRM2B (5). Therefore, RRM2 level is essential for DNA synthesis and cell proliferation (6).
The optimal intracellular concentrations of deoxyribonucleotides are critical for authentic DNA synthesis during DNA replication and repair (7). Defective RR often leads to cell cycle arrest, growth retardation, and high mutation rate (8). The high level of RRM2 protein and RR activity in human nasopharyngeal cancer cells results in ionizing radiation resistance through enhancing damage repair during G2 phase of the cell cycle (1). CHK1 is widely known as a DNA damage checkpoint signaling protein (9). RRM2 and CHK1 could be stimulated by DNA damage caused by ionizing radiation, UV, and camptothecins (CPT) (3, 10–13).
Embryo implantation into the maternal uterus is a crucial step for the successful establishment of mammalian pregnancy. Defects in implantation and trophoblast invasion are presently considered the major challenges for the successful establishment of pregnancy (14). Although many embryo implantation-specific factors have been identified during the implantation period, the underlying molecular mechanism is still unknown. Based on our microarray analysis, Rrm2 expression was significantly higher in mouse uterus at implantation sites than that at interimplantation sites.3 A previous microarray study also showed that Rrm2 expression was up-regulated by 2.55-fold at implantation sites compared with interimplantation sites in mouse uteri (15). However, the expression, regulation, and function of RRM2 in mouse uterus during embryo implantation and decidualization are still unknown. We hypothesized that RRM2 may play a role during embryo implantation and decidualization. In this study, we showed that RRM2 was strongly expressed in the decidua and up-regulated by progesterone and DNA damage. The specific inhibitors for RRM2 could effectively inhibit uterine decidualization in mice.
MATERIALS AND METHODS
Animal Treatments
Mature mice (Kunming White outbred strain) were housed in a temperature- and light-controlled environment (14 h light/10 h dark) with free access to regular food and water. All animal procedures were approved by the Institutional Animal Care and Use Committee of Shantou University.
Female mice were mated with fertile or vasectomized males of the same strain to induce pregnancy or pseudopregnancy (day 1 is the day of vaginal plug). From day 1 to 4, pregnancy was confirmed by recovering embryos from the oviducts or uteri. The implantation sites on day 5 were visualized through intravenous injection of 0.1 ml of 1% Chicago blue dye (Sigma) in saline.
To induce and maintain delayed implantation, pregnant mice were ovariectomized at 0830–0900 h on day 4 of pregnancy and treated with daily injection of progesterone (1 mg/mouse; Sigma) from day 5 to 7. Estradiol-17β (25 ng/mouse; Sigma) was given to progesterone-primed delayed mice to initiate implantation on day 7 of pregnancy. For the activation group, the mice were sacrificed to collect uteri 24 h after estrogen treatment. Delayed implantation was confirmed by flushing blastocysts from one horn of the uterus. The implantation sites of activated uterus were identified through intravenous injection of 0.1 ml of 1% Chicago blue dye.
Artificial decidualization was induced by intraluminally infusing 25 μl of sesame oil (Sigma) into one uterine horn on day 4 of pseudopregnancy, whereas the contralateral uninjected horn served as a control. The uteri were collected on day 8 of pseudopregnancy.
To determine effects of estrogen and progesterone on RRM2 expression, steroid hormonal treatments were initiated 2 weeks after ovariectomy. The ovariectomized mice were injected subcutaneously with estrodiol-17β (100 ng/mouse), progesterone (1 mg/mouse), or a combination of estrodiol-17β and progesterone, respectively. The control mice received vehicle (sesame oil, 0.1 ml/mouse) only. For RU486 treatment, ovariectomized mice were injected with RU486 (0.75 mg/mouse; Sigma) 1 h before progesterone treatment. Mice were sacrificed at different times after the hormone injections to collect uteri for RNA and protein extraction.
Hydroxyurea (HU) (30 mg/0.2 ml/mouse; Sigma) was dissolved in PBS and injected twice on day 4 (1800 and 2200 h). PBS was injected intraperitoneally as a control. After mice were sacrificed at 0900 on day 7, the weight of each implantation site was measured. At least 18 mice were used in each group. To determine the effects of hydroxyurea on decidualization, mice under the induction of artificial decidualization were treated with a daily injection of hydroxyurea (30 mg/0.2 ml/mouse) or PBS on days 5 and 6. Uteri were collected and weighed on day 8 of pseudopregnancy.
Trimidox (3 mg/0.15 ml/mouse; Cayman Chemical, Ann Arbor, MI) was dissolved in DMSO and injected intraperitoneally twice on day 4 (1800 and 2200 h). The weight of each implantation site was measured similarly to hydroxyurea treatment.
In Situ Hybridization
Total RNAs from mouse uteri on day 8 of pregnancy were reverse-transcribed and amplified with the corresponding primers. The amplified fragment of each gene was cloned into pGEM-T plasmid (pGEM-T vector system, Promega, Madison, WI) and verified by sequencing. Digoxigenin-labeled antisense or sense cRNA probes were transcribed in vitro using a digoxigenin RNA labeling kit (Roche Applied Science). Primers used for in situ hybridization are listed in Table 1.
TABLE 1.
Primers used in this study
| Gene name | Primer sequences | Accession number | Size | Application |
|---|---|---|---|---|
| bp | ||||
| Rrm1 | TACCTGGAGCCTTGGCACTT | BC016450 | 76 | Real-time PCR |
| CTCGCTGTTCTTCCTTGCCT | ||||
| Rrm1 | ACCACGAAGCACCCTGACTA | BC016450 | 334 | In situ hybridization |
| GAATCCCCACAGAAACCCTC | ||||
| Rrm2 | GATTTAGCCAAGAAGTTCAAGTTACAG | M14223 | 168 | Real-time PCR |
| TCACACAAGGCATAGTTTCAATAGC | ||||
| Rrm2 | CGTTGAGGATGAGCCGTTAC | M14223 | 431 | In situ hybridization |
| CCCAGTCAGCCTTCTTCTTCAC | ||||
| Rrm2 | TGAGAACTTGGTGGAGCG | M14223 | 469 | RT-PCR |
| CCTAACGGCGTTGGTGAT | ||||
| Rrm2 | TTTGAGCTGCCACATGGTGT | chr12:25392235–25392362 | 128 | ChIP |
| GGCTCCACTGACTCTCCCAG | ||||
| Rrm2b | AAAAGCAACAGAACAGCGAAAA | BC058103 | 319 | Real-time PCR |
| GCCAAGGTCACATCAAGGAGTAA | ||||
| Rrm2b | ACCATTTTGGATTTGTGC | BC058103 | 401 | In situ hybridization |
| GCTGCCATCATTTTATTTG | ||||
| Dtprp | AGCCAGAAATCACTGCCACT | NM_010088 | 119 | Real-time PCR |
| TGATCCATGCACCCATAAAA | ||||
| c-Myc | CAGGACTGTATGTGGAGCGGTTTC | NM_001177354 | 214 | Real-time PCR |
| TGTCGTTGAGCGGGTAGGGA | ||||
| Chk1 | CATGAAGCGGGCCATAGACT | NM_007691 | 106 | Real-time PCR |
| TTCCCTCCTGTGGCCATAGA | ||||
| E2f1 | CACTTTGGTCTCGAGGAGGG | BC052160 | 121 | Real-time PCR |
| AGGTGGGCATCTCCAGACAC | ||||
| Rpl7 | GCAGATGTACCGCACTGAGATTC | M29016 | 129 | Real-time PCR |
| ACCTTTGGGCTTACTCCATTGATA | ||||
| Gapdh | ACCACAGTCCATGCCATCAC | NM_008084 | 452 | RT-PCR |
| TCCACCACCCTGTTGCTGTA |
Frozen sections (10 μm) were mounted on 3-aminopropyltriethoxysilane (Sigma)-treated slides and fixed in 4% paraformaldehyde solution in PBS. Hybridization was performed at 55 °C for 16 h as described previously (16). Then sections were incubated in sheep anti-digoxigenin antibody conjugated to alkaline phosphatase (1:5,000; Roche Applied Science). The signal was visualized with the buffer containing 0.4 mm 5-bromo-4-chloro-3-indolyl phosphate and 0.4 mm nitro blue tetrazolium. Endogenous alkaline phosphatase activity was inhibited with 2 mm levamisole (Sigma). All of the sections were counterstained with 1% methyl green. The positive signal was visualized as a dark brown color. The sense probe for each gene was also hybridized and served as negative control. No detectable signals were observed with sense probes.
Immunohistochemistry
Immunohistochemistry was performed as described previously (17). Briefly, sections were incubated with goat anti-RRM1 (sc-11733, Santa Cruz Biotechnology, Inc., Santa Cruz, CA), goat anti-RRM2 (sc-10848, Santa Cruz Biotechnology, Inc.), or normal goat IgG in 10% horse serum overnight at 4 °C. After washing in PBS, the sections were incubated with biotinylated secondary antibody followed by an avidin-alkaline phosphatase complex and Vector Red according to the manufacturer's protocol (Vectastain ABC-AP kit; Vector Laboratories, Burlingame, CA). The signal was visualized as a red color. Endogenous alkaline phosphatase activity was inhibited with levamisole.
The staining intensity of RRM1 and RRM2 on mouse uterine sections was analyzed by Image-Pro Plus (version 6.0, Media Cybernetics), using a modified method basing on one described previously (18). Briefly, the positive staining in selected region was converted to a gray scale image, and the sum of integral optical density was counted. Eight random areas of interest for each section were selected for evaluation. The time required to perform the analysis process was greatly reduced by creating macros. The expression level score was calculated by dividing the sum of integral optical density by the area of the selected region. The multiple comparison was performed with one-way analysis of variance followed by Tukey's test. p < 0.05 was considered statistically significant.
RT-PCR
RNA (500 ng of total RNAs from each sample) was isolated using TRIzol reagent (Invitrogen), digested by RQ1 deoxyribonuclease I (Promega), and reverse transcribed into cDNA with the PrimeScript reverse transcriptase reagent kit (Perfect Real Time, TaKaRa (Dalian, China)). The resulting cDNA was subjected to PCR analysis. Amplified fragments were separated by electrophoresis on 2% agarose gels and visualized by ethidium bromide staining.
Isolation of Uterine Stromal Cells
Uterine stromal cells were isolated as described previously (19). Briefly, uterine horns from day 4 mice were split longitudinally, and digested in HBSS containing 1% (w/v) trypsin (Amresco, Solon, OH) and 6 mg/ml dispase (Roche Applied Science). The digested uteri were shaken gently to dislodge sheets of luminal epithelial cells. The remaining tissues were rinsed three times with HBSS and incubated in HBSS containing 0.15 mg/ml collagenase I (Invitrogen) at 37 °C for 30 min, followed by vigorously shaking until supernatants became turbid. The supernatants were then passed through a 70-μm wire gauze filter to eliminate epithelial sheets and centrifuged. The cell pellets were washed twice with HBSS and resuspended in complete medium consisting of DMEM-nutrient mixture F-12 Ham (DMEM/F-12; Sigma) with 10% charcoal-treated fetal bovine serum (cFBS; Invitrogen). Cells were plated onto 35-mm culture dishes at a concentration of 1 × 106 cells/dish or 2 × 105 cells/well for 24-well culture plates. After an initial culture for 30 min, the isolated stromal cells were further cultured in fresh medium with 2% cFBS.
MTT Assay
Stromal cells were cultured in 24-well plates and treated with 0, 0.01, 0.1, and 1 mm HU, respectively. The culture medium was changed with serum-free medium containing 0.5 mg/ml MTT (Sigma) 24 h later. Stromal cells were further cultured at 37 °C for 4 h. After removal of the medium, 500 μl of DMSO was added into each well to dissolve the formazan crystals. The absorbance at 570 nm was determined using a NanoDrop® ND-1000 spectrophotometer. Triplicate wells were assayed for each condition, and S.D. values were determined.
Flow Cytometry Assay
After the treatment with 1 mm HU for 24 h, cultured cells were collected in PBS and fixed overnight in 70% cold ethanol at 4 °C. Fixed cells were collected by centrifugation, washed with PBS, and then treated with RNase and stained with propidium iodide. Following filtration through nylon mesh to remove cell clumps, the cell suspension was analyzed on flow cytometry immediately. The percentage of stromal cells in G0/G1, S, and G2/M phases of the cell cycle was calculated from DNA histogram data using counter software.
In Vitro Decidualization
In vitro decidualization was performed as described previously with modification (20, 21). Briefly, mouse endometrial stromal cells isolated from day 4 of pregnancy were treated with 10 nm estradiol-17β and 1 μm progesterone with/without 0.1 mm hydroxyurea. Culture medium was changed every 2 days. Cells were collected 3 days later.
Steroid Hormonal Treatments in Vitro
Cultured stromal cells were treated with 10 nm estradiol-17β, 1 μm progesterone, or a combination of estrodiol-17β and progesterone, respectively. For further studies, cells were treated with cycloheximide (CHX (1 μg/ml); Sigma), RU486 (1 μm), Ly294002 (15 μm; Cell Signaling), or 10058-F4 (20 μm or 40 μm; Sigma) in the absence or presence of progesterone, respectively. Then cells were collected at different times for further quantitative analysis by real-time RT-PCR and Western blot.
Plasmid Construction, Transfection, and Luciferase Assay
For luciferase reporter experiments, the promoter segment of mouse Rrm2 was amplified by PCR from mouse genomic DNA. The forward primer was 5′-CCGGGGTACCGTAGAAGGGAGTGAAGCAAC-3′ containing a KpnI site (underlined sequence), whereas the reverse primer was 5′-TATTAAGCTTCGGCGGAGACGGCGAGGA-3′, which included a HindIII site (underlined sequence). The amplified fragment was digested by restriction enzymes and inserted into the pGL3-basic vector (Promega) upstream from the start codon of luciferase.
Transfections of uterine stromal cells were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. pRL-TK plasmid containing Renilla luciferase was co-transfected with Rrm2-pGL3 for data normalization. The medium was replaced with complete culture medium containing 2% cFBS 6 h later. Following an additional 12 h of culture, stromal cells were then treated with estrodiol-17β (10 nm) and progesterone (1 μm), respectively. Both estrodiol-17β and progesterone were dissolved in ethanol. The final concentration of ethanol was kept under 0.1% (v/v). Then cell lysate was collected for luciferase analysis after 24 h of culture. Firefly and Renilla luciferase activities were measured using the Dual-Luciferase reporter assay system (Promega), and each group was assayed in triplicate.
To determine the effect of c-MYC on the Rrm2 promoter, stromal cells were transfected with Rrm2-PGL3 and pRL-TK in combination with pcDNA 3.1(+) or c-Myc overexpression vectors, respectively. Culture medium was changed by complete medium with 2% cFBS 6 h later. Cells were collected for luciferase activity analysis in 12 h.
Real-time RT-PCR
Total RNAs from mouse uteri or cultured cells were isolated using TRIzol reagent (Invitrogen), digested by RQ1 deoxyribonuclease I (Promega), and reverse transcribed into cDNA with the PrimeScript reverse transcriptase reagent kit (Perfect Real Time, TaKaRa). For real-time RT-PCR, cDNA was amplified using a SYBR Premix Ex Taq kit (DRR041S, TaKaRa) on the Rotor-Gene 3000A system (Corbett Research, Mortlake, Australia). The conditions used for real-time PCR were as follows: 95 °C for 10 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 34 s. All reactions were run in triplicate. Gapdh or Rpl7 was used for normalization. Data from real-time PCR were analyzed using the 2−ΔΔCt method. Primers used for Real-time PCR were listed in Table 1.
Northern Blot
Total RNAs (5 μg) were resolved on 1.2% agarose gel, transferred onto nylon membranes. Membranes were UV-cross-linked and hybridized with digoxigenin-labeled cRNA probe for Rrm2 in the hybridization solution (BioDev, MK161–2, Beijing, China) at 68 °C for 16 h. Then membranes were washed in 2× SSC with 0.1% SDS for 5 min, twice in 0.2× SSC with 0.1% SDS at 68 °C for 15 min each, and in washing buffer (0.1 m maleic acid, 0.15 m NaCl, and 0.3% Tween 20) for 5 min. After blocking in 1% blocking reagent (Roche Applied Science) for 30 min, membranes were incubated with anti-digoxigenin antibody conjugated with alkaline phosphatase for 30 min. Following washing in washing buffer and detection buffer (0.1 m Tris, 0.1 m NaCl, pH 9.5), membranes were incubated in CDP-Star (Roche Applied Science) and exposed with x-ray film. For normalization, membranes were stripped and reprobed with digoxigenin-labeled Gapdh. For samples from uterus, 28S RNA was used as an internal reference.
Western Blot
Cultured cells were collected directly in lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Triton X-100, and 0.25% sodium deoxycholate), whereas uterine tissues were homogenized in lysis buffer. Protein concentration was measured with a BCA reagent kit (Applygen, Beijing, China). Samples were run on a 10% polyacrylamide gel and transferred onto nitrocellulose membranes. After they were blocked in 5% nonfat dry milk in PBST (0.1% Tween 20 in PBS) for 1 h, membranes were incubated overnight at 4 °C with each primary antibody for RRM1, RRM2, c-MYC (9402, Cell Signaling, Beverly, MA), or p-AKT (9272, Cell Signaling), respectively. After three washes in 5% milk in PBST for 10 min each, membranes were incubated in matched second antibody conjugated with horseradish peroxidase for 1 h followed by two washes in 5% nonfat milk in PBST, PBST, and PBS, respectively. The signals were developed with the ECL chemiluminescent kit (Amersham Biosciences).
Co-immunoprecipitation
The sample used for co-immunoprecipitation was the implantation site of mouse uterus on day 8 of pregnancy. Co-immunoprecipitation was performed with goat anti-RRM1 antibody using goat IgG as a control. The immunoprecipitates were resolved on a 10% polyacrylamide gel and transferred onto nitrocellulose membranes. Western blot was performed as above using anti-RRM1 and anti-RRM2 antibodies, respectively.
Chromatin Immunoprecipitation (ChIP)
ChIP was conducted as described previously with modification (22). Mouse decidua from day 8 of pregnancy was washed once with PBS and fixed with 1% formaldehyde for 10 min to covalently cross-link any DNA-protein complexes. The cross-link was quenched by adding 570 μl of 2.5 m glycine. Tissues were washed three times with PBS and homogenized with a glass homogenizer. After they were incubated in Mg-NI buffer (15 mm Tris-HCl, pH 7.9, 5 mm MgCl2, 60 mm KCl, 0.5 mm DTT, 15 mm NaCl, and 300 mm sucrose) and Mg-NI-Nonidet P-40 buffer, cell pellets were resuspended in lysis buffer and sonicated by repeating a program of 15 s on and 30 s off at 45% amplitude 10 times. After the samples were centrifuged at 13,000 rpm for 10 min to pellet cell debris, the soluble chromatin was harvested. Immunoprecipitations were performed with 1 μg of IgG (catalog no. 2729, Cell Signaling), anti-AcH3 (catalog no. 06-599, Millipore), or anti-c-MYC (sc-764X, Santa Cruz Biotechnology, Inc.) and incubated at 4 °C overnight. For immunoprecipitation, protein A slurry was added to pull down the DNA-protein complexes. Then complexes were washed and treated with proteinase K. Relative levels of transcription factor enrichment on the gene promoter were determined by real-time PCR and normalized to input DNA. Primers used in real-time PCR are listed in Table l.
DNA Damage Induction of Uterine Stromal Cells
Stromal cells isolated from mouse uterus on day 4 were treated with DMSO, caffeine (3 mm), UCN-01 (0.25 μm), or KU-55933 (10 μm) for 1 h, respectively. Then each group was divided into three subgroups, which were treated with vehicle, CPT (1 μm), or UV (20 J). Cells were collected for RNA extraction and protein purification at the corresponding time.
To explore the role of E2F1 on RRM2 expression, the overexpression vector, E2f1-pcDNA3.1, and siRNA targeted to E2f1 were transfected into mouse stromal cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions, respectively.
For luciferase reporter experiments, pRL-TK plasmid containing Renilla luciferase was cotransfected with E2f1-pcDNA 3.1 and Rrm2-pGL3 for data normalization. After 24 h of culture, cell lysate was collected for luciferase analysis. Firefly and Renilla luciferase activities were measured using the Dual-Luciferase reporter assay system (Promega), and each group was assayed in triplicate.
For RNA interference, the medium was replaced with complete culture medium containing 2% cFBS 6 h after transfection. Following an additional 12-h culture, cells were treated with vehicle or CPT (1 μm). Cells were collected for Western blot 36 h later.
To determine the role of CHK1 on RRM2 expression regulated by progesterone, stromal cells were pretreated with RU486, UCN-01, or RU486 plus UCN-01 for 1 h, followed by vehicle or progesterone treatment. Cells were collected for Western blot after a 24-h treatment with progesterone.
Statistical Analysis
All of the experiments were independently repeated at least three times. Specifically, for in vivo experiments, at least three separate mice were used for each treatment. For in vitro experiments, each treatment was repeated at least three times from the isolation and culture of uterine stromal cells. The significance of difference between two groups was assessed by Student's t test. The multiple comparisons were performed with one-way analysis of variance. Results are expressed as means ± S.E. of at least three separate experiments. p < 0.05 was considered statistically significant.
RESULTS
Localization of RRM1, RRM2, and RRM2b in Mouse Uterus during Pregnancy
Ribonucleotide reductase is composed of two regulatory subunits (RRM1) and two catalytic subunits (RRM2 or RRM2b). In situ hybridization and immunohistochemistry were performed to detect the localization of RRM1, RRM2, and RRM2b.
From day 1 to 4 of pregnancy, there was no visible Rrm1 mRNA and protein signal in the uterus (data not shown). On day 5, a specific Rrm1 mRNA signal was detected in the subluminal stroma surrounding the implanting blastocyst, and no signal was seen in the interimplantation sites (Fig. 1A). From day 6 to 8, both RRM1 mRNA and protein signals were highly detected in the decidua and increased as the decidua were developing. Rrm1 mRNA was also expressed in the embryos from day 6 to 8 (Fig. 1A). There was no detectable RRM1 immunostaining in the uteri from day 1 to 5 of pregnancy (data not shown).
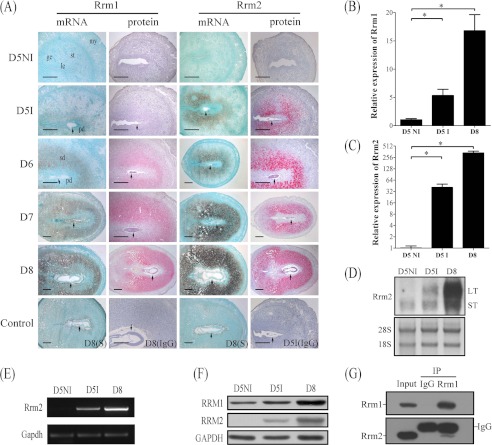
FIGURE 1.
Expression of RRM1 and RRM2 in mouse uterus during early pregnancy. A, both mRNA and protein localizations of RRM1 and RRM2 in mouse uterus from day 5 (D5) to day 8 (D8) were detected by in situ hybridization and immunostaining, respectively. Real-time RT-PCR was performed to quantify the mRNA level of Rrm1 (B) and Rrm2 (C) in mouse uterus on day 5 (D5NI, interimplantation site on day 5; D5I, implantation site on day 5) and day 8. D, Northern blot. Both long transcripts (LT) and small transcripts (ST) of Rrm2 were up-regulated at the implantation sites. 28S was used as internal reference. E, RT-PCR analysis of Rrm2 mRNA level in mouse pregnant uteri. Strong Rrm2 bands were observed in implantation sites on days 5 and 8 compared with the interimplantation site on day 5 of pregnancy. F, Western blot of RRM1 and RRM2 protein in mouse uterus on days 5 and 8. G, physical interaction between RRM1 and RRM2 in mouse decidua was detected by co-immunoprecipitation. ge, glandular epithelium; le, luminal epithelium; my, myometrium; st, stroma; pd, primary decidua; sd, secondary decidua; arrow, embryo. Bar, 300 μm. *, p < 0.05; error bars, S.E.
There were no detectable signals for RRM2 mRNA and protein in mouse uteri from day 1 to 4 (data not shown) and at the interimplantation sites on day 5. At the implantation sites, there were strong signals for RRM2 mRNA and protein in the subluminal stromal cells on day 5. The signal spread to the whole decidual area from day 6 to 8 (Fig. 1A). Quantitative data for RRM2 immunostaining were provided in supplemental Fig. 1.
Because no mRNA signal for Rrm2b was detected in mouse uterus from day 1 to 8 of pregnancy (data not shown), no further analysis was performed on RRM2b.
Quantitative Analysis of Rrm1 and Rrm2 mRNA Expression in Mouse Uterus during Pregnancy
Real-time RT-PCR was performed to quantify mRNA expression of Rrm1, Rrm2, and Rrm2b in mouse uterus during early pregnancy. Compared with interimplantation sites on day 5, the level of Rrm1 at the implantation sites on days 5 and 8 was up-regulated 6.6- and 20.8-fold, respectively (Fig. 1B). Rrm2 expression increased 18.6-fold at the implantation sites on day 5 and 153.3-fold on day 8 compared with interimplantation sites on day 5 (Fig. 1C). However, there was no significant change for Rrm2b mRNA expression between implantation sites and interimplantation sites on day 5 and day 8 of pregnancy (data not shown).
Northern blot was also performed to detect the transcripts of Rrm2 expressed in mouse uterus during pregnancy. At the interimplantation sites, only one transcript, the smaller one, was detected. However, both transcripts of Rrm2 were significantly up-regulated at the implantation sites on day 5 and day 8 compared with interimplantation sites (Fig. 1D). Rrm2 expression of mouse uteri on both days 5 and 8 was also confirmed by RT-PCR (Fig. 1E).
By Western blot, only one protein band of RRM2 was detected on days 5 and 8 at the corresponding position. Compared with interimplantation sites, both RRM1 and RRM2 protein were up-regulated at the implantation sites on days 5 and 8 (Fig. 1F).
Expression of RRM1 and RRM2 during Pseudopregnancy and under Delayed Implantation
To address whether the expression of RRM1 and RRM2 is dependent on the embryo, we examined the expression of RRM1 and RRM2 during pseudopregnancy. There were no detectable signals for RRM1 and RRM2 in mouse uteri from day 1 to 5 of pseudopregnancy (data not shown).
When embryo implantation was delayed by ovariectomy, there were no detectable mRNA signals for Rrm1 and Rrm2 in the uterus. After estrogen activated the dormant blastocyst to implant, both Rrm1 and Rrm2 signals were detected in the stromal cells of the implantation sites (Fig. 2A and supplemental Fig. 1). A high level of RRM2 immunostaining was detected in the stromal cells surrounding implanting blastocyst once delayed implantation was terminated. However, there was no detectable RRM1 immunostaining in both delayed and activated mouse uteri.
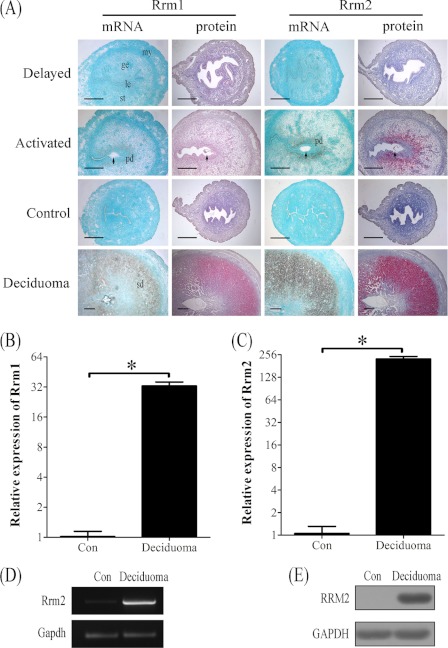
FIGURE 2.
Both mRNA and protein expressions of RRM1 and RRM2 at implantation sites in mouse uteri under delayed implantation and artificial decidualization, respectively. A, in situ hybridization and immunostaining of RRM1 and RRM2 under delayed implantation and artificial decidualization, respectively. B, real-time PCR analysis of Rrm1 under artificial decidualization. C, real-time PCR analysis of Rrm2 mRNA expression under artificial decidualization. D, RT-PCR analysis of Rrm2 mRNA level. Rrm2 mRNA level in mouse deciduoma was significantly higher compared with control. Gapdh was used for loading control. E, Western blot analysis of RRM2 protein. RRM2 protein level in deciduoma was significantly stronger than control. ge, glandular epithelium; le, luminal epithelium; my, myometrium; st, stroma; pd, primary decidua; sd, secondary decidua; arrow, embryo. Bar, 300 μm. *, p < 0.05; error bars, S.E.
Expression of RRM1 and RRM2 under Artificial Decidualization
Artificial decidualization was performed to see whether the expression of RRM1 and RRM2 in decidua was dependent on the presence of embryos. There were no detectable signals for both RRM1 and RRM2 in the uninjected control horn. Under artificial decidualization, both mRNA and protein levels of RRM1 and RRM2 were strongly detected in the decidualized cells (Fig. 2A and supplemental Fig. 1). Results from real-time RT-PCR also indicated that mRNA expression of Rrm1 and Rrm2 increased up to 32.8- and 224.7-fold, respectively (Fig. 2, B and C). By RT-PCR, Rrm2 expression was also significantly up-regulated in deciduoma compared with control (Fig. 2D). The high level of RRM2 protein in deciduoma was also confirmed by Western blot (Fig. 2E).
Interaction between RRM1 and RRM2 in Mouse Uterus during Pregnancy
Because both RRM1 and RRM2 are required for enzymatic activity, co-immunoprecipitation was performed to examine whether there is a physical interaction between RRM1 and RRM2 in mouse decidua on day 8 of pregnancy. Both goat IgG and goat anti-RRM1 were used to precipitate the protein complexes. Then Western blot was carried out to examine the immunoprecipitated complex with goat anti-RRM1 and goat anti-RRM2, respectively. There were no detectable bands for goat IgG group, whereas there was a strong RRM1 band with goat anti-RRM1 and a strong RRM2 band with goat anti-RRM2, respectively, indicating a physical binding between RRM1 and RRM2 in mouse decidua (Fig. 1G).
Effects of Hydroxyurea on Decidualization
Hydroxyurea is a specific inhibitor for ribonucleotide reductase activity (23). Hydroxyurea was intraperitoneally injected on day 4 of pregnancy to investigate the role of RRM2 during pregnancy. Because hydroxyurea has a very short half-life of not more than 3 h, mice were administrated by two injections of hydroxyurea at 1800 and 2200 h, respectively. Compared with control, hydroxyurea injection significantly reduced the weight of implantation sites on day 7 (Fig. 3, A and B).
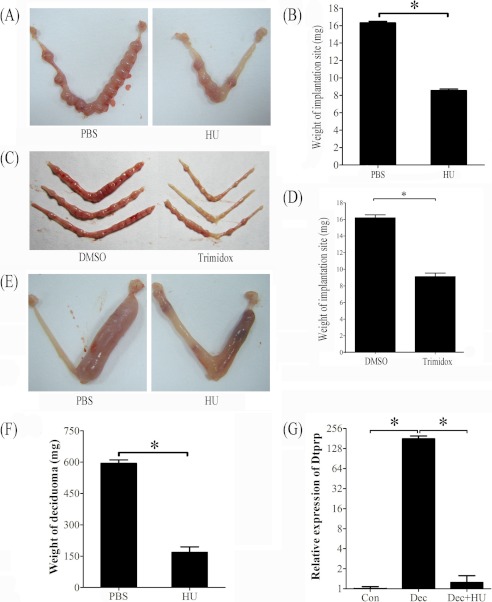
FIGURE 3.
Effects of hydroxyurea and trimidox on decidua and deciduoma. A, a representative photo showing the weight of implantation site was significantly decreased after pregnant mice were treated with hydroxyurea. B, the quantification of the weight of implantation sites after pregnant mice were treated with hydroxyurea. C, a representative photograph showing mouse uteri on day 7 after day 4 pregnant mice were treated with DMSO (control) or trimidox twice (1800 and 2200 h). D, the weight of implantation site was significantly decreased by trimidox treatment compared with control. E, a representative photo showing the weight of deciduoma was significantly reduced after pseudopregnant mice under artificial decidualization were treated with hydroxyurea. F, the quantification of deciduoma after pseudopregnant mice under artificial decidualization were treated with hydroxyurea. G, relative expression level of Dtprp mRNA in mouse uterus after pseudopregnant mice under artificial decidualization were treated with hydroxyurea. Con, control uterus; Dec, deciduoma; Dec + HU, deciduoma after HU injection. *, p < 0.05; error bars, S.E.
Trimidox, another specific inhibitor for RRM2 (24, 25), was also used to verify effects of RRM2 on decidualization. When pregnant mice on day 4 were treated with trimidox twice, the weight of the implantation site on day 7 was significantly reduced compared with control (Fig. 3, C and D).
To confirm the role of RRM2 during decidualization, hydroxyurea was also intraperitoneally injected into pseudopregnant mice twice on days 6 and 7 under artificial decidualization. Compared with PBS injection, the weight of deciduoma induced by sesame oil injection was significantly reduced by hydroxyurea injection (Fig. 3, E and F).
Because decidual/trophoblast prolactin-related protein (Dtprp) is a reliable marker for decidualization in mice (26), real-time RT-PCR was performed to examine the effects of hydroxyurea on Dtprp expression. Compared with uninjected control horn, Dtprp level was up-regulated 179.7-fold in deciduoma under artificial decidualization. Hydroxyurea significantly reduced Dtprp expression in deciduoma, which was similar to that in uninjected control horn (Fig. 3G).
Effect of Hydroxyurea on Decidualization in Vitro
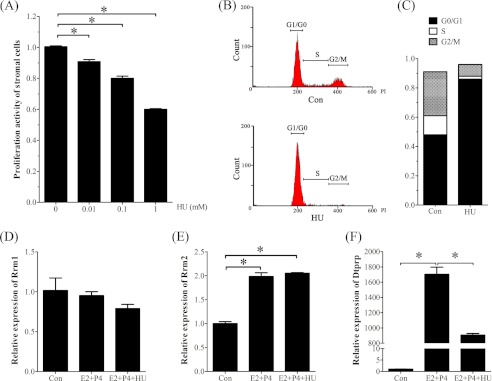
To investigate the regulation and role of RRM2 during decidualization, stromal cells isolated from mouse uteri on day 4 were induced for in vitro decidualization. The effective concentrations of hydroxyurea were determined by an MTT test. Compared with the control, the proliferation activity of stromal cells was significantly decreased by hydroxyurea in a dose-dependent manner (Fig. 4A). The proliferation activity of stromal cells treated with 1 mm hydroxyurea was decreased to 60% of control. To confirm the role of hydroxyurea, a flow cytometric assay was used to examine the proliferating stromal cells. The results also showed that the number of stromal cells in the S + G2/M phase was significantly decreased after a 24-h treatment with 1 mm hydroxyurea (Fig. 4, B and C).
FIGURE 4.
Effects of hydroxyurea on cell proliferation and decidualization in vitro. A, MTT assay of cell proliferation after stromal cells were treated with different concentrations of hydroxyurea. B, flow cytometry analysis of cell cycle after stromal cells were treated with hydroxyurea. C, histogram of the stromal cells under different cell cycles after stromal cells were treated with hydroxyurea. D, real-time RT-PCR of Rrm1 mRNA expression under in vitro decidualization. E, real-time RT-PCR of Rrm2 mRNA expression under in vitro decidualization. F, real-time RT-PCR of Dtprp mRNA expression under in vitro decidualization. Con, control; E2 + P4, estradiol-17β plus progesterone; E2 + P4 + HU, estradiol-17β plus progesterone plus hydroxyurea. *, p < 0.05; error bars, S.E.
Then a lower concentration of hydroxyurea (0.1 mm) was chosen to study its role on decidualization. Hydroxyurea had no obvious effects on Rrm1 mRNA expression (Fig. 4D). Under in vitro decidualization, the mRNA levels of Rrm2 and Dtprp were significantly stimulated (Fig. 4, E and F). Although hydroxyurea significantly decreased Dtprp expression, it had no effect on Rrm2 expression (Fig. 4E).
Regulation of Ovarian Steroid Hormones on RRM2 Expression
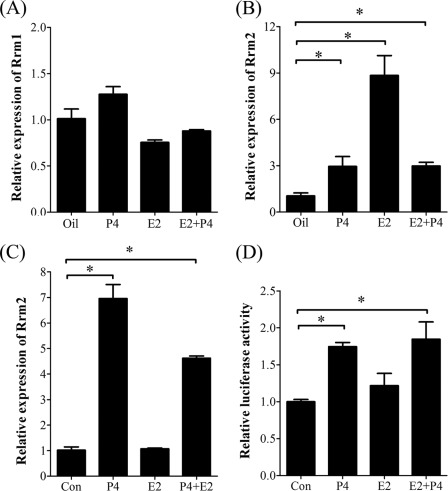
Estrogen and progesterone are essential for mouse embryo implantation. Ovariectomized mice were used to examine whether RRM2 expression was regulated by estrogen and progesterone. By real-time RT-PCR and Western blot analysis, there was no considerable change for RRM1 expression after ovariectomized mice were treated with steroid hormones compared with control (Figs. 5A and 6G). However, both E2 and progesterone could elevate Rrm2 expression in uteri of ovariectomized mice (Fig. 5B). In the in vitro cultured stromal cells, Rrm2 expression was significantly induced by progesterone (Fig. 5C). Through a luciferase activity assay, the promoter activity of Rrm2 was also induced by progesterone in cultured stromal cells (Fig. 5D).
FIGURE 5.
Steroid hormonal regulation on RRM1 and RRM2 expression. A, real-time RT-PCR of Rrm1 mRNA level after stromal cells were treated with steroid hormones. B, real-time RT-PCR of Rrm2 mRNA level after ovariectomized mice were treated with steroid hormones. C, real-time RT-PCR of Rrm2 mRNA level after stromal cells were treated with steroid hormones. D, the relative luciferase activity of Rrm2 promoter when stromal cells were treated with steroid hormones. *, p < 0.05; error bars, S.E.
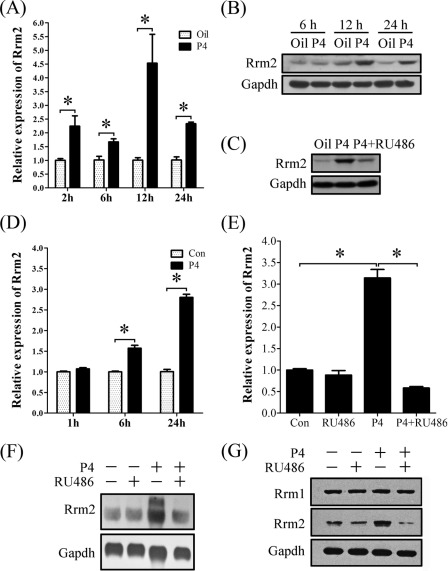
FIGURE 6.
Progesterone regulation on RRM2 expression in vivo and in vitro. A, real-time RT-PCR of Rrm2 expression after ovariectomized mice were treated with progesterone for different times. B, Western blot of RRM2 protein after ovariectomized mice were treated with progesterone for different times. C, protein expression of RRM2 in uteri of ovariectomized mice injected with oil, P4, or P4 plus RU486. D, real-time RT-PCR of Rrm2 expression after stromal cells were treated with progesterone for different times. E, real-time RT-PCR of Rrm2 expression after stromal cells were treated with RU486, progesterone, or both progesterone and RU486. F, Northern blot analysis of both transcripts of Rrm2 in stromal cells treated with RU486, progesterone, or both progesterone and RU486. G, Western blot analysis of RRM1 and RRM2 proteins in stromal cells treated with RU486, progesterone, or both progesterone and RU486. *, p < 0.05; error bars, S.E.
To further study progesterone regulation on RRM2 expression, ovariectomized mice were treated with progesterone in a time course. Although RRM2 expression was stimulated by progesterone at 3, 6, 12, and 24 h, respectively, the highest level was detected at 12 h after progesterone treatment (Fig. 6, A and B). Progesterone antagonist, RU486, significantly reduced progesterone-stimulated RRM2 expression (Fig. 6C). In vitro, Rrm2 expression was elevated from 6 h after progesterone treatment (Fig. 6D). After stromal cells were pretreated with RU486, no considerable increase of Rrm2 mRNA was detected in stromal cells with a 24-h treatment of progesterone compared with control (Fig. 6E).
To determine the regulation of progesterone on different transcripts of Rrm2, Northern blot was performed with total RNAs extracted from stromal cells that were treated by progesterone, RU486, or progesterone plus RU486. Only the smaller transcript was detected in control and RU486 treatment. Progesterone significantly stimulated the expression of both transcripts, whereas RU486 pretreatment effectively inhibited the stimulation of progesterone (Fig. 6F). Progesterone stimulation on RRM2 expression was further confirmed by Western blot, but only one band was detected (Fig. 6G).
Progesterone Induction on RRM2 Expression through Indirect Way
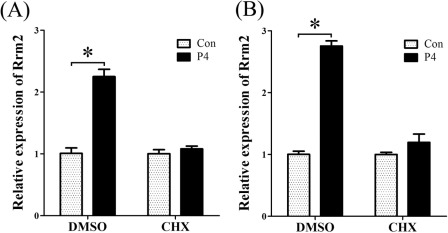
In order to verify whether progesterone induced RRM2 expression through a direct or indirect way, stromal cells were pretreated with cycloheximide 2 h before progesterone treatment. Without CHX pretreatment, Rrm2 mRNA was significantly induced at 6 and 12 h after progesterone treatment. Once stromal cells were pretreated with CHX, there was no detectable difference between control and progesterone treatment (Fig. 7, A and B), showing that progesterone induced RRM2 expression through an indirect way.
FIGURE 7.
Effects of cycloheximide on progesterone-induced RRM2 expression. Real-time RT-PCR was performed to examine Rrm2 expression in progesterone-treated stromal cells with or without pretreatment of CHX (1 μg/ml). After a 2-h pretreatment with CHX, stromal cells were treated with progesterone for 6 h (A) and 12 h (B), respectively. *, p < 0.05; error bars, S.E.
c-MYC Is Important Mediator of Progesterone Role in RRM2 Expression
Because the expression pattern of RRM2 during the peri-implantation period was similar to c-MYC, and c-MYC was stimulated by progesterone in stromal cells (27), we assumed that c-MYC, as an important transcription factor of cell proliferation, may participate in the regulation of RRM2 expression induced by progesterone. Both c-MYC and RRM2 protein at implantation sites were much higher than interimplantation sites on day 5 during pregnancy (Fig. 8A). Real-time RT-PCR and Western blot were also performed to examine the effect of progesterone on c-MYC expression. The results from time course treatments in vivo (Fig. 8, B and D) and in vitro (Fig. 8, C and E) showed that the up-regulation of c-MYC was elicited quickly in 2 h by progesterone.
FIGURE 8.
The expression of p-AKT, c-MYC, and RRM2 in mouse uterus and cultured stromal cells treated with progesterone. A, Western blot of p-AKT, c-MYC, and RRM2 proteins in mouse uterus on day 5. B, real-time PCR of c-Myc mRNA after ovariectomized mice were treated with progesterone for different times. C, real-time PCR of c-Myc mRNA after stromal cells were treated with progesterone for different times. D, Western blot of p-AKT, c-MYC, and RRM2 after ovariectomized mice were treated with progesterone for different times. E, Western blot of p-AKT, c-MYC, and RRM2 after stromal cells were treated with progesterone for different times. *, p < 0.05; error bars, S.E.
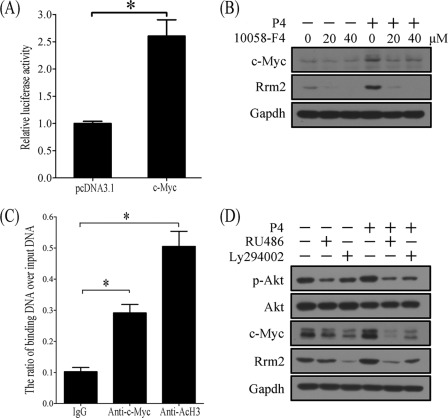
To study c-MYC regulation on RRM2 expression, stromal cells were transfected with Rrm2-PGL3 containing Rrm2 promoter and pRL-TK. Compared with control, c-Myc overexpression significantly up-regulated the promoter activity of Rrm2 (Fig. 9A). Progesterone stimulation of RRM2 expression in stromal cells was blocked by the pretreatment of 10058-F4, an inhibitor of c-MYC (Fig. 9B). Based on our prediction, there are five c-MYC binding sites in the Rrm2 promoter region. ChIP was performed to verify c-MYC regulation of Rrm2. For a positive control of active transcription, acetylated H3 binding was obviously enriched. Compared with control, c-MYC binding on the Rrm2 promoter was significantly enriched (Fig. 9C).
FIGURE 9.
The involvement of p-AKT and c-MYC in progesterone-induced RRM2 expression. A, the relative luciferase activity of Rrm2 promoter after c-Myc was overexpressed in stromal cells. B, Western blot of c-MYC and RRM2 in progesterone-treated stromal cells after pretreatment with 20 or 40 μm 10058-F4, an inhibitor of c-Myc. 10058-F4 can block the role of progesterone on c-Myc and RRM2. C, ChIP analysis. Uterine decidua from implantation sites on day 8 were used for chromatin immunoprecipitation with anti-c-Myc or anti-acetylated H3 (AcH3) antibody (a positive control for active transcription). IgG was used for a negative control. D, effects of RU486 and Ly294002 on progesterone-induced RRM2 expression. Stromal cells were pretreated with RU486 or Ly294002 for 1 h and then treated with progesterone. *, p < 0.05; error bars, S.E.
PI3K/AKT Mediated Role of Progesterone in c-MYC and RRM2
c-MYC has been shown to be up-regulated by prolactin through the PI3K/AKT pathway (28), we hypothesized that PI3K/AKT may also mediate the regulation of c-MYC and RRM2 expression in mouse uterine stromal cells responding to progesterone stimulation. The protein levels of p-AKT significantly increased at implantation sites on day 5 during pregnancy, whereas no visible difference of total AKT was detected (Fig. 8A). The results from time gradient treatments in vivo (Fig. 8D) and in vitro (Fig. 8E) showed that p-AKT was elicited quickly by progesterone treatment, which meant that p-AKT should locate at the upstream of RRM2 responding to progesterone stimulation.
To confirm our hypothesis, stromal cells were pretreated with RU486 (1 μm) or Ly294002 (PI3K inhibitor, 15 μm) 1 h before progesterone addition. Western blot was performed to detect the expression of p-AKT, c-MYC, and RRM2. Progesterone treatment significantly stimulated stromal cells to express p-AKT, c-MYC, and RRM2 in 12 h. A 1-h pretreatment with RU486 or Ly294002 could inhibit progesterone stimulation of RPM2 expression, which indicated that progesterone induced c-MYC and RRM2 expression through the PI3K/AKT pathway (Fig. 9D).
Up-regulation of RRM2 Was Mediated by ATR-directed DNA Damage Response to Replication Stress in Mouse Decidualization
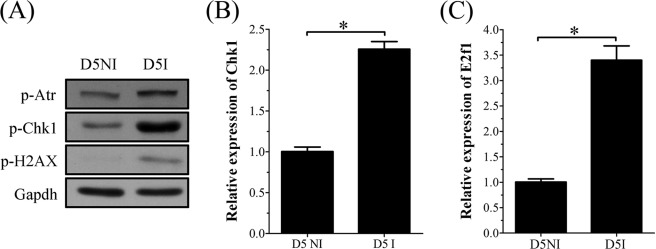
Uterine decidualization is characterized by extensive proliferation, differentiation, and endoreduplication. DNA damage frequently occurs during proliferation and causes replication stress by blocking DNA replication. Because RRM2 expression could be up-regulated by DNA damage caused by ionization irradiation or camptothecin (CPT), the topoisomerase I inhibitor, we hypothesized that the up-regulation of RRM2 in mouse decidualization may also be stimulated concurrently by DNA damage except for progesterone induction. To confirm the existence of DNA damage in mouse uterus during decidualization, Western blot was performed to analyze mouse uterus on day 5 of pregnancy to detect the expression of γ-H2AX, a sensitive marker for DNA damage. Compared with the interimplantation site, the p-H2AX level was significantly higher at the implantation site on day 5, suggesting that DNA damage does occur in day 5 uterus (Fig. 10A). We then further examined effects of DNA damage on RRM2 expression during decidualization. A previous study showed that CHK1 and E2F1 mediated the role of ATR/ATM on RRM2 expression responding to CPT-mediated DNA damage (29). Our results from real-time RT-PCR and Western blot also showed that the expression of p-ATR, CHK1, and E2F1 was also up-regulated at implantation sites on day 5 compared with interimplantation sites (Fig. 10, A–C). We assumed that DNA damage that occurred during decidualization might induce RRM2 expression through activating the ATR/ATM-CHK1-E2F1 pathway.
FIGURE 10.
Expression of DNA damage-related proteins at interimplantation site (D5NI) and implantation site (D5I) in mouse uterus on day 5. A, Western blot of p-ATR, p-CHK1, and p-H2AX. B, real-time RT-PCR of Chk1 mRNA. C, real-time RT-PCR of E2f1 mRNA. *, p < 0.05; error bars, S.E.
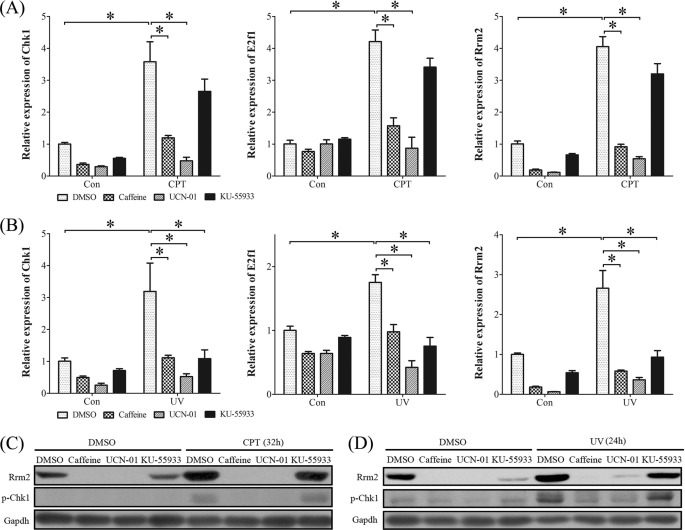
Besides intrinsic factors, many extrinsic factors can also cause DNA damage, including metabolic reactive oxygen species, ionizing radiation, UV light, genotoxic chemicals, and CPT. Then UV light and CPT were chosen to mimic DNA damage and replication stress, respectively. Compared with control, the mRNA levels of Chk1, E2f1, and Rrm2 in stromal cells were significantly elicited by CPT (1 μm) (Fig. 11A) and UV (20 J) (Fig. 11B), respectively. Pretreatments with caffeine (inhibitor of ATR/ATM, 3 mm) or UCN-01 (inhibitor of CHK1, 0.25 μm) significantly inhibited the stimulation of CPT and UV on mRNA expression of Chk1, E2f1, and Rrm2. KU-55933 (a specific inhibitor of ATM, 10 μm) could also block the action of UV but had less effect on CPT treatment. The conspicuous induction of p-CHK1 and RRM2 protein by CPT and UV was also detected in mouse stromal cells, which were inhibited by the pretreatment with caffeine, UCN-01, or KU-55933 (Fig. 11, C and D).
FIGURE 11.
Expression of CHK1, E2F1, and RRM2 in stromal cells treated with CPT or UV. A, real-time RT-PCR of Chk1, E2f1, and Rrm2 mRNA levels after CPT (1 μm)-treated stromal cells were pretreated with caffeine (inhibitor of ATR/ATM), KU-55933 (inhibitor of ATM), or UCN-01 (inhibitor of CHK1), respectively. B, real-time RT-PCR of Chk1, E2f1, and Rrm2 mRNA levels after UV (20 J)-treated stromal cells were pretreated with caffeine, KU-55933, or UCN-01, respectively. C, Western blot of RRM2 and p-CHK1 proteins after CPT-treated stromal cells were pretreated with caffeine, KU-55933, or UCN-01, respectively. D, Western blot of RRM2 and p-CHK1 proteins after UV-treated stromal cells were pretreated with caffeine, KU-55933, or UCN-01, respectively. *, p < 0.05; error bars, S.E.
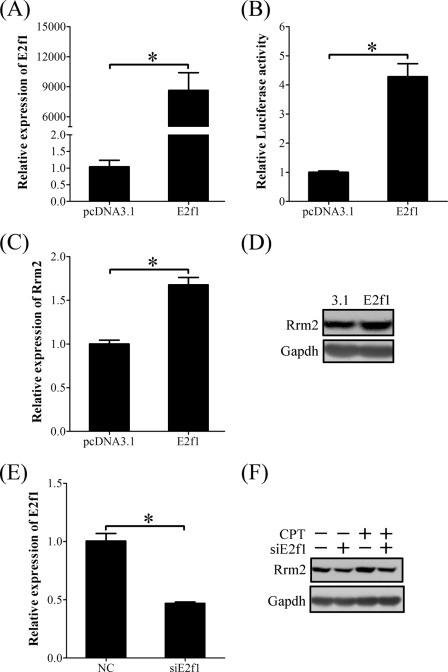
These results suggested that E2F1 and RRM2 should be downstream of the ATR/ATM-CHK1 pathway. In order to prove the action of E2F1 on RRM2 expression induced by DNA damage and replication stress, stromal cells isolated from mouse uterus were transfected with E2f1-pcDNA 3.1 vector. E2f1 overexpression significantly up-regulated the luciferase activity of the Rrm2 promoter (Fig. 12, A and B). Both mRNA and protein of RRM2 displayed obvious induction in cells transfected with E2f1 expression vector (Fig. 12, C and D).
FIGURE 12.
E2F1 regulation on RRM2 expression. A, real-time RT-PCR of E2f1 mRNA level in E2f1-overexpressing stromal cells. B, the relative luciferase activity of Rrm2 promoter in E2f1-overexpressing stromal cells. C, real-time RT-PCR of Rrm2 mRNA level in E2f1-overexpressing stromal cells. D, Western blot of RRM2 protein in E2f1-overexpressing stromal cells. E, real-time RT-PCR of E2f1 mRNA expression after stromal cells were transfected with E2f1 siRNA. F, Western blot of RRM2 protein after stromal cells were transfected with E2f1 siRNA (siE2f1) and then treated with CPT. *, p < 0.05; error bars, S.E.
We also investigated the effect of siRNA targeting E2f1 on RRM2 expression in the presence or absence of CPT. When E2f1 mRNA expression was knocked down to 50% by siRNA in stromal cells (Fig. 12E), the up-regulation of Rrm2 by CPT was partially suppressed (Fig. 12F). These results suggested that the role of CPT in RRM2 expression was mediated by E2F1.
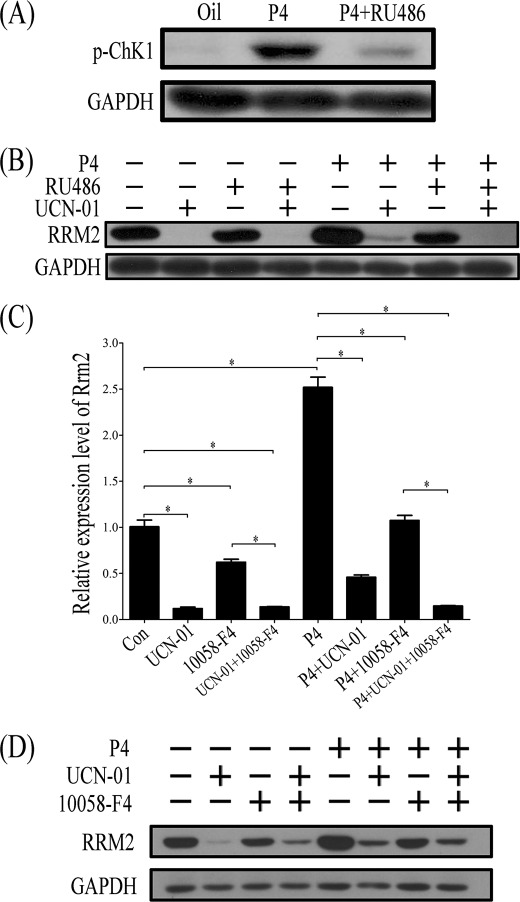
Progesterone is essential for decidualization. Our results suggested that both progesterone and replication stress were important factors to induce RRM2 expression. To further explore the relationship between progesterone and DNA damage, ovariectomized mice were used to examine effects of progesterone on CHK1. p-CHK1 was significantly up-regulated in mouse uterus after progesterone treatment, whereas RU486 could block the stimulation of progesterone on p-CHK1 expression (Fig. 13A).
FIGURE 13.
CHK1 regulation on RRM2 expression. A, Western blot of p-CHK1 protein after ovariectomized mice were treated with progesterone or progesterone plus RU486. B, Western blot of RRM2 protein after progesterone-treated stromal cells were pretreated with UCN-01, RU486, or UCN-01 plus RU486. C, real-time RT-PCR of Rrm2 mRNA expression after stromal cells were treated with progesterone (P4), UCN-01, 10058-F4, or a combination of them. D, Western blot analysis of RRM2 protein expression after stromal cells were treated with progesterone (P4), UCN-01, 10058-F4, or a combination of them. *, p < 0.05; error bars, S.E.
In cultured stromal cells, RRM2 expression was significantly stimulated by progesterone. However, the pretreatment with UCN-01 sharply reduced the progesterone induction on RRM2 (Fig. 13B). Therefore, progesterone may up-regulate RRM2 expression through stimulating cell proliferation and inducing DNA replication stress in mouse uterus during decidualization.
In order to study the interaction between CHK1 and c-MYC, progesterone-treated stromal cells were co-treated with UCN-01 and 10058-F4, respectively. After cells were treated with progesterone, both RRM2 mRNA and protein were significantly up-regulated. RRM2 stimulation by progesterone was significantly abrogated by either UCN-01 or 10058-F4 treatment. Rrm2 mRNA level stimulated by progesterone was further inhibited by the treatment with both UCN-01 and 10058-F4. However, progesterone-induced RRM2 protein level was not further reduced by both inhibitors (Fig. 13, C and D).
DISCUSSION
RRM2 Expression Pattern during Embryo Implantation and Decidualization
Embryo implantation is a complex developmental process including embryo attachment, subsequent invasion into the stroma, and then the proliferation and differentiation of the uterine stromal cells to decidua (30). As a proliferation-related molecule, RRM2 was strongly expressed in the decidua on days 6–8 of pregnancy, which was confirmed by real-time RT-PCR, in situ hybridization, immunostaining, and Western blot. A previous study also showed that Rrm2 was up-regulated 2.55-fold at implantation sites compared with interimplantation sites in mouse uteri (15). RRM2 was not detected during pseudopregnancy and under delayed implantation, but it was expressed in the estrogen-activated uterus, suggesting that RRM2 expression at implantation sites requires the presence of active blastocysts. Mammalian RR consists of two different subunits, RR1 and RR2, and both are required for the RR activity (6). Because RRM2b was not detected in mouse uterus during peri-implantation period, it is possible that RRM1 and RRM2 form the ribonucleotide reductase. Both RRM1 and RRM2 were strongly co-localized in the decidua from day 6 to 8 of pregnancy and in the deciduoma. Our co-immunoprecipitation data also confirmed a physical interaction between RRM1 and RRM2, suggesting that ribonucleotide reductase in mouse decidua mainly consists of RRM1 and RRM2.
The level of RRM1 appears to be constant throughout the cell cycle in proliferating cells, whereas RRM2 level peaks in S phase and plays an essential role in regulating the active RR for DNA synthesis and cell proliferation (4, 6, 31–32). Therefore, we focused on RRM2 for further analysis.
Progesterone Regulation of RRM2 Expression through PI3K/AKT/c-MYC Pathway
Embryo implantation and decidualization are tightly regulated by the steroid hormones, estrogen and progesterone (33). Although RRM2 is significantly induced by estrogen in ovariectomized mice, estrogen has no obvious effects on RRM2 expression in isolated endometrial stromal cells in our study. Additionally, progesterone alone can induce decidualization in ovariectomized wild-type or ERKO mice in response to intraluminal oil infusion in the absence of estrogen (34). The failure of decidualization was also observed in PR knock-out (PRKO) mice (35). Based on these data, progesterone is essential for mouse decidualization. In this study, progesterone stimulated RRM2 expression in ovariectomized mouse uterus and in cultured uterine stromal cells, which was attenuated by progesterone receptor antagonist RU486, indicating that progesterone action on RRM2 was mediated through the progesterone receptor. Therefore, we focused on RRM2 regulation by progesterone in our study.
A previous study indicates that the transcription factor c-MYC was strongly expressed in the decidual cells in mouse uterus from day 5 to 8 and was up-regulated by progesterone (27). It seems that c-MYC shared a similar expression pattern with RRM2 in our study. We assumed that c-MYC might transcriptionally regulate RRM2 expression. In this study, RRM2 expression was stimulated by c-MYC overexpression and down-regulated by c-MYC inhibitor. It has been shown that c-MYC can cause the amplification of the mouse Rrm2 gene and induce simultaneous Rrm2 replication forks, whereas Rrm2 normally replicates with a single fork (36). We showed that progesterone stimulated the expression of both c-MYC and RRM2 in ovariectomized mouse uterus and in cultured stromal cells, respectively. Because c-MYC could be up-regulated by prolactin through PI3K/AKT pathway (28), we studied the effect of PI3K/AKT on regulation of c-MYC and RRM2 expression in mouse uterine stromal cells responding to progesterone stimulation. Progesterone significantly induced the phosphorylation of AKT, whereas pretreatment of Ly294002 could block progesterone induction on c-MYC and RRM2. Thus, progesterone might encourage RRM2 expression mediated by the PI3K/AKT/c-MYC pathway to induce proliferation and decidualization.
RRM2 Expression and DNA Damage
In rodents, uterine decidualization is characterized by extensive proliferation, differentiation, and polyploidization of endometrial stromal cells at the site of embryo implantation (37). Recent studies showed that polyploidization can be induced by persistent DNA damage signaling in response to double strand break-inducing agent (zeocin), doxorubicin, bleomycin, and repeated UV irradiation, through activation of the ATR/ATM-CHK1/CHK2 signal (38). We supposed that the polyploidization during decidualization might also be mediated by DNA damage response. Histone H2AX phosphorylation, a sensitive marker for DNA double strand breaks (39), was used to detect DNA damage in the decidua. The level of H2AX phosphorylation at implantation sites was significantly higher than at interimplantation sites, indicating that DNA damage does occur in the decidua. A number of DNA damage-related genes such as damage-specific DNA-binding protein 1 (Ddb1) and growth arrest and DNA-damage-inducible 45 β (Gadd45b) and γ (Gadd45g) are also up-regulated in decidual cells after embryo implantation (40).3 Furthermore, human GADD45 and forkhead transcription factor FOXO1a, an inducer of DDB1, are induced upon differentiation of the stromal compartment in the middle to late secretory phase of the cycle as well as being expressed in decidualizing endometrial stromal cells in culture (41–43). These data suggested that decidualization in mice is accompanied by DNA damage.
DNA damage in budding yeast stimulates the transcription of the RNR genes and leads to high RR activity and dNTP pool (10–11, 44). In both PC3 cells and KB cells, RRM2 was up-regulated by UV (3). ATR and its downstream phosphorylation target, CHK1, are also generally activated in response to UV and agents that stop DNA replication forks (12, 45). In cancer cells, CPT can cause replication-associated DNA double strand breaks by collisions between replication forks and drug-stabilized Top1 cleavage complexes and up-regulate RRM2 mediated by CHK1 and E2F1 (13, 46). Because RRM2 was strongly expressed in mouse decidua, and the level of CHK1 and E2F1 expression was higher at implantation sites than at interimplantation sites, we assume that the high level of RRM2 expression may be caused by DNA damage through ATR/ATM-CHK1-E2F1 signaling, and RRM2 may take part in the formation of polyploidy in the decidua. When cultured stromal cells were treated with UV or CPT, RRM2, CHK1 phosphorylation, and E2F1 were up-regulated, indicating that the factors causing DNA damage could increase the level of RRM2, CHK1 phosphorylation, and E2F1. The stimulation of RRM2 expression caused by CPT or UV could be blocked by caffeine (inhibitor for ATR/ATM), UCN (inhibitor for CHK1), and KU-55933 (inhibitor for ATM), respectively. Our data indicated that the high level of RRM2 expression in the decidua should be partially stimulated by DNA damage through the ATR/ATM-CHK1-E2F1 pathway. DNA damage signaling through ATR/ATM-CHK1/CHK2 is also involved in polyploidization (38). The high level of CHK1 and RRM2 following DNA damage should be required for normal DNA replication and polyploidization during decidualization.
In this study, progesterone can stimulate the expression of both c-MYC and p-CHK1. Progesterone-induced RRM2 expression is inhibited by either 10058-F4 or UCN-01. When cells were treated with both 10058-F4 and UCN-01, progesterone-induced Rrm2 mRNA level is further reduced compared with 10058-F4 or UCN-01 alone. These data suggest that both AKT/c-MYC and ATR/ATM-CHK1-E2F1 pathways are involved in the regulation of RRM2 expression during mouse decidualization. How these two pathways coordinately regulate RRM2 expression during decidualization remains to be determined.
RRM2 Function during Decidualization
The integrity of the feto-maternal interface consisting of the maternal decidua and the invading placental trophoblast is critical for survival of the conceptus. The decidual process can confer protection against oxidative stress-induced cell damage (47). During decidualization, FOXO1 may enhance the ability of decidualized cells to prevent oxidative damage, whereas FOXO3a repression can disable the signaling pathway responsible for oxidative cell death (41). Human chorionic gonadotropin is one of the earliest signals secreted by the implanting embryo. Recombinant, but not urinary, human chorionic gonadotropin protected decidualizing human endometrial stromal cells from oxidative cell death through enhancing the expression of manganese superoxide dismutase and selectively limiting activation of the apoptotic machinery in decidualizing human stromal cells (47). Although rodents and humans have a similar hemochorial type of placenta and exhibit similarities in terms of hormonal regulation of gene expression in the uterus, they are different in many aspects. It is still not clear whether mouse decidualization has similar protective actions as in humans. In polyploid decidual cells in mice, 135 genes related to apoptosis are also specifically suppressed compared with non-polyploid cells (40). DNA damage-induced RRM2 elevation may also be involved in DNA repair and protection against oxidative stress-induced cell death.
In our study, RRM2 was strongly expressed in the decidua on days 5–8 of pregnancy, suggesting that it may play a role in cell proliferation during decidualization. Polyploid cells in mouse decidual tissue have been confirmed as a result of endoreduplication, and multinucleate cells arise from mitoses without subsequent cell division (48). RRM2 may also be an important mediator of polyploidization. When pregnant mice were treated with RRM2 inhibitors, hydroxyurea or trimidox, the weight of implantation sites was significantly reduced compared with control. Additionally, artificially induced decidualization was significantly inhibited after pseudopregnant mice were treated with hydroxyurea. Under in vitro decidualization, the decidualization marker, Dtprp expression, was also down-regulated by hydroxyurea treatment. These data indicated that RRM2 should be essential for decidualization in mice. A previous study also showed that decidualization in rats was affected through inhibiting cell proliferation by hydroxyurea (49). Because proliferation is the first step for uterine stromal cells to enter decidualization phase, RRM2, as an important proliferation-related gene, may be essential for successful embryo implantation and decidualization.
Supplementary Material
This work was supported by National Basic Research Program of China Grant 2011CB944402 and National Natural Science Foundation of China Grants 30930013 and 31071276.

This article contains supplemental Fig. 1.
W. Lei, X.-H. Feng, W.-B. Deng, H. Ni, Z.-R. Zhang, B. Jia, X.-L. Yang, T.-S. Wang, J.-L. Liu, R.-W. Su, X.-H. Liang, Q.-R. Qi, and Z.-M. Yang, unpublished data.
- RR
- ribonucleotide reductase
- HU
- hydroxyurea
- CHX
- cycloheximide
- CPT
- camptothecin
- cFBS
- charcoal-treated fetal bovine serum
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- p-AKT
- p-CHK1, and p-H2AX, phosphorylated AKT, CHK1, and H2AX, respectively.
REFERENCES
- 1. Hu Z. Z., Huang H., Cheema A., Jung M., Dritschilo A., Wu C. H. (2008) Integrated bioinformatics for radiation-induced pathway analysis from proteomics and microarray data. J. Proteomics Bioinform. 1, 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nordlund P., Reichard P. (2006) Ribonucleotide reductases. Annu. Rev. Biochem. 75, 681–706 [DOI] [PubMed] [Google Scholar]
- 3. Zhou B., Liu X., Mo X., Xue L., Darwish D., Qiu W., Shih J., Hwu E. B., Luh F., Yen Y. (2003) The human ribonucleotide reductase subunit hRRM2 complements p53R2 in response to UV-induced DNA repair in cells with mutant p53. Cancer Res. 63, 6583–6594 [PubMed] [Google Scholar]
- 4. Engström Y., Eriksson S., Jildevik I., Skog S., Thelander L., Tribukait B. (1985) Cell cycle-dependent expression of mammalian ribonucleotide reductase. Differential regulation of the two subunits. J. Biol. Chem. 260, 9114–9116 [PubMed] [Google Scholar]
- 5. Shao J., Zhou B., Zhu L., Qiu W., Yuan Y. C., Xi B., Yen Y. (2004) In vitro characterization of enzymatic properties and inhibition of the p53R2 subunit of human ribonucleotide reductase. Cancer Res. 64, 1–6 [DOI] [PubMed] [Google Scholar]
- 6. Eriksson S., Gräslund A., Skog S., Thelander L., Tribukait B. (1984) Cell cycle-dependent regulation of mammalian ribonucleotide reductase. The S phase-correlated increase in subunit M2 is regulated by de novo protein synthesis. J. Biol. Chem. 259, 11695–11700 [PubMed] [Google Scholar]
- 7. Reichard P. (1993) From RNA to DNA, why so many ribonucleotide reductases? Science 260, 1773–1777 [DOI] [PubMed] [Google Scholar]
- 8. Wang C., Liu Z. (2006) Arabidopsis ribonucleotide reductases are critical for cell cycle progression, DNA damage repair, and plant development. Plant Cell 18, 350–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wilsker D., Petermann E., Helleday T., Bunz F. (2008) Essential function of Chk1 can be uncoupled from DNA damage checkpoint and replication control. Proc. Natl. Acad. Sci. U.S.A. 105, 20752–20757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao H., Piwnica-Worms H. (2001) ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell Biol. 21, 4129–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang M., Elledge S. J. (1997) Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol. Cell Biol. 17, 6105–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cha R. S., Kleckner N. (2002) ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297, 602–606 [DOI] [PubMed] [Google Scholar]
- 13. Holm C., Covey J. M., Kerrigan D., Pommier Y. (1989) Differential requirement of DNA replication for the cytotoxicity of DNA topoisomerase I and II inhibitors in Chinese hamster DC3F cells. Cancer Res. 49, 6365–6368 [PubMed] [Google Scholar]
- 14. Herrler A., von Rango U., Beier H. M. (2003) Embryo-maternal signaling. How the embryo starts talking to its mother to accomplish implantation. Reprod. Biomed. Online 6, 244–256 [DOI] [PubMed] [Google Scholar]
- 15. Reese J., Das S. K., Paria B. C., Lim H., Song H., Matsumoto H., Knudtson K. L., DuBois R. N., Dey S. K. (2001) Global gene expression analysis to identify molecular markers of uterine receptivity and embryo implantation. J. Biol. Chem. 276, 44137–44145 [DOI] [PubMed] [Google Scholar]
- 16. Ma X. H., Hu S. J., Ni H., Zhao Y. C., Tian Z., Liu J. L., Ren G., Liang X. H., Yu H., Wan P., Yang Z. M. (2006) Serial analysis of gene expression in mouse uterus at the implantation site. J. Biol. Chem. 281, 9351–9360 [DOI] [PubMed] [Google Scholar]
- 17. Ni H., Sun T., Ding N. Z., Ma X. H., Yang Z. M. (2002) Differential expression of microsomal prostaglandin E synthase at implantation sites and in decidual cells of mouse uterus. Biol. Reprod. 67, 351–358 [DOI] [PubMed] [Google Scholar]
- 18. Xavier L. L., Viola G. G., Ferraz A. C., Da Cunha C., Deonizio J. M., Netto C. A., Achaval M. (2005) A simple and fast densitometric method for the analysis of tyrosine hydroxylase immunoreactivity in the substantia nigra pars compacta and in the ventral tegmental area. Brain Res. Brain Res. Protoc. 16, 58–64 [DOI] [PubMed] [Google Scholar]
- 19. Hu S. J., Ren G., Liu J. L., Zhao Z. A., Yu Y. S., Su R. W., Ma X. H., Ni H., Lei W., Yang Z. M. (2008) MicroRNA expression and regulation in mouse uterus during embryo implantation. J. Biol. Chem. 283, 23473–23484 [DOI] [PubMed] [Google Scholar]
- 20. Liang X. H., Zhao Z. A., Deng W. B., Tian Z., Lei W., Xu X., Zhang X. H., Su R. W., Yang Z. M. (2010) Estrogen regulates amiloride-binding protein 1 through CCAAT/enhancer-binding protein-β in mouse uterus during embryo implantation and decidualization. Endocrinology 151, 5007–5016 [DOI] [PubMed] [Google Scholar]
- 21. Li Q., Kannan A., Wang W., Demayo F. J., Taylor R. N., Bagchi M. K., Bagchi I. C. (2007) Bone morphogenetic protein 2 functions via a conserved signaling pathway involving Wnt4 to regulate uterine decidualization in the mouse and the human. J. Biol. Chem. 282, 31725–31732 [DOI] [PubMed] [Google Scholar]
- 22. Ai N., Hu X., Ding F., Yu B., Wang H., Lu X., Zhang K., Li Y., Han A., Lin W., Liu R., Chen R. (2011) Signal-induced Brd4 release from chromatin is essential for its role transition from chromatin targeting to transcriptional regulation. Nucleic Acids Res. 39, 9592–9604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saban N., Bujak M. (2009) Hydroxyurea and hydroxamic acid derivatives as antitumor drugs. Cancer Chemother. Pharmacol. 64, 213–221 [DOI] [PubMed] [Google Scholar]
- 24. Szekeres T., Gharehbaghi K., Fritzer M., Woody M., Srivastava A., van't Riet B., Jayaram H. N., Elford H. L. (1994) Biochemical and antitumor activity of trimidox, a new inhibitor of ribonucleotide reductase. Cancer Chemother. Pharmacol. 34, 63–66 [DOI] [PubMed] [Google Scholar]
- 25. Fritzer-Szekeres M., Salamon A., Grusch M., Horvath Z., Höchtl T., Steinbrugger R., Jäger W., Krupitza G., Elford H. L., Szekeres T. (2002) Trimidox, an inhibitor of ribonucleotide reductase, synergistically enhances the inhibition of colony formation by Ara-C in HL-60 human promyelocytic leukemia cells. Biochem. Pharmacol. 64, 481–485 [DOI] [PubMed] [Google Scholar]
- 26. Kimura F., Takakura K., Takebayashi K., Ishikawa H., Kasahara K., Goto S., Noda Y. (2001) Messenger ribonucleic acid for the mouse decidual prolactin is present and induced during in vitro decidualization of endometrial stromal cells. Gynecol. Endocrinol. 15, 426–432 [DOI] [PubMed] [Google Scholar]
- 27. Huet-Hudson Y. M., Andrews G. K., Dey S. K. (1989) Cell type-specific localization of c-MYC protein in the mouse uterus. Modulation by steroid hormones and analysis of the periimplantation period. Endocrinology 125, 1683–1690 [DOI] [PubMed] [Google Scholar]
- 28. Domínguez-Cáceres M. A., García-Martínez J. M., Calcabrini A., González L., Porque P. G., León J., Martín-Pérez J. (2004) Prolactin induces c-Myc expression and cell survival through activation of Src/Akt pathway in lymphoid cells. Oncogene 23, 7378–7390 [DOI] [PubMed] [Google Scholar]
- 29. Zhang Y. W., Jones T. L., Martin S. E., Caplen N. J., Pommier Y. (2009) Implication of checkpoint kinase-dependent up-regulation of ribonucleotide reductase R2 in DNA damage response. J. Biol. Chem. 284, 18085–18095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang H., Dey S. K. (2006) Roadmap to embryo implantation. Clues from mouse models. Nat. Rev. Genet. 7, 185–199 [DOI] [PubMed] [Google Scholar]
- 31. Björklund S., Skog S., Tribukait B., Thelander L. (1990) S phase-specific expression of mammalian ribonucleotide reductase R1 and R2 subunit mRNAs. Biochemistry 29, 5452–5458 [DOI] [PubMed] [Google Scholar]
- 32. Mann G. J., Musgrove E. A., Fox R. M., Thelander L. (1988) Ribonucleotide reductase M1 subunit in cellular proliferation, quiescence, and differentiation. Cancer Res. 48, 5151–5156 [PubMed] [Google Scholar]
- 33. Franco H. L., Jeong J. W., Tsai S. Y., Lydon J. P., DeMayo F. J. (2008) In vivo analysis of progesterone receptor action in the uterus during embryo implantation. Semin. Cell Dev. Biol. 19, 178–186 [DOI] [PubMed] [Google Scholar]
- 34. Paria B. C., Tan J., Lubahn D. B., Dey S. K., Das S. K. (1999) Uterine decidual response occurs in estrogen receptor-α-deficient mice. Endocrinology 140, 2704–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lydon J. P., DeMayo F. J., Funk C. R., Mani S. K., Hughes A. R., Montgomery C. A., Jr., Shyamala G., Conneely O. M., O'Malley B. W. (1995) Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev. 9, 2266–2278 [DOI] [PubMed] [Google Scholar]
- 36. Kuschak T. I., Taylor C., McMillan-Ward E., Israels S., Henderson D. W., Mushinski J. F., Wright J. A., Mai S. (1999) The ribonucleotide reductase R2 gene is a non-transcribed target of c-Myc-induced genomic instability. Gene 238, 351–365 [DOI] [PubMed] [Google Scholar]
- 37. Tan Y., Li M., Cox S., Davis M. K., Tawfik O., Paria B. C., Das S. K. (2004) HB-EGF directs stromal cell polyploidy and decidualization via cyclin D3 during implantation. Dev. Biol. 265, 181–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Davoli T., Denchi E. L., de Lange T. (2010) Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell 141, 81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bonner W. M., Redon C. E., Dickey J. S., Nakamura A. J., Sedelnikova O. A., Solier S., Pommier Y. (2008) GammaH2AX and cancer. Nat. Rev. Cancer 8, 957–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ma X., Gao F., Rusie A., Hemingway J., Ostmann A. B., Sroga J. M., Jegga A. G., Das S. K. (2011) Decidual cell polyploidization necessitates mitochondrial activity. PLoS One 6, e26774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kajihara T., Jones M., Fusi L., Takano M., Feroze-Zaidi F., Pirianov G., Mehmet H., Ishihara O., Higham J. M., Lam E. W., Brosens J. J. (2006) Differential expression of FOXO1 and FOXO3a confers resistance to oxidative cell death upon endometrial decidualization. Mol. Endocrinol. 20, 2444–2455 [DOI] [PubMed] [Google Scholar]
- 42. Labied S., Kajihara T., Madureira P. A., Fusi L., Jones M. C., Higham J. M., Varshochi R., Francis J. M., Zoumpoulidou G., Essafi A., Fernandez de Mattos S., Lam E. W., Brosens J. J. (2006) Progestins regulate the expression and activity of the forkhead transcription factor FOXO1 in differentiating human endometrium. Mol. Endocrinol. 20, 35–44 [DOI] [PubMed] [Google Scholar]
- 43. Gellersen B., Brosens I. A., Brosens J. J. (2007) Decidualization of the human endometrium. Mechanisms, functions, and clinical perspectives. Semin. Reprod. Med. 25, 445–453 [DOI] [PubMed] [Google Scholar]
- 44. Huang M., Zhou Z., Elledge S. J. (1998) The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94, 595–605 [DOI] [PubMed] [Google Scholar]
- 45. Ward I. M., Minn K., Chen J. (2004) UV-induced ataxia-telangiectasia-mutated and Rad3-related (ATR) activation requires replication stress. J. Biol. Chem. 279, 9677–9680 [DOI] [PubMed] [Google Scholar]
- 46. Hsiang Y. H., Lihou M. G., Liu L. F. (1989) Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 49, 5077–5082 [PubMed] [Google Scholar]
- 47. Kajihara T., Tochigi H., Uchino S., Itakura A., Brosens J. J., Ishihara O. (2011) Differential effects of urinary and recombinant chorionic gonadotropin on oxidative stress responses in decidualizing human endometrial stromal cells. Placenta 32, 592–597 [DOI] [PubMed] [Google Scholar]
- 48. Ansell J. D., Barlow P. W., McLaren A. (1974) Binucleate and polyploid cells in the decidua of the mouse. J. Embryol. Exp. Morphol. 31, 223–227 [PubMed] [Google Scholar]
- 49. Spencer F., Chi L., Zhu M. X. (2000) Hydroxyurea inhibition of cellular and developmental activities in the decidualized and pregnant uteri of rats. J. Appl. Toxicol. 20, 407–412 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.