Abstract
Post-harvest ozone application has recently been shown to inhibit the onset of senescence symptoms on fleshy fruit and vegetables; however, the exact mechanism of action is yet unknown. To characterize the impact of ozone on the post-harvest performance of kiwifruit (Actinidia deliciosa cv. ‘Hayward’), fruits were cold stored (0 °C, 95% relative humidity) in a commercial ethylene-free room for 1, 3, or 5 months in the absence (control) or presence of ozone (0.3 μl l−1) and subsequently were allowed to ripen at a higher temperature (20 °C), herein defined as the shelf-life period, for up to 12 days. Ozone blocked ethylene production, delayed ripening, and stimulated antioxidant and anti-radical activities of fruits. Proteomic analysis using 1D-SDS-PAGE and mass spectrometry identified 102 kiwifruit proteins during ripening, which are mainly involved in energy, protein metabolism, defence, and cell structure. Ripening induced protein carbonylation in kiwifruit but this effect was depressed by ozone. A set of candidate kiwifruit proteins that are sensitive to carbonylation was also discovered. Overall, the present data indicate that ozone improved kiwifruit post-harvest behaviour, thus providing a first step towards understanding the active role of this molecule in fruit ripening.
Keywords: Actinidia deliciosa, antioxidants, anti-radical activity, ethylene, ozone, post-harvest storage, protein carbonylation
Introduction
Fruit ripening is a complex, genetically programmed senescence process, and its elucidation relies more on a holistic than on a single process study approach (Ramina et al., 2008). Fleshy fruits have been classified as climacteric or non-climacteric, depending on whether or not a fruit exhibits a peak in respiration and ethylene production during ripening (Giovannoni, 2004). The major area of ripening research involves the post-harvest management, since horticultural industry faces huge economic losses every year as a result of ethylene-induced senescence (Martínez-Romero et al., 2007). Therefore, understanding the ripening programme is crucial to control ripening under post-harvest conditions and thus to reduce massive spoilage of fruits during storage (Michailides and Manganaris, 2009).
Ozone (O3), also known as triatomic oxygen, is a naturally occurring highly reactive form of oxygen. Ozone signalling in the induction of cell death and defence genes and interaction with other reactive oxygen species and ethylene is well documented in plants (Castagna et al., 2007). In addition, recent studies have demonstrated the benefits of post-harvest ozone application on fleshy fruits and vegetables. However, the specific mode of action of this molecule is not yet fully understood. It has been suggested that ozone exposure prevents microbial spoilage and some diseases, decreases respiratory activity (Zhang et al., 2005), promotes ethylene oxidation in storage rooms (Skog and Chu, 2001), stimulates sucrose degradation and reduces the emission of volatile esters (Perez et al., 1999), induces the production of antioxidant compounds with health-promoting properties (Allende et al., 2007; Artés-Hernández et al., 2007), and modulates protein and enzymatic profiles (Baur et al., 2004; Zhang et al., 2005).
Proteomics represent a rapidly developing large-scale approach to globally investigate complex biological processes such as fruit ripening. It has recently been used to describe proteome changes during ripening in many fruit species, including grape (Deytieux et al., 2007; Giribaldi et al., 2007, 2010; Negri et al., 2008; Martínez-Esteso et al., 2011), orange (Muccilli et al., 2009), strawberry (Bianco et al., 2009), peach (Nilo et al., 2010; Zhang et al., 2011), pear (Pedreschi et al., 2007; Pedreschi et al., 2008), and tomato (Rocco et al., 2006; Faurobert et al., 2007; Page et al., 2010). Proteins undergo various post-translational modifications that can affect its activity, conformation, folding, distribution, stability, and therefore its function (Rinalducci et al., 2008). Specifically, mild oxidants, which are usually produced during fruit ripening (Pedreschi et al., 2008), signal through chemical reactions with specific atoms of target proteins resulting in covalent protein modifications, including oxidation, tyrosine nitration, S-nitrosylation, glutathionylation, and disulfide formation. Importantly, protein oxidation is expected to be a major component of the oxidative signalling that regulates many physiological outputs (Møller et al., 2007; Wong et al., 2010). Amongst a variety of methods for assessing oxidative-based protein modifications, protein carbonylation has been used (Job et al., 2005). With the employment of redox proteomic approaches, several carbonylated proteins have been identified during the senescence of apple (Qin et al., 2009a) and peach (Qin et al., 2009b). However, there is still a lack of information about the proteomic hallmarks during fruit ripening, especially when taking into consideration the comparatively low number of horticultural crops with their genomes completed (Hertog et al., 2011).
Kiwifruit is an economically important crop being highly appreciated for its high levels of bioactive compounds (i.e. antioxidants) that are closely related to its ability to tolerate prolonged cold storage (Tavarini et al., 2008). Currently, the cultivation of Actinidia and the post-harvest behaviour of kiwifruit are considered to be very important in Greece. The economic importance is due to the fact that Greece is one of the largest producers of kiwifruit in the world and over one-half of Greece’s kiwifruit production is exported. Besides such socioeconomic considerations, kiwifruit provides an excellent experimental model for investigating the post-harvest action of ozone. Indeed, kiwifruit displays non-climacteric behaviour at temperatures ≤10 °C (e.g. during cold storage) and climacteric behaviour at temperatures in the order of 20 °C (e.g. during ripening in most storage/conditioning rooms following cold storage, herein referred to as the shelf life) (Antunes, 2007). In addition, recent evidence suggests that ozone application during cold storage could influence the post-harvest life of kiwifruit by suppressing the development of stem-end rot disease caused by Botrytis cinerea (Barboni et al., 2010; Minas et al., 2010). On the other hand, a recent microarray analysis combined with transgenic approaches revealed the expression of a surprisingly large number of genes of unknown function in ripened kiwifruit (Atkinson et al., 2011), testifying to the complexity of the kiwifruit ripening process. In addition, the availability of the Actinidia expressed sequence tag (EST) database (Crowhurst et al., 2008) offers an opportunity to identify kiwifruit proteins during ripening, which, along with gene expression analysis, could globally add to the growing understanding of molecular changes underpinning climacteric fruit ripening.
This study characterized the physiological impact of ozone on qualitative and antioxidant-related properties of kiwifruit subjected to shelf-life conditions (20 °C), following short (1 month), intermediate (3 month), and extended (5 month) low-temperature storage (0 °C, 95% relative humidity). Also, it documents, as far as is known for the first time, the proteome of kiwifruit during ripening after removal from cold storage.
Materials and methods
Fruit material, experimental system, and sampling procedure
Kiwifruits (cv. ‘Hayward’), grown under standard cultural practices, were harvested from a commercial orchard (Meliki, Northern Greece) at the physiologically mature stage (Antunes, 2007) (mean weight: 95 ± 5 g, tissue firmness: 60.5 ± 2.3 N, soluble solids content: 7.9 ± 0.1% w/v). Subsequently, fruits were randomly divided into 19 lots of 30 fruits each. One lot was analysed at the time of harvest. The other lots (9 + 9) were subjected to cold storage (0 °C, 95% relative humidity) in the absence (control) or presence of continuously supplied gaseous ozone (0.3 μl l−1) (ozone treatment). In both cases, a Swingtherm catalytic reactor (model BS 500, Fruit Control Equipments, Milano, Italy) was used to control ethylene concentration at a desired level in the air circulating inside the cold storage room. Fruit were removed from cold storage rooms after 1, 3, or 5 months and subsequently were transferred to shelf-life conditions (20 °C) for up to 12 days (d).
Each lot was randomly divided into three 10-fruit sub-lots. From every sub-lot, after removal of the peel, four transverse and cylindrical pieces of flesh (∼4.0 g), were obtained from the equatorial region of each fruit, resulting in triplicate bulk samples per treatment. The first piece was used directly for assessment of quality parameters, the second and the third piece for the determination of fruit antioxidant profile and the fourth piece for the proteomic analysis, as described below.
Ethylene production and respiration rate were evaluated every 2 d of shelf life whilst physicochemical (tissue firmness, soluble solids content, and titratable acidity) and antioxidant properties (total antioxidant activity, ascorbic acid, and phenolic contents) were analysed after 1, 6, and 12 d of shelf life. Based on the antioxidant profiles, the anti-radical activity of kiwifruit extracts was tested at 1-d shelf life following 1, 3, or 5 months of cold storage. Finally, a proteomic approach and a Western blot (Oxyblot) analysis were used to characterize kiwifruit proteins during 1, 6, or 12 d of shelf life after 5 months of cold storage from control and ozone-treated fruit with contrasting ethylene production patterns. A scheme summarizing the experimental system and the sampling procedure is depicted in Supplementary Fig. S1 (available at JXB online).
Fruit quality and ripening parameters
Fruit weight was monitored in 20 independent fruits at harvest as well as after 1, 3, and 5 months of cold storage plus 1 d at shelf life, and weight loss was expressed as percentage of the initial weight at harvest. Tissue firmness was recorded by puncture measurements with a Chatillon penetrometer (Chatillon and Sons, New York, NY, USA) fitted with a flat 8-mm diameter probe. After the removal of a 1-mm-thick disc of skin at the fruit equator, the tip was inserted in two opposite sides and tissue firmness was expressed in Newtons (N). Soluble solids content was assessed in juice using a refractometer (Atago PR-1, Atago Co Ltd., Tokyo, Japan) and titratable acidity by titration of 5 g juice with 10 mM NaOH to pH 8.2.
Ethylene and respiration rate
Ethylene production and respiration rate were monitored at 1, 2, 4, 6, 8, 10, and 12 d of shelf life after removal from 1, 3, and 5 months of cold storage, respectively. Ethylene measurements were performed with a gas chromatograph (model 3300, Varian Analytical Instruments, Walnut Creek, CA, USA), equipped with a stainless-steel column filled with Porapak (length 100 cm, diameter 0.32 cm) at 50 °C and a flame-ionization detector at 120 °C, as described (Manganaris et al., 2007). Respiration rate was calculated from CO2 production in the gas phase of the jars, measured by an infrared gas analyser (Combo 280, David Bishop Instruments, UK).
Phytochemical analyses
Fruit material was extracted according to Minas et al. (2010) and the extract was used for the determination of phenol content and antioxidant activity, with the ferric ion reducing antioxidant power (FRAP) and the 1,1-diphenyl-2-picryl hydrazyl (DPPH) methods in a UV-Vis spectrophotometer (model UV-1700, Shimadzu, Kyoto, Japan). The amount of total phenolics was determined according to the Folin-Ciocalteu’s procedure (Scalbert et al., 1989; Skerget et al., 2005). Briefly, 0.5 ml extract was mixed with 2.5 ml of 1:10 diluted Folin–Ciocalteu’s phenol reagent, followed by 2.0 ml of 7.5% (w/v) Na2CO3. After 5 min at 50 °C, the absorbance was measured at 760 nm. Phenol content was estimated from a standard curve of gallic acid and results were expressed as μmol of gallic acid equivalents (GAE) of 100 g fresh weight (GAE, μmol 100 g−1 FW). For FRAP assay, a sample containing 3 ml freshly prepared FRAP solution (0.3 M acetate buffer (pH 3.6) containing 10 mM 2,4,6-tripyridyl-s-triazine and 40 mM FeCl3.6H2O) and 100 μl extract was incubated at 37 °C for 4 min and the absorbance was measured at 593 nm (Benzie and Strain, 1996). The absorbance change was converted into a FRAP value by relating the change of absorbance at 593 nm of the test sample to that of the standard solution of L-ascorbic acid and the results were expressed as μmol of ascorbic acid equivalent antioxidant capacity (AEAC) of 100 g FW (AEAC, μmol 100 g−1 FW). DPPH radical scavenging activity was determined according to a modified method of Brand-Williams et al. (1995). Briefly, 50 μl extract was added into 2.95 ml of 100 μmol l−1 DPPH methanolic solution, agitated, and kept in the dark at 20 °C. The decrease in absorbance was measured at 517 nm after 30 min and the results were expressed as μmol of ascorbic acid equivalent antioxidant capacity (AEAC) of 100 g FW (AEAC, μmol 100 g−1 FW). Ascorbic acid was measured with the Merck RQflex reflectometer set (Merck, Darmstadt, Germany) (Pantelidis et al., 2007) according to the protocol for kiwifruit (Ascorbic Acid in Kiwifruit, Merck) and the results were expressed as μmol of ascorbic acid of 100 g FW (μmol 100 g−1 FW).
Anti-radical activity
The protective effect of kiwifruit phenols against hydroxyl radicals (HO) generated by Fenton’s reagent was estimated with the DNA nicking assay (Hu and Kitts, 2001). In brief, in a total volume of 20 μl, 0.5 μg pBR322 plasmid DNA was mixed with 4 μl of 180 μM FeCl3.6H2O, 4 μl of 18 μM ascorbic acid, 4 μl of 81 mM H2O2, and phenolic extract (0.002 μmol GAE). The reaction system was incubated in a water bath at 37 °C for 1 h. Then, the whole mixture was loaded onto a 1.5% (w/v) agarose gel and evaluated as described below.
Following the synthesis and measurement of peroxynitrite (ONOO−, 302 nm, ϵ = 1670 M−1 cm−1) (Uppu and Pryor, 1996), the protective activity of kiwifruit extracts toward ONOO−-induced DNA damage was performed according to Barr and Gedamu (2003). In a total volume of 25 μl, the ONOO− was added in a reaction mixture containing 50 mM sodium phosphate buffer (pH 7.0) 10 mM NaCl, 0.1 mM diethylenetriaminepentaacetic acid, 0.5 μg intact pBR322 plasmid DNA and phenolic extract (0.0025 μmol GAE). After incubation (5 min), the DNA bands were separated on a 1.5% (w/v) agarose gel.
The inhibitory effect of phenolic extracts on DNA breakage induced by peroxyl radical (ROO) was evaluated as described by Lim et al. (2001). Here, 0.5 μg pBR322 plasmid DNA was mixed with 0.0025 μmol GAE of phenol extract in phosphate-buffered saline. An aliquot (2 μl) of 2,2′-azobis(2-amidinopropane) dihydrochloride were added to initiate the reaction (total volume 25 μl). The mixture was incubated for 2 h at 37 °C before being separated on a 1.5% (w/v) agarose gel.
In all cases, ethidium bromide-stained DNA bands were visualized under UV light, and the Image J software (National Institutes of Health; http://rsb.info.nih.gov/ij/) was employed to quantify DNA strand breaks. A correction factor of 1.4 applied to the quantitation of the signal with the supercoiled (form I) plasmid due to its lower affinity for ethidium bromide (Dong et al., 2006). All assays were run in triplicate and averaged.
Preparation of protein extracts
Fruit tissues without seeds (∼10 g) were ground in liquid nitrogen using a mortar and pestle. Total proteins were extracted from the resulting powder at 4 °C in 10 ml buffer containing 500 mM potassium phosphate, 1 mM EDTA, 1 M NaCl, 0.5 mM Triton X-100, Complete Mini protease inhibitor cocktail tablets (Roche Molecular Biochemicals), 60 U DNase I (Roche Diagnostics), and 6 U RNase A (Sigma). After 10 min at 4 °C, 20 mM dithiothreitol (DTT, Sigma) was added and the protein extracts were stirred (20 min at 4 °C) and centrifuged (5000 g for 5 min at 4°C). Cold ethanol was added to the supernatant until 20% (v/v) final concentration, the mixtures vortexed and stored at –20 °C for 2 h. The extracts were then centrifuged (15,000 g, 15 min, 4 °C). After removal of the pellet each supernatant was clarified by adding an equal volume of 20:80 ice-cold trichloroacetic acid/acetone containing 1 mM DTT and kept at –20 °C overnight. After centrifugation (15,000 g, 15 min, 4 °C), the precipitated protein pellet was washed three times with ice-cold acetone containing 10 mM DTT, washed once with an ice-cold solution of ethanol/ethyl acetate (1:1 v/v), and subsequently was dried under N2 flow. The proteins were resolubilized with the thiourea/urea lysis buffer as described (Harder et al., 1999; Catusse et al., 2008) containing 4% (v/v) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, 2% (v/v) Triton X-100, and 20 mM DTT. Protein concentrations were measured according to Bradford (1976) using a Bio-Rad protein assay kit (Kit II, cat. n. 500-0002). Bovine serum albumin (Sigma) was used as a standard.
1D-SDS-PAGE and data analysis
Protein samples were separated by 11.5% one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (1D-SDS-PAGE) as described by Laemmli (1970) and stained with Coomassie Brilliant Blue R250 solution (Bio-Rad). Triplicate gels were used for each sample. Stained gels were scanned with a GS-800 Calibrated densitometer (Bio-Rad) and analysed with the Quantity One software (version 4.7, Bio-Rad). Protein zones were auto detected (sensitivity, 150.00: lane width, 0.8 cm: minimum density, 0.10%: noise filter, 4.00: shoulder sensitivity, 1.10: size scale, 5) and, when needed, protein zones were identified manually, background subtracted and matched between biological and technical repeats, and normalized quantity determination of the zone volumes was performed (mode, relative quantity of zones in lane) along with statistical analysis.
Western blot analysis of carbonylated proteins
Electroblotting and membrane treatments were carried out as described (Tanou et al., 2009). The membranes were then treated with 0.5 mM 2,4-dinitrophenylhydrazine (DNPH) and probed using a primary antibody against DNPH (AbD Serotec, Oxford, UK) (1:3000). Immunoreactive bands of carbonylated proteins were detected using the WesternDot 625 Western Blot Kit (Invitrogen, Molecular Probes, Eugene, Oregon, USA). The relative quantitative UV-fluorescence signal of each lane of the Western blots was estimated with the Quantity One software. Protein oxidation levels were also quantified based on the detection of the reaction of DNPH with protein carbonyls to form protein hydrazones using a Shimadzu UV-1601 spectrophotometer according to Levine et al. (1990). Three biological replicates were performed for each assay.
Protein identification by mass spectrometry and database analysis
Protein zones were excised from gels and digested with trypsin as described (Tanou et al., 2010). Gel pieces were rinsed with water and acetonitrile, reduced with DTT, alkylated with iodoacetamide (Sigma), and incubated in a microtube overnight at 37 °C in the presence of 3 μl of 0.5 pmol/μl trypsin (sequencing grade, Roche) in 25 mM NH4HCO3. The tryptic fragments were extracted, dried, and reconstituted with 0.1% (v/v) formic acid. The peptides were loaded on a C18 column (Atlantis dC18, 3 μm, 75 μm × 150 mm Nano Ease, Waters) and eluted with a 5–60% linear gradient with water/acetonitrile (98:2, v/v) containing 0.1% (v/v) formic acid (buffer A) and water/acetonitrile (20:80, v/v) containing 0.1% (v/v) formic acid (buffer B) over 30 min at a flow rate of 200 nl min−1. Tryptic peptides were analysed by nano-liquid chromatography tandem mass spectrometry (LC-MS/MS, Q-TOF-Ultima Global equipped with a nano-ESI source coupled with a Cap LC nanoHPLC, Waters Micromass Waters, Saint Quentin en Yvelines, France) in the data dependent acquisition mode, allowing the selection of three precursor ions per survey scan. Only doubly and triply charged ions were selected for fragmentation over a mass range of 400–1300 m/z. A spray voltage of 3.2 kV was applied. The MS/MS raw data were processed (smooth 3/2 Savitzky Golay and no deisotoping) using the ProteinLynx Global Server 2.05 software (Waters) and peak lists were exported in the micromass pkl format. Peak lists of precursor and fragment ions were matched automatically to proteins in the National Center for Biotechnology Information non-redundant database (version 2008.05.10 (6,512,701 sequences; 2,221,612,072 residues; taxonomy: Viridiplantae, 481,095 sequences)) and the GenBank viridiplantae (EST Viridiplantae version 2008.03.14 (91,083,810 sequences; 15,967,802,572 residues; taxonomy: Viridiplantae, 91,083,810 sequences)), using a local Mascot version 2.2 program (Matrix Science, London, http://www.matrixscience.com) with the following parameters: trypsin specificity, one missed cleavage, carbamidomethyl cysteine and oxidation of methionine, and 0.2 Da mass tolerance on both precursor and fragment ions. To validate protein identification, only matches with scores above 46 or 63 (identity threshold for individual ion scores for protein or EST databanks, respectively), a threshold value calculated by the Mascot algorithm with the search parameters, were considered. All identified proteins had a MASCOT score greater than the significance level corresponding to P < 0.05. Moreover, among the positive matches, only protein identifications based on at least two different peptide sequences of more than six amino acids with an individual score above 20 were accepted. To validate protein identification with one single peptide (e.g., as in protein zone 19 and protein zone 25; see Supporting information, Active Links 1 and 2, respectively), the threshold score was set above the identity threshold (value of 46) with at least a series of five consecutive y or b fragments in the spectrum. These additional validation criteria are a good compromise to limit the number of false-positive matches without missing real proteins of interest. In some cases, the peptide masses and sequences obtained were blasted manually against the current databases.
Statistical analysis
Statistical analysis was carried out using the software package SPSS version 17.0 (SPSS, Chicago, IL, USA), by one-way analysis of variance significance (P < 0.05). For physiological analysis the averages of each treatment for every shelf-life period or time point after cold storage were compared by the Duncan’s multiple range test or pairwise Student’s t-test, respectively (significance level 95%). For 1D-PAGE and Western blot zone analysis, individual means were compared using Duncan’s multiple range test (significance level 95%).
Results
The impact of ozone on the ripening characteristics of kiwifruit
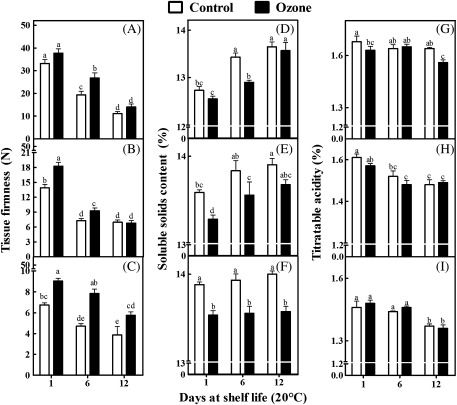
Kiwifruits exposed to ozone generally exhibited higher firmness retention compared with control fruits during shelf life (20 °C) after removal from 1, 3, and especially 5 months of cold storage (0 °C, 95% relative humidity) (Fig. 1A–C). An ozone-enriched cold storage atmosphere generally led to lower soluble solid contents in kiwifruits compared with control fruits (Fig. 1D–F), while titratable acidity generally remained unaffected by ozone (Fig. 1G–I). Also, ozone-treated kiwifruits exhibited lower weight loss compared with control fruits after 3 or 5 months of cold storage (Supplementary Fig. S2).
Fig. 1.
Tissue firmness (A–C), soluble solid content (SSC; D–F), and titratable acidity (TA; G–I) of kiwifruits (cv. ‘Hayward’) during shelf life (20 °C) for 1, 6, and 12 days after 1 (A,D,G), 3 (B,E,H), and 5 (C,F,I) months of cold storage (0 °C, 95% relative humidity) in the absence (control) or presence of ozone (0.3 μl l−1). Values are means ± SE (n = 30, n = 3, and n = 3 for fruit firmness, SSC, and TA, respectively). Bars for each shelf-life period with different letters are statistically significant (Duncan’s multiple range test, P < 0.05).
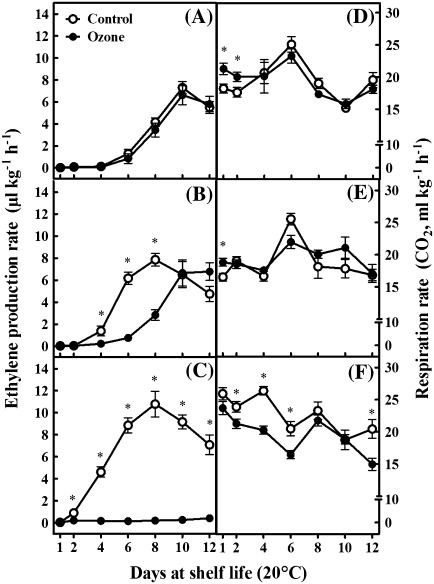
To investigate whether the ripening changes observed in response to ozone (Fig. 1) was ethylene-regulated, the production of this phytohormone was monitored in kiwifruits during shelf-life period. Ethylene production was not affected after 1 month of cold storage conducted in an ozone-enriched atmosphere compared with control fruits (Fig. 2A). However, ozone-treated fruits subjected to 3 months of cold storage exhibited a 2-d delay in the appearance of ethylene climacteric rise compared with untreated fruits (Fig. 2B). Intriguingly, ethylene remained at basal levels in ozone-treated kiwifruits exposed to a temperature of 20°C following 5 months of cold storage, without exhibiting the typical climacteric rise observed in control fruits (Fig. 2C), indicating that ozone blocked ethylene production in these conditions. It is noted that ethylene levels within the cold storage rooms were below the generally accepted levels for long-term kiwifruit storage (10 nl l−1), without differences among the cold storage conditions applied (data not shown).
Fig. 2.
Ethylene production (A–C) and respiration rate (D–F) of kiwifruits (cv. ‘Hayward’) during shelf life (20 °C) for 1, 2, 4, 6, 8, 10, and 12 days after 1 (A,D), 3 (B,E), and 5 (C,F) months of cold storage (0 °C, 95% relative humidity) in the absence (control) or presence of ozone (0.3 μl l−1). Values are means ± SE (n = 5). Asterisks indicate values that differ significantly at each time point (pairwise Student's t-test, P < 0.05).
A higher respiration rate was observed in ozone-treated fruits initially subjected to 1 month of cold storage and then to 1 or 2 d at 20 °C (Fig. 2D), as well as in fruits exposed to 3 months of cold storage followed by 1 d at 20 °C (Fig. 2E). In contrast, after 5 months of cold storage, ozone treatment caused a reduction in respiration rate compared with control fruits (Fig. 2F).
The impact of ozone on antioxidant and anti-radical ability of kiwifruit during ripening
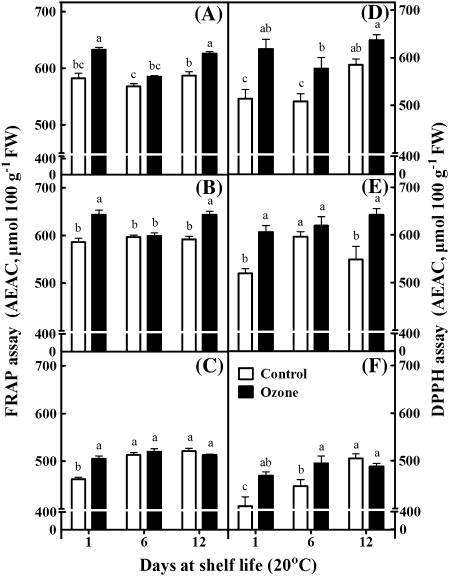
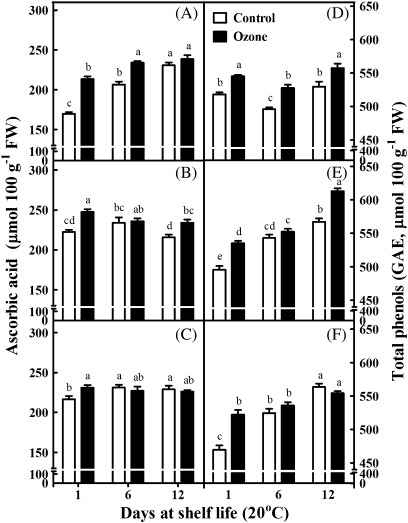
Fruits, as a multifunctional food, contain a large diversity of natural antioxidants (Sacchetti et al., 2005). In the present study, two different assay systems, namely the FRAP and DPPH assays, were employed to determine kiwifruit total antioxidant capacity (Fig. 3). The FRAP assay can measure only hydrophilic antioxidants. Therefore, compounds such as lipophilic carotenoids cannot be detected. On the other hand, the DPPH assay can detect antioxidants soluble in organic solvents such as alcohols or hydro-alcohol mixtures. However, this assay suffers from steric inaccessibility and a narrow linear range of absorbance versus concentration (Apak et al., 2007). The antioxidant activity, as evaluated with the FRAP assay, increased in kiwifruits after 12 d of shelf life following 1 month of cold storage (Fig. 3A). Also, the DPPH assay showed that fruit stored in an ozone-enriched cold atmosphere for 1 or 5 months and then exposed at 20 °C for 6 d exhibited a higher antioxidant activity than control fruits (Fig. 3D,F). Both assays indicated that the ozone treatment stimulated antioxidant capacity after 3 months of cold storage plus 12 d of shelf life (Fig. 3B,E). Notably, both antioxidant assays revealed that ozone treatment stimulated antioxidant synthesis after 1 d of shelf life (Fig. 3A–F). Furthermore, ozone-treated fruits generally exhibited higher levels of ascorbic acid content compared with control fruits, particularly after 1 d of shelf life (Fig. 4A–C). In addition, the total phenolic content was increased throughout the shelf-life period in fruits subjected to 1-month cold storage in an ozone-enriched atmosphere (Fig. 4D), as well as in fruits subjected to a 3-month exposure to ozone in cold conditions followed by 12 d of shelf life (Fig. 4E). As noted for total antioxidant capacity (Fig. 3A–F) and ascorbic acid content (Fig. 4A–C), ozone exposure for 1, 3, or 5 months in cold conditions and an additional 1 d of shelf life caused an induction in phenolic content (Fig. 4D–F).
Fig. 3.
Antioxidant activity, evaluated with the ferric ion reducing antioxidant power assay (A–C) and the 1,1-diphenyl-2-picryl hydrazyl assay (D–F), of kiwifruits (cv. ‘Hayward’) during shelf life (20 °C) for 1, 6, and 12 days after 1 (A,D), 3 (B,E), and 5 (C,F) months of cold storage (0 °C, 95% relative humidity) in the absence (control) or presence of ozone (0.3 μl l−1). Values are means ± SE (n = 6). Bars for each shelf-life period with different letters are statistically significant (Duncan’s multiple range test, P < 0.05).
Fig. 4.
Ascorbic acid (A–C) and total phenolic contents (D–F) of kiwifruits (cv. ‘Hayward’) during shelf life (20 °C) for 1, 6, and 12 days after 1 (A,D), 3 (B,E), and 5 (C,F) months of cold storage (0 °C, 95% relative humidity) in the absence (control) or presence of ozone (0.3 μl l−1). Values are means ± SE (n = 6). Bars for each shelf-life period with different letters are statistically significant (Duncan’s multiple range test, P < 0.05).
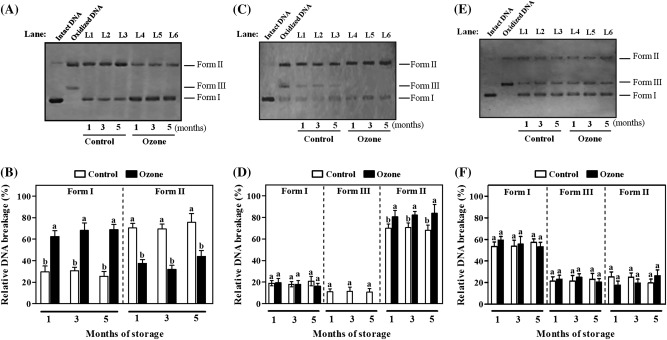
To strengthen the validity of the present results concerning the increased phenolic content in kiwifruit initially exposed to ozone and then subjected to 1 d of shelf life, the DNA nicking assay was employed to monitor the protective effect of kiwifruit’s phenolic extracts against injury by HO, ONOO−, and ROO (Fig. 5). Control experiments showed that incubation of intact plasmid DNA (Fig. 5A,C,E, Intact DNA, Form I) with HO produced by Fenton’s reaction mixture (Fe3/H2O2/ascorbic acid), authentic ONOO−, or ROO generated under thermal decomposition of dihydrochloride without kiwifruit extract caused open circular DNA (after a single-strand break) and linear DNA (after a double-strand break) (Fig. 5A,C,E, oxidized DNA, Forms II and III, respectively). The addition of extracts obtained from control fruits, which were stored for 1, 3, or 5 months in cold conditions plus 1 d of shelf-life period, substantially reduced radical-driven DNA strand breaking compared with positive controls (Fig. 5A,C,E, L1–L3). Both in-gel and quantitative analyses further showed that the corresponding phenolic-derived samples from ozone-treated kiwifruits (Fig. 5A,C,E, L4–L6) displayed higher ability to inhibit DNA damage induced by HO (Fig. 5A) or ONOO− (Fig. 5C), as inferred from a reduction in the intensity of oxidized Forms II (Fig. 5B) and III (Fig. 5D), respectively.
Fig. 5.
Inhibitory activity of kiwifruit (cv. ‘Hayward’) phenolic extracts against pBR322 plasmid DNA oxidation induced by hydroxyl radicals (HO) (A,B), peroxynitrite (ONOO−) (C,D), or peroxyl radicals (ROO) (E,F) during shelf life (20 °C) for 1 day after 1, 3, and 5 months of cold storage (0 °C, 95% relative humidity). (A,C,E) Forms of DNA in the presence of HO (A), ONOO− (C), or ROO (E) plus kiwifruit extracts: I, supercoiled DNA; II, open circular DNA; III, linear DNA. Lanes: Intact DNA, intact DNA; Oxidized DNA, DNA oxidized by HO (A), ONOO− (C), or ROO (E); L1–L6, kiwifruit extracts stored for the indicated times and sampled after 1 day. (B,D,F) Relative DNA breakage induced by HO (B), ONOO− (D), and ROO (F). The total intensity of the three DNA forms per lane is set to a value of 100. Values are means ± SE (n = 3). Bars for each shelf-life period with different letters are statistically significant (Duncan’s multiple range test, P < 0.05).
Nano-LC-MS/MS-based proteomic analysis in kiwifruit during ripening
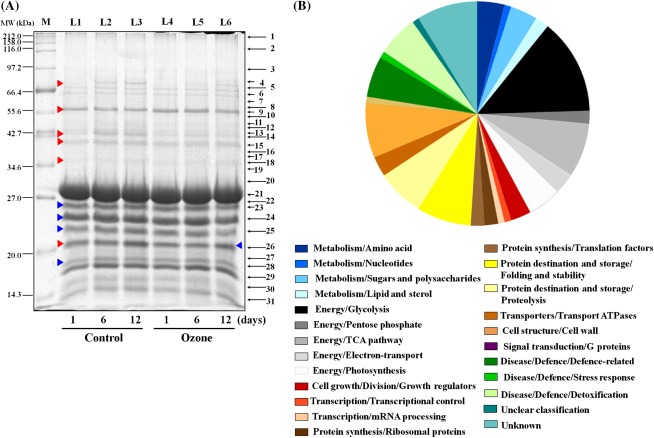
To complement this physiological study on the impact of ozone in kiwifruit post-harvest behaviour (Figs. 1–5), 1D-SDS-PAGE along with nano-LC-MS/MS was used to characterize the kiwifruit proteome during ripening at 20 °C. Experimental material from three independent biological replicates was collected at 1, 6, or 12 d shelf-life period following 5 months of cold storage in the absence or presence of ozone. This protocol allowed investigating the proteome of kiwifruits exhibiting contrasting ethylene patterns (Fig. 2C). A total of 31 protein zones were resolved by 1D-SDS-PAGE (1–31; marked in Fig. 6A) and variations in protein abundance within these zones upon comparing the fruit samples are shown in Supplementary Table S1. Interestingly, zones 4, 14, 15, 18, and 26 (Fig. 6A) were generally induced during ripening in both control and ozone-treated kiwifruits, whereas zones 4 and 14 showed decreased intensity of protein bands for fruit exposed to ozone compared with control fruit (Supplementary Table S1). In addition, protein zone 9 (Fig. 6A) remained unaffected during the ripening processes but was temporally and selectively modified by ozone (Supplementary Table S1). Protein zones (1–31, Fig. 6A) were excised and digested with trypsin and the resulting peptides were subjected to mass spectrometry analysis. Through this approach, a total of 102 different proteins were characterized following MASCOT searching, including two unidentified proteins (protein zones 3 and 30 in Fig. 6A). Table 1 summarizes the kiwifruit proteins identified by mass spectrometry analysis and a detailed list of these proteins is presented in Supplementary Table S2. An obvious limitation to the interpretation of the present results is the limited genomic background available in kiwifruit as only 19% of the present proteins were identified in Actinidia genus (Supplementary Table S2). Meanwhile, some of the identified proteins were present in more than one zone (Supplementary Table S2), indicating that these proteins are either products from different genes or arise from different post-translational protein modifications. The subcellular localization of these proteins was also evaluated based on database searches (Giribaldi et al., 2007). The largest portions of the identified kiwifruit proteins were localized in the cytosol (29%), chloroplast (17%), apoplast (14%), and mitochondria (9%) (Supplementary Table S2).
Fig. 6.
Characterization of kiwifruit (cv. ‘Hayward’) proteins during shelf life (20 °C) for 1, 6, and 12 days after cold storage (0 °C, 95% relative humidity) for 5 months in the absence (control) or presence of ozone (0.3 μl l−1). (A) Kiwifruit proteins (12 μg per lane) separated by 1D-SDS-PAGE and stained with Coomassie Brilliant Blue R250. For each treatment analysed, 1D-gels were run in triplicate. Protein zones were cut out, digested in gel using trypsin, and analysed by mass spectrometry and subsequent database searches. Lanes: M, molecular marker; L1–L6, kiwifruit proteins analysed at the indicated times. The localization of the protein zones (1–31) subjected to mass spectrometry analysis is reported on the right: the zones are labelled with the same numbers as in Table 1. Red and blue arrowheads indicate protein zones that were changed in abundance or carbonylated, respectively. (B) Functional categories of the identified kiwifruit proteins. The identified proteins were classified into various functional categories according to Bevan et al. (1998). The area for each category indicates the relative percentage of proteins in that category.
Table 1.
Identified kiwifruit proteins
Proteins are ontologically classified into functional categories (01–30) proposed by Bevan et al. (1998). Detailed information of protein identification is provided in Supplementary Table S2.
| Functional category | Protein name |
| 01 Metabolism (11) | |
| 01.01 | Ferredoxin-dependent glutamate synthase |
| 01.05 | β-Galactosidase |
| 01.01 | Alanine aminotransferase |
| 01.01 | Glutamate dehydrogenase |
| 01.05 | α-Galactosidase |
| 01.06 | Acyl-coenzyme A oxidase (2) |
| 01.01 | Methylcrotonoyl-CoA carboxylase |
| 01.05 | UDP-glucuronate decarboxylase |
| 01.05 | UDP-glucose-1-phosphate uridylyltransferase |
| 01.03 | Nucleoside diphosphate kinase |
| 02 Energy (32) | |
| 02.10 | Malate dehydrogenase (7) |
| 02.01 | Enolase (6) |
| 02.10 | Isocitrate dehydrogenase |
| 02.07 | Transaldolase (2) |
| 02.30 | Malate dehydrogenase (2) |
| 02.01 | Glyceraldehyde-3-phosphate dehydrogenase |
| 02.01 | Fructose-bisphosphate aldolase (5) |
| 02.30 | Fructose-bisphosphate aldolase (2) |
| 02.30 | Glyceraldehyde-3-phosphate dehydrogenase |
| 02.20 | NADPH quinone oxidoreductase |
| 02.20 | Fruit protein oxidoreductase NAD-binding (2) |
| 02.01 | Triosephosphate isomerase (2) |
| 03 Cell Growth (3) | |
| 03.26 | Agglutinin (2) |
| 03.26 | β−1,3−Glucanase |
| 04 Transcription (2) | |
| 04.05. 01.04 | Zinc finger (Ran-binding) family protein |
| 04.22 | Maturase |
| 05 Protein synthesis (4) | |
| 05.04 | Elongation factor |
| 05.01 | 60S ribosomal protein (2) |
| 05.04 | Eukaryotic translation initiation factor |
| 06 Protein destination and storage (15) | |
| 06.01 | HSP 70 (3) |
| 06.01 | HSP 70 ATP binding isoform |
| 06.01 | HSP 70 early responsive to dehydration |
| 06.13 | Leucine aminopeptidase (2) |
| 06.13 | Actinidin |
| 06.01 | HSP 70 dnaK protein |
| 06.01 | Cysteine protease |
| 06.01 | Protein disufide isomerase |
| 06.13 | Actinidin chain A |
| 06.13 | Chymopapain |
| 06.01 | Peptidyl-prolyl cis-trans isomerase |
| 06.13 | Cystatin |
| 07 Transporters (3) | |
| 07.22 | ATP synthase (F1) subunit β (2) |
| 07.22 | ATP synthase subunit E |
| 09 Cell structure (8) | |
| 09.01 | Pectinesterase (2) |
| 09.01 | Endo-β−1,4−glucanase |
| 09.01 | Xyloglucan endotransglycosylase |
| 09.01 | α-Expansin (2) |
| 09.01 | Pectinmethylesterase inhibitor (2) |
| 10 Signal transduction (1) | |
| 10.04.10 | Small GTP-binding protein |
| 11 Disease/defence (13) | |
| 11.06 | Glutathione reductase |
| 11.02 | Polygalacturonase inhibitor |
| 11.06 | Peroxidase |
| 11.06 | Lactoylglutathione lyase (3) |
| 11.02 | Kiwellin |
| 11.02 | Thaumatin (2) |
| 11.06 | 2-Cys peroxiredoxin |
| 11.02 | 2-Cys peroxiredoxin |
| 11.02 | Bet v 1 related allergen |
| 11.05 | Glycine-rich RNA binding protein |
| 12 Unclear classification (1) | |
| 12 | Phosphoinositide 5-phosphatase |
| 30-Unknown (9) | |
| 30 | Un-identified (2) |
| 30 | Hypothetical protein (6) |
| 30 | Serine-rich protein |
The identified kiwifruit proteins were then classified into functional categories (Fig. 6B), using gene classification resources proposed by Bevan et al. (1998). Identified kiwifruit proteins were mainly classified into the general categories of energy (31%), protein metabolism/modification (19%), plant defence (13%), and cell structure/cell wall (8%) (Fig. 6B). Several proteins associated with metabolism/amino acid (4%), metabolism/sugars and polysaccharides (4%), cell growth/division/growth regulators (3%), and transporters/transport ATPases (3%) were also identified. An over-representation of proteins specifically involved in glycolysis (14%), the tricarboxylic acid pathway (8%), photosynthesis (5%), and electron-transport (3%) was recorded in the general set of energy-related proteins, whereas the most common in the protein metabolism/modification category were proteins involved in protein destination and storage/folding and stability (8%) and protein destination and storage/proteolysis (7%). Identified proteins that belonging to plant defence include proteins involved in disease/defence/defence-related (6%) and disease/defence/detoxification (6%). The representative kiwifruit proteins in each functional class are listed in Table 1 and are discussed individually below.
Protein carbonylation profile in kiwifruit during ripening
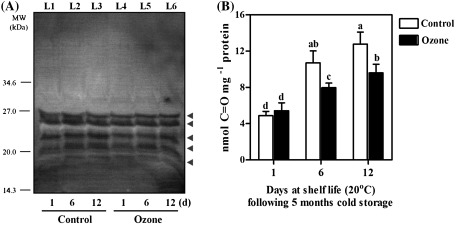
The strong impact of ripening and ozone on the antioxidant properties of kiwifruit extracts (Figs. 3–5) led to the investigation of whether the different conditions used to ripen the kiwifruits could be associated with specific features in protein oxidation patterns. To this end, kiwifruit proteins were sampled at the previously mentioned time points (Fig. 6A) and analysed in Western blot experiments based upon the specific detection of carbonylated proteins with anti-DNP antibodies. Analysis of kiwifruit proteins by 1D-SDS-PAGE followed by immunological detection using the Oxyblots revealed five carbonyl-reactive protein zones (Fig. 7A), indicating the involvement of oxidative stress in the kiwifruit ripening process. The carbonylated protein zones exhibited low molecular masses, ranging from ∼17.0 to 27.0 kDa. By matching immunopositive polypeptides (Fig. 7A) to corresponding zones on Coomassie Blue stained gels (Fig. 6A; zones 23, 24, 25, 26, and 28), 14 proteins were tentatively proposed as representing candidate carbonylated proteins (Supplementary Table S2). Quantitation of the immunoblot results by densitometry revealed an overall increased level of protein carbonylation during kiwifruit ripening (Supplementary Fig. S3). In agreement with this, quantitative measurements of protein carbonyl group content (Fig. 7B) confirmed that the extent of protein carbonylation increased during the ripening process. However, the protein carbonyl level at 6 and 12 d of shelf life following 5 months of cold storage was lower in ozone-treated fruits as compared with that of control fruits.
Fig. 7.
Protein carbonylation in kiwifruits (cv. ‘Hayward’) during shelf life (20 °C) for 1, 6, and 12 days after cold storage (0 °C, 95% relative humidity) for 5 months in the absence (control) or presence of ozone (0.3 μl l−1). (A) Oxidatively modified proteins detected with Oxyblot analysis (see Materials and methods). Kiwifruit proteins that were identified by mass spectrometry from carbonyl-reactive zones (arrowheads) are listed in Supplementary Table S2 as ‘CC; candidate carbonylated’ and also indicated in Fig. 6A. The gel is representative of three independent analyses. Lanes: L1–L6, kiwifruit proteins analysed at the indicated times. (B) Protein oxidation measured as carbonyl group content. Values are means ± SE (n = 3). Bars followed by the same letter are not statistically significant (Duncan’s multiple range test, P < 0.05).
Discussion
Ozone delayed ripening and blocked ethylene production in kiwifruit
Fruit ripening is a unique cellular process and provides an important contribution to human nutrition (Martínez-Romero et al., 2007). In the present study, kiwifruit ripening was monitored during shelf life at 20 °C, following cold storage (0 °C, 95% relative humidity) in the absence (control) or presence of ozone (0.3 μl l−1) by measuring their tissue firmness, soluble solids content, titratable acidity, ethylene production, and respiration rate. Results showed that kiwifruits soften rapidly after transfer from low-temperature storage conditions (0 °C) to ripening temperatures (20 °C), which is consistent with the characteristic softening behaviour of this fruit (Koukounaras and Sfakiotakis, 2007). However, it should be noted that ozone-treated kiwifruits displayed higher firmness retention, especially after 5 months of cold storage (Fig. 1A–C), than control fruits. This observation along with previous data (Tzortzakis et al., 2007a; Rodoni et al., 2010) suggests that ozone is efficient in promoting higher firmness retention. In addition, ozone-treated kiwifruits displayed lower soluble solids contents than control fruits. (Fig. 1D–F). Collectively, these data suggest that ozone is effective at suppressing ripening and senescence process, and therefore can extend kiwifruit post-harvest life.
Kiwifruit has a typical climacteric behaviour with ethylene playing a major role in the ripening process (Antunes and Sfakiotakis, 2002). Control fruits showed a climacteric behaviour by starting autocatalytic production of ethylene when placed at 20 °C, though the exact starting time may depend on the duration of cold storage (Fig. 2A–C). Results regarding the impact of ozone on ethylene production during fruit ripening are contradictory and limited (Skog and Chu, 2001; Palou et al., 2002; Tzortzakis et al., 2007a). The present study found that ozone delayed climacteric ethylene rise and totally blocked ethylene production in kiwifruits after intermediate (3 month) or extended (5 month) cold storage, respectively. Since the ethylene climacteric rise is tightly linked with kiwifruit softening (Antunes and Sfakiotakis 2002), the present data revealed a key post-harvest function of ozone that could be the main aetiological reason for the markedly retarded ripening process (Fig. 1). This regulatory ozone-driven mechanism may provide an excellent model for further studies addressing the role of ethylene signal transduction pathway and ripening phenomena in climacteric fruits. The present data also demonstrated' that kiwifruit are characterized by a time lag between the increase in respiration rate and the induction of ethylene production, confirming previous observations (Sfakiotakis et al., 1997). In addition, respiration rates are enhanced by ozone treatment after short (Fig. 2D) or intermediate (Fig. 2E) cold storage and subsequent transfer to 20 °C. This fact, which was also observed by Forney et al. (2007) in cold-stored vegetables, may indicate that kiwifruits experience physiological injury as a result of the oxidative stress caused by ozone exposure. However, the respiration rate is reduced in ozone-treated kiwifruits following extended cold storage compared with control fruits (Fig. 2F). This specific reduction in respiration may be related to the absence of climacteric peak of ethylene production in fruits exposed to ozone for 5 months (Fig. 2C), as previously suggested (Antunes and Sfakiotakis, 2002).
Ozone acts as an antioxidant and anti-radical elicitor in kiwifruits during ripening
As kiwifruit is a good source of phytochemicals (Tavarini et al., 2008), the present study evaluated the antioxidant potency of kiwifruit extracts during shelf-life period. An induction of antioxidant activity, as measured by both FRAP and DPPH assays, was observed in ozone-treated kiwifruits, particularly after 1 d of shelf life (Fig. 3). These data indicated that the exposure of kiwifruits to ozone stimulated their antioxidant activity, presumably because of its action as a signal molecule for the activation of antioxidant-stress responses in kiwifruit, which possibly experienced an oxidative stress during the cold storage period. Although a variety of negative and positive interactions have been reported between ozone with either ascorbic acid (Allende et al., 2007; Tavarini et al., 2008) or phenolic compounds (Allende et al., 2007; Artés-Hernández et al., 2007; Tzortzakis et al., 2007b) in fruits undergoing post-harvest ripening, the present study further showed that ozone application may increase ascorbic acid and phenol content in kiwifruits, especially after 1 d of shelf life (Fig. 4).
Because of the differential patterns of the antioxidant-related parameters between ozone-treated and untreated kiwifruits ripened for 1 d at 20 °C after 1, 3, or 5 months cold storage (Figs. 3 and 4), this study investigated the relative phenolic-dependent anti-radical activity of kiwifruit extracts with respect to their protective abilities to counteract pBR322 DNA strand scission against HO, ONOO−, or ROO. Phenolics extracted from untreated fruits were quite effective at protecting radical-derived DNA scission (Fig. 5A,C,E), thus confirming earlier observations that kiwifruit extracts can protect DNA from oxidation (Collins et al., 2001). Both in-gel assay (Fig. 5A,C) and quantitative analysis (Fig. 5B,D) revealed that the phenolic extracts of ozone-treated kiwifruits were very potent in inhibiting DNA breakage induced by HO or ONOO−, thereby lending evidence that ozone acts as a potent anti-radical elicitor against commonly encountered oxidant and nitrosant damaging agents.
Characterization of kiwifruit proteome during ripening
Despite increasing interest in plant proteomics (Agrawal et al., 2011; Job et al., 2011), no systematic analysis of the kiwifruit proteome has been published. To fill this gap, 1D-PAGE coupled with mass spectrometry analysis was applied to characterize the reference proteomic map in kiwifruit during ripening at 20 °C. This allowed the characterization of 102 kiwifruit proteins (Table 1; see also Supplementary Table S2), which is a substantial increase because on 25 August 2011 only 17 reviewed kiwi proteins were listed in the Uniprot/Swiss-prot database, of which only three were from kiwifruit (http://www.uniprot.org/uniprot/?query=kiwi&sort=score). The largest functional group of identified kiwifruit proteins is related to energy (Fig. 6B). This is a diverse group of proteins that includes proteins playing a role mainly in glycolysis and the tricarboxylic acid cycle. These proteins are expected to provide the necessary energy metabolites for the ongoing ripening process, as previously suggested in peach fruit (Qin et al., 2009b). The present analysis also showed the presence of various forms of malate dehydrogenase (Fig. 6A, protein zones 6, 14–17), enolase (Fig. 6A, protein zones 8–13), isocitrate dehydrogenase (Fig. 6A, protein zone 11), transaldolase (Fig. 6A, protein zone 14), glyceraldehyde-3-phosphate dehydrogenase (Fig. 6A, protein zones 15 and 17), fructose-bisphosphate aldolase (Fig. 6A, protein zones 15–20), NADPH quinone oxidoreductase (Fig. 6A, protein zone 18), and oxidoreductase NAD-binding (Fig. 6A, protein zones 22 and 23), suggesting that during ripening, specific energy-related isoforms of kiwifruit proteins are presented. Another enzyme that may be linked to the energy group is the mitochondrial ATP synthase (F1) subunit β, which was also identified (Fig. 6A, protein zones 8 and 9), possibly due to the increased demand for ATP during starch–sugar conversion in ripening kiwifruits. An interesting detection was the identification of maturase (Fig. 6A, protein zone 7). Chloroplast maturases evolved into general group II intron splicing factors, potentially mirroring a key step in the evolution of spliceosomal introns. The presence of the maturase in the kiwifruit proteome is indicative of ‘generation of precursor metabolites and energy’ as well as ‘metabolic compound salvage’ linking the splicing organellar introns to global signals that regulate gene expression in response to energy state (Mohr and Lambowitz, 2003), thus suggesting a role for this maturase as an effector protein during kiwifruit ripening.
The second major functional group consisted of proteins involved in protein metabolism and modification (Fig. 6B), including proteins with chaperone function such as HSP70 (Fig. 6A, protein zone 4). The accumulation of heat-shock proteins (HSPs) seems to be a common ripening regulatory mechanism in fruits (Giribaldi et al., 2007; Bianco et al., 2009; Muccilli et al., 2009; Manganaris et al., 2011) as these proteins play pivotal role in the degradation of damaged or misfolded peptides as occurs during fruit ripening (Faurobert et al., 2007). Also, the identification of 60S ribosomal protein (Fig. 6A, protein zones 21 and 31) and eukaryotic translation initiation factor 5A (Fig. 6A, protein zone 28) in kiwifruits during ripening is in accordance with the characterized role of these proteins in the regulation of pineapple fruit senescence (Moyle et al., 2005). Additionally, many proteins were identified, such as actinidin (Fig. 6A, protein zone 9), cysteine protease (Fig. 6A, protein zone 14), actinidin chain A (Fig. 6A, protein zone 21), chymopapain (Fig. 6A, protein zone 21), leucine aminopeptidase (Fig. 6A, protein zone 28), and cystatin (Fig. 6A, protein zone 31), which are responsible for proteolysis and nitrogen recycling and could be involved in the degradation of damaged or misfolded peptides during ripening (Faurobert et al., 2007). Finally, the present study identified the biotin-dependent methylcrotonoyl-CoA carboxylase, an enzyme involved in leucine catabolism in all living cells and which in plants is strongly induced during leaf senescence (Alban et al., 2000). Therefore, this enzyme might be involved in maintaining leucine homeostasis during kiwifruit ripening.
A third major functional group consisted of proteins playing a role in plant defence. Considering that both low-temperature storage and fruit ripening are associated with oxidative processes (Pedreschi et al., 2008), the presence of reactive oxygen species-scavenging proteins in kiwifruits, such as glutathione reductase (Fig. 6A, protein zone 10) and peroxidase (Fig. 6A, protein zone 15), could be required for satisfying the cellular oxidative conditions favourable for ripening. In addition, 2-Cys peroxiredoxin (Fig. 6A, protein zone 25) and small GTP-binding protein (Fig. 6A, protein zone 26) were also identified. 2-Cys peroxiredoxins constitute an ubiquitous group of peroxidases that exhibit dual physiological functions both as antioxidant and molecular chaperone in response to oxidative stimuli (Aran et al., 2009), whereas small GTP-binding protein acts as a molecular switch in oxidative transduction cascades (Shichrur and Yalovsky, 2006). The detection of such proteins in the present study supports their role in oxidative signalling during kiwifruit ripening. Further, the identification of nuclear glycine-rich RNA binding protein (Fig. 6A, protein zone 31) in kiwifruit is in agreement with the observation that this protein performs a RNA chaperone function during cold adaptation (Kim et al., 2010). Since methylglyoxal (a metabolite deriving from glycolysis) is highly cytotoxic, it is also interesting to note the present identification of various forms of lactoylglutathione lyase in kiwifruit (Fig. 6A, protein zones 17–19). This enzyme, also called glyoxalase I, catalyses the first step of the glyoxal pathway, in which methylglyoxal and glutathione are converted into S-lactoylglutathione, which is further converted into lactic acid (Jia et al., 2006). Thus the glyoxalase activity along with the identification of triosephosphate isomerase (Fig. 6A, protein zone 23), which is directly involved in the generation of methylglyoxal, likely reflects a mechanism to control the level of toxic methylglyoxal in kiwifruit, as previously proposed in pear fruit stored under long-term controlled-atmosphere conditions (Pedreschi et al., 2007).
Proteins belonging to the category of cell wall-modifying and structural proteins, such as pectinesterase (Fig. 6A, protein zones 9 and 10), endo-β-1,4-glucanase (Fig. 6A, protein zone 10), xyloglucan endotransglycosylase (Fig. 6A, protein zone 19), α-expansin (Fig. 6A, protein zones 24 and 25), and pectinmethylesterase inhibitor (Fig. 6A, protein zones 27 and 28), were also identified. The identification of cell wall-loosening proteins supports the pivotal role of cell-wall extensibility required for the softening process during kiwifruit ripening (Fig. 1C). Particularly relevant is also the identification of proteins related to metabolism of sugars/polysaccharides, such as β-galactosidase (Fig. 6A, protein zone 4) and α-galactosidase (Fig. 6A, protein zone 14). Both enzymes are able to release α- or β-linked D-galactose from polymers, oligosaccharides, or secondary metabolites and are responsible for most of cell-wall degradation and softening in kiwifruit (Ross et al., 1993). In addition, kiwifruit proteins involved in the regulation of growth, such as β-1,3-glucanase (Fig. 6A, protein zone 19) and agglutinin (Fig. 6A, protein zones 8 and 9), may be also involved in the softening process as previously suggested (Bogoeva et al., 2004; Deytieux et al., 2007).
It is noted that several of the presently identified kiwifruit proteins, such as β-1,3-glucanase, malate dehydrogenase, eukaryotic translation initiation factor 5A, enolase, expansin, glyceraldehyde-3-phosphate, glycine-rich RNA binding protein, small GTP-binding protein, HSPs, isocitrate dehydrogenase, malate dehydrogenase, pectinesterase, and triosephosphate isomerase (Table 1; see also Supplementary Table S2), have also been identified in a number of fruit species during their ripening (Rocco et al., 2006; Deytieux et al., 2007; Faurobert et al., 2007; Giribaldi et al., 2007, 2010; Pedreschi et al., 2007, 2008; Negri et al., 2008; Bianco et al., 2009; Muccilli et al., 2009; Page et al., 2010; Martínez-Esteso et al., 2011; Zhang et al., 2011), thus highlighting their general importance for the ripening process in fruits. In contrast to previous data obtained from studies with other climacteric fruits (Rocco et al., 2006; Faurobert et al., 2007; Page et al., 2010), the present study did not identify any proteins directly linked to ethylene metabolism in kiwifruit but cannot exclude the possibility that ethylene-related proteins were undetected due to the limited dynamic range afforded by classical gel-based proteomics, including 1D-SDS-PAGE analysis. Also, it has to be noted that a prerequisite for a separated protein to be identified is that it has either been sequenced in the organism being examined or that it shares a high sequence similarity with a sequenced protein from another organism (Blomqvist et al., 2008). It should be mentioned that the pattern of proteomic functional categories in kiwifruits (Fig. 6B) exhibits differences when compared with other fleshy fruit species. For example, the most abundant class of identified proteins in orange at ripening time belongs to sugar metabolism (Muccilli et al., 2009), whereas only few sugar-associated proteins were identified in kiwifruit (Table 1; see also Supplementary Table S2), possibly indicating that the presence of a group of proteins is a fingerprint of particular fruit species during ripening. It should be pointed out that the present data represent the first proteomic reference map of kiwifruit that, along with a recent microarray-based study of gene expression patterns in ripe kiwifruit (Atkinson et al., 2011), may provide useful information for ripening marker selection in Actinidia. Large-scale 2D resolution approaches of the Actinidia deliciosa proteome and quantitative profiling studies are a necessary next step for gaining a better understanding of the active role of ozone and the relevant importance of individual kiwifruit proteins in the ripening process.
Protein carbonylation is increased during kiwifruit ripening
The remarkable antioxidant and anti-radical responses observed in ozone-treated fruits (Figs. 3–5) prompted this investigation of whether ozone treatment can modify the protein carbonylation status in kiwifruit during ripening. Quantitative evaluation of the immunoblot results (Supplementary Fig. S3) together with the measurement of protein carbonyl group content (Fig. 7B) showed that protein carbonylation was progressively induced during ripening, thus confirming that protein oxidation is linked to fruit senescence (Qin et al., 2009a,b). The observed lower levels of protein carbonyls in ozone-treated fruits during ripening compared with control fruits (Fig. 7B) may be a consequence of the inhibition of ethylene biosynthesis (Fig. 2C) and the relatively high antioxidant and anti-radical activity in ozone-treated fruits (Figs. 3 and 4C,F and Fig. 5A–D, respectively) and may be associated with delayed senescence features in kiwifruit (Fig. 1). Since previous studies have indicated that protein oxidation is not necessarily a deleterious phenomenon (Job et al., 2005; Oracz et al., 2007) but has been involved in cellular signalling (Wong et al., 2010), the overall pattern of protein carbonylation in kiwifruit (Fig. 7) raises the hypothesis that this redox modification is an important signalling mechanism governing fruit activity during ripening.
In summary, the use of an ozone-enriched cold atmosphere markedly delayed kiwifruit ripening at 20 °C compared with conventional storage. The present data also underscores that the delay/block of climacteric rise of ethylene production is a key mechanism targeted by ozone in kiwifruit upon ripening. Furthermore, this study provides evidence that ozone application during cold storage acts as an antioxidant and anti-radical elicitor in kiwifruit. This report is the first draft of the Actinidia proteome during ripening using 1D-SDS-PAGE and mass spectrometry techniques. A number of proteins that are related to energy, protein metabolism, defence, cell structure, and amino acid or sugar/polysaccharide metabolism are identified. In addition, this investigation shows that ripening and ozone specifically regulate kiwifruit protein carbonylation. These results provide insights into kiwifruit metabolism during post-harvest life and also a starting point to develop ozone-based treatments strategies for controlling fruit ripening and quality in practice.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Table S1. Quantification of protein zones from 1D-gels of kiwifruit proteins.
Supplementary Table S2. Detailed information of the identified kiwifruit proteins.
Supplementary Fig. S1. Scheme summarizing the experimental set up.
Supplementary Fig. S2. Weight loss of kiwifruits (cv. Hayward) after cold storage in the absence or presence of ozone for 1, 3, or 5 months plus 1 day of shelf life.
Supplementary Fig. S3. Relative abundance (arbitrary units) of carbonyl-reactive signal of the immunoblot results in Fig. 7A.
Supporting information
Active Link 1. Mascot results for xyloglucan endotransglycosylase (protein zone 19) identifications based on one peptide sequences: http://139.124.36.211/mascot/cgi/peptide_view.pl?file=./data/20100517/F064815.dat&query=235&hit=1&index=gi%7c950299&px=1§ion=5&ave_thresh=45.
Active Link 2. Mascot results for α-expansin (protein zone 25) identifications based on one peptide sequences: http://139.124.36.211/mascot/cgi/peptide_view.pl?file=../data/20100517/F064843.dat&query=31&hit=1&index=gi%7c1815681&px=1§ion=5&ave_thresh=45.
Acknowledgments
ISM wishes to acknowledge the Hellenic State Scholarships Foundation (IKY) for financial support. We thank Samuel Granjeaud from TAGC (IFR 137, Marseille, France) for technical assistance on database searching.
References
- Agrawal GK, Job D, Zivy M, et al. Time to articulate a vision for the future of plant proteomics – a global perspective: an initiative for establishing the international plant proteomics organization (INPPO) Proteomics. 2011;11:1559–1568. doi: 10.1002/pmic.201000608. [DOI] [PubMed] [Google Scholar]
- Alban C, Job D, Douce R. Biotin metabolism in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 2000;51:17–47. doi: 10.1146/annurev.arplant.51.1.17. [DOI] [PubMed] [Google Scholar]
- Allende A, Marin A, Buendía B, Tomás-Barberán F, Gil MI. Impact of combined post-harvest treatments (UV-C light, gaseous O3, superatmospheric O2 and high CO2) on health promoting compounds and shelf-life of strawberries. Postharvest Biology and Technology. 2007;46:201–211. [Google Scholar]
- Antunes MDC. The role of ethylene in kiwifruit ripening and senescence. Stewart Postharvest Review. 2007;3:1–8. [Google Scholar]
- Antunes MDC, Sfakiotakis EM. Ethylene biosynthesis and ripening behaviour of ‘Hayward’ kiwifruit subjected to some controlled atmospheres. Postharvest Biology and Technology. 2002;26:167–179. [Google Scholar]
- Apak R, Güçlü K, Demirata B, Özyürek M, Çelik SE, Bektaşoğlu B, Berker KI, Özyurt D. Comparative evaluation of various total antioxidant capacity assays applied to phenolic compounds with the CUPRAC assay. Molecules. 2007;12:1496–1547. doi: 10.3390/12071496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran M, Ferrero DS, Pagano E, Wolosiuk RA. Typical 2-Cys peroxiredoxins- modulation by covalent transformations and noncovalent interactions. FEBS Journal. 2009;276:2478–2493. doi: 10.1111/j.1742-4658.2009.06984.x. [DOI] [PubMed] [Google Scholar]
- Artés-Hernandez F, Aguayo E, Artés F, Tomás-Barberán FA. Enriched ozone atmosphere enhances bioactive phenolics in seedless table grapes after prolonged shelf life. Journal of the Science of Food and Agriculture. 2007;87:824–831. [Google Scholar]
- Atkinson RG, Gunaseelan K, Wang MY, et al. Dissecting the role of climacteric ethylene in kiwifruit (Actinidia chinensis) ripening using an 1 aminocyclopropane-1-carboxylic acid oxidase knockdown line. Journal of Experimental Botany. 2011;62:3821–3835. doi: 10.1093/jxb/err063. [DOI] [PubMed] [Google Scholar]
- Barboni T, Cannac M, Chiaramonti N. Effect of cold storage and ozone treatment on physicochemical parameters, soluble sugars and organic acids in Actinidia deliciosa. Food Chemistry. 2010;121:946–951. [Google Scholar]
- Barr SD, Gedamu L. Role of peroxidoxins in Leishmania chagasi survival: evidence of an enzymatic defense against nitrosative stress. Journal of Biological Chemistry. 2003;278:10816–10823. doi: 10.1074/jbc.M212990200. [DOI] [PubMed] [Google Scholar]
- Baur S, Klaiber RG, Koblo A, Carle R. Effect of different washing procedures on phenolic metabolism of shredded packaged iceberg lettuce during storage. Journal of Agricultural and Food Chemistry. 2004;52:7017–7025. doi: 10.1021/jf048961a. [DOI] [PubMed] [Google Scholar]
- Benzie IFF, Strain JJ. The ferric reducing ability of plasma (FRAP) as a measure of ‘antioxidant power’: the FRAP assay. Analytical Biochemistry. 1996;39:70–76. doi: 10.1006/abio.1996.0292. [DOI] [PubMed] [Google Scholar]
- Bevan M, Bancroft I, Bent E, et al. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature. 1998;39:1485–1488. doi: 10.1038/35140. [DOI] [PubMed] [Google Scholar]
- Bianco L, Lopez L, Scalone AG, Di Carli M, Desiderio A, Benvenuto E, Perrotta G. Strawberry proteome characterization and its regulation during fruit ripening and in different genotypes. Journal of Proteomics. 2009;72:586–607. doi: 10.1016/j.jprot.2008.11.019. [DOI] [PubMed] [Google Scholar]
- Blomqvist LA, Ryberg M, Sundqvist C. Proteomic analysis of highly purified prolamellar bodies reveals their significance in chloroplast development. Photosynthesis Research. 2008;96:37–50. doi: 10.1007/s11120-007-9281-y. [DOI] [PubMed] [Google Scholar]
- Bogoeva VP, Radeva MA, Atanasova LY, Stoitsova SR, Boteva RN. Fluorescence analysis of hormone binding activities of wheat germ agglutinin. Biochimica et Biophysica Acta. 2004;6:213–218. doi: 10.1016/j.bbapap.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brand-Williams W, Cuvelier ME, Berset C. Use of a free radical method to evaluate antioxidant activity. LWT – Food Science and Technology. 1995;28:25–30. [Google Scholar]
- Castagna A, Ederli L, Pasqualini S, Mensuali-Sodi A, Baldan B, Donnini S, Ranieri A. The tomato ethylene receptor LE-ETR3 (NR) is not involved in mediating ozone sensitivity: causal relationships among ethylene emission, oxidative burst and tissue damage. New Phytologist. 2007;174:342–356. doi: 10.1111/j.1469-8137.2007.02010.x. [DOI] [PubMed] [Google Scholar]
- Catusse J, Strub J, Job C, Van Dorsselaer A, Job D. Proteome-wide characterization of sugarbeet seed vigor and its tissue specific expression. Proceedings of the National Academy of Sciences of the United States of America USA. 2008;105:10262–10267. doi: 10.1073/pnas.0800585105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins BH, Horska A, Hotten PM, Riddoch C, Collins AR. Kiwifruit protects against oxidative DNA damage in human cells and in vitro. Nutrition and Cancer. 2001;39:148–153. doi: 10.1207/S15327914nc391_20. [DOI] [PubMed] [Google Scholar]
- Crowhurst RN, Gleave AP, MacRae EA, et al. Analysis of expressed sequence tags from Actinidia: applications of a cross species EST database for gene discovery in the areas of flavor, health, color and ripening. BMC Genomics. 2008;9:351. doi: 10.1186/1471-2164-9-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deytieux C, Gény L, Lapaillerie D, Claverol S, Bonneu M, Donèche B. Proteome analysis of grape skins during ripening. Journal of Experimental Botany. 2007;58:1851–1862. doi: 10.1093/jxb/erm049. [DOI] [PubMed] [Google Scholar]
- Dong M, Vongchampa V, Gingipalli L, Cloutier JF, Kow YW, O’Connor T, Dedon PC. Development of enzymatic probes of oxidative and nitrosative DNA damage caused by reactive nitrogen species. Mutation Research. 2006;594:120–134. doi: 10.1016/j.mrfmmm.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Faurobert M, Mihr C, Bertin N, Pawlowski T, Negroni L, Sommerer N, Causse M. Major proteome variations associated with cherry tomato pericarp development and ripening. Plant Physiology. 2007;143:1327–1346. doi: 10.1104/pp.106.092817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forney CF, Song J, Hildebrand PD, Fan L, McRae KB. Interactive effects of ozone and 1-methylcyclopropene on decay resistance and quality of stored carrots. Postharvest Biology and Technology. 2007;45:341–348. [Google Scholar]
- Giovannoni JJ. Genetic regulation of fruit development and ripening. The Plant Cell. 2004;16:170–180. doi: 10.1105/tpc.019158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giribaldi M, Gény L, Delrot S, Schubert A. Proteomic analysis of the effects of ABA treatments on ripening Vitis vinifera berries. Journal of Experimental Botany. 2010;61:2447–2458. doi: 10.1093/jxb/erq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giribaldi M, Perugini I, Sauvage FX, Schubert A. Analysis of protein changes during grape berry ripening by 2-DE and MALDI-TOF. Proteomics. 2007;7:3154–3170. doi: 10.1002/pmic.200600974. [DOI] [PubMed] [Google Scholar]
- Harder A, Wildgruber R, Nawrocki A, Fey SJ, Larsen PM, Görg A. Comparison of yeast cell protein solubilization procedures for two-dimensional electrophoresis. Electrophoresis. 1999;20:826–829. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<826::AID-ELPS826>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Hertog MLATM, Rudell DR, Pedreschi R, Schaffer RJ, Geeraerd AH, Nicolaï BM, Ferguson I. Where systems biology meets postharvest. Postharvest Biology and Technology. 2011;62:223–237. [Google Scholar]
- Hu C, Kitts D. Evaluation of antioxidant activity of epigallocatehin gallate in biphasic model systems in vitro. Molecular and Cellular Biochemistry. 2001;218:147–155. doi: 10.1023/a:1007220928446. [DOI] [PubMed] [Google Scholar]
- Jia X, Hollung K, Therkildsen M, Hildrum KI, Bendixen E. Proteome analysis of early post-mortem changes in two bovine muscle types: M. longissimus dorsi and M. semitendinosis. Proteomics. 2006;6:936–944. doi: 10.1002/pmic.200500249. [DOI] [PubMed] [Google Scholar]
- Job D, Haynes PA, Zivy M. Plant proteomics. Proteomics. 2011;11:1557–8. doi: 10.1002/pmic.201190035. [DOI] [PubMed] [Google Scholar]
- Job C, Rajjou L, Lovigny Y, Belghazi M, Job D. Patterns of protein oxidation in Arabidopsis seeds and during germination. Plant Physiology. 2005;138:790–802. doi: 10.1104/pp.105.062778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Kim WY, Kwak KJ, Oh SH, Han YS, Kang H. Glycine-rich RNA-binding proteins are functionally conserved in Arabidopsis thaliana and Oryza sativa during cold adaptation process. Journal of Experimental Botany. 2010;61:2317–2325. doi: 10.1093/jxb/erq058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukounaras A, Sfakiotakis E. Effect of 1-MCP prestorage treatment on ethylene on ethylene and CO2 production and quality of ‘Hayward’ kiwifruit during shelf-life after short, medium and long term cold storage. Postharvest Biology and Technology. 2007;46:174–180. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levine RL, Garland D, Oliver CN, Amici A, Climent I, Lenz A-G, Ahn B-W, Shaltiel S, Stadtman ER. Determination of carbonyl content in oxidatively modified proteins. Methods in Enzymology. 1990;186:464–478. doi: 10.1016/0076-6879(90)86141-h. [DOI] [PubMed] [Google Scholar]
- Lim K, Hu C, Kitts DD. Antioxidant activity of a Rhus verniciflua stokes ethanol extract. Food and Chemical Toxicology. 2001;39:229–237. doi: 10.1016/s0278-6915(00)00135-6. [DOI] [PubMed] [Google Scholar]
- Manganaris GA, Rasori A, Bassi D, Ramina A, Tonutti P, Bonghi C. Comparative transcript profiling of apricot (Prunus armeniaca L.) fruit development and on-tree ripening. Tree Genetics and Genomes. 2011;7:609–616. [Google Scholar]
- Manganaris GA, Vasilakakis M, Diamantidis G, Mignani I. The effect of postharvest calcium application on tissue calcium concentration, quality attributes, incidence of flesh browning and cell wall physicochemical aspects of peach fruits. Food Chemistry. 2007;100:1385–1392. [Google Scholar]
- Martínez-Esteso MJ, Sellés-Marchart S, Lijavetzky D, Pedreño MA, Bru-Martínez R. A DIGE-based quantitative proteomic analysis of grape berry flesh development and ripening reveals key events in sugar and organic acid metabolism. Journal of Experimental Botany. 2011;62:2521–2569. doi: 10.1093/jxb/erq434. [DOI] [PubMed] [Google Scholar]
- Martínez-Romero D, Bailen G, Serrano M, Guillen F, Valverde JM, Zapata P, Castillo S, Valero D. Tools to maintain postharvest fruit and vegetable quality through the inhibition of ethylene action: a review. Critical Reviews in Food Science and Nutrition. 2007;47:543–560. doi: 10.1080/10408390600846390. [DOI] [PubMed] [Google Scholar]
- Michailides TJ, Manganaris GA. Harvesting and handling effects on postharvest decay. Stewart Postharvest Review. 2009;5:1–7. [Google Scholar]
- Minas IS, Karaoglanidis GS, Manganaris GA, Vasilakakis M. Effect of ozone application during cold storage of kiwifruit on the development of stem-end rot caused by Botrytis cinerea. Postharvest Biology and Technology. 2010;58:203–210. [Google Scholar]
- Mohr G, Lambowitz AM. Putative proteins related to group II intron reverse transcriptase/maturases are encoded by nuclear genes in higher plants. Nucleic Acids Research. 2003;31:647–652. doi: 10.1093/nar/gkg153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller IM, Jensen PE, Hansson A. Oxidative modifications to cellular components in plants. Annual Review of Plant Biology. 2007;58:459–481. doi: 10.1146/annurev.arplant.58.032806.103946. [DOI] [PubMed] [Google Scholar]
- Moyle R, Fairbairn DJ, Ripi J, Crowe M, Botella JR. Developing pineapple fruit has a small transcriptome dominated by metallothionein. Journal of Experimental Botany. 2005;56:101–112. doi: 10.1093/jxb/eri015. [DOI] [PubMed] [Google Scholar]
- Muccilli V, Licciardello C, Fontanini D, Russo MP, Cunsolo V, Saletti R, Reforgiato Recupero G, Foti S. Proteome analysis of Citrus sinensis L. (Osbeck) flesh at ripening time. Journal of Proteomics. 2009;73:134–152. doi: 10.1016/j.jprot.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Negri AS, Prinsi B, Rossoni M, Failla O, Scienza A, Cocucci M, Espen L. Proteome changes in the skin of the grape cultivar Barbera among different stages of ripening. BMC Genomics. 2008;9:378. doi: 10.1186/1471-2164-9-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilo R, Saffie C, Lilley K, et al. Proteomic analysis of peach fruit mesocarp softening and chilling injury using difference gel electrophoresis (DIGE) BMC Genomics. 2010;11:43. doi: 10.1186/1471-2164-11-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oracz K, Bouteau HE, Farrant JM, Cooper K, Belghazi M, Job C, Job D, Corbineau F, Bailly C. ROS production and protein oxidation as a novel mechanism for seed dormancy alleviation. The Plant Journal. 2007;50:452–465. doi: 10.1111/j.1365-313X.2007.03063.x. [DOI] [PubMed] [Google Scholar]
- Page D, Gouble B, Valot B, Bouchet JP, Callot C, Kretzschmar A, Causse M, Renard CM, Faurobert M. Protective proteins are differentially expressed in tomato genotypes differing for their tolerance to low-temperature storage. Planta. 2010;232:483–500. doi: 10.1007/s00425-010-1184-z. [DOI] [PubMed] [Google Scholar]
- Palou L, Crisosto CH, Smilanick JL, Adaskaveg JE, Zoffoli JP. Effects of continuous 0.3 ppm ozone exposure on decay development and physiological responses of peaches and table grapes in cold storage. Postharvest Biology and Technology. 2002;24:39–48. [Google Scholar]
- Pantelidis G, Vasilakakis M, Manganaris G, Diamantidis Gr. Antioxidant capacity, phenol, anthocyanin and ascorbic acid contents in raspberries, red currants, gooseberries and Cornelian cherries. Food Chemistry. 2007;102:777–783. [Google Scholar]
- Pedreschi R, Hertog M, Robben J, Noben J-P, Nicolaï B. Physiological implications of controlled atmosphere storage of ‘Conference’ pears (Pyrus communis L.): a proteomic approach. Postharvest Biology and Technology. 2008;50:110–116. [Google Scholar]
- Pedreschi R, Vanstreels E, Carpentier S, Hertog M, Lammertyn J, Robben J, Noben J-P, Swennen R, Vanderleyden J, Nicolaï BM. Proteomic analysis of core breakdown disorder in Conference pears (Pyrus communis L.) Proteomics. 2007;7:2083–2099. doi: 10.1002/pmic.200600723. [DOI] [PubMed] [Google Scholar]
- Perez AG, Sanz C, Rios JJ, Olias R, Olias JM. Effects of ozone treatment on postharvest strawberry quality. Journal of Agricultural and Food Chemistry. 1999;47:1652–1656. doi: 10.1021/jf980829l. [DOI] [PubMed] [Google Scholar]
- Qin G, Meng X, Wang Q, Tian S. Oxidative damage of mitochondrial proteins contributes to fruit senescence: a redox proteomics analysis. Journal of Proteome Research. 2009b;8:2449–2462. doi: 10.1021/pr801046m. [DOI] [PubMed] [Google Scholar]
- Qin G, Wang Q, Liu J, Li B, Tian S. Proteomic analysis of changes in mitochondrial protein expression during fruit senescence. Proteomics. 2009a;9:4241–4253. doi: 10.1002/pmic.200900133. [DOI] [PubMed] [Google Scholar]
- Ramina A, Tonutti P, Mc Glasson B. Ripening, nutrition, and postharvest physiology. In: Layne D, Bassi D, editors. The Peach Botany, Production and Uses. Oxfordshire, UK: CABI; 2008. pp. 550–574. [Google Scholar]
- Rinalducci S, Murgiano L, Zolla L. Redox proteomics: basic principles and future perspectives for the detection of protein oxidation in plants. Journal of Experimental Botany. 2008;59:3781–3801. doi: 10.1093/jxb/ern252. [DOI] [PubMed] [Google Scholar]
- Rocco M, D’Ambrosio C, Arena S, Faurobert M, Scaloni A, Marra M. Proteomic analysis of tomato fruits from two ecotypes during ripening. Proteomics. 2006;6:3781–3791. doi: 10.1002/pmic.200600128. [DOI] [PubMed] [Google Scholar]
- Rodoni L, Casadei N, Concellón A, Chaves Alicia AR, Vicente AR. Effect of short-term ozone treatments on tomato (Solanum lycopersicum L.) fruit quality and cell wall degradation. Journal of Agricultural and Food Chemistry. 2010;58:594–599. doi: 10.1021/jf9029145. [DOI] [PubMed] [Google Scholar]
- Ross GS, Redgwell RJ, MacRae EA. Kiwifruit β-galactosidase: isolation and activity against specific fruit cell-wall polysaccharides. Planta. 1993;189:499–506. [Google Scholar]
- Sacchetti G, Maietti S, Muzzoli M, Scaglianti M, Manfredini S, Radice M, Bruni R. Comparative evaluation of 11 essential oils of different origin as functional antioxidants, antiradicals and antimicrobials in foods. Food Chemistry. 2005;91:621–632. [Google Scholar]
- Scalbert A, Monties B, Janin G. Tannins in wood: comparison of different estimation methods. Journal of Agricultural and Food Chemistry. 1989;37:1324–1329. [Google Scholar]
- Sfakiotakis E, Antunes MD, Stavroulakis G, Niklis N, Ververidis P, Gerasopoulos D. Ethylene biosynthesis and its regulation in ripening ‘Hayward’ kiwifruit. In: Kanellis AK, Chang C, Kende H, Grierson D, editors. Biology and Biotechnology of the Plant Hormone Ethylene. Dordrecht: Kluwer Academic Publishers; 1997. pp. 47–56. [Google Scholar]
- Shichrur K, Yalovsky S. Turning ON the switch-RhoGEFs in plants. Trends in Plant Science. 2006;11:57–59. doi: 10.1016/j.tplants.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Skerget M, Kotnik P, Hadolin M, Hras A, Simonic M, Knez Z. Phenols, proanthocyanidins, flavones and flavonols in some plant materials and the antioxidant activities. Food Chemistry. 2005;89:191–198. [Google Scholar]
- Skog LJ, Chu CL. Effect of ozone on qualities of fruits and vegetables in cold storage. Canadian Journal of Plant Science. 2001;81:773–778. [Google Scholar]
- Tanou G, Job C, Belghazi M, Molassiotis A, Diamantidis G, Job D. Proteomic signatures uncover hydrogen peroxide and nitric oxide cross-talk signaling network in citrus plants. Journal of Proteome Research. 2010;9:5994–6006. doi: 10.1021/pr100782h. [DOI] [PubMed] [Google Scholar]
- Tanou G, Job C, Rajjou L, Arc E, Belghazi M, Diamantidis G, Molassiotis A, Job D. Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. The Plant Journal. 2009;60:795–804. doi: 10.1111/j.1365-313X.2009.04000.x. [DOI] [PubMed] [Google Scholar]
- Tavarini S, Degl’Innocenti E, Remorini D, Massai R, Guidi L. Antioxidant capacity, ascorbic acid, total phenols and carotenoids changes during harvest and after storage of Hayward kiwifruit. Food Chemistry. 2008;107:282–288. [Google Scholar]
- Tzortzakis N, Borland A, Singleton I, Barnes J. Impact of atmospheric ozone-enrichment on quality-related attributes of tomato fruit. Postharvest Biology and Technology. 2007a;45:317–325. [Google Scholar]
- Tzortzakis N, Singleton I, Barnes J. Deployment of low-level ozone-enrichment for the preservation of chilled fresh produce. Postharvest Biology and Technology. 2007b;43:261–270. [Google Scholar]
- Uppu RM, Pryor WA. Synthesis of peroxynitrite in a two-phase system using isoamyl nitrite and hydrogen peroxide. Analytical Biochemistry. 1996;236:242–249. [PubMed] [Google Scholar]
- Wong CM, Marcocci L, Liu L, Suzuki YJ. Cell signaling by protein carbonylation and decarbonylation. Antioxidants and Redox Signaling. 2010;12:393–404. doi: 10.1089/ars.2009.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lu Z, Yu Z, Gao X. Preservation fresh-cut celery by treatment of ozonated water. Food Control. 2005;16:279–283. [Google Scholar]
- Zhang L, Yu Z, Jiang L, Jiang J, Luo H, Fu L. Effect of post-harvest heat treatment on proteome change of peach fruit during ripening. Journal of Proteomics. 2011;74:1135–1149. doi: 10.1016/j.jprot.2011.04.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.