Abstract
The flavonoid-derived proanthocyanidins (PAs) are one class of the major defence phenolics in poplar leaves. Transcriptional activation of PA biosynthetic genes, resulting in PA accumulation in leaves, was detected following infection by the fungal Marssonina brunnea f.sp. multigermtubi using digital gene expression analysis. In order to study PA biosynthesis and its induction by fungi, a putative leucoanthocyanidin reductase gene, PtrLAR3, was isolated from Populus trichocarpa. Sequence comparison of PtrLAR3 with other known leucoanthocyanidin reductase proteins revealed high amino acid sequence similarity. Semi-quantitative reverse-transcription (RT) PCR and quantitative real-time PCR analysis demonstrated that PtrLAR3 was expressed in various tissues and the highest level of expression was observed in roots. Overexpression of PtrLAR3 in Chinese white poplar (Populus tomentosa Carr.) led to a significant plant-wide increase in PA levels. In vitro assays showed that crude leaf extracts from 35S:PtrLAR3 transformants were able to inhibit significantly the hyphal growth of M. brunnea f.sp. multigermtubi compared to the extracts from control plants. The transgenic 35S:PtrLAR3 poplar plants displayed a significant (P < 0.05) reduction in their disease symptoms compared with the control. RT-PCR analysis showed that PtrLAR3 expression was up-regulated in all transformants. These results suggested that constitutive expression of endogenous PtrLAR3 could be exploited to improve resistance to fungal pathogens in poplar.
Keywords: Marssonina brunnea, poplar, proanthocyanidin biosynthesis, PtrLAR3
Introduction
Proanthocyanidins (PAs), also known as condensed tannins, are the polyphenolic compounds synthesized via the flavonoid biosynthetic pathway. They occur in many plant species and play an important role in defence against herbivores and pathogens (Harborne and Grayer, 1993; Peters and Constabel, 2002; Tanner et al., 2003). The increase in PA concentration in forage crops can protect ruminants against pasture bloat (Dixon et al., 1996; Douglas et al., 1999; McMahon et al., 2000). PAs also act as potential antioxidants with beneficial effects for human health by protecting against free radical-mediated injury and cardiovascular diseases (Bagchi et al., 2000; Lin et al., 2002; Cos et al., 2004). Additionally, PAs also contribute to the taste of numerous fruits and beverages, such as fruit juices, tea, and wine (Dixon et al., 2005). Thus, it is important to understand the mechanisms of biosynthesis and regulation of PA polymers in planta.
PA biosynthesis is one branch of the flavonoid biosynthetic pathway that also produces anthocyanins and flavonols. The flavonoid pathway has been characterized both genetically and biochemically in several plant species (Shirley et al., 1992; Boss et al., 1996; Winkel-Shirley, 2001). The biosynthesis of PAs and anthocyanins shares the same up-stream flavonoid pathway, leading to the production of flavan-3,4-diols which are utilized as co-precursors for anthocyanin and PA polymer synthesis (Stafford, 1990; Springob et al., 2003). Biosynthesis of PA polymers is considered to be catalysed by leucoanthocyanidin reductase (LAR), converting leucocyanidin to catechin (Stafford, 1990). Leucoanthocyanidin is also the substrate for anthocyanidin synthase (ANS) to produce anthocyanidin, which is then converted to the other PA precursors, epicatechins, by the catalysation of anthocyanidin reductase (ANR). To date, LAR genes have been isolated from several plant species and the activity correlated with PA accumulation has been characterized (Stafford, 1990; Joseph et al., 1998; Marles et al., 2003; Pang et al., 2007; Paolocci et al., 2007). Tanner et al. (2003) purified firstly an LAR gene from Desmodium uncinatum and the recombinant protein was shown to catalyse the conversion of leucocyanidin, leucodelphinidin, or leucopelargonidin to the corresponding 2,3-trans-flavan-3-ol. This finding clearly established the role of LAR in PA biosynthesis. In Arabidopsis thaliana, the PA biosynthetic pathway has been best characterized, but this species accumulates only epicatechin-based starter units and lacks any obvious LAR orthologue (Abrahams et al., 2003). Two LAR orthologues from grapevine (Vitis vinifera L. cv. Shiraz) have been reported and they have different patterns of expression in skin and seeds (Bogs et al., 2005). Only VvLAR1 has been functionally characterized by enzymic assay of the recombinant protein and has been shown to possess LAR enzyme activity, but data have still to be provided for VvLAR2. In Medicago truncatula, there is an apparent lack of catechin in the PAs, but a single LAR gene has been cloned and characterized. Transgenic tobacco expressing MtLAR showed the lack of catechin or PA production, raising questions as to the actual role of LAR in M. truncatula (Pang et al., 2007). More recently, two LAR genes were reported in Lotus corniculatus, but only one of them, LcLAR1, was shown to encode a protein with demonstrable LAR activity (Paolocci et al., 2007).
Poplar (Populus spp.) is a widespread forest tree with significant economic and ecological importance worldwide, but the species is susceptible to different fungal diseases. Toxicity of PAs, usually estimated by the measurement of the reduction of in vitro mycelial growth, is well documented for several filamentous fungi, such as Aspergillus niger, Colletotrichum graminicola, Gloeophyllum trabeum, and Trichoderma viride (Harborne, 1980; Scalbert, 1991). The transcriptional response of hybrid poplar (Populus trichocarpa × Populus deltoides) to poplar leaf rust (Melampsora medusae) infection has been analysed using the Populus cDNA microarray and the genes encoding enzymes required for PA biosynthesis were up-regulated dramatically (Miranda et al., 2007). Phytochemical analysis has confirmed that PA levels increase in infected leaves late in infection, linking this pathway for the first time to the pathogen-defence response in poplar (Miranda et al., 2007).
With the completion of the P. trichocarpa genome sequence, a wide range of genomic and genetic resources are now available for this species (Tuskan et al., 2006); thus Populus has been selected as a model legume for biochemical, genetic, and genomic studies (Jansson and Douglas, 2007). In a previous study, Tsai et al. (2006) found that three LAR genes in Populus occurred in two distinct phylogenetic lineages, but showed little difference in their tissue distribution. During the PA increase, the transcript abundance of PtrLAR1 and PtrLAR3 was wound-stimulated by two- and ten-fold, pointing to differential regulation (Tsai et al., 2006).
The current report describes experiments to profile the transcriptional response of poplar leaves in a compatible interaction with the black spot fungus Marssonina brunnea f.sp. multigermtubi using Illumina’s digital gene expression (DGE) platform. A cDNA fragment encoding PtrLAR3 was isolated from P. trichocarpa by reverse-transcription PCR (RT-PCR). The PA content and PtrLAR3 transcription level in the different tissues were measured and the function of PtrLAR3 was determined, for which a chimeric PtrLAR3 gene was constitutively expressed under the control of the cauliflower mosaic virus 35S promoter. Transgenic plants were further evaluated for resistance to infection by the pathogenic fungus M. brunnea f.sp. multigermtubi. The results indicated that PA biosynthesis is mediated by overexpression of PtrLAR3 and leads to enhanced resistance to fungal pathogens in transgenic poplar plants.
Materials and methods
Plant materials and bacterial strains
Poplar plants were grown in the greenhouse at 25 °C under a 14/10 light/dark cycle with supplemental light (4500 lux). All tested tissues, including leaf, stem, root, and petiole, were harvested from greenhouse material, separated, and frozen in liquid nitrogen until further processing. To determine the defence mechanisms of triploid Populus tomentosa Carr. against black spot disease, a susceptible P. tomentosa Carr. clone (clone 51) was inoculated with M. brunnea f.sp. multigermtubi. Infected and control leaves were harvested at 3 days post inoculation (dpi) for RNA isolation and cDNA synthesis.
Escherichia coli DH5α was used as the host strain for transformation, genetic manipulation, and nucleotide sequencing. Agrobacterium tumefaciens EHA105 was used for the transformation of P. trichocarpa. Carr. (clone 73).
Cloning of PtrLAR3
Total RNA was isolated from frozen tissues of poplar plants using a RNA RNeasy Plant Mini Kit (Qiagen, Germany) following the manufacturer’s instructions. Leaves and petioles were excised from stems, including the fourth (young) and fifth (mature) internodes from the top of the stems (height 1 m). First-strand cDNA was synthesized from 2 μg DNase-treated RNA with RT-AMV transcriptase (TaKaRa, Dalian, China) in a total volume of 20 μl using oligo d(T)18 at 42 °C for 30 min.
The full open reading frame of PtrLAR3 was amplified with gene-specific primers (LAR3-F: 5′-ACATGAATGGTCATTCTCCA-3′; LAR3-R: 5′-TCATGCTGTAATAAATAAAG- 3′; Joint Genome Institute, http://genome.jgi-psf.org/poplar/poplar.info.html) by RT-PCR with 2 μl cDNA from roots. The PCR reaction was carried out with Pfu DNA polymerase (TaKaRa) in a total volume of 50 μl with an initial denaturing step at 94 °C for 3 min, 34 cycles of 94 °C for 45 s, 58 °C for 30 s, and 72 °C for 90s and a final extension step at 72 °C for 10 min. The amplification products were cloned into the plant binary vector pCXSN, which is a zero-background TA cloning system that provides simple and high-efficiency direct cloning of PCR-amplified DNA fragments (Chen et al., 2009). The resulting vector 35S:PtrLAR3, containing the PtrLAR3 open reading frame down-stream of the cauliflower mosaic virus 35S promoter and the hygromycin phosphotransferase gene (Hpt) as a plant-selectable marker conferring hygromycin resistance was transferred into A. tumefaciens EHA105 by the freeze–thaw method.
Transformation of P. tomentosa Carr. plants
Transgenic Chinese white poplar (P. tomentosa Carr.) plants were generated by Agrobacterium-mediated transformation as described previously (Jia et al., 2010). Recombinant Agrobacterium was used to infect poplar leaf discs and putative transgenic plants were selected on woody plant medium (WPM) (Lloyd and McCown, 1980) supplemented with 10 mg/l hygromycin. Rooted plantlets were acclimatized in pots at 25 °C in a 14/10 light/dark cycle and then transferred to the greenhouse for further studies.
DNA extraction and PCR analysis
Genomic DNA was extracted from leaves (300 mg) of untransformed control and hygromcyin-resistant plants using the modified cetyltrimethylammonium bromide extraction method as previously described (Jia et al., 2010). To determine the presence of transgenes, putative transgenic plants were screened preliminarily by PCR analysis (Luo et al., 2006). The following primers were designed for Hpt: forward: 5′-ATCGGACGATTGCGTCGTCGCATC-3′; reverse: 5′-GTGTCACGTTG CAAGACCTG-3′. The PCR conditions were an initial denaturing step at 94 °C for 3 min and 35 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min. The amplification products were resolved on a 1% (w/v) agarose gel and visualized after ethidium bromide staining.
Semi-quantitative RT-PCR and quantitative real-time PCR analysis
Total RNA was extracted from leaves, roots, stems, and petioles of poplar plants and treated with DNase I (TaKaRa) according to the manufacturer’s instructions. All RNA was purified and first-strand cDNA was synthesized as described above. The reverse-transcribed cDNA samples were used for quantitative real-time PCR, which was performed on a TaKaRa real-time-PCR detection system. 18S rRNA was used as an internal control. The RT-PCR conditions were an initial denaturation step at 94 °C for 3 min, 28 cycles of 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 1 min, and an extension step at 72 °C for 10 min. The amplification products were resolved by 1% (w/v) agarose gel electrophoresis and visualized with ethidium bromide under UV light.
Quantitative real-time PCR analysis was performed as described Tsai et al. (2006) in a 20-μl reaction volume containing 10 μl of SYBR Green master mix reagent (TaKaRa). The primers were designed using Primer 5.0 software: forward and reverse primers for PtrLAR3 amplification were PtrLAR3-F (5′-CGGTGATGGGACAGTTAAAG-3′) and PtrLAR3-R (5′-CATGGCAACAAAGCTCTCAA-3′). Each reaction was performed in duplicate and with three biological replicates along with no-template controls. The gene-quantification method was based on the relative expression of the target gene versus the reference gene (Actin) (Tsai et al., 2006).
Sequence comparisons and phylogenetic analysis
The deduced amino acid sequences were analysed using the program DNAMAN and the software MAGE version 4.0(Lynnonon Biosoft, Quebec, Canada). Alignment of the deduced amino acid sequences was performed using DNAMAN. The phylogenetic relationships of LARs were analysed with the neighbour-joining method using MAGE version 4.0.
Extraction and quantification of proanthocyanidins
For extraction of PAs, tissues were ground in liquid nitrogen and 1.0 g batches were extracted with 5 ml extraction solution (70% acetone and 0.5% acetic acid) by vortexing followed by sonication at 30 °C for 30 min. Following centrifugation at 2500 g for 10 min, residues were re-extracted twice, as above. Supernatants were pooled and then extracted with 3 ml chloroform, and the aqueous supernatant was re-extracted twice with chloroform and three times with hexane. Samples were freeze dried, resuspended in extraction solution to a final concentration of 3 g original sample ml−1, spun briefly, and transferred to another tube, and then the soluble PA content was determined using dimethylaminocinnamaldehyde (DMACA) reagent with catechin standards (Pang et al., 2007).
The insoluble PA content was tested using the butanol/HCl method (Porter et al., 1986). The residues from the above tissue extractions were dried in air for 2 days, and then 1 ml butanol/HCl reagent was added and the mixture was sonicated at room temperature for 60 min and centrifuged at 2500 g for 10 min. Supernatants were transferred to cuvettes for determination of absorption at 550 nm and were then boiled for 1 h. After cooling to room temperature, the A550 was recorded again and the first value subtracted from the second. Absorbance values were converted into PA equivalents using a standard curve (2.5, 5, 10, 20, and 40 mg) of procyanidin B1 (Indofine). Three independent experiments were performed for each sample.
Histochemical staining with DMACA
Histochemical analysis of PA accumulation in various tissues was performed as described by Li et al. (1996). In brief, plant tissues were decolourized in 5 ml of 30% acetic acid in ethanol for 12 h and washed with 75% ethanol. PAs were detected by staining tissues for 3 h with 1% (w/v) DMACA in ethanol and 6 M HCl (1:1, v/v). Images of stem and petiole sections were recorded using a Nikon microscope.
DGE library construction and sequencing
DGE experiments were performed as described by Zhang et al. (2010). Total RNA isolated from five M. brunnea f.sp. multigermtubi-infected and control leaves was pooled by treatment, respectively. Beads with oligo d(T)18 were used to isolate poly(A) mRNA. The first-strand cDNA was synthesized using a random hexamer-primer and reverse transcriptase (Invitrogen). The second-strand cDNA was synthesized using RNase H (Invitrogen) and DNA polymerase I (New England BioLabs). Then the cDNA libraries were prepared according to Illumina’s protocols. Briefly, one individual single-end cDNA library was constructed for each sample. Libraries were prepared from a 150–200-bp size-selected fraction following adapter ligation and agarose gel separation. The libraries were sequenced on the Illumina GA platform for 35 cycles. The paired-end libraries were sequenced for 44–75 bp.
Mapping short reads to the Populus genome and annotated gene set
The P. trichocarpa genome and annotated gene set were downloaded from the Populus public database (Joint Genome Institute) and 17,273 P. trichocarpa full-length cDNAs were collected from the reference genome sequence of Populus (Phytozome version 2.0). The cDNAs were aligned to the P. trichocarpa genome and those with identities higher than 80% were retained for further analysis. A nonredundant gene set of P. trichocarpa was created by merging the sequences of the Beijing Genomics Institute Gene Finder predicted genes and cDNAs and then removing the smaller one if two transcripts had at least 100 bp overlapping. After removing reads containing sequencing adapters and reads of low quality (reads containing >5 ambiguous residues), reads were aligned to the P. trichocarpa genome using Short Oligonucleotide Alignment Program (SOAP) (Li et al., 2008) allowing up to two mismatches. Reads that failed to be mapped progressively had one base trimmed off from the 39-end and mapped to the genome again until a match was found. For paired-ends reads, insert size between paired reads was set at 1 bp–10 kb to allow reads spanning introns of different sizes. A similar strategy was used to align reads to the nonredundant gene set, but the insert length range was restricted to 1 kb for paired-end read mapping.
In vitro assays for antifungal activity
To prepare a crude leaf extract, fresh poplar leaves (10 g) were homogenized under liquid N2 and extracted with 10 ml extraction buffer (pH 7.0) containing a mixture of protease inhibitors (1 mM PMSF, 1 mM N-ethylmaleimide, 5 mM EDTA, and 0.02 mM pepstatin A). Ground tissues were then centrifuged at 9000 g for 10 min at room temperature and extracts were collected from each sample. The supernatant was referred to as the crude leaf extract. In vitro antifungal activity assay was performed as described by Wu (1988). The pathogenic fungus M. brunnea f.sp. multigermtubi was employed for the assay of antifungal activity on potato/dextrose/agar (PDA). In brief, the assay used Petri dishes (90 mm diameter) containing 20 ml PDA to which 500 μl crude leaf extract was added when the medium had cooled to 50 °C. The plates were inoculated in the centre and incubated in the dark at 28 °C for 72 h, during which the hyphae grew outwards from the centre. Hyphal inhibition was checked daily and the hyphal growth was photographed using microscopy (Nikon) at 72 h.
Evaluation of transgenic plants for resistance against M. brunnea f.sp. multigermtubi
To test the resistance of transgenic poplar against fungal infections, the in vivo test was performed with M. brunnea f.sp. multigermtubi. Transgenic poplar plants were grown in a 14/10 light/dark and 25/23 °C cycle in the greenhouse. Hyphal fragments and spores were harvested and placed on the leaves of 3-month-old plants. The inoculated plants were incubated in a growth chamber for 4 d and the infected leaves were digitally photographed. Adobe Photoshop was used to calculate lesion area. Each experiment was performed twice, with at least three replicates per treatment, and contained wild-type controls.
Results
M. brunnea f.sp. multigermtubi infection induces PA biosynthesis in poplar leaves
In hybrid poplar, Melampsora medusae rust infection strongly induced genes for enzymes involved in flavonoid and proanthocyanidin biosynthesis late in the infection process (Miranda et al., 2007). To investigate the defence mechanisms of Chinese white poplar (P. tomentosa Carr.) for infection with black spot disease, total RNA was isolated from five plants infected with M. brunnea f.sp. multigermtubi and pooled together for the construction of DGE libraries as well as sequencing. In total, approximately 5.9 million filtered, high-quality reads with average length of 40 bp were sequenced from each sample and more than 20,000 contigs exhibited high similarity to a gene in the reference genome sequence of Populus (Phytozome version 2.0).
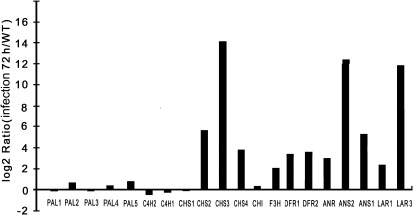
The expression patterns of flavonoid pathway up-stream genes (PtrPALs and PtrC4Hs) and structural genes involved in the biosynthesis of PAs (PtrCHSs, PtrCHI, PtrF3H, PtrDFRs, PtrANS, PtrANR, and PtrLARs) were examined in infected and control leaves. Almost all enzymes involved in the PA biosynthesis pathway are represented by more than one gene in poplar (Tsai et al., 2006). In the current study, at least one gene in each enzymic step was up-regulated, with the exception of cinnamate 4-hydroxylase (C4H); however, the PtrPAL paralogues PtrPAL1, PtrPAL3 and the PtrCHS paralogues PtrCHS1 were down-regulated. (Fig. 1 and Supplementary Table S1, available at JXB online).
Fig. 1.
Quantitative analysis of changes in transcript levels of flavonoid pathway genes in response to infection of poplar leaves with Marssonina brunnea f.sp. multigermtubi using digital gene expression analysis.
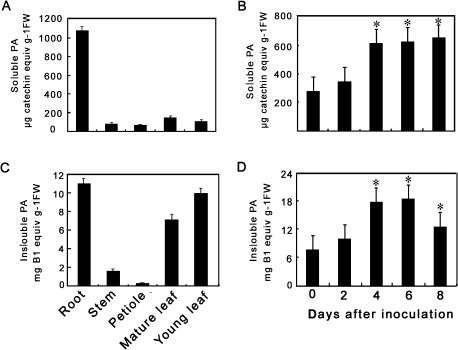
To correlate the gene expression data with PA accumulation, PA concentrations were examined in various tissues of P. tomentosa Carr (Fig. 2). The highest levels of soluble PAs (approximately 1.1 mg catechin equivalents (g fresh weight)−1) were detected in roots and much lower levels were found in stems, petioles, and leaves (Fig. 2A). Insoluble PAs were also extracted from the residues left after extraction of soluble PAs by repeated sonication in butanol/HCl (Porter et al., 1986), followed by heating to generate coloured anthocyanin from degradation of the PAs (Fig. 2C). The highest levels of insoluble PAs were detected in roots: about 11.4 mg procyanidin B1 equivalents (g fresh weight)−1. Insoluble PAs were also relatively high in leaves (young or mature), but very low in stems and petioles. This study then determined whether PAs accumulated in Chinese white poplar leaves following M. brunnea f.sp. multigermtubi infection. PAs were extracted from the leaves at various time points after inoculation. The results showed that a strong and statistically significant increase (P > 0.05) in PA levels in infected leaves at 4 days post inoculation (dpi) as compared with control leaves (Fig. 2B, D), which was consistent with the induction of PtrLARs and PtrANR, the genes specific to the PA biosynthesis pathway.
Fig. 2.
Levels of proanthocyanidins (PAs) in Populus tomentosa Carr. (A) Soluble PAs in different tissues from mature plants. (B) Insoluble PAs in different tissues from mature plants. (C) Soluble PAs in leaves at 0, 2, 4, 6, and 8 days after inoculation with Marssonina brunnea f.sp. multigermtubi. (D) Insoluble PAs in leaves at 0, 2, 4, 6, and 8 days after inoculation with M. brunnea f.sp. multigermtubi. Soluble PAs were determined by reaction with dimethylaminocinnamaldehyde reagent; insoluble PAs were determined by butanol/HCl hydrolysis and estimation of resulting anthocyanidin. Asterisks indicate significant differences using Student’s t-test (P < 0.05).
Isolation of PtrLAR3 cDNA encoding leucoanthocyanidin reductase from P. trichocarpa
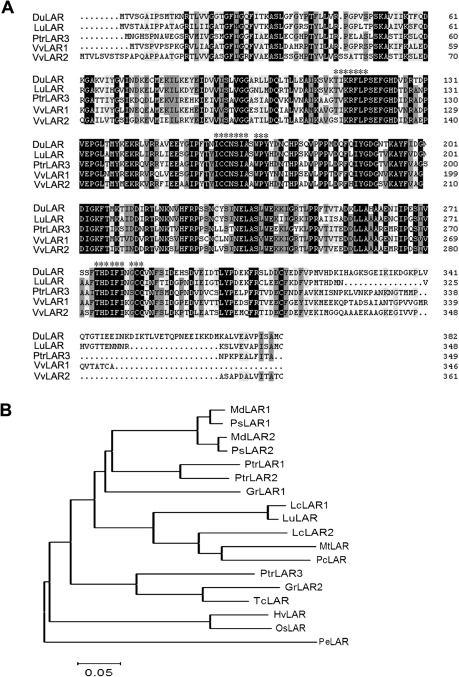
The final step of the PA biosynthesis pathway in poplar is catalysed by PtrLAR and PtrANR. The DGE analysis showed that, of the genes specific to the PA pathway, PtrLAR3 showed greater increase in expression level than PtrLAR1 or PtrANR in infected leaves (Fig. 1), indicating that PtrLAR3 may play major role in the accumulation of polymeric PAs against M. brunnea f.sp. multigermtubi infection. The isolation of a full-length PtrLAR3 cDNA would provide an important tool for analysing induction of PA synthesis at the molecular level. Based on the sequences deposited in the Populus genome database, the PtrLAR3 cDNA clone was cloned by RT-PCR from total RNA of P. trichocarpa leaves using gene-specific primers. The nucleotide sequence of PtrLAR3 was 1447-bp long and was predicted to encode a protein of 349 amino acid residues (Fig. 3A). Comparison of the PtrLAR3 sequence with databases showed high identity with LAR proteins from Vitis vinifera (VvLAR1, 59.49%; VvLAR2, 64.09%), Lotus uliginosus (LuLAR, 58.31%), and Desmodium uncinatum (DuLAR, 53.26%). The LAR-specific amino acid motifs RFLP, ICCN, and THD, as described previously (Tanner et al., 2003; Bogs et al., 2005), were found in the deduced PtrLAR3 protein, located at amino acid positions 113–123, 158–168, and 274–283, respectively. The conserved sequences of these motifs were identical in both LuLAR and DuLAR, with only two amino acid substitutions in VvLAR1 and one in VvLAR2 (Fig. 3A). The calculated isoelectric point and molecular mass of PtrLAR3 were 5.94 and 38.67 kD, respectively.
Fig. 3.
Comparison of PtrLAR3-deduced amino acid sequence with other LAR proteins. (A) Sequence alignment of the deduced PtrLAR3 sequence with other LAR sequences. Sequences are from Desmodium uncinatum (DuLAR, CAD79341), Lotus uliginosus (LuLAR, AAU45392), Populus tomentosa (PtrLAR3, EEF06163), Vitis vinifera (VvLAR1, AAZ82410; VvLAR2, AAZ82411). Identical amino acids are indicated by black letters on a white background, conservative amino acids by black on a dark gray background, and similar amino acids by black on a light gray background. Asterisks indicate the RFLP, ICCN, and THD motifs. Peptide sequences were aligned with the DNAMAN program. (B) Phylogenetic relationships of LAR proteins from P. tomentosa Carr. and selected species. Phylogenetic analysis was performed by the neighbour-joining method using MEGA version 4. Bar , 0.05 substitutions per site. GenBank accession numbers are: GrLAR1 (Gossypium raimondii, CAI56319); GrLAR2 (G. raimondii, CAI56325); HvLAR (Hordeum vulgare, CAI56320); LcLAR1 (Lotus corniculatus, ABC71326); LcLAR2 (L. corniculatus, ABC71328); LuLAR (Lotus uliginosus, AAU45392); MdLAR1 (Malus × domestica, AAX12185); MdLAR2 (Malus × domestica, AAX12186); MtLAR (Medicago truncatula, CAI56327); OsLAR (Oryza sativa, CAI56328); PcLAR (Phaseolus coccineus, CAI56322); PeLAR (Pinus taeda, CAI56321); PsLAR1 (Pyrus communis, ABB77696); PsLAR2 (Pyrus communis, ABB77697); PtrLAR1 (P. tomentosa, EEE89746); PtrLAR2 (P. tomentosa, EEF01056); PtrLAR3 (P. tomentosa, EEF06163); TcLAR (Theobroma cacao, ADD51358).
The neighbour-joining phylogenetic tree using the predicted amino acid sequences of LARs (Fig. 3B) showed that these proteins were clustered into two distinct groups, in which only PeLAR, from the gymnosperm Pinus taeda, belonged to one group, whereas LARs from angiosperm species belonged to another group. Of the angiosperm LARs, HvLAR (Hordeum vulgare) and OsLAR (Oryza sativa), of monocot species, formed a subgroup distinct from the dicot species. Among the dicotyledonous branches, the branch distribution revealed that PtrLAR3 formed an independent lineage in a cluster containing GsLAR2 from Gossypium arboretum and TcLAR from Theobroma cacao, suggesting that these proteins belonging to woody plants may have a close evolutionary relationship.
Tissue-specific expression of PtrLAR3
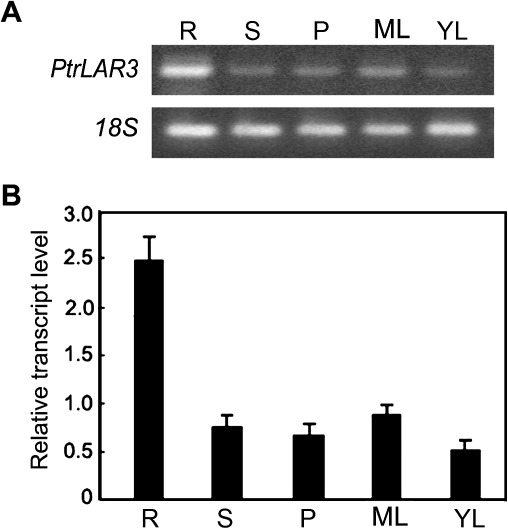
This study examined the expression of PtrLAR3 by semi-quantitative RT-PCR with total RNA from various tissues of P. trichocarpa (Fig. 4A). The highest mRNA level was observed in the roots, and relatively low but unambiguous expression was evident in the stems, leaves, and petioles. Quantitative real-time PCR analysis with different tissues further showed that the relative levels of the PtrLAR3 transcripts in the roots were more than twice as abundant as in the stems and were 3-fold higher than in the petioles (Fig. 4B). These results are consistent with those obtained for PA accumulation in different tissues of P. trichocarpa.
Fig. 4.
PtrLAR3 expression in Populus trichocarpa tissues. (A) Semi-quantitative RT-PCR analysis of PtrLAR3 expression in various tissues of P. trichocarpa. (B) Quantitative real-time PCR analysis of PtrLAR3 transcript levels in various tissues of P. trichocarpa. Poplar 18S expression was used as a control. Total RNA was isolated from roots (R), stems (S), petioles (P), mature leaves (ML), and young leaves (YL).
Molecular functional characterization of transgenic P. tomentosa Carr. plants overexpressing PtrLAR3
To investigate the function of PtrLAR3, the PtrLAR3 open reading frame under the control of the cauliflower mosaic virus 35S promoter was introduced into Chinese white poplar by A. tumefaciens-mediated transformation for constitutive expression. A total of 16 hygromycin-resistant putative transformants were obtained and grown in the greenhouse. None of the generated transgenic plants showed any phenotypic changes compared with wild-type plants (Supplementary Fig. S1). PCR analysis using gene-specific primers showed that an expected amplification product specific for Hpt was obtained from all transgenic lines tested, whereas no signal was detected from wild-type plants (Supplementary Fig. S2), confirming the integration of the transgene into the poplar genome. From all of the independent hygromycin-resistant transgenic lines harbouring the 35S:PtrLAR3 construct, three independent lines (2, 5, and 6) with high PtrLAR3 transcript levels were selected for further analysis.
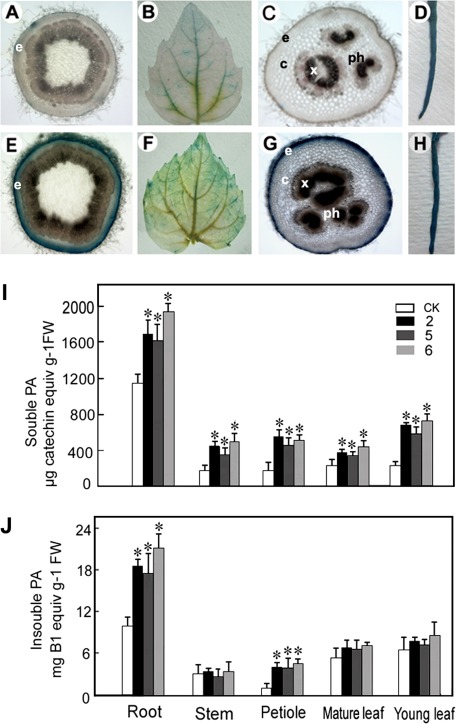
To determine PA localization in 35S:PtrLAR3 and control poplar plants, leaves, roots, petiole and stem sections were stained with DMACA, which reacts specifically with PAs and flavan-3-ols to form a blue chromophore (Li et al., 1996). As expected, DMACA stained the tissues of the empty-vector control lines (Fig. 5A–D) less intensely than the 35S:PtrLAR3 tissues (Fig. 5E–H). In stem sections, PAs were observed in the epidermis of control plants (Fig. 5A), while much higher concentration of PAs was detected in the epidermis in the PtrLAR3-overexpressing plants (Fig. 5E). In 35S:PtrLAR3 leaves, PAs were present at much higher concentrations compared with the control leaves (Fig. 5B and F). In petioles, staining was observed only in the epidermal cells in the controls (Fig. 5C), while in 35S:PtrLAR3 plants, strong staining was observed in the epidermal, cortex, phloem, and xylem cells (Fig. 5G). Most notably, DMACA staining was very abundant in the roots of both the control and the 35S:PtrLAR3 plants (Fig. 5D and H).
Fig. 5.
PA accumulation in different tissues of transgenic lines constitutively expressing PtrLAR3. PAs were localized by staining different tissues of control and PtrLAR3-overexpressor plants with the PA-specific stain dimethylaminocinnamaldehyde (DMACA; blue). (A–D) Control stem, leaf, petiole, and root, respectively. (E–H) PtrLAR3-overexpressing stem, leaf, petiole, and root, respectively. (I) Soluble PA levels in different tissues as determined by extraction and reaction with DMACA reagent. (J) Insoluble PA levels in different tissues. CK, empty-vector control; 2, 5, and 6, PtrLAR3-overexpressing lines. Asterisks indicate significant differences using Student’s t-test (P < 0.05).
To further confirm that PA biosynthesis can indeed be enhanced by PtrLAR3 overexpression, PA levels in various tissues of control and 35S:PtrLAR3 plants were determined. Three transgenic lines (2, 5, and 6) showed significantly increased contents (P < 0.05) of both soluble and insoluble PAs in their roots and petioles compared with those in the empty-vector controls (Fig. 5I, J). The increases of soluble PAs in the stems and leaves of all 35S:PtrLAR3 lines were significant (P < 0.05) but the increases of insoluble PAs were not (Fig. 5I, J), which was consistent with the results of DMACA staining. Taken together, in PtrLAR3-overexpressing plants, many of the same cell types that are competent in producing PAs are stimulated to produce much higher PA levels.
Overexpression of PtrLAR3 in P. tomentosa Carr. enhanced resistance to M. brunnea f.sp. multigermtubi
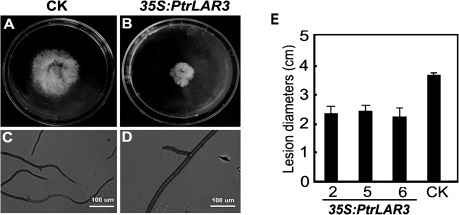
It is well established that PAs inhibit the in vitro growth of several filamentous fungi, thus preventing their rapid development in planta (Swain and Hillis, 1959). To determine whether the constitutive expression of PtrLAR3 conferred resistance to fungal pathogens, the antifungal activity of crude leaf extracts from transgenic poplar plants was tested against M. brunnea f.sp. multigermtubi. As shown in Fig. 6, in vitro mycelial growth of the pathogens was inhibited by crude extracts from transgenic 35S:PtrLAR3 plants, whereas no inhibition zone was found with the use of extracts from the empty-vector control line. Quantitative measurement revealed that leaf crude extracts from transgenic 35S:PtrLAR3 plants inhibited hyphal growth of M. brunnea f.sp. multigermtubi by up to 45% (Fig. 6E). Furthermore, the inhibition by PAs against M. brunnea f.sp. multigermtubi was evaluated microscopically: the crude extracts from the 35S:PtrLAR3 lines caused abnormal hyphal growth of M. brunnea f.sp. multigermtubi, seen as short hyphae, swollen tips, and less hyphal branches, in comparison to the controls (Fig. 6C, D).
Fig. 6.
In vitro antifungal activity of crude extracts from transgenic 35S:PtrLAR3 plants. (A, B) Inhibition of hyphal growth of Marssonina brunnea f.sp. multigermtubi on the plate supplied with crude leaf extracts from control plants (A) and PtrLAR3-overexpressing plants (B). (C, D) Microscopic observation of hyphal growth of M. brunnea f.sp. multigermtubi on plates without (C) and with (B) crude leaf extracts. Photomicrographs were taken after 72 h of incubation of M. brunnea f.sp. multigermtubi; bars, 100 μm. (E) Quantitative measurement of the inhibition of fungal growth of M. brunnea f.sp. multigermtubi with crude leaf extracts from control and PtrLAR3-overexpressing plants. CK, empty-vector control; 2, 5, and 6, PtrLAR3-overexpressing lines. Values are means of at least three replications. Error bars indicate standard deviations.
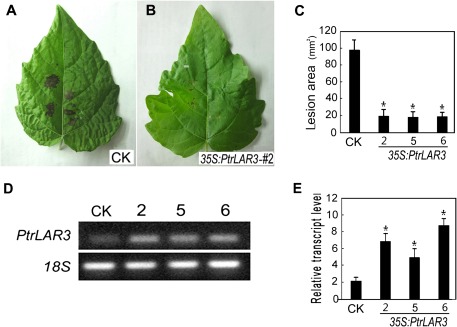
To assess the resistance of transgenic poplar plants overexpressing PtrLAR3 against black spot disease, excised leaves of transgenic and control lines were inoculated with agar plugs containing hyphae of M. brunnea f.sp. multigermtubi. Compared with the severe disease symptoms seen on the control leaves at 4 dpi (Fig. 7A), only slight necrotic lesions appeared on the leaves of all of the transgenic 35S:PtrLAR3 lines tested (Fig. 7B). Further quantification assays showed that lesions were significantly larger in the control plants (P < 0.05) than in 35S:PtrLAR3 lines (Fig. 7C), indicating that the transgenic lines constitutively expressing PtrLAR3 had increased resistance to fungal pathogens. RT-PCR analysis showed that PtrLAR3 was expressed in the transgenic plants at various levels (Fig. 7D). Further, quantitative experiments revealed that high-level transgene expression was found in the selected transgenic lines (Fig. 7E), compared to the empty-vector control line, suggesting that the level of disease resistance in transgenic plants could be positively correlated to PtrLAR3 mRNA levels.
Fig. 7.
Resistance of transgenic poplar plants inoculated with Marssonina brunnea f.sp. multigermtubi. Poplar leaves infected with M. brunnea f.sp. multigermtubi were photographed 4 days after inoculation. (A) Empty-vector control leaves. (B) Transgenic leaves from 35S:PtrLAR3 line 2. (C) Mean infected area of transgenic lines to the fungal pathogen; PtrLAR3 confers resistance to M. brunnea f.sp. multigermtubi in transgenic poplar plants. (D) Semi-quantitative reverse-transcription PCR analysis of PtrLAR3 expression in leaves of transgenic poplar plants; ethidium bromide-stained products amplified with PtrLAR3-specific primers from transgenic poplar cDNA and control cDNA. (E) Quantitative real-time PCR analysis of PtrLAR3 transcript levels in leaves of transgenic poplar plants. CK, empty-vector control; 2, 5, and 6, PtrLAR3-overexpressing lines. Values are means of three replications. Error bars indicate standard deviation. Asterisks indicate a statistically significant difference between control and transgenic plants (P < 0.05 by Student’s t-test).
Discussion
Worldwide, Populus is of increasing importance as commercial sources of fibre and fuel (Stettler et al., 1996). With the rapid development of artificial poplar forests, poplar diseases have increased. Marssonina leaf spot is one of the most common and devastating diseases of poplar, caused by the coelomycetous genus Marssonina (O’Riordain and Kavanagh, 1965; Li, 1984; Newcombe and Callan, 1998; Spiers, 1998). Marssonina causes very small, dark brown spots on leaves, petioles, and young green capsules and infected poplar trees exhibit premature leaf fall and reduction in photosynthetic capacity and growth. The fungus M. brunnea (e.g. M. brunnea f.sp. multigermtubi) has been considered the most important pathogen and occurs in most regions of China, resulting in serious economic losses in poplar plantations.
In poplar trees, PAs accumulate in leaves, roots, and a variety of other tissues, with levels as high as 35% dry weight (Salminen et al., 2004). In the present study, the higher PA contents were found in roots and leaves of P. tomentosa Carr., compared to stems and petioles (Fig. 2A, C). The flavonoid-derived PAs are considered as the major defence phenolics produced in poplar leaves. Damage to leaves by insect herbivory causes a rapid accumulation of PAs, both at the site of damage and distally in undamaged leaves (Peters and Constabel, 2002), suggesting that these compounds function in herbivore defence. The present study also found that PA accumulation was stimulated in leaves of Chinese white poplar by M. brunnea f.sp. multigermtubi at 3 dpi (Fig. 2B, D). This rapid PA accumulation is mediated by the activation of genes encoding enzymes involved in PA synthesis (Tsai et al., 2006). Therefore, the present study further determined the transcriptional response of P. tomentosa Carr. to infection by M. brunnea f.sp. multigermtubi using a DGE analysis. The results showed that flavonoid pathway genes were strongly induced, leading to PA accumulation in response to M. brunnea f.sp. multigermtubi (Fig. 1). The accumulation of PAs following mechanical wounding, fungal infection, or herbivore attack has been observed in previous studies. For instance, mechanical wounding, insect herbivory, and methyl jasmonate treatment all induced expression of dihydrofavonol-4-reductase, a key enzyme for PA synthesis, in aspen leaves resulting in PA accumulation (Peters and Constabel, 2002). In a hybrid cottonwood (Populus fremontii × angustifolia), transcript abundance of leaf-expressed flavonoid genes essential for synthesis of the PA precursor proanthocyanidin was wound stimulated (Tsai et al., 2006), and the most strongly wound-induced gene family members in P. fremontii × angustifolia also correspond to those genes identified in this study as being most highly induced by M. brunnea f.sp. multigermtubi in P. tomentosa Carr. leaves.
To date, genetic evidence for LAR function in late flavonoid biosynthetic pathway has been obtained for several plants (Tanner et al., 2003; Bogs et al., 2005; Routaboul et al., 2006; Pang et al., 2007; Paolocci et al., 2007). Interestingly, Arabidopsis does not possess an obvious LAR orthologue and has a PA composed of only epicatechin (Abrahams et al., 2003; Tanner et al., 2003). Furthermore, LAR expression is found in non-seed tissues of some species, such as L. corniculatus (Paolocci et al., 2007) and grape (Bogs et al., 2005), and these tissues accumulate high levels of PAs. PAs may play a multifunctional role in plants, including defence against microbes, protection from photo-damage, and storage of excess carbon (Hemming and Lindroth, 1995; Close and McArthur, 2002; Dixon, 2005). In the present study, the flavonoid structural gene family members (including CHS, CHI, F3H, DFR, and ANS) analysed in the DGE experiments were up-regulated within 72 h of pathogen infection in poplar, as were the PA-specific ANR and LAR, which participate in two separate pathways to PA biosynthesis in most species (Dixon et al., 2005).
A high level of accumulation of PtrLAR3 transcripts was observed in poplar leaves infected by M. brunnea f.sp. multigermtubi, indicating this gene may play an important role in pathogen defence in poplar. In order to investigate further, the present study overexpressed PtrLAR3 in poplar. As expected, PAs were present at much higher concentrations in roots as compared with leaves of control plants (Fig. 5). The quantitative assay using DMACA/HCl to detect soluble PAs and butanol/HCl to detect insoluble PAs revealed a significant increase in soluble PA concentrations (P < 0.05) in all tissues analysed, whereas insoluble PAs accumulated to higher levels in the roots and petioles of all transgenic lines, compared to the stems and leaves (Fig. 5). Interestingly, in a previous study, Desmodium LAR was ectopically expressed in tobacco but catechin was not accumulated (Tanner et al., 2003). Recently, transgenic tobacco plants constitutively overexpressing MtLAR from M. truncatula showed reduced anthocyanin content, but no catechin or increased levels of PAs were detected either in leaves or in flowers (Pang et al., 2007). The precise function of PtrLAR3 in PA biosynthesis in poplar requires further investigation through in vitro biochemical analysis.
Plants encounter numerous pathogens in the natural environment. Most plants produce a broad range of secondary metabolites that are toxic to pathogens and herbivores. PAs are important molecules for plant adaptation to the environment. In poplar leaves, the transcriptional activation of the PA biosynthetic genes leading to PA accumulation in leaves occurs after herbivore damage and mechanical wounding as well as after infection by the fungal biotroph Melampsora medusae (Peters and Constabel, 2002; Stevens and Lindroth, 2005; Miranda et al., 2007; Mellway et al., 2009). The present data demonstrated that PtrLAR3 overexpression resulted in PA accumulation in poplar leaves (Fig. 5), therefore, it was speculated that transgenic plants overexpressing PtrLAR3 would display increased resistance to fungal pathogens. In vitro assays using crude leaf extracts from transgenic 35:PtrLAR3 plants showed that hyphal growth of M. brunnea f.sp. multigermtubi was inhibited significantly, while no inhibition of hyphal growth was found with the use of extracts isolated from the empty-vector control plants (Fig. 6). This finding is agreement with PA toxicity to several filamentous fungi (Harborne, 1980; Scalbert, 1991). In vivo assays (detached leaves) showed that, after infection with M. brunnea f.sp. multigermtubi, there was a significant reduction in disease symptoms in transgenic leaves compared with the control (Fig. 7A–C), indicating that overexpression of PtrLAR3 in poplar significantly enhanced resistance to fungal pathogens.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Table S1. Comparison of changes in expression levels of PA biosynthetic genes leading to PA accumulation following Marssonina brunnea f.sp. multigermtubi infection using DGE analysis.
Supplementary Fig. S1. Photographs of PtrLAR3-overexpressing Populus tomentosa Carr. after 4 weeks of growth.
Supplementary Fig. S2. PCR analysis of transgenic poplar plants.
Acknowledgments
The authors thank Prof. Jingjiang Hu (Northwest A&F University, Shanxi, China) for providing M. brunnea f.sp. multigermtubi. This work was supported by the National Natural Science Foundation of China (3081576, 31171620), the National Key Project for Research on Transgenic Plant (2011ZX08010-003), the Natural Science Foundation Project of CQ CSTC (CSTC, 2009BA1004), and the Fundamental Research Funds for the Central Universities (XDJK2009B018).
References
- Abrahams S, Lee E, Walker AR, Tanner GJ, Larkin PJ, Ashton AR. The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. The Plant Journal. 2003;35:624–636. doi: 10.1046/j.1365-313x.2003.01834.x. [DOI] [PubMed] [Google Scholar]
- Bagchi D, Bagchi M, Stohs SJ, Das DK, Ray SD, Kuszynski CA, Joshi SS, Pruess HG. Free radicals and grape seed proanthocyanidin extract: importance in human health and disease prevention. Toxicology. 2000;148:187–197. doi: 10.1016/s0300-483x(00)00210-9. [DOI] [PubMed] [Google Scholar]
- Bogs J, Downey MO, Harvey JS, Ashton AR, Tanner GT, Robinson SP. Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape and grapevine leaves. Plant Physiology. 2005;139:652–663. doi: 10.1104/pp.105.064238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Davies C, Robinson SP. Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiology. 1996;111:1059–1066. doi: 10.1104/pp.111.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Songkumarn P, Liu J, Wang GL. A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiology. 2009;150:1111–1121. doi: 10.1104/pp.109.137125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close DC, McArthur C. Rethinking the role of many plant phenolics: protection from photodamage not herbivores? Oikos. 2002;9:166–172. [Google Scholar]
- Cos P, De Bruyne T, Hermans N, Apers S, Berghe DV, Vlietinck AJ. Proanthocyanidins in health care: current and new trends. Current Medicinal Chemistry. 2004;11:1345–1359. doi: 10.2174/0929867043365288. [DOI] [PubMed] [Google Scholar]
- Dixon RA. Engineering of plant natural product pathways. Current Opinion in Plant Biology. 2005:329–336. doi: 10.1016/j.pbi.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Dixon RA, Lamb CJ, Masoud S, Sewalt VJH, Paiva NL. Metabolic engineering: prospects for crop improvement through the genetic manipulation of phenylpropanoid biosynthesis and defense responses – a review. Gene. 1996;179:61–71. doi: 10.1016/s0378-1119(96)00327-7. [DOI] [PubMed] [Google Scholar]
- Dixon RA, Sharma SB, Xie D. Proanthocyanidins: a final frontier in flavonoid research? New Phytologist. 2005;165:9–28. doi: 10.1111/j.1469-8137.2004.01217.x. [DOI] [PubMed] [Google Scholar]
- Douglas GB, Stienezen M, Waghorn GC, Foote AG, Purchas RW. Effect of condensed tannins in birdsfoot trefoil (Lotus corniculatus) and sulla (Hedysarum coronarium) on body weight, carcass fat depth, and wool growth of lambs in New Zealand. New Zealand Journal of Agricultural Research. 1999;42:55–64. [Google Scholar]
- Harborne JB. Secondary plant products. In: EA Bell, BW Charlwood., editors. Encyclopedia of Plant Physiology. 1980. new series, vol. 8. Berlin: Springer-Verlag, pp. 329–420. [Google Scholar]
- Harborne JB, Grayer RJ. Flavonoids and insects. In: JB Harborne., editor. The Flavonoids: advances in research since 1986. London: Chapman and Hall; 1993. pp. 589–618. [Google Scholar]
- Hemming JDC, Lindroth RL. Intraspecific variation in aspen phytochemistry: effects on performance of gypsy moths and forest tent caterpillars. Oecologia. 1995;10:79–88. doi: 10.1007/BF00328428. [DOI] [PubMed] [Google Scholar]
- Jansson S, Douglas CJ. Populus: a model system for plant biology. Annual Review of Plant Biology. 2007;58:435–458. doi: 10.1146/annurev.arplant.58.032806.103956. [DOI] [PubMed] [Google Scholar]
- Jia ZC, Sun YM, Yuan L, Tian Q, Luo KM. The chitinase gene (Bbchit1) from Beauveria bassiana enhances resistance to Cytospora chrysosperma in Populus tomentosa Carr. Biotechnology Letters. 2010;32:1325–1332. doi: 10.1007/s10529-010-0297-6. [DOI] [PubMed] [Google Scholar]
- Joseph R, Tanner G, Larkin P. Proanthocyanidin synthesis in the forage legume Onobrychis vicifolia: a study of chalcone synthase, dihydroflavonol 4-reductase and leucoanthocyanidin 4-reductase in developing leaves. Australian Journal of Plant Physiology. 1998;25:271–278. [Google Scholar]
- Li CD. Study on two specifications of Marssonina populi. Journal of Nanjing Forestry University. 1984:10–16. [Google Scholar]
- Li RQ, Li YR, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- Li YG, Tanner GJ, Larkin PJ. The DMACA-HCl protocol and the threshold proanthocyanidin content for bloat safety in forage legumes. Journal of the Science of Food and Agriculture. 1996;70:89–101. [Google Scholar]
- Lin LC, Kuo YC, Chou CJ. Immunomodulatory proanthocyanidins from Ecdysanthera utilis. Journal of Natural Products. 2002;65:505–508. doi: 10.1021/np010414l. [DOI] [PubMed] [Google Scholar]
- Lloyd G, McCown B. Commercially feasible micropropagation of mountain laural (Kalmla latlfolia) by use of shoot tip cultures. Combined Proceedings of the International Plant Propagators’ Society. 1980;30:421–427. [Google Scholar]
- Luo KM, Zheng XL, Chen YQ, Xiao YH, Zhao DG, McAvoy R, Pei Y, Li Y. The maize Knotted1 gene is an effective positive selectable marker gene for Agrobacterium-mediated tobacco transformation. Plant Cell Reports. 2006;2:403–409. doi: 10.1007/s00299-005-0051-z. [DOI] [PubMed] [Google Scholar]
- Marles MAS, Ray H, Gruber MY. New perspectives on proanthocyanidin biochemistry and molecular regulation. Phytochemistry. 2003;64:367–383. doi: 10.1016/s0031-9422(03)00377-7. [DOI] [PubMed] [Google Scholar]
- McMahon LR, McAllister TA, Berg BP, Majak W, Acharya SN, Popp JD, Coulman BE, Wang Y, Cheng KJ. A review of the effects of forage condensed tannins on ruminal fermentation and bloat in grazing cattle. Canadian Journal of Plant Science. 2000;80:469–485. [Google Scholar]
- Mellway RD, Tran LT, Prouse MB, Campbell MM, Constabel CP. The wound-, pathogen-, and ultraviolet B-responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in poplar. Plant Physiology. 2009;150:924–941. doi: 10.1104/pp.109.139071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M, Ralph SG, Mellway R, White R, Heath MC, Bohlmann J, Constabel CP. The transcriptional response of hybrid poplar (Populus trichocarpa × P. deltoides) to infection by Melampsora medusae leaf rust involves induction of flavonoid pathway genes leading to the accumulation of proanthocyanidins. Molecular Plant–Microbe Interactions. 2007;20:816–831. doi: 10.1094/MPMI-20-7-0816. [DOI] [PubMed] [Google Scholar]
- Newcombe G. Association of Mmdl, a major gene for resistance to Melampsora medusae f.sp. deltoidae, with quantitative traits in poplar rust. Phytopathology. 1998;88:114–121. doi: 10.1094/PHYTO.1998.88.2.114. [DOI] [PubMed] [Google Scholar]
- O’Riordain F, Kavanagh T. Marssonina leaf spot of poplar. Irish Journal of Agricultural Research. 1965;4(2):233–35. [Google Scholar]
- Pang YZ, Peel GJ, Wright E, Wang ZY, Dixon RA. Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula. Plant Physiology. 2007;145:601–615. doi: 10.1104/pp.107.107326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolocci F, Robbins MP, Madeo L, Arcioni S, Martens S, Damiani F. Ectopic expression of a basic helix-loop-helix gene transactivates parallel pathways of proanthocyanidin biosynthesis: structure, expression analysis, and genetic control of leucoanthocyanidin 4-reductase and anthocyanidin reductase genes in Lotus corniculatus. Plant Physiology. 2007;143:504–516. doi: 10.1104/pp.106.090886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters DJ, Constabel CP. Molecular analysis of herbivore-induced condensed tannin synthesis: cloning and expression of dihydroflavonol reductase from trembling aspen (Populus tremuloides) The Plant Journal. 2002;32:701–712. doi: 10.1046/j.1365-313x.2002.01458.x. [DOI] [PubMed] [Google Scholar]
- Porter LJ, Hrstich LN, Chan BG. The conversion of procyanidins and prodelphinidins to cyanidin and delphinidin. Phytochemistry. 1986;2:223–230. [Google Scholar]
- Routaboul JM, Kerhoas L, Debeaujon I, Pourcel L, CabocheM Einhorn J, Lepiniec L. Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta. 2006;224:96–107. doi: 10.1007/s00425-005-0197-5. [DOI] [PubMed] [Google Scholar]
- Salminen JP, Roslin T, Karonen M, Sinkkonen J, Pihlaja K, Pulkkinen P. Seasonal variation in the content of hydrolyzable tannins, flavonoid glycosides and proanthocyanidins in oak leaves. Journal of Chemistry Ecology. 2004;30:1693–711. doi: 10.1023/b:joec.0000042396.40756.b7. [DOI] [PubMed] [Google Scholar]
- Scalbert A. Antimicrobial properties of tannins. Phytochemistry. 1991;30:3875–3883. [Google Scholar]
- Shirley BW, Hanley S, Goodman HM. Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. The Plant Cell. 1992;4:333–347. doi: 10.1105/tpc.4.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiers AG. Melampsora and Marssonina pathogens of poplars and willows in New Zealand. European Journal of Plant Pathology. 1998;28:233–240. [Google Scholar]
- Springob K, Nakajima J, Yamazaki M, Saito K. Recent advances in the biosynthesis and accumulation of anthocyanins. Natural Product Reports. 2003;20:288–303. doi: 10.1039/b109542k. [DOI] [PubMed] [Google Scholar]
- Stettler JA, Ward JV, Liss WJ, Frissell CA, Williams RN, Lichatowich JA, Coutant CC. A general protocol for restoration for restoration of regulated rivers. Regulated Rivers: Research & Management. 1996;12:391–414. [Google Scholar]
- Stevens MT, Lindroth RL. Induced resistance in the indeterminate growth of aspen (Populus tremuloides) Oecologia. 2005;145:298–306. doi: 10.1007/s00442-005-0128-y. [DOI] [PubMed] [Google Scholar]
- Swain T, Hillis WEJ. Phenolic constituents of Prunus domestica I. Quantitative analysis of phenolic constituents. Journal of the Science of Food and Agriculture. 1959;10:63. [Google Scholar]
- Tanner GJ, Francki KT, Abrahams S, Watson JM, Larkin PJ, Ashton AR. Proanthocyanidin biosynthesis in plants: purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. Journal of Biology Chemistry. 2003;278:31647–31656. doi: 10.1074/jbc.M302783200. [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Harding SA, Tschaplinski TJ, Lindroth RL, Yuan Y. Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New Phytologist. 2006;172:47–62. doi: 10.1111/j.1469-8137.2006.01798.x. [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, et al. The genome of black cottonwood Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Guo G, Hu X, et al. Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Research. 2010;20:646–54. doi: 10.1101/gr.100677.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.