Background: The IcmF family protein TssM is a conserved T6SS component.
Results: TssM exhibits ATPase activity, and its impaired ATP binding/hydrolysis activity causes loss of TssM-TssL-Hcp complex formation and Hcp secretion.
Conclusion: TssM functions as a T6SS energizer to recruit Hcp into the TssM-TssL complex and powers Hcp secretion.
Significance: This is the first demonstration of TssM ATPase activity and its role in protein secretion.
Keywords: ATPases, Membrane Proteins, Microbiology, Protein Secretion, Protein-Protein Interactions, ATPase, Agrobacterium tumefaciens, TssL, TssM, Type VI Secretion
Abstract
The type VI secretion system (T6SS) with diversified functions is widely distributed in pathogenic Proteobacteria. The IcmF (intracellular multiplication protein F) family protein TssM is a conserved T6SS inner membrane protein. Despite the conservation of its Walker A nucleotide-binding motif, the NTPase activity of TssM and its role in T6SS remain obscure. In this study, we characterized TssM in the plant pathogen Agrobacterium tumefaciens and provided the first biochemical evidence for TssM exhibiting ATPase activity to power the secretion of the T6SS hallmark protein, hemolysin-coregulated protein (Hcp). Amino acid substitutions in the Walker A motif of TssM caused reduced ATP binding and hydrolysis activity. Importantly, we discovered the Walker B motif of TssM and demonstrated that it is critical for ATP hydrolysis activity. Protein-protein interaction studies and protease susceptibility assays indicated that TssM undergoes an ATP binding-induced conformational change and that subsequent ATP hydrolysis is crucial for recruiting Hcp to interact with the periplasmic domain of the TssM-interacting protein TssL (an IcmH/DotU family protein) into a ternary complex and mediating Hcp secretion. Our findings strongly argue that TssM functions as a T6SS energizer to recruit Hcp into the TssM-TssL inner membrane complex prior to Hcp secretion across the outer membrane.
Introduction
Protein secretion systems play central roles in the export or import of macromolecules in both prokaryotic and eukaryotic cells. In Gram-negative bacteria, the translocation of proteins across the inner (or cytoplasmic) membrane (IM)2 is mediated by either the general secretory (Sec) pathway (1) or the twin-arginine translocation (Tat) pathway (2). In addition, at least six types of specialized protein secretion systems, type I to type VI (T1SS–T6SS) have evolved in Gram-negative bacteria for protein/DNA transport in response to specific environmental cues (3–9).
The most recently discovered T6SS is highly conserved and widely distributed in pathogenic Proteobacteria (9–13). T6SSs are involved in virulence or anti-virulence, biofilm formation, and cytotoxicity to eukaryotic or prokaryotic hosts, suggesting the diversified functions of this secretion system (14–17). Secretion of hemolysin-coregulated protein (Hcp) to the extracellular milieu is the hallmark of a functional T6SS. A unique feature of Hcp and another commonly observed secreted extracellular protein (or exoprotein), VgrG (valine-glycine repeat protein G), is that they both lack a recognizable Sec- or Tat-dependent signal peptide and are an integral part of the secretion apparatus because of their mutual requirement for secretion (18–20). Hcp forms a hexamer ring with a 40 Å internal pore (21) and is capable of stacking in a tube-like structure in vitro (22). Remarkably, the structure and sequence analogy of Hcp (23) and VgrG (19, 24) to tail tube gp19 and spike gp5/gp27 of the T4 bacteriophage, respectively, has led to a proposed Hcp-VgrG phage tail-like structure. Moreover, two of the Vibrio cholerae T6SS components, TssB (VipA) and TssC (VipB), following the general type VI secretion (Tss) nomenclature (25), form a cogwheel-like tubular complex, which is also structurally similar to the T4 tail sheath protein (26, 27). In addition, T6SS also encodes components with sequence similarity to T4 phage gp25, a baseplate protein with lysozyme activity (23), even though no lysozyme activity could be detected (28). Therefore, it is conceivable that T6SS may assemble into an inverted phage tail-like surface appendage similar to the T3S needle or T4S pilus (29). However, whether and how this postulated structure is assembled in vivo and the molecular mechanisms underlying Hcp/VgrG secretion across double membranes remain largely unknown.
Bacterial secretion systems generally require ATPases or proton motive force to energize assembly of the secretion machinery and/or substrate translocation (30–34). In T6SSs, two proteins with known ATPase activity or nucleotide binding site may function as energizers. The AAA+-type ATPase TssH (ClpV) mediates the remodeling of cogwheel-like TssB/TssC (VipA/VipB) tubules via ATP hydrolysis, which is crucial for Hcp secretion (26, 35). The IcmF family protein TssM is a conserved T6SS component and putative NTPase in which the importance of its Walker A nucleotide binding site for Hcp secretion remains controversial (20, 36). TssM is a polytopic IM protein interacting with another conserved T6SS protein, TssL, a bitopic IcmH/DotU family IM protein, to form an IM protein complex (36). However, the functional role of TssM and TssL in Hcp secretion remains unclear.
Here, we have characterized the biochemical functions of TssM and its interacting protein, TssL, for their mechanistic roles in Hcp secretion in T6SS of the plant pathogenic bacterium Agrobacterium tumefaciens. We provide compelling evidence that TssM exhibits ATPase activity and its ATP hydrolysis but not ATP binding activity is crucial for assembly of TssM-TssL-Hcp IM complex and energizes the secretion of Hcp from A. tumefaciens. The direct interaction of Hcp with the periplasmic domain of TssL further suggested that TssM might function as an energizer to trigger periplasmic Hcp secretion across the OM.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Primers
Table 1 lists all of the strains and plasmids used in this study. The primers used are listed in supplemental Table S1. A. tumefaciens TssM and TssL were named previously ImpL and ImpK, respectively (36). The plasmids were maintained by the addition of 50 μg/ml gentamycin and 200 μg/ml spectinomycin for A. tumefaciens and 100 μg/ml ampicillin, 100 μg/ml spectinomycin, 50 μg/ml chloramphenicol, and 25 μg/ml gentamycin for Escherichia coli.
TABLE 1.
Strains and plasmids used in this study
| Strain/Plasmid | Genotype/Description | Source |
|---|---|---|
| E. coli | ||
| EN2 | E. coli BL21 (DE3) dnaK deleted strain | Ref. 39 |
| BL21(DE3) | Overexpressing proteins driven by T7 promoter | Ref. 74 |
| A. tumefaciens | ||
| C58 | Wild type virulent strain containing nopaline-type Ti-plasmid pTiC58 | Eugene Nester |
| ΔtssM | In-frame deletion of tssM (impL) in C58 | Ref. 36 |
| ΔtssMΔtssL | In-frame deletion of tssM (impL) and tssL (impK) in C58 | This study |
| Plasmids | ||
| pRL662 | GmR, vector derived from pBBR1MCS-2, lac promoter expression vector | Ref. 75 |
| pTrc200 | SpR, pVS1 origin lacIq, trc promoter expression vector | Ref. 76 |
| pTrc200HA | SpR, HA sequence inserted between PstI and HindIII site of pTrc200 | This study |
| pET-22b(+) | ApR, overexpression vector to generate C-terminal His-tagged protein driven by T7 promoter | Novagen |
| pACYCDuet-1 | CmR, vector for coexpression of two target genes | Novagen |
| pTssM | GmR, pRL662 expressing TssM (ImpL) | Ref. 36 |
| pTssL | SpR, pTrc200 expressing TssL (ImpK) | This study |
| pTrc-TssL-His | SpR, pTrc200 expressing His-tagged TssL (ImpK) | This study |
| pTssMG144A/K145A | GmR, pRL662 expressing TssM (ImpL) with both G144A and K145A substitutions | Ref. 36 |
| pTssMD188A/G191A | GmR, pRL662 expressing TssM (ImpL) with both D188A and G191A substitutions | This study |
| pRL662-Hcp | GmR, pRL662 expressing Hcp | Ref. 38 |
| pTrc-Hcp | SpR, pTrc200 expressing Hcp | This study |
| pET-TssL-His | ApR, pET-22b(+) overexpressing His-tagged TssL (ImpK) | Ref. 36 |
| pET-TssL(Peri)-His | ApR, pET-22b(+) overexpressing periplasmic domain of His-tagged TssL (ImpK) | Ref. 36 |
| pET-TssL(Cyto)-HA | ApR, pET-22b(+) overexpressing cytoplasmic domain of HA-tagged TssL (ImpK) | This study |
| pTrc-TssM-HA | SpR, pTrc200 expressing HA-tagged TssM (ImpL) | This study |
| pET-TssM-His | ApR, pET-22b(+) overexpressing His-tagged TssM (ImpL) | This study |
| pET-TssMG144A/K145A-His | ApR, pET-22b(+) overexpressing His-tagged TssM (ImpL) with both G144A and K145A substitutions | This study |
| pET-TssM4PM-His | ApR, pET-22b(+) overexpressing His-tagged TssM (ImpL) with G139AG144A K145AT146A substitutions | This study |
| pACYC-TssL-TssM -His | CmR, pACYCDuet-1 co-overexpressing TssL (ImpK) and His-tagged TssM (ImpL) | This study |
| pACYC-TssL-TssMG144A/K145A-His | CmR, pACYCDuet-1 co-overexpressing TssL (ImpK) and His-tagged TssM (ImpL) with both G144A and K145A substitutions | This study |
| pACYC-TssL-TssMD188A/G191A-His | CmR, pACYCDuet-1 co-overexpressing TssL (ImpK) and His-tagged TssM (ImpL) with both D188A and G191A substitutions | This study |
| pET-Hcp-His | ApR, pET22b overexpressing His-tagged Hcp | Ref. 38 |
Plasmid Construction and Generation of In-frame Deletion Mutants
All in-frame deletion mutants were generated in A. tumefaciens strain C58 via double crossover using the suicide plasmid pJQ200KS (37) as described (36, 38). For plasmids used in protein purification, the plasmid pET-TssM-His was created by ligating the NdeI/XhoI-digested tssM PCR product (primers 1 and 2) into the same site of pET22b-(+). To construct plasmid pET-TssMG144A/K145A-His or pET-TssM4PM-His, the 1.5-kb NdeI/BamHI-digested tssM fragment from the plasmid pET-TssM-His was removed and replaced with the 1.5-kb NdeI/BamHI-digested tssM PCR product containing either the G144A/K145A mutation (primers 3a and 3b and primers 4a and 4b) or the G139A/G144A/K145A/T146A mutation (primers 5a and 5b and primers 6a and 6b), respectively. For plasmid pACYC-TssL-TssM-His, the tssL PCR product (primers 7a and 7b) was cloned into the NcoI/EcoRI sites of pACYCDUET-1 to create plasmid pACYC-TssL. The tssM PCR product (primers 8a and 8b) was cloned into the NdeI/XhoI sites of pACYC-TssL resulting in pACYC-TssL-TssM-His. The plasmids pACYC-TssL-TssMG144A/K145A-His (primers 3a and 3b and primers 4a and 4b) and pACYC-TssL-TssMD188A/G191A-His (primers 9a and 9b and primers 10a and 10b) were constructed following the procedure described for construction of pET-TssMG144A/K145A-His.
For plasmid pTssL, the tssL ORF was amplified (primers 21 and 22) and cloned into the NcoI/BamHI sites of pTrc200. For plasmids used in complementation or protein-protein interaction, the plasmid pTssMD188A/G191A was created by replacing the 1.5-kb XhoI/BamHI-digested tssM fragment of plasmid pRL662-TssM (36) with the XhoI/BamHI-digested PCR product (primers 11a and 11b and primers 12a and 12b). For plasmid pTrc-Hcp, the hcp ORF was amplified (primers 13 and 14) and cloned into the NcoI/XbaI sites of pTrc200. For plasmid pTrc-TssL(Cyto)-HA, the PCR product of tssL (primers 15 and 16) encoding amino acids 1–255 was digested by NdeI/BamHI and cloned into the same site of pTrc200HA. Then, the plasmid pTrc-TssL(Cyto)-HA was used as a template for amplifying the tssL(Cyto)-HA PCR product (primers 17 and 18). The PCR product was digested with NdeI/HindIII and ligated into the same site of pET22b-(+) to result in plasmid pET-TssL(Cyto)-HA. For plasmid pTrc-TssM-HA, the tssM ORF was amplified (primers 19 and 20) and cloned into the KpnI/XbaI sites of pTrc200HA. For plasmid pTssL-His, the tssL ORF with His6 sequences was amplified (primers 23 and 24) and cloned into the NcoI/XbaI sites of pTrc200.
TssM Protein Purification for ATP Binding and Hydrolysis Activity Assay
His-tagged wild type (WT) TssM protein or its variants were co-expressed with TssL, or His-tagged TssL was expressed alone in E. coli dnaK deletion mutant strain EN2 (39) and purified for ATP binding and the hydrolysis activity assay. The plasmids used in co-expressing WT TssM or its variants with TssL are indicated in each experiment. The strains were grown in Luria broth medium at 30 °C till A600 = 0.6. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mm, and growth continued for 4 h at 25 °C. The cells were suspended in lysis buffer (50 mm Tris-Cl, pH 7.5, 0.3 m NaCl, 0.1 mm EDTA, 1 mm DTT, 10% glycerol, 1 mm PMSF, and 0.5 mg/ml lysozyme) supplemented with 0.1 mg/ml DNase and 0.1 mg/ml RNase. After two passages through a French pressure cell at 16,000 p.s.i., the unbroken cells were removed by centrifugation at 20,000 × g for 15 min at 4 °C. The supernatant was further centrifuged at 150,000 × g for 1 h at 4 °C. The pellet, enriched with membrane proteins, was incubated in extraction buffer (50 mm Tris-Cl, pH 7.5, 0.1 mm EDTA, 0.1 m NaCl, 10% glycerol, 1 mm DTT, and 2% n-dodecyl-β-d-maltoside (DDM)) overnight and centrifuged at 150,000 × g for 1 h to obtain the supernatant containing the DDM-soluble membrane proteins. The DDM-solubilized proteins were further adjusted to final buffer condition (50 mm Tris-Cl, pH 7.5, 0.3 m NaCl, 10% glycerol, 0.1% DDM, and 10 mm imidazole) before preincubation with Ni2+-nitrilotriacetic acid (Ni-NTA) resin for 1 h. The precleared protein sample was loaded onto an NTA column, washed with washing buffer (50 mm Tris-Cl, pH 7.5, 0.15 m NaCl, 10% glycerol, 0.05% DDM, and 15 mm imidazole), and eluted with elution buffer (50 mm Tris-Cl, pH 7.5, 0.05 m NaCl, 10% glycerol, 0.05% DDM, and 200 mm imidazole). The eluted fractions were concentrated and exchanged to a buffer containing 50 mm Tris-Cl, pH 7.5, 0.05 m NaCl, 10% glycerol, and 0.05% DDM using either 100-kDa Microcon cut-offs (Millipore YM-100) for WT TssM and its variants or 30-kDa Microcon cut-offs for negative control TssL-His. The purified WT TssM and its variants were kept on ice until ATP binding or the ATPase activity assay within 3 days after purification to avoid protein degradation and loss of activity.
ATP Binding Assay
The TssM nucleotide binding activity was determined using a fluorescent ATP analog (TNP-ATP; Molecular Probes, Inc.) monitored by a Jasco FP-6500 spectrofluorometer. Purified TssM proteins were resuspended at 0.25 μm in reaction buffer (50 mm Tris-HCl, pH 7.5, 0.05 m NaCl, 0.05% DDM) in the presence or absence of 2 μm TNP-ATP and incubated at room temperature for 1 min. Fluorescence was excited at 410 nm, and emission was scanned in the range of 470 to 650 nm. All spectra were corrected by subtracting the background fluorescence emitted from buffer control.
ATP Hydrolysis Analysis
ATPase activity was determined using a malachite green assay (40, 41) with minor modifications. Briefly, 8 pmol of protein was resuspended in 120 μl of reaction buffer (50 mm Tris-HCl, pH 7.5, 1 mm DTT, 0.1% Nonidet P-40, 5 mm MgCl2, 25 μm zinc acetate, 5 mm potassium acetate, and 1 mm ATP) and incubated at 37 °C. The reaction mixture was mixed with 480 μl of malachite green reagent (0.03% malachite green hydrochloride, 1 n HCl, 1.05% ammonium molybdate, and 0.1% Triton X-100) to monitor the release of inorganic phosphate (Pi). 60 μl of 34% sodium citrate was added to stop the reaction, and further incubated at room temperature for 40 min before reading at 650 nm. Each sample was assayed at 0 and 120 min in triplicate, and the total Pi for each reaction was compared with a Pi standard curve prepared with K2HPO4. All readings were corrected by subtracting the background values from “no protein” controls.
In Vitro Pulldown Assay of TssL-TssM-HA Complex and Hcp-His
Hcp-His and TssL-TssM-HA complex were purified separately from E. coli BL21(DE3) before the in vitro pulldown assay. For Hcp-His protein purification, E. coli BL21(DE3) harboring Hcp-His plasmid (pET-Hcp-His) was grown at 37 °C until A600 = 0.5. IPTG was added at 0.4 mm final concentration, and growth continued for 2 h. The cell pellet resuspended in lysis buffer (20 mm Tris-Cl, pH 7.5, 0.5 m NaCl, 15 mm imidazole, 0.2 mg/ml DNase, 0.2 mg/ml RNase, and 1 mm PMSF) was lysed by a French pressure cell as described above. After removal of the unbroken cells and membranes, the cell lysate was loaded onto an Ni2+-NTA affinity column, washed with lysis buffer, and eluted with elution buffer as described above, except 250 mm imidazole was used. The eluted fraction was loaded onto a Superdex 200 column pre-equilibrated with buffer containing 20 mm Tris-Cl, pH 7.5, and 0.3 m NaCl. The fractions containing the Hcp-His peak were pooled and concentrated with 10-kDa Microcon cut-offs. The purified Hcp-His was stored at −20 °C until used for the in vitro pulldown assay with the TssL-TssM-HA complex.
For purification of the TssL-TssM-HA protein complex, the E. coli BL21(DE3) cells expressing both TssM-HA (pTssM-HA) and TssL (pET-TssL) were grown at 37 °C until A600 = 0.6 before the addition of IPTG (1 mm final concentration), and growth was continued for 4 h at 25 °C. The cell pellet was suspended in lysis buffer (50 mm Tris-Cl, pH 8.0, 0.3 m NaCl, 1 mm EDTA, 1 mm DTT, 1 mm PMSF, 0.5 mg/ml lysozyme, 0.2 mg/ml DNase, and 0.2 mg/ml RNase) and lysed by a French pressure cell as described above. The membrane pellet was resuspended in extraction buffer (50 mm Tris-Cl, pH 8.0, 0.15 m NaCl, 1 mm PMSF, and 1% DDM) and rocked overnight at 4 °C. The DDM-solubilized proteins were incubated with anti-HA-agarose beads (Sigma) at 4 °C for 2 h. After washing with 50 mm Tris-Cl, pH 8.0, 0.15 m NaCl, and 0.05% DDM, the beads containing the TssL-TssM-HA complex were further incubated with 0.04 unit/μl hexokinase and 1 mm glucose for 15 min at room temperature in buffer containing 50 mm Tris-Cl, pH 8.0, 0.15 m NaCl, 0.05% DDM, and 1 mm MgCl2. After washing with 50 mm Tris-Cl, pH 8.0, 0.15 m NaCl, 0.05% DDM, and 5 mm MgCl2 to remove hexokinase, the TssL-TssM-HA complex retained on the beads was used for a pulldown assay with purified Hcp-His. Then 2.5 μg of purified Hcp-His proteins was added to the 1 ml of anti-HA beads containing the TssL-TssM-HA complex and was aliquotted evenly before incubation with 1 mm ATP or non-hydrolyzable ATP analog (AMMNP; Sigma) overnight at 4 °C. The beads were washed three times with buffer (50 mm Tris-Cl, pH 8.0, 0.15 m NaCl, and 0.05% DDM); 2× Laemmli buffer was added, and the mixture was boiled for 10 min before SDS-PAGE followed by immunoblotting.
ATP Measurements
The ATP levels in untreated and arsenate-treated A. tumefaciens cells were measured by an ATP bioluminescence assay kit (CLSII; Roche Applied Science). Briefly, 100 μl of cells resuspended in 80% DMSO to A600 = 1 was mixed with 400 μl of ice-cold H2O. The amounts of ATP were measured for luminescence using a luminometer (Lumat LB9507; Berthold Technologies) by mixing 20 μl of the sample with 100 μl of luciferase reagent.
Spheroplast Preparation and Protease Susceptibility Assay
A. tumefaciens cells grown in AB-MES medium, pH 5.5, (42) at 25 °C for 6 h until A600 = 0.3 were treated with 25 mm sodium arsenate or H2O for 1 h. Cells were harvested by centrifugation and washed once with 50 mm Tris-Cl, pH 7.5. To prepare the spheroplast, the cell pellet was resuspended gently in buffer containing 50 mm Tris-Cl, pH 7.5, 20% sucrose, 2 mm EDTA, 0.2 mm DTT, and 0.5 mg/ml lysozyme and incubated on ice for 1 h. The spheroplasts were treated with Streptomyces griseus protease (Sigma) at a final concentration of 25, 50, 75, or 100 μg/ml for 10 min on ice in the presence of 10 mm MgCl2. The reaction was stopped by adding Laemmli buffer to 1× and immediately boiled for 10 min before SDS-PAGE followed by immunoblotting.
Pulldown Assay in A. tumefaciens
The membrane fraction from the A. tumefaciens ΔtssMΔtssL strain complemented with various plasmids (pTssL-His + pTssM, pTssMG144A/K145A, pTssMD188A/G191A, or pRL662 vector) was prepared as described above for protein purification. Membrane proteins were solubilized with 2% DDM in the presence of 50 mm Tris-Cl, pH 7.5, and 1 mm PMSF by rocking overnight at 4 °C. The solubilized membrane proteins were harvested and further diluted to 0.05% of DDM in buffer containing 50 mm Tris-Cl, pH 7.5, 0.3 m NaCl, and 10 mm imidazole. The protein samples were loaded onto NTA-resin and washed with 10× bed volumes of wash buffer (50 mm Tris-Cl, pH 7.5, 0.3 m NaCl, 15 mm imidazole, and 0.01% DDM). The proteins were eluted with elution buffer (50 mm Tris-Cl, pH 7.5, 0.15 m NaCl, 250 mm imidazole, and 0.01% DDM). The eluted fraction was precipitated with trichloroacetic acid overnight at 4 °C (38). The trichloroacetic acid pellet was resuspended in 1× Laemmli buffer and analyzed with SDS-PAGE followed by immunoblotting.
Hcp Secretion Assay
A. tumefaciens cells grown in AB-MES medium, pH 5.5 (42), at 25 °C for 6 h were used for Hcp secretion assays as described previously (36, 38).
Immunoblotting
Immunoblot analysis was performed as described previously (42) using primary polyclonal antibody against C-terminal TssM (C-TssM) (36), N-terminal TssL (N-TssL, amino acids 1–255), Hcp (38), ActC (43), or GroEL (44) followed by secondary antibody using horseradish peroxidase-conjugated goat anti-rabbit and detection using the Western Lightning System (PerkinElmer Life Sciences). Chemiluminescent signals were visualized on a high performance chemiluminescence film (GE Healthcare) or by using the BioSpectrum AC Imaging System (Ultra-Violet Products, Ltd.) to detect and quantify the photon intensity of protein signals.
RESULTS
TssM Exhibits ATPase Activity, Which Is Crucial for Hcp Secretion
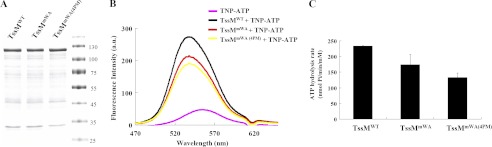
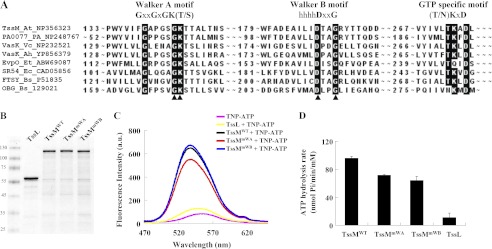
The conservation of the A. tumefaciens TssM Walker A motif in IcmF family proteins and the genetic evidence for its requirement in mediating Hcp secretion suggests that TssM functions as an ATPase (36). However, biochemical evidence for this assumption is lacking. Because TssM is a 128-kDa IM protein with three transmembrane domains (36), it has been a challenging task to purify and maintain the stability of full-length TssM fused with various fusion proteins or tags (data no shown). Nonetheless, we were able to obtain full-length TssM-His with high purity within 3 days after purification even though the proteolysis of full-length TssM could not be completely eliminated (Fig. 1A). The E. coli dnaK mutant strain EN2 (39) was used to overexpress and purify TssM-His in the presence of TssL, which stabilizes TssM in A. tumefaciens (36) to avoid contamination of the E. coli DnaK ATPase during purification. The His-tagged wild type TssM (TssMWT) and the Hcp secretion-deficient Walker A mutation variant, TssMG144A/K145A (TssMmWA), were purified in parallel by extraction from E. coli membranes with nonionic detergent DDM and preincubation with nickel resin to remove nonspecific binding proteins before purification on a nickel resin column. Purified WT TssM (TssMWT) possesses ATP binding activity, as evidenced by the increased fluorescence emission intensity upon binding to the fluorescent ATP analog TNP-ATP (45–47), and it exhibited ATP hydrolysis activity (Fig. 1, B and C). Surprisingly, the Walker A variant, TssMmWA, retained significant albeit lower ATP binding and hydrolysis activity as compared with TssMWT (Fig. 1, B and C).
FIGURE 1.
TssM binds and hydrolyzes ATP. A, DDM-extracted membrane proteins from E. coli strain EN2 co-expressing WT or variants of TssM-His (pET-TssM-His, pET-TssMG144A/K145A-His, or pET-TssMG139A/G144A/K145A/T146A-His) with TssL (pTssL) were purified by Ni-NTA resin. The purity of wild type TssM-His (TssMWT), TssMG144A/K145A-His (TssMmWA), and TssMG139A/G144A/K145A/T146A-His (TssMmWA(4PM)) used for ATP binding and hydrolysis assays was examined by Coomassie Blue-stained SDS-PAGE. The sizes of the molecular mass standards (in kDa; Fermentas Inc.) are indicated. B, ATP binding activity of WT and its variants. Fluorescence spectra of TNP-ATP bound to WT and its variants were detected in the range of 470 to 650 nm. The spectrum for each sample is indicated as WT TssM-His (black, TssMWT +TNP-ATP), TssMG144A/K145A-His (red, TssMmWA +TNP-ATP), TssMG139A/G144A/K145A/T146A-His (yellow, TssMmWA(4PM) +TNP-ATP), or TNP-ATP alone (pink). At least three independent experiments were performed for which representative data are shown. C, ATP hydrolysis activity of WT and its variants. The ATP hydrolysis activity was determined using a malachite green assay to measure the released free inorganic phosphate upon ATP hydrolysis. Average values for ATP hydrolysis activity from three independent experiments are shown with standard deviations.
To determine whether the detected ATP binding and hydrolysis activities of the Walker A variant derived from contaminating ATPases co-purified from E. coli, we generated an additional TssM variant expressing the full-length protein with mutations in two additional conserved amino acids in the Walker A motif of TssM (TssMmWA(4PM): G139A/G144A/K145A/T146A). Further reduction of TssMmWA(4PM) in ATP binding and hydrolysis activities suggested that TssMmWA indeed does not completely lose its activity in ATP binding and hydrolysis. Because this Walker A variant, TssMmWA, caused the loss of Hcp secretion from A. tumefaciens (36), we suggest that TssM exhibits ATPase activity and its full ATPase activity is required to mediate Hcp secretion.
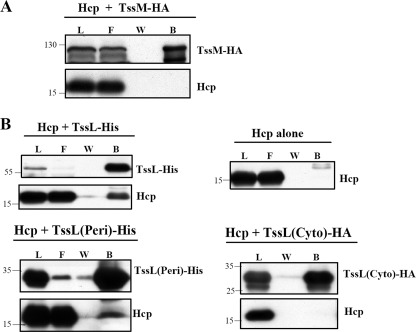
Hcp Interacts Directly with TssL but Not TssM
The importance of TssM ATPase activity in mediating Hcp secretion motivated us to investigate whether TssM interacts with Hcp and functions as an energizer to drive Hcp secretion across bacterial membrane. However, no evidence for an interaction between TssM and Hcp was detected by co-immunoprecipitation of TssM tagged with HA in E. coli co-expressing Hcp (Fig. 2A). Because TssM forms a protein complex with TssL (36), we then tested whether Hcp might interact with TssL. Indeed, TssL-His could pull down Hcp from E. coli that co-expressed both TssL-His and Hcp (Fig. 2B). As a control, no Hcp was detected in the bound fraction purified from E. coli expressing Hcp alone.
FIGURE 2.
Interaction studies of Hcp with TssL and TssM. A, the cell lysate of E. coli co-expressing Hcp with full-length TssM tagged with HA (TssM-HA) was subjected to co-immunoprecipitation using anti-HA beads. B, the cell lysate of E. coli expressing Hcp alone or in the presence of full-length TssL tagged with His (TssL-His), the C-terminal periplasmic domain of TssL-His (TssL(Peri)-His), or the N-terminal cytoplasmic domain of TssL (TssL(Cyto)-HA) were purified by either Ni2+-NTA affinity chromatography or co-immunoprecipitation using anti-HA beads. The load (L), flow-through (F) and washed (W) and bound (B) fractions were analyzed by immunoblotting with antisera against C-TssM, N-TssL, Hcp, or His6 (for TssL(Peri)-His), respectively. The sizes of the molecular mass standards (kDa) are indicated.
To dissect the Hcp interaction domain of TssL, we co-expressed Hcp with the N-terminal cytoplasmic domain of TssL (TssL(Cyto)-HA) or the C-terminal periplasmic domain of TssL (TssL(Peri)-His) in E. coli. Hcp co-eluted with the periplasmic domain of TssL-His in the bound fraction (Fig. 2B). In contrast, no Hcp was detected in the bound fractions purified from E. coli co-expressing TssL(Cyto)-HA with Hcp. From this data, we concluded that Hcp interacts specifically with the periplasmic domain of TssL.
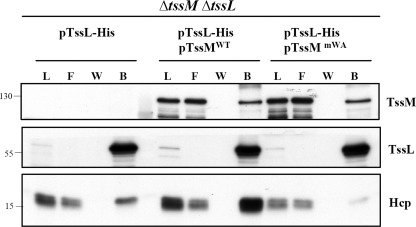
Full ATPase Activity of TssM Is Crucial for Assembly of TssM-TssL-Hcp Complex
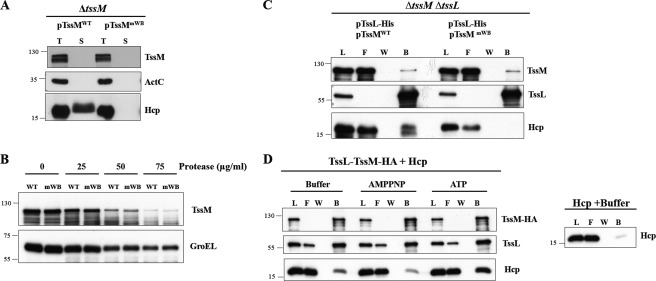
The previously shown TssM-TssL interaction (36) and our current discovery of a TssL-Hcp interaction in E. coli urged us to investigate whether Hcp interacts with the TssM-TssL IM protein complex in A. tumefaciens, and if so, whether the ATPase activity of TssM plays a role in this process. To answer this question, we performed a pulldown assay in the A. tumefaciens ΔtssMΔtssL double mutant expressing TssL-His either in the presence or absence of WT TssMWT or Walker A variant TssMmWA. The protein complexes were extracted by DDM from the membrane fraction followed by Ni2+-resin purification. TssL-His was able to pull down Hcp in the presence or absence of TssM in A. tumefaciens (Fig. 3). However, although TssL-His could pull down either TssMWT or TssMmWA at similar levels, only trace amounts of Hcp could be pulled down by TssL-His from A. tumefaciens expressing TssMmWA as compared with that expressing TssMWT (Fig. 3). These data suggested that the formation of the TssM-TssL complex is independent of TssM ATPase activity, whereas its full ATPase activity is important for the formation of the TssM-TssL-Hcp IM complex in A. tumefaciens.
FIGURE 3.
Full ATPase activity of TssM is crucial for assembly of TssM-TssL-Hcp complex. Membrane fractions isolated from the A. tumefaciens ΔtssMΔtssL double mutant expressing TssL-His alone or in the presence of WT TssM (TssMWT) or TssMG144A/K145A (TssMmWA) were solubilized with DDM. The DDM-solubilized membrane proteins were loaded onto Ni2+-NTA resin to purify TssL-His and its interacting partner, TssM. The load (L), flow-through (F) and washed (W) and bound (B) fractions were analyzed by immunoblotting with antisera against C-TssM, N-TssL, and Hcp, respectively. The sizes of the molecular mass standards (kDa) are indicated.
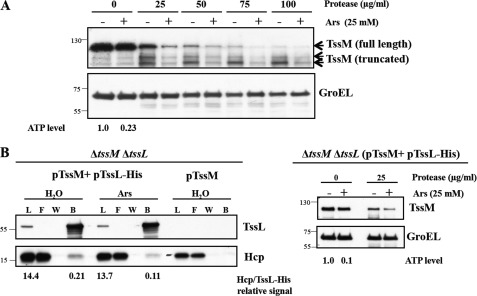
Cellular ATP Levels Modulate TssM Conformation and TssM-TssL-Hcp Complex Formation
It is plausible that ATP binding or hydrolysis of TssM may induce a structural transition for promoting the interaction of TssL and Hcp. To test this possibility, we monitored the conformational change of TssM by its susceptibility to protease digestion, an assay that has been used to detect the VirB10 and TonB conformational switch under energy-dissipated conditions (48, 49). A. tumefaciens cells were treated with either H2O or arsenate to dissipate intracellular ATP without affecting transmembrane potential (48), and the spheroplasts were prepared for the protease susceptibility assay. As expected from its topology (36), TssM was prone to protease digestion, and an increase in protease concentration caused the reduction of full-length TssM along with the presence of truncated TssM. Arsenate treatment was able to deplete about 80% of the cellular ATP level and increased TssM susceptibility to protease as compared with that of the H2O-treated condition (Fig. 4A). As a control, the cytoplasmic GroEL protein remained resistant to protease digestion in the presence or absence of arsenate, demonstrating that treatment with arsenate did not cause detectable nonspecific cell lysis or proteolysis. The result, that TssM was more resistant to protease digestion at high intracellular ATP levels, indicated that TssM undergoes a conformational switch between the ATP-bound and unbound state.
FIGURE 4.
Cellular ATP levels modulate TssM conformation and TssM-TssL-Hcp complex formation. A, spheroplasts prepared from arsenate-treated (Ars) or untreated (H2O) A. tumefaciens WT C58 cells were incubated with different protease concentrations. The level of susceptibility or resistance to protease was analyzed by immunoblotting with antisera against C-TssM and GroEL. Cytoplasmic GroEL serves as an internal control, as its protein level is not affected by energy depletion and resistance to protease. The cellular ATP levels were quantified and presented as the ratio relative to the H2O-treated control. B, detection of TssL-His-Hcp complex formation in energy-depleted condition. The A. tumefaciens ΔtssMΔtssL double mutant co-expressing TssM in the absence or presence of TssL-His was treated with arsenate or H2O followed by DDM extraction of isolated membrane fractions. The DDM-soluble membrane proteins were purified by Ni2+-NTA affinity chromatography (left panel). The formation of TssL-His-Hcp complexes was analyzed by immunoblotting with antisera against C-TssM and Hcp, respectively. The relative photon intensity of Hcp used for pulldown or bound to the same amount of purified TssL-His from H2O or arsenate-treated cells was quantified as described under “Experimental Procedures.” The conformational change of TssM in the ΔtssMΔtssL double mutant complemented with TssL-His and TssM was monitored by protease susceptibility, revealing the same susceptibility patterns as the WT C58 strain (right panel). The cellular ATP levels were quantified and presented as the ratio relative to the H2O-treated control.
We next determined whether cellular ATP levels affect the formation of the TssM-TssL-Hcp complex, which may be modulated by the conformational switch of TssM. The ΔtssMΔtssL mutant complemented with pTssM and pTssL-His plasmids was treated with either H2O or arsenate prior to the pulldown assay. Similar to the observation in the WT strain (Fig. 4A), TssM in the complemented strain revealed higher susceptibility to protease in an ATP-depleted condition (Fig. 4B, right panel). By quantitative immunoblot analysis, the relative protein signals of Hcp to TssL-His used for the pulldown assay from the cells treated with either H2O or arsenate were detected at a similar ratio (14.4 versus 13.7). In the ATP-depleted condition, we consistently detected only ∼50% of Hcp proteins that were pulled down by TssL-His as compared with that of Hcp bound by the same amount of TssL-His from H2O-treated cells (0.21 versus 0.11; Fig. 4B, left panel). As a control, no Hcp was detected in the bound fraction in the absence of TssL-His. Taken together, the cellular ATP levels regulate both a conformational switch in TssM and efficient assembly of the TssM-TssL-Hcp IM complex.
TssM Possesses a Walker B Motif That Is Crucial for ATP Hydrolysis
The retention of ATP binding and hydrolysis activities exhibited by the Walker A variant, TssMmWA (Fig. 1, B and C), motivated us to determine whether other motifs in addition to Walker A may contribute to these activities. By careful sequence inspection of TssM, we identified a putative Walker B motif that is highly conserved among T6SS orthologs and known GTPase family members. The identified Walker B motif contains a signature sequence, hhhhDXXG, to coordinate Mg2+, which is required for ATP hydrolysis. Interestingly, a distal (N/T)KXD, which provides specificity for guanine over other nucleotide bases (50, 51), was also identified (Fig. 5A).
FIGURE 5.
TssM possesses a Walker B motif required for its ATP hydrolysis. A, Walker A, Walker B, and potential GTP-specific motifs were revealed by amino acid sequence alignment of selected T6SS TssM orthologs and other GTPase family members. The conserved amino acid residues of each motif are highlighted; the arrowheads indicate the amino acids targeted for mutagenesis. The proteins used for alignments are indicated in the order of their protein name, organism, and GenbankTM accession number. At, A. tumefaciens C58; PA, P. aeruginosa PAO1; Vc, V. cholerae O1; Ah, A. hydrophila ATCC 7966; Et, E. tarda PPD130/91; Ec, E. coli CT18; Bs, Bacillus subtilis. B, DDM-extracted membrane proteins from E. coli EN2 strain expressing the WT or a variant of TssM-His (TssMWT: pACYC-TssL-TssM-His; TssMmWA: pACYC-TssL-TssMG144A/K145A-His; TssMmWB: and pACYC-TssL-TssMD188A/G191A-His) or TssL-His (TssL: pET-TssL-His) were purified in parallel using Ni-NTA resin. The purity of TssMWT, TssMmWA, TssMmWB, and TssL used for ATP binding and hydrolysis assays was examined by Coomassie Blue-stained SDS-PAGE. The sizes of the molecular mass standards (kDa; Fermentas) are indicated. C, ATP binding activity of TssM and its variants. Fluorescence spectra of TNP-ATP bound to TssM and its variants were detected in the range of 470 to 650 nm. The spectrum for each sample was indicated as WT TssM-His (black, TssMWT +TNP-ATP), TssMG144A/K145A-His (red, TssMmWA +TNP-ATP), TssMD188A/G191A-His (blue, TssMmWB +TNP-ATP), TssL-His (yellow, TssL +TNP-ATP), or TNP-ATP alone (pink). D, ATP hydrolysis activity of TssM and its variants. ATP hydrolysis activity was determined using the malachite green assay to measure the free inorganic phosphate released upon ATP hydrolysis. Average values for ATP hydrolysis activity from three independent experiments are shown with standard deviations.
Thus, we generated mutations in the putative Walker B motif by replacing both conserved Asp-188 and Gly-191 with alanine (Ala) using site-directed mutagenesis. The Walker B mutant (TssMmWB: TssMD188A/G191A) was overexpressed and purified in parallel with wild type TssMWT, Walker A mutant TssMmWA, in the presence of TssL for ATPase activity assays (Fig. 5, B–D). In addition, His-tagged TssL serving as a negative control (TssL) was purified in parallel. The mutations in the Walker B motif did not result in any loss of ATP binding activity, because the TssMmWB bound the fluorescence TNP-ATP as well as the TssMWT (Fig. 5C). In contrast, the hydrolysis activity of TssMmWB was reduced to a similar level to that of TssMmWA (Fig. 5D). The fact that much lower ATPase activity was detected from purified TssL as compared with that of TssMmWA and TssMmWB suggested that TssMmWB may not completely lose its ATPase activity. Because TssMmWB retained WT-like ATP binding affinity, its reduced ATP hydrolysis activity should be attributed to the mutations in its Walker B motif. Thus, we discovered that TssM possesses a Walker B motif that functions in ATP hydrolysis.
ATP Hydrolysis of TssM Drives TssM-TssL-Hcp Complex Formation and Powers Hcp Secretion
It is clear that the reduced ATP binding/hydrolysis activity of the Walker A variant, TssMmWA, caused inefficient assembly of the TssM-TssL-Hcp complex (Fig. 3) and a deficiency in Hcp secretion (36). However, it remained unanswered whether the structural transition and subsequent Hcp secretion were induced by the ATP binding or the hydrolysis activity of TssM. With the ATP binding/hydrolysis uncoupling of Walker B variant TssMmWB at hand, we asked whether ATP hydrolysis by TssM was required for Hcp secretion. We expressed both TssMWT and TssMmWB in the A. tumefaciens ΔtssM mutant and performed Hcp secretion assays. The Walker B variant resulted in a complete loss of Hcp secretion to the culture media (Fig. 6A), suggesting that TssM mediates Hcp secretion via its ATP hydrolysis activity. To investigate whether the observed conformational switch of TssM is induced by ATP binding or hydrolysis, we performed protease susceptibility assays to determine whether the mutation in the Walker B motif altered protease susceptibility. As shown in Fig. 6B, TssMmWB had a similar degree of protease resistance as TssMWT, suggesting that the reduced ATP hydrolysis activity did not compromise its ATP-regulated conformational switch. Taken together, we concluded that ATP binding per se is able to induce a conformational switch of TssM but is not sufficient to trigger secretion of Hcp.
FIGURE 6.
Effect of TssM ATP hydrolysis activity in Hcp secretion and the formation of TssM-TssL-Hcp complex and its conformational change. A, total proteins (T) and secreted proteins (S) isolated from the A. tumefaciens ΔtssM mutant harboring plasmids expressing either WT TssM (pTssMWT) or Walker B amino acid substitution mutant TssMD188A/G191A (pTssMmWB) were analyzed by immunoblotting using antisera against Hcp, C-TssM, and ActC, respectively. ActC serves as a nonsecreted protein control. The positions of the molecular mass markers (in kDa) are indicated. B, the A. tumefaciens ΔtssMΔtssL strain harboring plasmids expressing TssL-His (pTssL-His) with either WT TssM (pTssM) or the Walker B mutant (pTssMD188A/G191A) was subjected to the spheroplast protease susceptibility assay at different protease concentrations as indicated. The degree of susceptibility or resistance to protease in ΔtssMΔtssL (pTssL-His + pTssM) (WT) and ΔtssMΔtssL (pTssL-His + pTssMD188A/G191A) (mWB) was analyzed by immunoblotting using antisera against C-TssM and GroEL, respectively. C, DDM-soluble membrane proteins isolated from ΔtssMΔtssL (pTssL-His + pTssMWT) and ΔtssMΔtssL (pTssL-His + pTssMmWB) were purified by Ni2+-NTA affinity chromatography. The load (L), flow-through (F), and washed (W) and bound (B) fractions were analyzed by immunoblotting with antisera against N-TssL and Hcp, respectively. The positions of molecular mass markers (in kDa) are indicated. D, in vitro assay for TssM-TssL-Hcp complex formation in the presence of ATP or non-hydrolyzable ATP. TssL-TssM-HA protein complex purified from E. coli membrane was treated with hexokinase to remove cellular ATP and then incubated with purified Hcp-His in the presence or absence of ATP or non-hydrolyzable ATP (AMPPNP) followed by co-immunoprecipitation using anti-HA-agarose beads. The load, flow-through and washed and bound fractions were analyzed by immunoblotting with antisera against N-TssL, Hcp, and C-TssM, respectively. The right panel shows the background signal of nonspecific binding of Hcp-His to the beads in absence of TssL-TssM-HA complex.
Next, we investigated whether ATP binding is sufficient to recruit Hcp into the TssM-TssL complex for the assembly of a ternary complex. Thus, we performed the pulldown assay using the DDM-extracted membrane complex isolated from the A. tumefaciens ΔtssMΔtssL mutant expressing either the WT or Walker B variant (TssMmWB) in the presence of TssL-His. Although similar amounts of TssMWT or TssMmWB could be pulled down by TssL-His, no Hcp could be detected in the TssMmWB-TssL-His complex (Fig. 6C). This result suggested that the ATP hydrolysis activity of TssM is crucial for recruiting Hcp into the TssM-TssL IM complex in A. tumefaciens. Notably, the ATP binding-induced conformational change in TssM was not sufficient to trigger the formation of the TssM-TssL-Hcp complex.
We noticed that the membrane-associated Hcp protein amounts were lower in either the absence of TssM or the presence of TssMmWA or TssMmWB variants as compared with in the presence of TssMWT (Figs. 3 and 6C). Thus, the deficiency of Hcp to be pulled down by TssL in the mutants could be partly due to the lower abundance of membrane-associated Hcp. Therefore, we designed an in vitro pulldown assay to confirm the role of TssM ATP hydrolysis in this ternary complex formation when equal amounts of Hcp were present. We incubated purified Hcp-His with the purified TssL-TssM-HA complex to determine the formation of the ternary complex in the presence of ATP or the non-hydrolyzable ATP analog AMPPNP in vitro. The purified TssL-TssM-HA complex was pretreated with hexokinase to eliminate the contaminated ATP co-purified from E. coli (52). By co-immunoprecipitation using anti-HA-agarose beads, we found that much more Hcp-His was co-eluted with the TssL-TssM-HA complex in the presence of ATP than in the presence of AMPPNP or buffer controls (Fig. 6D). Because trace amounts of Hcp-His were nonspecifically bound to anti-HA beads in the absence of the TssL-TssM-HA complex, the low levels of Hcp co-immunoprecipitated with TssL-TssM-HA complex may be due to the nonspecific binding of Hcp-His to the beads under this experimental condition. Taken together, our results strongly argue that TssM is an energizer that hydrolyzes ATP to recruit Hcp into the TssM-TssL IM complex, which may be a prerequisite step for Hcp secretion across the OM.
DISCUSSION
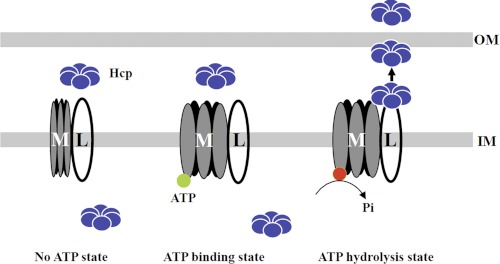
TssM and TssL are core T6SS components, which both play essential roles in Hcp secretion in the functional T6SSs identified to date (9). Despite the conservation of the Walker A motif in TssM (20, 36), its potential function as an NTPase has not been well demonstrated. Previously, we provided genetic evidence indicating the importance of the TssM Walker A motif in Hcp secretion from A. tumefaciens (36). In this work, we have further provided biochemical evidence that TssM exhibits ATPase activity capable of hydrolyzing ATP to recruit Hcp into the TssM-TssL IM complex and powers Hcp secretion across the OM. Based on our findings, we propose a general model of TssM-TssL complex-mediated Hcp secretion across the OM (Fig. 7). We reveal here that TssM undergoes a conformational change upon ATP binding. ATP binding, however, is not sufficient to trigger TssM-TssL-Hcp complex formation and Hcp secretion. Instead, the ATP-bound state is critical for subsequent ATP hydrolysis to recruit Hcp to the periplasmic domain of TssL to form a TssM-TssL-Hcp IM complex. Building this ternary complex could be a prerequisite for Hcp secretion across the OM that is powered by the ATP hydrolysis of TssM.
FIGURE 7.
Model of TssM-TssL complex-mediated Hcp secretion across the OM. TssM (M) and TssL (L) form a stable IM complex; their interaction is not affected by the ATP binding state of TssM. In the absence of ATP, no conformational change of TssM is induced, and the interaction of TssL with Hcp at the periplasm is prohibited. Upon ATP binding, a conformational change of TssM is induced, but this structural transition is not sufficient to recruit Hcp into the TssM-TssL IM complex. We postulate that ATP hydrolysis of TssM recruits Hcp into the periplasmic domain of TssL. Subsequently, energy derived from ATP hydrolysis drives the translocation of Hcp across the OM. Hexameric Hcp is depicted as interacting with TssL, but the oligomeric states and the origin of Hcp interacting with the TssM-TssL complex remains to be determined. The green circles depict ATP, and red circles depict ADP.
TssM belongs to the P-loop NTPase family, based on the two conserved motifs known as Walker A (GXXXXGKT) and Walker B box (hhhh(D/E)) (53). Interestingly, the Walker B motif of TssM has a signature of hhhhDXXG that belongs to GTPase family proteins (Fig. 5A). Apart from the Walker A and B motifs, TssM also contains a highly conserved (N/T)KXD motif belonging to the TRAFAC/SIMBI class of GTPases, which provides specificity for guanine over other nucleotide bases (50, 51). Hence, TssM has the hallmarks of a GTP-binding protein, and it is possible that TssM might also utilize GTP as substrate. Indeed, using a malachite green assay, we observed that TssM is capable of hydrolyzing GTP.3 However, whether GTP or ATP is the preferred nucleotide and whether GTP binding is physiologically relevant are beyond the scope of this study and await future investigation.
Demonstrating ATP binding and hydrolysis of TssM in vitro has been challenging because of the instability of the 128-kDa IM protein with three transmembrane domains (36). Nevertheless, we have succeeded in demonstrating these activities biochemically upon purifying full-length TssM-His from the membrane fraction of E. coli co-expressing TssL to stabilize TssM. Because of the instability of purified TssM variants with either N- or C-terminal truncations or a deletion of the entire Walker A motif (36) (data not shown), we purified His-tagged TssL to serve as a negative control for ATP binding/hydrolysis activity assays (Fig. 5, B and C). As compared with the lower ATP binding/hydrolysis activity contributed by TssMmWA(4PM) (Fig. 1, B and C) and background activity by purified TssL (Fig. 5), we suggest that the mutations in Walker A and B motifs (TssMmWA and TssMmWB) may not completely impair ATP binding and hydrolysis activity, respectively. Nevertheless, the concomitant reduction of ATP binding and ATP hydrolysis activities of TssMmWA, together with the WT-like ATP binding activity but impaired ATP hydrolysis activity of TssMmWB (Fig. 5), clearly indicated that Walker A and Walker B motifs contribute to the ATP binding and hydrolysis activity of TssM, respectively. Importantly, the loss of Hcp secretion caused by the TssMmWA and TssMmWB variants (36) (Fig. 6A) strongly suggest that both the Walker A and Walker B motifs of TssM are critical for the function of T6SS in A. tumefaciens. These data may also explain the contradictory conclusions on the importance of the Walker A motif of TssM orthologs in A. tumefaciens and Edwardsiella tarda (20, 36). Although the full ATPase activity of TssM is required to mediate Hcp secretion from A. tumefaciens, the presumably reduced ATPase activity conferred by the Walker A mutants of E. tarda TssM (EvpO) may remain sufficient for Hcp secretion.
The interaction of TssM and TssL in the IM is apparently independent of ATP binding/hydrolysis activity of TssM (Fig. 3), an observation that differs from the requirement of ATP binding of ExeA in ExeA-ExeB IM complex formation in Aeromonas hydrophila T2SS (54). The evidence that Hcp can only be recruited efficiently into the TssM-TssL complex in vivo and in vitro in an ATP hydrolysis-dependent manner (Figs. 3 and 6) strongly argues that TssM plays a direct role in regulating the dynamic interaction of TssL-Hcp via its ATPase activity. This ATP-dependent assembly of the TssM-TssL IM complex with Hcp in T6SS revealed both distinct and similar mechanisms with those in T2SS and T4SS. In T4SS, VirB11 and VirD4 energize the complex formation of VirB10-VirB7-VirB9 to bridge the IM and OM (48). In T2SS of A. hydrophila, the ATPase ExeA forms a complex with the bitopic subunit ExeB in the IM (54) and bridges the IM and OM through its binding to the peptidoglycan layer and OM secretin (55, 56). In the case of T6SS, it is the TssM-interacting protein TssL or another T6SS IM component, TagL (SciZ), that possess a conserved peptidoglycan-binding motif required for Hcp secretion (57).3 The interaction of the TssM ortholog with the OM protein TssJ (SciN) in enteroaggregative E. coli and E. tarda leads to the formation of a multiprotein complex linking both IM and OM proteins through peptidoglycan (20, 57, 58). TssJ is a bona fide T6SS OM protein present in many but not all T6SSs identified to date (59). Although no proteins homologous to TssJ or any putative OM protein were encoded in the A. tumefaciens T6SS gene cluster (9), we envision that the TssM-TssL IM complex may interact with peptidoglycan via the peptidoglycan-binding motif of TssL and bridge the OM prior to or during the process of Hcp secretion across the OM. It is plausible that TssJ, or other unknown OM components, may resemble the T2SS secretin ExeD, for which multimerization and localization into the OM is energized by the ExeA ATPase (60, 61). The functional and mechanistic similarity of trafficking ATPases in T2SS, T4SS, and T6SS argues for a convergent evolution of ATPase in the bacterial protein secretion systems.
Our observation that TssM is more resistant to protease digestion at higher cellular ATP levels (Fig. 4A), whereas the Walker B variant TssMmWB remains protease-resistant at the WT level (Fig. 6B), suggests an ATP binding-induced conformational change. This protease-resistant conformational transition induced by ATP binding has also been observed in other trafficking ATPases such as E. coli HlyB (62), Salmonella typhimurium MalK (63), and E. coli SecA (64), suggesting a common and critical feature for ATPases. In addition to ATPases, VirB10 (48) and TonB (49) function as energy sensors of the complex in which they undergo a protease-susceptible conformational change when sensing ATP or proton. T2SS ExeB shares sequence and structure similarities with TonB and is protected by ExeA from proteolytic degradation (65). Interestingly, TssL possesses the common features of an energy sensor, including its interaction with ATPase, a bitopic membrane topology, and a proline-rich region in the C-terminal periplasmic domain (48), suggesting a possible function for TssL as an energy sensor undergoing a conformational change when sensing ATP-bound TssM. The evidence that TssL is able to interact with Hcp in the absence of TssM but the efficient recruitment of Hcp into TssM-TssL complex requires the ATP hydrolysis activity of TssM (Figs. 3 and 6D) indeed supports the ATP-hydrolysis driven conformation transition of TssL in this ternary complex. The periplasmic domain of TssM may undergo a conformational change when binding to TssL via its N-terminal cytoplasmic domain (36). It is possible that the periplasmic domain of TssL is only accessible for interaction with Hcp during ATP hydrolysis but may be locked or masked in the absence of ATP hydrolysis conferred by TssM. The TssL periplasmic domain could become wide open when not interacting with TssM and therefore accessible to interact with Hcp. However, we could not detect any protease susceptibility difference in TssL under energy-depleted condition or in the absence of TssM (data no shown). Future studies using fluorescence resonance energy transfer (66) to detect possible TssL conformational change in real time may allow us to investigate further exactly how TssL senses the input of energy from the cytoplasmic domain of TssM and responds by interacting with Hcp at the periplasm.
Notably, the interaction of Hcp with the C-terminal periplasmic domain but not the N-terminal cytoplasmic domain of TssL in E. coli (Fig. 2) suggested that the assembly of TssM-TssL-Hcp occurs in the periplasmic space of A. tumefaciens. Hcp localizes mostly in the cytosol but also is present in the cell periphery, including both membranes and the periplasm, based on our previous immunoelectron microscopy (38) and biochemical fractionation studies.4 Moreover, the localization of Hcp in the periplasm is also supported by the presence of Hcp in the periplasmic fraction of various bacteria (67–70) and the detection of alkaline phosphatase activity of the P. aeruginosa Hcp1-PhoA fusion protein in E. coli (28). However, it remains unclear how Hcp translocates across the IM to the periplasm, as no typical Sec/Tat signal peptide is found in Hcp. A recent study on the export of SodA, a protein lacking a classical signal peptide, to the periplasm in a SecA-dependent but SecB-independent manner in Rhizobium leguminosarum (71) suggests the existence of one or more novel protein translocation mechanisms. Therefore, Hcp may be translocated across the IM to the periplasm via an unconventional pathway and represent an intermediate form of Hcp in the process of building the nanotube-like structure toward the bacterial surface. Even though Hcp can form a hexamer when expressed in E. coli (21, 68) or a nanotube stacked in head-to-tail pattern in vitro (22), the nanotube-like structure of Hcp has not been observed in vivo. We postulated that TssM energizes the hexameric form of Hcp into the TssM-TssL IM complex at the periplasmic face, where it assembles into a nanotube-like structure with VgrG at the tip to allow the tube protruding out of the OM into the extracellular space. Future detailed biochemical and microscopy analysis of the TssM-TssL-Hcp subcomplex assembly in vivo and in vitro will illustrate the mechanistic roles of TssM and TssL in Hcp assembly and/or secretion.
TssH and TssM, the T6SS ATPases, are required for Hcp secretion (20, 21, 26, 38). The AAA+ ATPase TssH (ClpV) interacts with TssB (VipA) and TssC (VipB), and its ATP hydrolysis is crucial for threading the TssB/TssC cogwheel-like tubule, which is similar to the T4 bacteriophage tail sheath structure (26, 72). By analyzing the TssB/TssC tubule variants, Pietrosiuk et al. (35) further document the necessity of TssH-triggered TssB/TssC tubule disassembly for T6SS activity. However, TssH does not interact directly with the secreted exoproteins Hcp and VgrG (26), which presumably resemble the internal phage tail tube structure assembled underneath tail sheath (73). TssM also does not interact directly with Hcp but hydrolyzes ATP to drive the assembly of the TssM-TssL-Hcp complex, which may be critical for Hcp secretion across the OM (Figs. 3, 6, and 7). Therefore, we propose that the T6SS uses two energizers, with TssM powering the assembly of the Hcp nanotube and TssH providing energy for contracting the tail sheath, to push Hcp secretion across the OM. The molecular mechanisms underlying the temporal and spatial coordination of TssM and TssH for T6SS assembly and secretion await future investigation.
Supplementary Material
Acknowledgments
We thank Benaroudj Nadia for kindly providing the EN2 strain, Kai Westphal for help with the malachite green assay, Yun-Long Tsai for constructing the plasmid pTrc200HA, and members of the Lai laboratory for helpful discussion. We also thank Jer-Sheng Lin, Chih-Feng Wu, and Hung-Yi Wu for critical reading of the manuscript. We also acknowledge Ban-Yang Chang for providing GroEL antisera and the DNA Sequencing Core Laboratory located in the Institute of Plant and Microbial Biology, Academia Sinica for DNA sequencing.
This work was supported by National Science Council Grant NSC 98-2311-B-001-002-MY3 and a joint grant from the German Academic Exchange Service (DAAD) and the Taiwan National Science Council (PPP Grant 0970029248P) (to F. N. and E.-M. L.).

This article contains supplemental Table S1.
L. Ma and E. Lai, unpublished results.
J. Lin and E. Lai, unpublished results.
- IM
- inner membrane
- OM
- outer membrane
- T6SS
- type VI secretion system
- Hcp
- hemolysin-coregulated protein
- VgrG
- valine-glycine repeat protein G
- IPTG
- isopropyl-β-d-thiogalactopyranoside
- DDM
- n-dodecyl-β-d-maltoside
- Ni-NTA
- nickel-nitrilotriacetic acid
- TNP-ATP
- 2′,3′-O-(2,4,6-trinitrophenyl) adenosine 5′-triphosphate
- AMPPNP
- adenosine 5′-(β,γ-imido)triphosphate
- Tss
- type VI secretion.
REFERENCES
- 1. Driessen A. J., Nouwen N. (2008) Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77, 643–667 [DOI] [PubMed] [Google Scholar]
- 2. Brüser T. (2007) The twin-arginine translocation system and its capability for protein secretion in biotechnological protein production. Appl. Microbiol. Biotechnol. 76, 35–45 [DOI] [PubMed] [Google Scholar]
- 3. Galán J. E., Wolf-Watz H. (2006) Protein delivery into eukaryotic cells by type III secretion machines. Nature 444, 567–573 [DOI] [PubMed] [Google Scholar]
- 4. Cianciotto N. P. (2005) Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 13, 581–588 [DOI] [PubMed] [Google Scholar]
- 5. Delepelaire P. (2004) Type I secretion in Gram-negative bacteria. Biochim. Biophys. Acta 1694, 149–161 [DOI] [PubMed] [Google Scholar]
- 6. Henderson I. R., Navarro-Garcia F., Desvaux M., Fernandez R. C., Ala'Aldeen D. (2004) Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68, 692–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Juhas M., Crook D. W., Hood D. W. (2008) Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell. Microbiol. 10, 2377–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bingle L. E., Bailey C. M., Pallen M. J. (2008) Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 11, 3–8 [DOI] [PubMed] [Google Scholar]
- 9. Cascales E. (2008) The type VI secretion toolkit. EMBO Rep. 9, 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Filloux A., Hachani A., Bleves S. (2008) The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154, 1570–1583 [DOI] [PubMed] [Google Scholar]
- 11. Boyer F., Fichant G., Berthod J., Vandenbrouck Y., Attree I. (2009) Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pukatzki S., McAuley S. B., Miyata S. T. (2009) The type VI secretion system: translocation of effectors and effector domains. Curr. Opin. Microbiol. 12, 11–17 [DOI] [PubMed] [Google Scholar]
- 13. Records A. R. (2011) The type VI secretion system: a multipurpose delivery system with a phage-like machinery. Mol. Plant Microbe Interact. 24, 751–757 [DOI] [PubMed] [Google Scholar]
- 14. Jani A. J., Cotter P. A. (2010) Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe 8, 2–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schwarz S., Hood R. D., Mougous J. D. (2010) What is type VI secretion doing in all those bugs? Trends Microbiol. 18, 531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leung K. Y., Siame B. A., Snowball H., Mok Y. K. (2011) Type VI secretion regulation: cross-talk and intracellular communication. Curr. Opin. Microbiol. 14, 9–15 [DOI] [PubMed] [Google Scholar]
- 17. Bernard C. S., Brunet Y. R., Gueguen E., Cascales E. (2010) Nooks and crannies in type VI secretion regulation. J. Bacteriol. 192, 3850–3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pukatzki S., Ma A. T., Sturtevant D., Krastins B., Sarracino D., Nelson W. C., Heidelberg J. F., Mekalanos J. J. (2006) Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pukatzki S., Ma A. T., Revel A. T., Sturtevant D., Mekalanos J. J. (2007) Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U.S.A. 104, 15508–15513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zheng J., Leung K. Y. (2007) Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 66, 1192–1206 [DOI] [PubMed] [Google Scholar]
- 21. Mougous J. D., Cuff M. E., Raunser S., Shen A., Zhou M., Gifford C. A., Goodman A. L., Joachimiak G., Ordoñez C. L., Lory S., Walz T., Joachimiak A., Mekalanos J. J. (2006) A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ballister E. R., Lai A. H., Zuckermann R. N., Cheng Y., Mougous J. D. (2008) In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc. Natl. Acad. Sci. U.S.A. 105, 3733–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leiman P. G., Basler M., Ramagopal U. A., Bonanno J. B., Sauder J. M., Pukatzki S., Burley S. K., Almo S. C., Mekalanos J. J. (2009) Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U.S.A. 106, 4154–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hachani A., Lossi N. S., Hamilton A., Jones C., Bleves S., Albesa-Jové D., Filloux A. (2011) Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. J. Biol. Chem. 286, 12317–12327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shalom G., Shaw J. G., Thomas M. S. (2007) In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153, 2689–2699 [DOI] [PubMed] [Google Scholar]
- 26. Bönemann G., Pietrosiuk A., Diemand A., Zentgraf H., Mogk A. (2009) Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 28, 315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bönemann G., Pietrosiuk A., Mogk A. (2010) Tubules and donuts: a type VI secretion story. Mol. Microbiol. 76, 815–821 [DOI] [PubMed] [Google Scholar]
- 28. Lossi N. S., Dajani R., Freemont P., Filloux A. (2011) Structure-function analysis of HsiF, a gp25-like component of the type VI secretion system, in Pseudomonas aeruginosa. Microbiology 157, 3292–3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hayes C. S., Aoki S. K., Low D. A. (2010) Bacterial contact-dependent delivery systems. Annu. Rev. Genet. 44, 71–90 [DOI] [PubMed] [Google Scholar]
- 30. Shiue S. J., Kao K. M., Leu W. M., Chen L. Y., Chan N. L., Hu N. T. (2006) XpsE oligomerization triggered by ATP binding, not hydrolysis, leads to its association with XpsL. EMBO J. 25, 1426–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Atmakuri K., Cascales E., Christie P. J. (2004) Energetic components VirD4, VirB11, and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol. Microbiol. 54, 1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Possot O. M., Letellier L., Pugsley A. P. (1997) Energy requirement for pullulanase secretion by the main terminal branch of the general secretory pathway. Mol. Microbiol. 24, 457–464 [DOI] [PubMed] [Google Scholar]
- 33. Akeda Y., Galán J. E. (2005) Chaperone release and unfolding of substrates in type III secretion. Nature 437, 911–915 [DOI] [PubMed] [Google Scholar]
- 34. van der Wolk J. P., de Wit J. G., Driessen A. J. (1997) The catalytic cycle of the Escherichia coli SecA ATPase comprises two distinct preprotein translocation events. EMBO J. 16, 7297–7304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pietrosiuk A., Lenherr E. D., Falk S., Bönemann G., Kopp J., Zentgraf H., Sinning I., Mogk A. (2011) Molecular basis for the unique role of the AAA+ chaperone ClpV in type VI protein secretion. J. Biol. Chem. 286, 30010–30021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ma L. S., Lin J. S., Lai E. M. (2009) An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its walker a motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J. Bacteriol. 191, 4316–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Quandt J., Hynes M. F. (1993) Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127, 15–21 [DOI] [PubMed] [Google Scholar]
- 38. Wu H. Y., Chung P. C., Shih H. W., Wen S. R., Lai E. M. (2008) Secretome analysis uncovers an Hcp family protein secreted via a type VI secretion system in Agrobacterium tumefaciens. J. Bacteriol. 190, 2841–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ratelade J., Miot M. C., Johnson E., Betton J. M., Mazodier P., Benaroudj N. (2009) Production of recombinant proteins in the lon-deficient BL21(DE3) strain of Escherichia coli in the absence of the DnaK chaperone. Appl. Environ. Microbiol. 75, 3803–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Henkel R. D., VandeBerg J. L., Walsh R. A. (1988) A microassay for ATPase. Anal. Biochem. 169, 312–318 [DOI] [PubMed] [Google Scholar]
- 41. Camberg J. L., Sandkvist M. (2005) Molecular analysis of the Vibrio cholerae type II secretion ATPase EpsE. J. Bacteriol. 187, 249–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lai E. M., Kado C. I. (1998) Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J. Bacteriol. 180, 2711–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu A. C., Shih H. W., Hsu T., Lai E. M. (2008) A citrate-inducible gene, encoding a putative tricarboxylate transporter, is down-regulated by the organic solvent DMSO in Agrobacterium tumefaciens. J. Appl. Microbiol. 105, 1372–1383 [DOI] [PubMed] [Google Scholar]
- 44. Chang B. Y., Chen K. Y., Wen Y. D., Liao C. T. (1994) The response of a Bacillus subtilis temperature-sensitive sigA mutant to heat stress. J. Bacteriol. 176, 3102–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cho Y. K., Ríos S. E., Kim J. J., Miziorko H. M. (2001) Investigation of invariant serine/threonine residues in mevalonate kinase. Tests of the functional significance of a proposed substrate binding motif and a site implicated in human inherited disease. J. Biol. Chem. 276, 12573–12578 [DOI] [PubMed] [Google Scholar]
- 46. Hormaeche I., Alkorta I., Moro F., Valpuesta J. M., Goni F. M., De La Cruz F. (2002) Purification and properties of TrwB, a hexameric, ATP-binding integral membrane protein essential for R388 plasmid conjugation. J. Biol. Chem. 277, 46456–46462 [DOI] [PubMed] [Google Scholar]
- 47. Yao H., Hersh L. B. (2006) Characterization of the binding of the fluorescent ATP analog TNP-ATP to insulysin. Arch. Biochem. Biophys. 451, 175–181 [DOI] [PubMed] [Google Scholar]
- 48. Cascales E., Christie P. J. (2004) Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc. Natl. Acad. Sci. U.S.A. 101, 17228–17233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Larsen R. A., Thomas M. G., Postle K. (1999) Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol. Microbiol. 31, 1809–1824 [DOI] [PubMed] [Google Scholar]
- 50. Bourne H. R., Sanders D. A., McCormick F. (1991) The GTPase superfamily: conserved structure and molecular mechanism. Nature 349, 117–127 [DOI] [PubMed] [Google Scholar]
- 51. Brown E. D. (2005) Conserved P-loop GTPases of unknown function in bacteria: an emerging and vital ensemble in bacterial physiology. Biochem. Cell Biol. 83, 738–746 [DOI] [PubMed] [Google Scholar]
- 52. Motojima F., Yoshida M. (2003) Discrimination of ATP, ADP, and AMPPNP by chaperonin GroEL: hexokinase treatment revealed the exclusive role of ATP. J. Biol. Chem. 278, 26648–26654 [DOI] [PubMed] [Google Scholar]
- 53. Walker J. E., Saraste M., Runswick M. J., Gay N. J. (1982) Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases, and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1, 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schoenhofen I. C., Stratilo C., Howard S. P. (1998) An ExeAB complex in the type II secretion pathway of Aeromonas hydrophila: effect of ATP-binding cassette mutations on complex formation and function. Mol. Microbiol. 29, 1237–1247 [DOI] [PubMed] [Google Scholar]
- 55. Li G., Miller A., Bull H., Howard S. P. (2011) Assembly of the type II secretion system: identification of ExeA residues critical for peptidoglycan binding and secretin multimerization. J. Bacteriol. 193, 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Howard S. P., Gebhart C., Langen G. R., Li G., Strozen T. G. (2006) Interactions between peptidoglycan and the ExeAB complex during assembly of the type II secretin of Aeromonas hydrophila. Mol. Microbiol. 59, 1062–1072 [DOI] [PubMed] [Google Scholar]
- 57. Aschtgen M. S., Gavioli M., Dessen A., Lloubès R., Cascales E. (2010) The SciZ protein anchors the enteroaggregative Escherichia coli type VI secretion system to the cell wall. Mol. Microbiol. 75, 886–899 [DOI] [PubMed] [Google Scholar]
- 58. Felisberto-Rodrigues C., Durand E., Aschtgen M. S., Blangy S., Ortiz-Lombardia M., Douzi B., Cambillau C., Cascales E. (2011) Towards a structural comprehension of bacterial type VI secretion systems: characterization of the TssJ-TssM complex of an Escherichia coli pathovar. PLoS Pathog. 7, e1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Aschtgen M. S., Bernard C. S., De Bentzmann S., Lloubès R., Cascales E. (2008) SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J. Bacteriol. 190, 7523–7531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ast V. M., Schoenhofen I. C., Langen G. R., Stratilo C. W., Chamberlain M. D., Howard S. P. (2002) Expression of the ExeAB complex of Aeromonas hydrophila is required for the localization and assembly of the ExeD secretion port multimer. Mol. Microbiol. 44, 217–231 [DOI] [PubMed] [Google Scholar]
- 61. Schoenhofen I. C., Li G., Strozen T. G., Howard S. P. (2005) Purification and characterization of the N-terminal domain of ExeA: a novel ATPase involved in the type II secretion pathway of Aeromonas hydrophila. J. Bacteriol. 187, 6370–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Koronakis E., Hughes C., Milisav I., Koronakis V. (1995) Protein exporter function and in vitro ATPase activity are correlated in ABC domain mutants of HlyB. Mol. Microbiol. 16, 87–96 [DOI] [PubMed] [Google Scholar]
- 63. Schneider E., Wilken S., Schmid R. (1994) Nucleotide-induced conformational changes of MalK, a bacterial ATP binding cassette transporter protein. J. Biol. Chem. 269, 20456–20461 [PubMed] [Google Scholar]
- 64. Shinkai A., Mei L. H., Tokuda H., Mizushima S. (1991) The conformation of SecA, as revealed by its protease sensitivity, is altered upon interaction with ATP, presecretory proteins, everted membrane vesicles, and phospholipids. J. Biol. Chem. 266, 5827–5833 [PubMed] [Google Scholar]
- 65. Howard S. P., Meiklejohn H. G., Shivak D., Jahagirdar R. (1996) A TonB-like protein and a novel membrane protein containing an ATP-binding cassette function together in exotoxin secretion. Mol. Microbiol. 22, 595–604 [DOI] [PubMed] [Google Scholar]
- 66. Heyduk T. (2002) Measuring protein conformational changes by FRET/LRET. Curr. Opin. Biotechnol. 13, 292–296 [DOI] [PubMed] [Google Scholar]
- 67. Mougous J. D., Gifford C. A., Ramsdell T. L., Mekalanos J. J. (2007) Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat. Cell Biol. 9, 797–803 [DOI] [PubMed] [Google Scholar]
- 68. Jobichen C., Chakraborty S., Li M., Zheng J., Joseph L., Mok Y. K., Leung K. Y., Sivaraman J. (2010) Structural basis for the secretion of EvpC: a key type VI secretion system protein from Edwardsiella tarda. PLoS One 5, e12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Miyata S. T., Kitaoka M., Brooks T. M., McAuley S. B., Pukatzki S. (2011) Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect. Immun. 79, 2941–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. de Bruin O. M., Duplantis B. N., Ludu J. S., Hare R. F., Nix E. B., Schmerk C. L., Robb C. S., Boraston A. B., Hueffer K., Nano F. E. (2011) The biochemical properties of the Francisella pathogenicity island (FPI)-encoded proteins IglA, IglB, IglC, PdpB, and DotU suggest roles in type VI secretion. Microbiology 157, 3483–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Krehenbrink M., Edwards A., Downie J. A. (2011) The superoxide dismutase SodA is targeted to the periplasm in a SecA-dependent manner by a novel mechanism. Mol. Microbiol. 82, 164–179 [DOI] [PubMed] [Google Scholar]
- 72. Aksyuk A. A., Leiman P. G., Kurochkina L. P., Shneider M. M., Kostyuchenko V. A., Mesyanzhinov V. V., Rossmann M. G. (2009) The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria. EMBO J. 28, 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Leiman P. G., Arisaka F., van Raaij M. J., Kostyuchenko V. A., Aksyuk A. A., Kanamaru S., Rossmann M. G. (2010) Morphogenesis of the T4 tail and tail fibers. Virol. J. 7, 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. (1990) Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185, 60–89 [DOI] [PubMed] [Google Scholar]
- 75. Vergunst A. C., Schrammeijer B., den Dulk-Ras A., de Vlaam C. M., Regensburg-Tuïnk T. J., Hooykaas P. J. (2000) VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290, 979–982 [DOI] [PubMed] [Google Scholar]
- 76. Schmidt-Eisenlohr H., Domke N., Baron C. (1999) TraC of IncN plasmid pKM101 associates with membranes and extracellular high-molecular-weight structures in Escherichia coli. J. Bacteriol. 181, 5563–5571 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.