Abstract
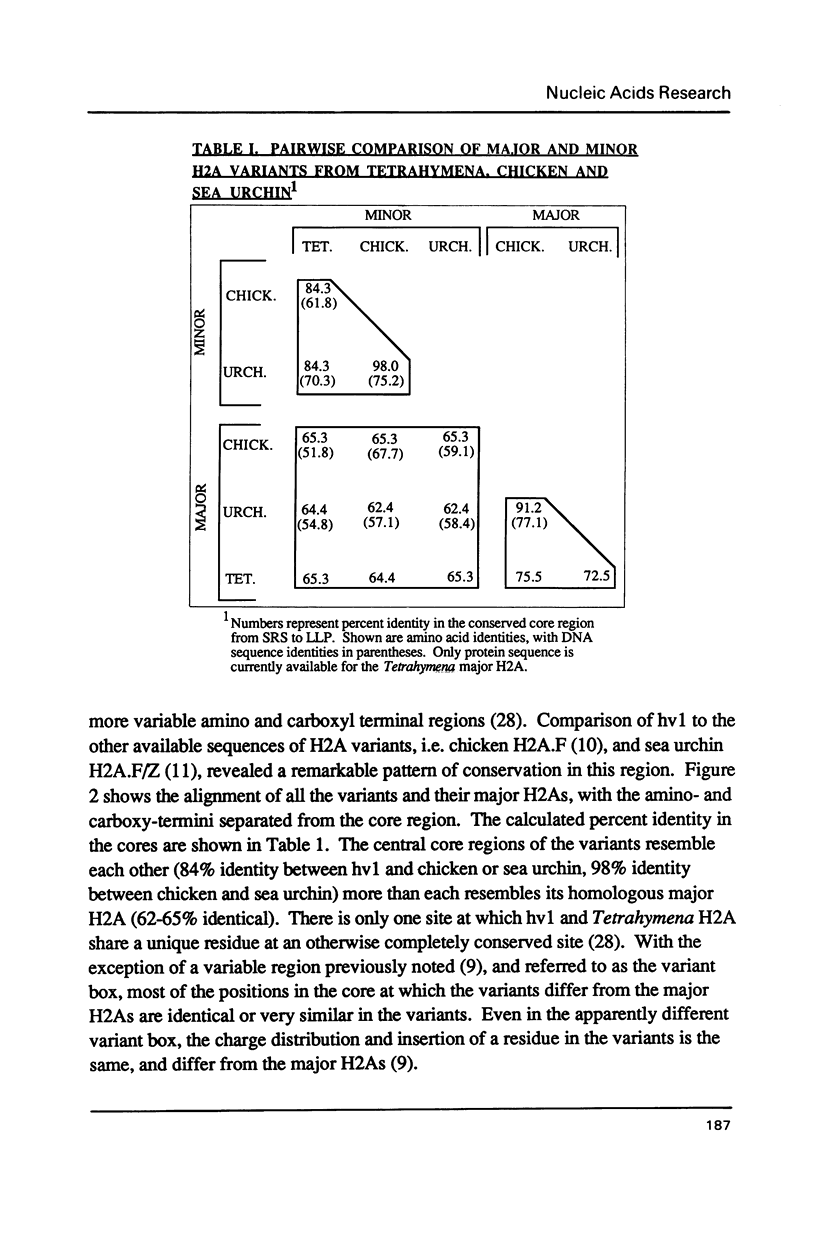
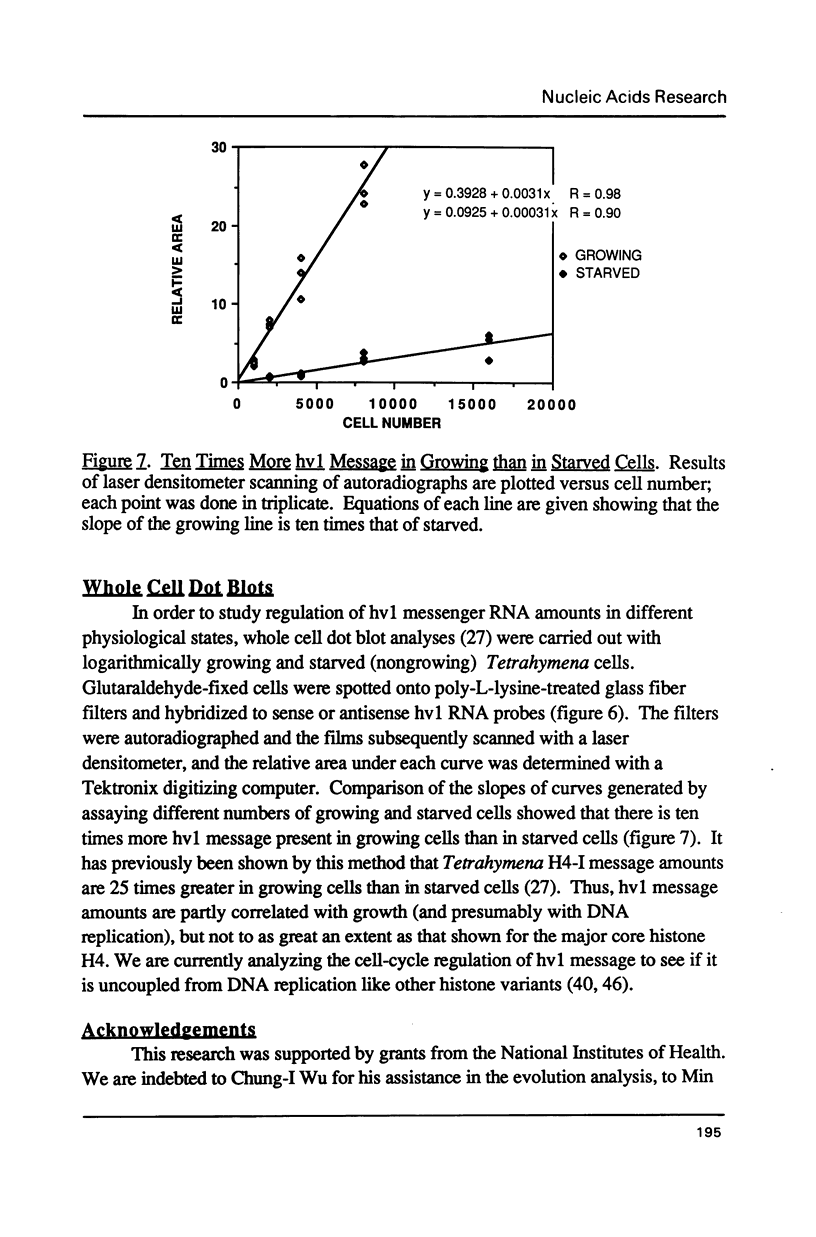
hv1 is a histone H2A variant found in the transcriptionally active Tetrahymena macronucleus, but not in the transcriptionally inert micronucleus. hv1 also contains antigenic determinants conserved in the histone complements of representatives of all four eukaryotic kingdoms. A cDNA clone encoding hv1 has been isolated and sequenced. Comparison of the derived protein sequence of hv1 with that of the chicken variant H2A.F and the sea urchin variant H2A.F/Z reveals remarkable homology in all but the extreme amino- and carboxy-termini and a small region in the conserved core. Putative regions of conserved antigenicity are discussed. Evidence is presented that suggests that hv1 is a single-copy, intron-containing gene that encodes a polyadenylated message. Unusual features in the 3' flanking sequence and in codon usage are also described. Evidence is also presented showing that hv1 message amounts are ten-fold greater in growing cells than in starved cells.
Full text
PDF

















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allis C. D., Glover C. V., Bowen J. K., Gorovsky M. A. Histone variants specific to the transcriptionally active, amitotically dividing macronucleus of the unicellular eucaryote, Tetrahymena thermophila. Cell. 1980 Jul;20(3):609–617. doi: 10.1016/0092-8674(80)90307-4. [DOI] [PubMed] [Google Scholar]
- Allis C. D., Richman R., Gorovsky M. A., Ziegler Y. S., Touchstone B., Bradley W. A., Cook R. G. hv1 is an evolutionarily conserved H2A variant that is preferentially associated with active genes. J Biol Chem. 1986 Feb 5;261(4):1941–1948. [PubMed] [Google Scholar]
- Allis C. D., Ziegler Y. S., Gorovsky M. A., Olmsted J. B. A conserved histone variant enriched in nucleoli of mammalian cells. Cell. 1982 Nov;31(1):131–136. doi: 10.1016/0092-8674(82)90412-3. [DOI] [PubMed] [Google Scholar]
- Ball D. J., Slaughter C. A., Hensley P., Garrard W. T. Amino acid sequence of the N-terminal domain of calf thymus histone H2A.Z. FEBS Lett. 1983 Apr 5;154(1):166–170. doi: 10.1016/0014-5793(83)80896-5. [DOI] [PubMed] [Google Scholar]
- Bannon G. A., Bowen J. K., Yao M. C., Gorovsky M. A. Tetrahymena H4 genes: structure, evolution and organization in macro- and micronuclei. Nucleic Acids Res. 1984 Feb 24;12(4):1961–1975. doi: 10.1093/nar/12.4.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannon G. A., Calzone F. J., Bowen J. K., Allis C. D., Gorovsky M. A. Multiple, independently regulated, polyadenylated messages for histone H3 and H4 in Tetrahymena. Nucleic Acids Res. 1983 Jun 25;11(12):3903–3917. doi: 10.1093/nar/11.12.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar Y. M., McCullar M. K., Chronister R. B. Immunocytochemical localization of a histone H2A variant in the mammalian nucleolar chromatin. Cell Biol Int Rep. 1984 Nov;8(11):971–979. doi: 10.1016/0309-1651(84)90195-4. [DOI] [PubMed] [Google Scholar]
- Blackburn E. H., Karrer K. M. Genomic reorganization in ciliated protozoans. Annu Rev Genet. 1986;20:501–521. doi: 10.1146/annurev.ge.20.120186.002441. [DOI] [PubMed] [Google Scholar]
- Brown D. T., Wellman S. E., Sittman D. B. Changes in the levels of three different classes of histone mRNA during murine erythroleukemia cell differentiation. Mol Cell Biol. 1985 Nov;5(11):2879–2886. doi: 10.1128/mcb.5.11.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush D., Dodgson J. B., Choi O. R., Stevens P. W., Engel J. D. Replacement variant histone genes contain intervening sequences. Mol Cell Biol. 1985 Jun;5(6):1307–1317. doi: 10.1128/mcb.5.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- D'Andrea R., Harvey R., Wells J. R. Vertebrate histone genes: nucleotide sequence of a chicken H2A gene and regulatory flanking sequences. Nucleic Acids Res. 1981 Jul 10;9(13):3119–3128. doi: 10.1093/nar/9.13.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst S. G., Miller H., Brenner C. A., Nocente-McGrath C., Francis S., McIsaac R. Characterization of a cDNA clone coding for a sea urchin histone H2A variant related to the H2A.F/Z histone protein in vertebrates. Nucleic Acids Res. 1987 Jun 11;15(11):4629–4644. doi: 10.1093/nar/15.11.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrner K., Yarger J., Hereford L. Yeast histone mRNA is polyadenylated. Nucleic Acids Res. 1980 Dec 11;8(23):5725–5737. doi: 10.1093/nar/8.23.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fusauchi Y., Iwai K. Tetrahymena histone H2A. Isolation and two variant sequences. J Biochem. 1983 Jun;93(6):1487–1497. doi: 10.1093/oxfordjournals.jbchem.a134286. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A., Yao M. C., Keevert J. B., Pleger G. L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9(0):311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Harvey R. P., Whiting J. A., Coles L. S., Krieg P. A., Wells J. R. H2A.F: an extremely variant histone H2A sequence expressed in the chicken embryo. Proc Natl Acad Sci U S A. 1983 May;80(10):2819–2823. doi: 10.1073/pnas.80.10.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H., Nomoto M., Iwai K. Tetrahymena histone H4. Complete amino acid sequences of two variants. J Biochem. 1984 Nov;96(5):1449–1456. [PubMed] [Google Scholar]
- Helms C., Graham M. Y., Dutchik J. E., Olson M. V. A new method for purifying lambda DNA from phage lysates. DNA. 1985 Feb;4(1):39–49. doi: 10.1089/dna.1985.4.39. [DOI] [PubMed] [Google Scholar]
- Hentschel C. C., Birnstiel M. L. The organization and expression of histone gene families. Cell. 1981 Aug;25(2):301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- Horowitz S., Bowen J. K., Bannon G. A., Gorovsky M. A. Unusual features of transcribed and translated regions of the histone H4 gene family of Tetrahymena thermophila. Nucleic Acids Res. 1987 Jan 12;15(1):141–160. doi: 10.1093/nar/15.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard E. A., Blackburn E. H. Reproducible and variable genomic rearrangements occur in the developing somatic nucleus of the ciliate Tetrahymena thermophila. Mol Cell Biol. 1985 Aug;5(8):2039–2050. doi: 10.1128/mcb.5.8.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985 Jan;2(1):13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- Kolodrubetz D., Rykowski M. C., Grunstein M. Histone H2A subtypes associate interchangeably in vivo with histone H2B subtypes. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7814–7818. doi: 10.1073/pnas.79.24.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel B. E., Roth S. Y., Cook R. G., Allis C. D., Davie J. R. Changes in the histone H2A variant H2A.Z and polyubiquitinated histone species in developing trout testis. Biochemistry. 1987 Jul 14;26(14):4417–4421. doi: 10.1021/bi00388a034. [DOI] [PubMed] [Google Scholar]
- Nomoto M., Imai N., Saiga H., Matsui T., Mita T. Characterization of two types of histone H2B genes from macronuclei of Tetrahymena thermophila. Nucleic Acids Res. 1987 Jul 24;15(14):5681–5697. doi: 10.1093/nar/15.14.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okayama H., Berg P. High-efficiency cloning of full-length cDNA. Mol Cell Biol. 1982 Feb;2(2):161–170. doi: 10.1128/mcb.2.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Old R. W., Woodland H. R. Histone genes: not so simple after all. Cell. 1984 Oct;38(3):624–626. doi: 10.1016/0092-8674(84)90256-3. [DOI] [PubMed] [Google Scholar]
- Pederson D. S., Shupe K., Gorovsky M. A. Changes in chromatin structure accompany modulation of the rate of transcription of 5S ribosomal genes in Tetrahymena. Nucleic Acids Res. 1984 Nov 26;12(22):8489–8507. doi: 10.1093/nar/12.22.8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pustell J., Kafatos F. C. A convenient and adaptable package of computer programs for DNA and protein sequence management, analysis and homology determination. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):643–655. doi: 10.1093/nar/12.1part2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rykowski M. C., Wallis J. W., Choe J., Grunstein M. Histone H2B subtypes are dispensable during the yeast cell cycle. Cell. 1981 Aug;25(2):477–487. doi: 10.1016/0092-8674(81)90066-0. [DOI] [PubMed] [Google Scholar]
- Sogin M. L., Elwood H. J., Gunderson J. H. Evolutionary diversity of eukaryotic small-subunit rRNA genes. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1383–1387. doi: 10.1073/pnas.83.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sures I., Lowry J., Kedes L. H. The DNA sequence of sea urchin (S. purpuratus) H2A, H2B and H3 histone coding and spacer regions. Cell. 1978 Nov;15(3):1033–1044. doi: 10.1016/0092-8674(78)90287-8. [DOI] [PubMed] [Google Scholar]
- Wenkert D., Allis C. D. Timing of the appearance of macronuclear-specific histone variant hv1 and gene expression in developing new macronuclei of Tetrahymena thermophila. J Cell Biol. 1984 Jun;98(6):2107–2117. doi: 10.1083/jcb.98.6.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudt L. P., Pastink A., Kempers-Veenstra A. E., Jansen A. E., Mager W. H., Planta R. J. The genes coding for histone H3 and H4 in Neurospora crassa are unique and contain intervening sequences. Nucleic Acids Res. 1983 Aug 25;11(16):5347–5360. doi: 10.1093/nar/11.16.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Allis C. D., Richman R., Cook R. G., Gorovsky M. A. An intervening sequence in an unusual histone H1 gene of Tetrahymena thermophila. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8674–8678. doi: 10.1073/pnas.83.22.8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R. S., Bonner W. M. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell. 1981 Dec;27(2 Pt 1):321–330. doi: 10.1016/0092-8674(81)90415-3. [DOI] [PubMed] [Google Scholar]
- Wu R. S., Panusz H. T., Hatch C. L., Bonner W. M. Histones and their modifications. CRC Crit Rev Biochem. 1986;20(2):201–263. doi: 10.3109/10409238609083735. [DOI] [PubMed] [Google Scholar]
- Yu S. M., Gorovsky M. A. In situ dot blots: quantitation of mRNA in intact cells. Nucleic Acids Res. 1986 Oct 10;14(19):7597–7615. doi: 10.1093/nar/14.19.7597. [DOI] [PMC free article] [PubMed] [Google Scholar]






