Abstract
Three genes encoding anthocyanidin reductase (ANR) in apple (Malus×domestica Borkh.), designated MdANR1, MdANR2a, and MdANR2b, have been cloned and characterized. MdANR1 shows 91% identity in coding DNA sequences with MdANR2a and MdANR2b, while MdANR2a and MdANR2b are allelic and share 99% nucleotide sequence identity in the coding region. MdANR1 and MdANR2 genes are located on linkage groups 10 and 5, respectively. Expression levels of both MdANR1 and MdANR2 genes are generally higher in yellow-skinned cv. Golden Delicious than in red-skinned cv. Red Delicious. Transcript accumulation of MdANR1 and MdANR2 genes in fruits gradually decreased throughout fruit development. Ectopic expression of apple MdANR genes in tobacco positively and negatively regulates the biosynthesis of proanthocyanidins (PAs) and anthocyanin, respectively, resulting in white, pale pink-coloured, and white/red variegated flowers. The accumulation of anthocyanin is significantly reduced in all tobacco transgenic flowers, while catechin and epicatechin contents in transgenic flowers are significantly higher than those in flowers of wild-type plants. The inhibition of anthocyanin synthesis in tobacco transgenic flowers overexpressing MdANR genes is probably attributed to down-regulation of CHALCONE ISOMERASE (CHI) and DIHYDROFLAVONOL-4-REDUCTASE (DFR) genes involved in the anthocyanin pathway. Interestingly, several transgenic lines show no detectable transcripts of the gene encoding leucoanthocyanidin reductase (LAR) in flowers, but accumulate higher levels of catechin in flowers of transgenic plants than those of wild-type plants. This finding suggests that the ANR gene may be capable of generating catechin via an alternative route, although this mechanism is yet to be further elucidated.
Keywords: Anthocyanin, anthocyanidin reductase, flavonoid, Malus, proanthocyanidin
Introduction
Proanthocyanidins (PAs), also known as condensed tannins, are phenolic polymers of condensed flavan-3-ols and are among the major flavonoid compounds found in higher plants (Winkel-Shirley, 2001). PAs are powerful antioxidants, and thus provide multiple health benefits to humans, including anti-inflammatory effects, immunity enhancement, as well as lowering risks of cardiovascular diseases and certain cancers (Santos-Buelga and Scalbert, 2000). PAs can also protect ruminants against pasture bloat disease and enhance ruminant nutrition (McMahon et al., 2000). Moreover, PAs can interact with proteins, particularly saliva proteins such as α-amylase, resulting in the astringent and bitter sensations in many fruits and fruit juices (Vidal et al., 2003; Obreque-Slier et al., 2010; Renard et al., 2011). Therefore, it is important to further our understanding of PA biosynthesis, and to engineer PAs in fruit and forage crops to improve their nutritional and health protective values.
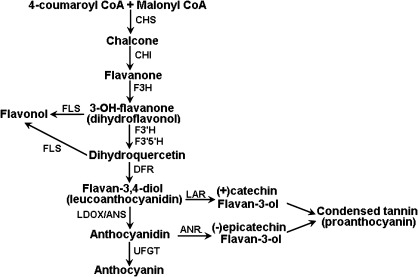
The biosynthesis pathway of PAs is a branch of the anthocyanin biosynthesis pathway, and begins with the synthesis of flavan-3-ol units, such as catechin and epicatechin (Fig. 1). Catechin is derived from leucocyanidin by leucoanthocyanidin reductase (LAR), while epicatechins are synthesized from cyanidin by anthocyanidin reductase (ANR). The functionality of LAR has been characterized in Desmodium uncinatum (Tanner et al., 2003) and grapevine (Bogs et al., 2005). ANR has been initially identified in Arabidopsis, and it is encoded by the BANYULS (BAN) gene (Xie et al., 2003). ANR utilizes cyanidin as a substrate, rather than leucocyanidin, which is consistent with the fact that leucoanthocyanidin dioxygenase (LDOX) is essential for PA synthesis in Arabidopsis (Abrahams et al., 2003). Ectopic expression of BAN in tobacco flower petals and Arabidopsis leaves results in loss of anthocyanins and accumulation of condensed tannins, suggesting that there is an interaction between anthocyanidin and PA pathways (Xie et al., 2003). However, there are no reports on how overexpression of either BAN or other PA pathway genes influence expression of genes involved in the anthocyanin pathway.
Fig. 1.
A general schematic diagram of the flavonoid biosynthetic pathway. CHI, chalcone isomerase; CHS, chalcone synthase; F3H, flavonoid 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; FLS, flavonol synthase; DFR or BAN, dihydroflavonol reductase; LAR, leucoanthocyanidin reductase; ANS, anthocyanidin synthase; ANR, anthocyanidin reductase; UFGT, glucose transferase.
Besides the two structural genes BAN and ANR, MYB-regulatory genes have also been identified as being involved in PA biosynthesis. For example, expression of BAN in Arabidopsis seed coat is co-regulated by at least three MYB transcription factorss, TT2, TT8, and TTG1 (Nesi et al., 2001; Baudry et al., 2004). In persimmon fruits, expression levels of DkMYB4 are positively correlated with accumulation of DkANR transcripts, but have little influence on accumulation of DkLAR transcripts (Akagi et al., 2009). In contrast, grapevine VvMYBPA1 can activate expression of both VvANR and VvLAR genes in grape berries and leaves (Bogs et al., 2007).
Apple (Malus×domestica Borkh.) is one of the most widely cultivated fruit crops grown around the world. Since red coloration is an important contributor to apple fruit quality, the molecular mechanism underlying apple red coloration has recently been extensively studied. For example, red coloration of the flesh of apple fruit is due to increased MdMYB10 transcripts and subsequent accumulation of anthocyanins throughout the plant (Espley et al., 2007). Two alleles of MdMYB10, MdMYBA and MdMYB1, play important roles in red coloration of the skin of apple fruit (Takos et al., 2006; Ban et al., 2007). In addition, apple fruits are also well known to be rich in PAs (Souquet, 1996). Thus, apple is an important edible source of PAs with known nutrient and health benefits for humans (Hammerstone et al., 2000). However, few studies have been reported on genes involved in the PA pathway in apple. In fact, the PA pathway is in competition with anthocyanin synthesis, especially via the ANR gene as it utilizes the same substrate, anthocyanidin. Moreover, functional characterization of ANR and LAR genes has so far only been reported in Medicago truncatula (Tanner et al., 2003), Arabidopsis (Xie et al., 2003), and Vitis vinifera (Bogs et al., 2005). Many questions related to the mechanism of how plants synthesize PAs still remain unanswered. Therefore, investigation of MdANR genes in apple will not only provide valuable insights into the molecular mechanism of PA accumulation in plants, but will also aid in our understanding of coloration of fruit.
In this study, isolation, genetic mapping, and functional characterization of the gene family encoding ANR in apple is reported. Expression profiles of apple ANR genes in fruit were also investigated at different stages of development. Moreover, the functionality of different members of the apple ANR gene family is characterized via ectopic expression in tobacco. The results demonstrate that ectopic expression of apple ANR genes in tobacco flowers inhibits expression of both CHALCONE ISOMERASE (CHI) and DIHYDROFLAVONOL-4-REDUCTASE (DFR) genes, resulting in loss of anthocyanin. These findings will aid in future attempts to manipulate flavonoid biosynthesis in apple as well as in other plants.
Materials and methods
Plant material
Apple leaves, flowers, and fruits at different developmental stages were collected, and whole fruits were used for gene expression studies. Wild-type and T2 transgenic plants of tobacco (Nicotiana tabacum cv. Petite Havana SR1) were grown in a greenhouse, and flowers at the full-bloom stage were harvested for the analysis of gene expression and flavonoid compounds. All the samples were frozen in liquid nitrogen upon collection, and stored at –80 °C until needed.
Isolation of genes encoding ANR in apple
A full-length cDNA of MdANR (GenBank accession no. DQ099803) was obtained from an apple expressed sequence tag (EST) database (http://titan.biotec.uiuc.edu/apple/), and a pair of primers (5′-CACGACCAAACCTGTTCCTT-3′/5′-GTTGCAACCCCTGTC AACTT-3′) was designed to screen an apple bacterial artificial chromosome (BAC) library (cv. GoldRush) according to a previously described PCR-based screening protocol (Xu et al., 2001). The PCR program consisted of 34 cycles of 30 s at 94 °C, 30 s at 58 °C, 60 s at 72 °C, and a final extension for 5 min at 72 C. Positive BAC clones were then subjected to Southern blot analysis, and three clones representing different gene copies were selected and subjected to subcloning. BAC DNA was extracted from a 300 ml culture using the Plasmid Midi kit (QIAGEN, Valencia, CA, USA), and the subcloning of BAC DNA was carried out according to a previously reported protocol (Han et al., 2007). The primer walking strategy was then used to sequence positive subclones to recover full genomic DNA sequences encoding ANR in apple.
Southern blot hybridization of BAC and genomic DNA
Genomic DNA (30 μg) from leaves of cv. GoldRush and 25 ng of BAC DNA from each positive clone were digested with BamHI, separated on a 1.0% agarose gel, and then transferred onto nylon membranes (Amersham Biosciences, Pittsburgh, PA, USA) using the capillary transfer method. A pair of primers (5′-TTCTCTTATGGCCGGTCCTT-3′/5′-CGTCCTCCACATGTGCAATG-3′) was designed to DNA probes using cDNA from leaves of cv. GoldRush as a template. Hybridization was carried out using the DIG Easy Hyb kit (Roche, Indianapolis, IN, USA) according to the manufacturer's instructions. Blots were exposed to a Lumi-Film X-ray film (Hyperfilm™, Amersham) at room temperature for 25 min.
Mapping of MdANR genes onto the integrated apple genetic and physical map
Two markers were developed based on insertion/deletion (InDel) polymorphism to tag MdANR1 and MdANR2 genes, and their primer sequences were 5′-CTGCCGCATGATGATCTTCAC-3′/5′-GAATCTCTTCGTGGACCACTCC-3′ and 5′-ATCTGCTTCTGGTCGGTACA-3′/5′-TTGGCCTTGAAGTACTCCAC-3′, respectively. The two InDel markers were used for screening an F1 population derived from a cross between ‘Co-op 16’ and ‘Co-op 17’. PCR products were separated on 2% (w/v) agarose gels. The linkage analysis was conducted using JoinMap version 4.0 according to a previously reported strategy (Han et al., 2011).
Quantitative real-time PCR (qRT-PCR) analysis
Total RNA from leaf and flower tissues were extracted using an RNAqueous Kit (Ambion, Austin, TX, USA) according to the manufacturer's instructions. Total RNA from apple fruits was isolated according to the protocol described by Gasic et al. (2004). A total of 3 μg of RNA was treated with DNase I (Invitrogen) to remove contaminating DNA, and then subjected to cDNA synthesis using the SuperScript III RT kit (Invitrogen) according to the manufacturer's instructions.
The SYBR Green real-time PCR assay was carried out in a total volume of 15 μl, containing 12.5 μl of 2× SYBR Green I Master Mix (Applied Biosystems), 0.2 μM (each) of specific primers, and 100 ng of template cDNA. The amplification program consisted of one cycle of 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. The fluorescent product was detected at the last step of each cycle. Following amplification, melting temperatures of PCR products were analysed to determine the specificity of the PCR product. Melting curves were obtained by slow heating at 0.5 °C s−1, from 60 °C to 90 °C while continuously monitoring the fluorescence signal. A negative control without a cDNA template was run with each analysis to normalize the data, and evaluate the overall specificity. The qRT-PCRs were carried out in 96-well plates in a 7300 Real Time PCR System (Applied Biosystems). All samples were run in triplicate. An apple actin gene was used as an endogenous control gene. Differences in cycle threshold (Ct) between target and actin genes were used to determine the relative transcript level of the target gene, and calculated as 2 exp–(Cttarget–Ctactin). Real-time PCR primer sequences are listed on Supplementary Table S1 available at JXB online.
Expression vector construction and tobacco transformation
Two pairs of primers, 5′-TGACTCTAGAATGGCCACCCAACAACCCATCT-3′/5′-ATACGAGCTCCTAGTTCTGCAGCAGC CCCTTT-3′ and 5′-TGACTCTAGAATGGCCACCCAACAAC CCA TCT-3′/5′-ATACGAGCTCTTTTATTTTCTTTTTTCTCAT C-3′, were designed to amplify the whole coding sequences of MdANR1 and MdANR2, respectively, using cDNA from leaves of cv. GoldRush as templates. The forward and reverse primers contained XhoI/SacI sites at the 5' end, respectively. The PCR amplification was conducted using proofreading DNA polymerase Platinum® Pfx (Invitrogen), and PCR products, digested with XhoI and SacI, were ligated into XhoI/SacI-digested pBI121 binary vector. The binary vector containing each of the MdANR genes was sequenced. As a result, three constructs carrying coding regions of MdANR1, MdANR2a, and MdANR2b, respectively, were generated.
Each construct was immobilized into Agrobacterium tumefaciens strain GV3101, and co-cultivated with tobacco leaf sections following a previously reported protocol (Horsch et al., 1985), with some modification. Briefly, leaf sections (∼0.5 cm2) of 10-day-old seedlings were incubated with Agrobacterium suspension (A600=0.5) for 30 min at 28 °C with shaking at 100 rpm. Leaf explants were transferred to a solid medium supplemented with B5 vitamins, 3% (w/v) sucrose, 1 mg l−1 6-benzyladenine (BA), 0.1 mg l−1 α-naphthaleneacetic acid (NAA), 0.8% tissue culture agar, pH 5.6, and 100 mg l−1 kanamycin. Explants were incubated in the dark for 2 weeks at 25 °C, and then transferred to a growth chamber at 25 °C under a 16 h photoperiod. Finally, kanamycin-resistant plantlets were transferred to soil mix, acclimatized, and grown in the greenhouse.
Flavonoid analysis
Anthocyanins and flavonols were extracted from 50 mg of finely-ground tissues in 1 ml of 1% HCl/methanol (v/v) at room temperature in the dark with continuous shaking for 1 h, and centrifuged at 13, 000 rpm for 15 min. A total of 100 μl of supernatant was transferred to a fresh tube, acid-hydrolysed by adding 30 μl of 3 N HCl, and incubated at 70 °C for 1 h in a Thermo Hybaid MBS 0.25s thermocycler (Thermo Scientific). PAs were extracted using 1 ml of 70% (v/v) acetone containing 0.1% (w/v) ascorbate, and incubated at room temperature for 24 h in darkness. The extract was centrifuged at 13, 000 rpm for 15 min at room temperature, and the supernatant was transferred to a new 1.5 ml microfuge tube. An aliquot of 200 μl of extract was dried at 35 °C and resuspended in 100 μl of 1% (v/v) HCl/methanol and 100 μl of 200 mM sodium acetate (pH 7.5).
Flavonoids were identified using liquid chromatography–tandem mass spectrometry (LC-MS/MS) and their contents were calculated by comparison with commercial standards, including kaempferol, quercetin, cyanidin, catechin, and epicatechin (Sigma). The LC-MS/MS analysis was performed on a 5500 QTRAP mass spectrometer (AB Sciex) which was equipped with a 1200 Agilent HPLC. An Analyst (version 1.5.1, Applied Biosystems) was used for data acquisition and processing. A Phenomenex column (3u C6-Phenly 11A, 4.6×50 mm) was used for separation. The HPLC flow rate was set at 0.3 ml min−1. HPLC mobile phases consisted of A (0.1% formic acid in H2O) and B (0.1% formic acid in acetonitrile). The autosampler was kept at 5 °C. The gradient for catechin and epicatechin was as follows: 0 min, 90% A; 10 min, 50% A; 13–18 min, 0% A; and 18.1–25 min, 90% A. The gradients for cyanidin, kaempferol, and quercetin were as follows: 0 min, 70% A; 7–12.5 min, 0% A; and 13–20 min, 70% A. The injection volumes were 20 μl or 10 μl for anthocyanin or PA analysis, respectively. Multiple reaction monitoring was used to quantify catechin and epicatechin (m/z 291.0→139.2), cyanidin (m/z 287.2→213.2), kaempferol (m/z 287.1→153.2), and quercetin (m/z 303.1→153.1). The electrospray voltage was set to 5500 V, the heater was set at 600°C, the curtain gas was 35 psi, and both GS1 and GS2 were at 60 psi. All samples were run three times.
Results
Three members of a gene family encode anthocyanin reductase in apple
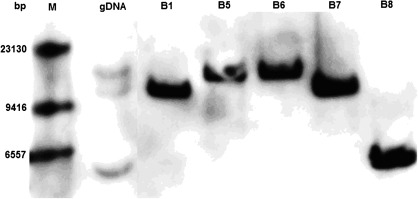
To identify all gene copies encoding ANR present in the apple genome, positive BAC clones and genomic DNA from ‘GoldRush’ leaves were subjected to Southern blot analysis. The genomic DNA consisted of three bands (Fig. 2), indicating that there are three copies of MdANR genes in the apple genome. The three BAC clones, designated B5, B6, and B7, yielded high, middle, and low bands, respectively (Fig. 1), and were thus selected to recover genomic DNA sequences of MdANR genes in apple.
Fig. 2.
Southern blot of BAC clones containing MdANR genes. Genomic DNA (gDNA) from apple ‘GoldRush’ was used to determine the number of gene copies of MdANR genes in the apple genome. The five positive BAC clones were designated as B1, B5, B6, B7, and B8. The digoxigenin (DIG)-labelled DNA probes correspond to cDNA fragments of the last two exons.
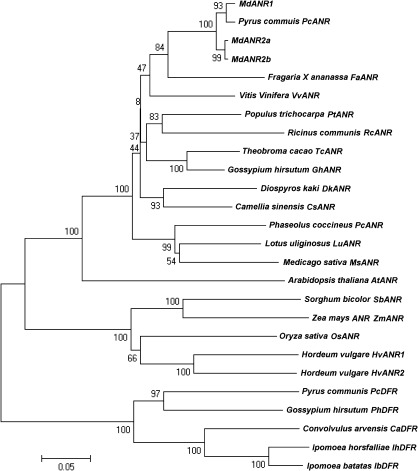
Three ANR genes, designated MdANR1 (GenBank accession no. JN035299), MdANR2a (GenBank accession no. JN035300), and MdANR2b (GenBank accession no. JN035301), were identified in apple. All three MdANR genes are composed of six exons and five introns, and contain an open reading frame of 1332 bp encoding a putative protein of 339 amino acids. Phylogenetic analysis was performed using coding DNA sequences of genes encoding ANR as well as closely related genes encoding DFR in plants. The results indicated that all three apple MdANR genes were clustered together with plant ANR genes and separated from plant DFR genes, indicating that all three genes encoded ANR (Fig. 3).
Fig. 3.
Phylogenetic tree derived from nucleotide sequences of genes encoding ANR and DFR in plants. The scale bar represents 0.05 substitutions per site, and the numbers next to the nodes are bootstrap values from 1000 replicates. The GenBank accession numbers are as follows: PcANR (DQ251189), FaANR (DQ664192), VvANR (XM_002271336), PtANR (XM_002317234), TcANR (GU324348), GhANR (EF187443), DkANR (AB195284), CsANR (AY641729), LuANR (EF197823), RcANR (XM_002518532), PcANR (BN000164), MsANR (HM754630), AtANR (NM_104854), HvANR1 (AK373696), HvANR2 (AK365124), OsANR (NM_001060512), SbANR (XM_002447113), ZmANR (BT064433), PcDFR (AY227730), IhDFR (GQ180935), IbDFR (EU402466), CaDFR (EU189076), and GhDFR (FJ713480).
MdANR1 shows ∼79% and ∼95% nucleotide sequence identities in genomic and coding regions, respectively, with either MdANR2a or MdANR2b. MdANR2a and MdANR2b share 97.0% and 99.9% nucleotide sequence identities in genomic and coding regions, respectively. MdANR2a and MdANR2b were deemed allelic, as described later. Differences in nucleotide sequences between the two genomic fragments of MdANR2a and MdANR2b are mainly attributed to several small InDels. The amino acid sequence of MdANR1 has 97% identity with sequences of both MdANR2a and MdANR2b. The deduced amino acid sequences of MdANR2a and MdANR2b are almost identical, except for two sequences that are different.
Mapping of MdANR1 and MdANR2 onto the apple genetic linkage map
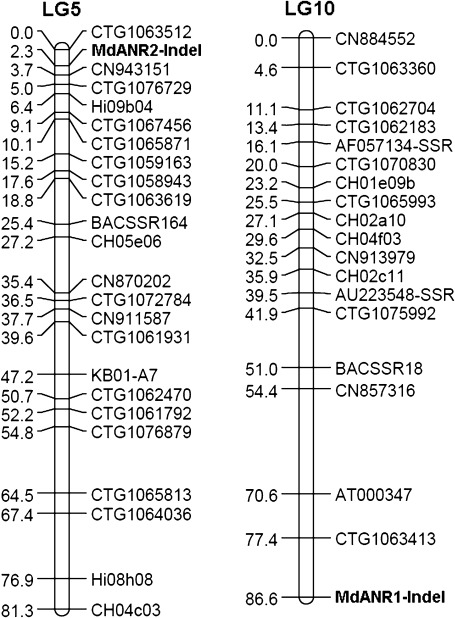
The genomic DNA sequence of MdANR1 from apple cv. GoldRush was BLASTed against the genome sequence database of apple cv. Golden Delicious (http://www.rosaceae.org/gb/gbrowse/malus_x_domestica/), and an InDel, ∼2.2 kb downstream of the stop codon, was identified. Based on this InDel, a marker was developed, designated as MdANR1-indel, and used for screening a segregating population derived from a ‘Coop 17’בCoop 16’ cross (Supplememtary Fig. S1 at JXB online). As a result, the apple MdANR1 gene was anchored onto linkage group 10 (Fig. 4).
Fig. 4.
Mapping of MdANR1 and MdANR2 genes onto apple genetic linkage groups 10 and 5, respectively.
Similarly, alignment of genomic DNA sequences of MdANR2a and MdANR2b revealed an InDel in the last intron, and a marker, designated MdANR2-indel, was developed based on this Indel. The MdANR2-indel was subsequently used to screen the ‘Coop 17’בCoop 16’ mapping population, and all progeny segregated into three genotypes (Supplementary Fig. S2 at JXB online). The first genotype had a high band corresponding to the MdANR2a allele, and the second genotype had a low band corresponding to the MdANR2b allele. However, the third genotype carried two bands that corresponded to both alleles. This result clearly demonstrated that MdANR2a and MdANR2b were alleles of the MdANR2 gene. Analysis of the genetic map indicated that the MdANR2 gene was located on linkage group 5 (Fig. 4).
Expression profiles of MdANR1 and MdANR2 genes in apple
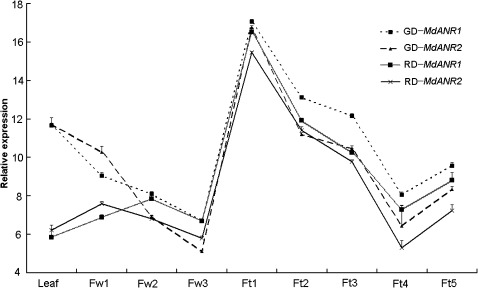
Two genotypes, a red-skinned cv. Red Delicious and a yellow-skinned cv. Golden Delicious, were selected to investigate expression profiles of genes encoding ANR in apple. qRT-PCR analysis indicated that MdANR1 and MdANR2 genes were expressed in all analysed tissues, including leaf, flower, and fruit (Fig. 5). Transcript accumulation of both MdANR1 and MdANR2 genes was highest in young fruitlets [9 days after pollination (DAP)] of both ‘Red Delicious’ and ‘Golden Delicious’, and then declined as fruitlets continued to develop. Transcript levels of both MdANR1 and MdANR2 genes in fruits reached their lowest levels at 104 DAP, but then slightly increased at fruit maturity.
Fig. 5.
Real-time PCR analysis of MdANR1 and MdANR2a/b gene expression in various floral and fruit tissues of ‘Red Delicious’ and ‘Golden Delicious’. Differential expression of MdANR1 and MdANR2 between the two apple cultivars Red Delicious and Golden Delicious was compared. Abbreviations for stages of development are as follows: Fw1, flower buds at the pink stage; Fw2, flower buds at the balloon stage; Fw3, flowers at full bloom; Ft1, fruitlets 9 DAP; Ft2, fruitlets 16 DAP; Ft3, fruitlets 44 DAP; Ft4, fruits at 104 DAP; Ft5, fruits at 145 DAP; RD, ‘Red Delicious’; GD, ‘Golden Delicious’. All expression data were normalized to the apple actin gene, and values are means of three technical replicates.
Overall, transcript accumulation of both MdANR1 and MdANR2 was higher in ‘Golden Delicious’ than in ‘Red Delicious’. Transcript levels of both MdANR1 and MdANR2 were relatively lower in flowers than in both leaves and fruits at early and middle stages of development. Accumulation of transcripts of both MdANR1 and MdANR2 were lower in flowers of cv. Golden Delicious, and continued to decline throughout flower development; however, transcripts of both genes remained relatively constant in flowers of cv. Red Delicious throughout flower development.
Functional analysis of MdANR genes in tobacco
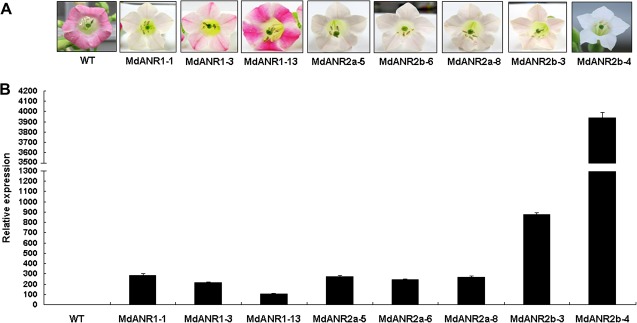
Coding region sequences encoding MdANR1, MdANR2a, and MdANR2b were separately transferred into tobacco under control of the Cauliflower mosaic virus (CaMV) 35S promoter, and several transgenic lines were generated for each construct. Flower colours of all T2 transgenic lines were different from those of wild-type plants, indicating that all three genes encoding ANR in apple were functional (Fig. 6a). Moreover, T2 transgenic lines produced flowers of different pigmentation patterns. For example, transgenic lines carrying MdANR1 produced either pale or white variegation with red pendant flowers. All three transgenic lines carrying MdANR2a produced pale-pink flowers, and two transgenic lines overexpressing MdANR2b produced either pale pink-coloured or pure white flowers, respectively.
Fig. 6.
Ectopic expression of MdANR1 and MdANR2 genes in tobacco. (A) Tobacco flowers of wild-type (WT) and T2 transgenic lines. Transgenic lines containing MdANR1, MdANR2a, and MdANR2b genes were pre-fixed with MdANR1, MdANR2a, and MdANR2b, respectively. (B) Expression profiles of MdANR genes in flowers of transgenic tobacco lines.
Following LC-MS/MS analysis, it was revealed that flowers of all transgenic lines accumulated lower levels of anthocyanin than wild-type flowers (Table 1). Anthocyanin contents in either white- or pale pink-coloured transgenic flowers were too low to be detected. White and red variegated transgenic flowers accumulated certain amounts of anthocyanin, but these levels were significantly lower than those of wild-type flowers. Moreover, flowers of all transgenic lines accumulated higher levels of both catechin and epicatechin than did wild-type flowers. Transgenic line MdANR2b-4 producing pure white flowers accumulated the highest levels of epicatechin, while transgenic lines MdANR1-3 and MdANR1-13, producing white and red variegated flowers, respectively, had lower levels of epicatechin when compared with other transgenic lines. Flowers of all transgenic lines, except for MdANR2a-6, accumulated higher levels of quercetin when compared with flowers of the wild-type control. Moreover, flowers of all transgenic lines, except for MdANR2b-3 and MdANR2b-4, produced lower levels of kaempferol when compared with flowers of wild-type control.
Table 1.
Flavonoid contents in transgenic and wild-type tobacco flowersa
| Flower | Flavonol (μg g−1) |
Anthocyanin (ng g−1) | Proanthocyanidin (ng g−1) |
||
| Kaempferol | Quercetin | Cyanidin | Catechin | Epicatechin | |
| Wild type | 60.57±0.83 | 71.80±1.51 | 1722±64.37 | 4.80±0.53 | 6.47±0.76 |
| MdANR1-1 | 53.73±0.64 | 87.60±4.42 | N/A | 9.07±0.99 | 9.93±1.01 |
| MdANR1-3 | 32.53±0.96 | 78.17±5.58 | 759±5.77 | 5.33±0.23 | 8.40±0.87 |
| MdANR1-13 | 43.57±1.81 | 86.57±1.80 | 1063±18.38 | 5.47±0.64 | 7.20±0.20 |
| MdANR2a-5 | 42.10±1.67 | 99.67±4.89 | N/A | 9.53±0.58 | 9.60±0.20 |
| MdANR2a-6 | 26.30±1.47 | 68.40±6.62 | N/A | 6.07±0.42 | 8.80±0.20 |
| MdANR2a-8 | 42.20±2.73 | 83.13±6.65 | N/A | 10.33±1.48 | 11.93±1.97 |
| MdANR2b-3 | 68.77±3.37 | 92.90±4.03 | N/A | 6.80±0.20 | 8.73±0.61 |
| MdANR2b-4 | 145.33±10.21 | 493.67±21.92 | N/A | 5.60±0.20 | 16.73±0.50 |
All data correspond to mean values ±SD of three biological replicates. N/A, not available.
Following qRT-PCR analysis, it was revealed that transgenic line MdANR2b-4, producing pure white flowers, exhibited the highest levels of transgene expression. MdANR1-3 and MdANR1-13 lines, producing white and red variegated flowers, respectively, exhibited relatively lower levels of transgene expression in their flowers when compared with those of other transgenic lines (Fig. 6b). Taken together, ectopic expression of MdANR genes in tobacco promoted the biosynthesis of PAs in flowers, but inhibited anthocyanin accumulation.
Influence of overexpressing MdANR genes on expression of other flavonoid pathway genes in transgenic tobacco flowers
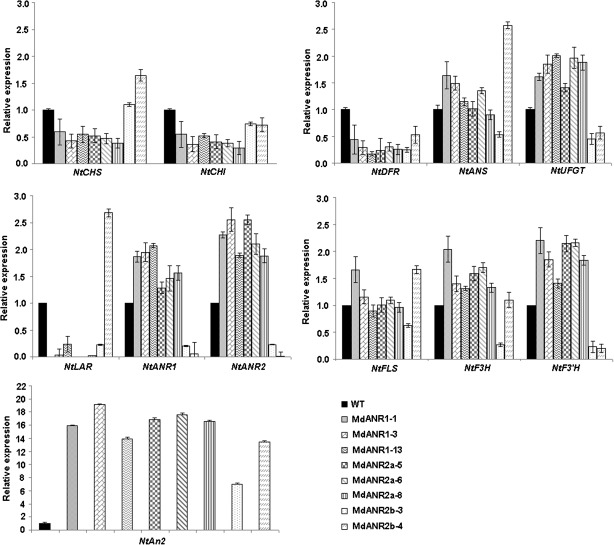
qRT-PCR analysis was conducted to investigate the coordinate interaction of MdANR genes with other flavonoid pathway genes in transgenic tobacco flowers, including structural genes such as NtCHS, NtCHI, NtF3H, NtF3′H, NtFLS, NtDFR, NtUFGT, NtLAR, NtANR1, and NtANR2, and the regulator gene NtAn2. Expression of all flavonoid structural genes in flowers of all different transgenic lines carrying apple MdANR genes showed significant differences (Fig. 7). Overexpression of MdANR genes in tobacco greatly influenced expression of flavonoid structural genes in these flowers. For example, expression of NtANR, NtUFGT, and NtF3'H genes was up-regulated in flowers of transgenic plants carrying either MdANR1 or MdANR2a, but these were down-regulated in transgenic flowers overexpressing MdANR2b. Flowers of transgenic lines such as MdANR1-1, MdANR2a-5, and MdANR2a-6 showed no detectable expression of the NtLAR gene. Of all genes investigated, the two genes NtCHI and NtDFR showed lower levels of expression in flowers of all transgenic lines. Moreover, transcript accumulation of the R2R3 MYB gene NtAn2 was significantly higher in flowers of all transgenic plants than those of the wild-type control.
Fig. 7.
Expression profiles of flavonoid-related structural biosynthetic genes in flowers of transgenic tobacco lines carrying MdANR genes. All mRNA transcripts expressed in transgenic flowers were quantified relative to those expressed in wild-type tobacco flowers.
Discussion
ANR and LAR catalyse the synthesis of flavan-3-ol, an initiating monomer of condensed tannin or PA synthesis, from cyanidin and leucoanthocyanidin, respectively. ANR is not only important for PA synthesis, but it also influences the synthesis of anthocyanin by competing with UFGT (UDP-glucose:flavonoid 3-O-glucosyltransferase) activity by which it converts anthocyanidin to anthocyanin (Bog et al., 2005). Thus, ANR plays an important role in both anthocyanin and PA synthesis in plants. To date, genes encoding ANR have been isolated and characterized in Arabidopsis and grapevines (Xie et al., 2003; Bogs et al., 2005). However, it is not clear how ANR genes interact with other flavonoid structural genes to coordinate the biosynthesis of anthocyanin. Herein, a gene family encoding ANR in apple has been isolated and functional analysis has been conducted. The results will provide insights into the interaction between MdANR genes and other genes involved in flavonoid biosynthesis.
Duplication of MdANR genes is related to the polyploid origin of the apple genome
Two MdANR genes, MdANR1 and MdANR2, have been identified in the apple genome. MdANR1 and MdANR2 genes share 91% nucleotide sequence identity in their coding regions, and are located on linkage groups 10 and 5, respectively. The apple is a diploid (2n=34), but it has an allopolyploid origin, and has a wide genetic diversity (Korban and Tartarini, 2009; Velasco et al., 2010, Zhang et al., 2011). Recently, integrated physical and genetic maps of the apple have demonstrated that linkage groups 5 and 10 are of homologous chromosome pairs (Han et al., 2011). Moreover, regulation of genes during early stages of fruit development has been investigated (Soria-Guerra et al., 2011). Interestingly, two FLAVONOID 3′ HYDROXYLASE (F3′H) genes, MdF3′H1 and MdF3′H2, have been identified in the apple genome (Han et al., 2010). MdF3′H1 and MdF3′H2 share 91% nucleotide sequence identity in their coding regions, and are located on linkage groups 6 and 14, respectively. The apple linkage groups 6 and 14 are also homologous chromosome pairs (Han et al., 2011). Thus, it is clear that duplication of both ANR and F3′H genes in apple must be simultaneously derived from whole-genome duplication during the process of speciation (Xu and Korban, 2004). Moreover, the two apple MdANR genes are functional, and are expressed in fruits throughout fruit development. Multiple genes expressed in fruits may be responsible for the presence of high contents of PAs in apple (Souquet, 1996). In addition, coloration patterns of flowers of transgenic tobacco overexpressing MdANR1 are different from those of transgenic flowers overexpressing MdANR2. These differences may be attributed to transgene positional effects. However, it cannot be ruled out that MdANR1 and MdANR2 genes may have also functionally diverged during the course of evolutionary development of apple.
Incomplete inhibition of anthocyanin production results in novel floral pigmentation patterns
Floral colours and pigmentation patterns, such as white and red variegation, are of important ornamental value. Novel floral pigmentation patterns have been obtained by genetic modification of the anthocyanin biosynthesis pathway in several ornamental plants such as petunia, lisianthus, and torenia that naturally produce patterned flowers, with some white and coloured petal areas (Napoli et al., 1990; Davies, 2009). Variegated pigmentation patterns are pre-determined by morphological signals within petals that control levels of gene inhibition, and in turn those signals would interact with environmental signals that influence pigmentation (van der Krol et al., 1990). Tobacco plants do not produce flowers that are naturally patterned or variegated. However, variegated flower phenotypes have been observed in tobacco plants overexpressing an Arabidopsis transposable element, Tag1-R (Liu et al., 2001). Yet, no patterned tobacco flowers have been reported following inhibition of anthocyanin biosynthesis.
It has been reported that anthocyanin biosynthesis can be inhibited by overproduction of enzymes that compete for the substrate (Joung et al., 2003). For example, introduction of a Medicago CHALCONE REDUCTASE (CHR) gene into petunia altered flower coloration from deep purple to pale purple, but did not yield any white-coloured flowers (Davies et al., 1998). The ANR enzyme is known to compete with the UFGT enzyme to convert anthocyanidin to epicatechin. In a previous study, it has been demonstrated that ectopic expression of an Arabidopsis BAN gene in tobacco can significantly inhibit the biosynthesis of anthocyanin, resulting in white-coloured flowers (Xie et al., 2003). Here, it IS further found that overexpression of apple ANR genes in tobacco not only produces white flowers or pale pink-coloured flowers, but has also resulted in recovery of flowers with novel pigmentation patterns, with some petal areas that are white and others that are red.
To determine whether or not transgene copy number had an effect on coloration of transgenic flowers, Southern blot analysis was conducted for those transgenic lines carrying the MdANR1 gene. All three transgenic lines producing different coloured flowers contained a single copy of the MdANR1 transgene (Supplementary Fig. S3 at JXB online). Thus, differences in flower pigmentation could not be attributed to transgene copy number. Moreover, two transgenic lines of MdANR1 produced white and red variegated flowers and showed lower levels of transgene expression when compared with other transgenic lines of MdANR2 producing either white or pale pink-coloured flowers. In addition, previous studies demonstrated that introducing an enzyme to compete with anthocyanin biosynthetic enzymes could successfully lower pigment levels, but this was not sufficient for depleting pigmentation in flowers and resulting in white-coloured flowers (Davies et al., 1998; Xie et al., 2003). Interestingly in this study, a single transgenic line, MdANR2b-4, produced pure white flowers, yet showed very high levels of MdANR2b expression. Taken together, these findings suggested that low levels of expression of MdANR genes might be partially responsible for incomplete inhibition of anthocyanin synthesis in transgenic tobacco flowers.
The apple ANR gene may also have a redundant function in converting anthocyanidin into catechin
It is well documented that LAR and ANR catalyse the conversion of leucocyanidin and cyanidin into catechin and epicatechin, respectively (Bogs et al., 2005; Szankowski et al., 2009). In this study, flowers of all transgenic tobacco lines overexpressing either MdANR1 or MdANR2 genes have accumulated higher levels of both catechin and epicatechin compared with those of wild-type tobacco plants. However, overexpression of an MdANR gene has contributed to significant suppression of the function of the LAR native gene, as expression levels of the LAR gene in most transgenic tobacco lines has been too low to be detected. Thus, other genes in anthocyanin biosynthesis, besides LAR, must be capable of promoting accumulation of catechin.
Incubation of BAN proteins of M. truncatula and Arabidopsis with anthocyanidins, cyanidin, and pelargonidin has resulted in the formation of epicatechin as the major product and catechin as a minor product (Xie et al., 2003). Similarly, findings in this study also suggest that ANR or BAN could synthesize both epicatechin and catechin. This in turn raises a question as to whether the synthesis of catechin results from epimerization of epicatechin or whether it is a product that is catalysed by ANR. It has been reported that incubating either epicatechin or catechin with either MtANR or AtANR in the presence of NADPH does not result in epimerization into catechin or epicatechin, respectively (Xie et al., 2004). LDOX is essential for PA synthesis in Arabidopsis (Abrahams et al., 2003), thus suggesting that ANR cannot efficiently use leucoanthocyanidin as a substrate to produce catechin. Thus, it is likely that BAN could directly convert anthocyanidin into catechin. In addition, it is worth noting that catechin might be formed through chemical epimerization of epicatechin (Xie et al., 2004). Further studies are needed to clarify whether the catechin detected in this study is attributed to the epimerization of epicatechin to catechin. It has been reported that high temperatures and alkalization are two main factors that induce the epimerization of epicatechin into catechin (Kofink et al., 2007). However, in this study, PA extraction has been conducted under pH 7.5 and 35 °C, which is not high. Moreover, the accumulated catechin, presumed to be formed through chemical epimerization, is observed as a minor byproduct (Xie et al., 2003). In this study, the amounts of epicatechin and catechin accumulating in flowers of some transgenic tobacco lines overexpressing an MdANR gene are almost equal; thus, formation of catechin in these lines cannot be fully explained by chemical epimerization alone.
Overexpression of MdANR genes causes co-suppression of other structural genes involved in flavonoid biosynthesis
Overexpression of the CHALCONE SYNTHASE (CHS) gene results in co-suppression of homologous genes in petunia, and this co-suppression is related to an RNA silencing mechanism (Baulcombe, 2004). In this study, overexpression of MdANR genes has resulted in co-suppression of NtLAR, NtDFR, and NtCHI genes in transgenic tobacco lines. Of these three genes, NtLAR and NtDFR are related to MdANR. Collectively, MdANR, NtLAR, and NtDFR belong to the reductase–epimerase–dehydrogenase (RED) superfamily although they share low identity in their DNA coding sequences. However, co-suppression of NtLAR and NtDFR may not be related to RNA silencing for the following reasons. First, ANR genes from apple and tobacco share ∼52% identity in their DNA coding sequences, and overexpression of MdANR does not result in co-suppression of NtANR genes in tobacco. Secondly, ANR is more closely related to DFR than to LAR; however, overexpression of an MdANR gene suppresses expression of NtLAR more severely than that of NtDFR. Therefore, the mechanism of co-suppression is rather complicated, and requires further investigation.
LAR converts leucoanthocyanidin into catechin and it competes with anthocyanidin synthase (ANS)/LDOX for the substrate to produce an alternative initiating unit for PA biosynthesis. It is known that ANR and LAR are NAPDH-dependent reductases. Thus, overexpression of MdANR in this study will offer little opportunity for NtLAR to accept NAPDH, resulting in low levels of expression of NtLAR in flowers of transgenic tobacco lines. Interactions among DFR, CHS, and CHI have been previously identified in Arabidopsis (Burbulis and Winkel-Shirley, 1999). Therefore, it is likely that co-suppression of NtLAR and NtDFR may be due to interactions among enzymes involved in the flavonoid biosynthetic pathway.
Variegated patterns of flower coloration of transgenic tobacco overexpressing MdANR are related to low levels of expression of both NtCHI and NtDFR
In this study, expression of structural and regulatory genes involved in the flavonoid biosynthesis pathway has been investigated in flowers of transgenic tobacco lines overexpressing MdANR genes. Of the flavonoid structural genes, NtCHI and NtDFR have demonstrated significantly lower levels of expression in flowers of all transgenic tobacco lines compared with those of wild-type plants. It is well known that expression of CHI and DFR genes is controlled by regulatory genes such as R2R3 MYB transcription factors (Grotewold, 2006). More recently, an R2R3 MYB regulator from tobacco (NtAn2) has been isolated and reported to be a key gene controlling anthocyanin production in reproductive tissues of tobacco (Pattanaik et al., 2010). Interestingly, in this study, flowers of all transgenic tobacco lines have demonstrated significantly higher levels of expression of NtAn2 than those of wild-type tobacco. Thus, it seems that overexpression of MdANR may affect the regulatory role of the NtAn2 gene. However, it is not yet clear as to whether or not the ANR protein can interact with R2R3 MYB proteins, thus leading to their enhanced transcriptional activities.
Transgenic tobacco lines carrying MdANR genes accumulate up to 10 ng g−1 more PAs than wild-type plants, and most show slight changes in flavonol accumulation compared with wild-type plants. However, most MdANR transgenic lines have not accumulated anthocyanins, whereas wild-type plants have accumulated >1700 ng g−1 anthocyanins. These results suggest that ANR genes may coordinately interact with other flavonoid genes to regulate flavonoid biosynthesis. Overall, overexpression of MdANR genes in tobacco results in a significant decrease in the total amount of flavonoids accumulated in flowers. The loss of anthocyanin in tobacco transgenic flowers overexpressing MdANR genes is probably attributed to suppression of the two genes NtCHI and NtDFR.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Segregation for the MdANR1 gene in an F1 apple population.
Figure S2. Segregation for the MdANR2 gene in an F1 apple population.
Figure S3. Copy numbers of MdANR1 genes in transgenic tobacco lines determined by Southern blot hybridization.
Table S1. Primer sequences for qRT-PCR.
Acknowledgments
This work was supported by funds received from the National Program on Key Basic Research Project of China (973 Program) (for YH) under grant no. 2011CB100600 and funds provided by USDA-NIFA-SCRI grant AG 2009-51181-06023 (SSK).
References
- Abrahams S, Lee E, Walker AR, Tanner GJ, Larkin PJ, Ashton AR. The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. The Plant Journal. 2003;35:624–636. doi: 10.1046/j.1365-313x.2003.01834.x. [DOI] [PubMed] [Google Scholar]
- Akagi T, Ikegami A, Tsujimoto T, Kobayashi S, Sato A, Kono A, Yonemori K. DkMyb4 is a Myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit. Plant Physiology. 2009;151:2028–2045. doi: 10.1104/pp.109.146985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, Moriguchi T. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant and Cell Physiology. 2007;48:958–970. doi: 10.1093/pcp/pcm066. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. The Plant Journal. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- Burbulis IE, Winkel-Shirley B. Interactions among enzymes of the arabidopsis flavonoid biosynthetic pathway. Proceedings of the National Academy of Sciences, USA. 1999;96:12929–12934. doi: 10.1073/pnas.96.22.12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs J, Downey MO, Harvey JS, Ashton AR, Tanner GJ, Robinson SP. Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiology. 2005;139:652–663. doi: 10.1104/pp.105.064238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs J, Jaffé FW, Takos AM, Walker AR, Robinson SP. The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiology. 2007;143:1347–1361. doi: 10.1104/pp.106.093203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KM. Modifying anthocyanin production in flowers. In: Gould K, Davies K, Winefield C, editors. Anthocyanins: biosynthesis, functions, and applications. New York: Springer; 2009. pp. 49–84. [Google Scholar]
- Davies KM, Bloor SJ, Spiller GB, Deroles SC. Production of yellow colour in flowers: redirection of flavonoid biosynthesis in petunia. The Plant Journal. 1998;13:259–266. [Google Scholar]
- Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. The Plant Journal. 2007;49:414–427. doi: 10.1111/j.1365-313X.2006.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasic K, Hernandez A, Korban SS. RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Molecular Biology Reporter. 2004;22:437a–437g. [Google Scholar]
- Grotewold E. The genetics and biochemistry of floral pigments. Annual Review of Plant Biology. 2006;57:761–780. doi: 10.1146/annurev.arplant.57.032905.105248. [DOI] [PubMed] [Google Scholar]
- Hammerstone JF, Lazarus SA, Schmitz HH. Procyanidin content and variation in some commonly consumed foods. Journal of Nutrition. 2000;130:2086S–2092S. doi: 10.1093/jn/130.8.2086S. [DOI] [PubMed] [Google Scholar]
- Han Y, Bendik E, Sun FJ, Gasic K, Korban SS. Genomic isolation of genes encoding starch branching enzyme II (SBEII) in apple: toward characterization of evolutionary disparity in SbeII genes between monocots and eudicots. Planta. 2007;226:1265–1276. doi: 10.1007/s00425-007-0555-6. [DOI] [PubMed] [Google Scholar]
- Han Y, Vimolmangkang S, Soria-Guerra RE, Rosales-Mendoza S, Zheng D, Lygin AV, Korban SS. Ectopic expression of apple F3′H genes contributes to anthocyanin accumulation in the Arabidopsis tt7 mutant grown under nitrogen stress. Plant Physiology. 2010;153:806–820. doi: 10.1104/pp.109.152801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Zheng D, Vimolmangkang S, Khan MA, Beever JE, Korban SS. Integration of physical and genetic maps in apple confirms whole-genome and segmental duplications in the apple genome. Journal of Experimental Botany. 2011;62:5117–5130. doi: 10.1093/jxb/err215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1230. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Joung J-Y, Mangai Kasthuri G, Park J-Y, Kang W-J, Kim H-S, Yoon B-S, Joung H, Jeon J-H. An overexpression of chalcone reductase of Pueraria montana var. lobata alters biosynthesis of anthocyanin and 5′-deoxyflavonoids in transgenic tobacco. Biochemical and Biophysical Research Communications. 2003;303:326–331. doi: 10.1016/s0006-291x(03)00344-9. [DOI] [PubMed] [Google Scholar]
- Kofink M, Papagiannopoulos M, Galensa R. (–)-Catechin in cocoa and chocolate: occurrence and analysis of an atypical flavan-3-ol enantiomer. Molecules. 2007;12:1274–1288. doi: 10.3390/12071274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korban SS, Tartarini S. Apple structural genomics. In: Folta KM, Gardiner SE, editors. Genetics and genomics of Rosacea. New York: Springer-Science; 2009. pp. 85–119. [Google Scholar]
- Liu D, Galli M, Crawford NM. Engineering variegated floral patterns in tobacco plants using the arabidopsis transposable element tag1. Plant and Cell Physiology. 2001;42:419–423. doi: 10.1093/pcp/pce053. [DOI] [PubMed] [Google Scholar]
- McMahon LR, McAllister TA, Berg BP, Majak W, Acharya SN, Popp JD, Coulman BE, Wang Y, Cheng KJ. A review of the effects of forage condensed tannins on ruminal fermentation and bloat in grazing cattle. Canadian Journal of Plant Science. 2000;80:469–485. [Google Scholar]
- Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. The Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. The Plant Cell. 2001;13:2099–2114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obreque-Slíer E, Peña-Neira A, López-Solís R. Enhancement of both salivary protein-enological tannin interactions and astringency perception by ethanol. Journal of Agricultural and Food Chemistry. 2010;58:3729–3735. doi: 10.1021/jf903659t. [DOI] [PubMed] [Google Scholar]
- Pattanaik S, Kong Q, Zaitlin D, Werkman JR, Xie CH, Patra B, Yuan L. Isolation and functional characterization of a floral tissue-specific R2R3 MYB regulator from tobacco. Planta. 2010;231:1061–1076. doi: 10.1007/s00425-010-1108-y. [DOI] [PubMed] [Google Scholar]
- Renard CMGC, Le Quéré JM, Bauduin R Symoneaux R, Le Bourvellec C, Baron A. Modulating polyphenolic composition and organoleptic properties of apple juices by manipulating the pressing conditions. Food Chemistry. 2011;124:117–125. [Google Scholar]
- Santos-Buelga C, Scalbert A. Proanthocyanidins and tannin-like compounds—nature, occurrence, dietary intake and effects on nutrition and health. Journal of the Science of Food and Agriculture. 2000;80:1094–1117. [Google Scholar]
- Soria-Guerra R, Rosales-Mendoza S, Gasic K, Band M, Wisniewski ME, Korban SS. Gene expression is highly regulated in early developing fruit of apple. Plant Molecular Biology Reporter. 2011;29:885–897. [Google Scholar]
- Souquet J. Polymeric proanthocyanidins from grape skins. Phytochemistry. 1996;43:509–512. [Google Scholar]
- Szankowski I, Flachowsky H, Li H, Halbwirth H, Treutter D, Regos I, Hanke MV, Stich K, Fischer TC. Shift in polyphenol profile and sublethal phenotype caused by silencing of anthocyanidin synthase in apple (Malus sp.) Planta. 2009;229:681–692. doi: 10.1007/s00425-008-0864-4. [DOI] [PubMed] [Google Scholar]
- Takos AM, Jaffé FW, Jacob SR, Bogs J, Robinson SP, Walker AR. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiology. 2006;142:1216–1232. doi: 10.1104/pp.106.088104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner GJ, Francki KT, Abrahams S, Watson JM, Larkin PJ, Ashton AR. Proanthocyanidin biosynthesis in plants: purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. Journal of Biological Chemistry. 2003;278:31647–31656. doi: 10.1074/jbc.M302783200. [DOI] [PubMed] [Google Scholar]
- van der Krol AR, Mur LA, De Lange P, Gerats AGM, Mol JNM, Stuitje AR. Antisense chalcone synthase genes in petunia: visualization of variable transgene expression. Molecular and General Genetics. 1990;220:204–212. [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, et al. The genome of the domesticated apple (Malus × domestica Borkh.) Nature Genetics. 2010;42:833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- Vidal S, Francis L, Guyot S, Marnet N, Kwiatkowski M, Gawel R, Cheynier V, Waters EJ. The mouth-feel properties of grape and apple proanthocyanidins in a wine-like medium. Journal of the Science of Food and Agriculture. 2003;83:564–573. [Google Scholar]
- Winkel-Shirley B. It takes a garden. How work on diverse plant species has contributed to an understanding of flavonoid metabolism. Plant Physiology. 2001;127:1399–1404. [PMC free article] [PubMed] [Google Scholar]
- Xie D-Y, Sharma SB, Dixon RA. Anthocyanidin reductases from Medicago truncatula and. Arabidopsis thaliana. Archives of Biochemistry and Biophysics. 2004;422:91–102. doi: 10.1016/j.abb.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA. Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science. 2003;299:396–399. doi: 10.1126/science.1078540. [DOI] [PubMed] [Google Scholar]
- Xu M, Korban SS. Somatic variation plays a key role in the evolution of the Vf gene family residing in the Vf locus that confers resistance to apple scab disease. Molecular Phylogenetics and Evolution. 2004;32:57–65. doi: 10.1016/j.ympev.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Xu M, Song J, Cheng Z, Jiang J, Korban SS. A bacterial artificial chromosome (BAC) library of Malus floribunda 821 and contig construction for positional cloning of the apple scab resistance vf gene. Genome. 2001;44:1104–1113. [PubMed] [Google Scholar]
- Zhang Q, Li J, Zhao Y, Korban SS, Han Y. 2011. Evaluation of genetic diversity in Chinese wild apple species along with apple cultivars using SSR markers. Plant Molecular Biology Reporter. DOI 10.1007/s11105-011-0366-6 (in press) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.