Abstract
Flavonols, phenylalanine-derived secondary metabolites, have protective and regulatory functions in plants. In Arabidopsis thaliana, they are consecutively glycosylated at their 3-OH and 7-OH groups. UGT78D1 and UGT78D2 are the major flavonol 3-O-glycosyltransferases in Arabidopsis leaves. The ugt78d1 ugt78d2 double mutant, which was strongly compromised in the initial 3-O-glycosylation, showed a severe and specific repression of flavonol biosynthesis, retaining only one-third of the wild-type level. This metabolic phenotype was associated with a repressed transcription of several flavonol biosynthetic genes including the committed step chalcone synthase [(CHS) or TRANSPARENT TESTA 4 (TT4)]. Furthermore, the committed step of the upstream, general phenylpropanoid pathway, phenylalanine ammonia-lyase (PAL), was down-regulated in its enzyme activity and in the transcription of the flavonol-related PAL1 and PAL2. However, a complete blocking of flavonoid biosynthesis at CHS released PAL inhibition in a tt4 ugt78d1 ugt78d2 line. PAL activity was even enhanced in the flavonol synthase 1 mutant, which compromises the final formation of flavonol aglycones. The dependence of the PAL feedback inhibition on flavonols was confirmed by chemical complementation of tt4 ugt78d1 ugt78d2 using naringenin, a downstream flavonoid intermediate, which restored the PAL repression. Although aglycones were not analytically detectable, this study provides genetic evidence for a novel, flavonol-dependent feedback inhibition of the flavonol biosynthetic pathway and PAL. It was conditioned by the compromised flavonol-3-O-conjugation and a decrease in flavonol content, yet dependent on a residual, flavonol synthase 1 (FLS1)-related capacity to form flavonol aglycones. Thus, this regulation would not react to a reduced metabolic flux into flavonol biosynthesis, but it might prevent the accumulation of non-glycosylated, toxic flavonols.
Keywords: Feedback inhibition, flavonoids, flavonols, flavonol synthase, phenylalanine ammonia-lyase, phenylpropanoids, UDP-carbohydrate-dependent glycosyltransferase
Introduction
The phenylalanine-derived flavonoids are important plant secondary metabolites. In Arabidopsis thaliana, flavonoids comprise flavonols, anthocyanins, and proanthocyanidins. Flavonols have been the subject of intense research interest because they were shown to have diverse roles in plants such as protection from oxidative damage and UV radiation, inhibition of auxin transport, control of pollen function, and signalling to symbiotic organisms (Mo et al., 1992; Li et al., 1993; Peer et al., 2004; Harrison, 2005; Taylor and Grotewold, 2005; Wasson et al., 2006; Santelia et al., 2008; Buer et al., 2010). In addition, flavonols taken in as food and feed ingredients have also been implicated as being either protective, regulatory, or cytotoxic molecules in mammals (Lin and Weng, 2006).
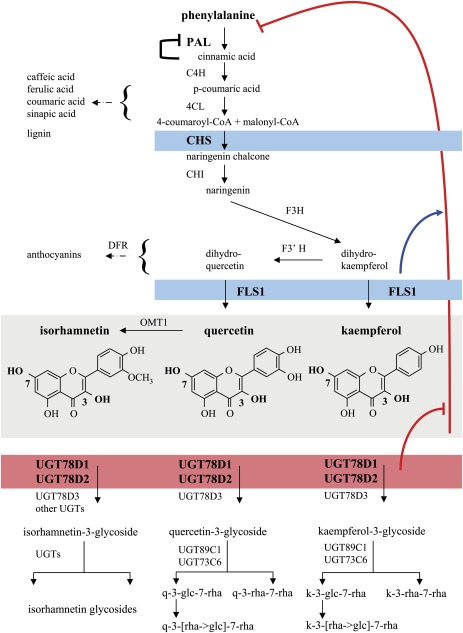
The biosynthesis of flavonols as well as the general phenylpropanoid pathway supplying the flavonoid precursor p-coumaroyl-CoA have been well characterized in many plant species including A. thaliana (Dixon and Steele, 1999; Harborne and Williams, 2000, 2001; Winkel-Shirley, 2001) (Fig. 1). Phenylalanine ammonia-lyase (PAL) catalyses the committed step of the phenylpropanoid pathway at the link between primary and secondary metabolism. Different PAL isoforms have been proposed to have different metabolic roles in A. thaliana. PAL1 and PAL2 are involved in flavonoid and lignin biosynthesis, while PAL4 appears to be specific for the lignin branch (Raes et al., 2003; Rohde et al., 2004; Huang et al., 2010). Chalcone synthase [(CHS) or TT4 (TRANSPARENT TESTA 4)] catalyses the committed step of flavonoid biosynthesis and is encoded by a single gene in A. thaliana (Winkel-Shirley, 2006). The tt4 mutant is completely devoid of flavonoids, which provides a good tool for analysing the consequences of a complete loss of flavonoids in plants. Although six Arabidopsis flavonol synthase (FLS) isoforms were identified, only FLS1 contributed considerably to the last step of the formation of the flavonol aglycones (Owens et al., 2008; Stracke et al., 2009). FLS3 had a very low activity, which could be only tracked down in an fls1 background (Preuß et al., 2009).
Fig. 1.
Phenylpropanoid and flavonol biosynthesis pathways in Arabidopsis thaliana including the FLS1-dependent feedback inhibition. The pathways leading from the central precursor phenylalanine to flavonols as well as to other phenylpropanoids such as anthocyanins, sinapic acid derivatives, and lignin are displayed (Winkel-Shirley, 2001; Tohge et al., 2005; Yonekura-Sakakibara et al., 2007). Phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate-CoA ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3'-hydroxylase (F3'H), flavonol synthase (FLS1), O-methyltransferase (OMT1), dihydroflavonol 4-reductase (DFR), flavonol 3-O-rhamnosyltransferase (UGT78D1), flavonoid 3-O-glucosyltransferase (UGT78D2), flavonol 3-O-arabinosyltransferase (UGT78D3), flavonol 7-O-rhamnosyltransferase (UGT89C1), and flavonol 7-O-glucosyltransferase (UGT73C6). Known (cinnamic acid) and the newly discovered metabolic feedback inhibitors (flavonols) are indicated. The inhibition (red) of PAL (and downstream flavonoid-specific genes) is provoked by a compromised 3-O-glycosylation in the ugt78d1 ugt78d2 double mutant. This regulation was shown to depend on (blue) the function of CHS or FLS1, which are necessary to generate the flavonol aglycones. k-3-glc-7-rha, kaempferol 3-O-glucoside-7-O-rhamnoside (f1); k-3-[rha→glc]-7-rha, kaempferol-3-O-[rhamnosyl (1→2 glucoside)]-7-O-rhamnoside (f2); k-3-rha-7-rha, kaempferol 3-O-rhamnoside-7-O-rhamnoside (f3). The structurally equivalent quercetin (q) glycosides are abbreviated in an analogous way.
Flavonol aglycones are further glycosylated in a complex manner in plant cells. Glycosylation is frequently involved in the biosynthesis of secondary metabolites to modify their stability, solubility, or localization, and thereby the biological properties of the conjugated molecules (Li et al., 2001; Meßner et al., 2003; Bowles et al., 2006). In the case of the hydrophobic flavonols, glycosylation renders them more water soluble and less toxic, and it may enable flavonol transport and compartmentation (Cos et al., 2001; Bowles et al., 2006; Winkel-Shirley, 2006; Moreira et al., 2007; Xiao et al., 2009). However, the physiological significance of the complex glycosylation pattern found in plants remains unclear. Glycosylation is achieved by UDP-carbohydrate-dependent glycosyltransferases (UGTs), which transfer sugars to their aglycone substrates by the formation of a glycosidic bond (Li et al., 2001). Flavonols are usually glycosylated at their 3-OH and 7-OH positions in A. thaliana (Veit and Pauli, 1999; Bloor and Abrahams, 2002; Jones et al., 2003; Tohge et al., 2005; Yonekura-Sakakibara et al., 2007, 2008). Based on the in vitro substrate preferences of recombinant UGT proteins and the flavonol glycoside profiles of the Arabidopsis mutants deficient for glycosylation at these two positions, the 3-O-glycosylation is considered to be the first step of conjugation followed by 7-O-glycosylation. Five UGTs involved in flavonol glycosylation have been identified: UGT78D1, UGT78D2, UGT78D3, UGT73C6, and UGT89C1 (Fig. 1) (Jones et al., 2003; Tohge et al., 2005; Yonekura-Sakakibara et al., 2007, 2008). In leaves, UGT78D1 and UGT78D2 are the major 3-O-glycosyltransferases, whereas UGT78D3 makes only a minor contribution to the overall 3-O-glycosylation (Jones et al., 2003; Tohge et al., 2005; Yonekura-Sakakibara et al., 2008). UGT73C6 contributes to 7-O-glucosylation in leaves, but only trace amounts of flavonols are 7-O-glucosylated. Instead, 7-O-rhamnosylation catalysed by UGT89C1 is the major form of 7-O-conjugation (Yonekura-Sakakibara et al., 2007).
The phenylpropanoid and flavonoid biosynthesis pathways are subject to multiple levels of control. They are dependent on developmental stages and tissues as well as on exogenous stimuli, in particular on irradiation, nutrient deprivation, and temperature (Kubasek et al., 1992; Taylor and Grotewold, 2005; Kaffarnik et al., 2006; Winkel-Shirley, 2006; Olsen et al., 2009; Götz et al., 2010). Both post-transcriptional modification and transcriptional controls have been implicated in these different types of regulation. Several MYB-related and bZIP-type transcription factors regulating flavonol biosnythesis have been identified, which themselves are subject to transcriptional control (Franken et al., 1991; Pairoba and Walbot, 2003; Ulm et al., 2004; Ramsay and Glover, 2005; Tohge et al., 2005; Quattrocchio et al., 2006; Winkel-Shirley, 2006; Stracke et al., 2007; Dubos et al., 2008; Lillo et al., 2008; Olsen et al., 2009; Stracke et al., 2010). In particular, PAL, as the committed step into the phenylpropanoid pathway, has been shown to be metabolically regulated through negative feedback by cinnamic acid on PAL transcription and on enzyme activity (Lamb, 1979; Bolwell et al., 1986; Mavandad et al., 1990; Blount et al., 2000).
Here, an Arabidopsis ugt78d1 ugt78d2 double mutant was characterized that strongly compromised initial 3-O-glycosylation of flavonols. However, instead of an accumulation of flavonol aglycones or alternative conjugations by UGT78D3 or other unknown 3-O-glycosyltransferase in this line, the biosynthesis of flavonols was strongly repressed. Other branches of the phenylpropanoid pathway were not or only marginally affected. This reduced biosynthesis of flavonols was correlated with a repression of both flavonol biosynthetic genes and PAL genes, and of PAL enzyme activity. Further genetic and molecular analyses suggested a novel feedback repression of PAL that was directly related to the strongly compromised initial flavonol-3-O-conjugation, yet dependent on the capacity for flavonol aglycone formation.
Materials and methods
Plant lines and growth conditions
Mutant lines were originally obtained from NASC and ABRC stock centres and have been described elsewhere (Jones et al., 2003; Tohge et al., 2005; Yonekura-Sakakibara et al., 2007, 2008; Bashandy et al., 2009; Stracke et al., 2009). ugt78d1 (SAIL_568F08), ugt78d2 (SALK_049338), ugt73c6 (SAIL_525H07), ugt89c1 (SALK_071113), and tt4 (SALK_020583) mutants have the Columbia wild-type (Col-0) background, while fls1 (RIKEN_PST16145) has been generated in the Nössen accession. Double and triple mutant combinations were verified by PCR-based genotyping of the knockout insertions. Overexpression lines were established in Col-0 using vectors pAlligator2 and pK2GW7 (Karimi et al., 2002; Bensmihen et al., 2004) to constitutively express UGT78D1 and UGT78D2, respectively. Plants were grown in a growth chamber at 22°C employing a 16/8 h photoperiod and 100–120 μmol m−2 s−1 irradiance. Plants were usually grown on soil, except for flavonol quantification in roots and for β-glucuronidase (GUS) reporter analyses, for which plants were cultured on half-strength Murashige and Skoog medium (M5524, Sigma GmbH, Germany) containing 1.5% sucrose and 0.6% phyto-agar.
For naringenin feeding experiments, seeds were surface sterilized and planted on half-strength MS agar medium supplemented with 1% sucrose; seedlings were allowed to grow for 12 d with an 11 h photoperiod. Then seedlings were transferred to 250 ml beakers containing half-strength MS medium with 1% sucrose and 100 μM naringenin (Roth, Karlsruhe, Germany). After a 45 h incubation under continuous light with gentle shaking, the plantlets were rinsed with water and the rosettes were harvested for further analyses.
Histochemical GUS assay
Genomic fragments upstream of the start codon of both UGT78D isoforms (–1 bp to –1469 bp for UGT78D1 or to –1481 bp for UGT78D2) were amplified from Col-0 genomic DNA by PCR and introduced into vector pBGWFS7 (Karimi et al., 2002), forming UGT78Dpro:GUS-GFP fusions, which were transformed into Col-0 plants. Histochemical analysis of the GUS reporter gene was performed as described before (Deruère et al., 1999).
Extraction of flavonol metabolites for HPLC analysis
Different plant materials were collected for flavonol extraction. Extracts were prepared by incubating ground (in liquid N2) plant material with methanol (1 ml per 100 mg) for 1 h with moderate rotation at 4°C. Except for the naringenin feeding experiment, naringenin was used as internal standard for flavonol quantification. The extracts were then clarified by centrifugation at 14 000 g for 10 min. One-third volume of distilled water was added to the supernatant, and centrifugation as above removed chlorophyll and lipids. For flavonol aglycone analysis, 100 μl of cleared extract was hydrolysed by adding an equal volume of 2 N HCl and incubating at 70°C for 40 min. Then 100 μl of methanol was added to prevent the precipitation of aglycones. Samples were centrifuged at 14 000 rpm for 15 min. A total of 20 μl of the cleared extract was analysed by high-pressure liquid chromatography (HPLC).
HPLC analysis of flavonols and sinapate esters
Samples were analysed using a reverse phase HPLC system at a flow rate of 1 ml min−1 with a 45 min linear gradient starting at 100% solution A to 100% B; 100% B was maintained for 5 min. Solvent A was 2% formic acid with 0.1% ammonium formate, and solvent B was 88% methanol with 0.1% ammonium formate. According to previous work, absorbance at 280 nm was used for detection (Turunen et al., 1999). The flavonol aglycones and sinapate esters were identified by the diode array spectra and retention time in comparison with authentic standards. In addition to the diode array spectra, the published flavonol glycoside patterns of the ugt78d1 and ugt78d2 single mutants were used for flavonol glycoside identification (Jones et al., 2003; Tohge et al., 2005).
Quantitative RT-PCR
Total RNA was isolated with Trizol Reagent (Invitrogen, Darmstadt, Germany) according to the manufacturer’s instructions. The first-strand cDNAs were synthesized using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Darmstadt, Germany). Data were collected in accordance with the 7500 real-time PCR system (Applied Biosystems). The experiments were repeated three times with independently cultured plant materials. S16 (At5g18380) and TUB9 (At4g20890) were used as reference genes; their stability was tested using geNorm and shown to be suitable as reference genes under these experimental condition (Vandesompele et al., 2002). The gene expression was compared between genotypes by the paired t-test. The paired testing takes into account the dependency of the data by the experimental design. The mean difference of the log10 intensities, which is identical to the log fold change, is tested against zero. In addition, t-test-based 95% confidence intervals (CIs) were calculated for the mean difference of the log intensities. The results were back-transformed by taking the power to base 10 to obtain the fold change and a 95% CI for the fold change. For all calculations, the R software was employed (R-Development-Core-Team, 2009).
The absolute expression levels of the PAL and 4CL isoforms analysed were different in Col-0 leaves. The Ct values of PAL1, PAL2, PAL3, PAL4, 4CL1, 4CL2, and 4CL3 were ∼25, 30, 34, 35, 32, 39, and 32 cycles, respectively, under the quantitative RT-PCR conditions used, indicating low (≥30) or undetectable transcript levels (≥34) for some isoforms.
Photometric determination of anthocyanins
Photometric determination of anthocyanins was performed according to a method published previously (Mehrtens et al., 2005). Anthocyanins of 3-week-old Arabidopsis leaves were extracted at 4°C for 1 h with moderate shaking after adding 250 μl of acidic methanol (1% HCl, w/v) to 50 mg of plant material, which had been homogenized on ice. The homogenate was clarified by centrifugation (14 000 rpm at room temperature for 5 min). Anthocyanins were quantified based on the absorption at 530 nm and 657 nm using Qanthocyanins=(A530–0.25×A657)×M−1, where Qanthocyanins is a corrected absorption value linearly correlated with the amount of anthocyanins, A530 and A657 are the absorptions at the indicated wavelengths, and M is the mass of the plant material used for extraction.
PAL activity assay
The assay method was modified from Olsen et al. (2008). Approximately 100 mg of fresh plant material was thoroughly homogenized with 3 ml of extraction buffer (containing 100 mM TRIS-HCl, pH 8.8, 12 mM 2-mercaptoethanol) using a mortar and pestle on ice. After centrifugation at 14 000 g for 10 min at 4 °C the supernatant was desalted using a Sephadex G25 column. The assays were performed at 37°C for 2 h in a mixture containing 500 μl of protein extract, 50 μl of 100 mM L-phenylalanine, and 450 μl of 100 mM TRIS-HCl (pH 8.8). The reaction was terminated by adding 50 μl of 5 M HCl and centrifuged at 14 000 g for 15 min. Control reactions were run without adding L-phenylalanine. The UV absorbance was recorded at 290 nm. Assays were performed in five technical repeats in each of the three independent experiments.
LC-ESI-MSn metabolite analysis
A 100 mg aliquot of freeze-dried Arabidopsis leaf powder was extracted with 500 μl of methanol containing 0.1 μg μl−1 4-methylumbelliferyl-β-D-glucuronide as an internal standard. Methanol was removed in a rotary vacuum concentrator and the extract was re-dissolved in 35 μl of water for analysis by liquid chromatography/electrospray ionization multistage mass spectrometry (LC-ESI-MSn). Metabolites were identified by their retention times, mass spectra, and product ion spectra in comparison with the data determined for authentic reference materials. Relative metabolite quantification was performed using the DataAnalysis 3.1 and QuantAnalysis 1.5 software (Bruker Daltonics, Bremen, Germany) normalizing all results to the internal standard.
Statistics
Statistical analyses of quantitative RT-PCR data were performed by the paired t-test using the R software package (R-Development-Core-Team, 2009). Other statistical analyses were performed using SAS 9.1 (SAS/STAT User’s Guide, Version 9.1, Cary NC, USA).
Results
Strongly reduced total flavonol content in leaves of the ugt78d1 ugt78d2 double mutant
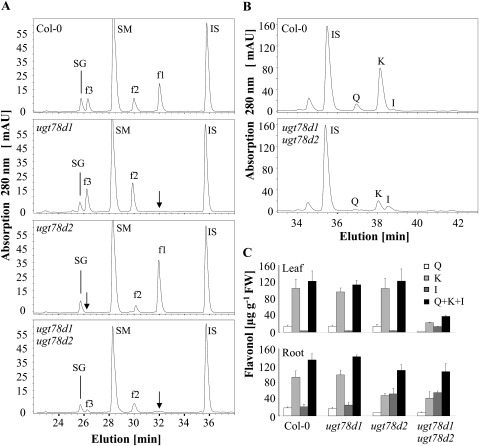
Three major flavonol glycosides, kaempferol 3-O-rhamnoside-7-O-rhamnoside (f1), kaempferol 3-O-glucoside-7-O-rhamnoside (f2), and kaempferol-3-O-[rhamnosyl (1→2 glucoside)]-7-O-rhamnoside (f3), were present in leaves of A. thaliana accession Columbia grown under non-stressed conditions (Bloor and Abrahams, 2002) (Fig. 2A). In ugt78d1 single mutant leaves, f1 was not detectable, whereas f2 and f3 accumulated to higher levels (Jones et al., 2003). However, in ugt78d2 single mutant leaves, f2 and f3 were strongly decreased, while f1 was increased (Tohge et al., 2005) (Fig. 2A). To quantify the total flavonol content in these ugt mutants, methanolic leaf extracts were hydrolysed and the released flavonol aglycones were analysed by HPLC. Although the flavonol glycoside pattern was altered in the ugt78d1 and ugt78d2 single mutants, both mutants maintained wild-type, total flavonol content (Fig. 2C). These observations suggest that UGT78D1 and UGT78D2 compete for flavonol aglycones as substrates, and the loss of flavonol 3-O-glycosylation in either single mutant is fully compensated by the remaining active flavonol 3-O-UGTs. However, the flavonol glycoside profile in leaves of the ugt78d1 ugt78d2 double mutant plants was severely altered; f2 and f3 were strongly reduced and f1 was not detectable. Furthermore, neither other forms of flavonol derivatives nor any flavonol aglycones were detected (Fig. 2A; data not shown). LC-MS analysis could also not reveal any flavonol aglycones (see Supplementary Table S1 available at JXB online). However, in contrast to the ugt single mutants, the total flavonol content was reduced; the ugt78d1 ugt78d2 double mutant contained less than one-third of the wild-type flavonol level. Kaempferol and quercetin contents were decreased to a greater extent, with only 21% and 18% of the wild-type content, respectively (Fig. 2B, C). However, isorhamnetin, a low abundant flavonol in wild-type plants, accumulated ∼3.5-fold in the ugt78d1 ugt78d2 double mutant (Fig. 2B, C). LC-MS analyses detected the strongly increased level of a compound corresponding to the molecular mass of an isorhamnetin-glucoside-rhamnoside, which may at least partly account for the increased isorhamnetin level in ugt78d1 ugt78d2 (see Supplementary Table S1). Enhanced isorhamnetin glucosides had also been described for the ugt78d2 single mutant, indicating the existence of an unknown 3-O-glucosyltransferase (Yonekura-Sakakibara et al., 2008).
Fig. 2.
Determination of flavonols from Col-0, ugt78d1, ugt78d2, and ugt78d1 ugt78d2 by HPLC analyses. (A) Representative HPLC diagrams of the flavonol glycoside profiles from leaves of 3-week-old plants. IS, internal standard (naringenin); SM, sinapoyl malate; SG, sinapoyl glucose; f1, f2, and f3 are as described in the legend of Fig. 1. Arrows indicate missing or repressed glycosides. (B) Representative HPLC diagrams of flavonol aglycone profiles from leaf extracts after acid hydrolysis. Q, quercetin; K, kaempferol; I, isorhamnetin. (C) Total flavonol aglycone quantification after acid hydrolysis of the methanolic extracts from leaves and roots. Means ±SD determined from three independent experiments are shown.
In summary, the loss of the major flavonol 3-O-glycosylation resulted in a strong reduction of total flavonol content. On the other hand, enhanced 3-O-glycosylation in UGT78D1 or UGT78D2 overexpression lines did not raise total flavonol levels (see Supplementary Fig. S1 at JXB online).
The expression pattern of UGT78D1 and UGT78D2 matches organ-specific reduction in flavonols
The reduction of total flavonol content was organ specific and different in inflorescences, stems, and leaves from that in roots. Similar to leaves, the total flavonol content was strongly reduced in both inflorescences and stems of ugt78d1 ugt78d2 plants as compared with the wild-type counterpart (data not shown). In roots, however, the total flavonol content of the double mutant was maintained at higher levels similar to the ugt78d2 single mutant (Fig. 2C).
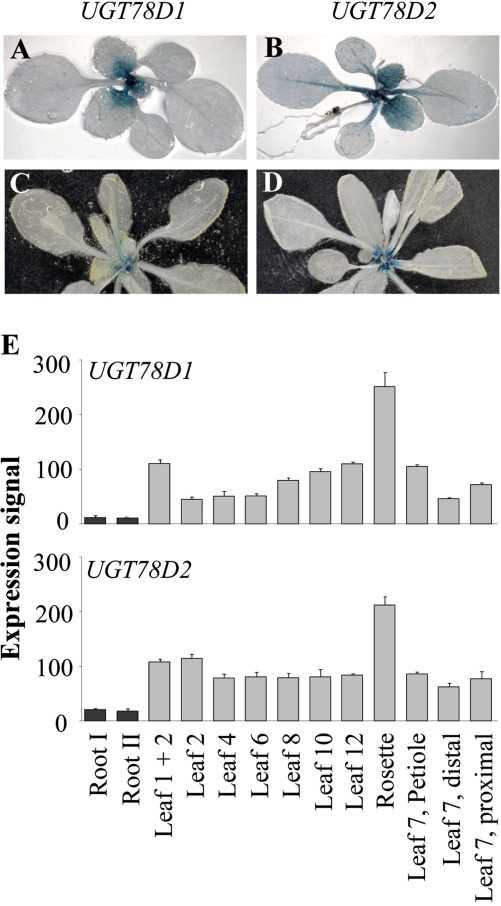
The expression pattern of both UGT78D1 and UGT78D2 positively correlated with the organ-dependent reduction in total flavonol content in the ugt double mutant. Two-week-old transgenic lines expressing the GUS reporter gene under the control of either the UGT78D1 or UGT78D2 promoter indicated that both genes were well expressed at the basal part of the young leaves (Fig. 3A, B). The expression of UGT78D2 was also observed along the central vein of the leaves as well as in the cotyledons (Fig. 3B). In 3-week-old plants, the expression of both UGT78D1 and UGT78D2 was lower compared with that of 2-week-old plants and predominantly at the basal part of leaf petioles (Fig. 3C, D). The expression of both UGT78D1 and UGT78D2 was very low in roots (data not shown; Jones et al., 2003). These findings were further corroborated by publicly available microarray data using the Arabidopsis eFP Browser (Schmid et al., 2005; Winter et al., 2007) (Fig. 3E).
Fig. 3.
Expression pattern of UGT78D1 and UGT78D2 indicated by staining transgenic lines harbouring promoter–GUS fusions. (A, B) Two-week-old plants. (C, D) Three-week-old plants. (E) Expression values of both UGT78D1 and UGT78D2 genes obtained using the Arabidopsis eFP Browser (bar.utoronto.ca) (Schmid et al., 2005; Winter et al., 2007) in roots, defined rosette leaves, complete rosettes, as well distal and proximal halves of leaf 7.
Flavonoid biosynthetic genes are transcriptionally down-regulated in ugt78d1 ugt78d2
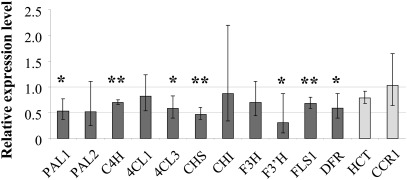
To examine whether the reduced flavonol content in leaves was accompanied by reduced transcript levels of the flavonoid biosynthetic genes, their mRNA abundance was assessed by quantitative RT-PCR in both ugt78d1 ugt78d2 double mutant and wild-type plants. In ugt78d1 ugt78d2, the transcripts of CHS, F3'H, FLS1, and DFR were significantly lowered to ∼50% in comparison with the wild-type counterpart (Fig. 4). However, the transcripts of CHI and F3H were not clearly different from wild-type levels. Furthermore, genes in the general phenylpropanoid pathway upstream of the flavonoid branch including PAL1, PAL2, PAL3, PAL4, 4CL1, 4CL2, 4CL3, and C4H were analysed. PAL1 and 4CL3 transcripts were significantly reduced to almost half in the double mutant compared with the wild type, whereas the reduction of C4H was less pronounced and 4CL1 mRNA levels were not altered. PAL2 showed only a tendency for repression, but it was more variable and also expressed to a lower extent than PAL1 in the wild type (Fig. 4). Eventually, the transcription of PAL3, PAL4, and 4CL2 in leaves was too low to allow an unambiguous evaluation in both the wild type and the mutant (Materials and methods). Thus, the committed steps of the phenylpropanoid pathway (PAL1) and of flavonoid biosynthesis (CHS) along with several intermediate steps were suppressed at the transcriptional level in ugt78d1 ugt78d2 to about one-half of wild-type expression. Neither PAL1 (and PAL2) nor CHS transcript levels were affected in either one of the single ugt mutants (see Supplementary Fig. S2 at JXB online).
Fig. 4.
Transcription of genes involved in phenylpropanoid and flavonoid biosynthetic pathways. Quantitative RT-PCR analyses were used to assess transcript levels of the indicated genes in the ugt78d1 ugt78d2 double mutant relative to Col-0 (Supplementary Table S2 at JXB online). Data were obtained from three independent experiments. Expression levels were normalized using S16 and TUB9 transcripts. Fold changes with 95% CIs are displayed. Asterisks indicate significance of the comparison with wild-type plants, *P < 0.05; **P < 0.01. The lower mean value of PAL2 indicated a tendency for repression of this gene which is expressed only to a low extent (P=0.0658).
Transcriptional control of biosynthetic genes
Several families of transcription factors, in particular R2R3-MYB, bHLH (basic helix–loop–helix), and WD40 proteins, as well as bZIP, WRKY, and MADS-box proteins interact and establish transcriptional networks to control the general phenylpropanoid and flavonoid biosynthetic genes (Borevitz et al., 2000; Zimmermann et al., 2004; Ramsay and Glover, 2005; Tohge et al., 2005; Quattrocchio et al., 2006; Stracke et al., 2007, 2010; Dubos et al., 2008; Hichri et al., 2011). Among them, Arabidopsis MYB11, MYB12, MYB111, or MYBL2 are flavonol-specific activators or repressors which are able to regulate flavonol biosynthesis independently from other cofactors (Czemmel et al., 2009). Since many of these transcription factors are controlled at the transcriptional level, several genes were analysed for deregulation in the ugt double mutant. However, neither MYB111, MYB12, MYBL2, PAP1/MYB75, PAP2/MYB90, nor HY5 (bZIP) transcript levels were significantly changed in parallel with the observed repression of the flavonoid biosynthetic genes (Supplementary Fig. S3 at JXB online). PAP2 was even induced, thus seemingly counteracting the suppression of the flavonoid branch.
Recently, miRNA156 was shown to affect the anthocyanin/flavonol ratio by targeting SPL9, a regulator of flavonoid-related transcriptional complexes (Gou et al., 2011). RNA interference could be also involved in the repression of flavonol biosynthesis by directly affecting the related genes. However, none of the repressed biosynthetic genes was listed as a confirmed or predicted target of known microRNAs (miRNAs) in A. thaliana (mpss.udel.edu/at/target.php). Furthermore, a blast search for known A. thaliana miRNAs derived from miRBase (www.mirbase.org) did not reveal any significant, high scoring hits. An independent search for homologous regions (20-mers allowing up to three mismatches) only identified imperfect overlaps within at most four genes (Y. Wang and G. Haberer, personal communication). Thus, a coordinated regulation of the repression based on direct RNA interference targeting the biosynthetic genes is unlikely.
Reduced PAL enzyme activity in ugt78d1 ugt78d2
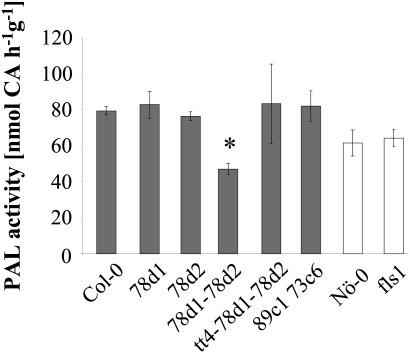
The decreased transcript levels of PAL1 in ugt78d1 ugt78d2 prompted examination of whether PAL activity was also reduced. PAL activity was assayed in total protein extracts from ugt78d1, ugt78d2, ugt78d1 ugt78d2, and wild-type leaves. Both ugt78d1 and ugt78d2 single mutants retained wild-type levels of PAL activity, whereas the ugt78d1 ugt78d2 double mutant showed a significant reduction to ∼60% (Fig. 5). However, the endogenous levels of the PAL substrate phenylalanine were not changed in the double mutant compared with the wild type (Supplementary Fig. S4 at JXB online).
Fig. 5.
Quantification of PAL activity of ugt78d1, ugt78d2, ugt78d1 ugt78d2, tt4 ugt78d1 ugt78d2, ugt89c1 ugt73c6, fls1, and wild-type (Col-0, Nö-0) plants. Crude protein extracts from leaves of 3-week-old plants were assayed for PAL activity. CA, cinnamic acid. The mean values ±SD of three independent biological samples (pools of 4–6 plants) are displayed. Col-0-based (dark) and Nö-0-derived (white bars) lines are indicated. *P < 0.05 (based on multiple comparisons of different mutants with the corresponding wild type using Dunnett’s t-tests). The experiment was repeated three times with similar results.
The influence of reduced PAL activity on non-flavonol branches of phenylpropanoid metabolism
In addition to the flavonol pathway, the repression of PAL activity in ugt78d1 ugt78d2 might also affect other phenylpropanoid-dependent branches; that is, the biosynthesis of anthocyanins, sinapate esters, and lignin. In another scenario affecting a single branch of the phenylpropanoid pathway, suppression of lignin biosynthesis has led to metabolic overflow and enhanced flavonoid production (Besseau et al., 2007).
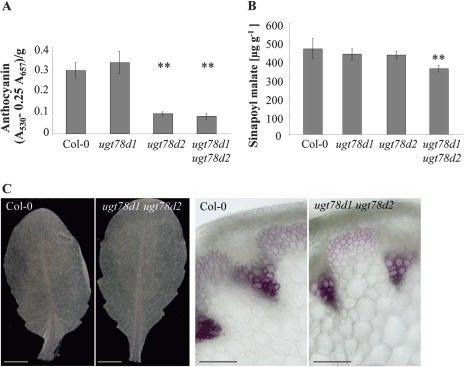
ugt78d1 accumulated essentially wild-type levels of total anthocyanins, while anthocyanin accumulation was already strongly suppressed in the ugt78d2 single mutant without any inhibitory effect on PAL expression (Figs 5, 6A) (Tohge et al., 2005). Since ugt78d1 ugt78d2 had low amounts of anthocyanin comparable with ugt78d2 (Fig. 6A), the repression of PAL did not further affect this downstream branch. However, DFR transcription was down-regulated in ugt78d1 ugt78d2 (Fig. 4).
Fig. 6.
Phenylpropanoid compounds of ugt78d1 ugt78d2 and Col-0 plants. (A) Photometric determination of the anthocyanin content in acidic methanolic leaf extracts of 3-week-old Arabidopsis plants: A530, absorption at 530 nm; A657, absorption at 657 nm. The mean values ±SD of three independent biological samples (pools of 4–6 plants) are displayed. **P < 0.01; paired t-tests. (B) Quantification of sinapoyl malate. The mean values ±SD of six independent biological samples (pools of 4–6 plants) are displayed. **P < 0.01; paired t-tests. This experiment was repeated three times with similar results. (C) Phloroglucinol-HCl-stained rosette leaves (left, bar=3 mm) from 3-week-old plants and hand cross-sections of inflorescence stems (directly below the third internode) (right, bar=100 μm) from 5-week-old plants.
Sinapate esters, particularly sinapoyl malate, are abundantly present in Arabidopsis leaves. The double mutant, in contrast to the single mutants, had a slightly, yet significantly, reduced sinapoyl malate content compared with the wild type (Fig. 6B). Sinapoyl glucose was also slightly reduced in the ugt double mutant (data not shown).
To test whether the lignin biosynthetic branch was affected, two key genes involved in lignin biosynthesis were analysed by quantitative RT-PCR. The single HCT (hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase) and the isoform CCR1 (cinnamoyl-CoA reductase Class I) are important in initiating the biosynthesis of monolignol precursors (and of other cinnamic acid derivatives) (HCT) and catalysing the first step of the monolignol-specific pathway (CCR1) in A. thaliana (Jones et al., 2001; Lavergeat et al., 2001; Raes et al., 2003; Besseau et al., 2007). Both HCT and CCR1 transcripts were not significantly affected in ugt78d1 ugt78d2 leaves (Fig. 4). In addition, changes in upstream steps contributing to lignin biosynthesis such as 4CL1, 4CL2, PAL3, and PAL4 (Raes et al., 2003) did not indicate transcriptional up-regulation (see above; Fig. 4). Thus, the biosynthesis of lignin precursors was essentially not altered at the transcriptional level. Furthermore, total lignin was examined histochemically using phloroglucinol-HCl staining of leaves and stems. The total flavonol content in inflorescence stems of ugt78d1 ugt78d2 was also reduced to a similar extent to that in leaves (data not shown). However, lignin deposits in both leaves and stems of ugt78d1 ugt78d2 were similar to wild-type levels (Fig. 6C). Finally, metabolites associated with the lignin biosynthesis pathway were assessed by LC-MS analysis. No immediate monolignol precursor was detected in the leaf extracts. Nevertheless, LC-MS analysis showed that the ugt78d1 ugt78d2 double mutant accumulated wild-type levels of the monolignol precursor-related derivatives p-coumaroyl-glucoside, p-coumaroyl-glucose-ester, and caffeoyl-glucose-ester (Supplementary Table S1 at JXB online). Thus, these results indicated that lignin biosynthesis was not significantly affected in the ugt double mutant.
In summary, these analyses demonstrated that there were at most marginal effects on non-flavonol-specific branches of phenylpropanoid biosyntheses in ugt78d1 ugt78d2; notably, there were no compensatory up-regulations.
The reduction of PAL expression of ugt78d1 ugt78d2 is dependent on flavonol aglycone formation
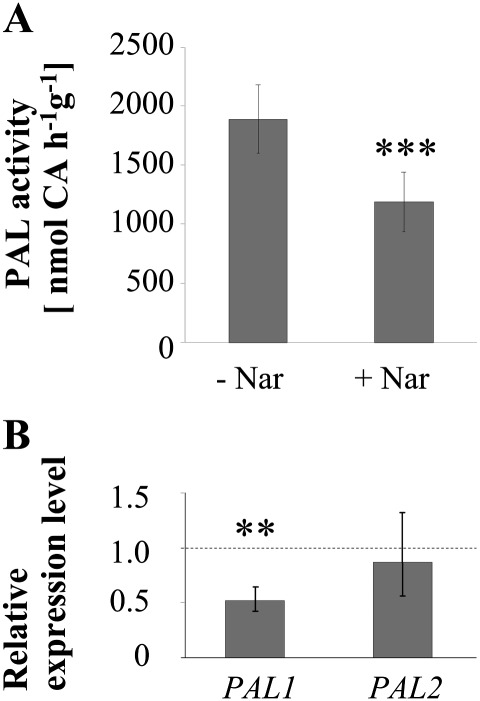
To examine whether flavonoids themselves could play a role in the flavonol-specific reduction in PAL expression, the tt4 ugt78d1 ugt78d2 triple mutant was employed. The triple mutant tt4 ugt78d1 ugt78d2 was blocked at the committed step into flavonoid biosynthesis in the ugt double mutant background (Fig. 1). In contrast to the ugt78d1 ugt78d2 line, tt4 ugt78d1 ugt78d2 as well as the parent tt4 mutant had wild-type PAL activity (Fig. 5; data not shown) and the transcripts of both PAL1 and PAL2 even showed a tendency towards being up-regulated in the triple mutant (Supplementary Fig. S5 at JXB online). Thus, the suppression of PAL expression in ugt78d1 ugt78d2 was released either by the loss of flavonoids or by the loss of CHS activity. To distinguish these two possibilities, a chemical complementation approach was employed. The feeding of naringenin—an intermediate downstream of CHS/TT4, but upstream of FLS—to tt4 ugt78d1 ugt78d2 could re-implement the inhibition of PAL expression. Naringenin feeding led to the repression of PAL activity and to the transcriptional down-regulation of PAL1 similar to the extent observed for the glycosyltransferase double mutant (Fig. 7). The transcription of PAL2 which was expressed to a much lower extent was not significantly affected under this condition (Fig. 7B). Flavonols had been formed after administration of naringenin, confirming the uptake and processing of the compound by tt4 ugt78d1 ugt78d2 (Supplementary Fig. S6 at JXB online). Thus, the repression of PAL in the 3-O-glycosylation-compromised ugt78d1 ugt78d2 double mutant was flavonoid-dependent and related to naringenin or downstream naringenin-dependent steps. To narrow down the key regulatory step further, additional mutants defective in reactions either directly upstream or downstream of 3-O-glycosylation were examined for effects on PAL expression.
Fig. 7.
Chemical complementation of tt4 ugt78d1 ugt78d2 with naringenin. (A) PAL activity of tt4 ugt78d1 ugt78d2 with and without feeding of naringenin. The mean values ±SD of six independent biological samples (pools of 4–6 plants) are displayed. CA, cinnamic acid. ***P < 0.001; paired t-tests. The experiment was repeated twice with similar results. (B) Expression of PAL1 and PAL2 genes in the tt4 ugt78d1 ugt78d2 triple mutant after naringenin feeding relative to the non-fed triple mutant by quantitative RT-PCR analysis. Fold changes with 95% CIs are displayed. Asterisks indicate significance of the difference from non-fed mutant plants: **P < 0.01. Data obtained from six biological sample samples (pools of 4–5 seedlings) are shown. The experiment was repeated twice with similar results.
The fls1 mutant severely blocks flavonol biosynthesis at its final step (i.e. the formation of the aglycone moiety) and is almost devoid of flavonols (Fig. 1) (Stracke et al., 2009). However, PAL activity was not reduced, and transcription of PAL1 and PAL2 was even enhanced in the fls1 background (Fig. 5; Supplementary Fig. S5 at JXB online).
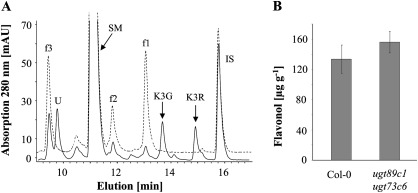
Flavonol 3-O-glycosylation is followed by 7-O-glycosylation. To examine whether a compromised flavonol 7-O-glycosylation would also lead to a reduction in flavonol content and a concomitant repression of PAL activity, the ugt89c1 ugt73c6 double mutant was generated (Fig. 1). The flavonol glycoside profile of ugt89c1 ugt73c6 leaves indicated a strongly suppressed 7-O-conjugation. The contents of the three abundant, 7-O-conjugated wild-type flavonol glycosides (f1, f2, and f3) were strongly decreased, whereas two flavonol 3-O-monoglycosides, kaempferol 3-O-glucoside and kaempferol 3-O-rhamnoside, accumulated (Fig. 8A). Yet, ugt89c1 ugt73c6 contained wild-type levels of total flavonols and had unchanged PAL enzyme activity (Figs 5, 8B).
Fig. 8.
Flavonol determination in ugt89c1 ugt73c6 and Col-0 leaves. (A) Representative HPLC diagrams of ugt89c1 ugt73c6 (solid line) and Col-0 (dashed line). Flavonols were extracted from the leaves of 3-week-old plants. IS, internal standard (naringenin); SM, sinapoyl malate; U, unknown peak; K3G, kaempferol 3-O-glucoside; K3R, kaempferol 3-O-rhamnoside. The identities of K3G and K3R were determined through comparison with authentic standards by HPLC analysis and then confirmed by LC-MS analysis (Supplementary Fig. S7 at JXB online). (B) Total flavonol aglycone quantification after acid hydrolysis of the leaf extracts. Each bar represents the mean ±SD as determined in three independent experiments. Abbreviations: CA, cinnamic acid; I, isorhamnetin; K, kaempferol; K3G, kaempferol 3-O-glucoside; K3R, kaempferol 3-O-rhamnoside; Q, quercetin; SM, sinapoyl malate; UGT, UDP-carbohydrate-dependent glycosyltransferase.
Discussion
Inhibition of flavonol biosynthesis upon a compromised 3-O-glycosylation
Flavonol biosynthesis is under developmental and environmental control in accordance with the diverse regulatory and protective functions of these secondary metabolites (Rohde et al., 2004; Winkel-Shirley, 2006; Hichri et al., 2011). Since flavonols constitute only one, usually minor, branch of phenylpropanoid-derived metabolites, their synthesis is also intensely regulated and integrated with other parts of the phenylpropanoid pathway. Flavonoid biosynthesis is regulated at the transcriptional level by ternary complexes comprising MYB, bHLH, and WD40 proteins, while MYB proteins that act independent of other cofactors specifically control the synthesis of flavonols. However, these transcription factors are subject to transcriptional control themselves (Baudry et al., 2004; Stracke et al., 2007, 2010; Hichri et al., 2011). A block in the lignin-directed branch led to a metabolic redirection and the accumulation of flavonoids (Besseau et al., 2007).
Here it is shown that a strongly compromised 3-O-glycosylation of flavonols resulted in a repression of flavonol biosynthesis itself, with ∼30% of residual total flavonols, which was specifically associated with the repression of transcripts of flavonol biosynthetic genes as well as the flavonol-related PAL genes and PAL enzyme activity of the general phenylpropanoid pathway (see below). However, it was crucial to observe that the repression of PAL was completely released upon blocking the committed step (tt4/chs) or the very last step of flavonol aglycone biosynthesis (fls1).
Specificity of inhibition of the flavonol branch
Flavonol biosynthesis is mostly dependent on PAL1 and PAL2; pal1 pal2 double mutants are almost devoid of flavonoids; that is, both anthocyanins and flavonols (Rohde et al., 2004; Huang et al., 2010). The pal1 pal2 mutant still contained ∼50% of PAL enzyme activity in leaves due to the activity of other isoforms (Rohde et al., 2004). The reduced flavonol biosynthesis in the ugt double mutant was specifically down-regulating the flavonol-related PAL1 and PAL2 transcription by ∼50% associated with a reduction in PAL activity by ∼40%. However, in comparison with the double pal1 pal2 knockout situation, this seemingly moderate reduction in the ugt78d1 ugt78d2 line is likely to represent a major and specific suppression of flavonol-related PAL activity.
Similar to PAL, four active 4CL genes have been identified in the genome of A. thaliana within the general phenylpropanoid pathway. The isoforms 4CL1 and 4CL2 are considered to be important for lignin biosynthesis, whereas 4CL3 is involved in flavonoid biosynthesis (Ehlting et al., 1999; Raes et al., 2003; Costa et al., 2005). Consistent with a specific repression of flavonoids, only the transcription of the flavonol-related 4CL3 was down-regulated in ugt78d1 ugt78d2 (Fig. 4).
Furthermore, the ugt78d1 ugt78d2-dependent suppression of the flavonol branch did not strongly affect other branches of the general phenylpropanoid pathway. In particular, lignin biosynthesis appeared not to be affected. This was in contrast to the reverse situation provoked by a repression of lignin biosynthesis. In this scenario, a redirection of the metabolic flux and an increased production of flavonoids have been found (Besseau et al., 2007). These differing consequences from repressing lignin versus flavonoid biosynthesis may well be due to the fact that the metabolic flux into the lignin branch is far higher than into flavonoids. The specificity of flavonol-related PAL suppression is also corroborated by the finding that the concentration of phenylalanine was not changed in the ugt double mutant. In contrast, in a pal1 or pal2 knockout line, it was enhanced 3-fold (Rohde et al., 2004).
Multiple control of PAL activity
PAL, the committed step of the general phenylpropanoid pathway, is subject to multiple levels of control at both transcription and enzyme activity. Importantly, at least in A. thaliana, a high correlation between transcription and enzyme activity has been found (Olsen et al., 2008). There is strong evidence that the flavonol biosynthetic enzymes and the upstream enzymes of the general phenylpropanoid pathway are organized as metabolons (Winkel, 2004). Thereby, the specific association of different PAL isoforms could be controlled, which might also depend on post-translational modifications of PALs such as phosphorylation (Allwood et al., 2002).
In addition to widely studied transcriptional networks in response to diverse stimuli such as light, UV-B irradiation, temperature, or nutrient deprivation (Ulm et al., 2004; Quattrocchio et al., 2006; Olsen et al., 2009), the phenylpropanoid biosynthetic pathway is known to be metabolically regulated by phenylpropanoid intermediates. The transcription of CHS is inhibited by cinnamic acid, but stimulated by p-coumaric acid (Loake et al., 1991, 1992). PAL activity and transcription of PAL genes have been shown to be negatively regulated by its product trans-cinnamic acid either via exogenous application of the compound or via blocking the downstream, cinnamic acid-metabolizing C4H activity (Lamb, 1979; Bolwell et al., 1986; Mavandad et al., 1990; Blount et al., 2000) (Fig. 1). Isolated tomato PAL isoforms were differentially sensitive in vitro to various phenylpropanoid compounds including cinnamic acid, suggesting an allosteric inhibition (Sarma et al., 1998). However, a further mechanistic explanation for these feedback inhibitions, in particular concerning the transcriptional suppression, has not yet been found.
This work provides genetic evidence for an additional, novel feedback regulation, which is not dependent on these known control mechanisms to repress PAL activity and flavonol biosynthesis. Suppression of flavonol biosynthesis provoked by a compromised 3-O-glycosylation led to the repression of flavonol biosynthesis along with the inhibition of flavonol biosynthetic genes including PAL1 (PAL2). However, a complete or almost complete block of flavonol biosynthesis in tt4 or fls1 mutant backgrounds released this suppression, although it eliminated the metabolite flux into the flavonol branch even more drastically than the ugt double mutant. Thus, these data suggest that the formation of free flavonol aglycones is required for this feedback inhibition (Fig. 1). Currently, a detailed mechanism for this regulation cannot be provided. However, in the case of the hydrophobic flavonol aglycones, two major, non-exclusive scenarios can be envisaged.
First, the proposed flavonol metabolon could be directly influenced in situ through binding of nascent, non-glycosylated flavonols to PAL or other components, which could transmit the repression by direct inhibition or by inducing secondary suppressive modifications. Such a scenario would be favoured as soon as the subsequent glycosylation is compromised, as evidenced in the ugt double mutant. Only minute, non-accumulating amounts of flavonol aglycones would be required, which could escape detection. In addition, the specificity of the regulation confining the repression basically to the flavonol branch and flavonol-related PAL activity could be achieved in this model due to the in situ interaction. At the protein level, flavonols had been shown to inhibit PAL activity only in an unspecific manner or only weakly at rather high aglycone concentrations (Sato and Sankawa, 1983; Sarma et al., 1998). Therefore, these in vitro studies may not directly translate to the situation of the metabolon in planta, but support such a possible interaction. Since PAL repression and in particular pal1 knockout have been demonstrated to lead to a suppression of CHS and DFR transcription (Rohde et al., 2004), the flavonol-dependent repression of PAL activity could promote a correlated down-regulation of the transcription of these genes, with PAL being a major coordinating node through an as yet unknown link to transcription.
Secondly, since flavonols have been also localized to the nucleus (Saslowsky et al., 2005), the transcriptional repressions could also be dependent on this property. Flavonols could interact with and block known transcription factors. Such a scenario could also be mediated by a dual function of flavonol-related enzymes binding to flavonols in situ and subsequently being translocated to the nucleus. Indeed, flavonol biosynthetic enzymes, specifically CHS and CHI, had been detected in the nucleus (Saslowsky et al., 2005). Thereby, the flavonol biosynthetic enzymes could regulate nuclear transcription, for example through interactions with the canonical flavonol-specific transcription factors. There are precedents for such nuclear functions of metabolic enzymes, such as the signalling dependent on hexokinase 1 in plants or on lactate dehydrogenase A and glyceraldehyde-3-phosphate dehydrogenase in mammalian cells (Kim and Dang, 2005; Cho et al., 2006).
Interestingly, the regulation of transcriptional activators such as PAP1, MYB111, and HY5 as well as the repressor MYBL2 was not found in the ugt double mutant as has been shown in other cases, where their genes were themselves regulated at the transcriptional level resulting in a concomitant up- or down-regulation of flavonol biosynthesis (Ulm et al., 2004; Ramsay and Glover, 2005; Tohge et al., 2005; Quattrocchio et al., 2006; Stracke et al., 2007, 2010; Olsen et al., 2009). Another, alternative regulation involving small RNAs appeared to be unlikely as well, since none of the regulated genes is a target of a known Arabidopsis miRNA nor do these genes share extended homologous regions among themselves, which could lead to a coordinated suppression of PAL and flavonol-related genes through RNA interference.
Physiological significance of the flavonol-dependent feedback inhibition
Flavonols are known to bind to proteins including histones or DNA polymerases and thus have the potential to interfere with gene regulation (Solimani, 1997; Mizushina et al., 2003; Ramadass et al., 2003; Peer et al., 2004). Such metabolite–protein interactions could be part of the mechanism of feedback control (see above), but they also provide an explanation for the potentially detrimental effects of the hydrophobic flavonol aglycones through (un)specific interactions with cellular proteins. Therefore, the accumulation of aglycones upon specifically blocking their initial 3-O-glycosylation is to be prevented by the flavonol-dependent feedback inhibition. This situation resembles a recently described regulation of chlorophyll biosynthesis, which also averts the accumulation of detrimental products. Repression of the final biosynthetic step catalysed by chlorophyll synthase, which conjugates the chlorophyllide core molecule through esterification with phytol, did not lead to the accumulation of precursors. Instead, analogous to the situation found here upon blocking flavonol conjugation, a specific repression of upstream enzyme activities in the chlorophyll branch as well as the committed step of biosynthesis of the porphyrin ring was found. This was also accompanied by a repression of several transcripts encoding these enzymes (Shalygo et al., 2009).
In conclusion, the flavonol-dependent feedback inhibition may serve as a means to avoid the accumulation of hydrophobic and potentially toxic flavonol aglycones upon a compromised flavonol-3-O-glycosylation. Genetic evidence is provided that residual flavonol aglycone formation is required for this feedback inhibition of the general phenylpropanoid pathway. In contrast, a complete block of flavonol biosynthesis and thus elimination of the metabolic flux does not lead to a repression of the flavonol biosynthetic capacity.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Total flavonol content in UGT78D1 and UGT78D2 overexpression lines in the Col-0 background.
Figure S2. Relative quantification of PAL1, PAL2, and CHS transcripts in the ugt78d1 and ugt78d2 single mutants compared with the wild type.
Figure S3. Transcript levels of transcription factor genes in the ugt double mutant versus the wild type.
Figure S4. Phenylalanine content in wild-type Col-0 and the ugt double mutant.
Figure S5. Transcription of PAL1 and PAL2 genes in fls1 and tt4 ugt78d1 ugt78d2 mutants relative to the wild type.
Figure S6. Flavonols detected in tt4 ugt78d1 ugt78d2 plantlets grown in liquid culture after naringenin feeding.
Figure S7. Confirmation of kaempferol 3-O-monoglucoside and kaempferol 3-O-monorhamnoside by LC-MS analysis.
Table S1. Additional phenylpropanoid metabolites detected in the ugt78d1 ugt78d2 double mutant and Col-0 leaves by LC-MS analysis.
Table S2. Primers used for real-time quantitative RT-PCR.
Acknowledgments
We would like to thank Ralf Stracke (Universität Bielefeld) for providing tt4 and ugt89c1 ugt73c6 seeds, Hagen Scherb (Helmholtz Zentrum München) for help with statistical analyses, Klaus Mayer and Georg Haberer (Helmholtz Zentrum München) for analysing potential RNA interference, Jörg Durner and Christian Lindermayr for critical reading, and Birgit Geist and Susanne Stich for excellent technical assistance. VSP was supported by a Kekulé fellowship of the Verband der Chemischen Industrie.
Glossary
Abbreviations
- CA
cinnamic acid
- I
isorhamnetin
- K
kaempferol
- K3G
kaempferol 3-O-glucoside
- K3R
kaempferol 3-O-rhamnoside
- Q
quercetin
- SM
sinapoyl malate
- UGT
UDP-carbohydrate-dependent glycosyltransferase
References
- Allwood EG, Davies DR, Gerrish C, Bolwell GP. Regulation of CDPKs, including identification of PAL kinase, in biotically stressed cells of French bean. Plant Molecular Biology. 2002;49:533–544. doi: 10.1023/a:1015502117870. [DOI] [PubMed] [Google Scholar]
- Bashandy T, Taconnat L, Renou JP, Meyer Y, Reichheld JP. Accumulation of flavonoids in an ntra ntrb mutant leads to tolerance to UV-C. Molecular Plant. 2009;2:249–258. doi: 10.1093/mp/ssn065. [DOI] [PubMed] [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. The Plant Journal. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- Bensmihen S, To A, Lambert G, Kroj T, Giraudat J, Parcy F. Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Letters. 2004;561:127–131. doi: 10.1016/S0014-5793(04)00148-6. [DOI] [PubMed] [Google Scholar]
- Besseau S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M. Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. The Plant Cell. 2007;19:148–162. doi: 10.1105/tpc.106.044495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloor SJ, Abrahams S. The structure of the major anthocyanin in Arabidopsis thaliana. Phytochemistry. 2002;59:343–346. doi: 10.1016/s0031-9422(01)00460-5. [DOI] [PubMed] [Google Scholar]
- Blount JW, Korth KL, Masoud SA, Rasmussen S, Lamb C, Dixon RA. Altering expression of cinnamic acid 4-hydroxylase in transgenic plants provides evidence for a feedback loop at the entry point into the phenylpropanoid pathway. Plant Physiology. 2000;122:107–116. doi: 10.1104/pp.122.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolwell GP, Cramer CL, Lamb CL, Schuch W, Dixon RA. L-Phenylalanine ammonia-lyase from Phaseolus vulgaris: modulation of the levels of active enzyme by trans-cinnamic acid. Planta. 1986;169:97–107. doi: 10.1007/BF01369780. [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. The Plant Cell. 2000;12:2383–2394. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles D, Lim EK, Poppenberger B, Vaistij FE. Glycosyltransferases of lipophilic small molecules. Annual Review of Plant Biology. 2006;57:567–597. doi: 10.1146/annurev.arplant.57.032905.105429. [DOI] [PubMed] [Google Scholar]
- Buer CS, Imin N, Djordjevic MA. Flavonoids: new roles for old molecules. Journal of Integrative Plant Biology. 2010;52:98–111. doi: 10.1111/j.1744-7909.2010.00905.x. [DOI] [PubMed] [Google Scholar]
- Cho YH, Yoo SD, Sheen J. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell. 2006;127:579–589. doi: 10.1016/j.cell.2006.09.028. [DOI] [PubMed] [Google Scholar]
- Cos P, Calomme M, Sindambiwe JB, De Bruyne T, Cimanga K, Pieters L, Vlietinck AJ, Vanden Berghe D. Cytotoxicity and lipid peroxidation-inhibiting activity of flavonoids. Planta Medica. 2001;67:515–519. doi: 10.1055/s-2001-16472. [DOI] [PubMed] [Google Scholar]
- Costa MA, Bedgar DL, Moinuddin SG, et al. Characterization in vitro and in vivo of the putative multigene 4-coumarate:CoA ligase network in Arabidopsis: syringyl lignin and sinapate/sinapyl alcohol derivative formation. Phytochemistry. 2005;66:2072–2091. doi: 10.1016/j.phytochem.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Czemmel S, Stracke R, Weisshaar B, Cordon N, Harris NN, Walker AR, Robinson SP, Bogs J. The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiology. 2009;151:1513–1530. doi: 10.1104/pp.109.142059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deruère J, Jackson K, Garbers C, Soll D, Delong A. The RCN1-encoded A subunit of protein phosphatase 2A increases phosphatase activity in vivo. The Plant Journal. 1999;20:389–399. doi: 10.1046/j.1365-313x.1999.00607.x. [DOI] [PubMed] [Google Scholar]
- Dixon RA, Steele CL. Flavonoids and isoflavonoids—a gold mine for metabolic engineering. Trends in Plant Science. 1999;4:394–400. doi: 10.1016/s1360-1385(99)01471-5. [DOI] [PubMed] [Google Scholar]
- Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. The Plant Journal. 2008;55:940–953. doi: 10.1111/j.1365-313X.2008.03564.x. [DOI] [PubMed] [Google Scholar]
- Ehlting J, Buttner D, Wang Q, Douglas CJ, Somssich IE, Kombrink E. Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. The Plant Journal. 1999;19:9–20. doi: 10.1046/j.1365-313x.1999.00491.x. [DOI] [PubMed] [Google Scholar]
- Franken P, Niesbach-Klosgen U, Weydemann U, Marechal-Drouard L, Saedler H, Wienand U. The duplicated chalcone synthase genes C2 and Whp (white pollen) of Zea mays are independently regulated; evidence for translational control of Whp expression by the anthocyanin intensifying gene in. EMBO Journal. 1991;10:2605–2612. doi: 10.1002/j.1460-2075.1991.tb07802.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz M, Albert A, Stich S, Heller W, Scherb H, Krins A, Langebartels C, Seidlitz HK, Ernst D. PAR modulation of the UV-dependent levels of flavonoid metabolites in Arabidopsis thaliana (L.) Heynh. leaf rosettes: cumulative effects after a whole vegetative growth period. Protoplasma. 2010;243:95–103. doi: 10.1007/s00709-009-0064-5. [DOI] [PubMed] [Google Scholar]
- Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. The Plant Cell. 2011;23:1512–1522. doi: 10.1105/tpc.111.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborne JB, Williams CA. Advances in flavonoid research since 1992. Phytochemistry. 2000;55:481–504. doi: 10.1016/s0031-9422(00)00235-1. [DOI] [PubMed] [Google Scholar]
- Harborne JB, Williams CA. Anthocyanins and other flavonoids. Natural Product Reports. 2001;18:310–333. doi: 10.1039/b006257j. [DOI] [PubMed] [Google Scholar]
- Harrison MJ. Signaling in the arbuscular mycorrhizal symbiosis. Annual Review of Microbiology. 2005;59:19–42. doi: 10.1146/annurev.micro.58.030603.123749. [DOI] [PubMed] [Google Scholar]
- Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. Journal of Experimental Botany. 2011;62:2465–2483. doi: 10.1093/jxb/erq442. [DOI] [PubMed] [Google Scholar]
- Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou YH, Yu JQ, Chen Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiology. 2010;153:1526–1538. doi: 10.1104/pp.110.157370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Ennos AR, Turner S. Cloning and characterization of irregular xylem4 (irx4): a severly lignin-deficient mutant of Arabidopsis. The Plant Journal. 2001;26:205–216. doi: 10.1046/j.1365-313x.2001.01021.x. [DOI] [PubMed] [Google Scholar]
- Jones P, Messner B, Nakajima J, Schäffner AR, Saito K. UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. Journal of Biological Chemistry. 2003;278:43910–43918. doi: 10.1074/jbc.M303523200. [DOI] [PubMed] [Google Scholar]
- Kaffarnik F, Seidlitz HK, Obermaier J, Sandermann H, Jr, Heller W. Environmental and developmental effects on the biosynthesis of UV-B screening pigments in Scots pine (Pinus sylvestris L.) needles. Plant, Cell and Environment. 2006;29:1484–1491. doi: 10.1111/j.1365-3040.2006.01518.x. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Kim JW, Dang CV. Multifaceted roles of glycolytic enzymes. Trends in Biochemical Sciences. 2005;30:142–150. doi: 10.1016/j.tibs.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Kubasek WL, Shirley BW, McKillop A, Goodman HM, Briggs W, Ausubel FM. Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. The Plant Cell. 1992;4:1229–1236. doi: 10.1105/tpc.4.10.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb CJ. Regulation of enzyme levels in phenylpropanoid biosynthesis: characterization of the modulation by light and pathway intermediates. Archives of Biochemistry and Biophysics. 1979;192:311–317. doi: 10.1016/0003-9861(79)90097-3. [DOI] [PubMed] [Google Scholar]
- Lauvergeat V, Lacomme C, Lacombe E, Lasserre E, Roby D, Grima-Pettenati J. Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry. 2001;57:1187–1195. doi: 10.1016/s0031-9422(01)00053-x. [DOI] [PubMed] [Google Scholar]
- Li J, Ou-Lee TM, Raba R, Amundson RG, Last RL. Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. The Plant Cell. 1993;5:171–179. doi: 10.1105/tpc.5.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Baldauf S, Lim EK, Bowles DJ. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. Journal of Biological Chemistry. 2001;276:4338–4343. doi: 10.1074/jbc.M007447200. [DOI] [PubMed] [Google Scholar]
- Lillo C, Lea US, Ruoff P. Nutrient depletion as a key factor for manipulating gene expression and product formation in different branches of the flavonoid pathway. Plant, Cell and Environment. 2008;31:587–601. doi: 10.1111/j.1365-3040.2007.01748.x. [DOI] [PubMed] [Google Scholar]
- Lin JK, Weng MS. Flavonoids as nutraceuticals. In: Grotewold E, editor. The science of flavonoids. New York: Springer Science+BusinessMedia, Inc; 2006. pp. 213–238. [Google Scholar]
- Loake GJ, Choudhary AD, Harrison MJ, Mavandad M, Lamb CJ, Dixon RA. Phenylpropanoid pathway intermediates regulate transient expression of a chalcone synthase gene promoter. The Plant Cell. 1991;3:829–840. doi: 10.1105/tpc.3.8.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loake GJ, Faktor O, Lamb CJ, Dixon RA. Combination of H-box [CCTACC(N)7CT] and G-box (CACGTG) cis elements is necessary for feed-forward stimulation of a chalcone synthase promoter by the phenylpropanoid-pathway intermediate p-coumaric acid. Proceedings of the National Academy of Sciences, USA. 1992;89:9230–9234. doi: 10.1073/pnas.89.19.9230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavandad M, Edwards R, Liang X, Lamb CJ, Dixon RA. Effects of trans-cinnamic acid on expression of the bean phenylalanine ammonia-lyase gene family. Plant Physiology. 1990;94:671–680. doi: 10.1104/pp.94.2.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiology. 2005;138:1083–1096. doi: 10.1104/pp.104.058032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meßner B, Thulke O, Schäffner AR. Arabidopsis glucosyltransferases with activities toward both endogenous and xenobiotic substrates. Planta. 2003;217:138–146. doi: 10.1007/s00425-002-0969-0. [DOI] [PubMed] [Google Scholar]
- Mizushina Y, Ishidoh T, Kamisuki S, Nakazawa S, Takemura M, Sugawara F, Yoshida H, Sakaguchi K. Flavonoid glycoside: a new inhibitor of eukaryotic DNA polymerase alpha and a new carrier for inhibitor-affinity chromatography. Biochemical and Biophysical Research Communications. 2003;301:480–487. doi: 10.1016/s0006-291x(02)03083-8. [DOI] [PubMed] [Google Scholar]
- Mo Y, Nagel C, Taylor LP. Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proceedings of the National Academy of Sciences, USA. 1992;89:7213–7217. doi: 10.1073/pnas.89.15.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira MR, Kanashiro A, Kabeya LM, Polizello AC, Azzolini AE, Curti C, Oliveira CA, T-do Amaral A, Lucisano-Valim YM. Neutrophil effector functions triggered by Fc-gamma and/or complement receptors are dependent on B-ring hydroxylation pattern and physicochemical properties of flavonols. Life Sciences. 2007;81:317–326. doi: 10.1016/j.lfs.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Olsen KM, Lea US, Slimestad R, Verheul M, Lillo C. Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. Journal of Plant Physiology. 2008;165:1491–1499. doi: 10.1016/j.jplph.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Olsen KM, Slimestad R, Lea US, Brede C, Lovdal T, Ruoff P, Verheul M, Lillo C. Temperature and nitrogen effects on regulators and products of the flavonoid pathway: experimental and kinetic model studies. Plant, Cell and Environment. 2009;32:286–299. doi: 10.1111/j.1365-3040.2008.01920.x. [DOI] [PubMed] [Google Scholar]
- Owens DK, Alerding AB, Crosby KC, Bandara AB, Westwood JH, Winkel BS. Functional analysis of a predicted flavonol synthase gene family in Arabidopsis. Plant Physiology. 2008;147:1046–1061. doi: 10.1104/pp.108.117457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairoba CF, Walbot V. Post-transcriptional regulation of expression of the Bronze2 gene of Zea mays L. Plant Molecular Biology. 2003;53:75–86. doi: 10.1023/B:PLAN.0000009267.76482.ce. [DOI] [PubMed] [Google Scholar]
- Peer WA, Bandyopadhyay A, Blakeslee JJ, Makam SN, Chen RJ, Masson PH, Murphy AS. Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. The Plant Cell. 2004;16:1898–1911. doi: 10.1105/tpc.021501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuß A, Stracke R, Weisshaar B, Hillebrecht A, Matern U, Martens S. Arabidopsis thaliana expresses a second functional flavonol synthase. FEBS Letters. 2009;583:1981–1986. doi: 10.1016/j.febslet.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Baudry A, Lepiniec L, Grotewold E. The regulation of flavonoid biosynthesis. In: Grotewold E, editor. The science of flavonoids. New York: Springer Science+BusinessMedia, Inc; 2006. pp. 97–122. [Google Scholar]
- R-Development-Core-Team . A language and environment for statistical computing. Foundation for Statistical Computing, Vienna, Austria; 2009. http://www.R-project.org. [Google Scholar]
- Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiology. 2003;133:1051–1071. doi: 10.1104/pp.103.026484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadass P, Meerarani P, Toborek M, Robertson LW, Hennig B. Dietary flavonoids modulate PCB-induced oxidative stress, CYP1A1 induction, and AhR–DNA binding activity in vascular endothelial cells. Toxicological Sciences. 2003;76:212–219. doi: 10.1093/toxsci/kfg227. [DOI] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ. MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends in Plant Science. 2005;10:63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Rohde A, Morreel K, Ralph J, et al. Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. The Plant Cell. 2004;16:2749–2771. doi: 10.1105/tpc.104.023705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santelia D, Henrichs S, Vincenzetti V, et al. Flavonoids redirect PIN-mediated polar auxin fluxes during root gravitropic responses. Journal of Biological Chemistry. 2008;283:31218–31226. doi: 10.1074/jbc.M710122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma AD, Sreelakshmi Y, Sharma R. Differential expression and properties of phenylalanine ammonia-lyase isoforms in tomato leaves. Phytochemistry. 1998;49:2233–2243. doi: 10.1016/s0031-9422(98)00336-7. [DOI] [PubMed] [Google Scholar]
- Saslowsky DE, Warek U, Winkel BS. Nuclear localization of flavonoid enzymes in Arabidopsis. Journal of Biological Chemistry. 2005;280:23735–23740. doi: 10.1074/jbc.M413506200. [DOI] [PubMed] [Google Scholar]
- Sato T, Sankawa U. Inhibition of phenylalanine ammonia-lyase by flavonoids. Chemical and Pharmaceutical Bulletin. 1983;31:149–155. [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nature Genetics. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Shalygo N, Czarnecki O, Peter E, Grimm B. Expression of chlorophyll synthase is also involved in feedback-control of chlorophyll biosynthesis. Plant Molecular Biology. 2009;71:425–436. doi: 10.1007/s11103-009-9532-8. [DOI] [PubMed] [Google Scholar]
- Solimani R. The flavonols quercetin, rutin and morin in DNA solution: UV-vis dichroic (and mid-infrared) analysis explain the possible association between the biopolymer and a nucleophilic vegetable-dye. Biochimica et Biophysica Acta. 1997;1336:281–294. doi: 10.1016/s0304-4165(97)00038-x. [DOI] [PubMed] [Google Scholar]
- Stracke R, De Vos RC, Bartelniewoehner L, Ishihara H, Sagasser M, Martens S, Weisshaar B. Metabolomic and genetic analyses of flavonol synthesis in Arabidopsis thaliana support the in vivo involvement of leucoanthocyanidin dioxygenase. Planta. 2009;229:427–445. doi: 10.1007/s00425-008-0841-y. [DOI] [PubMed] [Google Scholar]
- Stracke R, Favory JJ, Gruber H, Bartelniewoehner L, Bartels S, Binkert M, Funk M, Weisshaar B, Ulm R. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant, Cell and Environment. 2010;33:88–103. doi: 10.1111/j.1365-3040.2009.02061.x. [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. The Plant Journal. 2007;50:660–677. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor LP, Grotewold E. Flavonoids as developmental regulators. Current Opinion in Plant Biology. 2005;8:317–323. doi: 10.1016/j.pbi.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. The Plant Journal. 2005;42:218–235. doi: 10.1111/j.1365-313X.2005.02371.x. [DOI] [PubMed] [Google Scholar]
- Turunen M, Heller W, Stich S, Sandermann H, Sutinen ML, Norokorpi Y. The effects of UV exclusion on the soluble phenolics of young Scots pine seedlings in the subarctic. Environmental Pollution. 1999;106:219–228. doi: 10.1016/s0269-7491(99)00070-6. [DOI] [PubMed] [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Mate Z, Adam E, Oakeley EJ, Schäfer E, Nagy F. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2004;101:1397–1402. doi: 10.1073/pnas.0308044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002 doi: 10.1186/gb-2002-3-7-research0034. 3, RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit M, Pauli GF. Major flavonoids from Arabidopsis thaliana leaves. Journal of Natural Products. 1999;62:1301–1303. doi: 10.1021/np990080o. [DOI] [PubMed] [Google Scholar]
- Wasson AP, Pellerone FI, Mathesius U. Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. The Plant Cell. 2006;18:1617–1629. doi: 10.1105/tpc.105.038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel BS. Metabolic channeling in plants. Annual Review of Plant Biology. 2004;55:85–107. doi: 10.1146/annurev.arplant.55.031903.141714. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. The biosynthesis of flavonoids. In: Grotewold E, editor. The science of flavonoids. New York: Springer Science+BusinessMedia, Inc; 2006. pp. 71–95. [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An ‘Electronic Fluorescent Pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007 doi: 10.1371/journal.pone.0000718. 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Cao H, Wang Y, Zhao J, Wei X. Glycosylation of dietary flavonoids decreases the affinities for plasma protein. Journal of Agricultural and Food Chemistry. 2009;57:6642–6648. doi: 10.1021/jf901456u. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Niida R, Saito K. Identification of a flavonol 7-O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. Journal of Biological Chemistry. 2007;282:14932–14941. doi: 10.1074/jbc.M611498200. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, Saito K. Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene–metabolite correlations in. Arabidopsis. The Plant Cell. 2008;20:2160–2176. doi: 10.1105/tpc.108.058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. The Plant Journal. 2004;40:22–34. doi: 10.1111/j.1365-313X.2004.02183.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.