Background: The length of the O-antigen polysaccharide is determined by polysaccharide co-polymerases(PCPs).
Results: We have determined structures of several chimeric PCPs that lead to different polysaccharide lengths and determined their oligomerization state.
Conclusion: The polysaccharide length distribution depends primarily on the residues on the outer surface of the PCP oligomer.
Significance: Our data provide support for the octameric assembly of WzzB PCPs.
Keywords: Cell Surface, Crystal Structure, Lipopolysaccharide (LPS), Mutagenesis, Protein Chimeras, Protein-Protein Interactions, O-antigen, Polysaccharide Length Control
Abstract
The surface O-antigen polymers of Gram-negative bacteria exhibit a modal length distribution that depends on dedicated chain length regulator periplasmic proteins (polysaccharide co-polymerases, PCPs) anchored in the inner membrane by two transmembrane helices. In an attempt to determine whether structural changes underlie the O-antigen modal length specification, we have determined the crystal structures of several closely related PCPs, namely two chimeric PCP-1 family members solved at 1.6 and 2.8 Å and a wild-type PCP-1 from Shigella flexneri solved at 2.8 Å. The chimeric proteins form circular octamers, whereas the wild-type WzzB from S. flexneri was found to be an open trimer. We also present the structure of a WzzFepE mutant, which exhibits severe attenuation in its ability to produce very long O-antigen polymers. Our findings suggest that the differences in the modal length distribution depend primarily on the surface-exposed amino acids in specific regions rather than on the differences in the oligomeric state of the PCP protomers.
Introduction
Bacterial surface is lined with complex carbohydrate polymers that form a protective barrier against the external environment. For pathogenic bacteria, these polymers are an essential component of the survival toolkit within the host and frequently are critical determinants of pathogen-host interactions. In particular, the LPS O-antigen (OAg)2 is emerging as an important pathogenesis factor contributing to virulence of a number of human pathogens (1–7). The O-antigen is the most chemically diverse component of the outer membrane lipopolysaccharide and varies among even the same species of bacteria. The mature O-antigen chain is a polymer made of the repeating units, with each O-unit containing 4–6 sugar residues (8). The proteins involved in the biosynthesis of the nucleotide sugars, glycosyl transfers, and OAg processing machinery are encoded on one or more gene clusters (9, 10). The OAg processing machinery is responsible for the assembly of the polymer in the periplasmic space and for the transport to the cell surface (11). In a system referred to as Wzy-dependent synthesis, the individual O-units are synthesized at the cytoplasmic side of the inner membrane, then flipped to the periplasm by a flippase Wzx, and assemble into a high molecular mass polysaccharide by Wzy polymerase (9). The assembly is completed by the ligase WaaL, which transfers the mature O-antigen to the sugar moiety of the outer core of the lipid A molecule (9).
The number of the O-units in a given O-antigen polymer exposed on bacterial surface falls within a narrow range distribution, resulting in polymers of a well defined modal length. Much like the chemical composition of the individual sugar moieties making up the polymer, the length of the polysaccharide is species- and serotype-specific (10, 12). Early genetic studies identified a single gene wzzB (previously cld or rol) to be responsible for defining the specific number of the repeat units in a given O-antigen polymer (13). Subsequent biochemical investigations have found that the product of wzzB gene is a homo-oligomeric protein embedded in the inner membrane through two transmembrane helices and containing a large periplasmic domain (14). Shortly after, the presence of similar structural features was discovered in seemingly unrelated proteins involved in trafficking a range of complex polysaccharides in both Gram-negative and Gram-positive bacteria (14–16). These functionally related proteins were named polysaccharide co-polymerases (PCPs) (17) and are further subdivided into various subfamilies depending on the type of the polysaccharide biosynthetic pathway they are part of and the presence of a cytosolic tyrosine kinase domain (11, 17).
Currently, atomic-level structural data are available only for periplasmic domains of a few family members. Crystallographic studies of the WzzB from Salmonella typhimurium (WzzST), FepE (WzzFepE) from Escherichia coli O157:H7, and WzzE from E. coli revealed that these proteins adopt a very similar three-dimensional fold despite a very limited sequence similarity (18). The oligomeric chain length regulators resemble elongated bell-shaped structures, with the protomers composed of an α-β base domain and a long α-helix extending about 100 Å away from the inner membrane into the periplasm (18). In the crystal structures, soluble domains of WzzB, WzzFepE, and WzzE were found in different oligomeric organizations composed of five, nine, and eight protomers, respectively. The size of the oligomers was addressed independently also by electron microscopy and small angle scattering. The study by electron microscopy of the full-length WzzB and WzzFepE reconstituted in proteoliposomes led Larue et al. (19) to suggest a hexameric structure for these oligomers. A small angle x-ray scattering study of the full-length E. coli (O86:H2) WzzB solubilized in n-dodecyl-β-maltoside suggested on the other hand the presence of tetramers in solution (20), although higher order oligomers were observed when the salt concentration was increased. The contradictory results of these studies performed away from the natural environment within the bacterial membrane leaves uncertain the native oligomeric state of the PCPs within the inner membrane.
Despite the current functional and structural data, little is understood about the precise molecular mechanism underlying the chain length control. The specific modal length appears to be associated with each chain length regulator because when expressed in a closely related bacterial species, the PCP imposes its own characteristic modal length distribution despite the drastically different chemical structure of the carbohydrate polymer (14, 21, 22). Previously, we have used a structure-guided chimeric protein approach to establish which regions of the chain length regulator are responsible for the control of modal length. By swapping various structural elements between two closely WzzB molecules yet imposing distinctly different modal length distributions, Shigella flexneri WzzBSF and S. typhimurium WzzBST, we were able to produce a number of functional chimeric proteins exhibiting a range of intermediate modal lengths (22). These findings posed a series of further questions, one of which was whether the observed differences in phenotypes could be the result of significant local conformational differences between the wild-type protein and the corresponding chimeric chain length regulators.
To address this question, we determined the structures of the periplasmic domain of the S. flexneri WzzBSF and three chimeric chain length regulator proteins generated from parental WzzBSF and WzzBST and compared them with the parental structures. The structural alignments demonstrated little differences in the main-chain conformations, suggesting that a given modal length is determined by the side chains of residues in selected regions. Most of these residues were found to be surface-exposed on the external face of the oligomer. The chimeras form oligomers with a variable number of protomers, bell-shaped octamers similar to the oligomers of WzzE, pentamers similar to the previously observed E. coli WzzB, and open-face trimers. The size of the octamers corresponds very well to the electron microscopy data, and we surmise that this is the most likely arrangement of WzzB in the bacterial cell. Lastly, we determined the structure of a WzzFepE loop deletion mutant, which confers strikingly different modal length from the wild-type protein, and discovered that it forms a nonameric structure very similar to its wild-type counterpart. All of these observations suggest that the modality is dictated predominantly by the nature of the external surface amino acids rather then by structural alterations of the chain length regulator itself.
EXPERIMENTAL PROCEDURES
Construction of Expression Vectors Encoding Periplasmic Domains of Chimeric Proteins
All plasmids were purified from E. coli DH5α using the plasmid mini prep kit (Sigma-Aldrich). The sequences corresponding to amino acids (Glu-54–Lys-293) were PCR-amplified from the corresponding plasmids pSK10 and pSK11 (22) using the following primer set: 5′-GAG CAG GGA TCC GAA AAA TGG ACA TCC ACG GCG (8) and 3′-GAC CAG GAA TTC TTA TTT CGG ACT ATC GCG ACG AAT AGG. The resultant PCR product was cut with BamHI and EcoRI and ligated into the pET-based histidine expression vector pFO4. The sequence corresponding to the periplasmic domain of WzzB from S. flexneri was amplified from the genomic DNA and was subcloned in pET-based expression vector as described above. All constructs were verified by sequencing and confirmed to contain a periplasmic domain of the chimeric WzzB with a noncleavable N-terminal hexahistidine tag. WzzFepE (Δ258:266::GSG) mutant was constructed using the overlap PCR method (40). The sequence (Lys-64–Pro-330) corresponding to the periplasmic domain was then subcloned into the pET-based expression vector containing N-terminal hexahistidine tag.
Expression and Purification of the Chimeric Proteins
E. coli BL21 (DE3) (Invitrogen) transformed with the above expression plasmids were grown to mid-log phase, cooled to 20 °C, and induced with 0.1 mm isopropyl-d-thiogalactoside. Growth was continued for 20 h before the cells were collected by centrifugation at 4,000 × g for 20 min at 4 °C. Cell pellets were resuspended and washed twice with ice-cold wash buffer A (200 mm NaCl, 10 mm imidazole, 40 mm Tris, pH 8.0, supplemented with 1 mm p-aminobenzamidine and 100 μm leupeptin protein inhibitors). Cells were broken by sonication for a total of 6 × 10-s bursts. Cell debris was removed by centrifugation for 45 min at 15,000 × g in the cold, and the supernatant was incubated with 0.5 ml of 50% nickel-nitrilotriacetic acid-agarose beads for 2 h at 4 °C pre-equilibrated with wash buffer A. The beads were washed with 10 column volumes of buffer A containing 20 mm imidazole and 10 column volumes of buffer B (200 mm NaCl, 30 mm Tris at pH 8.0) containing 40 mm imidazole, and chimeric WzzB were eluted in buffer A containing 250 mm imidazole. The purity of the eluted protein was analyzed on 12% sodium dodecyl sulfate (SDS)-polyacrylamide gels and stained with Coomassie Blue, and the buffer was exchanged into 200 mm NaCl, 40 mm Tris, pH 8.0, using a 10DG desalting column (Bio-Rad, Mississauga, Ontario, Canada). The protein was concentrated 10-fold with a Centricon concentrator (30,000 molecular weight cut-off) and was further purified by gel filtration on a Superdex S200 column (GE Healthcare) in buffer B. The purified protein was concentrated to 10 mg/ml in the same buffer, flash-frozen in liquid nitrogen, and stored at −80 °C. Se-Met-containing proteins were obtained using a similar protocol by growing BL21(DE3) on minimal medium containing l-selenomethionine. Protein concentrations were determined with the Bio-Rad protein assay, using bovine serum albumin as a standard, and dynamic light scattering experiments were carried out using a DynaPro plate reader (Wyatt Technologies, Santa Barbara, CA) to assess sample polydispersity prior to crystallization.
Crystallization
The initial crystallization conditions for WzzBSF were identified using Qiagen JCSG Core suite screens (Qiagen, Mississauga, Ontario, Canada) and optimized using the hanging drop vapor diffusion method. The best WzzBSF crystals were grown by equilibration of 1 μl of protein (at 6 mg/ml) in buffer 30 mm Hepes, pH 7.5, 200 mm NaCl, with 1 μl of reservoir solution containing 20% PEG 4000, 0.1 m Hepes, pH 7.5, and 0.1 m MgCl2 and equilibrating over the reservoir solution. The crystals belonged to P21 with a = 81.5, b = 62.6, c = 90.7 Å with three molecules in the asymmetric unit and Vm of 2.75 Å3 Da−1. For each chimeric protein, a number of commercial crystallization kits from Qiagen, Hampton Research, and Molecular Dimensions were screened using the sitting drop vapor diffusion method at 298 K, mixing 400 nl of concentrated protein solution (10 mg/ml) with 400 nl of the corresponding reservoir solution. For SF4, a number of initial hits in various high salt conditions of the MbClass screen (Qiagen) could be seen 5 days later. Most of these were further optimized in Linbro 24-well plates using the hanging drop vapor diffusion method, and the conditions that led to the best diffracting crystal were 3.0 m sodium formate, pH 6.0 (HNaO2). The crystals belonged to the space group I422 with a = 115.7, c = 210.9 Å with two molecules in the asymmetric unit and Vm of 3.14 Å3 Da−1. A similar crystal form was obtained for SF5 with initial crystals grown in 1.0 m sodium citrate (unbuffered) and was further optimized by a hanging drop vapor diffusion by equilibration of 1 μl of protein concentrated to 8 mg/ml in buffer B with 1 μl of reservoir solution. The best diffracting crystals were grown in a condition containing 2.0 m sodium citrate + 5% (v/v) glycerol and, as SF4, belonged to space group I422 with a = 116.9, c = 219.1 Å, and Vm of 3.34 Å3 Da3. Native periplasmic WzzFepE (Δ258:266::GSG) was concentrated to 18 mg/ml in the buffer containing 0.1 m Hepes, pH 7.5, 200 mm NaCl, and 2% w/v glycerol. The crystals were grown by equilibration of 1 μl of protein mixed with 1 μl of reservoir solution (0.5 m KCl, 0.05 m MOPS, pH 7.0, 12% (w/v) PEG 4000, and 20% w/v glycerol over 500 μl of reservoir solution) and belonged to space group C2 with a = 208.3, b = 141.23, and c = 136.9 Å, with nine molecules in the asymmetric unit and Vm of 3.82 Å Da−1.
X-ray Data Collection, Structure Solution, and Refinement
The crystal, a Se-Met-labeled wild-type S. flexneri WzzBSF, was flash-cooled in a nitrogen stream to 100 K, and diffraction data were collected to 2.8 Å resolution at the CMCF-1 beamline, Canadian Light Source. Ten of the expected 12 selenium atoms were found using ShelxD (23). Initial phasing was performed by Phaser (24), and the dual-space fragment extension procedure was used to improve the initial phases (25). The initial model was built using the autobuild option in Phenix and subsequently was completed manually using the program Coot. Iterative cycles of refinement and manual building were carried out using Phenix (26) and COOT (27), respectively. The final Rwork and Rfree were 24.5 and 30.3%, respectively.
Diffraction data for a Se-Met-labeled SF4 crystal flash-cooled in a nitrogen stream at 100 K were collected to 1.6 Å at 31-ID beamline, Advanced Photon Source, Argonne National Laboratory. Data integration and scaling were performed with the program suite HKL3000 (28). Initial phases were determined by single-wavelength anomalous dispersion at the selenium peak energy (λ = 0.9794 Å). Eight of the 10 expected selenium atoms in the asymmetric unit were located with the program SHELXD (23), and the initial phases were calculated and improved using the program MLPHARE (29). The model was built by ARP/wARP (30) using the data set to 1.6 Å resolution and completed manually using the program COOT and refined with Phenix. All measured reflections were used in the refinement. The water molecules were added, and the translation-libration-screw model for anisotropic temperature factors was applied (31). The refinement converged to an Rwork value of 21.8% and an Rfree value of 23.8%. The N-terminal histidine tag was not visible in the electron density map and was not included in the final model. Data sets for native SF5 crystals were collected at the CMCF-1 beamline of the Canadian Light Source to 2.6 Å resolution. SF5 structure was solved by molecular replacement with a monomer of SF4 as a search model using the program Phaser. Residues Lys-126–Gln-129 were also poorly resolved and were not built in either subunit. The refinement was carried out using Phenix with translation-libration-screw model for anisotropic temperature factors. The final Rwork values were 23.5 and 29.6%, respectively. In each structure, the tips of several side chains, mostly lysines, arginines, and glutamates, were also disordered and were not included in the models. Both SF4 and SF5 have good stereochemistry as analyzed by the program PROCHECK (32).
Data sets for the native WzzFepE (Δ258:266::GSG) mutant were collected to 2.8 Å resolution at the CMCF-1 beamline, Canadian Light Source, and its structure was solved by molecular replacement with a nonamer of a WT WzzFepE (18) as a search model using the program Phaser. Direct methods (OASIS as part of the CCP4 suite) were used to improve the phases obtained by the molecular replacement (33), and the whole model was rebuilt by and subsequently refined using Phenix. The density for residues 257–266 was very ambiguous, and they were not modeled. Rwork and Rfree converged to 24.2 and 28.9%, respectively. Data collection and refinement statistics are given in Table 1.
TABLE 1.
X-ray data and refinement statistics
| SF4 | SF5 | WzzBSF | WzzFepE (Δ258:266::GSG) | |
|---|---|---|---|---|
| Space group | I422 | I422 | P21 | C2 |
| a, b, c (Å) | 115.7, 115.7, 210.9 | 116.9, 116.9, 219.1 | 81.5, 62.2, 90.7 | 208.3, 141.3, 136.9 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 94.7, 90 | 90, 107.0, 90 |
| Wavelength (Å) | 0.96406 | 0.97949 | 0.97912 | 0.97937 |
| Resolution (Å) | 50–1.6A | 50–2.8A | 50–2.8A | 50–2.8A |
| Completeness (High resolution) (%) | 98.8 (93.1) | 99.4 (100) | 99.9 (100) | 99.9 (100) |
| Rsym (%) | 6.3 (23.5) | 7.88 (39.0) | 7.2 (44) | 11.3 (91.6) |
| I/σ | 63.3 (11.1) | 12.2 (4.3) | 22.6 (3.6) | 13.14 (1.57) |
| Rwork (No. of reflections) | 21.8 | 23.5 | 24.5 | 24.2 |
| Rfree (No. of reflections) | 23.8 | 29.6 | 30.3 | 28.9 |
| No. of observed reflections | 972,196 | 186,262 | 84,954 | 351929 |
| No. of unique reflections | 127,447 | 23,649 | 22,670 | 93614 |
| B-factor (No. of atoms) | ||||
| Protein | 25.2 (3,514) | 29.5 (3,494) | 48.2 (4,895) | 59.403 (16883) |
| Solvent | 33.6 (790) | 24.4 (91) | 33.4 (44) | - |
| Ramachandran plot | ||||
| Favored (%) | 96.4 | 92.6 | 90.3 | 98.0% |
| Allowed regions (%) | 3.4 | 6.2 | 6.9 | 2.0% |
| Generously allowed (%) | 0.0 | 0.2 | 2.3 | 0.7% |
| Disallowed regions (%) | 0.2 | 0.5 | 0.5 | 0.0% |
| r.m.s.d.a | ||||
| Bond lengths (Å) | 0.016 | 0.009 | 0.010 | 0.009 |
| Bond angles (°) | 1.43 | 1.14 | 1.39 | 1.21 |
| Protein Data Bank (PDB) code | 4E29 | 4E2C | 4E2H | 4E2L |
a r.m.s.d., root mean square deviation.
RESULTS
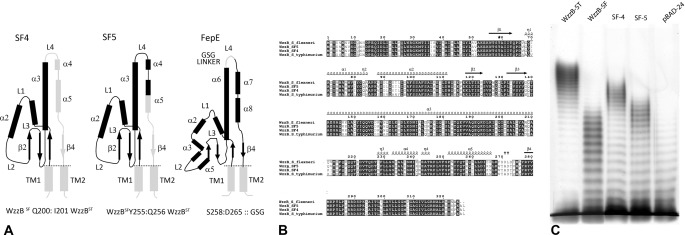
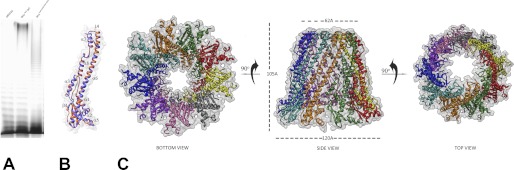
In our previous work, we reported construction of several chimeric chain length regulators based on two parental WzzBs, WzzBSF and WzzBST, which share 70% amino acid identity and yet lead to distinctly different O-antigen modal length distributions (22). The chimeric WzzBs confer the O-antigen modal lengths that are in between those of the two parental WzzBs. We have previously reported the structure of the WzzBST (18). In an effort to understand the structural basis for the observed phenotypic outcomes, we first determined the crystal structure of the periplasmic domain of WzzBSF (amino acids 52–293) and subsequently of two chimeras, which we refer to as SF4, WzzBSF(1–200)-WzzBST(201:327), and SF5, WzzBSF(1–255)-WzzBST(256:327). Both chimeras were composed of larger N-terminal portions of WzzBSF and shorter C-terminal regions of WzzBST (Fig. 1A). The two chimeric proteins share 96% sequence identity (Fig. 1B) and are over 80% identical to the WzzBSF and WzzBST parents (Fig. 1B). Remarkably, these minor differences in the amino acid sequence are sufficient to cause pronounced differences in modal lengths of the resulting O-antigen chains; WzzBSF leads to a modal length of ∼13 repeating units, WzzBST leads to ∼20 repeating units, SF4 leads to ∼17 repeating units, and SF5 leads to ∼15 repeating units (Fig. 1C).
FIGURE 1.
WzzB chimeras and WzzFepE. A, topology of the chimeric WzzB and WzzFepE proteins used for crystallographic analysis indicating the segments used to construct each of the hybrid chain length regulators. The segments in black were taken from WzzBSF, whereas those shaded in gray were from WzzBST (α, α-helix; β, β-strand; L, loop; TM, transmembrane domain). B, sequence alignment of the chimeric WzzB proteins and the corresponding parental WzzBSF (gi 24052434) and WzzBST (gi 154322) proteins. C, silver-stained polyacrylamide gel showing the O-antigen LPS profile of E. coli EVV33 (pMF19) expressing corresponding chimeric proteins. Lane 1, parental WzzBST encoded by pWzzB-ST; lane 2, parental WzzBSF encoded by pWzzB-SF; lane 3, SF4; lane 4; SF5; and lane 5, pBAD24 empty vector control.
Structure of WzzBSF
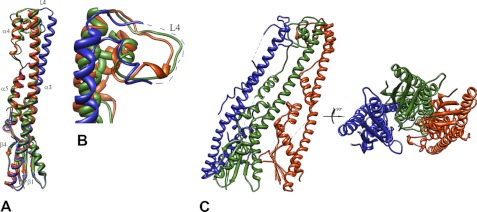
The crystals of WzzBSF were obtained using PEG 4000 as a precipitating agent, and the structure was solved using the experimental phases derived from a single anomalous dispersion experiment to eliminate model bias. Unlike the WzzBST (18), this protein forms open trimers in the crystal (Fig. 2A). The same organization was also previously found for the periplasmic domain of WzzBEC from E. coli W3110, determined at a low resolution (18).3 Interestingly, all three WzzBSF subunits within the trimer are bent to a different degree near their midpoints of the central, long helix α3, resulting in movements of the top parts of the molecules (loop L4) by several Å, with the largest tilt in the C subunit (Fig. 2, B and C). The observed different bending of the central helix may stem from crystal packing differences and a nonidentical environment of the inner and outer protomers in the trimer. This indicates a certain amount of flexibility of the central helix that may play a functional role of the chain regulators in vivo. In the pentameric structure of WzzBST reported previously by our group (18), these conformational changes could not be discerned as the top parts of all the subunits in the pentamer were disordered. The two outside protomers pack tightly to the central protomer along their entire lengths in a manner very similar to that observed previously in the closed oligomers of WzzFepE and WzzE and the open WzzBEc trimer (18). The intersubunit interface area, as calculated by the PISA server (34), is ∼1950 Å2. This interprotomer packing observed in the pentamers of WzzBST is an exception in that the contacts are made predominantly by the base domains with the intersubunit interface area being only 570 Å2 (18).
FIGURE 2.
Graphic representation of WzzBSF. A, superposition of the three protomers of the periplasmic domain of S. flexneri WzzBSF (subunit A is yellow; subunit B is blue; and subunit C is green) reveals the different degree of bending of the helix α3. B, a close-up view of the top part of the molecule showing pronounced shifts in the L4 region. C, two orientations of the WzzBSF trimer (α, α-helix; β, β-strand; L, loop regions). This and subsequent figures were prepared using the program UCSF Chimera (42).
Structures of SF4 and SF5 Chimeras
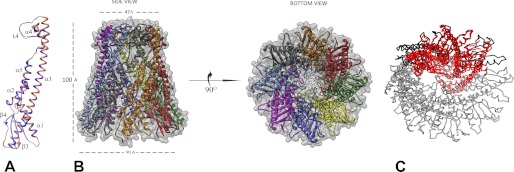
Both of the chimeric proteins were crystallized under high salt conditions and produced similar crystal forms, both belonging to the I422 space group but with dimensions along the c-axis differing by 8 Å (Table 1). Each crystal contained two protomers in the asymmetric unit (Fig. 3A), which assembled into the bell-shaped octamers through 4-fold crystallographic symmetry (Fig. 3B). This oligomerization state is different from the pentameric assembly previously observed for wild-type WzzBST and the open trimers of WzzBSF (Table 2) but very similar to the oligomeric form of WzzE. The bell-shaped oligomer has the height of 95 Å and an outer diameter of 90 Å at the base and 55 Å at the top. These dimensions are similar to those seen for the WzzE octamer (95, 113, and 67 Å, respectively (18)) and significantly larger then the pentameric WzzBST (18). In both chimeric proteins, the loop region facing the internal cavity of the oligomer (loop L1, residues 124–131) is poorly ordered, and consequently, this region was not modeled.
FIGURE 3.
Crystal structure of SF4 chimeric chain length regulator. A, structural alignment of the two crystallographically independent subunits (subunit A is orange, and subunit B is blue). B, a side view and an end-on view of the graphic representation of the octamer obtained by the application of the crystallographic symmetry. C, structural superposition of the WzzBSF trimer (red) on the octamer of SF4. The trimer has smaller radius of curvature than the octamer.
TABLE 2.
Summary of the PCP-1 structures crystallized to date
| Protein | Crystallization condition | Space group | Oligomer | Unit cell parameters | Resolution |
|---|---|---|---|---|---|
| Å | |||||
| WzzBST | 0.1 m Bis-Tris, pH 5.5, 0.2 m ammonium acetate, 25% (w/v) PEG 3350 | C2 | Pentamer | 88.0, 112.7, 170.9 Å, 90, 97.3, 90° | 3.1 |
| ST1 WzzBST(1–95)-WzzBSF(96–325) | 18% PEG 4000, 0.1 m Bis-Tris, pH 5.5 | P21 | Trimer | 83, 61, 92 Å, 90, 95, 90 | 3.3, streaky spots |
| ST5 WzzBST(1–255)-WzzBSF(256–325) | 22% PEG 3350, sodium citrate, pH 5.5 | P212121 | Pentamer | 67, 121, 210 Å, 90, 90, 90° | 3.1, streaky spots |
| SF4 WzzBSF(1:200)-WzzBST(201:327) | 3.0 m sodium formate, pH 6.0 | I422 | Octamer | 115.7, 115.7, 210.9 Å 90, 90, 90° | 1.8 |
| SF5 WzzBSF(1–255) WzzBST(256–327) | 2.0 m sodium citrate, 5% w/v glycerol | I422 | Octamer | 116.9, 116.9,219.1 Å 90, 90, 90° | 2.8 |
| WzzSF | 20% PEG 4000, 0.1 m Hepes, pH 7.5, and 0.1 m MgCl2 | P21 | Trimer | 81.5, 62.2, 90.7 Å, 90, 94.7, 90° | 2.8 |
| WzzB O157:H7 | 10 mm HEPES, pH 7.0, 150 mm NaCl with 2 ml reservoir (18% (w/v) PEG 3350, 5% (v/v) glycerol, 25 mm di-ammonium hydrogen citrate | P21 | Trimer | 81.3, 62.6, 91.7, 90, 91.4, 90° | 3.7 |
| WzzFepE O157:H7 | 30 mmm Hepes, pH 7.0, 1.4 m sodium citrate | P6322 | Nonamer | 139.7, 139.7, 276.7 Å, 90, 90, 120° | 2.7 |
| WzzFepE O157:H7 | 1.5 m sodium citrate, pH 6.5 | P3221 | Nonamer | 160.7, 160.7, 276.9 Å 90, 90, 120° | 3.1 |
| WzzFepE O157:H7 (Δ258:266: GSG) | 0.5 m KCl, 0.05 m MOPS, pH 7.0, 12% (w/v) PEG 4000, and 20% w/v glycerol | C2 | Nonamer | 208.3, 141.3, 136.9 Å, 90, 107.0, 90° | 2.8 |
| WzzE | 18% PEG 3350, 0.05 mm sodium citrate | P21 | Octamer | 107.4, 108.3, 122.6 Å, 90, 102.2, 90° | 2.4 |
The tightly packed protomers form numerous side-to-side contacts via ∼15 hydrogen bonds, five salt bridges, and numerous van der Waals contacts, which extend along the entire length of each protomer. In the base domain, the contacts are made between strand β4 and helix α3 of one protomer with helix α2′ and the bottom of strand β1′ of the other. The long central helix α3 also contributes to the intermolecular contacts forming numerous interactions with the residues in helices α4′ and α5′ of the neighboring subunit. A significant fraction of residues in loop L4 at the top of the oligomer is also engaged in multiple contacts with the residues in the L4 regions of the neighboring protomers. Overall, the surface area of the intersubunit interface seen in the chimeric chain length regulators constitutes 1935 and 1906 Å2 (or SF4 and SF5, respectively).
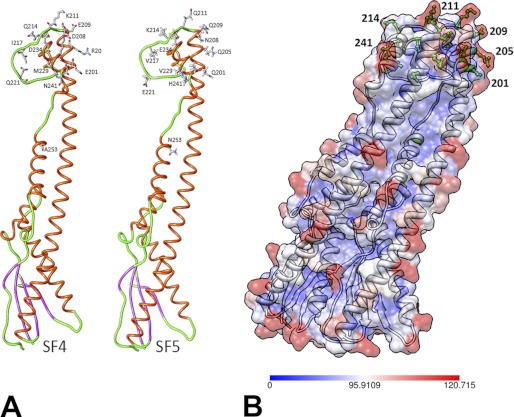
In the previous structural studies of class-1 polysaccharide co-polymerases, the regions of the protomers most distant from the membrane, comprising the top of the central helix and the following residues, were disordered to various extents in all of the crystal structures (18). Our subsequent functional studies demonstrated that this part of the molecule plays an essential role in several O-antigen chain length regulators (22). In the structures of both SF4 and SF5, most of this segment could be reliably modeled (Fig. 4A), providing the first view of the stable conformation of the region most distant from the membrane and capping the top of the oligomer. The conformation of this loop differs slightly between the two molecules in the asymmetric unit and is also somewhat different in the two chimeras, reflecting a degree of inherent flexibility of this loop.
FIGURE 4.
Differences between SF4 and SF5. A, residues differing between the SF4 and SF5 are shown explicitly in stick representation. Helices are colored red, strands are magenta, and loops are green. B, surface accessibility analysis of the two WzzB chimeras with the most buried residues colored in blue, whereas the most solvent-accessible residues colored in red. The most surface-accessible residues differing between the two chimeras are colored in green and are depicted in ball-and-stick representation.
Further structural comparisons revealed that the chimeric proteins are very similar, with the root mean square deviation for all Cα atoms 0.4 Å. The amino acids differing between the two chimeric proteins are predominantly distributed throughout the top of the molecule and are found at the tip of the central long helix α3, in the loop L4 region, and within helix α4. Out of the 13 amino acid differences, most either represent conservative substitutions (I217V, D234E, R205Q, M228L) or are associated with a loss or a gain of a charge (E201Q, D208N, E209Q, K211Q, Q214K, Q221E). One substitution (K237E) leads to a charge reversal, and two are nonconservative substitutions (Q241L, A253N) (Fig. 4A). Six out of these 13 residues (residues 205, 214, 217, 221, 228, and 234) are involved in intersubunit contacts, yet these substitutions do not affect significantly the protomer association. In all of the proteins examined, the internal cavity is rich in positively charged regions, whereas the external surface contains patches of both positive and negative charges. The profile of the solvent-accessible surface revealed that some of the polar residues, which differ between the two chimeras, are highly solvent-exposed (Fig. 4B). Mainly, these are found at the tip of the central long helix α3 (205 and 209), in the first part of the loop L4 region (211 and 214), and within the helix α4 (241) (Fig. 4B).
Structure of WzzFepE Loop L4 Mutant
In our previous work, we found the loop L4 region to be essential for OAg chain length determination as various modifications to this part of the protein resulted in drastic reductions of modality regulated by WzzFepE and completely abolished the function of WzzB (22). We have further investigated this notion and sought to determine whether a more subtle mutation of the loop L4 would be sufficient to alter the modality. To this end, we substituted the first part of L4 in WzzFepE, composed of the amino acids Ser-258–Asp-265, by a Gly-Ser-Gly linker (Fig. 1A). This modification, which affected only the tip of the L4 loop, rendered the chain length regulator to produce the OAg modality comparable with what was observed by the other loop L4 mutants (Fig. 5A). To determine whether this replacement led to conformational changes in the WzzFepE structure that could account for a significantly reduced modal length, we have solved the crystal structure of the periplasmic domain of this WzzFepE(GSG) mutant at 2.8 Å resolution. The mutant was found to crystallize in the C2 space group with nine molecules in the asymmetric unit forming a circular, closed nonamer, the same oligomeric assembly as was previously seen for the wild-type protein (18). As in the wild-type WzzFepE, the electron density for the GSG linker replacing loop L4 could not be unequivocally traced in the electron density map, indicating its flexibility, but the main chain of the wild-type and mutant WzzFepE were nearly identical with a root mean square deviation of 0.37 Å for all Cα atoms (Fig. 5B).
FIGURE 5.
Structural analysis of WzzFepE(ΔSer-258–Asp-265::GSG). A, silver-stained polyacrylamide gel showing the LPS profiles of vector only (lane1), wild-type WzzFepE (lane 2), and WzzFepE(ΔSer-258–Asp-265::GSG) expressed in E. coli K-12 containing the pMF19 to allow the expression of O16 LPS (lane 3). B, structure of the WzzFepE(ΔSer-258–Asp-265::GSG) monomer (orange) overlaid with a structure of the wild-type WzzFepE monomer demonstrating identical main-chain confirmations. C, views of the WzzFepE(ΔSer-258–Asp-265::GSG) nonamer from the bottom, side, and top.
DISCUSSION
What is the oligomeric state of WzzBs in the periplasm? In the previous crystallographic studies (18), the periplasmic domains of the three different PCPs exhibited different oligomerization states, with WzzFepE being a nonamer, WzzE being an octamer, and WzzBST being a pentamer. Although the protomer-protomer packing in WzzFepE and WzzE was similar and the contacts extended along the entire length of the protomers, those of WzzBST interacted only through their base domains. The observations by crystallography of a variety of oligomeric sizes as well as circular and open oligomers, by electron microscopy of circular oligomers interpreted as hexamers (19), and by cross-linking studies of higher molecular weight oligomers for the Wzz from S. flexneri (14, 35) raise the question of what the in vivo oligomerization state of the polysaccharide co-polymerases really is. In solution, the purified soluble periplasmic domains of PCPs form smaller oligomers, likely dimers as judged by size exclusion chromatography (Ref. 18 and present work, data not shown). Moreover, while pursuing these studies, we have obtained other crystal forms of these and other chimeric WzzB proteins. Although the poor quality of these crystals did not allow for the high resolution structure determination, the molecular replacement solutions indicated a different oligomeric assembly from that seen for either WzzB chimera SF4 or WzzB chimera SF5. The periplasmic domain of ST1 chimeric protein (WzzBST(1–95)-WzzBSF(96–325) was found to be a trimer, whereas that of ST5 chimeric protein (WzzBST(1–255)-WzzBSF(256–325) was found to be a pentamer (Table 2). Although these soluble domains exhibit a propensity to assemble into the oligomers of various compositions, comparing crystallization conditions of the crystals (Table 2), we conclude that the number of subunits and the assembly into the open or the closed forms is not strictly dependent on the given crystallization condition. Both high salt and high polyethylene glycol conditions appear to be conducive to the formation of closed and open oligomers (Table 2).
Because we have observed the circular octameric oligomers of WzzB chimeras in the crystals, we have reanalyzed the electron micrographs of Larue et al. (19) of the two-dimensional crystals of various detergent-solubilized O-antigen chain length regulators. Their estimate of the largest diameter of WzzB from the electron microscopy reconstructions was ∼100 Å. This was smaller than the dimensions of the WzzFepE nonamer, slightly smaller than the WzzE octamer, and larger than the WzzBST pentamer. The authors derived a model of a hexamer based on the crystal structure of WzzE that measured 100 Å at the base, 95 Å in length, and 55 Å at the top and concluded that this model had the size compatible with the EM reconstruction. Here, we show that the WzzB chimeras SF4 and SF5 form octamers in the crystal with dimensions of 95 Å across the base, 100 Å in length, and 50 Å across the top of the octameric ring. These octamers are therefore in a very good agreement with the EM data. Moreover, removing two protomers from this octamer would result in a smaller oligomer size than constructed by Larue et al. (19), too small to fit the EM data. To further investigate this, we have analyzed the curvature of the open trimers of WzzBSF. Superposition of this trimer on the SF4 octamer shows that it displays higher curvature (smaller radius) (Fig. 3C). We have computationally extended the trimer following its curvature and found that this accommodates a hexamer. However, the dimensions of this artificially constructed hexamer would be ∼70 Å, which is inconsistent with the EM studies of the full-length proteins (19) and with the number of protomers in a circular oligomers observed in the crystal structures. Clearly, a crystal structure of the full-length WzzB is needed to clarify the controversy surrounding the real oligomerization state of these molecules and to determine whether the size of the oligomer is indeed playing a role in the determination of the length of the carbohydrate polymer. However, the fact that the size of the octamers is in agreement with that derived for the full-length proteins resuspended in proteoliposomes supports the view that octamers represent the physiologically relevant assembly of WzzB. Interestingly, the cytosolic kinase domain from a PCP-IIa family member Wzc was also found to assemble as octamers (36). Wza, an outer membrane lipoprotein, proposed to form a trans-envelope complex with Wzc, also adopts the octameric organization in the crystals (37).
The EM images of full-length WzzFepE reconstituted in lipid bilayers indicated particles of ∼100 Å diameter at the base (19), as compared with 115 Å of the nonamer in the crystal, which the authors suggested would be more consistent with a hexamer. Here, we report the crystal structure of a WzzFepE mutant crystallized under significantly different crystallization conditions than the wild type and in a different space group (C2 versus P6322 or P3221) and yet forming a nonamer with the same overall dimensions as that of the wild-type WzzFepE oligomer. Although we cannot exclude the possibility that the presence of the transmembrane regions might affect the number of protomers within the oligomer through their interaction with the surrounding lipid bilayers, we surmise that the same oligomeric arrangement with nine protomers is also likely for the full-length protein. We have remeasured the distances between the particles in the negatively stained EM micrographs (Fig. 4F in Ref. 19) using the specified calibrated pixel size of 3.84 Å and obtained the distance between the centers of the neighboring subunits of 115 Å for a full-length WzzFepE, which is very similar to the size of the nonamer observed in the crystals.
Another interesting finding that emerged from the structural analysis of WzzFepE (Δ258:266::GSG) mutant is that it exhibits the same main-chain conformation and oligomeric state, yet leads to a drastically different modal length distribution. We previously considered that the oligomer size might be a significant factor in the OAg modal length determination mechanism, with the larger oligomers producing polymers containing more repeating O-units (12, 18). Kintz and Goldberg (41) recently reported that a residue involved in maintaining the oligomer stability of WzzFepE homologue from Pseudomonas aeruginosa (Wzz2) is partially responsible for the specification of a given O-antigen chain length. The structure of our WzzFepE mutant, which leads to much shorter O-antigen chains and yet forms the same oligomer as the wild-type WzzFepE, strongly suggests that the oligomeric size alone is not a sole factor of the modal length determination. Instead, our finding reiterates the importance of the flexible region at the top of the molecule in the mechanism of the OAg chain length control. This is interesting in light of the reported observation that OAg modality can be reproduced in vitro with only Wzy polymerase and WzzB chain regulator present (38). Loop L4 region is positioned too far from the inner membrane to form physical protein-protein interactions with the Wzy polymerase, and therefore, L4 is likely to serve as one of the sugar-binding faces interacting with the polar O-antigen subunits. Indeed, most of the differences between SF4 and SF5 chimeric proteins, which produce the O-antigens with distinctly different modal lengths, also fall in this region (Fig. 4A). As subtle as the differences are, they would be sufficient to confer differential affinities to the O-antigen-PCP interaction. Recently, it was shown that the Wzy polymerase-O-antigen interactions in Pseudomonas are very sensitive to even conservative amino acid changes in specific positions as even Arg-to-Lys mutation led to a complete loss of the polymerase function, which was attributed to the loss of sugar binding (39). Clearly, the top of the molecule is not the only site of O-antigen-PCP interaction. In our previous study, we established the importance of the 269–274 segment. This region is the same in SF4 and SF5 and contains only one negatively charged residue (Asp-273); however, in WzzBSF, it contains three negatively charged residues (Asp-270, Asp-271, and Asp-274). This patch would thus be expected to form a different network of interactions with the incoming repeat units, perhaps leading to a weaker affinity between the O-antigen and the chain length regulator.
In summary, our work provides another structural insight into the PCP-1 machinery involved in the secretion of high molecular weight sugar polysaccharides at the atomic resolution. It demonstrates previously unseen oligomerization states for the WzzB periplasmic domains, of which the octameric assembly appears to be more physiologically relevant than the previously reported pentameric arrangement, and is in agreement with the EM data reported for the full-length proteins. The structures of the chimeric domains show for the first time the structural details of the top region of the oligomer, which is involved in PCP function. Our structural analysis of the WzzFepE mutant also brings forth two important points. First, it shows that the mutation of the L4 region does not exert its phenotypic effect through the structural effects on the individual protomers. Second, the finding that the periplasmic domains of both wild-type and mutant FepE were found to be nonamers despite the wide differences of the crystallization conditions provides the evidence that this arrangement may indeed represent the preferred physiological state of the WzzFepE O-antigen chain length regulator.
Acknowledgments
We thank Dr. Xiang Ruan and Dr. Miguel Valvano for assistance with the LPS extraction and analysis and Yunge Li for invaluable advice on crystal optimization. We thank S. Labiuk for help with data collection. The data collection described in this study was performed at the Canadian Light Source, which is supported by the Natural Sciences and Engineering Research Council, National Research Council, Canadian Institutes of Health Research, and the University of Saskatchewan.
This work was supported by Grant MOP-89787 from the Canadian Institutes of Health Research (to M. C.).
The atomic coordinates and structure factors (codes 4E29, 4E2C, 4E2H, and 4E2L) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
A. Tocilj, A. Matte, C. Munger, and M. Cygler, unpublished results.
- OAg
- O-antigen
- PCP
- polysaccharide co-polymerases
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Murray G. L., Attridge S. R., Morona R. (2003) Regulation of Salmonella typhimurium lipopolysaccharide O-antigen chain length is required for virulence: identification of FepE as a second Wzz. Mol. Microbiol. 47, 1395–1406 [DOI] [PubMed] [Google Scholar]
- 2. Murray G. L., Attridge S. R., Morona R. (2006) Altering the length of the lipopolysaccharide O-antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. J. Bacteriol. 188, 2735–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Duerr C. U., Zenk S. F., Chassin C., Pott J., Gütle D., Hensel M., Hornef M. W. (2009) O-antigen delays lipopolysaccharide recognition and impairs antibacterial host defense in murine intestinal epithelial cells. PLoS Pathog. 5, e1000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Paixão T. A., Roux C. M., den Hartigh A. B., Sankaran-Walters S., Dandekar S., Santos R. L., Tsolis R. M. (2009) Establishment of systemic Brucella melitensis infection through the digestive tract requires urease, the type IV secretion system, and lipopolysaccharide O-antigen. Infect. Immun. 77, 4197–4208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. West N. P., Sansonetti P., Mounier J., Exley R. M., Parsot C., Guadagnini S., Prévost M. C., Prochnicka-Chalufour A., Delepierre M., Tanguy M., Tang C. M. (2005) Optimization of virulence functions through glucosylation of Shigella LPS. Science 307, 1313–1317 [DOI] [PubMed] [Google Scholar]
- 6. Saldías M. S., Ortega X., Valvano M. A. (2009) Burkholderia cenocepacia O-antigen lipopolysaccharide prevents phagocytosis by macrophages and adhesion to epithelial cells. J. Med. Microbiol. 58, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 7. Clay C. D., Soni S., Gunn J. S., Schlesinger L. S. (2008) Evasion of complement-mediated lysis and complement C3 deposition are regulated by Francisella tularensis lipopolysaccharide O-antigen. J. Immunol. 181, 5568–5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Samuel G., Reeves P. (2003) Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 338, 2503–2519 [DOI] [PubMed] [Google Scholar]
- 9. Valvano M. A. (2003) Export of O-specific lipopolysaccharide. Front. Biosci. 8, s452–s471 [DOI] [PubMed] [Google Scholar]
- 10. Raetz C. R., Whitfield C. (2002) Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cuthbertson L., Mainprize I. L., Naismith J. H., Whitfield C. (2009) Pivotal roles of the outer membrane polysaccharide export and polysaccharide co-polymerase protein families in export of extracellular polysaccharides in Gram-negative bacteria. Microbiol. Mol. Biol. Rev. 73, 155–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morona R., Purins L., Tocilj A., Matte A., Cygler M. (2009) Sequence-structure relationships in polysaccharide co-polymerase (PCP) proteins. Trends Biochem. Sci. 34, 78–84 [DOI] [PubMed] [Google Scholar]
- 13. Batchelor R. A., Haraguchi G. E., Hull R. A., Hull S. I. (1991) Regulation by a novel protein of the bimodal distribution of lipopolysaccharide in the outer membrane of Escherichia coli. J Bacteriol. 173, 5699–5704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Daniels C., Morona R. (1999) Analysis of Shigella flexneri Wzz (Rol) function by mutagenesis and cross-linking: Wzz is able to oligomerize. Mol. Microbiol. 34, 181–194 [DOI] [PubMed] [Google Scholar]
- 15. Rosenow C., Esumeh F., Roberts I. S., Jann K. (1995) Characterization and localization of the KpsE protein of Escherichia coli K5, which is involved in polysaccharide export. J. Bacteriol. 177, 1137–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paulsen I. T., Beness A. M., Saier M. H., Jr. (1997) Computer-based analyses of the protein constituents of transport systems catalyzing export of complex carbohydrates in bacteria. Microbiology 143, 2685–2699 [DOI] [PubMed] [Google Scholar]
- 17. Morona R., Van Den Bosch L., Daniels C. (2000) Evaluation of Wzz/MPA1/MPA2 proteins based on the presence of coiled-coil regions. Microbiology 146, 1–4 [DOI] [PubMed] [Google Scholar]
- 18. Tocilj A., Munger C., Proteau A., Morona R., Purins L., Ajamian E., Wagner J., Papadopoulos M., Van Den Bosch L., Rubinstein J. L., Féthière J., Matte A., Cygler M. (2008) Bacterial polysaccharide co-polymerases share a common framework for control of polymer length. Nat. Struct. Mol. Biol. 15, 130–138 [DOI] [PubMed] [Google Scholar]
- 19. Larue K., Kimber M. S., Ford R., Whitfield C. (2009) Biochemical and structural analysis of bacterial O-antigen chain length regulator proteins reveals a conserved quaternary structure. J. Biol. Chem. 284, 7395–7403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tang K. H., Guo H., Yi W., Tsai M. D., Wang P. G. (2007) Investigation of the conformational states of Wzz and the Wzz·O-antigen complex under near-physiological conditions. Biochemistry 46, 11744–11752 [DOI] [PubMed] [Google Scholar]
- 21. Franco A. V., Liu D., Reeves P. R. (1998) The Wzz (Cld) protein in Escherichia coli: amino acid sequence variation determines O-antigen chain length specificity. J. Bacteriol. 180, 2670–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kalynych S., Ruan X., Valvano M. A., Cygler M. (2011) Structure-guided investigation of lipopolysaccharide O-antigen chain length regulators reveals regions critical for modal length control. J. Bacteriol. 193, 3710–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schneider T. R., Sheldrick G. M. (2002) Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 24. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yao D. Q., Huang S., Wang J. W., Gu Y. X., Zheng C. D., Fan H. F., Watanabe N., Tanaka I. (2006) SAD phasing by OASIS-2004: case studies of dual-space fragment extension. Acta Crystallogr. D Biol. Crystallogr. 62, 883–890 [DOI] [PubMed] [Google Scholar]
- 26. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 28. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. (2006) HKL-3000: the integration of data reduction and structure solution: from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 29. Otwinowski Z. (1991) Maximum likelihood refinement of heavy atom parameters in isomorphous replacement and anormalous scattering in Proceedings of the CCP4 Study Weekend (Wolf W., Evans P. R., Leslie A. G., eds), pp. 80–88, Daresbury Laboratory, Warrington, UK [Google Scholar]
- 30. Perrakis A., Morris R., Lamzin V. S. (1999) Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 31. Winn M. D., Isupov M. N., Murshudov G. N. (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 32. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) Procheck: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 33. He Y., Yao D. Q., Gu Y. X., Lin Z. J., Zheng C. D., Fan H. F. (2007) OASIS and molecular-replacement model completion. Acta Crystallogr. D Biol. Crystallogr. 63, 793–799 [DOI] [PubMed] [Google Scholar]
- 34. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 35. Purins L., Van Den Bosch L., Richardson V., Morona R. (2008) Coiled-coil regions play a role in the function of the Shigella flexneri O-antigen chain length regulator WzzpHS2. Microbiology 154, 1104–1116 [DOI] [PubMed] [Google Scholar]
- 36. Bechet E., Gruszczyk J., Terreux R., Gueguen-Chaignon V., Vigouroux A., Obadia B., Cozzone A. J., Nessler S., Grangeasse C. (2010) Identification of structural and molecular determinants of the tyrosine-kinase Wzc and implications in capsular polysaccharide export. Mol. Microbiol. 77, 1315–1325 [DOI] [PubMed] [Google Scholar]
- 37. Dong C., Beis K., Nesper J., Brunkan-Lamontagne A. L., Clarke B. R., Whitfield C., Naismith J. H. (2006) Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature 444, 226–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Woodward R., Yi W., Li L., Zhao G., Eguchi H., Sridhar P. R., Guo H., Song J. K., Motari E., Cai L., Kelleher P., Liu X., Han W., Zhang W., Ding Y., Li M., Wang P. G. (2010) In vitro bacterial polysaccharide biosynthesis: defining the functions of Wzy and Wzz. Nat. Chem. Biol. 6, 418–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Islam S. T., Gold A. C., Taylor V. L., Anderson E. M., Ford R. C., Lam J. S. (2011) Dual conserved periplasmic loops possess essential charge characteristics that support a catch-and-release mechanism of O-antigen polymerization by Wzy in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 286, 20600–20605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Horton R. M., Hunt H. D., Ho S. N., Pullen J. K., Pease L. R. (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77, 61–68 [DOI] [PubMed] [Google Scholar]
- 41. Kintz E. N., Goldberg J. B. (2011) Site-directed mutagenesis reveals key residue for O-antigen chain length regulation and protein stability in Pseudomonas aeruginosa Wzz2. J. Biol. Chem. 286, 44277–44284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]