Background: Tyrosine kinases and phosphatases modulate TGF-β signaling.
Results: PTP1B deficiency attenuates TGF-β-induced Smad phosphorylation correlating with suppression of growth inhibition and apoptosis, an effect that is reverted by genistein, and p65 or NOX1 knockdown.
Conclusion: Lack of PTP1B impairs Smad activation in a NOX1 and NF-κB-dependent manner.
Significance: New insights are opened into the role of phosphatases in TGF-β signaling.
Keywords: Hepatocyte, NADPH Oxidase, NF-κB, Protein Phosphatase, Transforming Growth Factor-β (TGF-β), PTP1B
Abstract
Transforming growth factor-β (TGF-β) plays a dual role in hepatocytes, mediating both tumor suppressor and promoter effects. The suppressor effects of the cytokine can be negatively regulated by activation of survival signals, mostly dependent on tyrosine kinase activity. The aim of our work was to study the role of the protein-tyrosine phosphatase 1B (PTP1B) on the cellular responses to TGF-β, using for this purpose immortalized neonatal hepatocytes isolated from both PTP1B+/+ and PTP1B−/− mice. We have found that PTP1B deficiency conferred resistance to TGF-β suppressor effects, such as apoptosis and growth inhibition, correlating with lower Smad2/Smad3 activation. Both responses were recovered in the presence of the general tyrosine kinase inhibitor genistein. PTP1B−/− cells showed elevated NF-κB activation in response to TGF-β. Knockdown of the NF-κB p65 subunit increased cell response in terms of Smads phosphorylation and apoptosis. Interestingly, these effects were accompanied by inhibition of Smad7 up-regulation. In addition, lack of PTP1B promoted an altered NADPH oxidase (NOX) expression pattern in response to TGF-β, strongly increasing the NOX1/NOX4 ratio, which was reverted by genistein and p65 knockdown. Importantly, NOX1 knockdown inhibited nuclear translocation of p65, promoted Smad phosphorylation, and decreased Smad7 levels. In summary, our results suggest that PTP1B deficiency confers resistance to TGF-β through Smad inhibition, an effect that is mediated by NOX1-dependent NF-κB activation, which in turn, increases the level of the Smad inhibitor Smad7 and participates in a positive feedback loop on NOX1 up-regulation.
Introduction
Transforming growth factor-β (TGF-β) is an important regulatory cytokine in the liver. Among its physiological roles, it exerts essential suppressor functions through regulating hepatocyte proliferation and cell death (1, 2). However, TGF-β may also modulate protumorigenic processes, such as epithelial to mesenchymal transition, which mediates cell migration and survival, and also cell invasion, immune regulation or microenvironment modification (3). Indeed, malignancy of liver tumors can be related to their so-called TGF-β gene signature: tumors expressing antiapoptotic and metastatic genes (late signature) display a higher invasive phenotype and increased tumor recurrence compared with those that show suppressor genes (early signature) (4).
TGF-β final effects in the cell can be modulated by activation of survival signals dependent on different protein kinases, such as the epidermal growth factor receptor (EGFR)4 or the Ras/ERK pathway. These kinases are generally regulated by its phosphorylation status, which is tightly controlled by mechanisms involving inactivation by protein phosphatases (5). Among them, protein-tyrosine phosphatase 1B (PTP1B) is a widely expressed member of the nonreceptor PTPs, which is located mainly associated with intracellular membranes such as the endoplasmic reticulum (6). This phosphatase plays important roles regulating insulin signaling and susceptibility to apoptosis because its deficiency confers resistance to cell death triggered by different stimuli (7, 8). These cellular effects are dependent on the modulation of several prosurvival pathways directly controlled by PTP1B, such as the EGFR, IGF-1R, or the IR (9). In addition, several reports have linked PTP1B to NF-κB regulation (10, 11). Importantly, the NF-κB pathway has been also described as an inhibitor of the TGF-β suppressor effects in hepatocytes (12).
To exert its proapoptotic activity in hepatocytes, TGF-β requires the production of reactive oxygen species (13, 14), mainly originated by the NADPH oxidase NOX4, which is necessary for apoptosis execution in hepatocytes sensitive to the cytokine (15). However, the role of NADPH oxidases in TGF-β-mediated signaling in liver is complex. In fact, recent results indicate that TGF-β might induce another member of the NOX family (NOX1) in hepatoma cells, with prosurvival properties (16). Thus, NOX proteins might play a dual role in the intracellular signaling induced by TGF-β in liver cells, being the balance in the expression of different members of the family determinant for the final effects of the cytokine. Importantly, we have described that EGFR or Ras/ERK activation would counteract the suppressor effects of the cytokine, inhibiting NOX4 induction or modifying the balance between NOX1 and NOX4 (16–19). Moreover, we have also previously proposed a relationship between NF-κB and NADPH oxidases mediating prosurvival signals in response to TGF-β (20).
On that basis, the aim of our work was to study the role of PTP1B on the cellular responses to TGF-β, using for this purpose immortalized neonatal hepatocytes isolated from both PTP1B+/+ and PTP1B−/− mice. We have found that PTP1B deficiency conferred resistance to TGF-β suppressor effects through Smad signaling inhibition. Interestingly, NOX1-dependent NF-κB activation mediates this response, which correlates with increased levels of Smad7 and a positive feedback loop of NOX1 up-regulation.
EXPERIMENTAL PROCEDURES
Cell Culture Conditions
Isolation and immortalization of neonatal hepatocytes from PTP1B+/+ and PTP1B−/− mice was described previously (21). For cell culture, cells were grown in DMEM (Lonza, Basel, Switzerland) supplemented with 10% fetal bovine serum and maintained in a humidified atmosphere of 37 °C, 5% CO2. For experiments, cells at 70% confluence were serum-deprived for 4 h and treated with 2 ng/ml TGF-β (Merck). When indicated, the following products were added 30 min before TGF-β and maintained during the experiment: EGF (20 ng/ml; a gift from Serono Laboratories, Madrid, Spain), the tyrosine kinase inhibitor genistein (50 μm, from Sigma), IGF-1R inhibitor II (Merck) or anti-mouse IGF-1 antibody (R&D Systems).
Analysis of Cell Number
Cell number was analyzed after crystal violet staining (0.2% in 2% ethanol), as described previously (14).
Analysis of Cell Cycle
The percentage of cells in each phase of the cell cycle was determined by flow cytometry and analyzed using the software ModFit LTTM (Verity Software House) as described previously (22).
Proliferation Measurement by [3H]Thymidine Incorporation
Proliferation was measured after 48 h of incubation with 1 μCi/ml, 1 μm thymidine (Hartmann Analytic GmbH, Braunschweig, Germany), as described previously (23). Radioactivity was measured in a scintillation counter 1209 Rackbeta (Wallac, Turku, Finland) and expressed as a percentage of control.
Measurement of Intracellular Redox State
The oxidation-sensitive fluorescent probe 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA; Invitrogen) was used to analyze the total intracellular content of reactive oxygen species as described previously (16). Fluorescence was measured in a Microplate Fluorescence Reader Fluostar Optima and expressed as percentage of control after correction with protein content.
Analysis of Apoptotic Nuclei
Cells were fixed with 4% paraformaldehyde in PBS for 30 min at room temperature, stained for 5 min with 2 ng/ml DAPI solution, and mounted with Vectashield (Vector Laboratories, Burlingame, CA). Cells were visualized in an Olympus BX-60 with the appropriate filter, and fragmented nuclei were counted as positive in at least 10 different fields.
Analysis of Caspase-3 Activity
Caspase-3 activity was analyzed fluorometrically upon incubation of 20 μg of cell lysates with 6.6 μg/ml Ac-DEVD-AMC (BD Pharmingen) for 2 h at 37 °C as described previously (16). Protein concentration of cell lysates was determined using the Bio-Rad protein assay kit. Results are calculated as units of caspase-3 activity/μg of protein per h.
Analysis of Gene Expression
RNeasy Mini kit (Qiagen) was used for total RNA isolation. Reverse transcription (RT) was carried out using the High Capacity Reverse Transcriptase kit (Applied Biosystems), with 500 ng of total RNA from each sample for complementary DNA synthesis. Semiquantitative PCRs were performed using specific primers for mouse samples: Smad7, 5′-TGTTCAGGTGGCCGGATCTCAG-3′ and 5′-GATGCCACAGCCGATCTTGCTC-3′; Smad4, 5′-CAAGTAATCGCGCATCAACG-3′ and 5′-GCAGGATGATTGGAAATGGG-3′; Smad3, 5′-GCCATTCCATTCCCGAGAAC-3′ and 5′-CACTGAGGCACTCCGCAAAG-3′; Smad2, 5′-CCCAGCAGGAATTGAGCCAC-3′ and 5′-ACACTGTTGCAGGGTGCCAG-3′; TGF-βRII, 5′-CGCCAACAACATCAACCACAAC-3′ and 5′-CTGTGAACAATGGGCATCTTGG-3′; TGF-βRI, 5′-AGACAATGGGACATGGACGCAG-3′ and 5′-TGCATAGATGTCAGCGCGTTTG-3′; PTP1B, 5′-TGGCCTGACTTTGGAGTCCC-3′ and 5′-CTCCAGTGTGCGTTTGGGTG-3′; 18s, 5′-GCGAAAGCATTTGCCAAGAA-3′ and 5′-CATCACAGACCTGTTATTGC-3′.
PCR products were obtained after 30–35 cycles of amplification at annealing temperatures of 57–62 °C and analyzed by 1.5% agarose gel electrophoresis. Expression of 18s was analyzed as a loading control, as indicated. The −RT channel contained RNA that had not been treated with the RT mixture and is shown as a specificity control.
For real-time quantitative PCR, expression levels were determined in duplicate in an ABI Prism7700 System, using the SYBR Green PCR Master Mix (Applied Biosystems). Reactions were performed with the following primers mouse specific primers: NOX1, 5′-TCCTTCGCTTTTATCGCTCC-3′ and 5′-TCGCTTCCTCATCTGCAATTC-3′; NOX4, 5′-TCCAAGCTCATTTCCCACAG-3′ and 5′-CGGAGTTCCATTACATCAGAGG-3′; IGF-1, 5′- TCATGTCGTCTTCACACCTCTTC-3′ and 5′-CCACACACGAACTGAAGAGCAT-3′; IGF-1R, 5′-GTGGGGGCTCGTGTTTCTC-3′ and 5′-GATCACCGTGCAGTTTTCCA-3′; Smad2, 5′-TCACAGACCCATCAAACTCG-3′ and 5′-ACTCAGCAAACACTTCCCC-3′; Smad3, 5′-CCGAGAACACTAACTTCCCT-3′ and 5′-CATCTTCACTCAGGTAGCCAG-3′; 18s, 5′-GCGAAAGCATTTGCCAAGAA-3′ and 5′-CATCACAGACCTGTTATTGC-3′.
Western Blot Analysis
Total protein extracts, nuclear extracts, and Western blotting procedures were carried out as described previously (16, 20). The antibodies used were: mouse anti-β-actin (clone AC-15); rabbit anti-phospho-Akt (Ser473), rabbit anti-Akt, rabbit anti-phospho-p44/42 MAPK (Thr202/Tyr204), anti-p44/42 MAPK, rabbit anti-PTP1B, rabbit anti-p65; rabbit anti-IκBα, mouse anti-Smad2, rabbit anti-phospho-Smad2 (Ser465/Ser467), rabbit anti-Smad3, rabbit anti-phospho-Smad3 (Ser423/Ser425), rabbit anti-Smad4, goat anti-Smad2/3, mouse anti-Smad7, rabbit anti-PCNA. All antibodies were from Cell Signaling Technology, except anti-β-actin from Sigma, anti-PTP1B and anti-Smad3 from Millipore, and anti-Smad2/3, anti-Smad7, anti-p65, anti-IκBα, and anti-phospho-IGF-IR (Tyr1165/Tyr1166) from Santa Cruz Biotechnology. Antibodies were used at 1:1000 dilution, except β-actin (1:3000). Protein concentration was measured with BCA Protein Assay kit (Pierce).
Immunocytochemistry Studies
Fluorescence microscopy studies were performed as described previously (22). Cells were fixed with 4% paraformaldehyde in PBS for 30 min at room temperature and incubated with primary antibody (1:50) diluted in 1% BSA for 2 h at room temperature. After several washes with PBS, the samples were incubated with Alexa Fluor 488-conjugated anti-rabbit for 1 h at room temperature (1:200) and mounted in Vectashield. Representative images were taken with a Spot 4.3 digital camera and software and edited in Adobe Photoshop. Cells were visualized in an Olympus BX-60 with the appropriate filters.
Knockdown Assays
For transient siRNA transfection, cells were transfected at 70% confluence using TransIT-siQuest (Mirus, Madison, WI) at 1:300 dilution in Complete medium, according to the manufacturer's recommendation, with a final siRNA concentration of 50 nm during 8 h. After 16 h of incubation in Complete medium, cells were trypsinized and seeded for experiments. Oligonucleotides were obtained from Sigma-Genosys. The oligonucleotide sequences were as follows: unsilencing, 5′-GUAAGACACGACUUAUCC-3′; p65, 5′-UUACUCGGCAGAUCUUGAG-3′; NOX1, 5′-GAGGAGAGCUUGGGUGAAA-3′; IGF-1, 5′-GCUGCAAAGGAGAAGGAAA-3′; IGF-1R, 5′-CAGAAGUGGAGCAGAAUAA-3′; EGFR, 5′-GAACAACAGAGCTGAGAAA-3′; Smad2, 5′-ACTCAGAATTGCAATACT-3′; Smad3, 5′-CTACCTGAGTGAAGATGGA-3′.
The unsilencing siRNA used was selected from previous works (16). Specific oligonucleotides with maximal knockdown efficiency were selected among three different sequences for each gene.
RESULTS
Immortalized Neonatal Hepatocytes Isolated from PTP1B-deficient Mice Are Resistant to TGF-β
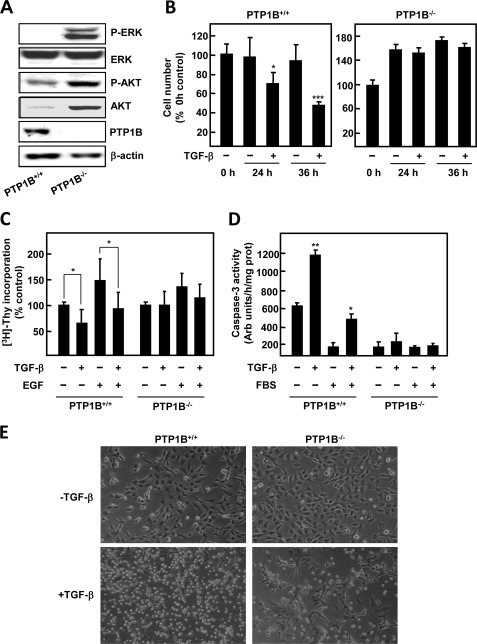
To investigate the role of the tyrosine phosphatase PTP1B in TGF-β signaling in hepatocytes, we first compared basal activation of several kinases in wild-type (PTP1B+/+) and PTP1B-deficient (PTP1B−/−) immortalized hepatocytes. PTP1B−/− cells showed high phosphorylation of ERK1/2 and AKT; total levels of the latter were also elevated in these cells (Fig. 1A). Next, we wanted to compare the response to TGF-β of wild-type and PTP1B-deficient cells. Crystal violet staining revealed that, indeed, cell number of wild-type hepatocytes was reduced to half upon 36 h of treatment, whereas PTP1B−/− hepatocytes remained unaffected (Fig. 1B). As reduction of cell number by TGF-β has been reported to be a combinatory effect of apoptosis induction and growth inhibition, we decided to measure both parameters in response to the cytokine. On the one side, TGF-β was able to inhibit basal and EGF-induced DNA synthesis of wild-type hepatocytes (Fig. 1C and Table 1), whereas basal cell growth of PTP1B−/− hepatocytes was not affected by the cytokine. EGF-induced increase of cell number in PTP1B-deficient cells was attenuated by TGF-β, but effects were modest and did not reach statistical significance (Fig. 1C and Table 1). On the other side, we studied apoptosis induction by measuring caspase-3 activity, which was induced by TGF-β treatment either in the presence or in the absence of serum only in wild-type cells (Fig. 1D). We further confirmed these results by phase contrast microscopy, which revealed a great number of wild-type hepatocytes with morphological characteristics of apoptosis after 36 h of incubation with the cytokine (Fig. 1E). By contrast, under similar experimental conditions most PTP1B−/− cells remained attached to the plate (Fig. 1E). In summary, these results indicate that PTP1B−/− hepatocytes are resistant to the suppressor effects of TGF-β, namely growth inhibition and apoptosis triggering.
FIGURE 1.
Immortalized neonatal hepatocytes generated from PTP1B-deficient mice are resistant to TGF-β in terms of cell viability. A, representative Western blot of total lysates from both wild-type (PTP1B+/+) and PTP1B-deficient (PTP1B−/−) immortalized hepatocytes. β-Actin was used as loading control. B, left, PTP1B+/+ cells; right, PTP1B−/− cells. Cell number was measured by crystal violet staining; cells were serum-depleted for 4 h and treated for the indicated times with 2 ng/ml TGF-β. Data were calculated relative to zero time and represent the mean ± S.E. (error bars) of three independent experiments. Student's t test was calculated versus zero time for each cell type: *, p < 0.05; ***, p < 0.001. C, DNA synthesis analysis by [3H]thymidine incorporation. When indicated, cells were pretreated for 30 min with 20 ng/ml EGF and then incubated for an additional 40 h in the absence or presence of 2 ng/ml TGF-β. Data were calculated as a percentage of untreated cells for each cell type and represent the mean ± S.E. of three independent experiments. Student's t test was calculated as TGF-β-treated versus nontreated cells for each experimental condition (EGF-pretreated or nonpretreated for each cell type): *, p < 0.05. D, PTP1B+/+ and PTP1B−/− hepatocytes were treated for 24 h with 2 ng/ml TGF-β the presence or absence of FBS, and caspase-3 activity was determined. Data were expressed as arbitrary units/h and per μg of protein and were calculated as percentage of untreated cells and represent the mean ± S.E. of three independent experiments. Student's t test was calculated as TGF-β-treated versus nontreated cells for each experimental condition (FBS-pretreated or nonpretreated for each cell type): *, p < 0.05; **, p < 0.01. E, representative phase contrast photographs of PTP1B+/+ and PTP1B−/− hepatocytes at 36 h of TGF-β treatment.
TABLE 1.
PTP1B−/− hepatocytes are resistant to growth inhibition induced by TGF-β
PTP1B+/+ and PTP1B−/− hepatocytes were serum-depleted for 4 h and then treated for an additional 48 h with 20 ng/ml EGF, 2 ng/ml TGF-β, or both. Numbers represent the mean ± S.E. of three independent determinations of the percentage of cells in each phase of the cell cycle, analyzed by flow cytometry.
| Treatment | PTP1B+/+ |
PTP1B−/− |
||||
|---|---|---|---|---|---|---|
| G0/G1 | S | G2/M | G0/G1 | S | G2/M | |
| Control | 62.40 ± 6.47 | 27.94 ± 11.13 | 9.64 ± 4.95 | 51.57 ± 3.80 | 46.38 ± 2.43 | 5.2 ± 4.66 |
| TGF-β | 71.46 ± 4.78 | 20.58 ± 8.24 | 7.95 ± 3.99 | 54.12 ± 6.41 | 50,86 ± 8.56 | 6.71 ± 1.52 |
| EGF | 48.16 ± 2.01 | 45.16 ± 4.36 | 10.01 ± 3.80 | 39.14 ± 3.80 | 53.79 ± 2.00 | 10.62 ± 2.05 |
| EGF + TGF-β | 62.34 ± 16.66 | 23.29 ± 15.29 | 11.88 ± 0.97 | 31.38 ± 11.01 | 61.50 ± 13.85 | 6.77 ± 3.86 |
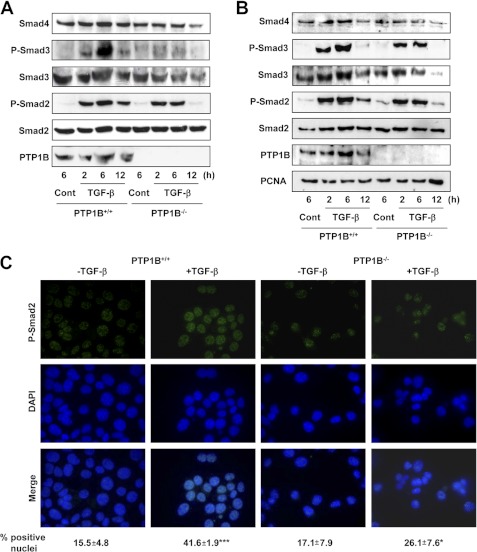
PTP1B−/− Hepatocytes Show Diminished Smad Phosphorylation and Translocation, Both Recovered in Presence of Genistein
Taking into consideration that PTP1B-deficient hepatocytes show resistance to the diverse TGF-β effects, we next decided to study the molecules involved in the signaling of the cytokine. First, we confirmed that these hepatocytes expressed all of the components required for TGF-β-mediated signal transduction, such as receptors and Smad proteins (supplemental Fig. 1). For these reasons, we next decided to check the phosphorylation status of Smad2 and Smad3, both in total (Fig. 2A) and nuclear (Fig. 2B) extracts. Interestingly, we found that duration and strength of Smad2 and Smad3 phosphorylation are greatly reduced both in total or nuclear extracts in PTP1B−/− hepatocytes compared with wild type. Consequently, phospho-Smad2 translocation to the nucleus is attenuated in these cells (Fig. 2C). A direct correlation between the TGF-β sensitivity and phospho-Smad2 levels was also confirmed using wild-type cells stably overexpressing PTP1B, which showed higher apoptotic rate and phosphorylated Smad2 in response to the cytokine compared with control cells (supplemental Fig. 2).
FIGURE 2.
PTP1B−/− hepatocytes show decreased Smad2/3 phosphorylation and translocation to the nucleus in upon TGF-β treatment. Cells were serum-depleted for 4 h and treated with TGF-β for the indicated times. A, Western blot of total lysates. B, Western blot of nuclear extracts. Total Smad2 and PCNA were used as loading control, respectively, in A and B. C, phospho-Smad2 immunocytochemistry (top row), DAPI (middle row), and merge of both (bottom row) at 2 h. Below, a quantification of p-Smad2-positive nuclei in at least 10 different fields for experiment (n = 3) is shown. Student's t test was calculated as TGF-β-treated versus nontreated cells for each cell type: *, p < 0.05; ***, p < 0.001. A representative experiment of three is shown in each case.
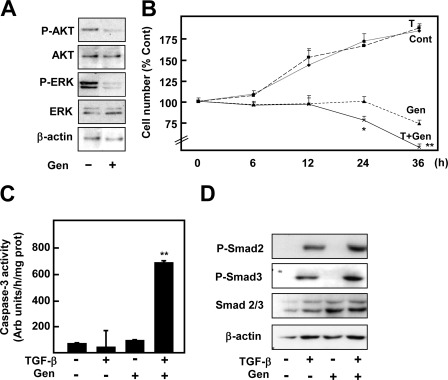
As stated above, PTP1B deficiency led to constitutive kinase phosphorylation in the absence of stimuli (Fig. 1A), which might counteract Smad signaling (24). Looking for the possible kinase responsible for resistance to TGF-β in PTP1B-deficient hepatocytes, we first decided to incubate cells in the presence of different kinase inhibitors: PD98059 for ERK1/2, LY294002 for PI3K/AKT, and PP2 for Src. We found that, although the inhibitors caused some effects on basal viability, none of them was able to sensitize PTP1B−/− hepatocytes to TGF-β treatment (data not shown). Importantly, when a general tyrosine kinase inhibitor, genistein, was added to the culture medium the response to TGF-β was recovered (Fig. 3). First of all, we found that 6 h of genistein treatment was sufficient to inhibit the AKT and ERK basal phosphorylation detected in PTP1B−/− cells (Fig. 3A). In addition, we observed that combined genistein and TGF-β treatment induced a great loss of viability which was accompanied by caspase-3 activation (Fig. 3, B and C). Interestingly, the combined treatment also increased Smad2 and Smad3 phosphorylation (Fig. 3D). Thus, resistance to TGF-β in PTP1B-deficient cells correlates with reduced Smad2 and Smad3 phosphorylation, which is recovered in the presence of a general tyrosine kinase inhibitor.
FIGURE 3.
Genistein pretreatment recovers the response to TGF-β in terms of cell death and Smad phosphorylation in PTP1B−/− hepatocytes. Except when indicated, PTP1B−/− hepatocytes were incubated for 30 min with 50 μm genistein (Gen) and then treated or not with TGF-β. A, Western blot of total lysates at 6 h of genistein treatment. B, cell number at the indicated times. C, caspase-3 activity at 16 h. D, Western blot of total lysates at 2 h. β-Actin was used as loading control in A and D. In B and C data represent the mean ± S.E. (error bars) of four independent experiments. Student's t test was calculated as TGF-β-treated versus nontreated cells: *, p < 0.05; **, p < 0.01.
As a result, we decided to investigate the possible implication of different tyrosine kinase receptors in this response. We found that IGF-1 levels were approximately 30 times higher in PTP1B-deficient hepatocytes, correlating with strong receptor activation (supplemental Fig. 3A). However, neither inhibition nor down-regulation of the IGF-1/IGF-1R pathway (supplemental Fig. 3), EGFR (supplemental Fig. 4) nor the IR individually (data not shown) nor different combinations of siRNA (data not shown) could recover the response to TGF-β.
Activation of NF-κB Pathway Is Responsible for TGF-β Resistance in PTP1B−/− Hepatocytes
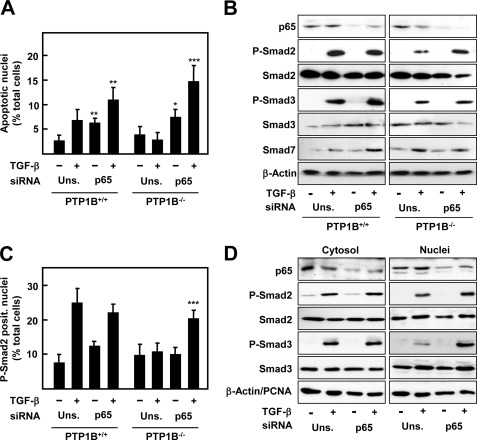
In addition to its direct effect on tyrosine kinases, several reports have related genistein-proapototic effects with NF-κB pathway inhibition, which also can counteract TGF-β signaling (12, 25). For this reason, we decided to investigate a possible implication of NF-κB in our model. First, we measured the nuclear translocation of the p65 subunit, as a hallmark of NF-κB activation, in response to TGF-β in both PTP1B+/+ and PTP1B−/− hepatocytes. As shown in Fig. 4, PTP1B−/− cells showed an increased p65 nuclear localization, which correlated with lower levels of inhibitory protein IκBα upon TGF-β treatment. Interestingly, and as described in other experimental systems, genistein inhibited IκBα degradation and p65 translocation (Fig. 4, B and C). To test the role of the NF-κB pathway in the resistance to TGF-β of PTP1B−/− cells, we decided to perform a siRNA-targeted knockdown against p65. Thus, silencing p65 sensitized both PTP1B+/+ and PTP1B−/− to TGF-β proapoptotic effects, as measured by appearance of fragmented nuclei at 30 h (Fig. 5A). Western blot analysis determined that phosphorylated levels of both Smad2 and Smad3 in PTP1B−/− cells were increased when p65 was knocked down, correlating to lower levels of the inhibitory Smad7 (Fig. 5B). Accordingly, nuclear phospho-Smad2 and 3 were also significantly augmented upon p65 silencing (Fig. 5, C and D). On the contrary, Smad levels, or phosphorylation, in PTP1B+/+ cells did not seem to be affected by p65 knockdown. Together, these results indicate that an increased NF-κB activation upon TGF-β treatment is responsible for the resistance to the cytokine in PTP1B-deficient hepatocytes.
FIGURE 4.
PTP1B−/− hepatocytes show an increased NF-κB pathway activation in response to TGF-β. PTP1B+/+ and PTP1B−/− cells were serum-depleted for 4 h, incubated for 30 min with 50 μm genistein (Gen) when indicated, and then treated or not with TGF-β for an additional 2 h. A, representative p65 inmunocytochemistry. B, percentage of p65-positive nuclei in at least 10 different fields for the experiment (n = 3). Student's t test was calculated as PTP1B+/+ versus PTP1B−/− cells (left) or genistein-pretreated versus nonpretreated cells (right). *, p < 0.05; **, p < 0.01. Error bars, S.E. C, representative Western blot of IκBα. β-Actin was used as loading control.
FIGURE 5.
p65 knockdown in PTP1B−/− hepatocytes recovers the apoptotic response to TGF-β and Smad2 and Smad3 phosphorylation and nuclear translocation, coincident with lower Smad7 induction. PTP1B+/+ and PTP1B−/− cells were transfected with either an unspecific siRNA (Uns.) or p65 siRNA (p65) and then treated or not with TGF-β. A, percentage of apoptotic nuclei at 30 h. B, Western blot of total lysates at 2 h. C, percentage of p-Smad2-positive nuclei in at least 10 different fields for experiment (n = 3) at 2 h. D, Western blot of cytosolic/nuclear extracts of PTP1B−/− treated for 2 h with TGF-β. β-Actin was used as loading control for total, and cytosolic extracts and PCNA were used as loading control for nuclear extracts in B and D. Data represent the mean ± S.E. (error bars) of three independent experiments. Student's t test was calculated as unsilencing siRNA versus p65 siRNA for each experimental condition in each cell type: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
NOX1-dependent Signaling Accounts for TGF-β Resistance in PTP1B−/− Hepatocytes by Activating NF-κB Pathway
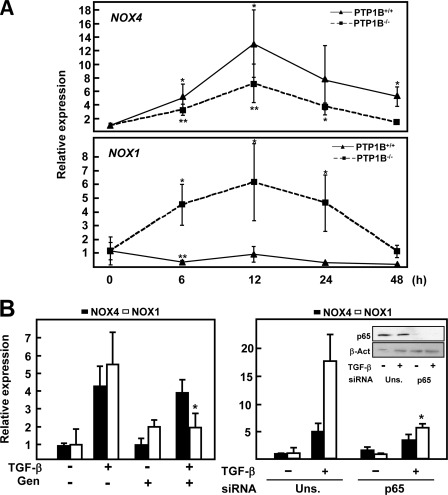
As described above, NOX-derived reactive oxygen species are important regulators of TGF-β-induced cell death in hepatocytes and may be contemplated as proapoptotic or antiapoptotic, depending mainly on the NOX enzyme induced (16). In addition, we have previously proposed a relationship between NF-κB and NADPH oxidases in response to TGF-β (20). For these reasons, we decided to investigate whether they could be implicated in the resistance to TGF-β observed in PTP1B-deficient cells. When we comparatively measured NOX1 and NOX4 expression, we found their expression regulated differently: first, PTP1B-deficient cells showed a decreased but still significant NOX4 induction compared with wild-type cells; second, although NOX1 is repressed by TGF-β in PTP1B+/+ cells, it is strongly induced in PTP1B−/− hepatocytes (Fig. 6A). In addition, basal expression of NOX1 was significantly higher in PTP1B−/− cells (data not shown). These results suggest that PTP1B-deficient cells show an imbalance in the expression of NOX enzymes upon TGF-β treatment, leading to a more prosurvival phenotype. Accordingly, we decided to measure their levels in genistein-dependent or p65 knockdown-dependent sensitization to TGF-β. NOX1 up-regulation by TGF-β was impeded by both genistein and p65 knockdown, whereas NOX4 expression was not affected (Fig. 6B).
FIGURE 6.
PTP1B−/− hepatocytes show a prosurvival pattern of NADPH oxidase induction in response to TGF-β. A, NOX1 and NOX4 expression at the indicated times of TGF-β treatment was determined by real-time PCR. Data were calculated with respect to zero time for each cell type. B, left, cells were treated for 30 min with genistein in the presence or the absence of TGF-β for an additional 12 h. Right, PTP1B+/+ and PTP1B−/− cells were transfected with either an unspecific siRNA (Uns.) or p65 siRNA (p65) and then treated or not with TGF-β for 12 h. Data represent the mean ± S.E. (error bars) of at least three independent experiments. Student's t test was calculated versus zero time (A), genistein-pretreated versus nonpretreated cells (B, left), or TGF-β-treated cells for each siRNA (B, right). *, p < 0.05; **, p < 0.01.
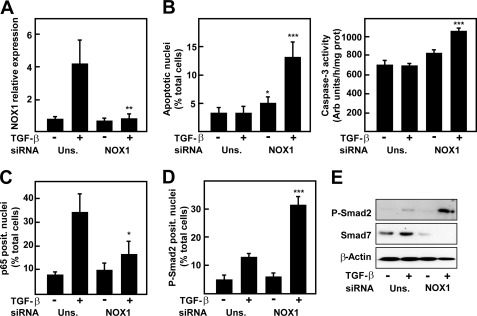
For these reasons, we wanted to test the effects of NOX1-targeted knockdown on TGF-β-dependent cell death in PTP1B−/− cells. NOX1 silencing, which was effective in blocking an TGF-β-induced increase in NOX1 (Fig. 7A), recovered the apoptotic response to TGF-β in terms of percentage of apoptotic nuclei and caspase-3 activity, confirming the prosurvival role of this protein in hepatocytes (Fig. 7B). It is worthy to note that NOX1 silencing also decreased p65 nuclear translocation (Fig. 7C) while increasing Smad2 phosphorylation and nuclear localization (Fig. 7, D and E). Importantly, we observed an inhibition of Smad7 protein level, as it occurred when p65 was silenced (Fig. 7E). These results imply that NOX1 activation is upstream NF-κB activation; however, down-regulation of NOX1 when p65 was knocked down suggests the existence of a positive feedback loop on NOX1 expression levels dependent on NF-κB.
FIGURE 7.
NOX1 knock-down in PTP1B−/− hepatocytes recovers the apoptotic response to TGF-β and Smad2 phosphorylation, inhibiting p65 nuclear translocation. PTP1B−/− cells were transfected with either an unspecific siRNA (Uns.) or NOX1 siRNA (NOX1) and then treated or not with TGF-β. A, real-time PCR of NOX1 at 12 h. B, cell death measured as percentage of apoptotic nuclei at 30 h (left) or caspase-3 activity at 16 h (right). C, percentage of p65-positive nuclei. D, percentage of phospho-Smad2-positive nuclei. E, Western blot of total lysates. β-Actin was used as loading control. Data represent the mean ± S.E. (error bars) of two experiments with duplicate or triplicate wells. Student's t test was calculated versus unspecific siRNA: *, p < 0.05; **, p < 0.01;***, p < 0.001).
DISCUSSION
TGF-β plays a dual role in the control of liver pathophysiology, exerting both suppressor and tumor promoter functions. We and others have described previously that suppressor functions of TGF-β in hepatocytes, mainly growth inhibition and apoptosis induction, can be inhibited by different survival signals through activation of the EGFR, PI3K/AKT, and/or ERK1/2 hyperactivation (26–28) that are frequently observed in tumoral hepatocytes (18, 19). In most cases, such prosurvival signals are dependent on growth factor or cytokine receptors, whose activation status is controlled mainly by phosphorylation/dephosphorylation cycles triggered by protein kinases and phosphatases. In this context, the tyrosine phosphatase PTP1B seemed a good candidate for the modulation of the suppressor effects of TGF-β because it controls several receptor tyrosine kinases, such as the EGFR, the IR, or IGF-1R (29). As hypothesized, PTP1B-deficient hepatocytes showed great resistance to the suppressor effects of TGF-β, correlating with high ERK1/2 and AKT phosphorylations (Fig. 1). In addition, these cells were less responsive to exogenous EGF in terms of growth, which might reflect EGFR activation under basal conditions (Fig. 1C and Table 1).
TGF-β-induced growth inhibition and apoptosis are controlled in a Smad-dependent manner (30). We discarded that the expression of essential components of the TGF-β canonical pathway might be altered in PTP1B−/− cells (supplemental Fig. 1), but we found a diminished Smad2 and 3 phosphorylation and nuclear translocation upon TGF-β treatment compared with wild-type cells (Fig. 2). TGF-β pathway is strongly regulated at the level of Smad activation and nuclear translocation by different means. One mechanism involves Smad sequestration in the cytosol by proteins acting as repressors such as SnoN (31, 32) or AKT (33, 34). However, these proteins do not seem to participate in our system, because neither SnoN knockdown nor AKT inhibition with LY294002 was able to recover response to TGF-β in PTP1B-deficient cells (data not shown). Moreover, we found that although IGF-1 levels were about 30 times higher in PTP1B-deficient hepatocytes, correlating with strong receptor activation (supplemental Fig. 3), knockdown or inhibition of IGF-1R, IGF-1, or IR suppressed autocrine growth of PTP1B-deficient cells but was not able to recover the response to TGF-β in terms of cell death (supplemental Fig. 3 and data not shown). Knockdown experiments using specific siRNAs against EGFR, or combinations of EGFR, IR, IGF-1, or IGF-1R were also ineffective (supplemental Fig. 4 and data not shown). All of these negative results indicate that the impairment of TGF-β-induced suppressor effects due to the kinase activation status of the cell related to PTP1B deficiency cannot be overcome by simple inhibition of two or three kinases because it is probably the sum of signals from multiple origins. For these reasons, the apoptotic response was only observed in the presence of genistein, a general tyrosine kinase inhibitor (Fig. 3).
Several articles had demonstrated NF-κB-dependent inhibition of the proapoptotic effects of TGF-β both in normal and transformed hepatocytes (12, 35, 36). Indeed, we have described that TGF-β can induce not only proapoptotic but also prosurvival signals in hepatocytes. Interestingly, these signals are dependent on NF-κB activity (20, 37). In our model, results obtained with p65-targeted knockdown confirm the inhibitory effect of NF-κB on TGF-β signaling in both PTP1B+/+ and PTP1B−/− hepatocytes (Figs. 4 and 5). Thus, we hypothesize that kinase hyperactivation due to PTP1B lack provokes an early or excessive NF-κB activation, which completely blocks the response to the cytokine in PTP1B-deficient cells.
The existence of a connection between PTP1B and NOX proteins has been proposed previously (38, 39). However, such a relationship was always contemplated as unidirectional toward the regulation of PTP1B activity by NOX-derived reactive oxygen species. In fact, it is well known that protein phosphatases are regulated mainly by oxidation/reduction of a cysteine localized in their active center (40). In contrast, what we propose herein involves a differential NOX regulation at the expression level depending on the presence of PTP1B (Fig. 6). NOX1 regulation by TGF-β seems to be Smad-independent (because TGF-β-induced changes are not affected by either Smad2 or Smad3 siRNAs) (supplemental Fig. 5), but NF-κB-dependent (Fig. 6B). Indeed, a direct dependence of NOX1 transcriptional levels on NF-κΒ has been described previously in vascular smooth muscle cells (41). As mentioned above, NOX1 and NOX4 play opposite roles in the control of cell survival and death in response to TGF-β. In this regard, we have reported that NOX4 is up-regulated in response to the cytokine in normal and tumoral hepatocytes (15, 16), whereas NOX1 remains unchanged or is even down-regulated in fetal hepatocytes treated with TGF-β (15, 20). In agreement with these previous results, in this study we demonstrate that NOX4 up-regulation and NOX1 down-regulation also occur in neonatal wild-type hepatocytes upon TGF-β treatment (Fig. 5B). Hence, it was reasonable to hypothesize that the results obtained in PTP1B−/− cells, which increased both NOX1 and NOX4, suggest a change in the balance between cell death and survival signaling toward a more resistant phenotype associated with NOX1 up-regulation. Accordingly, the proapoptotic conditions achieved with the combined treatment of genistein and TGF-β or p65 silencing impeded NOX1 increase, strongly suggesting a role for this protein in our model (Fig. 6B). NOX1 knockdown experiments (Fig. 7B) confirmed the crucial role of this protein in the survival of hepatocytes to proapoptotic insults dependent on TGF-β or growth factor deprivation as we have described previously in tumoral hepatocytes (16, 23, 42). Indeed, results presented here indicate that one of the mechanisms for NOX1 to promote survival is via NF-κB activation because its knockdown reduced p65 nuclear translocation (Fig. 7C). Such a relationship between NOX1 and NF-κB to promote survival upon TGF-β treatment had been suggested previously by our group, because we described that a NADPH oxidase apocynin-inhibitable (NOX1 or NOX2 in hepatocytes) was responsible for NF-κB activation in fetal hepatocytes, which promoted cell survival (20). Indeed, several reports in other cellular systems described NF-κB activation by NOX1 in response to potentially harmful stimuli such as advanced glycation end products (43) or proinflammatory cytokines such as TNF-α or IL-1β (44). Moreover, such prosurvival signaling involving nuclear p65 and NOX1 has been suggested to be important for colon cancer development (45, 46).
Our results suggest that NOX1 would be located upstream of NF-κB in the signaling cascade, but the kinase/kinases regulated by PTP1B responsible for the initial NOX1 activation/up-regulation upon TGF-β treatment remain unknown. It is possible to hypothesize that, because NOX1 has been described to participate in the signaling cascades of several tyrosine kinase receptors and intracellular kinases, NOX1 knockdown blunts several signaling pathways simultaneously leading to NF-κB activation.
However, it is well known that the TGF-β pathway is tightly controlled by a mechanism of autoinhibitory feedback loop involving Smad7 up-regulation (47), which binds to the receptor I and interferes with the phosphorylation of Smad2 and Smad3 (48). Western blotting experiments performed at 2 h of treatment revealed that Smad7 induction was greater in PTP1B−/− cells (Fig. 5E and data not shown). Furthermore, we observed the existence of a correlation among the situations in which the apoptotic response and Smad phosphorylation upon TGF-β treatment were recovered and lowered Smad7 induction (Figs. 5B and 7E). Thus, we propose here that higher Smad7 early induction dependent on NF-κB activation may be inhibiting Smad2 and Smad3 phosphorylation in PTP1B-deficient cells. Indeed, it has been described previously that NF-κB can counteract TGF-β pathway signaling by direct binding of the transcription factor to the Smad7 promoter in a response observed within the first minutes upon stimulation (49, 50).
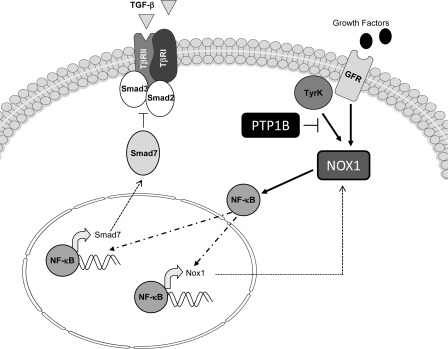
In summary, our results suggest that lack of PTP1B expression impairs TGF-β-induced Smad activation in a NOX1- and NF-κB-dependent manner, which could involve changes in Smad7 levels (Fig. 8). Further work is necessary to study the potential role of this novel pathway in pathological conditions implicating kinase hyperactivation, such as liver cancer.
FIGURE 8.
Proposed model for the effect of PTP1B in TGF-β-dependent suppressor signaling in neonatal hepatocytes. PTP1B-inhibitable signals from intracellular tyrosine kinases or growth factor receptors activate NOX1, which in turn, promotes p65 nuclear translocation. NF-κB increases Smad7 levels by either transcriptional activation or protein stability regulation, which inhibits Smad2 and Smad3 phosphorylation and up-regulates NOX1, to activate an inhibitory positive feedback loop.
Acknowledgments
We thank Dr. E. Castaño from the Scientific and Technical Services, University of Barcelona. We also thank Henrik Westerberg for editing the grammar and spelling in the manuscript.
This work was supported by Ministerio de Ciencia e Innovación, Spain, Grants BFU2009-07219 and ISCIII-RTICC RD06/0020 (to I. F.) and SAF2009-08114 (to A. M. V.) and by Agència de Gestió d'Ajuts Universitari i de Recerca-Generalitat de Catalunya Grant 2009SGR-312 (to I. F.).

This article contains supplemental Figs. 1–5.
- EGFR
- epidermal growth factor receptor
- IGF-1R
- insulin-like growth factor 1 receptor
- IR
- insulin receptor
- NOX
- NADPH oxidase
- PCNA
- proliferating cell nuclear antigen
- PTP1B
- protein-tyrosine phosphatase 1B.
REFERENCES
- 1. Carr B. I., Hayashi I., Branum E. L., Moses H. L. (1986) Inhibition of DNA synthesis in rat hepatocytes by platelet-derived type β transforming growth factor. Cancer Res. 46, 2330–2334 [PubMed] [Google Scholar]
- 2. Oberhammer F. A., Pavelka M., Sharma S., Tiefenbacher R., Purchio A. F., Bursch W., Schulte-Hermann R. (1992) Induction of apoptosis in cultured hepatocytes and in regressing liver by transforming growth factor β1. Proc. Natl. Acad. Sci. U.S.A. 89, 5408–5412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Massagué J. (2008) TGFβ in cancer. Cell 134, 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Coulouarn C., Factor V. M., Thorgeirsson S. S. (2008) Transforming growth factor-β gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology 47, 2059–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hunter T. (1998) The role of tyrosine phosphorylation in cell growth and disease. Harvey Lect. 94, 81–119 [PubMed] [Google Scholar]
- 6. Frangioni J. V., Beahm P. H., Shifrin V., Jost C. A., Neel B. G. (1992) The nontransmembrane tyrosine phosphatase PTP-1B localizes to the endoplasmic reticulum via its 35-amino acid C-terminal sequence. Cell 68, 545–560 [DOI] [PubMed] [Google Scholar]
- 7. Miranda S., González-Rodríguez A., Revuelta-Cervantes J., Rondinone C. M., Valverde A. M. (2010) Beneficial effects of PTP1B deficiency on brown adipocyte differentiation and protection against apoptosis induced by pro- and anti-inflammatory stimuli. Cell. Signal. 22, 645–659 [DOI] [PubMed] [Google Scholar]
- 8. González-Rodriguez A., Escribano O., Alba J., Rondinone C. M., Benito M., Valverde A. M. (2007) Levels of protein-tyrosine phosphatase 1B determine susceptibility to apoptosis in serum-deprived hepatocytes. J. Cell. Physiol. 212, 76–88 [DOI] [PubMed] [Google Scholar]
- 9. Valverde A. M., González-Rodríguez A. (2011) IRS2 and PTP1B: two opposite modulators of hepatic insulin signalling. Arch. Physiol. Biochem. 117, 105–115 [DOI] [PubMed] [Google Scholar]
- 10. Sangwan V., Paliouras G. N., Cheng A., Dubé N., Tremblay M. L., Park M. (2006) Protein-tyrosine phosphatase 1B deficiency protects against Fas-induced hepatic failure. J. Biol. Chem. 281, 221–228 [DOI] [PubMed] [Google Scholar]
- 11. Chacón P. J., Arévalo M. A., Tébar A. R. (2010) NGF-activated protein-tyrosine phosphatase 1B mediates the phosphorylation and degradation of IκBα coupled to NF-κB activation, thereby controlling dendrite morphology. Mol. Cell. Neurosci. 43, 384–393 [DOI] [PubMed] [Google Scholar]
- 12. Arsura M., FitzGerald M. J., Fausto N., Sonenshein G. E. (1997) Nuclear factor-κB/Rel blocks transforming growth factor β1-induced apoptosis of murine hepatocyte cell lines. Cell Growth Differ. 8, 1049–1059 [PubMed] [Google Scholar]
- 13. Herrera B., Alvarez A. M., Sánchez A., Fernández M., Roncero C., Benito M., Fabregat I. (2001) Reactive oxygen species (ROS) mediates the mitochondrial-dependent apoptosis induced by transforming growth factor β in fetal hepatocytes. FASEB J. 15, 741–751 [DOI] [PubMed] [Google Scholar]
- 14. Sánchez A., Alvarez A. M., Benito M., Fabregat I. (1996) Apoptosis induced by transforming growth factor-β in fetal hepatocyte primary cultures: involvement of reactive oxygen intermediates. J. Biol. Chem. 271, 7416–7422 [DOI] [PubMed] [Google Scholar]
- 15. Carmona-Cuenca I., Roncero C., Sancho P., Caja L., Fausto N., Fernández M., Fabregat I. (2008) Up-regulation of the NADPH oxidase NOX4 by TGF-β in hepatocytes is required for its pro-apoptotic activity. J. Hepatol. 49, 965–976 [DOI] [PubMed] [Google Scholar]
- 16. Sancho P., Bertran E., Caja L., Carmona-Cuenca I., Murillo M. M., Fabregat I. (2009) The inhibition of the epidermal growth factor (EGF) pathway enhances TGF-β-induced apoptosis in rat hepatoma cells through inducing oxidative stress coincident with a change in the expression pattern of the NADPH oxidases (NOX) isoforms. Biochim. Biophys. Acta 1793, 253–263 [DOI] [PubMed] [Google Scholar]
- 17. Carmona-Cuenca I., Herrera B., Ventura J. J., Roncero C., Fernández M., Fabregat I. (2006) EGF blocks NADPH oxidase activation by TGF-β in fetal rat hepatocytes, impairing oxidative stress, and cell death. J. Cell. Physiol. 207, 322–330 [DOI] [PubMed] [Google Scholar]
- 18. Caja L., Sancho P., Bertran E., Fabregat I. (2011) Dissecting the effect of targeting the epidermal growth factor receptor on TGF-β-induced apoptosis in human hepatocellular carcinoma cells. J. Hepatol. 55, 351–358 [DOI] [PubMed] [Google Scholar]
- 19. Caja L., Sancho P., Bertran E., Iglesias-Serret D., Gil J., Fabregat I. (2009) Overactivation of the MEK/ERK pathway in liver tumor cells confers resistance to TGF-β-induced cell death through impairing up-regulation of the NADPH oxidase NOX4. Cancer Res. 69, 7595–7602 [DOI] [PubMed] [Google Scholar]
- 20. Murillo M. M., Carmona-Cuenca I., Del Castillo G., Ortiz C., Roncero C., Sánchez A., Fernández M., Fabregat I. (2007) Activation of NADPH oxidase by transforming growth factor-β in hepatocytes mediates up-regulation of epidermal growth factor receptor ligands through a nuclear factor-κB-dependent mechanism. Biochem. J. 405, 251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gonzalez-Rodriguez A., Clampit J. E., Escribano O., Benito M., Rondinone C. M., Valverde A. M. (2007) Developmental switch from prolonged insulin action to increased insulin sensitivity in protein-tyrosine phosphatase 1B-deficient hepatocytes. Endocrinology 148, 594–608 [DOI] [PubMed] [Google Scholar]
- 22. Caja L., Ortiz C., Bertran E., Murillo M. M., Miró-Obradors M. J., Palacios E., Fabregat I. (2007) Differential intracellular signalling induced by TGF-β in rat adult hepatocytes and hepatoma cells: implications in liver carcinogenesis. Cell. Signal. 19, 683–694 [DOI] [PubMed] [Google Scholar]
- 23. Sancho P., Fabregat I. (2010) NADPH oxidase NOX1 controls autocrine growth of liver tumor cells through up-regulation of the epidermal growth factor receptor pathway. J. Biol. Chem. 285, 24815–24824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Itoh S., ten Dijke P. (2007) Negative regulation of TGF-β receptor/Smad signal transduction. Curr. Opin. Cell Biol. 19, 176–184 [DOI] [PubMed] [Google Scholar]
- 25. Banerjee S., Li Y., Wang Z., Sarkar F. H. (2008) Multi-targeted therapy of cancer by genistein. Cancer Lett. 269, 226–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Valdés F., Murillo M. M., Valverde A. M., Herrera B., Sánchez A., Benito M., Fernández M., Fabregat I. (2004) Transforming growth factor-β activates both pro-apoptotic and survival signals in fetal rat hepatocytes. Exp. Cell Res. 292, 209–218 [DOI] [PubMed] [Google Scholar]
- 27. Murillo M. M., del Castillo G., Sánchez A., Fernández M., Fabregat I. (2005) Involvement of EGF receptor and c-Src in the survival signals induced by TGF-β1 in hepatocytes. Oncogene 24, 4580–4587 [DOI] [PubMed] [Google Scholar]
- 28. Fabregat I., Herrera B., Fernández M., Alvarez A. M., Sánchez A., Roncero C., Ventura J. J., Valverde A. M., Benito M. (2000) Epidermal growth factor impairs the cytochrome c/caspase-3 apoptotic pathway induced by transforming growth factor β in rat fetal hepatocytes via a phosphoinositide 3-kinase-dependent pathway. Hepatology 32, 528–535 [DOI] [PubMed] [Google Scholar]
- 29. Stuible M., Tremblay M. L. (2010) In control at the ER: PTP1B and the down-regulation of RTKs by dephosphorylation and endocytosis. Trends Cell Biol. 20, 672–679 [DOI] [PubMed] [Google Scholar]
- 30. Heldin C. H., Landström M., Moustakas A. (2009) Mechanism of TGF-β signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr. Opin. Cell Biol. 21, 166–176 [DOI] [PubMed] [Google Scholar]
- 31. Krakowski A. R., Laboureau J., Mauviel A., Bissell M. J., Luo K. (2005) Cytoplasmic SnoN in normal tissues and nonmalignant cells antagonizes TGF-β signaling by sequestration of the Smad proteins. Proc. Natl. Acad. Sci. U.S.A. 102, 12437–12442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mayoral R., Valverde A. M., Llorente Izquierdo C., González-Rodríguez A., Boscá L., Martín-Sanz P. (2010) Impairment of transforming growth factor β signaling in caveolin-1-deficient hepatocytes: role in liver regeneration. J. Biol. Chem. 285, 3633–3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Remy I., Montmarquette A., Michnick S. W. (2004) PKB/Akt modulates TGF-β signalling through a direct interaction with Smad3. Nat. Cell Biol. 6, 358–365 [DOI] [PubMed] [Google Scholar]
- 34. Conery A. R., Cao Y., Thompson E. A., Townsend C. M., Jr., Ko T. C., Luo K. (2004) Akt interacts directly with Smad3 to regulate the sensitivity to TGF-β-induced apoptosis. Nat. Cell Biol. 6, 366–372 [DOI] [PubMed] [Google Scholar]
- 35. Arsura M., Mercurio F., Oliver A. L., Thorgeirsson S. S., Sonenshein G. E. (2000) Role of the IκB kinase complex in oncogenic Ras- and Raf-mediated transformation of rat liver epithelial cells. Mol. Cell. Biol. 20, 5381–5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Arsura M., Panta G. R., Bilyeu J. D., Cavin L. G., Sovak M. A., Oliver A. A., Factor V., Heuchel R., Mercurio F., Thorgeirsson S. S., Sonenshein G. E. (2003) Transient activation of NF-κB through a TAK1/IKK kinase pathway by TGF-β1 inhibits AP-1/SMAD signaling and apoptosis: implications in liver tumor formation. Oncogene 22, 412–425 [DOI] [PubMed] [Google Scholar]
- 37. Franco D. L., Mainez J., Vega S., Sancho P., Murillo M. M., de Frutos C. A., Del Castillo G., López-Blau C., Fabregat I., Nieto M. A. (2010) Snail1 suppresses TGF-β-induced apoptosis and is sufficient to trigger EMT in hepatocytes. J. Cell Sci. 123, 3467–3477 [DOI] [PubMed] [Google Scholar]
- 38. Chen K., Kirber M. T., Xiao H., Yang Y., Keaney J. F., Jr. (2008) Regulation of ROS signal transduction by NADPH oxidase 4 localization. J. Cell Biol. 181, 1129–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mahadev K., Motoshima H., Wu X., Ruddy J. M., Arnold R. S., Cheng G., Lambeth J. D., Goldstein B. J. (2004) The NAD(P)H oxidase homolog Nox4 modulates insulin-stimulated generation of H2O2 and plays an integral role in insulin signal transduction. Mol. Cell. Biol. 24, 1844–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Salmeen A., Barford D. (2005) Functions and mechanisms of redox regulation of cysteine-based phosphatases. Antioxid. Redox Signal. 7, 560–577 [DOI] [PubMed] [Google Scholar]
- 41. Manea A., Tanase L. I., Raicu M., Simionescu M. (2010) Transcriptional regulation of NADPH oxidase isoforms, Nox1 and Nox4, by nuclear factor-κB in human aortic smooth muscle cells. Biochem. Biophys. Res. Commun. 396, 901–907 [DOI] [PubMed] [Google Scholar]
- 42. Sancho P., Fabregat I. (2011) The NADPH oxidase inhibitor VAS2870 impairs cell growth and enhances TGF-β-induced apoptosis of liver tumor cells. Biochem. Pharmacol. 81, 917–924 [DOI] [PubMed] [Google Scholar]
- 43. San Martin A., Foncea R., Laurindo F. R., Ebensperger R., Griendling K. K., Leighton F. (2007) Nox1-based NADPH oxidase-derived superoxide is required for VSMC activation by advanced glycation end-products. Free Radic. Biol. Med. 42, 1671–1679 [DOI] [PubMed] [Google Scholar]
- 44. Miller F. J., Jr., Filali M., Huss G. J., Stanic B., Chamseddine A., Barna T. J., Lamb F. S. (2007) Cytokine activation of nuclear factor κB in vascular smooth muscle cells requires signaling endosomes containing Nox1 and ClC-3. Circ. Res. 101, 663–671 [DOI] [PubMed] [Google Scholar]
- 45. Fukuyama M., Rokutan K., Sano T., Miyake H., Shimada M., Tashiro S. (2005) Overexpression of a novel superoxide-producing enzyme, NADPH oxidase 1, in adenoma and well differentiated adenocarcinoma of the human colon. Cancer Lett. 221, 97–104 [DOI] [PubMed] [Google Scholar]
- 46. Wang R., Dashwood W. M., Nian H., Löhr C. V., Fischer K. A., Tsuchiya N., Nakagama H., Ashktorab H., Dashwood R. H. (2011) NADPH oxidase overexpression in human colon cancers and rat colon tumors induced by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP). Int. J. Cancer 128, 2581–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nakao A., Afrakhte M., Morén A., Nakayama T., Christian J. L., Heuchel R., Itoh S., Kawabata M., Heldin N. E., Heldin C. H., ten Dijke P. (1997) Identification of Smad7, a TGFβ-inducible antagonist of TGFβ signalling. Nature 389, 631–635 [DOI] [PubMed] [Google Scholar]
- 48. Hayashi H., Abdollah S., Qiu Y., Cai J., Xu Y. Y., Grinnell B. W., Richardson M. A., Topper J. N., Gimbrone M. A., Jr., Wrana J. L., Falb D. (1997) The MAD-related protein Smad7 associates with the TGFβ receptor and functions as an antagonist of TGFβ signaling. Cell 89, 1165–1173 [DOI] [PubMed] [Google Scholar]
- 49. Nagarajan R. P., Chen F., Li W., Vig E., Harrington M. A., Nakshatri H., Chen Y. (2000) Repression of transforming growth factor-β-mediated transcription by nuclear factor κB. Biochem. J. 348, 591–596 [PMC free article] [PubMed] [Google Scholar]
- 50. Bitzer M., von Gersdorff G., Liang D., Dominguez-Rosales A., Beg A. A., Rojkind M., Böttinger E. P. (2000) A mechanism of suppression of TGF-β/SMAD signaling by NF-κB/RelA. Genes Dev. 14, 187–197 [PMC free article] [PubMed] [Google Scholar]