Background: microRNA172 (miR172) accumulation is regulated by ambient temperature.
Results: FCA promotes miR172 processing by directly binding to the primary miR172 transcripts at warm temperature.
Conclusion: FCA regulates miR172 accumulation at the processing step in response to changes in ambient temperature.
Significance: The FCA-miR172 regulon is crucial for understanding thermosensory flowering in Arabidopsis.
Keywords: Arabidopsis, Development, MicroRNA, Plant Biochemistry, Plant Molecular Biology, FCA, Flowering
Abstract
Ambient temperature fluctuates diurnally and seasonally. It profoundly influences the timing of flowering in plants. The floral integrator FLOWERING LOCUS T (FT) mediates ambient temperature signals via the thermosensory pathway in Arabidopsis flowering. microRNA172 (miR172), which promotes flowering by inducing FT, also responds to changes in ambient temperature. However, it is largely unknown how miR172 integrates ambient temperature signals into the flowering genetic network. Here, we show that Arabidopsis RNA-binding protein FCA promotes the processing of primary microRNA172 transcripts (pri-miR172) in response to changes in ambient temperature. Ambient temperature regulates miR172 biogenesis primarily at the pri-miR172 processing step. miR172 abundance is elevated at 23 °C but not at 16 °C. miR172 accumulation at 23 °C requires functional FCA. FCA binds to the flanking sequences of the stem-loop within the pri-miR172 transcripts via the RNA recognition motif. FCA also binds to the primary transcripts of other temperature-responsive miRNAs, such as miR398 and miR399. Notably, levels of FCA mRNAs and proteins increase at 23 °C but remain low at 16 °C, supporting the role of FCA in temperature perception. Our data show that FCA regulation of miR172 processing is an early event in the thermosensory flowering pathway. We propose that the FCA-miR172 regulon provides an adaptive strategy that fine tunes the onset of flowering under fluctuating ambient temperature conditions.
Introduction
Ambient temperature influences diverse aspects of plant developmental processes (1, 2). Temperature extremes, such as cold and heat, initiate signaling cascades and metabolic adaptation responses that enhance plant survival. These temperature effects may be associated with recent concerns about changes in flowering time of many plant species in the temperate zones, possibly due to global warming (3, 4).
Flowering is a major developmental trait strongly influenced by ambient temperature. Molecular genetic studies in Arabidopsis have shown that even small changes in ambient temperature significantly affect flowering time (1, 2). Arabidopsis flowering is delayed at low temperatures but accelerated at high temperatures (5, 6). The effects of ambient temperature on flowering are mediated by the floral integrator FT2 via the thermosensory flowering pathway (5, 6). Recent studies have shown that various flowering genes, including FCA, FVE, SHORT VEGETATIVE PHASE (SVP), FLOWERING LOCUS M (FLM), EARLY FLOWERING 3 (ELF3), and TERMINAL FLOWER 1 (TFL1), are involved in this flowering pathway (5–8). It has also been reported that the histone H2A variant H2A.Z also plays a role in temperature perception in flowering (9). However, it is largely unknown how the components of the thermosensory flowering pathway are functionally interrelated.
FCA was originally identified as a component of the autonomous flowering pathway (10, 11). FCA regulates FLOWERING LOCUS C (FLC) expression by modulating proximal polyadenylation site selection of noncoding antisense RNAs (12–14). FCA also regulates its own expression by modulating alternative RNA cleavage and polyadenylation of the primary FCA transcripts (15, 16). It functions through physical interactions with FY, a WD repeat-containing protein that is closely related to the yeast RNA 3′-end processing protein factor Pfs2p (17). In addition, RNA 3′-end processing activity of FCA is required for silencing of multiple RNAs and subsequent DNA methylation in a locus-dependent manner (18–20), suggesting that molecular mechanisms underlying the FCA function are more complicated than previously expected.
MicroRNA172 (miR172) is one of the most extensively studied miRNAs in plants, particularly in flowering transition and floral development (21–26). It represses a small group of genes encoding APETALA2 (AP2)-like transcription factors, including AP2, TARGET OF EAT 1 (TOE1), TOE2, TOE3, SCHLAFMÜTZE (SMZ), and SCHNARCHZAPFEN (SNZ). It promotes photoperiodic flowering by inducing FT in a CONSTANS (CO)-independent manner (22–24). Interestingly, miR172 accumulation is ambient temperature-responsive (27). Its abundance is lower at 16 °C than at 23 °C. Overexpression of miR172 causes temperature-insensitive flowering, suggesting that miR172 may function in thermosensory flowering. Of particular interest is the observation that miR172 abundance is reduced significantly in fca mutants (22), raising the possibility that miR172 is involved in the FCA-mediated thermosensory flowering.
In this work, we demonstrate that FCA regulates the processing of primary transcripts of miR172 and other temperature-responsive miRNAs, such as miR398 and miR399 (27). FCA binds to the flanking sequences of the stem-loop in the primary miRNA transcripts. FCA activity is modulated by ambient temperature at both the transcriptional and protein levels. These observations support that FCA serves as a platform for miRNA processing by incorporating temperature signals into the miRNA processing machinery.
EXPERIMENTAL PROCEDURES
Plant Materials and Growth Conditions
Arabidopsis thaliana lines used were in a Col-0 background unless specified otherwise. Plants were grown in soil or on 1/2× Murashige and Skoog (MS)-agar plates (hereafter referred to as MS-agar plates) in a controlled culture room at 23 °C under long days (16 h light and 8 h dark). To examine the effects of ambient temperatures on gene expression, plants were grown at either 16 or 23 °C for the indicated times. To examine the kinetic patterns of miRNA accumulation, plants were grown at either 23 °C for 1 week or at 16 °C for 2 weeks. The two plant groups were considered to be in similar developmental stages as evaluated by comparing overall sizes and shapes of plants, leaf morphology and numbers, and formation of abaxial trichomes (28, 29).
Generation of miR172-overproducing Transgenic Plants
The miR172b gene sequence covering a 1410-bp transcription unit was amplified from a Col-0 cDNA pool by RT-PCR as described previously (22). The miR172b gene construct was transformed into Col-0 plants and several flowering time mutants, such as fca-9 and fve-3. Approximately 30 independent transgenic plants that exhibit early flowering were isolated from each transformation. Homozygotic transgenic plants having a single copy T-DNA insertional event were identified by herbicide selection for two additional generations and analysis of segregation ratios.
Analysis of Gene Transcript Levels
Gene transcript levels were determined by quantitative real time RT-PCR (qRT-PCR). qRT-PCRs were carried out in 96-well blocks with an Applied Biosystems 7500 Real-Time PCR System (Foster City, CA) using the SYBR Green I Master Mix in a reaction volume of 25 μl. All qRT-PCRs were carried out in biological triplicates using RNA samples extracted from three independent plant materials treated under identical growth conditions and gene-specific primer pairs listed in supplemental Table S1. Data processing and determination of reaction specificities were carried out as described previously (30).
miRNA Northern Blot Analysis
Total RNA samples were extracted from appropriate plant materials by the TRIzol method according to the procedure provided by the supplier (Invitrogen) but with a few modifications as described previously (22). miRNA Northern blot analyses were carried out using the ULTRA-Hyb Oligo solution according to the procedure provided by the manufacturer (Ambion, Houston, TX). Oligonucleotide probes were 5′-end-labeled with [γ-32P]ATP and T4 polynucleotide kinase (Takara, Shiga, Japan). An oligonucleotide complementary to 5 S rRNA was also processed in the same manner and used as quality control for RNA preparations. Quantitation of bands on the blots was carried out by densitometry of images using the Labwork image acquisition and analysis software (Media Cybernetics, San Diego, CA) installed in the system. At least three representative blots were counted and averaged for each quantitation.
Preparation of MBP-FCA Fusion Protein
The FCA gene was subcloned into the pMAL-c2X Escherichia coli expression vector (New England Biolabs, Ipswich, MA) containing a maltose-binding protein (MBP)-coding sequence. Recombinant MBP and MBP-FCA proteins were synthesized in the E. coli Rosetta2 (DE3) pLysS strain (Novagen, Madison, WI) and partially purified as described previously (30).
In Vitro Pulldown Assays
DNA fragments corresponding to MIR172 gene transcripts were amplified by PCR from genomic DNA and subcloned into the pGEM-7Zf (+) E. coli expression vector (Promega, Madison, WI) under the control of the SP6 and T7 promoters. The vector constructs were used as templates for in vitro transcription in the presence of [α-32P]UTP to generate 32P-labeled MIR172 RNAs. The in vitro binding of FCA to RNAs was performed as described previously (31).
RNA in Vitro Pulldown Assay
Biotin-labeled MIR172 RNAs were in vitro transcribed using the Biotin RNA Labeling Mix (Roche Applied Science) and T7 RNA polymerase (Roche Applied Science), treated with RNase-free DNase I (New England Biolabs), and purified with an RNeasy Mini kit (Qiagen, Valencia, CA). Nuclear extract was obtained from 35S:FCA transgenic plants overexpressing the MYC-FCA fusion gene as described (32). Two micrograms of biotin-labeled RNAs and nuclear extract were mixed in pulldown buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 2 mm DTT, 0.05% Nonidet P-40) containing protease inhibitor mixture (Sigma-Aldrich) and incubated for 6 h at 4 °C, and then 30 μl of washed streptavidin-agarose beads (Roche Applied Science) were added to each binding reaction and further incubated for 2 h at 4 °C. Beads were washed briefly five times, boiled in SDS buffer, and then subjected to Western blots against anti-MYC antibody (Millipore, Billerica, MA).
Electrophoretic Mobility Shift Assay (EMSA)
EMSA assays were performed as described previously (31) using recombinant MBP-FCA fusion protein. The 32P-labeled RNAs were incubated for 30 min at room temperature with 1 μg of MBP-FCA fusion protein in binding buffer (10 mm Tris-HCl, pH 7.6, 50 mm NaCl, 1 mm EDTA, 5 mm DTT, 5% glycerol) supplemented with 100 ng of poly(dI-dC) in the presence or absence of competitor RNAs. The reaction mixtures were resolved on a 4% non-denaturing polyacrylamide gel at 100 V for 1 h at room temperature. The gels were dried on Whatman 3MM paper and exposed to x-ray films.
RNA Immunoprecipitation (RIP) Assay
RIP assays were carried out essentially as described previously (33). Two-week-old 35S:FCA transgenic plants grown on MS-agar plates were used for preparation of nuclear extracts. To collect protein-RNA complexes, an anti-MYC antibody (Millipore) and protein A-agarose beads (Millipore) were added to the nuclear extracts. After elution of protein-RNA complexes, DNA and proteins were removed from the protein-RNA eluent by using RNase-free DNase I (New England Biolabs) and proteinase K (Roche Applied Science), and RNA molecules were purified from the eluent using the RNeasy Mini kit (Qiagen). First strand cDNA was synthesized using the Moloney murine leukemia virus reverse transcriptase (Promega) and then subjected to qRT-PCRs.
RESULTS
miR172 in Thermosensory Flowering
Ambient temperature influences flowering time through the FT-mediated thermosensory pathway (5). The miR172 signaling pathway promotes flowering primarily by inducing FT gene (22–24). In addition, miR172 abundance is affected by changes in ambient temperature (27), raising a possibility that miR172 is linked with thermosensory flowering.
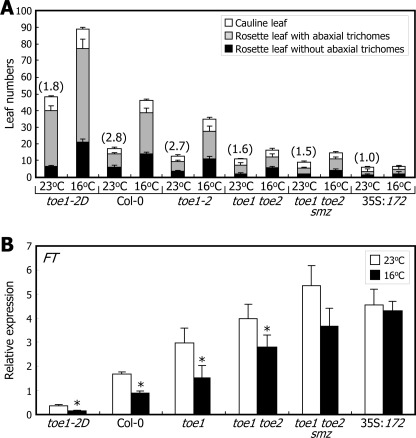
In support of this hypothesis, effects of ambient temperature on flowering initiation were reduced significantly in Arabidopsis plants overproducing miR172 (35S:172) (Fig. 1A). The temperature effects were also reduced in the miR172 target gene mutants, particularly in higher order mutants, such as toe1 toe2 double and toe1 toe2 smz triple mutants. Similarly, the sensitivity of FT transcription to ambient temperature was diminished in the 35S:172 transgenic plants and toe1 toe2 smz triple mutants (Fig. 1B), indicating that miR172 mediates ambient temperature signals in flowering time control.
FIGURE 1.
Roles of miR172 and its targets in thermosensory flowering. A, flowering phenotypes and developmental phase transitions. Plants were grown in soil at either 23 or 16 °C until flowering under long days. Leaf numbers in individual leaf developmental phases were counted and averaged using 30 plants for each plant genotype. Bars indicate S.E. The numbers in parentheses refer to the ratios of total leaf numbers at 16 and 23 °C (16/23 °C). B, levels of FT mRNA. Ten-day-old whole plants grown at either 23 or 16 °C were harvested at ZT16 for total RNA extraction. mRNA levels were examined by qRT-PCR (t test; *, p < 0.05). Error bars indicate S.E.
Ambient Temperature Regulation of miR172 Abundance
We set up a series of experiments to determine whether miR172 is involved in ambient temperature regulation of flowering.
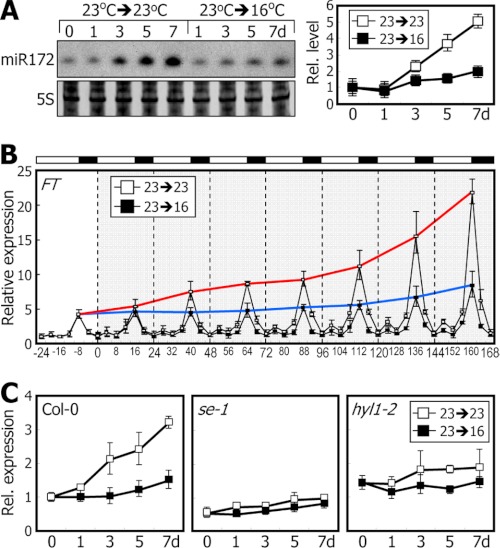
We first measured miR172 levels at different zeitgeber time (ZT) points in Col-0 plants grown at different temperatures. The miR172 level is very low during the early vegetative phase but is rapidly elevated during the late adult vegetative phase (29, 34). For accurate measurements of miR172 abundance, it was important to slow down the rate of plant growth. For this reason, all miR172 Northern blot analyses were carried out using plants grown on sucrose-deficient media. The miR172 levels were higher in plants grown at 23 °C than at 16 °C at all ZT points (supplemental Fig. S1). To examine systematically the temperature effects, 7-day-old plants grown at 23 °C were transferred to 16 °C, and kinetic levels of miR172 and FT mRNA were measured. Whereas the miR172 levels increased gradually in plants grown at 23 °C, the rate of increase was significantly lower in plants grown at 16 °C (Fig. 2A). Kinetic patterns of FT expression were also similar to those of miR172 accumulation (Fig. 2B) but disappeared in serrate-1 (se-1) (35) and hyponastic leaves 1-2 (hyl1-2) (36) mutants having defects in miRNA biogenesis (Fig. 2C). We also observed a correlation between the temporal accumulation patterns of FT mRNA and miR172 during the transition from 16 to 23 °C (supplemental Fig. S2).
FIGURE 2.
miR172 level is elevated at 23 °C. Levels of miR172 and FT mRNA levels were examined by Northern blot analysis and qRT-PCR, respectively. Error bars indicate S.E. d, days; Rel., relative. A and B, kinetic measurements of miR172 abundance (A) and FT mRNA levels (B) after transfer from 23 to 16 °C. Seven-day-old plants grown at 23 °C were either maintained at 23 °C or transferred to 16 °C. Whole plants were harvested at different ZT points for up to 7 days. In B, the red and blue lines indicate peaks of FT expression in plants grown at 23 and 16 °C, respectively. C, FT mRNA levels in se-1 and hyl1-2 mutants. Seven-day-old plants grown at 23 °C were either maintained at 23 °C or transferred to 16 °C. Whole plants were harvested at ZT16 for up to 7 days.
We constructed transgenic plants overexpressing TOE1-MYC and miR172-resistant TOE1 (rTOE1)-MYC fusion genes, resulting in 35S:TOE1 and 35S:rTOE1 transgenic plants, respectively (supplemental Fig. S3, A, B, C, and D), and the levels of TOE1 protein and FT mRNA were measured in the transgenic plants grown at different temperatures. The results showed that although the temperature effects on the levels of TOE1 protein and FT mRNA were relatively strong in the 35S:TOE1 plants the effects disappeared in the 35S:rTOE1 plants (supplemental Fig. S3, E and F). Our observations strengthen the possibility that miR172 and its targets are responsible for the thermosensory regulation of FT expression.
FCA Is Essential for miR172 Accumulation at 23 °C
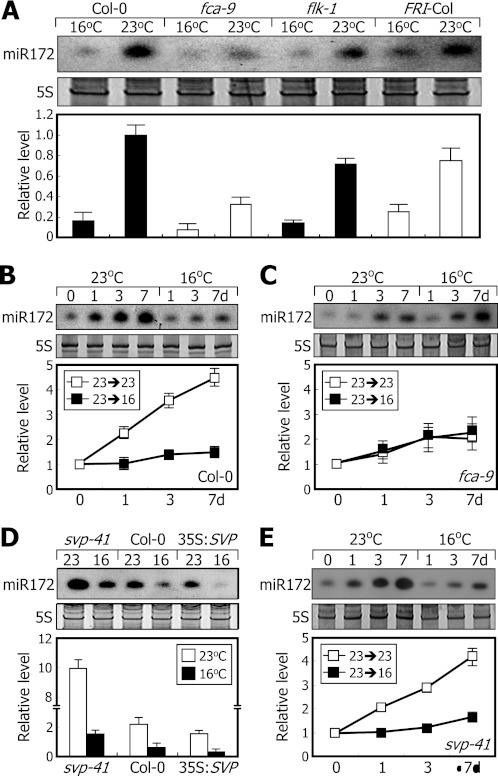
It has been shown that miR172 levels are relatively low in fca mutant (22). We therefore investigated whether FCA is required for miR172 accumulation at 23 °C. At 16 °C, miR172 levels were lower in the fca-9 and flk-1 mutants (37) as observed in Col-0 plant (Fig. 3A). The levels were also lower in FRI-Col plants in which FRIGIDA gene was introgressed into Col-0 plants (38). At 23 °C, whereas the miR172 levels were significantly elevated in Col-0, flk-1, and FRI-Col plants, they were elevated only slightly in the fca-9 mutant (Fig. 3A). We also examined the kinetic patterns of miR172 accumulation in the mutants that were grown at 23 °C and then exposed to 16 °C. The miR172 levels were lower in the fca-9 mutant even at 23 °C (Fig. 3, B and C), indicating that miR172 accumulation at 23 °C requires FCA but is independent of FLC.
FIGURE 3.
FCA is required for miR172 accumulation at 23 °C. miR172 levels were examined by Northern blot analysis. Error bars indicate S.E. A, relative miR172 levels in flowering time mutants. Ten-day-old plants grown at either 23 or 16 °C were harvested at ZT16 for extraction of total RNA. B and C, kinetic measurements of miR172 levels in Col-0 plant (B) and fca-9 mutant (C). Seven-day-old plants grown at 23 °C were further grown at either 23 or 16 °C for up to 7 days (d). Whole plants were harvested at ZT16 for extraction of total RNA. D, miR172 levels in 35S:SVP transgenic and svp-41 mutant plants. Ten-day-old plants grown at either 23 or 16 °C were harvested at ZT16 for extraction of total RNA. E, kinetic measurements of miR172 levels in svp-41 mutant. Preparation of plant materials and extraction of total RNA were carried out as described in B and C.
The SVP gene plays a role in the thermosensory flowering pathway (7). We found that at 23 °C miR172 levels were significantly higher in svp-41 mutant but detectably lower in SVP-overexpressing transgenic plants (35S:SVP) than in Col-0 plants when grown (Fig. 3D). When plants were exposed to 16 °C, miR172 levels were still reduced in the svp-41 mutant and 35S:SVP transgenic plants as observed in Col-0 plants. In addition, mutation of the SVP gene does not influence miR172 accumulation at both 16 and 23 °C (Fig. 3, B and E). These observations indicate that although the SVP gene has a negative effect on miR172 accumulation (27) thermosensory regulation of miR172 accumulation is mostly independent of SVP.
FCA Regulates Pri-miR172 Processing
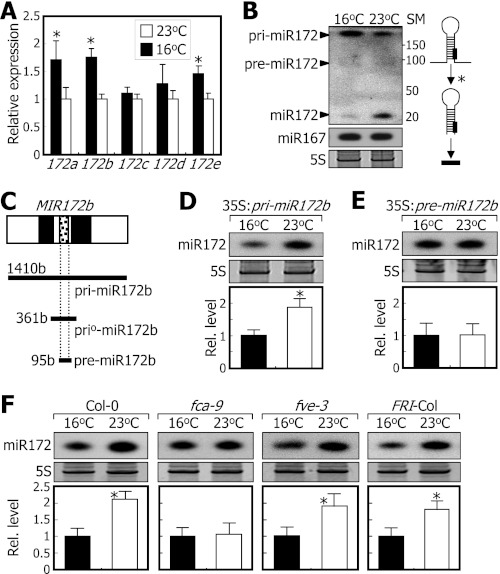
The next question was how ambient temperature regulates miR172 accumulation. Levels of primary transcripts of MIR172 genes (pri-miR172 transcripts) were slightly higher in plants grown at 16 °C than in plants grown at 23 °C (Fig. 4, A and B) unlike the high accumulation of miR172 at 23 °C. The pre-miR172 levels were equally low at both temperatures (Fig. 4B). These data indicate that ambient temperature regulates primarily the processing step from pri-miR172 to pre-miR172 rather than the transcription of MIR172 genes.
FIGURE 4.
FCA mediates temperature regulation of miR172 processing. Ten-day-old whole plants grown on MS-agar plates at either 23 or 16 °C were used for extraction of total RNA. miR172 levels were examined by Northern blot analysis. Error bars indicate S.E. A, effects of ambient temperature on MIR172 gene expression. Transcript levels of the MIR172 genes were analyzed by qRT-PCR (t test; *, p < 0.05). B, relative levels of pri-miR172 and pre-miR172 transcripts and mature miR172. miR167 was included for comparison. The asterisk indicates the pri-miR172-to-pre-miR172 processing step (right panel). The numbers indicate molecular sizes (in bases) of size marker (SM) RNAs. C, MIR172b gene constructs used. Black boxes are introns, and white boxes are exons. b, bases. D, miR172 levels in 35S:pri-miR172b transgenic plants. E, miR172 levels in 35S:pre-miR172b transgenic plants. F, miR172 levels in flowering time mutants overexpressing pri-miR172b construct driven by the cauliflower mosaic virus 35S promoter. Rel., relative.
To eliminate any temperature effects on MIR172 gene transcription in the assays, we produced 35S:pri-miR172b and 35S:pre-miR172b transgenic plants in which the MIR172b gene sequences consisting of 1410 and 95 bases, respectively, were overexpressed (Fig. 4C). In the 35S:pri-miR172b transgenic plants, the miR172 level was higher at 23 °C than at 16 °C (Fig. 4D). In contrast, the miR172 levels were similar at both temperatures in the 35S:pre-miR172b transgenic plants (Fig. 4E). Consistent with this, flowering of the 35S:pri-miR172b transgenic plants was delayed by 30–50% at 16 °C than at 23 °C, but the delayed flowering at 16 °C disappeared in the 35S:pre-miR172b transgenic plants (supplemental Fig. S4). These observations indicate that ambient temperature modulates the pri-miR172-to-pre-miR172 processing step.
We also transformed the pri-miR172b gene construct into fca-9 and fve-3 mutants, and miR172 levels were measured in the transgenic plants grown at different temperatures. Whereas the temperature effects on miR172 processing were maintained in the fve-3 background, the temperature effects disappeared in the fca-9 background (Fig. 4F). The lack of temperature effects in the miR172-overproducing fca-9 plants, unlike the small effects on miR172 processing in the parental fca-9 mutant (Fig. 3A), is certainly because the miR172 levels are extremely high in the miR172-overproducing fca-9 plant (supplemental Fig. S5). These observations indicate that FCA plays a primary role in ambient temperature regulation of miR172 processing.
FCA Binds to Pri-miR172 Transcripts
FCA is an RNA-binding protein that has two copies of RNA recognition motifs in the N-terminal region (15, 16). We therefore anticipated that it would regulate pri-miR172 processing by directly binding to the pri-miR172 transcripts.
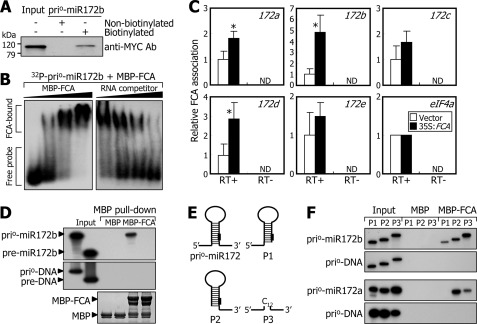
To test this hypothesis, we carried out RNA binding assays using nuclear extracts prepared from transgenic plants overexpressing the FCA-MYC fusion (35S:FCA). A modified version of the pri-miR172b RNA having ∼100 nucleotides at both sides of the stem-loop, designated prio-miR172b (Fig. 4C and supplemental Fig. S6A), was prepared to facilitate the RNA binding assays. The prio-miR172b RNAs were biotinylated by in vitro transcription. The RNA binding assays revealed that FCA binds efficiently to the biotinylated prio-miR172b RNAs (Fig. 5A). The assays also showed that the FCA-RNA interactions occur via the RNA recognition motifs (supplemental Fig. S6B).
FIGURE 5.
FCA binds to flanking sequences of pri-miR172 stem loop. A, in vitro pulldown assays on FCA binding to prio-miR172b RNA. Nuclear extracts were prepared from transgenic plants overexpressing the MYC-FCA gene fusion (35S:FCA) grown for 2 weeks on MS-agar plates. The in vitro transcribed and biotinylated prio-miR172b RNAs were used. The MYC-FCA fusion protein was detected immunologically using an anti-MYC antibody (Ab). kDa, kilodaltons. B, EMSA. Increasing amounts of recombinant MBP-FCA protein (0, 0.1, 0.5, and 2.0 μg) were added to the assay mixtures (left panel). Increasing amounts of unlabeled prio-miR172b RNA (0×, 5×, 25×, and 100×) were included in the assays to verify binding specificities (right panel). C, RIP assays. Total proteins extracted from 35S:FCA transgenic plants were immunoprecipitated with an anti-MYC antibody. RNA samples eluted from the protein-RNA complexes were analyzed by qRT-PCR (t test; *, p < 0.05). Error bars indicate S.E. RT, reverse transcription; ND, not detected. D, in vitro binding of FCA to prio-miR172b and pre-miR172b RNAs. Coprecipitation of radiolabeled prio-miR172b RNAs with MBP-FCA fusion proteins was detected by in vitro pulldown assays using amylose resin (top panel). PCR-amplified DNA fragments corresponding to the prio- and pre-miR172b RNAs were included in the assays (middle panel). Part of a Coomassie-stained gel is shown (bottom panel). E, deletion forms of prio-miR172 RNA. In the P3 construct, the flanking sequences were connected via a stretch of 12 C nucleotides. F, in vitro binding of FCA to deletion forms of prio-miR172a and prio-miR172b RNAs. In vitro pulldown assays were carried out as described in D.
We further examined the FCA-prio-miR172b RNA interactions by EMSA (Fig. 5B). The results showed that FCA binds to the prio-miR172b RNA in a dose-dependent manner, and the FCA binding was reduced in the presence of competitor RNAs (Fig. 5B). Similarly, FCA also bound to the prio-miR172a RNA (supplemental Fig. S6C). In addition, RIP assays using nuclear extracts of the 35S:FCA transgenic plants revealed that native FCA is strongly associated with pri-miR172a, pri-miR172b, and pri-miR172d transcripts in vivo (Fig. 5C).
FCA Binds to Flanking Sequences of Stem-Loop in Pri-miR172 Transcripts
We next asked what structural element(s) of pri-miR172 transcripts is responsible for FCA binding. Intriguingly, a set of in vitro pulldown assays showed that FCA binds to both the prio-miR172b and pre-miR172b RNAs under low stringency washing conditions containing 0.1% Triton X-100 (supplemental Fig. S6, D and E). However, under high stringency washing conditions containing 0.3% Triton X-100, FCA bound only to the prio-miR172b RNA (Fig. 5D, top panel). Meanwhile, FCA did not bind to double-stranded DNAs corresponding to the prio-miR172b and pre-miR172b RNAs (Fig. 5D, middle panel). FCA also bound to the prio-miR172a RNA (supplemental Fig. S6F), but it did not bind to the prio-miR172c RNA (supplemental Fig. S6G). These observations show that FCA binds primarily to some, if not all, of the primary miR172 transcripts, which is also consistent with the in vivo interaction of native FCA with pri-miR172 transcripts (Fig. 5C).
To determine the sequence element of the pri-miR172 transcripts required for FCA binding, we generated several deletion forms of the prio-miR172a and prio-miR172b RNAs (Fig. 5E). A set of in vitro pulldown assays revealed that FCA binds to the flanking sequences at both sides of the prio-miR172b RNA with a preference to the 3′-flanking sequence (Fig. 5F). Notably, FCA also bound to the P3 deletion forms missing the stem-loop sequences. Together with the FCA binding to pri-miR172 transcripts but not to pre-miR172 transcripts, these data demonstrate that FCA binds in a non-sequence-specific manner to the single-stranded flanking sequences of the stem-loops of at least pri-miR172a and pri-miR172b transcripts. The non-sequence-specific FCA binding to single-stranded RNA molecules has also been reported previously (11).
FCA Binds to Primary Transcripts of miR398 and miR399
Our data indicated that FCA binds to the pri-miR172 transcripts and promotes pri-miR172 processing. Besides miR172, several additional miRNAs, including miR156, miR398, and miR399, have been reported to be ambient temperature-responsive (27). Therefore, a question was whether FCA also regulates the processing of these miRNAs.
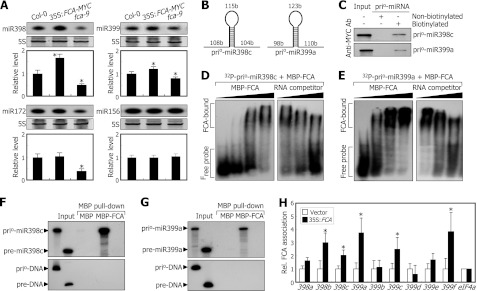
We first measured the levels of miR156, miR398, and miR399 in fca-9 mutant and 35S:FCA transgenic plants. miRNA Northern blot analysis revealed that the levels of miR398 and miR399 were reduced detectably in the fca-9 mutant (Fig. 6A), suggesting that FCA may also regulate the processing of miR398 and miR399. However, the levels of miR156 in the fca-9 mutant and 35S:FCA transgenic plants were similar to those in Col-0 plants.
FIGURE 6.
FCA promotes processing of pri-miR388 and pri-miR399 transcripts. The assays were carried out as described in Fig. 5. A, levels of miR156, miR172, miR398, and miR399. Two-week-old plants grown at 23 °C on MS-agar plates were harvested at ZT16. miRNA levels were examined by Northern blot analysis (t test; *, p < 0.01). Error bars indicate S.E. B, prio-miR398c and prio-miR399a RNAs assayed. b, bases. C, in vitro pulldown assays on FCA binding to prio-miR398c and prio-miR399a RNAs. The in vitro transcribed and biotinylated prio-miR398c and prio-miR399a RNAs were used. kDa, kilodaltons. D and E, EMSAs on FCA binding to prio-miR398c (D) and prio-miR399a (E) RNAs. F and G, in vitro binding of FCA to prio-miR398c (F) and prio-miR399a (G) RNAs. H, RIP assays on in vivo binding of FCA to pri-miR398 and pri-miR399 transcripts. Rel., relative; Ab, antibody.
To examine whether FCA binds to the primary transcripts of miR398 and miR399, we carried out RNA binding assays using in vitro transcribed and biotinylated prio-miR398c and prio-miR399a RNAs, which were prepared in a manner similar to that of prio-miR172 RNAs (Fig. 6B), and nuclear extracts prepared from the 35S:FCA transgenic plants. RNA binding assays showed that FCA binds to both the pri-miR398c and prio-miR399a RNAs (Fig. 6C). EMSAs also confirmed the FCA binding to the prio-miR398c and prio-miR399a RNAs in a dose-dependent manner (Fig. 6, D and E). In addition, in vitro pulldown assays under high stringency washing conditions showed that FCA binds to the prio-miR398c and prio-miR399a RNAs but not to their precursor miRNA forms (Fig. 6, F and G). RIP assays using nuclear extracts of the 35S:FCA transgenic plants confirmed that FCA interacts with the pri-miR398 and pri-miR399 transcripts in vivo (Fig. 6H). Together, these observations indicate that FCA mediates the pri-miRNA processing of various miRNAs involved in ambient temperature responses.
Notably, FCA activity is not restricted to temperature-responsive miRNAs. It was found that the levels of miR159, miR164, and miR167, which are uninfluenced by ambient temperature (27), are reduced obviously in the fca-9 mutant (supplemental Fig. S7A). In addition, FCA associated with pri-miR159 and pri-miR164 transcripts in vivo (supplemental Fig. S7B).
FCA Activity Is Temperature-regulated at Both Transcriptional and Protein Levels
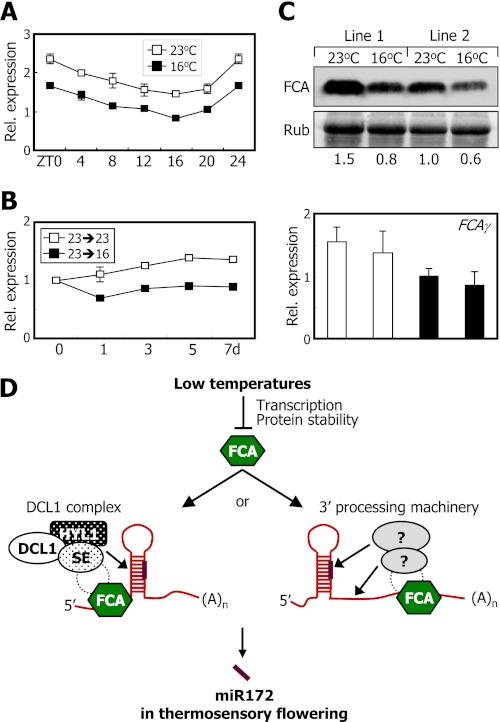
The next question was how ambient temperature signals are incorporated into the FCA-miR172 module. We measured the relative levels of FCAγ mRNA, an alternatively spliced variant encoding functional FCA protein (15, 16), in plants grown at either 23 or 16 °C. qRT-PCR analysis showed that the levels of FCAγ mRNA were higher in plants grown at 23 °C than at 16 °C at all ZT points (Fig. 7A). Kinetic patterns of FCAγ expression also revealed that the levels of FCAγ mRNA were slightly lower in plants grown at 16 °C (Fig. 7B). Other FCA transcript isoforms also decreased by ∼50% in plants grown at 16 °C (supplemental Fig. S8).
FIGURE 7.
FCA regulates miR172 processing in thermosensory flowering. In A–C, mRNA levels were determined by qRT-PCR (t test; *, p < 0.05). Error bars indicate S.E. A, effects of ambient temperature on the accumulation of FCAγ mRNA. qRT-PCRs were carried out using the plant materials described in supplemental Fig. S1. B, kinetic measurements of FCAγ mRNA levels after transfer from 23 to 16 °C. Relative levels of FCAγ mRNAs were determine using the cDNA samples of Col-0 plants described in Fig. 2C. C, effects of ambient temperature on FCA protein levels. The 35S:FCA transgenic plants were grown on MS-agar plates at either 23 or 16 °C for 10 days before harvesting whole plants. FCA protein was detected immunologically using an anti-MYC antibody (top panel). The numbers refer to the relative protein levels (middle panel). Relative levels of FCAγ mRNAs were determined by qRT-PCR (bottom panel). Rub, ribulose-bisphosphate carboxylase/oxygenase. D, schematic model of FCA function in thermosensory regulation of miR172 processing. Rel., relative.
To examine whether ambient temperature also affects the levels of FCA proteins, 35S:FCA transgenic plants were grown at different temperatures, and relative FCA protein levels were compared by Western blot analysis using an anti-MYC antibody. It was found that whereas FCA transcript levels were similar at both temperatures the FCA protein levels were reduced meaningfully in plants grown at 16 °C (Fig. 7C), showing that FCA activity is regulated by ambient temperature at both the gene transcription and protein levels. In support of the notion that FCA activity is regulated by ambient temperature, miR172 abundance was significantly higher in the 35S:FCA transgenic plants than in control plants grown at 16 °C (supplemental Fig. S9A). In addition, the 35S:FCA transgenic plants flowered earlier than control plants under low temperatures (supplemental Fig. S9, B and C).
We found that FCA binds to pri-miR172 transcripts to promote its processing. However, FCA has no ribonuclease activity (11). Therefore, another question was how FCA binding induces pri-miR172 processing. One possibility would be that FCA recruits its functional partner FY for the thermosensory regulation of miR172 processing. We found that fy mutation does not influence miR172 accumulation at both 16 and 23 °C (supplemental Fig. S10), suggesting that protein partners other than FY are necessary for the FCA-mediated miR172 processing.
DISCUSSION
Controlled RNA metabolism, which is mediated by an extensive set of RNA metabolic enzymes, RNA-binding proteins, and miRNAs, plays a role in diverse plant developmental processes and responses to environmental stimuli (39, 40). FCA is one of the best characterized RNA-binding proteins in plants. It has been well documented that FCA regulates alternative cleavage and polyadenylation of several primary transcripts, including its own mRNA precursors and noncoding antisense RNA transcripts (14, 16). It is also required for silencing RNAs in a locus-dependent manner and promoting subsequent asymmetric DNA methylation (18, 19). In addition, a recent whole-genome tiling array-based analysis has suggested that FCA, in collaboration with the RNA-binding protein FPA, plays a role in limiting intergenic transcription by modulating RNA 3′-end processing and transcriptional termination (20).
In this study, we found a distinct role for FCA in miRNA processing. FCA binds to the pri-miR172 transcripts and promotes their processing in response to ambient temperature changes. We found that FCA activity is regulated by ambient temperature at both gene transcriptional and protein levels. miR172, which acts as a positive riboregulator of FT expression, is ambient temperature-responsive (27). miR172 production is promoted at 23 °C but remains low at 16 °C, which is in good correlation with the kinetic patterns of FT gene transcription. The coordinated accumulation patterns of miR172 and FT mRNA under changing ambient temperature could be explained by the thermosensory regulation of miR172 processing by FCA.
Our data demonstrate that miR172 accumulation is regulated posttranscriptionally at the processing step from pri-miR172 to pre-miR172 and that FCA is required for the processing step. A critical question is how FCA regulates miR172 processing. Some clues can be inferred from studies on miRNA biogenesis in animal systems. It has been shown recently that several RNA-binding proteins, collectively known as heterogeneous nuclear ribonucleoproteins, are involved in miRNA processing and maturation (41, 42). Lin-28 binds to the stem-loop of the pri-let-7 transcripts and inhibits the miRNA processing steps mediated by Drosha, a member of the nuclear RNase III superfamily (43). In contrast, the heterogeneous nuclear ribonucleoprotein A1 binds to pri-miR-18a and facilitates the Drosha-mediated miRNA processing (44).
Similarly, FCA would regulate pri-miR172 processing through physical interaction with components of the miRNA processing machinery. miRNA processing steps are coordinated by DICER-LIKE 1 (DCL1), a nuclear RNase III enzyme that is assisted by the double-stranded RNA-binding protein HYPONASTIC LEAVES 1 (HYL1) and the C2H2 zinc finger protein SERRATE (SE) (45, 46). The DCL1-HYL1-SE heteropolymeric complex (DCL1 complex) regulates precise processing of pri-miRNA transcripts (47). In this view, FCA may recruit a core component of the DCL1 complex for miR172 processing (Fig. 7D).
It has been well documented that FCA regulates mRNA 3′-end processing in collaboration with FY (17). It is therefore possible that FCA modulates the 3′-end processing step of pri-miR172 transcripts and induces structural changes in the secondary structures required for pri-miR172 processing (Fig. 7D). It will be interesting to examine whether the 3′-end termination of the pri-miR172 transcripts is affected in fca mutants. Identification of FCA-interacting partners would also help elucidate how FCA regulates the 3′-end pri-miRNA processing step. Alternatively, miR172 accumulation would be regulated at least in part at the transcriptional level (27) in which FCA may also be involved. We found that although the overall levels of miR172 were reduced in fca-9 mutant miR172 accumulation was still sensitive to low temperatures (Fig. 3A), supporting the transcriptional control of miR172 accumulation. It has been shown that FCA acts as a chromatin-remodeling factor (12–14, 18–20). Therefore, it will be worth investigating whether FCA regulates the transcription of MIRNA genes through chromatin modifications.
We found that FCA regulates the pri-miRNA-to-pre-miRNA processing step of at least three miRNAs in Arabidopsis. FCA binds to their pri-miRNA transcripts with higher affinity than to their pre-miRNA transcripts. FCA is capable of binding even to prio-miR172 RNA deletions lacking the stem-loop structures in vitro, showing that it binds primarily to the flanking sequences of the stem-loop structures rather than the stem-loops themselves. This observation is also consistent with the previous observation that RNA recognition motif-containing RNA-binding proteins, such as FCA, bind to single-stranded RNAs in a non-sequence-specific manner but with a preference for U- and G-rich sequences (11, 48). There were no obvious sequence similarities in the flanking sequences of the stem-loops of pri-miR172a, pri-miR172b, pri-miR398c, and pri-miR399a transcripts. In addition, FCA also bound to the primary transcripts of miRNAs that are not involved in responses to changes in ambient temperature. Bases on these observations, one plausible explanation would be that FCA binding to single-stranded RNA sequences is assisted by its interacting partner.
Our data show that FCA is involved in the thermosensory regulation of miR172 processing. The FCA-miR172 regulatory pathway may not be a major pathway that regulates the FT-mediated thermosensory flowering. It has been reported that although fca mutants are late flowering and unresponsive to high temperature for their thermal induction fca flc double mutants are fully responsive to changes in ambient temperature (6), indicating that the lack of thermosensory effects on flowering in fca mutants is primarily due to high levels of FLC expression.
We found that although mutation of SVP gene eliminates the effects of ambient temperature on flowering time (7) miR172 accumulation was still influenced by ambient temperature in the svp mutants (Fig. 3, D and E). However, the rate of elevation of miR172 levels at 23 °C was significantly higher in the svp mutants in comparison with that in Col-0 plants. Considering the role of SVP as a transcription factor (7), our observations suggest that SVP may regulate the transcription of MIR172 genes in response to changes in ambient temperatures.
One of the major findings in this work is that the expression of FCA gene itself is regulated by ambient temperature. It has been known that the levels of FCA transcript isoforms are maintained by the autoregulatory mechanism mediated by FCA protein (15, 16). Therefore, although ambient temperature influences the FCA activity both at the transcription level and at the protein level, the thermosensory effects on FCA activity are not sufficient to explain those on miR172 accumulation and thermosensory flowering. It is therefore likely that the FCA-miR12 regulon identified in this work would be one of multiple signaling pathways, such as those mediated by FLC, FVE, and SVP (5–7), that regulate thermosensory flowering. In this regard, it is concluded that the FCA-miR172 regulon does not play a major role in thermosensory flowering. Instead, it may function as a fine tuning mechanism for the onset of flowering under fluctuating ambient temperature conditions.
It is notable that the fca-9 mutation reduces the accumulation of miRNAs, such as miR159, miR164, and miR167, which are not sensitive to temperature changes, and FCA binds to the primary transcripts of these miRNAs. It has been reported that the levels of these miRNAs are influenced by various endogenous and environmental stimuli, including growth hormones, light, and abiotic stresses (49, 50). Therefore, it is probable that FCA also functions in miRNA-mediated growth hormonal and light signaling pathways and stress responses.
Supplementary Material
Acknowledgment
We thank the Arabidopsis Biological Resource Center (ABRC, Ohio State University) for the SALK T-DNA lines.
This work was supported by the Leaping Research Program (Grant 20110016440) provided by the National Research Foundation of Korea; the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center Number PJ008103) provided by the Rural Development Administration; the Plant Signaling Network Research Center (Grant 20110001099); the Agricultural Research and Development Promotion Center (Grant 309017-03), Korea Ministry for Food, Agriculture, Forestry and Fisheries; and the Creative Research Initiatives of the National Research Foundation for the Ministry of Education, Science and Technology (Grant R16-2008-106-01000-0 to J. H. A.).

This article contains supplemental Figs. S1–S10 and Table S1.
- FT
- FLOWERING LOCUS T
- AP2
- APETALA2
- CO
- CONSTANS
- DCL1
- DICER-LIKE 1
- ELF3
- EARLY FLOWERING 3
- FLC
- FLOWERING LOCUS C
- FLM
- FLOWERING LOCUS M
- FRI
- FRIGIDA
- HYL1
- HYPONASTIC LEAVES 1
- MBP
- maltose-binding protein
- miRNA
- microRNA
- miR172
- microRNA172
- MS
- Murashige and Skoog
- pri-miR172
- primary microRNA172
- qRT-PCR
- quantitative real time RT-PCR
- RIP
- RNA immunoprecipitation
- SE
- SERRATE
- SMZ
- SCHLAFMÜTZE
- SNZ
- SCHNARCHZAPFEN
- SVP
- SHORT VEGETATIVE PHASE
- TFL1
- TERMINAL FLOWER 1
- TOE
- TARGET OF EAT
- ZT
- zeitgeber time
- rTOE1
- resistant TOE1.
REFERENCES
- 1. Penfield S. (2008) Temperature perception and signal transduction in plants. New Phytol. 179, 615–628 [DOI] [PubMed] [Google Scholar]
- 2. Samach A., Wigge P. A. (2005) Ambient temperature perception in plants. Curr. Opin. Plant Biol. 8, 483–486 [DOI] [PubMed] [Google Scholar]
- 3. Atkin O. K., Tjoelker M. G. (2003) Thermal acclimation and the dynamic response of plant respiration to temperature. Trends Plant Sci. 8, 343–351 [DOI] [PubMed] [Google Scholar]
- 4. Fitter A. H., Fitter R. S. (2002) Rapid changes in flowering time in British plants. Science 296, 1689–1691 [DOI] [PubMed] [Google Scholar]
- 5. Blázquez M. A., Ahn J. H., Weigel D. (2003) A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat. Genet. 33, 168–171 [DOI] [PubMed] [Google Scholar]
- 6. Balasubramanian S., Sureshkumar S., Lempe J., Weigel D. (2006) Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2, e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee J. H., Yoo S. J., Park S. H., Hwang I., Lee J. S., Ahn J. H. (2007) Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 21, 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Strasser B., Alvarez M. J., Califano A., Cerdán P. D. (2009) A complementary role for ELF3 and TFL1 in the regulation of flowering time by ambient temperature. Plant J. 58, 629–640 [DOI] [PubMed] [Google Scholar]
- 9. Kumar S. V., Wigge P. A. (2010) H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140, 136–147 [DOI] [PubMed] [Google Scholar]
- 10. Koornneef M., Hanhart C. J., van der Veen J. H. (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66 [DOI] [PubMed] [Google Scholar]
- 11. Macknight R., Bancroft I., Page T., Lister C., Schmidt R., Love K., Westphal L., Murphy G., Sherson S., Cobbett C., Dean C. (1997) FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89, 737–745 [DOI] [PubMed] [Google Scholar]
- 12. Liu F., Marquardt S., Lister C., Swiezewski S., Dean C. (2010) Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327, 94–97 [DOI] [PubMed] [Google Scholar]
- 13. Liu F., Quesada V., Crevillén P., Bäurle I., Swiezewski S., Dean C. (2007) The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol. Cell 28, 398–407 [DOI] [PubMed] [Google Scholar]
- 14. Hornyik C., Terzi L. C., Simpson G. G. (2010) The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev. Cell 18, 203–213 [DOI] [PubMed] [Google Scholar]
- 15. Macknight R., Duroux M., Laurie R., Dijkwel P., Simpson G., Dean C. (2002) Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell 14, 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Quesada V., Macknight R., Dean C., Simpson G. G. (2003) Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time. EMBO J. 22, 3142–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Simpson G. G., Dijkwel P. P., Quesada V., Henderson I., Dean C. (2003) FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113, 777–787 [DOI] [PubMed] [Google Scholar]
- 18. Bäurle I., Smith L., Baulcombe D. C., Dean C. (2007) Widespread role for the flowering-time regulators FCA and FPA in RNA-mediated chromatin silencing. Science 318, 109–112 [DOI] [PubMed] [Google Scholar]
- 19. Bäurle I., Dean C. (2008) Differential interactions of the autonomous pathway RRM proteins and chromatin regulators in the silencing of Arabidopsis targets. PLoS One 3, e2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sonmez C., Bäurle I., Magusin A., Dreos R., Laubinger S., Weigel D., Dean C. (2011) RNA 3′ processing functions of Arabidopsis FCA and FPA limit intergenic transcription. Proc. Natl. Acad. Sci. U.S.A. 108, 8508–8513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen X. (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jung J. H., Seo Y. H., Seo P. J., Reyes J. L., Yun J., Chua N. H., Park C. M. (2007) The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19, 2736–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mathieu J., Yant L. J., Mürdter F., Küttner F., Schmid M. (2009) Repression of flowering by the miR172 target SMZ. PLoS Biol. 7, e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yant L., Mathieu J., Dinh T. T., Ott F., Lanz C., Wollmann H., Chen X., Schmid M. (2010) Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22, 2156–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wollmann H., Mica E., Todesco M., Long J. A., Weigel D. (2010) On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 137, 3633–3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grigorova B., Mara C., Hollender C., Sijacic P., Chen X., Liu Z. (2011) LEUNIG and SEUSS co-repressors regulate miR172 expression in Arabidopsis flowers. Development 138, 2451–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee H., Yoo S. J., Lee J. H., Kim W., Yoo S. K., Fitzgerald H., Carrington J. C., Ahn J. H. (2010) Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res. 38, 3081–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Telfer A., Bollman K. M., Poethig R. S. (1997) Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124, 645–654 [DOI] [PubMed] [Google Scholar]
- 29. Wu G., Park M. Y., Conway S. R., Wang J. W., Weigel D., Poethig R. S. (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seo P. J., Kim M. J., Park J. Y., Kim S. Y., Jeon J., Lee Y. H., Kim J., Park C. M. (2010) Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 61, 661–671 [DOI] [PubMed] [Google Scholar]
- 31. Jiao X., Trifillis P., Kiledjian M. (2002) Identification of target messenger RNA substrates for the murine deleted in azoospermia-like RNA-binding protein. Biol. Reprod. 66, 475–485 [DOI] [PubMed] [Google Scholar]
- 32. Gendrel A. V., Lippman Z., Martienssen R., Colot V. (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2, 213–218 [DOI] [PubMed] [Google Scholar]
- 33. Terzi L. C., Simpson G. G. (2009) Arabidopsis RNA immunoprecipitation. Plant J. 59, 163–168 [DOI] [PubMed] [Google Scholar]
- 34. Jung J. H., Seo P. J., Kang S. K., Park C. M. (2011) miR172 signals are incorporated into the miR156 signaling pathway at the SPL3/4/5 genes in Arabidopsis developmental transitions. Plant Mol. Biol. 76, 35–45 [DOI] [PubMed] [Google Scholar]
- 35. Yang L., Liu Z., Lu F., Dong A., Huang H. (2006) SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 47, 841–850 [DOI] [PubMed] [Google Scholar]
- 36. Han M. H., Goud S., Song L., Fedoroff N. (2004) The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. U.S.A. 101, 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lim M. H., Kim J., Kim Y. S., Chung K. S., Seo Y. H., Lee I., Kim J., Hong C. B., Kim H. J., Park C. M. (2004) A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell 16, 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee I., Amasino R. M. (1995) Effect of vernalization, photoperiod, and light quality on the flowering phenotype of Arabidopsis plants containing the FRIGIDA gene. Plant Physiol. 108, 157–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim Y. K., Heo I., Kim V. N. (2010) Modifications of small RNAs and their associated proteins. Cell 143, 703–709 [DOI] [PubMed] [Google Scholar]
- 40. Lorković Z. J. (2009) Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci. 14, 229–236 [DOI] [PubMed] [Google Scholar]
- 41. Michlewski G., Guil S., Semple C. A., Cáceres J. F. (2008) Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol. Cell 32, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rybak A., Fuchs H., Smirnova L., Brandt C., Pohl E. E., Nitsch R., Wulczyn F. G. (2008) A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 10, 987–993 [DOI] [PubMed] [Google Scholar]
- 43. Newman M. A., Thomson J. M., Hammond S. M. (2008) Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14, 1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guil S., Cáceres J. F. (2007) The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 14, 591–596 [DOI] [PubMed] [Google Scholar]
- 45. Voinnet O. (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687 [DOI] [PubMed] [Google Scholar]
- 46. Xie Z., Khanna K., Ruan S. (2010) Expression of microRNAs and its regulation in plants. Semin. Cell Dev. Biol. 21, 790–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dong Z., Han M. H., Fedoroff N. (2008) The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl. Acad. Sci. U.S.A. 105, 9970–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cléry A., Blatter M., Allain F. H. (2008) RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 18, 290–298 [DOI] [PubMed] [Google Scholar]
- 49. Siré C., Moreno A. B., Garcia-Chapa M., López-Moya J. J., San Segundo B. (2009) Diurnal oscillation in the accumulation of Arabidopsis microRNAs, miR167, miR168, miR171 and miR398. FEBS Lett. 583, 1039–1044 [DOI] [PubMed] [Google Scholar]
- 50. Sunkar R., Chinnusamy V., Zhu J., Zhu J. K. (2007) Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 12, 301–309 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.