Background: The microstructure of nacre is controlled by the proteins in them.
Results: When PfN23 was knocked down, the shell formation in adults and larvae was suppressed.
Conclusion: The basic protein PfN23 is important for the control of crystal growth in nacre.
Significance: This might provide a valuable complementary to the classic view that acidic proteins control nacre formation.
Keywords: Biomaterials, Biomineralization, Calcium-binding Proteins, Crystal Structure, Extracellular Matrix Proteins, Calcium Carbonate, Pearl Oyster
Abstract
The fine microstructure of nacre (mother of pearl) illustrates the beauty of nature. Proteins found in nacre were believed to be “natural hands” that control nacre formation. In the classical view of nacre formation, nucleation of the main minerals, calcium carbonate, is induced on and by the acidic proteins in nacre. However, the basic proteins were not expected to be components of nacre. Here, we reported that a novel basic protein, PfN23, was a key accelerator in the control over crystal growth in nacre. The expression profile, in situ immunostaining, and in vitro immunodetection assays showed that PfN23 was localized within calcium carbonate crystals in the nacre. Knocking down the expression of PfN23 in adults via double-stranded RNA injection led to a disordered nacre surface in adults. Blocking the translation of PfN23 in embryos using morpholino oligomers led to the arrest of larval development. The in vitro crystallization assay showed that PfN23 increases the rate of calcium carbonate deposition and induced the formation of aragonite crystals with characteristics close to nacre. In addition, we constructed the peptides and truncations of different regions of this protein and found that the positively charged C-terminal region was a key region for the function of PfN23 Taken together, the basic protein PfN23 may be a key accelerator in the control of crystal growth in nacre. This provides a valuable balance to the classic view that acidic proteins control calcium carbonate deposition in nacre.
Introduction
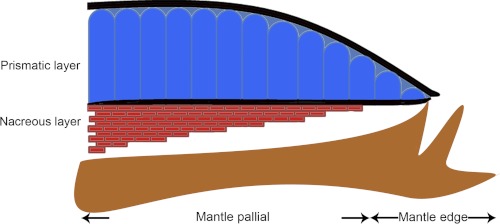
Pearls share an identical structure with the nacre of oyster shell (1, 2). Nacre (mother of pearl) is an organo-mineral assemblage that is dominated by calcium carbonate with a minor organic matrix complex, which produces exceptional hardness, strength, and osteoinductive activity compared with the pure mineral (3, 4). The shell of the pearl oyster, Pinctada fucata, which is most widely used to produce saltwater pearls, consists of two different forms of calcium carbonate, i.e. aragonite in the inner nacreous layer and calcite in the outer prismatic layer (Fig. 1) (5–7). Calcium carbonate crystals have three water-free polymorphs, which are, from stable to increasingly unstable, calcite, aragonite, and vaterite. Many studies of nacre formation have been conducted in the disciplines of biology, physics, chemistry, crystallography, and materials science, but our fundamental understanding of nacre is still very limited (8–13).
FIGURE 1.
Diagram of the cross-section of the shell and the mantle of P. fucata.
Pioneering studies have shown that the proteins in the shell have a dominant role in directing the highly organized nanocomposites in the layered structure of nacre, although they account for less than 5% of the shell weight (14, 15). Several proteins have been identified in the molluscan shell, and their functions in controlling calcium carbonate deposition have been extensively investigated (1, 16–25). Acidic matrix proteins are believed to be the major components with control over crystal growth, because of their cation-binding capacity (26). Models of the processes of nacre formation have been improved by our increasing knowledge of these proteins (27–29). Mollusks build their shells using a hydrophobic silk gel, highly acidic aspartic acid-rich proteins in a highly structured chitinous framework (30). In contrast to most acidic proteins, proteins with a basic isoelectric point were not expected to be components of the molluscan shell matrix, and they have been far less studied in the nacre (18, 27, 31, 32).
Three suppression subtractive hybridization libraries were previously constructed to isolate genes expressed during important developmental stages in shell formation (33). We retrieved expression sequence tags from these libraries to identify genes involved in nacre formation. In this study, we identified and characterized a basic protein, PfN23, in the nacre of the shell. The nature and function of PfN23 in nacre formation were elucidated by in vitro and in vivo analyses. Our results demonstrated that this basic protein may accelerate calcium carbonate precipitation and induce the formation of the nacre microstructure.
EXPERIMENTAL PROCEDURES
Animals
The oysters, P. fucata, were purchased from the Marine Biology Research Station at Daya Bay, CAS, Shenzhen, China. Animals were maintained in glass aquaria filled with aerated artificial seawater (Sude Instant Sea Salt, 3% at 20 °C) for 3 days prior to experimentation.
Primers
Refer to the supplemental material for primer details.
Identification of the Complete PfN23 Sequence
Total RNA was extracted from the mantle tissue of P. fucata using an SV total RNA isolation kit (Promega), according to the manufacturer's instructions. Rapid amplification of cDNA ends (RACE)3 was performed using a SMARTerTM RACE cDNA amplification kit (Clontech). RACE3 and RACE5 primers were used with the primers supplied with the 3′- and 5′-RACE kits, respectively. The full-length sequence was confirmed using the primers Full1 and Full2.
The deduced amino acid sequence of the gene was determined bioinformatically using the ExPASy Translate tool. The signal peptide was predicted using the SignalP 3.0. Disordered region was predicted using GlobPlot2. Analysis of hydrophobicity was done using the method of Kyte and Doolittle in ProtScale.
Gene Expression Analysis by RT-PCR
Using the methods described above, total RNA was isolated from the mantle edge, mantle pallial, gonad, foot, gill, hemocytes, viscus, and adductor muscles of adult individual P. fucata. Equal quantities (1 μg) of total RNA from different tissues were reverse-transcribed into cDNA using Quant Reverse Transcriptase (Tiangen), according to the manufacturer's instructions. RT-PCR was conducted using the primer pairs RT-1/RT-2 and GaRT-1/GaRT-2 to amplify PfN23 and GAPDH (included as a positive control for cDNA preparations) gene fragments, respectively. To avoid false-positive results and to detect cross-contamination of the samples, a negative control was performed in the absence of the cDNA template. All PCR products were subcloned and verified by sequencing.
Preparation of Polyclonal Antibodies against the Recombinant PfN23
To remove the signal peptide coding sequence from the open reading frame of PfN23, primers Exp1 and Exp2 were used to clone the PfN23, which was then subcloned into the expression vector PET28a. Recombinant PfN23 with a His6 tag at the N terminus was overexpressed in Escherichia coli (Transsetta DE3) with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside at 10 °C for 24 h. The cell sample was homogenized and purified using a combination of His-trap FF crude column and Superdex 75 10/300 GL column (GE Healthcare), according to the manufacturer's instructions. Eluted fractions containing purified recombinant PfN23 detected by SDS-PAGE were concentrated and desalted by ultrafiltration (Amicon, Ultra-15, 3 kDa) against 20 mm Na3PO4, 0.5 m NaCl buffer, pH 7.4. Polyclonal antibodies against PfN23 were raised in New Zealand rabbits following standard immunization procedures, and they were affinity-purified using an antibody purification kit (ZKCY-BIO), according to the manufacturer's instructions. The titer was determined using a standard enzyme-linked immunosorbent assay. The specificity of the antibodies was also tested using Western blots against the purified protein, polyhistidine, and the EDTA-soluble matrix extracted from the entire shell of P. fucata.
Matrix Extraction
EDTA-soluble and EDTA-insoluble matrices from different shell layers were prepared as described by Kong et al. (24), with some modifications. During the extraction of the soluble matrix, the supernatant was collected by centrifugation at 13,000 rpm for 30 min at 4 °C and then desalted by dialysis and ultrafiltration (Amicon, Ultra-15, 3 kDa). The amount of the protein was measured using a BCA assay kit (Pierce), according to the manufacturer's instructions.
Detection of PfN23 in Shell Extracts
The presence of native PfN23 in different extracts of the prismatic layer and the nacreous layer of the shell of P. fucata was tested by ELISA and Western blotting. During the ELISA, known amounts (0.1–5 ng) of PfN23 were used as a standard. The tests were repeated three times in triplicate. Protein samples from different EDTA shell extracts were separated by SDS-PAGE and transferred to a PVDF membrane (Millipore). Anti-PfN23 antibody was used at dilutions of 1:1000, and HRP-conjugated goat anti-rabbit secondary antibody (Calbiochem) was used at 1:10,000. The dot was finally incubated with Luminata Crescendo Western HRP substrate (Millipore) and exposed to x-ray film for 1 min. Control experiments were performed without the first antibody step.
Immunolocalization of PfN23 with Gold Particles
The immunogold-labeling assay was conducted as described by Marin et al. (34), with some modifications. The anti-PfN23 antibodies were used at dilutions of 1:200. Diluted (1:400) goat anti-rabbit antibodies coupled to 5-nm gold particles (Sigma) were used as secondary antibodies. Blank experiments were performed without the first antibody step. Samples were sputter-coated with carbon and analyzed using an FEI Sirion2000 scanning electron microscope (SEM) in the back-scattering electron mode.
In Vivo Silencing of PfN23
The RNAi assay was performed as described in Suzuki et al. (1), with some modifications. Primers RNAi-P1 and RNAi-P2 were used to amplify specific sequences from the first strand cDNA. In the GFP, pEGFP-C1 (Clontech) was used as the template. The PCR products were purified using a Wizard PCR PrepDNA purification system (Promega). A RiboMAXTM large scale RNA production system (T7) kit (Promega) was used to synthesize and purify the dsRNA. RNase-free DNase I (Takara) was used to digest the template DNA. The dsRNA was diluted to 40 μg/100 μl or 80 μg/100 μl using PBS and then injected into the adductor muscle of 2-year-old individuals with a shell length of 5–6 cm. Five individuals were used for each treatment. Total RNA from the mantle tissue of each oyster was extracted 6 days after injection and used to synthesize the first strand cDNA, as described above. Real time quantitative PCR was used to quantify the expression levels of each gene, where β-actin was used as an internal reference. quantitative PCR was conducted using the Mx3000PTM (Stratagene) with an SYBRH Premix Ex TaqTMII kit (Takara), according to the manufacturer's instructions (see supplemental material for primer details). The cycling parameters were 95 °C for 30 s (1 cycle), 95 °C for 5 s, 55 °C for 30 s, and 72 °C for 30 s (40 cycles). Dissociation curves were analyzed to determine the purity of the product and the specificity of amplification. In the control, the expression levels of the PBS injected group were normalized to a relative value of 1.0.
Six days after injection, the shells of the injection groups were thoroughly washed with Milli-Q water and air-dried. Shells were cut into pieces and then mounted on the scanner with the inner nacreous surface face-up, sputter-coated with 10-nm-thick gold, and analyzed using an FEI Quanta 200 scanning electron microscope.
Morpholinos were obtained from Gene Tools LLC (Philomath, OR). Antisense PfN23 morpholinos, i.e. morpholino A (5′-CAG TCA ACA ACG CCA CCC CCT TCA T-3′, Mo A) and morpholino B (5′-TAT TCC TAA ATG TCT TGT TCT TGT C-3′, Mo B), were used in the experiments. A 25-base random sequence mixture (Mo R) was used as a control.
The insemination process was as described in Fujimura et al. (35). Fertilized eggs were incubated at 25 °C. We then soaked 100–200 larvae at the two-cell to four-cell stages in a total volume of 1 ml of seawater containing 5 or 10 μm morpholino and 6 μm Endo-Porter-Aqueous (Gene Tools LLC). Observations of each colony were performed by microscopy and a polarizing microscope (Leica, DMR). The percentages of surviving and abnormally developed larvae were estimated independently by up to three different persons, which produced identical values with only a 10–20% deviation. Larvae were harvested after 32 h to analyze the effects of the morpholinos. Proteins were extracted from the larvae using a tissue protein extraction kit (CWBIO) and analyzed by Western blotting, as described above. The expression of the β-actin control was detected using an anti-β-actin antibody (1:1000, CWBIO).
Chitin and Calcium Carbonate Crystal Binding Assay
Chitin binding assays were performed as described by Inoue et al. (36), and the calcium carbonate crystal binding assay was conducted as described by Suzuki et al. (1), with some modifications. BSA or PfN23 (30 μg each) was incubated with 5 mg of chitin (Wako), calcite (Sigma), or aragonite (synthesized as described by Kitano et al. (37)) that had been previously equilibrated with 0.5% ammonium bicarbonate for 1 h at 4 °C. After removal of the solution by centrifugation, the insoluble mixture was successively washed twice with 200 μl each of distilled water and 0.2 m NaCl. Each washing was desalted by ultrafiltration (Millipore, 3-kDa cutoff) and lyophilized before freeze-dried proteins were resuspended directly in 15 μl of Laemmli sample buffer. The final insoluble residue was boiled in 30 μl of 2% (w/v) SDS containing 20% (v/v) 2-mercaptoethanol for 10 min, before the supernatant was separated by centrifugation at 12,000 rpm for 5 min. Each washing or supernatant was subjected to SDS-PAGE on a 12% gel under reducing conditions. After electrophoresis, the gel was stained with Coomassie Brilliant Blue.
Protein Labeling with Rhodamine and the Crystal Binding Assay
PfN23 and BSA were labeled with rhodamine using a HOOKTM-dye labeling kit (GE Healthcare), according to the manufacturer's instructions. The protein crystal binding assay was performed as described previously (38).
Inhibition of Calcium Carbonate Precipitation Assay
The effect of PfN23 on the rate of calcium carbonate precipitation was tested according to the method of Suzuki et al. (20), with some modifications. The sample solution (10 μl) was mixed with 100 μl of 100 mm sodium bicarbonate, pH 8.5. After the addition of 100 μl of 100 mm calcium chloride to the mixed solution, the formation of calcium carbonate precipitate was monitored by recording changes in the turbidity every minute for 5 min based on the absorbance at 570 nm measured using a spectrophotometer (Bio-Rad 680).
In Vitro Calcium Carbonate Crystallization Assay
Saturated calcium bicarbonate solution was prepared following the method of Xu et al. (39), with some modifications. CO2 gas was bubbled through the mixture of calcium carbonate and Milli-Q deionized water for 6 h, and the excess solid CaCO3 was removed by filtration. An aragonitic crystallizing solution was produced by the addition of 50 mm magnesium chloride to the first solution. Crystallization experiments were conducted by adding samples to the freshly prepared crystallization solution on a siliconized glass slide. After 24 h, the crystallization solution was removed, and crystals were characterized using an FEI Quanta 200 scanning electron microscope and Raman spectroscopy, which was recorded at an excitation wavelength of 514 nm. The spectra were scanned three times for 20 s in the range of 100–1400 cm−1 using a Renishaw RM2000 spectrometer.
Bombyx mori silk cocoons (Chenye, Sichuan, China) were cut into ∼2 mm length and boiled for 45 min in 0.02 m Na2CO3 to extract the glue-like sericin proteins from the structural fibroin proteins. The fibroin extract was then rinsed three times in Milli-Q water and dissolved in 9.3 m LiBr solution at 60 °C for 4 h. The supernatant was separated by centrifugation at 10,000 rpm for 30 min and dialyzed (cutoff of 3.5 kDa) against Milli-Q water for 48 h at 4 °C. The final concentration of silk protein was 8 mg/ml, as determined using a BCA assay kit (Pierce). Then 100 μl of silk solution was placed on glass slides (14 mm diameter) to produce a 40-μm film thickness. The films were allowed to dry overnight. The calcium carbonate crystallization assay was performed on the films, as described above.
RESULTS
Isolation and Bioinformatic Analysis of PfN23
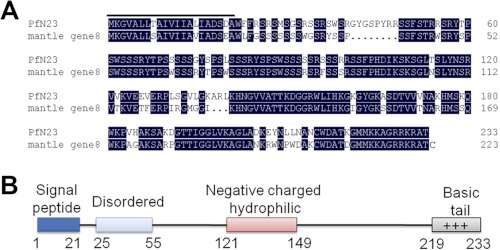
Based on the expression sequence tag in the suppression subtractive hybridization library of the D-shape stages (33) in P. fucata, we obtained a 1220-bp transcript including a 5′-untranslated region of 77 bp, an open reading frame of 702 bp encoding a deduced 233 amino acid protein, and a 3′-untranslated region of 518 bp (GenBankTM accession number JN995665). The deduced mature protein had a calculated molecular mass of 23.4 kDa, and it was referred to as PfN23. A BLASTx search against the GenBankTM nr data base indicated that PfN23 was similar to mantle gene 8 of P. fucata, and there were no similar genes in other species (Fig. 2A). However, mantle gene 8 had not been previously characterized before, so the function of PfN23 was unknown. The sequence analysis showed that PfN23 was a basic protein with a theoretical isoelectric point of 11.17. PfN23 was rich in Ser (22.2%) and positively charged residues (Arg + Lys, 19.8%), and it consisted of a signal peptide, a disordered region, a negatively charged and hydrophilic region, and a C-terminal basic region (Fig. 2B).
FIGURE 2.
Deduced amino acid sequence and bioinformatic analysis of PfN23. A, deduced amino acid sequence of PfN23 was aligned with mantle gene 8. The putative signal peptide in N-terminal is highlighted. B, schematic representation of PfN23 showing the predicted signal sequence, a disordered region, a negatively charged and hydrophilic region, and a C-terminal basic tail.
PfN23 in the Nacre
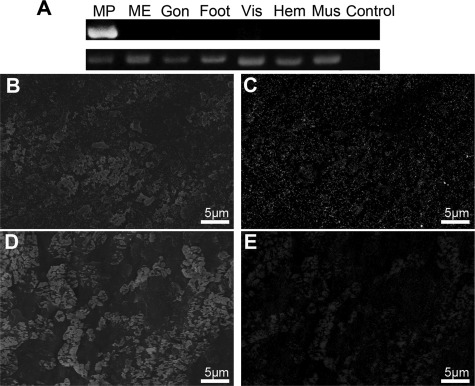
The specific expression of PfN23 was tested in a variety of P. fucata target tissues using RT-PCR (Fig. 3A). PfN23 expression was found to be specific to the mantle pallial which corresponded to the nacreous aragonite layer, although it was absent in the mantle edge that corresponded to the prismatic calcite layer. Biochemical analyses were performed to investigate whether native PfN23 was present in either the nacreous layer and/or the prismatic layer. Polyclonal antibodies raised against a recombinant form of PfN23 were used for the immunodetection of native PfN23 in EDTA extracts of shell using an ELISA. PfN23 was detected in the EDTA-soluble matrix of the shell that is known to be responsible for calcium carbonate formation (supplemental Fig. S1A). To further analyze the presence of PfN23 in different shell layers, EDTA extracts of the nacreous and prismatic layers were separated by SDS-PAGE followed by Western blotting. PfN23 was detected exclusively in the EDTA-soluble fractions of the nacre (supplemental Fig. S1B). In the control experiment, no staining was observed when anti-PfN23 antibodies were not used. The immunodetection assay confirmed the gene expression analysis that PfN23 expression corresponded to nacreous layer formation. To further elucidate the microstructural distribution of native PfN23 in the shell, the prismatic and nacreous layers were immunogold-labeled using anti-PfN23 antibodies as the primary antibody and 5 nm gold-labeled antibodies as the secondary antibody. Scanning in the back-scattered electron mode using scanning electron microscope could detect the high atomic number gold elements as white spots. PfN23 was detected on the whole surface of the nacreous layer with no concentrated areas (Fig. 3, B and C). In the control experiment, no staining was observed when anti-PfN23 antibodies were not used (Fig. 3, D and E).
FIGURE 3.
Detection of native PfN23. A, tissue-specific gene expression of PfN23 by RT-PCR analysis. Total RNA extracted from mantle edge (ME), mantle pallial (MP), gonad (Gon), foot, viscus (Vis), hemocytes (Hem), and adductor muscle (Mus) was used for RT-PCR. RT-PCR without a template was used as a negative control (Control). The housekeeping gene GAPDH was used as a positive control. B–E, detection of native PfN23 in shells by immunogold staining of the nacreous layer surface. B, SEM image of immunogold staining of the nacreous layer surface. D, SEM image of the control nacreous layer surface. C and E, back-scattered electron mode scanning electron microscope (SE-BSE) image of the same area shown in (B and D), respectively. Scale bars, 5 μm in B–E.
In Vivo Effect of PfN23 during Shell Formation
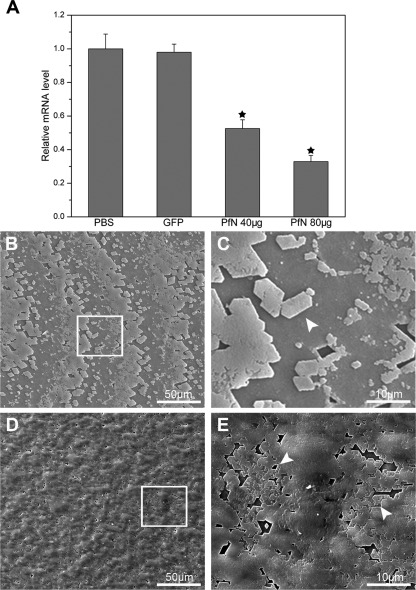
To further explore the role of PfN23 in nacre formation, a combination of RNAi and morpholino soaking assays was conducted. Specifically designed dsRNA of PfN23 was injected into the muscle of P. fucata to investigate the effects of PfN23 on the shell formation in adults. Compared with the PBS-injected groups, the PfN23 expression levels decreased to ∼50% in the 40-μg injected groups and 30% in the 80-μg injected groups. In contrast, the expression level of PfN23 was unchanged in the GFP dsRNA-injected groups (Fig. 4A). We also tested the expression levels of other matrix proteins i.e. aspein, N19, pearlin, prismalin-14, and prisilkin-39. The expression of these matrix proteins was not affected by the dsRNA of PfN23 (supplemental Fig. S2). The surface structure of the nacreous layer in each injected group was observed under SEM. In the GFP dsRNA-injected groups, the surface of the nacreous layer showed a normal stair-like growth pattern (Fig. 4B). Small flat tablets could be seen clearly on the surface (Fig. 4C, arrowhead). In the PfN23 dsRNA low dosage-injected group (40 μg), the surface also had a stair-like growth pattern (Fig. 4D). However, the flat tablets were covered with randomly accumulated crystals (Fig. 4E, arrowhead). The deposited crystals did not form the typical rectangular or hexagonal flat tablets. In the high dosage (80 μg) dsRNA-injected group, this abnormal phenomenon was more obvious (supplemental Fig. S3, A and B). In contrast, the morphology of the prismatic layers was not changed in the PfN23 and GFP dsRNA injected groups (data not shown).
FIGURE 4.
Effects of PfN23 on the nacre growth. A, expression levels of PfN23 were decreased by RNAi. For the control, the expression levels of the PBS-injected group were measured to a relative value of 1.0. The star represents a significant (p < 0.001) difference compared with the PBS-injected groups. B, SEM image of inner surface of GFP dsRNA injected group showing the stair-like growth pattern. D, SEM image of the 40 μg of dsRNA of PfN23 injected group. The growth pattern was disturbed. C and E, enlargement of the box shown in B and D, respectively. Arrowheads, the flat tablets on the surface of nacre. Scale bars, 50 μm in B and D and 10 μm in C and E.
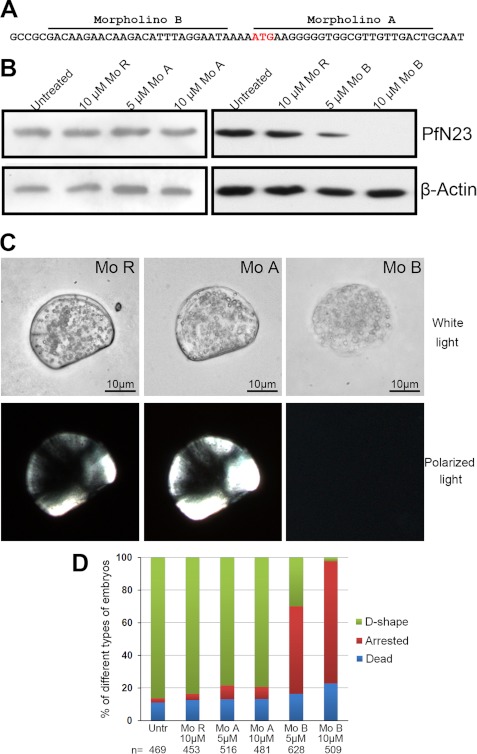
The gene was expressed during the D-shape stage of development (33), so the function of PfN23 was clarified using morpholinos. Two translational-blocking morpholinos were designed to block the translation of PfN23 (Fig. 5A), i.e. morpholino A (Mo A) and morpholino B (Mo B). A random morpholino (Mo R) was used as a control. The two-cell to four-cell stage embryos were soaked with 5 or 10 μm morpholinos to knock down the translation of PfN23. We conducted a Western blot 32 h after treatment to investigate PfN23 expression. As shown in Fig. 5B, the expression of PfN23 was not decreased compared with untreated groups when the Mo R or Mo A was used. However, Mo B decreases the expression of PfN23, and little protein was detected in the 10 μm-treated group. Larvae from all the groups could develop to the trochophore stage within 10 h. After 32 h, the larvae in the untreated, Mo R-treated, and Mo A-treated groups could develop to the D-shape stage, leading to a capital D-like morphology (Fig. 5C). In the Mo B-treated groups, the development of most larvae was arrested. In addition, no shell coverings were present on the arrested larvae (Fig. 5C). In the control or Mo A-treated groups, most larvae (∼80%) developed to the D-shape stage. When treated with 5 μm Mo B, only 29.8% (187/628) of larvae developed to the D-shape stage, whereas 53.8% (338/628) of larvae were arrested before they were covered with a shell. Compared with the 5 μm treatment, fewer larvae (2.4%, 12/509) developed to the D-shape stage, whereas more larvae (74.9%, 381/509) were arrested in the 10 μm Mo B-treated groups (Fig. 5D).
FIGURE 5.
Depletion of PfN23 using antisense morpholino oligomers. A, positions of the antisense morpholino oligomers and the ATG initiation codon are indicated. B, Western blot analysis using anti-PfN23 antibody showing that Mo B blocked translation of PfN23 mRNA. β-Actin was used as a loading control. C, phenotypes of morpholino oligomer-treated larvae 32 h after fertilization. The samples were analyzed under white polarized light to evaluate the formation of shells. D, frequency of the phenotypes obtained with different concentrations of Mo. The number n indicates the total number of analyzed embryos. Mo R, random morpholino oligomers, Mo A, morpholino A, and Mo B, morpholino B. Scale bars, 10 μm in C.
In Vitro Effect of PfN23 in Calcium Carbonate Crystallization
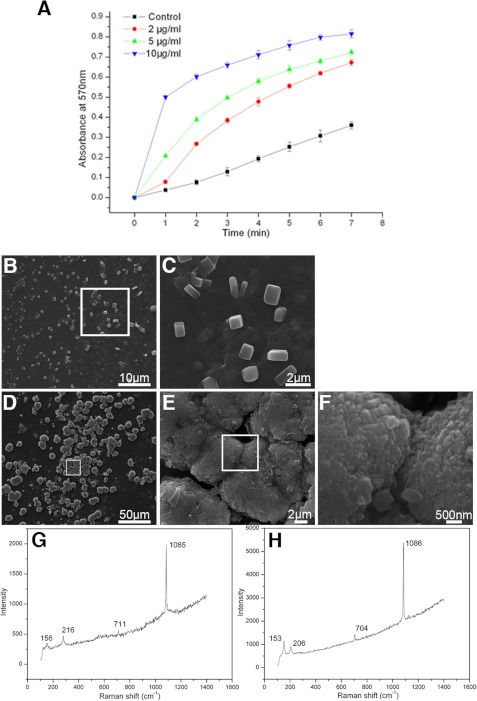
To further analyze the function of PfN23 in calcium carbonate crystallization, we tested its binding properties with the main component of shell, i.e. calcite, aragonite, and chitin, and found PfN23 could bind to calcite and aragonite (supplemental Fig. S4, A and B). We then tested its function during calcium carbonate crystallization. A series of precipitation experiments was performed to analyze the effect of PfN23 on the rate of calcium carbonate precipitation. Compared with the control, PfN23 increased the rate of precipitation, and this effect occurred in a concentration-dependent manner (Fig. 6A).
FIGURE 6.
Effects of PfN23 on calcium carbonate crystallization. A, accelerative activity of PfN23 during calcium carbonate precipitation. ■, BSA (10 μg/ml) was used as negative control; ●, 2 μg/ml PfN23; ▴, 5 μg/ml PfN23; ▾, 10 μg/ml PfN23 was present. B–E, effects of PfN23 on the crystals growth were tested on the silk film. B, 50 μg/ml BSA was used as negative control. D, 50 μg/ml PfN23 was added to the system. C, E, and F, SEM images of the enlargement of the box in B, D, and E, respectively. G and H, Raman spectra of the crystals formed in B and D, respectively. Scale bars, 50 μm in D, 10 μm in B, 2 μm in C and E, and 500 nm in F.
To investigate the effect of PfN23 on the growth of calcium carbonate crystals, in vitro crystallization experiments with or without magnesium were conducted. In the magnesium-free system, the effect of PfN23 on the growth of calcite was analyzed. In the control experiments, the precipitated crystals were all typical rhombohedra of calcite in the presence of BSA. PfN23 could induce the formation of vaterite in the calcite formation system. The percentages of the rhombohedral and round vaterite crystals in the deposited crystals were about 25% at 10 μg/ml PfN23, 44% at 30 μg/ml PfN23, and 65% at 50 μg/ml PfN23. (supplemental Fig. S5). Magnesium was introduced into the precipitation system to induce the formation of aragonite, which is the main component of the nacreous layer. Compared with calcite, the different concentrations of PfN23 had little effect on aragonite formation. Even at a high concentration of 50 μg/ml, the typical aragonite crystals were deposited with a normal needle-shaped morphology (supplemental Fig. S6). According to the models proposed by Weiner and co-workers (30), aragonite is formed in the silk-gel phase of the nacre. Given that PfN23 is a protein in the EDTA-soluble matrix of the shell, these findings suggested that a silk gel was required for the functionality of PfN23. We tested this hypothesis by assaying the in vitro crystallization on a silk film. With 50 μg/ml BSA, typical calcite crystals were deposited on the film (Fig. 6, B and C). The crystals were confirmed to be calcite by Raman spectra (Fig. 6G). However, compared with the crystals (∼10 μm) formed without the silk film (supplemental Fig. S5A), these crystals were smaller (∼2 μm), and they had rounder edges. With 50 μg/ml of PfN23, the deposited crystals were not separate particles and instead they amalgamated to form tablets that were similar to the microstructure of nacre (Fig. 6D). In addition, these tablets were composed of small sheets of crystals (Fig. 6, E and F). The Raman spectra showed that these crystals were aragonite (Fig. 6H). These results suggested that PfN23 could induce the formation of nacreous tablets in the silk-gel phase.
Functions of Different Regions of PfN23
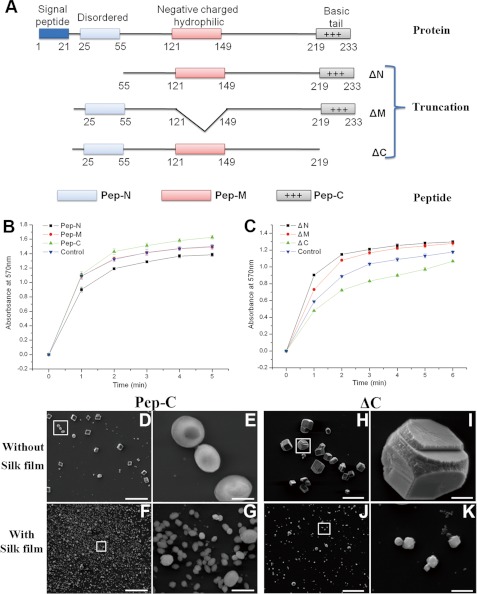
We constructed the peptides and truncations of the predicted disordered region (Peptide, Pep-N; mutation, ΔN), negatively charged and hydrophilic region (Peptide, Pep-M; mutation, ΔM), and C-terminal basic region (Peptide, Pep-C; mutation, ΔC) to evaluate the function of these regions (Fig. 7A). The Pep-N and Pep-M had little effect on the morphology of the deposited crystals in the in vitro crystallization experiments (supplemental Fig. S7, A–H). One point to mention is that the Pep-N could inhibit the growth of calcium carbonate crystals, and Pep-M had little effect on the rate of the growth of calcium carbonate crystals (Fig. 7B). Compared with these two regions, the peptide of C-terminal basic region (Pep-C) could promote the growth of calcite and induce the formation of vaterite (Fig. 7, B, D, and E, and supplemental Fig. S8A). The percentages of the round shaped vaterite in the deposited crystals were about 27% at 10 μg/ml Pep-C, 35% at 30 μg/ml Pep-C, and 44% at 50 μg/ml Pep-C. With silk gel, this peptide could induce the formation of small hexagonal vaterite crystals, but these crystals were not connected together (Fig. 7, F and G, and supplemental Fig. S8B). In addition, these crystals were quite like those deposited in the presence of the predicted disordered region or negatively charged and hydrophilic region truncations (supplemental Fig. S7, K, L, O, and P). When this basic region was deleted, the protein could not promote the crystallization or induce vaterite formation (Fig. 7, C, H, and I, and supplemental Fig. S8C). Without silk film, the crystals formed with ΔC (Fig. 7, H and I) looked like those formed with ΔN (supplemental Fig. S7, I and J). With silk gel, this basic region truncation had little effect on the crystal deposition (Fig. 7, J and K, and supplemental Fig. S8D). Taken together, this positively charged region may be a key region for the function of PfN23, and the other regions would also be involved to achieve the full function of PfN23.
FIGURE 7.
Functions of different regions of PfN23. A, schematic representation of truncations and peptides. Numbers indicate the position of amino acid. B and C, accelerative activities of truncations and peptides during calcium carbonate precipitation. Changes in the turbidity of the assayed solutions are shown. B, activities of peptides. ■, 10 μg/ml Pep-N; ●, 10 μg/ml Pep-M; ▴, 10 μg/ml Pep-C was present; ▾, 10 μg/ml BSA was used as a negative control. C, activities of truncations. ■, 10 μg/ml ΔN; ●, 10 μg/ml ΔM; ▴, 10 μg/ml ΔC was present; ▾, 10 μg/ml BSA was used as a negative control. D–K, in vitro crystallization experiments in the presence of truncations and peptides. 10 μg/ml Pep-C was added with (F) or without (D) the silk film. 10 μg/ml ΔC was added with (J) or without (H) the silk film. E, G, I, and K, enlargement of boxes in D, F, H, and J, respectively. Scale bars, 50 μm in D, F, H, and J and 5 μm in E, G, I, and K.
DISCUSSION
Matrix proteins are known to play crucial roles in nacre formation. Much progress has been made in the analysis of biogenic minerals, which facilitates their applications in the biomaterial and biomedical fields (10). Acidic aspartic acid-rich proteins that may interact with calcium atoms were believed to regulate crystal formation in nacre (40, 41). However, no basic proteins with a serine-rich sequence have been characterized that might induce the formation of nacre-like micro-tablets.
In this study, a serine-rich basic protein, PfN23, was cloned from a previously constructed suppression subtractive hybridization library (33). A combination of genetic and biochemical analyses showed that PfN23 was found in the EDTA-soluble fraction of nacre. Depending on their solubility in a decalcifying EDTA solution, proteins in nacre are usually classified into two groups. Proteins in the EDTA-soluble fraction are believed to localize within calcium carbonate crystals and control crystal morphology, phase switching, and orientation. In contrast, proteins in the EDTA-insoluble fraction are hypothesized to localize around calcium carbonate crystallites where they are responsible for framework formation (14, 17, 42). The exact position of PfN23 in the nacre was further confirmed using an immune gold assay, where positive signals for PfN23 were found in the tablets of nacre but not in the frames (Fig. 3C). This suggests a role for the protein in the control of aragonite formation in nacre.
We analyzed the in vivo function of PfN23 in P. fucata adults and larvae. When dsRNA was injected, a specific reduction of the PfN23 transcription levels was achieved. SEM images of the surface of nacre revealed an aberrant texture that could be attributed to the effects of PfN23 silencing. Taking that PfN23 increased the rate of calcium carbonate precipitation (Fig. 6). Thus, the driving force for crystallization was repressed when PfN23 was silenced. Most of the control molecules found in the crystallization space inhibit precipitation. The saturated solution was stabilized to a point where these inhibitors could not repress the crystallization process, and calcium carbonate crystals then randomly accumulated on the surface of the nacre. To our knowledge, the use of morpholino oligomers to study nacre formation in the developmental stages has not been described previously. Only a small number of microinjection studies have been performed with mollusks to analyze the development of larvae (43–45). Here, we present a convenient assay to deliver morpholinos to embryos of P. fucata by soaking. A translation-blocking morpholino was successfully delivered to knock down the expression of PfN23, and the larvae were arrested before they were covered with a shell. This appearance may also be due to the absence of the driving force for crystallization. Thus, the larvae could not deposit calcium carbonate to form external D-shaped shells. The calcium carbonate crystallization was controlled by two antagonistic mechanisms, inhibition and acceleration (34). If either one of the two mechanisms was inhibited, the formation of nacre would be disturbed.
In vitro crystallization experiments were conducted to evaluate the function of PfN23 during calcium carbonate crystallization. PfN23 increased the rate of calcium carbonate precipitation and induced the formation of vaterite. Other than calcite and aragonite, vaterite is one of the three nonhydrated crystalline polymorphs of calcium carbonate. Vaterite is highly unstable and when exposed to water it can readily recrystallize to calcite. PfN23 facilitates the formation of unstable vaterite, although other factors were needed to induce the nacre formation. The formation of aragonitic tablets was not induced solely by PfN23. According to the classical model of nacre formation (30), the nacre tablets are constructed in the silk gel that is embedded with a highly structured chitinous framework. Silk is usually the major protein fraction in nacre. It is rich in Gly and Ala or only Gly. The silk gel where nacre is built can inhibit the in vitro crystallization of calcium carbonate (46). Most proteins found in the nacre are known to inhibit calcium carbonate crystallization. Other than these repressors of nacre formation, proteins that might promote the rate of precipitation need to be considered. When in vitro crystallization experiments were conducted on the silk film, we found that the deposited aragonite crystals amalgamated to display characteristics similar to those observed in nacre. However, the shape of these crystals was not the same as the rectangular or hexagonal flat ones found in nacre. Another piece of the model of nacre formation is that crystallization processes occur in a highly structured chitinous framework that may be responsible for the shape of the tablets.
In conclusion, this study demonstrated that a basic protein, PfN23, has a key role in the formation of nacre. PfN23 has the potential for the in vitro reconstruction of nacre in the silk gel. Further studies on the mechanisms of nacre formation would shed new light on the synthesis of nacre in artificial systems.
Supplementary Material
Acknowledgments
We thank Wenguang Liu for technical support in the larvae culture and Yanchao Han for advice on the design of morpholino oligomers.
This work was supported by National Basic Research Program of China Grant 2010CB126405, National High Technology Research and Development Program of China 2010AA09Z405, National Natural Science Foundation of China Grants 31172382, 40876068, and U0831001 (Joint Fund with Guangdong), Specialized Research Fund for the Doctoral Program of Higher Education of China Grant 20090002110055, and Independent Research Projects of Tsinghua University Grant 20111080964.

This article contains supplemental Figs. S1–S8.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) JN995665.
- RACE
- rapid amplification of cDNA end
- SEM
- scanning electron microscope
- dsRNA
- double-stranded RNA
- Mo
- morpholino.
REFERENCES
- 1. Suzuki M., Saruwatari K., Kogure T., Yamamoto Y., Nishimura T., Kato T., Nagasawa H. (2009) An acidic matrix protein, Pif, is a key macromolecule for nacre formation. Science 325, 1388–1390 [DOI] [PubMed] [Google Scholar]
- 2. Li X., Huang Z. (2009) Unveiling the formation mechanism of pseudo-single crystal aragonite platelets in nacre. Physical. Rev. Lett. 102, 75502. [DOI] [PubMed] [Google Scholar]
- 3. Addadi L., Weiner S. (1997) Biomineralization. A pavement of pearl. Nature 389, 912–915 [Google Scholar]
- 4. Weiner S., Traub W. (1984) Macromolecules in mollusc shells and their functions in biomineralization. Philos. Trans. R. Soc. Lond. B Biol. Sci. 304, 425–434 [Google Scholar]
- 5. Simkiss K. (1977) Biomineralization and detoxification. Calcif. Tissue Res. 24, 199–200 [DOI] [PubMed] [Google Scholar]
- 6. Wilkinson B. H. (1979) Mineralization, paleooceanography, and the evolution of calcareous marine organisms. Geology 7, 524–527 [Google Scholar]
- 7. Lowenstam H. A., Abbott D. P. (1975) Vaterite. A mineralization product of the hard tissues of a marine organism (Ascidiacea). Science 188, 363–365 [DOI] [PubMed] [Google Scholar]
- 8. Feng Q. (2011) in Principles of Calcium-based Biomineralization Molecular Biomineralization (Müller W. E. G., ed) pp. 141–197, Springer-Verlag, Berlin: [DOI] [PubMed] [Google Scholar]
- 9. Marelli B., Ghezzi C. E., Alessandrino A., Barralet J. E., Freddi G., Nazhat S. N. (2012) Silk fibroin-derived polypeptide-induced biomineralization of collagen. Biomaterials 33, 102–108 [DOI] [PubMed] [Google Scholar]
- 10. Marin F., Luquet G., Marie B., Medakovic D. (2007) in Current Topics in Developmental Biology (Gerald P. S., ed) pp. 209–276, New York: [DOI] [PubMed] [Google Scholar]
- 11. Kniep R. (2010) in Fluorapatite-Gelatine-Nanocomposites. Self-organized Morphogenesis, Real Structure, and Relations to Natural Hard Materials. Biomineralization I (Naka K., ed) pp. 73–125, Springer, Berlin [Google Scholar]
- 12. Tang Z., Kotov N. A., Magonov S., Ozturk B. (2003) Nanostructured artificial nacre. Nat. Mater. 2, 413–418 [DOI] [PubMed] [Google Scholar]
- 13. Weiner S., Addadi L. (2011) Crystallization pathways in biomineralization. Annu. Rev. Materials Res. 41, 21–40 [Google Scholar]
- 14. Belcher A. M., Wu X. H., Christensen R. J., Hansma P. K., Stucky G. D., Morse D. E. (1996) Control of crystal phase switching and orientation by soluble mollusk-shell proteins. Nature 381, 56–58 [Google Scholar]
- 15. Contri M. B., Boraldi F., Taparelli F., De Paepe A., Ronchetti I. P. (1996) Matrix proteins with high affinity for calcium ions are associated with mineralization within the elastic fibers of pseudoxanthoma elasticum dermis. Am. J. Pathol. 148, 569–577 [PMC free article] [PubMed] [Google Scholar]
- 16. Miyamoto H., Miyashita T., Okushima M., Nakano S., Morita T., Matsushiro A. (1996) A carbonic anhydrase from the nacreous layer in oyster pearls. Proc. Natl. Acad. Sci. U.S.A. 93, 9657–9660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sudo S., Fujikawa T., Nagakura T., Ohkubo T., Sakaguchi K., Tanaka M., Nakashima K., Takahashi T. (1997) Structures of mollusc shell framework proteins. Nature 387, 563–564 [DOI] [PubMed] [Google Scholar]
- 18. Shen X., Belcher A. M., Hansma P. K., Stucky G. D., Morse D. E. (1997) Molecular cloning and characterization of lustrin A, a matrix protein from shell and pearl nacre of Haliotis rufescens. J. Biol. Chem. 272, 32472–32481 [DOI] [PubMed] [Google Scholar]
- 19. Samata T., Hayashi N., Kono M., Hasegawa K., Horita C., Akera S. (1999) A new matrix protein family related to the nacreous layer formation of Pinctada fucata. FEBS Lett. 462, 225–229 [DOI] [PubMed] [Google Scholar]
- 20. Suzuki M., Murayama E., Inoue H., Ozaki N., Tohse H., Kogure T., Nagasawa H. (2004) Characterization of Prismalin-14, a novel matrix protein from the prismatic layer of the Japanese pearl oyster (Pinctada fucata). Biochem. J. 382, 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yano M., Nagai K., Morimoto K., Miyamoto H. (2006) Shematrin. A family of glycine-rich structural proteins in the shell of the pearl oyster Pinctada fucata. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 144, 254–262 [DOI] [PubMed] [Google Scholar]
- 22. Yano M., Nagai K., Morimoto K., Miyamoto H. (2007) A novel nacre protein N19 in the pearl oyster Pinctada fucata. Biochem. Biophys. Res. Commun. 362, 158–163 [DOI] [PubMed] [Google Scholar]
- 23. Shechter A., Glazer L., Cheled S., Mor E., Weil S., Berman A., Bentov S., Aflalo E. D., Khalaila I., Sagi A. (2008) A gastrolith protein serving a dual role in the formation of an amorphous mineral containing extracellular matrix. Proc. Natl. Acad. Sci. U.S.A. 105, 7129–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kong Y., Jing G., Yan Z., Li C., Gong N., Zhu F., Li D., Zhang Y., Zheng G., Wang H., Xie L., Zhang R. (2009) Cloning and characterization of Prisilkin-39, a novel matrix protein serving a dual role in the prismatic layer formation from the oyster Pinctada fucata. J. Biol. Chem. 284, 10841–10854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Glazer L., Shechter A., Tom M., Yudkovski Y., Weil S., Aflalo E. D., Pamuru R. R., Khalaila I., Bentov S., Berman A., Sagi A. (2010) A protein involved in the assembly of an extracellular calcium storage matrix. J. Biol. Chem. 285, 12831–12839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gotliv B. A., Addadi L., Weiner S. (2003) Mollusk shell acidic proteins. In search of individual functions. ChemBioChem 4, 522–529 [DOI] [PubMed] [Google Scholar]
- 27. Mann K., Siedler F., Treccani L., Heinemann F., Fritz M. (2007) Perlinhibin, a cysteine-, histidine-, and arginine-rich miniprotein from abalone (Haliotis laevigata) nacre, inhibits in vitro calcium carbonate crystallization. Biophys. J. 93, 1246–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Levi-Kalisman Y., Falini G., Addadi L., Weiner S. (2001) Structure of the nacreous organic matrix of a bivalve mollusk shell examined in the hydrated state using cryo-TEM. J. Struct. Biol. 135, 8–17 [DOI] [PubMed] [Google Scholar]
- 29. Weiss I. M., Tuross N., Addadi L., Weiner S. (2002) Mollusc larval shell formation. Amorphous calcium carbonate is a precursor phase for aragonite. J. Exp. Zool. 293, 478–491 [DOI] [PubMed] [Google Scholar]
- 30. Addadi L., Joester D., Nudelman F., Weiner S. (2006) Mollusk shell formation. A source of new concepts for understanding biomineralization processes. CHEM-EUR. J. 12, 981–987 [DOI] [PubMed] [Google Scholar]
- 31. Treccani L., Mann K., Heinemann F., Fritz M. (2006) Perlwapin, an abalone nacre protein with three four-disulfide core (whey acidic protein) domains, inhibits the growth of calcium carbonate crystals. Biophys. J. 91, 2601–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weiss I. M., Kaufmann S., Mann K., Fritz M. (2000) Purification and characterization of perlucin and perlustrin, two new proteins from the shell of the mollusc Haliotis laevigata. Biochem. Biophys. Res. Commun. 267, 17–21 [DOI] [PubMed] [Google Scholar]
- 33. Fang D., Xu G. R., Hu Y. L., Pan C., Xie L. P., Zhang R. Q. (2011) Identification of genes directly involved in shell formation and their functions in pearl oyster, Pinctada fucata. PLoS One 6, e21860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marin F., Luquet G., Marie B., Medakovic D. (2008) Molluscan shell proteins. Primary structure, origin, and evolution. Curr. Top. Dev. Biol. 80, 209–276 [DOI] [PubMed] [Google Scholar]
- 35. Fujimura T., Wada K., Iwaki T. (1995) Development and morphology of the pearl oyster larvae, Pinctada fucata. Venus Jpn. J. Malacol. 54, 25–48 [Google Scholar]
- 36. Inoue H., Ohira T., Ozaki N., Nagasawa H. (2004) A novel calcium-binding peptide from the cuticle of the crayfish, Procambarus clarkii. Biochem. Biophys. Res. Commun. 318, 649–654 [DOI] [PubMed] [Google Scholar]
- 37. Kitano Y., Park K., Hood D. W. (1962) Pure aragonite synthesis. J. Geophys. Res. 67, 4873–4874 [Google Scholar]
- 38. Ma Z., Huang J., Sun J., Wang G., Li C., Xie L., Zhang R. (2007) A novel extrapallial fluid protein controls the morphology of nacre lamellae in the pearl oyster, Pinctada fucata. J. Biol. Chem. 282, 23253–23263 [DOI] [PubMed] [Google Scholar]
- 39. Xu G. F., Yao N., Aksay I. A., Groves J. T. (1998) Biomimetic synthesis of macroscopic scale calcium carbonate thin films. Evidence for a multistep process. J. Am. Chem. Soc. 120, 11977–11985 [Google Scholar]
- 40. Borbas J. E., Wheeler A. P., Sikes C. S. (1991) Molluscan shell matrix phosphoproteins. Correlation of degree of phosphorylation to shell mineral microstructure and to in vitro regulation of mineralization. J. Exp. Zool. 258, 1–13 [Google Scholar]
- 41. Lowenstam H. (1981) Minerals formed by organisms. Science 211, 1126–1131 [DOI] [PubMed] [Google Scholar]
- 42. Mann S., Archibald D. D., Didymus J. M., Douglas T., Heywood B. R., Meldrum F. C., Reeves N. J. (1993) Crystallization at Inorganic-organic Interfaces. Biominerals and biomimetic synthesis. Science 261, 1286–1292 [DOI] [PubMed] [Google Scholar]
- 43. Chan X. Y., Lambert J. D. (2011) Patterning a spiralian embryo. A segregated RNA for a Tis11 ortholog is required in the 3a and 3b cells of the Ilyanassa embryo. Dev. Biol. 349, 102–112 [DOI] [PubMed] [Google Scholar]
- 44. Henry J. Q., Perry K. J., Martindale M. Q. (2010) β-Catenin and early development in the gastropod, Crepidula fornicata. Integr. Comp. Biol. 50, 707–719 [DOI] [PubMed] [Google Scholar]
- 45. Rabinowitz J. S., Chan X. Y., Kingsley E. P., Duan Y., Lambert J. D. (2008) Nanos is required in somatic blast cell lineages in the posterior of a mollusk embryo. Curr. Biol. 18, 331–336 [DOI] [PubMed] [Google Scholar]
- 46. Falini G., Albeck S., Weiner S., Addadi L. (1996) Control of aragonite or calcite polymorphism by mollusk shell macromolecules. Science 271, 67–69 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.