Background: AT-hook motif-containing proteins are associated with chromatin modifications.
Results: The Arabidopsis AT-hook protein AHL22 regulates H3 acetylation and methylation in FT chromatin by binding to an intragenic AT-rich DNA sequence.
Conclusion: AHL22 acts as a chromatin remodeling factor that regulates FT expression in flowering induction.
Significance: Learning how the FT gene is regulated is critical for understanding gene regulatory mechanisms of flowering.
Keywords: Arabidopsis, Chromatin Modification, Chromosomes/Nonhistone Chromosomal Proteins, Histone Deacetylase, Nuclear Matrix, Arabidopsis, FT Chromatin, Flowering, Histone H3 Acetylation, Histone H3 Methylation
Abstract
Coordination of the onset of flowering with developmental status and seasonal cues is critical for reproductive success in plants. Molecular genetic studies on Arabidopsis mutants that have alterations in flowering time have identified a wide array of genes that belong to distinct genetic flowering pathways. The flowering time genes are regulated through versatile molecular and biochemical mechanisms, such as controlled RNA metabolism and chromatin modifications. Recent studies have shown that a group of AT-hook DNA-binding motif-containing proteins plays a role in plant developmental processes and stress responses. Here, we demonstrate that the AT-hook protein AHL22 (AT-hook motif nuclear localized 22) regulates flowering time by modifying FLOWERING LOCUS T (FT) chromatin in Arabidopsis. AHL22 binds to a stretch of the AT-rich sequence in the FT locus. It interacts with a subset of histone deacetylases. An Arabidopsis mutant overexpressing the AHL22 gene (OE-AHL22) exhibited delayed flowering, and FT transcription was significantly reduced in the mutant. Consistent with the delayed flowering and FT suppression in the OE-AHL22 mutant, histone 3 (H3) acetylation was reduced and H3 lysine 9 dimethylation was elevated in the FT chromatin. We propose that AHL22 acts as a chromatin remodeling factor that modifies the architecture of FT chromatin by modulating both H3 acetylation and methylation.
Introduction
The timing of flowering initiation is regulated through coordinated interactions of developmental programs, such as gibberellic acid, and seasonal cues, including photoperiod, exposure to prolonged low temperature (vernalization), and ambient temperature (1–3). The developmental and environmental signals converge to regulate floral integrators, such as FT,2 SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), and LEAFY (LFY) (2). The FT and SOC1 integrators are also regulated by the floral repressor FLOWERING LOCUS C (FLC) that incorporates vernalization and autonomous signals into the flowering genetic network (1, 2).
Expression of flowering time genes is modulated through various molecular and biochemical mechanisms in addition to the ordinary gene transcriptional regulation. Examples include controlled RNA metabolism, which is governed primarily by RNA-binding proteins and microRNAs, and epigenetic regulation, which is mediated mainly by histone modifications and DNA methylation (4–7). Several RNA-binding proteins have been shown to regulate RNA processing and selection of polyadenylation sites in their own genes and the FLC gene (4, 8). MicroRNAs regulate post-transcriptionally flowering time genes. miR156 suppresses a subset of genes encoding SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) transcription factors that promote flowering (5). MiR172 induces degradation of gene transcripts encoding a small group of APETALA2 (AP2)-like transcription factors that act as floral repressors (5).
Expression of flowering time genes is also regulated by epigenetic mechanisms that include post-translational modifications of histone and nonhistone proteins. Regulation of flowering initiation by histone modifications has been studied extensively in FLC chromatin. Molecular characterization of FLC repressors and activators in recent years has shown that at least three regulatory systems, vernalization, FRIGIDA (FRI), and autonomous pathway components, regulate FLC activity by modifying the FLC chromatin. It is known that specific lysine (K) residues in the N-terminal region of H3 are either methylated or acetylated (6). H3 trimethylation at Lys-4 and acetylation are associated with active FLC expression. In contrast, H3 deacetylation and methylation at Lys-9 and Lys-27 repress the FLC expression (6).
Nuclear matrix is a network of filamentous proteins and somewhat analogous to cellular cytoskeleton (9). Organization of the nuclear matrix is regulated in a temporal and spatial manner during the cell cycle (10, 11). It also contributes to dynamic chromatin reorganization occurring during DNA metabolism and gene expression (9–11). The matrix attachment region (MAR), which is also called the scaffold attachment region, is a stretch of AT-rich DNA sequence (ATR) that guides binding of genomic DNA to the nuclear matrix (10, 11). Therefore, MAR acts as a structural determinant of chromatin organization and recruits multiple MAR-binding factors that facilitate remodeling of the chromatin structure in regulating gene expression (12).
Various MAR-binding factors have been identified in yeast, animals, and plants (11, 12). A major group of the MAR-binding factors possesses a protein motif, called AT-hook that consists of 9–12 residues (13). In animals, many AT-hook proteins have been identified in diverse protein groups, and their roles have been demonstrated in different aspects of gene regulation (13, 14). In plants, a series of AT-hook proteins plays a role in developmental processes, such as flowering transition and stress responses (15–21). One example is the AT-hook motif nuclear localized 22 (AHL22) protein that belongs to the AHL family consisting of 29 members in Arabidopsis (22). Overexpression of the AHL22 gene delays flowering, and FT expression is reduced in the transgenic plants (19). In contrast, silencing of four AHL genes (AHL22, AHL18, AHL27, and AHL29) promotes flowering, suggesting that the AHL22 gene, and some other AHL genes as well, act as a floral repressors, possibly by modulating FT expression.
Here, we show that the AHL22 protein binds to an ATR sequence element within the FT locus, which has previously been predicted as a intragenic MAR (10), and regulates FT expression by recruiting a subset of histone deacetylases, such as HDA1/HDA19, HDA6, and HDA9. H3 acetylation was significantly reduced in the AHL22-overexpressing OE-AHL22 mutant. We also found that H3K9 dimethylation in the FT chromatin was elevated in the mutant, suggesting that AHL22 may also interact with histone methyltransferases. Our observations indicate that the FT chromatin is coordinately regulated through H3 acetylation and methylation during floral transition.
EXPERIMENTAL PROCEDURES
Plant Materials
Arabidopsis thaliana lines used were in Columbia (Col-0) background. Arabidopsis plants were grown in a controlled culture room at 23 °C under long days (LDs, 16-h light/8-h dark). To produce transgenic plants overexpressing Arabidopsis genes, the gene sequences were subcloned into the binary pB2GW7 vector under control of the cauliflower mosaic virus (CaMV) 35S-promoter (Invitrogen). The loss-of-function mutants ahl22-1 and ahl22-2 (SALK-018866 and SALK-143279, respectively) were isolated from a pool of T-DNA insertion lines deposited in the Arabidopsis Biological Resource Center (ABRC, Ohio State University).
Isolation of OE-AHL22 Mutant
The OE-AHL22 mutant was isolated from an Arabidopsis mutant pool that has been produced by randomly integrating the activation tagging vector pSKI015 that contains the CaMV 35S-enhancer element into the genome of Col-0 plants (23). The presence of a single T-DNA insertion event in the OE-AHL22 mutant was verified by genomic Southern blot analysis using the 35S-enhancer sequence as probe. The flanking genomic sequences of the T-DNA insertion site were determined by a plasmid rescue method (15).
Analysis of Transcript Levels
Transcript levels were examined by either Southern blot hybridization of semi-quantitative RT-PCR products or by quantitative real-time RT-PCR (qRT-PCR). RNA sample preparations, PCR conditions, and data processing have been described previously (23).
qRT-PCR was carried out in 96-well blocks with the Applied Biosystems 7500 Real-time PCR System using the SYBR Green I master mix in a volume of 20 μl. The two-step thermal cycling profile used was denaturation for 15 s at 94 °C and extension for 1 min at 68 °C. The comparative ΔΔCT method was used to evaluate the relative quantities of each amplified product in the samples. The threshold cycle (CT) was automatically determined for each reaction by the system set with default parameters. The specificity of amplifications was determined by melt curve analysis of the amplified products using the standard method installed in the system. The eIF4A (eukaryotic initiation factor 4A) gene (At3g13920) was included in the reactions as internal control for normalizing the variations in the cDNA amounts used. The RT-PCR and qRT-PCR primers used are listed in supplemental Table S1.
Flowering Time Measurements
Plants were grown in soil under LDs until flowering. Flowering times were determined by counting the number of rosette leaves at bolting. Fifteen to 20 plants were counted and averaged for each measurement.
AHL22 Binding to FT DNA
Binding of AHL22 to FT DNA was examined using recombinant the maltose-binding protein (MBP)-AHL22 fusion protein essentially as described previously (24) but with some modifications. The recombinant MBP-AHL22 fusion protein was produced in Escherichia coli strain BL21-codon Plus (DE3)-RIL (Stratagene, La Jolla, CA). After induction for 5 h at room temperature, E. coli cells were harvested and resuspended in lysis buffer A (20 mm Tris-HCl, pH 7.4, 200 mm NaCl, 1 mm EDTA, and 10 mm β-mercaptoethanol) containing protease inhibitor mixture (Sigma) and 1 mm PMSF. The cells were lysed by French press (8500 psi, three times). The cell lysates were sonicated for 30 s twice and centrifuged at 20,000 × g for 20 min. The supernatants were stored at −80 °C until use.
For purification of the fusion protein, 1 ml of cell lysates was mixed with amylose resin (New England Biolabs, Ipswich, MA) and incubated at 4 °C for 2 h. The resin was washed 3 times with fresh lysis buffer A. Bound proteins were eluted with 1× SDS-PAGE loading buffer, separated on 10% SDS-PAGE, and transferred to polyvinylidene fluoride membrane. The air-dried membrane blot was immersed in binding buffer (25 mm HEPES, pH 8.00, 60 mm KCl, 1 mm EDTA, 1 mm DTT, and 6 m guanidine hydrochloride) and gently shaken for 10 min at 4 °C. Renaturation of the bound proteins and MAR-binding assays were carried out as described previously (24).
Chromatin Immunoprecipitation Assays (ChIP)
ChIP assays were performed as described previously (25) using 2-week-old plants grown under LDs on ½ × Murashige & Skoog (MS)-agar plates (hereafter, referred to as MS-agar plates). Briefly, rosette leaves were vacuum-infiltrated with 1% formaldehyde for cross-linking and ground in liquid nitrogen after quenching the cross-linking process. Chromatin preparations were sonicated into 0.5–1-kb fragments. Specific antibodies against MYC (catalog number 05-419), H3Ac (catalog number 06-599), H3K9me2 (catalog number 07-212), and H3K27me3 (catalog number 07-449) (Millipore, Billerica, MA) were added to the chromatin solution, which was precleared with salmon sperm DNA/Protein A-agarose beads. The precipitates were eluted from the beads. Cross-links were reversed, and residual proteins were removed by incubation with proteinase K. DNA was recovered using the QIAquick spin column (Qiagen, Valencia, CA). Quantitative PCR was used to determine the amounts of genomic DNA enriched in the chromatin samples. The primers were designed to amplify DNA fragments of 100–200 bp (supplemental Table S1).
Electrophoretic Mobility Shift Assays (EMSA)
EMSAs were carried out as described previously (25) using recombinant MBP-AHL22 fusion protein. Double-stranded DNA fragments were end-labeled with [γ-32P]ATP using T4 polynucleotide kinase. The radiolabeled DNA fragments were incubated for 30 min at room temperature with 1 μg of the MBP-AHL22 fusion protein in binding buffer (10 mm Tris-HCl, pH 7.6, 50 mm NaCl, 1 mm EDTA, 5 mm DTT, 5% glycerol) supplemented with 100 ng of poly(dI-dC) in the presence or absence of competitor DNA fragments. The reaction mixtures were resolved on 4% nondenaturing polyacrylamide gel. The gel was dried on Whatman 3MM paper and exposed to x-ray films.
Subcellular Localization Assays
A full-size AHL22 cDNA was fused in-frame to the 3′ end of a green fluorescence protein (GFP)-coding sequence in the p2FGW7 vector (Invitrogen), and the fusion construct was transformed into Col-0 plants. Lateral roots were subject to fluorescence microscopy.
For bimolecular fluorescence complementation (BiFC) assays, a full-size AHL22 cDNA was fused in-frame to either the 5′ end of a DNA sequence encoding the N-terminal half of EYFP in the pSATN-nEYFP-C1 vector (E3081) or to the 3′ end of a DNA sequence encoding the C-terminal half of EYFP in the pSATN-cEYFP-C1 vector (E3082). The pSAT vectors were kindly provided by Stanton Gelvin (Purdue University). The expression constructs were cotransformed into Arabidopsis protoplasts by a polyethylene glycol-calcium transfection method (26). YFP signals were analyzed 14–18 h after transfection by fluorescence microscopy using the Zeiss LSM510 confocal microscope (Carl Zeiss, Yena, Germany).
In Vitro Pulldown Assays
HDAC cDNAs were amplified by RT-PCR and subcloned into the pGBKT7 vector, which contains the SP6 RNA polymerase promoter upstream of the multiple cloning sequence. [35S]-Labeled HDAC proteins were prepared by in vitro transcription/translation using the TnT SP6 wheat germ extract-coupled system (Promega, Madison, WI). The MBP-mAHL22 gene fusion was subcloned into the pMAL-c2X E. coli expression vector, and recombinant MBP-mAHL22 protein was prepared as described with the recombinant MBP-AHL22 protein above. In vitro pulldown assays were carried out as described previously (27) using 5 μl of 35S-labeled HDAC polypeptides and 5 μg of MBP alone or MBP fusion proteins.
Histochemical Staining
A promoter region consisting of a ∼2-kbp sequence upstream of the transcription start site of the AHL22 gene was transcriptionally fused to a β-glucuronidase (GUS)-coding sequence, and the pAHL22-GUS fusion was transformed into Col-0 plants. The pFT-GUS construct has been described previously (28). Plant sample processing and histochemical detection of GUS activities were carried out as described previously (25).
RESULTS
Pleiotropic Phenotypes of OE-AHL22 Mutant
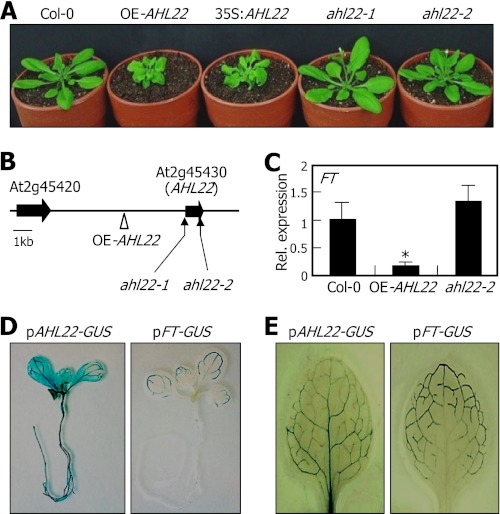
The AHL22-overexpressing OE-AHL22 mutant exhibited delayed flowering with small, curled rosette leaves (Fig. 1A and supplemental Fig. S1A). It was also featured by having short siliques and altered floral structure (supplemental Fig. S1, B and C). We mapped the site of T-DNA insertion by a plasmid rescue method (15). It was found that the T-DNA element was inserted adjacent to the At2g45430 locus in the mutant (Fig. 1B). Genomic Southern blot hybridization confirmed that there was a single T-DNA insertion event in the mutant (supplemental Fig. S2A). Gene expression assays showed that the At2g45430 gene was activated significantly in the mutant (supplemental Fig. S2B), suggesting that activation of the At2g45430 gene correlates with the OE-AHL22 phenotypes. To examine the co-relationship, the At2g45430 gene was overexpressed driven by the CaMV 35S-promoter in Col-0 plants. The resulting 35S:AHL22 transgenic plants recapitulated the OE-AHL22 phenotypes (Fig. 1A), indicating that the At2g45430 activation underlies the OE-AHL22 phenotypes. The At2g45430 gene has previously been named AT-hook motif nuclear localized 22 (AHL22) (19, 22).
FIGURE 1.
Phenotypic and molecular characterization of OE-AHL22 and ahl22 mutants. A, phenotypic comparison. Plants were grown in soil for 4 weeks under LDs before taking photographs. The AHL22 gene was overexpressed driven by the CaMV 35S-promoter in Col-0 plants, resulting in 35S:AHL22 transgenic plants. B, mapping of the T-DNA insertion events. The AHL22 gene does not have introns. C, FT transcript levels. Transcript levels were determined by qRT-PCR. Biological triplicates were averaged and statistically treated using a Student's t test (*, p < 0.01). Bars indicate S.E. of the mean. D and E, expression domains of AHL22 and FT genes. Whole-mount staining of 8-day-old seedlings (D) and staining of the first rosette leaves of 12-day-old seedlings (E) were displayed.
Two AHL22-deficient mutants, ahl22-1 and ahl22-2, were isolated from the T-DNA insertion pool deposited in the ABRC. The knock-out mutants did not show discernible phenotypes (Fig. 1A), possibly because of extensive functional redundancy between AHL22 and other AHL genes (19).
The most prominent phenotype of the OE-AHL22 mutant was late flowering, as has been observed in Arabidopsis mutant overexpressing ESCAROLA (ESC)/ORESARA7 (ORE7)/AHL27 gene (15, 17). The early-flowering phenotype of multiple ahl mutants also supports the role of the AHL22 gene and probably other AHL genes in flowering time control (19). Expression analysis of flowering time genes showed that the FT gene (At1g65480) is significantly suppressed in the OE-AHL22 mutant (Fig. 1C and supplemental Fig. S3A). LFY and AP1 genes, which act downstream of the FT gene (1, 3), were also suppressed in the mutant. In contrast, expression of the FT gene was slightly but reproducibly elevated in the ahl22 mutants (Fig. 1C), suggesting that the late-flowering phenotype of the OE-AHL22 mutant is at least in part caused by FT suppression.
To investigate the potential linkage between the late-flowering phenotype of the OE-AHL22 mutant and FT gene, we compared the spatial expression patterns of the FT and AHL22 genes. The pAHL22-GUS construct, in which a promoter region consisting of ∼2-kb upstream of the transcription start site of the AHL22 gene was fused transcriptionally to the GUS-coding sequence, was transformed into Col-0 plants. The pFT-GUS fusion has been constructed in a similar manner (28). Histochemical assays revealed that in 8-day-old seedlings, whereas GUS activity was detected broadly in the hypocotyls, roots, and vascular bundles of the leaves in the pAHL22-GUS transgenic plants, it was detected primarily in the vascular bundles of the leaves in the pFT-GUS transgenic plants (Fig. 1D). Close examination of GUS distribution patterns in the leaves of 12-day-old seedlings revealed that GUS activity was detected in the vascular bundles of the basal leaf area in the pAHL22-GUS transgenic plants (Fig. 1E). In contrast, it was detected in the vascular bundles of the distal leaf area in the pFT-GUS transgenic plants, as has been observed (28).
The AHL22 gene was highly expressed in earlier growth stages, but its expression decreased drastically during the 2–3 week period after germination (supplemental Fig. S3B), when Arabidopsis plants experience a transition from the juvenile to adult vegetative growth stages (1). In contrast, the FT gene exhibited a reversed expression kinetics. Together with FT suppression in the OE-AHL22 mutant, the opposite spatial and temporal expression patterns of the AHL22 and FT genes further support the notion that FT suppression is related with the AHL22-mediated late flowering.
Binding of AHL22 to FT DNA
AHL proteins possess MAR-binding activity (20, 22). Intragenic MARs are intimately associated with gene regulation (11). We therefore hypothesized that AHL22 might bind to intragenic MARs in the FT locus.
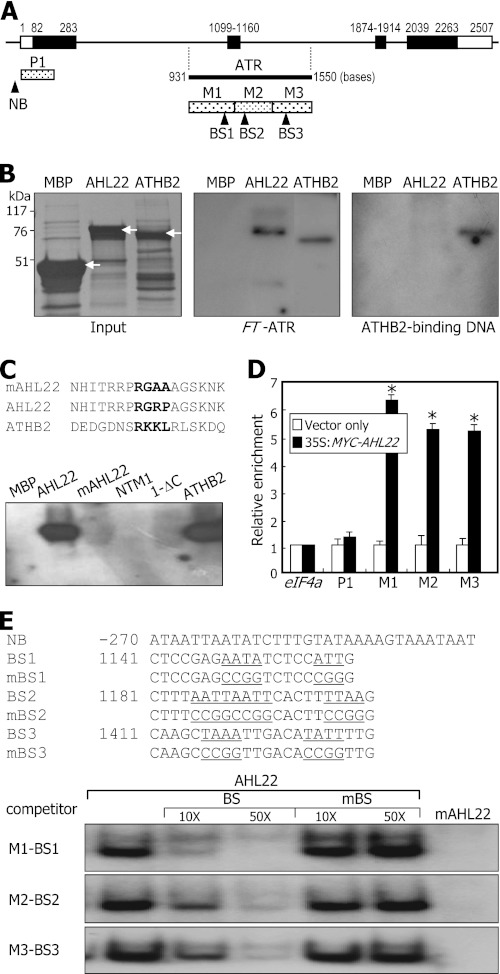
In silico mapping of the Arabidopsis genome sequence revealed that an ATR sequence element consisting of ∼620 nucleotides, which covers parts of introns 1 and 2 and exon 2 of the FT gene (Fig. 2A), has previously been identified as a putative intragenic MAR (10). An ATR sequence was also predicted in the AP1 locus (supplemental Fig. S4A).
FIGURE 2.
Binding of AHL22 to FT-ATR. A, location of ATR in FT locus. Black bars indicate exons, and white bars indicate untranslated regions. The FT-ATR was dissected into 3 sequence regions, M1 to M3. P1 is a control DNA fragment. Putative BSs of AHL22 were selected according to the rule proposed previously (40). NB, nonbinding sequence. B, AHL22 binding to FT-ATR. Recombinant AHL22 and ATHB2 proteins were prepared as MBP fusions in E. coli cells (left panel, white arrows). The same amounts of proteins shown on the protein gel and 32P-labeled DNA fragments were used in the in vitro binding assays (middle panel). The ATHB2-binding sequence was also assayed (right panel). C, effects of core sequence mutations on AHL22 binding to FT-ATR. The core sequence of the AT-hook motif (RGRP) was mutated to RGAA, resulting in mAHL22 (upper panel). NTM1 and ATHB2 transcription factors were included as controls in the assays (lower panel). D, ChIP assays on AHL22 binding to FT-ATR. The 35S:MYC-AHL22 transgenic plants grown on MS-agar plates for 2 weeks were used. Primer pairs specific to M1, M2, and M3 sequences were used. Three measurements were averaged and statistically treated (t test, *p < 0.01). Bars indicate mean ± S.E. E, EMSA on AHL22 binding to FT-ATR. The BS sequences were mutated to verify specific binding, resulting in mutated BS (mBS) sequences (upper panel). Increasing amounts of unlabeled BS or mutated BS oligonucleotides were added to the assay mixtures (lower panel).
We decided to examine whether AHL22 binds to the ATR sequence in the FT locus. Recombinant AHL22 protein was prepared as a MBP-AHL22 fusion in E. coli cells. The FT-ATR fragment was prepared by genomic PCR and end-labeled with [γ-32P]ATP. Southwestern analysis showed that AHL22 indeed binds to the FT-ATR (Fig. 2B). AHL22 also bound to the intragenic ATR sequence in the AP1 locus and the intergenic ATR sequence in the LFY locus (supplemental Fig. S4B).
The homeobox motif-containing ATHB2 transcription factor, which does not have the AT-hook (29), also bound to the FT-ATR. However, the ATHB2-binding DNA fragment, which is a distinct 9-bp dyad-symmetric sequence (CAAT(G/C)ATTG) (30), specifically bound only to ATHB2, but not to AHL22, suggesting that multiple regulatory factors binds to the FT-ATR. Sequence comparison identified a putative AT-hook-like sequence in the ATHB2 protein (Fig. 2C, upper panel). We did not examine whether the sequence motif is responsible for the binding of ATHB2 to the FT-ATR. Notably, the Arabidopsis high mobility group A protein (At1g14900), which has four AT-hook motifs in the C-terminal region, did not bind to the FT-MAR (supplemental Fig. S5), showing that not all AT-hook proteins bind to the FT-MAR.
To further examine the AHL22 binding to the FT-ATR, a mutated AHL22 protein (mAHL22) was synthesized by mutating the core RGRP sequence to RGAA within the AT-hook motif. The mAHL22 protein did not bind to the FT-ATR (Fig. 2C, lower panel), indicating that the interaction is mediated by the AT-hook, as has been shown with AHL1 (22). Although ATHB2 bound to the FT-ATR, additional control transcription factors, such as NTM1 and its activated form ΔC that contain the NAC DNA-binding domain (23), did not exhibit any detectable affinity for the FT-ATR, further supporting that AHL22 binding to the FT-ATR is specific.
We next examined whether AHL22 binds to the FT-ATR in vivo, ChIP assays were carried out using 35S:MYC-AHL22 transgenic plants that overexpress the MYC-AHL22 gene fusion, in which a MYC-coding sequence was fused in-frame to the 5′ end of the AHL22 gene. Chromatin preparations extracted from the transgenic seedlings were probed with an anti-MYC antibody. Primer sets were designed so that PCR products of ∼200 bp, such as M1, M2, and M3 that cover different regions of the FT-ATR (Fig. 2A), were synthesized. P1 is a control DNA sequence region that covers the 5′ untranslated region and part of exon 1, to which the FT repressor TEMPRANILLO 1 binds (31). The assays using the primer sets covering the M1, M2, and M3 regions showed clear enrichment of the FT-ATR sequence, whereas those covering the P1 region did not show any enrichment of the FT-ATR sequence (Fig. 2D). In addition, AHL22 binding to the M1 and M3 regions was further elevated in 35-day-old plants compared with that in 10-day-old plants (supplemental Fig. S6), which is certainly due to the developmental stage-dependent activation of FT chromatin (1). These observations demonstrate that AHL22 binding to the FT-ATR occurs in vivo.
We also carried out EMSA using DNA sequences bearing AAT, ATT, TAA, and TTA within the FT-ATR as probes (Fig. 2E, upper panel). The DNA fragments of ∼20 nucleotides containing the consensus motifs were end-labeled, and their binding to recombinant MBP-AHL22 fusion protein was assayed. It was found that AHL22 bound strongly to the binding sequences (BSs) that are homologous to the FT-ATR (Fig. 2E, lower panel). In addition, the AHL22 binding was significantly reduced in the presence of excess unlabeled BS fragments but only slightly reduced in the presence of mutated DNA fragments (mBSs), supporting the specific binding of AHL22 to the BS sequences. In contrast, we did not detect any detectable binding of mAHL22 to the BS sequences.
Effects of AT-hook Mutation on AHL22 Function in Flowering
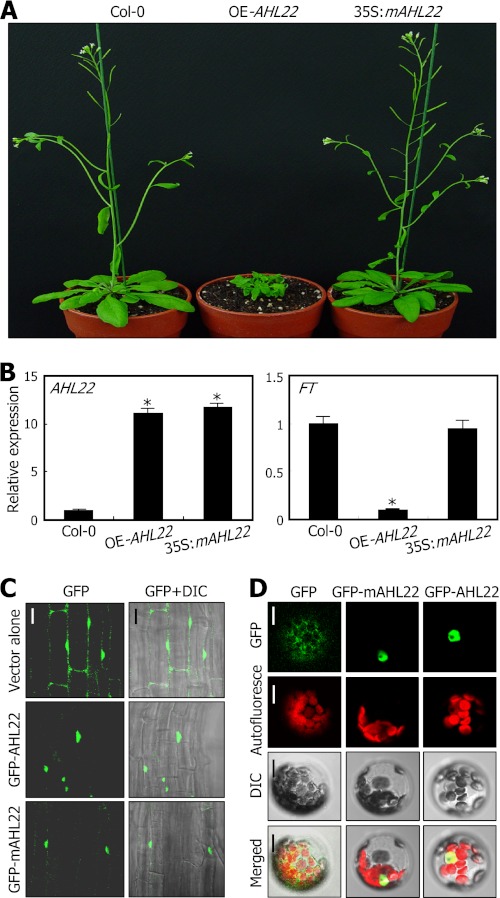
We next examined whether AHL22 binding to the FT-ATR is physiologically important in flowering. To examine this, the mAHL22 gene was overexpressed in Arabidopsis. Unlike the late-flowering 35S:AHL22 transgenic plants, the 35S:mAHL22 transgenic plant did not exhibit late flowering (Fig. 3A). qRT-PCR assays revealed that the mAHL22 transcript level in the 35S:mAHL22 transgenic plant was similar to that in the OE-AHL22 mutant (Fig. 3B, left panel). In contrast, the FT transcript level was not reduced in the 35S:mAHL22 transgenic plant, which was in contrast to the significant suppression of the FT gene in the OE-AHL22 mutant (Fig. 3B, right panel). These observations indicate that AHL22 binding to the FT-ATR is linked with the AHL22-mediated delaying flowering.
FIGURE 3.
Effects of AHL22 mutation on FT expression and flowering. A, flowering phenotypes of 35S:mAHL22 transgenic plants. B, relative transcript levels of FT and AHL22 genes. Two-week-old whole plants grown on MS-agar plates were used for extraction of total RNA. Transcript levels were determined by qRT-PCR. Biological triplicates were averaged and statistically treated (t test, *, p < 0.01). Bars indicate mean ± S.E. C and D, subcellular localization of AHL22 proteins. The GFP-AHL22 and GFP-mAHL22 gene fusions were either transformed into Col-0 plants (C) or transiently expressed in Arabidopsis protoplasts (D). In C, root samples were visualized by differential interference contrast microscopy (DIC) and fluorescence microscopy. Scale bar, 40 μm. In D, Arabidopsis protoplasts were examined by confocal microscopy. Scale bar, 10 μm.
Based on the specific binding of AHL22 to the FT-ATR, it was predicted that AHL22 would be localized in the nucleus. We examined the subcellular localization of AHL22 using transgenic plants overexpressing a GFP-AHL22 fusion, in which a GFP-coding sequence was fused in-frame to the 5′ end of the AHL22 gene. The assays showed that AHL22 is localized exclusively in the nucleus (Fig. 3C). The mAHL22 protein was also localized in the nucleus, indicating that the AT-hook motif is not essential for the nuclear localization of AHL22.
The subcellular distribution of the AHL22 and mAHL22 proteins was further examined by transiently expressing the GFP-AHL22 and GFP-mAHL22 fusions in Arabidopsis protoplasts. Both the AHL22 and mAHL22 proteins were localized exclusively in the nucleus (Fig. 3D), demonstrating that the AHL22 protein is localized in the nucleus, where it binds to the FT-ATR.
AHL22 Suppression of FT in Flowering
Our data showed that AHL22 suppresses FT expression by binding directly to the FT-ATR. Therefore, a question was whether the AHL22 suppression of FT is physiologically important in flowering.
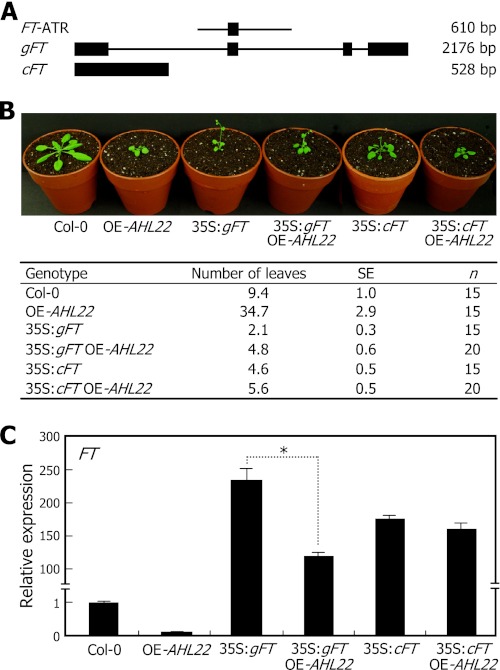
To address the question, we produced two independent transgenic plants: one overexpressing the genomic FT gene sequence (gFT) and the other overexpressing FT cDNA (cFT). The gFT gene consisting of 2180 bp included four exons and three introns (Fig. 4A). It also included the FT-ATR. In contrast, the cFT gene consisting of 528 bp lacks intact FT-ATR, and thus AHL22 is unable to bind to the FT cDNA. The 35S:gFT and 35S:cFT transgenic plants were also genetically crossed with the late-flowering OE-AHL22 mutant, resulting in 35S:gFT OE-AHL22 and 35S:cFT OE-AHL22 plants.
FIGURE 4.
AHL22 suppression of FT gene in flowering. A, FT gene constructs used. Black boxes indicate exons. B, flowering phenotypes of transgenic plants overexpressing either gFT or cFT sequences. Four-week-old plants grown in soil under LDs were photographed (upper panel). Flowering times were measured by counting rosette leaf numbers at bolting (lower panel). Fifteen to 20 plants were counted and averaged for each plant genotype. Values are mean ± S.E. C, FT transcript levels. Whole plants grown on MS-agar plates for 10 days under LDs were used for extraction of total RNA. Transcript levels were determined by qRT-PCR. Biological triplicates were averaged and statistically treated (t test, *, p < 0.01). Bars indicate mean ± S.E.
Both the 35S:gFT and 35S:cFT transgenic plants flowered very early at rosette leaf numbers (RLN) of 2.1 ± 0.3 and 4.6 ± 0.5, respectively (Fig. 4B). The FT transcript levels were accordingly elevated drastically in the transgenic plants (Fig. 4C). The 35S:gFT OE-AHL22 and 35S:cFT OE-AHL22 plants also exhibited early flowering (Fig. 4B). However, counting of the RLN revealed that the early-flowering phenotype of the 35S:gFT transgenic plants was detectably repressed in the 35S:gFT OE-AHL22 plants, which flowered at a RLN of 4.8 ± 0.6. In contrast, the early-flowering phenotype of the 35S:cFT transgenic plants were suppressed only slightly in the 35S:cFT OE-AHL22 transgenic plants, which flowered at a RLN of 5.6 ± 0.5.
Consistent with the changes in flowering times, the FT transcript level was detectably reduced in the 35S:gFT OE-AHL22 plants compared with that in the 35S:gFT transgenic plants (Fig. 4C). In contrast, the FT transcript level in the 35S:cFT OE-AHL22 plants was largely unchanged compared with that in the 35S:cFT transgenic plants, which is certainly because the FT cDNA is not targeted by AHL22. These observations demonstrate that AHL22 regulates flowering time by modulating FT expression.
AHL22 Regulation of H3 Acetylation and Methylation in FT Chromatin
Recent studies have shown that some AT-hook proteins function in chromatin remodeling in both animals and plants (13). MAR-binding factors play a role in gene regulation by mediating chromatin modifications (11, 14). Our data showed that AHL22 interacts with the FT-ATR, which has been suggested to act as an intragenic MAR (10). We therefore examined whether AHL22 repression of FT transcription is mediated by histone modifications.
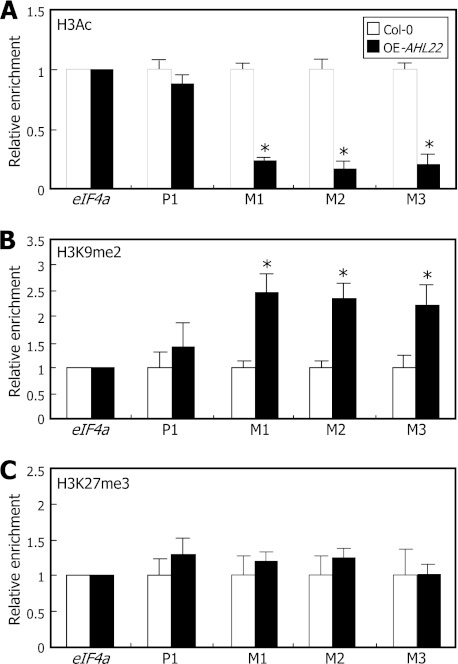
We carried out ChIP assays on the FT chromatin using the primer sets used in the ChIP assays on AHL22 binding to FT-ATR (Fig. 2D). The ChIP assays revealed that H3 acetylation (H3Ac), which is a mark for active gene expression (32), was reduced ∼70% in the FT-ATR in the OE-AHL22 mutant compared with that in Col-0 plants (Fig. 5A). In contrast, H3 dimethylation at Lys-9 (H3K9me2), a repressive mark for gene expression (33), increased ∼2-fold in the mutant (Fig. 5B). These observations indicate that AHL22 modulates FT chromatin within the FT-ATR by modulating H3 acetylation and Lys-9 dimethylation. In contrast, H3 trimethylation at Lys-27 (H3K27me3), which is another repressive mark for gene expression (34), was not changed to a discernible level in the mutant (Fig. 5C), suggesting that H3K27me3 is not involved in the AHL22-mediated modifications of the FT chromatin.
FIGURE 5.
Modifications of FT chromatin by AHL22. Relative levels of H3 modifications in FT chromatin were examined by ChIP assays using an anti-H3Ac (A), -H3K9me2 (B), or -H3K27me3 (C) antibody. PCR primer pairs specific to M1, M2, and M3 sequences, as shown in Fig. 2A, were used. Plants grown on MS-agar plants for 12 days under LDs were used for chromatin preparations. Three measurements were averaged and statistically treated (t test, *, p < 0.01). Bars indicate mean ± S.E.
Interactions of AHL22 with HDACs
Histone deacetylases (HDACs) are a group of enzymes that remove acetyl groups from acetylated Lys residues of histone proteins (35). We found that H3 acetylation is reduced in the FT chromatin of the OE-AHL22 mutant. We therefore asked whether the AHL22-mediated modifications of FT chromatin are related to HDACs.
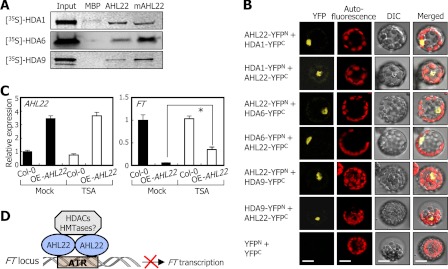
We first carried out in vitro pulldown assays using recombinant MBP-AHL22 fusion protein and in vitro translated HDAC polypeptides to examine whether AHL22 interacts with HDAC enzymes. It was found that AHL22 strongly interacted with HDA1/HDA19, HDA6, and HDA9 (Fig. 6A), which are involved in flowering timing and floral architecture (35). The three HDAC proteins did not bind to MBP alone, supporting the specific interaction between the HDAC enzymes and AHL22 protein.
FIGURE 6.
Interaction of AHL22 with HDACs. A, in vitro pulldown assays. [35S]-Labeled HDAC polypeptides were prepared by in vitro translation. Recombinant MBP-AHL22 and MBP-mAHL22 proteins prepared in E. coli cells were used. Input represents 5% of the HDAC protein used in each assay. B, BiFC assays in Arabidopsis protoplasts. The cYFP and nYFP fusions were cotransfected into Arabidopsis protoplasts and visualized by differential interference contrast (DIC) microscopy and fluorescence microscopy. Scale bar, 10 μm. C, effects of TSA on FT expression in OE-AHL22 mutant. Transcript levels were determined by qRT-PCR. Biological triplicates were averaged and statistically treated using a Student's t test (*, p < 0.01). Bars indicate mean ± S.E. D, schematic model of AHL22 function in flowering.
We also carried out BiFC assays to further examine the AHL22-HDAC interactions. The nYFP- and cYFP-coding sequences were fused in-frame to the 5′ and 3′ ends of the AHL22 and HDAC gene sequences, and the fusion constructs were coexpressed transiently in Arabidopsis protoplasts. Strong reconstituted YFP signals were detected in the nuclei of cells coexpressing the AHL22-nYFP and HDAC-cYFP fusions and the AHL22-cYFP and HDAC-nYFP fusions (Fig. 6B), confirming that AHL22 interacts with the HDAC enzymes in the nucleus.
Dynamic dimer formation regulates the binding specificity and affinity of transcription regulators to their target DNA or interacting partners (36). We therefore examined whether AHL22 forms homodimers by in vitro pulldown assays using recombinant MBP-AHL22 proteins and [35S]methionine-labeled AHL22 polypeptides. We found that the two AHL22 forms interact with each other (supplemental Fig. S7A). In addition, BiFC assays in Arabidopsis protoplasts showed that the two AHL22 forms interact with each other in vivo (supplemental Fig. S7B). Notably, the mAHL22 protein also interacts with AHL22 in both in vitro pulldown assays and BiFC assays (supplemental Fig. S7, A and B), indicating that the AT-hook motif is not required for the interactions. These observations support that the AHL22 proteins form multimers, probably homodimers.
We next examined whether HDAC activity is important for FT regulation by employing trichostatin A (TSA) that selectively inhibits class I and II mammalian HDAC enzymes (35). Arabidopsis plants were grown for 10 days on MS-agar plates containing 0.5 μm TSA, and FT transcript levels were examined. The FT transcript level was elevated at least 7-fold in the TSA-treated OE-AHL22 mutant (Fig. 6C). In contrast, the AHL22 transcription was not affected by TSA under identical conditions, indicating that HDACs participate in the AHL22-mediated suppression of FT transcription.
Altogether, our data demonstrate that the AHL22 protein suppresses FT transcription by binding to the FT-ATR and recruiting a subset of HDAC enzymes. A plausible working scenario is that the AHL22-HDAC complexes deacetylate acetylated histones in the FT chromatin (Fig. 6D). AHL22 also regulates H3 dimethylation at Lys-9, suggesting that histone methyltransferases are also involved in the AHL22-mediated modification of the FT chromatin.
DISCUSSION
The AHL proteins are characterized by having two conserved structural components: the AT-hook motif that binds to AT-rich stretches of DNA and the plant and prokaryotic conserved domain that mediates nuclear localization (22). Several AHL proteins have been functionally studied in diverse aspects of plant growth and developmental processes and stress responses in Arabidopsis. AGF1/AHL25 is critical for the negative feedback regulation of GA3 oxidase gene (16). SOB3/AHL29 and ESC/ORE7/AHL27 are known to regulate hypocotyl growth (18). It also acts as a negative regulator of leaf senescence (17). In addition, GIK/AHL21 plays a role in organ patterning and differentiation (20). Furthermore, it has been reported that AHL22 is involved in flowering induction and hypocotyl elongation (19). Meanwhile, overexpression of the AHL20 gene suppresses plant innate immune responses (21). AHL15, AHL19, and AHL27 have also been implicated in defense responses (21).
It is notable that AHL1 binds to MARs via the AT-hook motif (22). MARs are specific stretches of DNA sequences that are important for the structural organization of chromatin fibers by anchoring chromatin loops to nuclear matrix (10). A genome-scale study of gene expression patterns in conjunction with screening of potential intragenic MARs has shown that Arabidopsis genes possessing intragenic MARs tend to be less expressed irrespective of plant tissues and organs and differentially regulated throughout the plant growth stages (10, 11). It has been known that MARs link AHL proteins with chromatin modifications. For example, ESC/ORE7/AHL27 influences chromatin architecture by modulating the distribution of H2B (17). In addition, AHL21 represses the AUXIN RESPONSE FACTOR 3 (ARF3) gene by inducing H3 dimethylation at Lys-9 in the gene promoter during floral development (20).
In this work, we demonstrated that AHL22 suppresses FT expression by binding to the FT-ATR and recruiting a subset of HDAC enzymes, HDA1/HDA19, HDA6, and HDA9. The early-flowering phenotype of the 35S:gFT transgenic plants was compromised in the OE-AHL22 background (35S:gFT OE-AHL22). Consistent with the changes in flowering time, the FT transcript level was reduced in the 35S:gFT OE-AHL22 plants compared with that in the 35S:gFT transgenic plants. In contrast, the early-flowering phenotype of the 35S:cFT transgenic plants was reduced only slightly in the OE-AHL22 background (35S:cFT OE-AHL22), which is obviously because the FT cDNA does not have intact FT-ATR to which AHL22 binds.
More work is required to determine whether AHL22 is a bona fide MAR-binding factor and the FT-ATR is an intragenic MAR. The FT-ATR has been predicted as an intragenic MAR (10). A major group of MAR-binding factors possesses the AT-hook motif (13). AHL1 is associated with the nuclear matrix (22). It is therefore likely that AHL22 acts as a MAR-binding factor in Arabidopsis.
Our data strongly support that AHL22 regulates FT expression by modulating histone acetylation and methylation through physical interactions with HDACs and presumably methyltransferase enzymes. Whereas the level of H3Ac was reduced, that of H3K9me2 was elevated in the FT chromatin of the late-flowering OE-AHL22 mutant, whereas H3K27 trimethylation may not be involved in FT control (Fig. 5). The physiological significance of the AHL22-HDAC interactions in FT regulation and thus flowering time control is further supported by the effects of the HDAC inhibitor TSA on FT expression in the OE-AHL22 mutant. However, the role of the AHL22-HDAC interactions in flowering time control likely is more complicated than the proposed working model (Fig. 6D). AHL proteins are functionally redundant, and at least several AHL proteins are apparently involved in flowering time control (18, 19, 21), suggesting that different AHL proteins may interact with different HDAC enzymes. This view entails that various combinations of AHL-HDAC complexes would bind to the FT-ATR, depending on developmental and environmental signals.
We observed that LFY and AP1 genes, in addition to FT, are also suppressed in the OE-AHL22 mutant (supplemental Fig. S3A). In addition, AHL22 bound to the intragenic and intergenic ATR sequences in the AP1 and LFY loci, respectively. It is therefore likely that the AHL22-HDAC complexes also regulate the chromatin status of LFY and AP1 genes and other flowering time genes in addition to the FT gene. This signaling complexity may explain the relatively small changes of FT transcript levels and the timing of flowering initiation observed in the 35S:gFT OE-AHL22 plants compared with those in the 35S:gFT transgenic plants. Further work is necessary to determine how much the AHL22-HDAC regulation of FT chromatin contributes to the role of FT in flowering time control. In addition, the role of endogenous and environmental factors in regulating AHL22 activity should also be investigated.
Coordinated histone modifications mediate epigenetic regulation of gene expression in plants. One of the most extensively studied is epigenetic regulation of the floral repressor FLC. It has been known that epigenetic regulation of the FLC gene is mediated by complex networks of histone acetylation and methylation events. Activation of the FLC expression is achieved through several active chromatin modifications, such as acetylation of core histone tails, H3K4 methylation, and H3K36 dimethylation and trimethylation (6). In contrast, repressive histone modifications, including histone deacetylation, H3K4 demethylation, H3K9 and H3K27 methylation, and histone arginine methylation, repress the FLC expression (6).
Histone modifications in FT chromatin have recently been studied. Whereas H3K4 trimethylation in the FT chromatin is associated with FT activation, H3K27 trimethylation is associated with FT repression. It has been found that the H3K27 methyltransferase CURLY LEAF (CLF) represses FT expression (37). In addition, the chromodomain-containing LIKE HETEROCHROMATIN PROTEIN 1 (LHP1) protein binds to H3K27me3 in the FT chromatin and maintains the repressive state of FT expression (38). However, there is little known about the role of histone acetylation/deacetylation in the FT chromatin. We found that AHL22 binds to an AT-rich DNA sequence in the FT locus and reduces H3 acetylation. We also found that H3 dimethylation at Lys-9 is elevated in the FT chromatin of the OE-AHL22 mutant. It seems that the FT chromatin is regulated through coordinated actions of histone acetylation and methylation, although not precisely in the way that epigenetic modifications control the FLC gene.
HDAC enzymes play a role in global gene repression during developmental processes and stress adaptation responses in plants (35). AHL22 physically interacts with HDA1/HDA19, HDA6, and HDA9, which are homologous to yeast RPD3 (reduced potassium deficiency 3) and belong to the type I HDAC subfamily (35). In Saccharomyces pombe, mutations in the subunits of class I HDAC complexes affect H3K9 methylation (39), indicating that Lys-9 deacetylation is a prerequisite for subsequent H3 methylation. We found that H3K9 dimethylation was elevated in the FT chromatin of the OE-AHL22 mutant. Based on the previous and our own data, we believe that AHL22 regulates the FT chromatin in a similar manner to the yeast RPD3: H3 deacetylation by HDAC enzymes may precede H3K9 dimethylation to suppress FT expression. In this view, it is envisaged that AHL22 may also interact with histone methyltransferases (Fig. 6D).
Supplementary Material
Acknowledgment
We thank the Arabidopsis Biological Resource Center for the SALK T-DNA lines.
This work was supported by Leaping Research Program Grant 20110016440 provided by the National Research Foundation of Korea, the Next-Generation BioGreen 21 program (Plant Molecular Breeding Center No. PJ008103) provided by the Rural Development Administration, and Plant Signaling Network Research Center Grant 20110001099, National Research Foundation of Korea Grant 20110027355, the Agricultural R & D Promotion Center Grant 309017-03, and the Korea Ministry for Food, Agriculture, Forestry and Fisheries.

This article contains supplemental Figs. S1–S7 and Table S1.
- FT
- FLOWERING LOCUS T
- AHL22
- AT-hook motif nuclear localized 22
- ATR
- AT-rich sequence
- BiFC
- bimolecular fluorescence complementation
- ESC
- ESCAROLA
- FLC
- FLOWERING LOCUS C
- FRI
- FRIGIDA
- GUS
- β-glucuronidase
- H3
- histone 3
- H3Ac
- H3 acetylation
- H3K9me2
- H3 dimethylation at Lys-9
- H3K27me3
- H3 trimethylation at Lys-27
- HDAC
- histone deacetylase
- LFY
- LEAFY
- MAR
- matrix attachment region
- MBP
- maltose-binding protein
- MS
- Murashige & Skoog
- qRT-PCR
- quantitative real-time RT-PCR
- SOC1
- SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1
- TSA
- trichostatin A
- YFP
- yellow fluorescent protein
- BS
- binding sequence
- LD
- long day
- CaMV
- cauliflower mosaic virus
- gFT
- genomic FT
- cFT
- cDNA FT
- RLN
- rosette leaf number.
REFERENCES
- 1. Amasino R. (2010) Seasonal and developmental timing of flowering. Plant J. 61, 1001–1013 [DOI] [PubMed] [Google Scholar]
- 2. Simpson G. G., Dean C. (2002) Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289 [DOI] [PubMed] [Google Scholar]
- 3. Blázquez M. A., Ahn J. H., Weigel D. (2003) A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat. Genet. 33, 168–171 [DOI] [PubMed] [Google Scholar]
- 4. Simpson G. G. (2004) The autonomous pathway. Epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr. Opin. Plant Biol. 7, 570–574 [DOI] [PubMed] [Google Scholar]
- 5. Jones-Rhoades M. W., Bartel D. P., Bartel B. (2006) MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 57, 19–53 [DOI] [PubMed] [Google Scholar]
- 6. He Y. (2009) Control of the transition to flowering by chromatin modifications. Mol. Plant 2, 554–564 [DOI] [PubMed] [Google Scholar]
- 7. Yaish M. W., Colasanti J., Rothstein S. J. (2011) The role of epigenetic processes in controlling flowering time in plants exposed to stress. J. Exp. Bot. 62, 3727–3735 [DOI] [PubMed] [Google Scholar]
- 8. Liu F., Marquardt S., Lister C., Swiezewski S., Dean C. (2010) Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327, 94–97 [DOI] [PubMed] [Google Scholar]
- 9. Pederson T. (2000) Half a century of “the nuclear matrix.” Mol. Biol. Cell 11, 799–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rudd S., Frisch M., Grote K., Meyers B. C., Mayer K., Werner T. (2004) Genome-wide in silico mapping of scaffold/matrix attachment regions in Arabidopsis suggests correlation of intragenic scaffold/matrix attachment regions with gene expression. Plant Physiol. 135, 715–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tetko I. V., Haberer G., Rudd S., Meyers B., Mewes H. W., Mayer K. F. (2006) Spatiotemporal expression control correlates with intragenic scaffold matrix attachment regions (S/MARs) in Arabidopsis thaliana. PLoS Comput. Biol. 2, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang T. Y., Han Z. M., Chai Y. R., Zhang J. H. (2010) A minireview of MAR-binding proteins. Mol. Biol. Rep. 37, 3553–3560 [DOI] [PubMed] [Google Scholar]
- 13. Aravind L., Landsman D. (1998) AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 26, 4413–4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reeves R. (2010) Nuclear functions of the HMG proteins. Biochim. Biophys. Acta 1799, 3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weigel D., Ahn J. H., Blázquez M. A., Borevitz J. O., Christensen S. K., Fankhauser C., Ferrándiz C., Kardailsky I., Malancharuvil E. J., Neff M. M., Nguyen J. T., Sato S., Wang Z. Y., Xia Y., Dixon R. A., Harrison M. J., Lamb C. J., Yanofsky M. F., Chory J. (2000) Activation tagging in Arabidopsis. Plant Physiol. 122, 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matsushita A., Furumoto T., Ishida S., Takahashi Y. (2007) AGF1, an AT-hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase. Plant Physiol. 143, 1152–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lim P. O., Kim Y., Breeze E., Koo J. C., Woo H. R., Ryu J. S. (2007) Overexpression of a chromatin architecture-controlling AT-hook protein extends leaf longevity and increases the post-harvest storage life of plants. Plant J. 52, 1140–1153 [DOI] [PubMed] [Google Scholar]
- 18. Street I. H., Shah P. K., Smith A. M., Avery N., Neff M. M. (2008) The AT-hook-containing proteins SOB3/AHL29 and ESC/AHL27 are negative modulators of hypocotyl growth in Arabidopsis. Plant J. 54, 1–14 [DOI] [PubMed] [Google Scholar]
- 19. Xiao C., Chen F., Yu X., Lin C., Fu Y. F. (2009) Overexpression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol. Biol. 71, 39–50 [DOI] [PubMed] [Google Scholar]
- 20. Ng K. H., Yu H., Ito T. (2009) AGAMOUS controls GIANT KILLER, a multifunctional chromatin modifier in reproductive organ patterning and differentiation. PLoS Biol. 7, e1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lu H., Zou Y., Feng N. (2010) Overexpression of AHL20 negatively regulates defenses in Arabidopsis. J. Integr. Plant Biol. 52, 801–808 [DOI] [PubMed] [Google Scholar]
- 22. Fujimoto S., Matsunaga S., Yonemura M., Uchiyama S., Azuma T., Fukui K. (2004) Identification of a novel plant MAR DNA-binding protein localized on chromosomal surfaces. Plant Mol. Biol. 56, 225–239 [DOI] [PubMed] [Google Scholar]
- 23. Kim Y. S., Kim S. G., Park J. E., Park H. Y., Lim M. H., Chua N. H., Park C. M. (2006) A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18, 3132–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Parviz F., Hall D. D., Markwardt D. D., Heideman W. (1998) Transcriptional regulation of CLN3 expression by glucose in Saccharomyces cerevisiae. J. Bacteriol. 180, 4508–4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang S. D., Seo P. J., Yoon H. K., Park C. M. (2011) The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23, 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yoo S. D., Cho Y. H., Sheen J. (2007) Arabidopsis mesophyll protoplasts. A versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 [DOI] [PubMed] [Google Scholar]
- 27. Hong S. Y., Kim O. K., Kim S. G., Yang M. S., Park C. M. (2011) Nuclear import and DNA binding of the ZHD5 transcription factor is modulated by a competitive peptide inhibitor in Arabidopsis. J. Biol. Chem. 286, 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Takada S., Goto K. (2003) Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell 15, 2856–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schena M., Davis R. W. (1992) HD-Zip proteins, members of an Arabidopsis homeodomain protein superfamily. Proc. Natl. Acad. Sci. U.S.A. 89, 3894–3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sessa G., Morelli G., Ruberti I. (1993) The Athb-1 and -2 HD-Zip domains homodimerize forming complexes of different DNA binding specificities. EMBO J. 12, 3507–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Castillejo C., Pelaz S. (2008) The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 18, 1338–1343 [DOI] [PubMed] [Google Scholar]
- 32. Santos-Rosa H., Schneider R., Bannister A. J., Sherriff J., Bernstein B. E., Emre N. C., Schreiber S. L., Mellor J., Kouzarides T. (2002) Active genes are trimethylated at K4 of histone H3. Nature 419, 407–411 [DOI] [PubMed] [Google Scholar]
- 33. Litt M. D., Simpson M., Gaszner M., Allis C. D., Felsenfeld G. (2001) Correlation between histone lysine methylation and developmental changes at the chicken β-globin locus. Science 293, 2453–2455 [DOI] [PubMed] [Google Scholar]
- 34. Kirmizis A., Bartley S. M., Kuzmichev A., Margueron R., Reinberg D., Green R., Farnham P. J. (2004) Silencing of human polycomb target genes is associated with methylation of histone H3K27. Genes Dev. 18, 1592–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hollender C., Liu Z. (2008) Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 50, 875–885 [DOI] [PubMed] [Google Scholar]
- 36. Vinson C., Acharya A., Taparowsky E. J. (2006) Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochim. Biophys. Acta 1759, 4–12 [DOI] [PubMed] [Google Scholar]
- 37. Jiang D., Wang Y., Wang Y., He Y. (2008) Repression of flowering locus C and flowering locus T by the Arabidopsis polycomb repressive complex 2 components. PLoS One 3, e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Turck F., Roudier F., Farrona S., Martin-Magniette M. L., Guillaume E., Buisine N., Gagnot S., Martienssen R. A., Coupland G., Colot V. (2007) Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 3, e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Silverstein R. A., Richardson W., Levin H., Allshire R., Ekwall K. (2003) A new role for the transcriptional corepressor SIN3. Regulation of centromeres. Curr. Biol. 13, 68–72 [DOI] [PubMed] [Google Scholar]
- 40. Metcalf C. E., Wassarman D. A. (2006) DNA binding properties of TAF1 isoforms with two AT-hooks. J. Biol. Chem. 281, 30015–30023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.