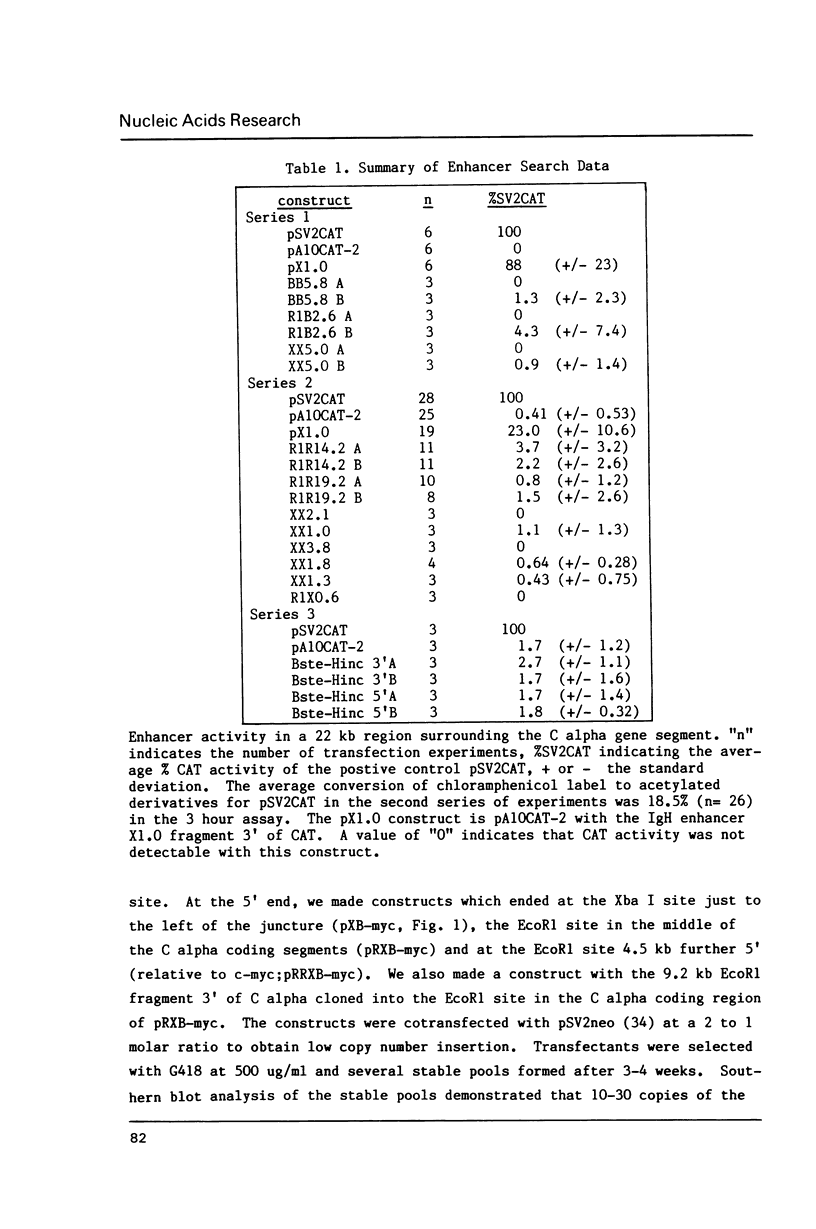
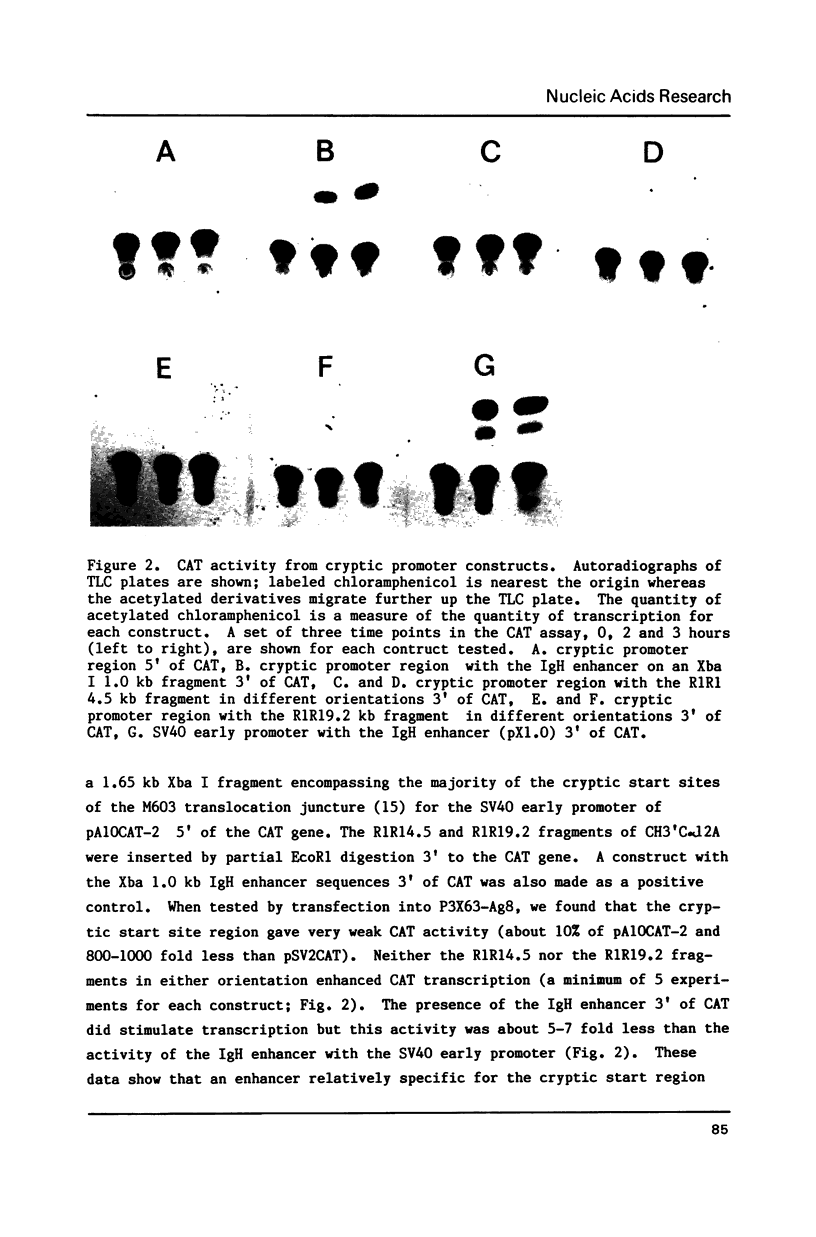
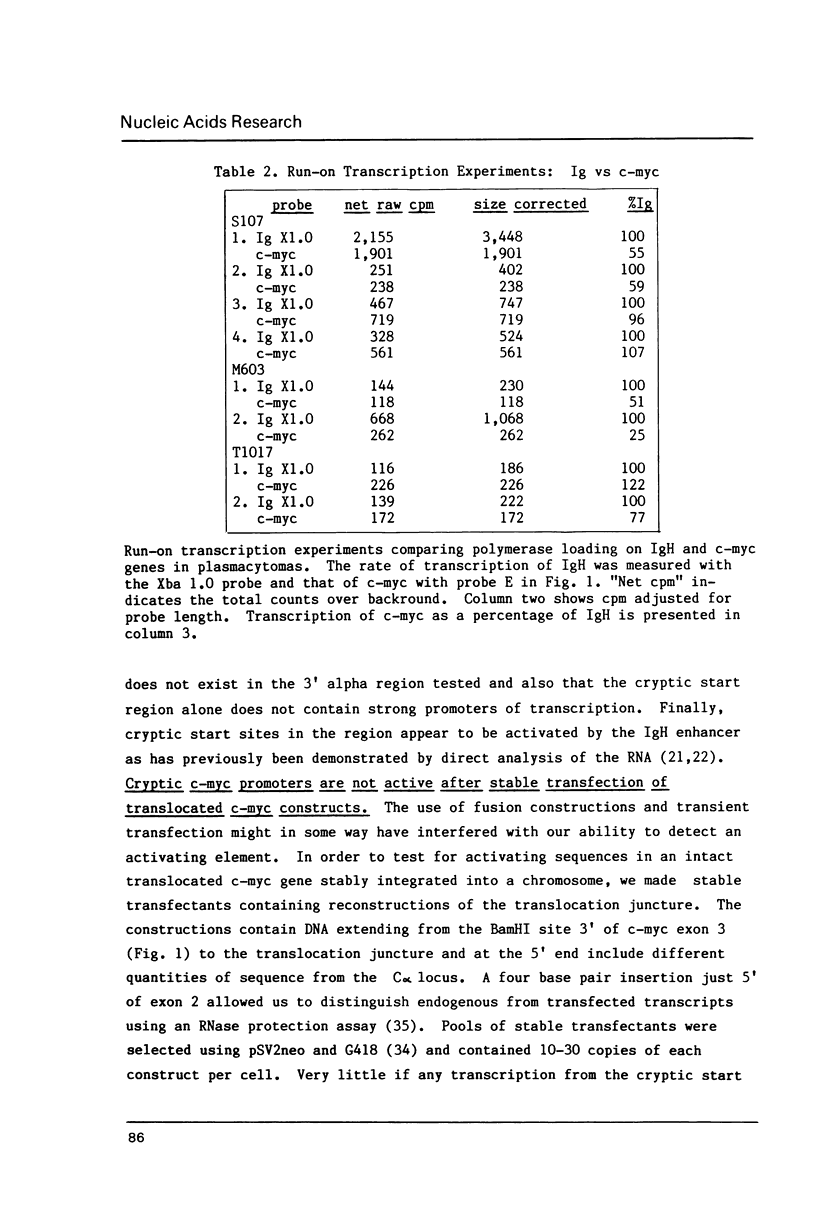
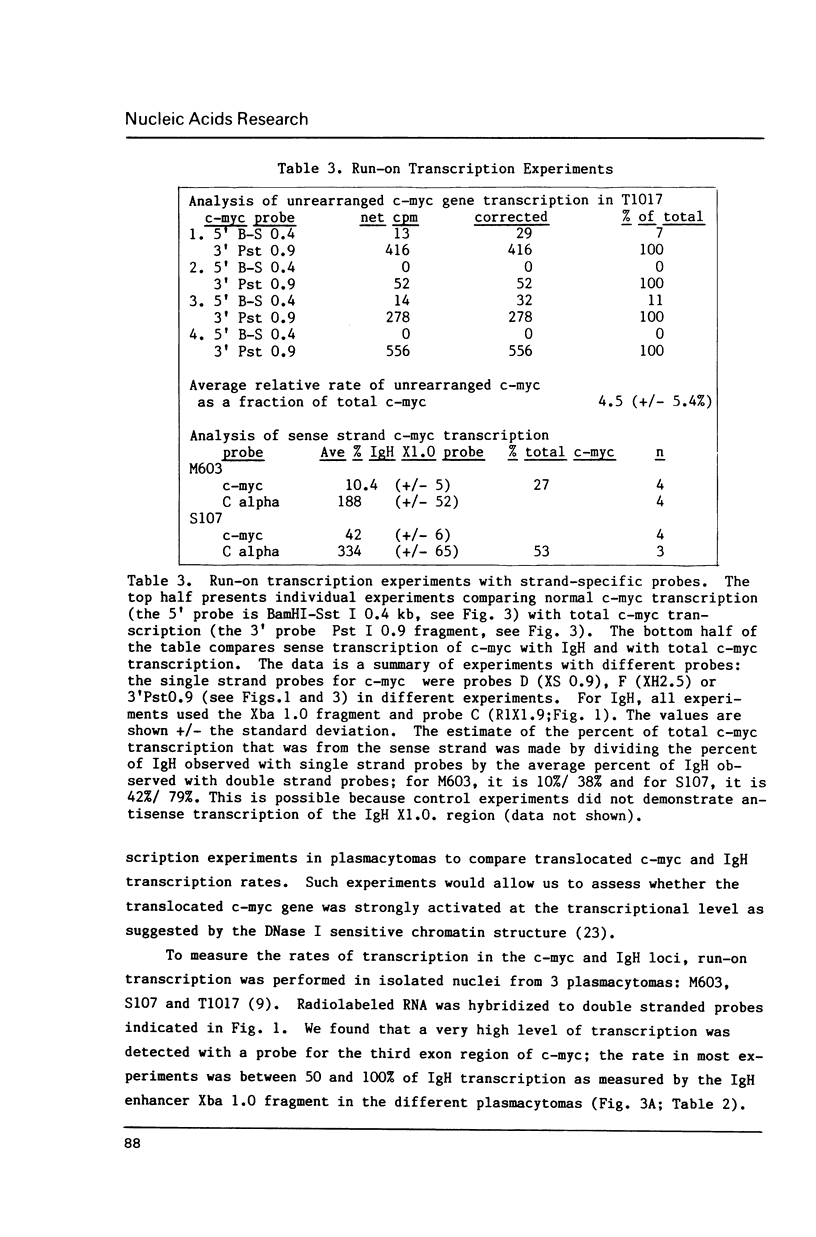
Abstract
We have studied the transcriptional activation of translocated c-myc genes in murine plasmacytomas in which the translocation juncture occurs within the first intron of c-myc and juxtaposes c-myc with the immunoglobulin C alpha gene segment. It has been widely suggested that a novel transcriptional enhancer element located near the C alpha gene segment might activate the translocated c-myc gene. We have carried out an extensive search for such an element and find no significant transcriptional enhancer activity in a 22 kb region encompassing the translocation junction, C alpha gene segment and regions 3' of C alpha. We also find that the cryptic promoter region of the translocated c-myc gene is a very weak promoter of transcription. Despite this evidence against the presence of strong transcriptional regulatory elements, the translocated c-myc gene locus is transcribed at high rates that are 25-greater than 100% of that measured for the highly active immunoglobulin genes in murine plasmacytomas. These data suggest the presence of a novel type of strong activator of transcription in the murine heavy chain locus.
Full text
PDF



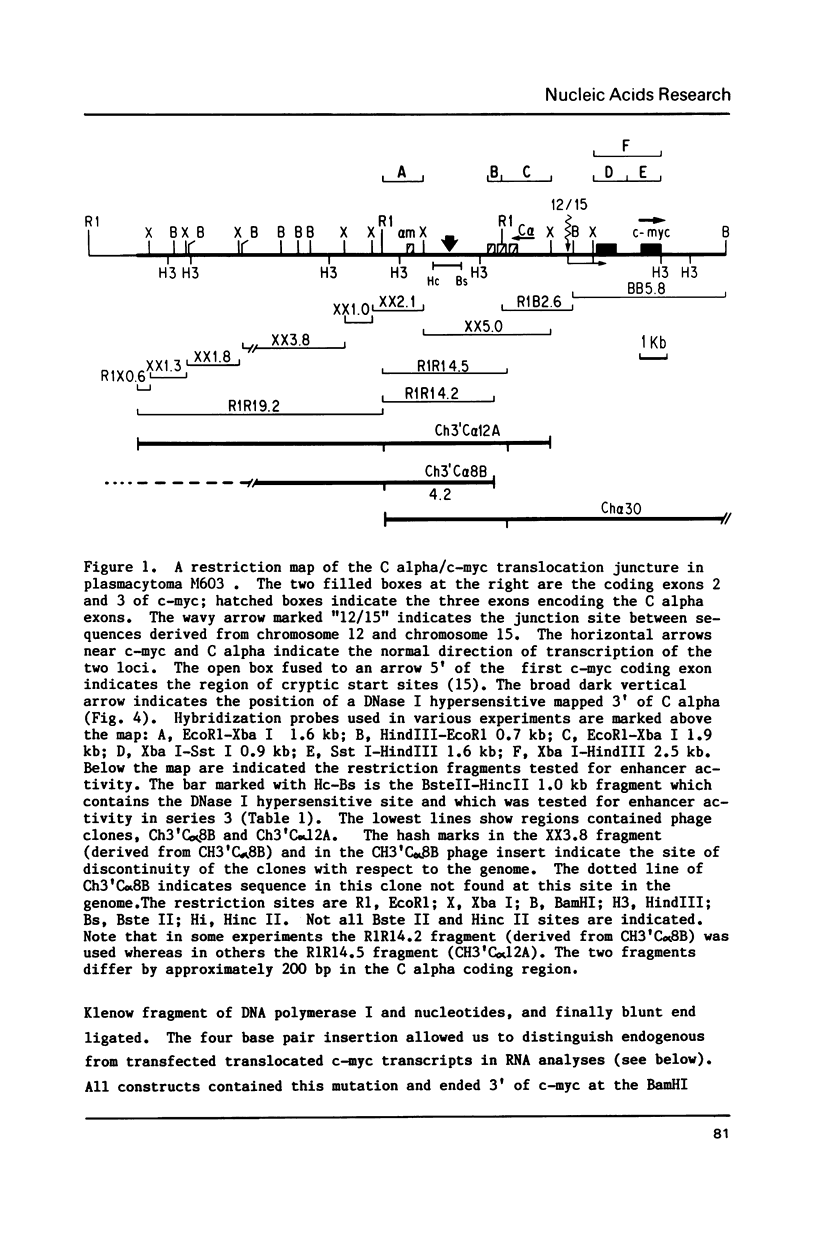











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams J. M., Gerondakis S., Webb E., Mitchell J., Bernard O., Cory S. Transcriptionally active DNA region that rearranges frequently in murine lymphoid tumors. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6966–6970. doi: 10.1073/pnas.79.22.6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams J. M., Harris A. W., Pinkert C. A., Corcoran L. M., Alexander W. S., Cory S., Palmiter R. D., Brinster R. L. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985 Dec 12;318(6046):533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Banerji J., Olson L., Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983 Jul;33(3):729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Novel promoter upstream of the human c-myc gene and regulation of c-myc expression in B-cell lymphomas. Mol Cell Biol. 1986 Oct;6(10):3481–3489. doi: 10.1128/mcb.6.10.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard O., Cory S., Gerondakis S., Webb E., Adams J. M. Sequence of the murine and human cellular myc oncogenes and two modes of myc transcription resulting from chromosome translocation in B lymphoid tumours. EMBO J. 1983;2(12):2375–2383. doi: 10.1002/j.1460-2075.1983.tb01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabi F., Neuberger M. S. Chromosome translocation activates heterogeneously initiated, bipolar transcription of a mouse c-myc gene. EMBO J. 1985 Mar;4(3):667–674. doi: 10.1002/j.1460-2075.1985.tb03681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calza R. E., Eckhardt L. A., DelGiudice T., Schildkraut C. L. Changes in gene position are accompanied by a change in time of replication. Cell. 1984 Mar;36(3):689–696. doi: 10.1016/0092-8674(84)90349-0. [DOI] [PubMed] [Google Scholar]
- Corcoran L. M., Cory S., Adams J. M. Transposition of the immunoglobulin heavy chain enhancer to the myc oncogene in a murine plasmacytoma. Cell. 1985 Jan;40(1):71–79. doi: 10.1016/0092-8674(85)90310-1. [DOI] [PubMed] [Google Scholar]
- Cory S., Gerondakis S., Adams J. M. Interchromosomal recombination of the cellular oncogene c-myc with the immunoglobulin heavy chain locus in murine plasmacytomas is a reciprocal exchange. EMBO J. 1983;2(5):697–703. doi: 10.1002/j.1460-2075.1983.tb01487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews S., Barth R., Hood L., Prehn J., Calame K. Mouse c-myc oncogene is located on chromosome 15 and translocated to chromosome 12 in plasmacytomas. Science. 1982 Dec 24;218(4579):1319–1321. doi: 10.1126/science.7146913. [DOI] [PubMed] [Google Scholar]
- Croce C. M., Erikson J., ar-Rushdi A., Aden D., Nishikura K. Translocated c-myc oncogene of Burkitt lymphoma is transcribed in plasma cells and repressed in lymphoblastoid cells. Proc Natl Acad Sci U S A. 1984 May;81(10):3170–3174. doi: 10.1073/pnas.81.10.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M., Kent R. B., Sonenshein G. E. Transcriptional activation of immunoglobulin alpha heavy-chain genes by translocation of the c-myc oncogene. 1983 Sep 29-Oct 5Nature. 305(5933):443–446. doi: 10.1038/305443a0. [DOI] [PubMed] [Google Scholar]
- Dunnick W., Baumgartner J., Fradkin L., Schultz C., Szurek P. Methylation of plasmacytoma c-myc genes. Gene. 1985;39(2-3):287–292. doi: 10.1016/0378-1119(85)90325-7. [DOI] [PubMed] [Google Scholar]
- Eick D., Piechaczyk M., Henglein B., Blanchard J. M., Traub B., Kofler E., Wiest S., Lenoir G. M., Bornkamm G. W. Aberrant c-myc RNAs of Burkitt's lymphoma cells have longer half-lives. EMBO J. 1985 Dec 30;4(13B):3717–3725. doi: 10.1002/j.1460-2075.1985.tb04140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrlander P. D., Piechaczyk M., Marcu K. B. Chromatin structure of the murine c-myc locus: implications for the regulation of normal and chromosomally translocated genes. EMBO J. 1985 Dec 1;4(12):3195–3202. doi: 10.1002/j.1460-2075.1985.tb04065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrlander P. D., Sümegi J., Yang J. Q., Wiener F., Marcu K. B., Klein G. Activation of the c-myc oncogene by the immunoglobulin heavy-chain gene enhancer after multiple switch region-mediated chromosome rearrangements in a murine plasmacytoma. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3746–3750. doi: 10.1073/pnas.82.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feo S., Harvey R., Showe L., Croce C. M. Regulation of translocated c-myc genes transfected into plasmacytoma cells. Proc Natl Acad Sci U S A. 1986 Feb;83(3):706–709. doi: 10.1073/pnas.83.3.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerondakis S., Cory S., Adams J. M. Translocation of the myc cellular oncogene to the immunoglobulin heavy chain locus in murine plasmacytomas is an imprecise reciprocal exchange. Cell. 1984 Apr;36(4):973–982. doi: 10.1016/0092-8674(84)90047-3. [DOI] [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983 Jul;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor P. D., Morrison S. L. Myeloma mutant with a novel 3' flanking region: loss of normal sequence and insertion of repetitive elements leads to decreased transcription but normal processing of the alpha heavy-chain gene products. Mol Cell Biol. 1986 Jun;6(6):1903–1916. doi: 10.1128/mcb.6.6.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C. V., Jacob P. E., Ringold G. M., Lee F. Expression and regulation of Escherichia coli lacZ gene fusions in mammalian cells. J Mol Appl Genet. 1983;2(1):101–109. [PubMed] [Google Scholar]
- Hayday A. C., Gillies S. D., Saito H., Wood C., Wiman K., Hayward W. S., Tonegawa S. Activation of a translocated human c-myc gene by an enhancer in the immunoglobulin heavy-chain locus. 1984 Jan 26-Feb 1Nature. 307(5949):334–340. doi: 10.1038/307334a0. [DOI] [PubMed] [Google Scholar]
- Honjo T. Immunoglobulin genes. Annu Rev Immunol. 1983;1:499–528. doi: 10.1146/annurev.iy.01.040183.002435. [DOI] [PubMed] [Google Scholar]
- Iguchi-Ariga S. M., Itani T., Kiji Y., Ariga H. Possible function of the c-myc product: promotion of cellular DNA replication. EMBO J. 1987 Aug;6(8):2365–2371. doi: 10.1002/j.1460-2075.1987.tb02513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakkis E., Prehn J., Calame K. An active chromatin structure acquired by translocated c-myc genes. Mol Cell Biol. 1986 Apr;6(4):1357–1361. doi: 10.1128/mcb.6.4.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G., Gruss P. Enhancer elements. Cell. 1983 Jun;33(2):313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Klein S., Sablitzky F., Radbruch A. Deletion of the IgH enhancer does not reduce immunoglobulin heavy chain production of a hybridoma IgD class switch variant. EMBO J. 1984 Nov;3(11):2473–2476. doi: 10.1002/j.1460-2075.1984.tb02158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher L. A., Ruddle F. H. Chromosome mediated gene transfer. Annu Rev Biochem. 1981;50:533–554. doi: 10.1146/annurev.bi.50.070181.002533. [DOI] [PubMed] [Google Scholar]
- Köhler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975 Aug 7;256(5517):495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Laimins L. A., Khoury G., Gorman C., Howard B., Gruss P. Host-specific activation of transcription by tandem repeats from simian virus 40 and Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6453–6457. doi: 10.1073/pnas.79.21.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder P., Battey J., Lenoir G., Moulding C., Murphy W., Potter H., Stewart T., Taub R. Translocations among antibody genes in human cancer. Science. 1983 Nov 18;222(4625):765–771. doi: 10.1126/science.6356357. [DOI] [PubMed] [Google Scholar]
- Lombardi L., Newcomb E. W., Dalla-Favera R. Pathogenesis of Burkitt lymphoma: expression of an activated c-myc oncogene causes the tumorigenic conversion of EBV-infected human B lymphoblasts. Cell. 1987 Apr 24;49(2):161–170. doi: 10.1016/0092-8674(87)90556-3. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercola M., Goverman J., Mirell C., Calame K. Immunoglobulin heavy-chain enhancer requires one or more tissue-specific factors. Science. 1985 Jan 18;227(4684):266–270. doi: 10.1126/science.3917575. [DOI] [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B. Intragenic pausing and anti-sense transcription within the murine c-myc locus. EMBO J. 1986 Nov;5(11):2859–2865. doi: 10.1002/j.1460-2075.1986.tb04580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen D. A., Chou J., MacKrell A. J., Casadaban M. J., Steiner D. F. Expression of a preproinsulin-beta-galactosidase gene fusion in mammalian cells. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5198–5202. doi: 10.1073/pnas.80.17.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K., ar-Rushdi A., Erikson J., DeJesus E., Dugan D., Croce C. M. Repression of rearranged mu gene and translocated c-myc in mouse 3T3 cells X Burkitt lymphoma cell hybrids. Science. 1984 Apr 27;224(4647):399–402. doi: 10.1126/science.6424234. [DOI] [PubMed] [Google Scholar]
- Nishikura K., ar-Rushdi A., Erikson J., Watt R., Rovera G., Croce C. M. Differential expression of the normal and of the translocated human c-myc oncogenes in B cells. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4822–4826. doi: 10.1073/pnas.80.15.4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter R. D., Brinster R. L. Germ-line transformation of mice. Annu Rev Genet. 1986;20:465–499. doi: 10.1146/annurev.ge.20.120186.002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechaczyk M., Yang J. Q., Blanchard J. M., Jeanteur P., Marcu K. B. Posttranscriptional mechanisms are responsible for accumulation of truncated c-myc RNAs in murine plasma cell tumors. Cell. 1985 Sep;42(2):589–597. doi: 10.1016/0092-8674(85)90116-3. [DOI] [PubMed] [Google Scholar]
- Prehn J., Mercola M., Calame K. Translocation affects normal c-myc promoter usage and activates fifteen cryptic c-myc transcription starts in plasmacytoma M603. Nucleic Acids Res. 1984 Dec 11;12(23):8987–9007. doi: 10.1093/nar/12.23.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J., Wall R. Immunoglobulin heavy chain genes: demethylation accompanies class switching. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7497–7501. doi: 10.1073/pnas.78.12.7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Stanton L. W., Watt R., Marcu K. B. Translocation, breakage and truncated transcripts of c-myc oncogene in murine plasmacytomas. Nature. 1983 Jun 2;303(5916):401–406. doi: 10.1038/303401a0. [DOI] [PubMed] [Google Scholar]
- Stavnezer-Nordgren J., Sirlin S. Specificity of immunoglobulin heavy chain switch correlates with activity of germline heavy chain genes prior to switching. EMBO J. 1986 Jan;5(1):95–102. doi: 10.1002/j.1460-2075.1986.tb04182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J., Sirlin S., Abbott J. Induction of immunoglobulin isotype switching in cultured I.29 B lymphoma cells. Characterization of the accompanying rearrangements of heavy chain genes. J Exp Med. 1985 Mar 1;161(3):577–601. doi: 10.1084/jem.161.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart T. A., Pattengale P. K., Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984 Oct;38(3):627–637. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- Storb U., Arp B. Methylation patterns of immunoglobulin genes in lymphoid cells: correlation of expression and differentiation with undermethylation. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6642–6646. doi: 10.1073/pnas.80.21.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studzinski G. P., Brelvi Z. S., Feldman S. C., Watt R. A. Participation of c-myc protein in DNA synthesis of human cells. Science. 1986 Oct 24;234(4775):467–470. doi: 10.1126/science.3532322. [DOI] [PubMed] [Google Scholar]
- Thomas K. R., Capecchi M. R. Introduction of homologous DNA sequences into mammalian cells induces mutations in the cognate gene. Nature. 1986 Nov 6;324(6092):34–38. doi: 10.1038/324034a0. [DOI] [PubMed] [Google Scholar]
- Wabl M. R., Burrows P. D. Expression of immunoglobulin heavy chain at a high level in the absence of a proposed immunoglobulin enhancer element in cis. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2452–2455. doi: 10.1073/pnas.81.8.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. F., Calame K. The endogenous immunoglobulin heavy chain enhancer can activate tandem VH promoters separated by a large distance. Cell. 1985 Dec;43(3 Pt 2):659–665. doi: 10.1016/0092-8674(85)90238-7. [DOI] [PubMed] [Google Scholar]
- Yancopoulos G. D., DePinho R. A., Zimmerman K. A., Lutzker S. G., Rosenberg N., Alt F. W. Secondary genomic rearrangement events in pre-B cells: VHDJH replacement by a LINE-1 sequence and directed class switching. EMBO J. 1986 Dec 1;5(12):3259–3266. doi: 10.1002/j.1460-2075.1986.tb04637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaller D. M., Eckhardt L. A. Deletion of a B-cell-specific enhancer affects transfected, but not endogenous, immunoglobulin heavy-chain gene expression. Proc Natl Acad Sci U S A. 1985 Aug;82(15):5088–5092. doi: 10.1073/pnas.82.15.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]