Abstract
MYB-type transcription factors play a diverse role in plant development and response to abiotic stress. This study isolated a rice R2R3-type MYB gene, OsMYB2, and functionally characterized its role in tolerance to abiotic stress by generating transgenic rice plants with overexpressing and RNA interference OsMYB2. Expression of OsMYB2 was up-regulated by salt, cold, and dehydration stress. OsMYB2 was localized in the nucleus with transactivation activity. No difference in growth and development between the OsMYB2-overexpressing and wild-type plants was observed under normal growth conditions, but the OsMYB2-overexpressing plants were more tolerant to salt, cold, and dehydration stresses and more sensitive to abscisic acid than wild-type plants. The OsMYB2-overexpressing plants accumulated greater amounts of soluble sugars and proline than wild-type plants under salt stress. Overexpression of OsMYB2 enhanced up-regulation of genes encoding proline synthase and transporters. The OsMYB2-overexpressing plants accumulated less amounts of H2O2 and malondialdehyde. The enhanced activities of antioxidant enzymes, including peroxidase, superoxide dismutase, and catalase, may underlie the lower H2O2 contents in OsMYB2-overexpressing plants. There was greater up-regulation of stress-related genes, including OsLEA3, OsRab16A, and OsDREB2A, in the OsMYB2-overexpressing plants. Microarray analysis showed that expression of numerous genes involving diverse functions in stress response was altered in the OsMYB2-overexpressing plants. These findings suggest that OsMYB2 encodes a stress-responsive MYB transcription factor that plays a regulatory role in tolerance of rice to salt, cold, and dehydration stress.
Keywords: Abiotic stress, MYB, OsMYB2, oxidative stress, proline, rice, soluble sugars
Introduction
Plants are sessile organisms and are frequently exposed to variable environmental stress that adversely affect plant growth and agricultural production. To cope with the stress, plants have evolved efficient mechanisms to sense and rapidly adapt to stressed conditions. As the phytohormone abscisic acid (ABA) plays a crucial role in the adaptive response of plants to abiotic stresses, the pathways leading to the adaptation to stress can be divided into two major categories: ABA-dependent and ABA-independent pathways (Xiang et al., 2008). During the response and adaptation to abiotic stress, there are many changes in biochemical and physiological processes. These include accumulation of osmolytes and cryoprotectants such as sugar and proline (Xin and Browse, 1998) to facilitate osmo-regulation and prevent oxidative damage due to disruption of reactive oxygen species (ROS) homeostasis (Suzuki and Mittler, 2006). In addition, many genes are activated, leading to accumulation of numerous proteins involved in resistance to abiotic stress, such as late embryogenesis abundant (LEA; Ma et al., 2010). Activation of the genes can protect plants from oxidative damage due to osmotic stress, ionic toxicity, and oxidative stress (Bartels, 2005). The expression of stress-induced genes is largely regulated by specific transcription factors (Hu et al., 2008). Among these transcription factors, members of the APETELA2 (AP2), bZIP, NAC, and MYB families have been well characterized for their regulatory roles in the response of plants to abiotic stress (Hu et al., 2008; Takasaki et al., 2010). There have been a number of studies demonstrating that transgenic plants overexpressing genes encoding transcription factors can greatly enhance their tolerance to various abiotic stresses such as salinity, cold, and drought (Dubouzet et al., 2003; Vannini et al., 2004; Nakashima et al., 2007; Xiang et al., 2008; Song et al., 2011).
MYB transcription factors occur widely in animals, plants, and fungi (Lippold et al., 2009). They were first identified as oncogenes in animals, where their function is linked to control of the cell cycle (Ito et al., 2001). MYB proteins contain one, two, or three imperfect repeats (51–53 amino acids) in their DNA-binding domain, and they are further classified into three subfamilies, type MYBR2R3, type MYBR1R2R3, and MYB-related, depending on the number of repeats in their MYB domains (Stracke et al., 2001; Chen et al., 2006). Among the MYB proteins in plants, the MYB family with the two-repeat (R2R3) is the most common one (Stracke et al., 2001). There are 126 and 109 R2R3-type MYB proteins in Arabidopsis and rice, respectively (Chen et al., 2006). The involvement of Arabidopsis MYB proteins in the regulation of secondary metabolism, control of cellular morphogenesis, and regulation of the meristem and the cell cycle has been demonstrated (Kranz et al., 1998).
More recently, R2R3-type MYB proteins are reported to be involved in responses of plants to environmental stress. For instance, AtMYC2 and AtMYB2 proteins play important roles as transcription factors in ABA-dependent gene expression under drought and salt stress (Abe et al., 2003). Denekamp and Smeekens (2003) reported that AtMYB102 is a key component to integrate signalling pathways in responses of Arabidopsis to wounding, osmotic stress, and ABA. Several studies also revealed that MYB proteins in Arabidopsis (AtMYB44, AtMYB60, and AtMYB61) are involved in regulation of stomatal aperture in response to drought stress (Cominelli et al., 2005; Liang et al., 2005; Jung et al., 2008). In addition, AtMYB15 was found to negatively regulate the induction of cold-responsive genes in an ABA-independent way (Agarwal et al., 2006). Lippold et al. (2009) reported that AtMYB41 controls the short-term transcriptional responses to osmotic stress.
Three MYB proteins have been reported to be involved in response of rice to abiotic stress. For instance, overexpression of OsMYB4 significantly confers tolerance to chilling and freezing stress in transgenic Arabidopsis (Vannini et al., 2004; Pasquali et al., 2008). Ma et al. (2009) reported that OsMYB3R-2 participates in the cold signalling pathway by targeting the cell cycle and a putative DREB/CBF. Moreover, a recent study revealed that OsMYBS3 is essential for conferring tolerance of rice plants to cold stress (Su et al., 2010). Among these characterized MYB proteins, OsMYB4 is the only R2R3-type protein, while OsMYB3R-2 and MYBS3 are R1R2R3-type and MYB-related proteins, respectively. Furthermore, studies on rice MYB proteins have mainly focused on their roles in response to cold stress, while little is known about the role of MYB proteins in response of rice to other abiotic stress, such as salt and dehydration stress.
This study isolated a R2R3-MYB transcription factor, designated OsMYB2, in rice. The role of OsMYB2 in response of rice plants to salt, cold, and dehydration stress was characterized by generating transgenic plants with overexpressing and RNA interference (RNAi) OsMYB2.
Materials and methods
Plant material, growth conditions, and stress treatments
Seeds of rice cultivar Zhonghua 10 (Oryza sativa L.) were surface-sterilized by incubation for 3 min in 75% ethanol, followed by 10 min in 0.1% HgCl2, and then washed thoroughly with sterile water. The sterilized seeds were germinated on half-strength Murashige and Skoog (1/2 MS) agar (0.6%, w/v, agar; pH 5.8) in darkness for 2 days. Thereafter the germinated seedlings were grown in a greenhouse at 28/25 °C (day/night) with a 14-h photoperiod. Two-week-old seedlings were treated with varying chemicals and abiotic stresses after the MS agar was washed off. Chemical treatments were conducted by exposing the seedlings to 1/2 MS medium containing 100 μM abscisic acid (ABA), 100 μM indoleacetic acid, 100 μM salicylic acid, or 10 μM brassinosteroids for 5 h and sampled for further analysis. For treatment with salt stress, 2-week-old seedlings were submerged into 1/2 MS medium containing 200 mM NaCl and sampled at varying periods after treatments. For cold and dehydration treatments, the 2-week-old seedlings were exposed to 2 °C and 20% PEG solution, respectively, and sampled at 0, 5, and 10 h after treatments.
Subcellular localization and transactivation assay
The whole coding sequence of OsMYB2 was ligated with XbaI and KpnI-digested pBI221 vector to generate pBI221-OsMYB2-GFP containing an OsMYB2-GFP fusion construct under the control of cauliflower mosaic virus 35S (CaMV 35S) promoter. The construct was confirmed by sequencing and used for transient transformation of onion (Allium cepa) epidermis via a gene gun (Bio-Rad). After 24 h of incubation, GFP fluorescence in transformed onion cells was observed under a confocal microscope (Zeiss, Germany).
To determine the transactivation activity, the open reading frame of OsMYB2 was generated by PCR amplification, cloned into KpnI and XbaI sites, and fused in-frame to the GAL4 DNA binding domain in the vector pGBKT7 (Invitrogen). The fused construct of pGBKT7-OsMYB2 was transformed into AH109 cells by the lithium acetate-mediated method. The transformed yeast strain was plated on SD/–Trp medium at 28 °C for 2 days. Yeast transformants from SD medium lacking Trp were then transferred and streaked onto solid SD agar lacking Trp/His/Ade (SD/–Trp/–His/–Ade) to score the growth response after 3 days. For the colony-lift filter assay (X-gal assay), the yeast was transferred to Whatman filter paper plus X-gal for transcription activation activity analysis within 8 h. Transcription factor OsNAC5 was used as a positive control.
Vector construction and plant transformation
The full-length cDNA of OsMYB2 were amplified from rice with the primers 5′-CGCGGATCCATGGACATGGCGCACGAGAG-3′ (BamHI site underlined) and 5′-CGGGGTACCTCACACGGCGGCCTGGGTGG-3′ (KpnI site underlined). The product was ligated into pGEM-T Easy vector (Promega) and sequenced. Then the OsMYB2 fragment digested from pGEM-T Easy-OsMYB2 was cloned into the KpnI-BamHI sites of a pUN1301 vector to obtain the pUN1301-OsMYB2 construct. OsMYB2 was driven by an ubiquitin promoter in the construct and a GUS marker was carried in the vector pUN1301 as described previously (Ge et al., 2004). The pUN1301-OsMYB2 construct was electroporated into Agrobacterium tumefaciens EHA105 and then introduced into rice embryonic calli by A. tumefaciens EHA105-meditated methods (Xu et al., 2005). OsMYB2 transgenic rice plants were selected in 1/2 MS medium containing 75 mg l−1 hygromycin (Roche, Germany).
The RNAi plasmid was constructed as described by Wang et al. (2004). Briefly, a 314-bp fragment of OsMYB2 was amplified using the primers 5′-GGGGTACCACTAGTGAGCTGTCGAGCACCACG-3′ (KpnI and SpeI sites underlined) and 5′-CGGGATCCGAGCTCTGGTCGTCCTCCATGCTC-3′ (BamHI and SacI sites underlined). Gene transformation protocols were used as described above.
Southern blot
Genomic DNA extracted from 14-d-old seedlings was digested with EcoRI, electrophoresed on a 0.8% agarose gel, and blotted onto a nylon membrane (Hybond–XL; Amersham Pharmacia Biotech) under alkaline conditions. α-32P-dCTP-labeled GUS amplified from pUN1301 was used as a probe for hybridization. The membrane was exposed to X-ray film (Eastern Kodak) at –80 °C for 5 d.
Determination of tolerance to salt, cold, and dehydration stress
Two-week-old seedlings of both wild-type and OsMYB2 transgenic rice were submerged into 1/2 MS medium supplemented with 200 mM NaCl for 2 days. Then the plants were transferred into the incubation solution without NaCl for an additional 4 days. For cold stress, 2-week-old seedlings of wild-type and OsMYB2 transgenic rice were subjected to treatment at 2 °C for 3 days and then transferred into a greenhouse at a temperature of 28/25 °C (day/night) with a 14-h photoperiod for 1 week. For dehydration stress, wild-type and OsMYB2 transgenic plants were exposed to 1/2 MS medium containing 20% PEG for 2 days and recovered in normal growth conditions for 1 week.
Wild-type and transgenic plants grown in soil were used to determine the stress tolerance. For treatment with salt stress, 40-d-old plants grown in pots were watered with 100 mM NaCl for 10 days and then recovered for another 8 days. For dehydration treatment, plants grown in pots for 60 days were subjected to drought stress for 7 days by withholding water and then were re-watered for 10 days. For cold stress, plants grown in pots under normal conditions for 30 days were exposed to 2 °C for 4 days and then recovered for another 6 days.
Determination of proline and soluble sugars
Proline contents in rice leaves were determined by the method described previously (Bates et al., 1973; Song et al., 2011). Total soluble sugar content was measured following the methods used previously (Baily, 1958; Song et al., 2011).
Determination of H2O2 and malondialdehyde
Hydrogen peroxide was measured as described previously (Alexieva et al., 2001) with some modifications. Briefly, 1 g leaf sample was ground with 0.1% trichloroacetic acid and centrifuged at 10,000 g for 20 min at 4 °C. The supernatant was used to measure H2O2 spectrophotometrically. The reaction mixture consisted of 1 ml of the extracted supernatant, 1 ml of K2PO4 buffer, and 2 ml of 1 M KI. The reaction was developed for 1 h in darkness and the absorbance was measured at 390 nm.
Malondialdehyde (MDA) content in rice leaves was determined following the protocols described by Song et al. (2011). Briefly, rice leaves were homogenized in 5 ml of 10% trichloroacetic acid containing 0.25% thiobarbituric acid. The mixture was incubated in water at 95 °C for 30 min and the reaction was stopped in an ice bath. The mixture was centrifuged at 10,000 g for 20 min and the absorbance of the supernatant was measured at 450, 532, and 600 nm.
Determination of peroxidase, superoxide dismutase, and catalase activity
Two-week-old plants were exposed to 200 mM NaCl for 24 h. Rice leaves (approx. 0.50 g) were ground thoroughly with a cold mortar and pestle in 50 mM potassium phosphate buffer (pH 7.8) containing 1% polyvinylpyrrolidone. The homogenate was centrifuged at 15000 g for 20 min at 4 °C. The supernatant was crude enzyme extraction. The activities of peroxidase (POD; EC 1.11.1.7), superoxide dismutase (SOD; EC 1.15.1.1), and catalase (CAT; EC 1.11.1.6) were measured using the protocols described by Miao et al. (2010).
RNA isolation and real-time reverse-transcription PCR
Total RNA was isolated from leaves using TRIzol reagent (Invitrogen) and treated with RNase-free DNase I (Promega). The total RNA was reverse-transcribed into first-strand cDNA with M-MLV reverse transcriptase (Promega). Real-time reverse-transcription (RT)-PCR was performed in an optical 96-well plate with a real-time Mx3000P PCR system. Each reaction contained 7.5 μl of 2×SYBR Green Master Mix reagent, 0.5 μl cDNA samples, and 0.6 μl of 10 μM gene-specific primers in a final volume of 15 μl. The thermal cycle used was 95 °C for 5 min and 40 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min. The primers were: for OsMYB2, 5′-GGGCTGAAACGCACAGGCAAGA-3′ and 5′-CTGCTTGGCGTGCTTCTGC-3′; for J033099M14, 5′-CTCAAATCAAGGCGTCAACTAAGA-3′ and 5′-TTTGTCAATATATACGTGGCATATACCA-3′; for J033031H21, 5′-CGCCCCTCCCCGTATCT-3′ and 5′-AGGAATGCGGCAACAAGTG-3′; for 03g44230, 5′-AGGGACGATGGAGTTCTAAAGCT-3′ and 5′-G GGATTCCAAAGGCAAAAAGA-3′; for 07g01090, 5′-GAGGAGGCTACCTGACT GTCAAC-3′ and 5′-GCTCATGAAGTCGCCAAGGA-3′ (Xiang et al., 2007); for OsLEA3, 5′-CGGCAGCGTCCTCCAAC-3′, 5′-CGGTCATCCCCAGCGTG-3′; for OsDREB2A, 5′-GCTGCACATCAGCACCTTCA-3′, 5′-TCCTGCACCTCAGGGACTAC-3′; for OsRab16A, 5′-CACACCACAGCAAGAGCTAAGTG-3′ and 5′-TGGTGCTCC ATCCTGCTTAAG-3′; and for actin, 5′-ACCACAGGTATTGTGTTGGACTC-3′ and 5′-AGAGCATATCCTTCATAGATGGG-3′. The relative quantification (Delta-Delta CT) was used to evaluate quantitative variation between the replicates. The amplification of actin (accession no. AB047313) was used as an internal control to normalize data.
Microarray analysis
Total RNA was isolated from wild-type and OsMYB2-overexpressing plants using TRIzol reagent. All processes for labelling, hybridization, and washing were performed through Affymetrix custom service (Capitalbio) by the protocols given at http://www.affymetrix.com/support/technical/mannual/exprssion-mannual.affx. Normalization was performed according to the standard Affymetrix protocols to allow the comparison of the samples. Expression of genes with changes of more than 3-fold was taken as significantly different. Two biological replicates were used in the present study.
Statistics
All data were analysed by analysis of variance using SAS statistics program. Statistical differences are referred to as significant when P < 0.05.
Results
Isolation and characterization of OsMYB2
OsMYB2 (accession no. AK120551) cDNA was isolated from a salt-stressed rice microarray. The gene comprises 1645 nucleotides with a 990-bp open reading frame. It encodes a putative protein of 329 amino acids with a calculated molecular mass of 36.0 kD and a pI of 4.59. A Blastp search indicated that it is an R2R3-Myb protein with two imperfect repeat sequences in its MYB domain (Supplementary Fig. S1, available at JXB online).
A phylogenetic tree based on the full-length amino acid sequences of rice MYB proteins was constructed (Supplementary Fig. S1). The resulting trees contained six clusters, which were named as C6, C12, C17, C20, C25, and Cr28 according to the classification by Zhang et al. (2011). OsMYB4 (LOC_Os04g43680) clustered with LOC_Os02g41510, LOC_Os10g33810 in C25. OsMYB3R-2 (LOC_Os01g62410) clustered in C6, which was composed of four R1R2R3-MYB proteins. OsMYBS3 (AAN63154) is a MYB-related protein and clustered in Cr28. OsMYB2 (LOC_Os3g20090) clustered with another five members in C12, representing the functional cluster of the regulation of stamen development. According to the phylogenetic tree, OsMYB2 protein had the highest similarity with LOC_Os07g48870, a R2R3-MYB protein with unknown function.
OsMYB2 is located in the nucleus and has transactivation activity in yeast
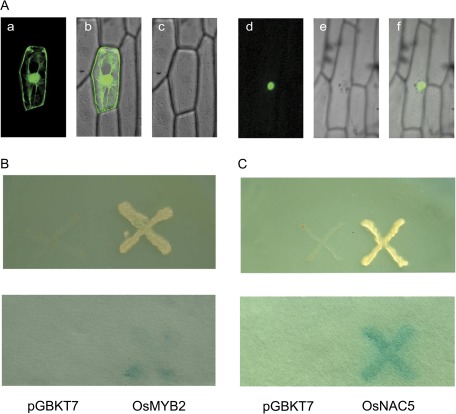
To examine the subcellular localization of OsMYB2, the open reading frame of OsMYB2 was fused to the 5′-terminus of the GFP reporter gene under the control of the CaMV 35S promoter. The recombinant constructs of the OsMYB2-GFP fusion gene and GFP alone were introduced into onion (A. cepa) epidermal cells by particle bombardment. The results showed that the OsMYB2-GFP fusion protein was specifically localized in the nucleus, whereas GFP alone showed ubiquitous distribution in the whole cell (Fig. 1A).
Fig. 1.
Subcellular localization and transactivation analysis of OsMYB2. (A) Nuclear localization of OsMYB2. Confocal images of onion epidermis cells under the GFP channel showing the constitutive localization of GFP (a) and nuclear localization of OsMYB2-GFP (d). The confocal images (b and e) are of the same cells in (a) and (d) with transmitted light. The merged images (c and f) are of (a) and (b) and (d) and (e), respectively. GFP or OsMYB2-GFP fusion was driven by the control of the cauliflower mosaic virus 35S promoter. Onion epidermal peels were bombarded with DNA-coated gold particles and GFP expression was visualized 24 h later. (B) Transactivation assay of OsMYB2 in the yeast strain AH109. Fusion protein of the GAL4 DNA-binding domain and OsMYB2 were expressed in yeast strain AH109. The vector pGBKT7 was expressed in yeast as a control. The culture solution of the transformed yeast was dropped onto SD plates without tryptophan, histidine, or adenine. The plates were incubated for 3 days (upper) and then subjected to β-galactosidase assay (lower). (C) Transactivation assay of OsNAC5 in the yeast strain AH109.
To test whether OsMYB2 has transcription activity, OsMYB2 was fused in-frame to GAL4 DNA-binding domain in the pGBKT7 vector and the fusion constructs pBD-OsMYB2 were transformed into the yeast strain AH109. The transcription factor OsNAC5, which has been shown to have transactivation activity and localize in nucleus, was used as a positive control during this assay (Takasaki et al., 2010; Song et al., 2011). As shown in Fig. 1B and C, only the transformants containing pBD-OsMYB2 and pBD-OsNAC5 grew normally on SD/–Trp/–His/–Ade medium exclusively and exhibited the activity of β-galactosidase reporter gene upon addition of X-gal on Whatman filter paper. Thus these results confirm that OsMYB2 is a transcription activator.
Expression profile of OsMYB2
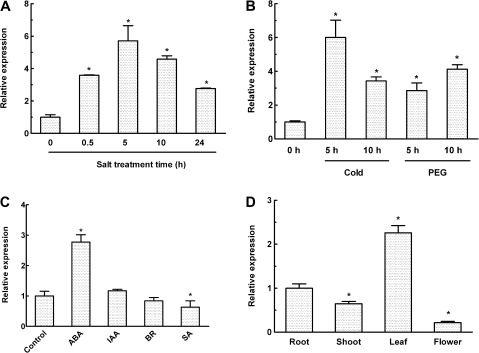
The response of OsMYB2 expression to salt, cold, and dehydration stress was monitored by real-time RT-PCR. An increase in the OsMYB2 transcript was observed after 30 min of exposure to salt stress. The salt stress-induced increase in the OsMYB2 transcript peaked after 5 h of salt stress, and thereafter the transcript declined gradually under salt stress (Fig. 2A). A similar increase in the OsMYB2 transcript was also observed when rice seedlings were exposed to low temperature (2 °C) or osmotic stress (20% PEG) (Fig. 2B). In addition, treatment of rice seedlings with ABA also led to an increase in expression of OsMYB2 (Fig. 2C). In contrast, exogenous application of salicylic acid reduced the expression of OsMYB2, while no effect of indoleacetic acid and brassinosteroids on the OsMYB2 transcript was observed. OsMYB2 was detected in roots, shoots, leaves, and flowers under non-stressed conditions, with the expression being greatest in leaves, followed by roots and shoots (Fig. 2D). The strong induction of this gene by abiotic stress prompted this study to check its promoter sequence (1500 bp upstream from the transcription start site) by searching the promoter sequence against the PLACE database (http://www.dna.affrc.go.jp/PLACE/). The promoter of OsMYB2 contains stress-responsive related cis-elements, such as ABRE and MYB and MYC recognition sites (Supplementary Fig. S2).
Fig. 2.
Real-time reverse-transcription (RT) PCR analysis for the expression of OsMYB2 in rice. (A and B) Time course of OsMYB2 expression during salt (A) and cold and PEG (B) treatments. (C) Expression of OsMYB2 under various hormone treatments. (D) Expression of OsMYB2 in different tissues. Total RNAs were prepared from 14-d-old seedlings of wild-type rice after the above treatments and then reverse-transcribed. The resultant cDNAs were used as templates for real-time RT-PCR and actin was used as an internal control. Data are mean±SE of three biological replicates. Asterisks indicate statistically significant differences (P < 0.05) from controls (0 h in A and B, root in D).
Overexpression of OsMYB2 enhanced tolerance to salt, cold, and dehydration stress
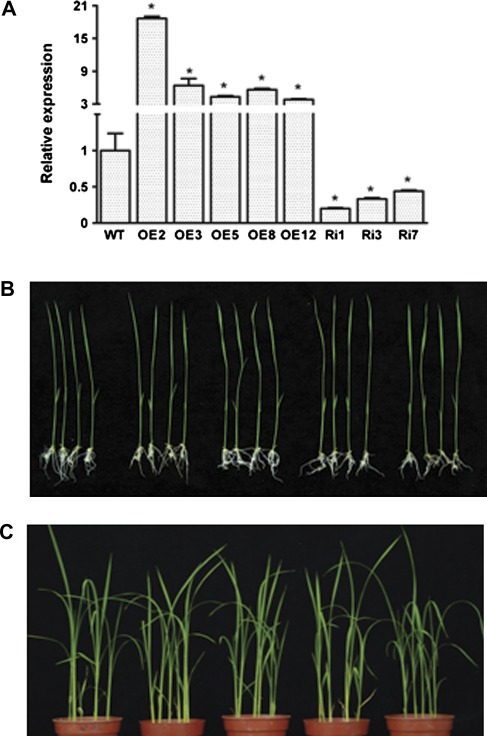
To study the function of OsMYB2, an overexpressing construct and an RNAi construct, under the control of an ubiquitin promoter and CaMV 35S respectively, were transformed into rice ‘Zhonghua 10’ and several transgenic lines were obtained. The transgenic rice lines were confirmed by hygromycin selection, GUS staining, and real-time RT-PCR. Compared with the untransformed wild-type rice, the abundance of the OsMYB2 transcript was higher in the OsMYB2-overexpressing lines (OE2, OE3, OE5, OE8, and OE12) and lower in the OsMYB2-underexpressing lines (Ri1, Ri3, and Ri7) than that of wild-type plants (Fig. 3A). To examine whether the phenotypes of transgenic lines differ from their wild-type, homozygous T3 progeny of the transgenic lines and the wild type were grown on 1/2 MS medium and in soil in the greenhouse. No differences in phenotypes among wild-type, overexpressing, and RNAi plants were observed when grown under normal, non-stressed conditions (Fig. 3B, C). T3 progeny of the transgenic lines (OE2, OE3, Ri1, and Ri3) were chosen based on their expression levels of OsMYB2 and seed availability to further study the physiological function of OsMYB2. Southern blotting with a GUS gene probe was performed to confirm integration of exogenous OsMYB2 into the rice genome, using DNA digested with EcoRI. Different hybridized patterns to the GUS probe were observed, indicating that OsMYB2 was integrated into the rice genome and that the four transgenic lines were independent (Supplementary Fig. S3).
Fig. 3.
Molecular characterization and phenotypes of OsMYB2 transgenic rice. (A) OsMYB2 expression in wild-type and transgenic rice. Total RNAs from 14-d-old wild-type and transgenic rice plants were isolated, reverse-transcribed, and analysed by real-time reverse-transcription PCR. Actin was used as an internal control. Error bars are based on three replicates. (B) The phenotypes of the T3 generation of wild-type and transgenic plants after growing on 1/2 MS medium for 14 days. (C) The phenotypes of the T3 generation of wild-type and transgenic plants after growing in soil for 30 days. Data are mean±SE of three biological replicates. Asterisks indicate statistically significant differences (P < 0.05) between wild-type (WT) and transgenic lines (OE and Ri).
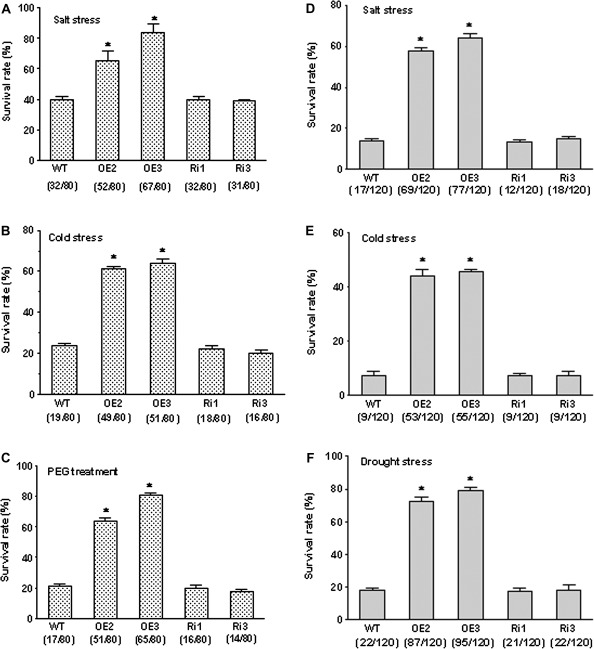
The involvement of OsMYB2 in salt, cold, and osmotic stress was investigated by exposing wild-type and the transgenic plants grown in hydroponic solution with NaCl, low temperature, and PEG. There was no difference between transgenic and wild-type plants when grown under normal, non-stressed conditions in hydroponic solution (Fig. 4A). Phenotypically, most OsMYB2-overexpressing seedlings remained green and showed continuous growth, whereas both wild-type and RNAi seedlings showed severe leaf rolling and wilting after exposure to the salt, cold, and osmotic stress (Fig. 4B-D). In addition, the two transgenic rice lines overexpressing OsMYB2 grown in soil also exhibited greater tolerance to NaCl, cold, and drought stress than wild-type and RNAi plants (Fig. 4E-J). This study also determined the survival rate for wild-type and transgenic plants grown in both hydroponic solution and soil challenged with salt, cold, and osmotic stress. As shown in Fig. 5A-C, the survival rate of overexpressing lines was significantly higher than that of wild-type and RNAi seedlings when exposed to salt stress (200 mM NaCl for 2 d), cold stress (5 °C for 3 d), and osmotic stress (20% PEG6000 for 2 d). The survival rates of the two overexpressing lines higher than those of wild-type and RNAi plants were also observed when rice seedlings grown in soil were challenged by salt, cold, and drought stress (Fig. 5D-F).
Fig. 4.
Effect of salt, cold, and dehydration stress on wild-type (WT) and transgenic (OE and Ri) rice plants. (A–D) Phenotypes of wild-type and transgenic rice plants grown on 1/2 MS medium for 14 days and then under normal conditions (A), salt stress (200 mM NaCl for 2 days and normal conditions for 4 days) (B), cold stress (2 °C for 3 days and normal conditions for 7 days) (C), and treatment with 20% PEG for 2 days and recovery for 1 week (D). (E–J) Phenotypes of wild-type and transgenic rice plants grown in soil under normal conditions (E, G, I), subjected to 100 mM NaCl for 10 days and then recovered for another 8 days (F), exposed to 2 °C for 4 days and then recovered for another 6 days (H), and exposed to drought stress for 7 days and then re-watered for 10 days (J). Age of seedlings: 40 d ((E and F), 30 d (G and H) and 60 d (I and J).
Fig. 5.
Effect of salt, cold, and dehydration stress on survival rates of wild-type and transgenic rice plants, corresponding to the plants and treatments as shown in Fig. 4. (A–C) Plants grown on 1/2 medium. (D–F) Plants grown in soil. Data are mean±SE of three replicates with total seedling number of 80 for all stress treatments. Values in parentheses are the numbers of survived seedlings/total seedlings used to calculate the survival rate. Asterisks indicate statistically significant differences (P < 0.05) between wild-type (WT) and transgenic lines (OE and Ri).
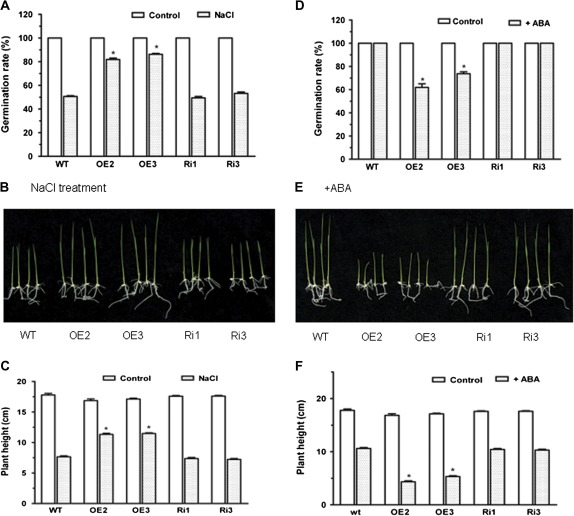
Overexpression of OsMYB2 altered sensitivity of seed germination and growth to salt stress and ABA
Given that expression of OsMYB2 was sensitive to ABA (Fig. 2C), this study further investigated the effects of salt stress and ABA on seed germination of wild-type, OsMYB2-overexpressing, and RNAi seeds. Exposure of both wild-type and transgenic seeds to NaCl reduced their germination rate (Fig. 6A). However, germination of OsMYB2-overexpressing seeds was less inhibited by NaCl than that of wild-type and RNAi seeds. For example, seed germination rate of the two OsMYB2-overexpressing lines (OE2, OE3) was 81% and 86% when incubated in the presence of 100 mM NaCl, while germination rate for wild-type and RNAi (Ri1, Ri3) seeds was found to be 51%, 49%, and 53% under the identical conditions, respectively. The effect of NaCl on seedling growth was also examined. In the saline medium containing 150 mM NaCl, the OsMYB2-overexpressing plants exhibited faster growth and their shoots were significantly longer than wild-type plants (Fig. 6B, C).
Fig. 6.
Responses of seed germination and seedling growth to treatment with NaCl and abscisic acid (ABA). (A–C) Responses to NaCl treatment: (A) germination rates: T3 generation seeds were soaked in distilled water for 1 day and then allowed to germinate on sterile-water-saturated filter paper with 100 mM NaCl for 5 days; (B) phenotypes: seeds were allowed to germinate in darkness for 2 days, and then transferred to 1/2 MS medium containing 150 mM NaCl under 28/25 °C (day/night) with a 14-h photoperiod for 12 days; (C) shoot heights of seedlings under normal and 150 mM NaCl conditions. (D–F) Responses to ABA treatment: (D) germination rates: T3 generation seeds were soaked in distilled water for 1 day and then allowed to germinate on sterile-water-saturated filter paper with 4 μM ABA for 5 days; (E) phenotypes: seeds were allowed to germinate in darkness for 2 days, and then transferred to 1/2 MS medium containing 4 μM ABA under 28/25 °C (day/night) with a 14-h photoperiod for 12 days; (F) shoot heights of seedlings under normal and 4 μM ABA conditions. Data are mean±SE of three replicates. Asterisks indicate statistically significant differences (P < 0.05) between wild-type (WT) and transgenic lines (OE and Ri). (This figure is available in colour at JXB online.)
In contrast to salt stress, seed germination of OsMYB2-overexpressing lines was more sensitive to ABA than that of wild-type and RNAi lines, such that wild-type and RNAi lines had higher seed germination rate than OsMYB2-overexpressing lines when ABA was present in the incubation medium (Fig. 6D). Like seed germination, growth of OsMYB2-overexpressing seedlings was more inhibited by ABA than that of wild-type and RNAi seedlings, as shown by a shorter length of OsMYB2-overexpressing seedlings than wild-type and RNAi seedlings when grown in the presence of ABA (Fig. 6E, F). No difference in shoot length of wild-type and the transgenic plants grown in control medium was found.
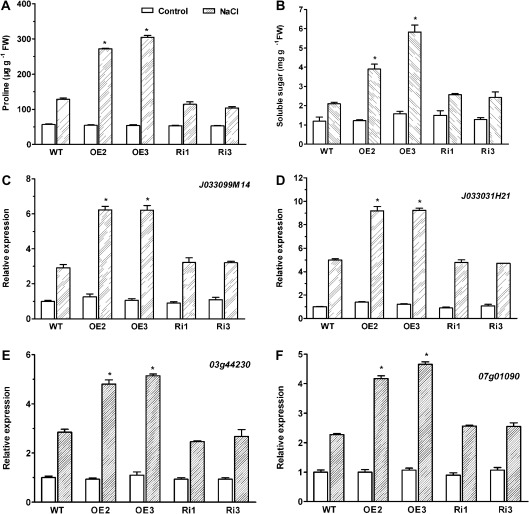
OsMYB2-overexpressing plants accumulated greater amounts of proline and soluble sugars
Accumulation of proline and soluble sugars to facilitate osmoregulation is a common adaptive mechanism for tolerance of plants to abiotic stress. To test whether the enhanced tolerance of OsMYB2-overexpressing plants to salt stress is related to the capacity to accumulate proline and soluble sugars, the effect of salt stress on contents of proline and soluble sugars in wild-type and transgenic plants was investigated. There was no significant difference in proline contents between wild-type and transgenic plants under non-stressed, control conditions (Fig. 7A). An increase in proline content was observed upon exposure to salt stress in both wild-type and transgenic plants. However, the increase in proline content in the OsMYB2-overexpressing plants was significantly higher than in wild-type plants. Similar to proline, no significant difference in content of soluble sugars between wild-type and transgenic plants was observed under control conditions (Fig. 7B). However, the two OsMYB2-overexpressing lines accumulated greater amounts of soluble sugars than wild-type plants when these plants were exposed to 200 mM NaCl for 2 d.
Fig. 7.
Effect of salt stress on contents of proline and soluble sugars and gene expression in wild-type and transgenic rice plants. (A and B) Wild-type and transgenic rice seedlings of 14-d-old were exposed to 200 mM NaCl for 2 days and then collected for determination of proline (A) and soluble sugars (B) contents. (C–F) Expression levels of putative proline synthase genes (J033099M14 and J033031H21; C and D) and transporter genes (03g44230 and 07g01090; E–F) in transgenic and wild-type plants. Total RNA was extracted from the 14-d-old rice seedlings grown under control and salt stress (200 mM NaCl) conditions for 24 hours. The transcript levels were measured by real-time reverse-transcription PCR. Actin was used as an internal control. Data are mean±SE of three replicates. Asterisks indicate statistically significant differences (P < 0.05) between wild-type (WT) and transgenic lines (OE and Ri). The accession numbers of the sequences of J033099M14, J033031H21, 03g44230, and 07g01090 are AK102633, AK101230, AK067118, and AK0666298.
To further elucidate the mechanism by which OsMYB2-overexpressing plants accumulate greater amounts of proline than wild-type plants under salt stress, the effects of salt stress on expression of genes responsible for proline biosynthesis and proline transport were investigated. As shown in Fig. 7C and D, treatment with salt stress led to a greater increase in transcripts of proline biosynthesis genes (Δ-1-pyrroline-5-carboxylate synthase genes, J033099M14 and J033031H21) in the OsMYB2-overexpressing lines than in the wild-type and RNAi lines. A similar greater up-regulation of the two genes encoding putative proline transport (03g44230 and 07g01090) in the two OsMYB2-overexpressing lines than in the wild-type and RNAi lines was also observed in response to salt stress (Fig. 7E, F).
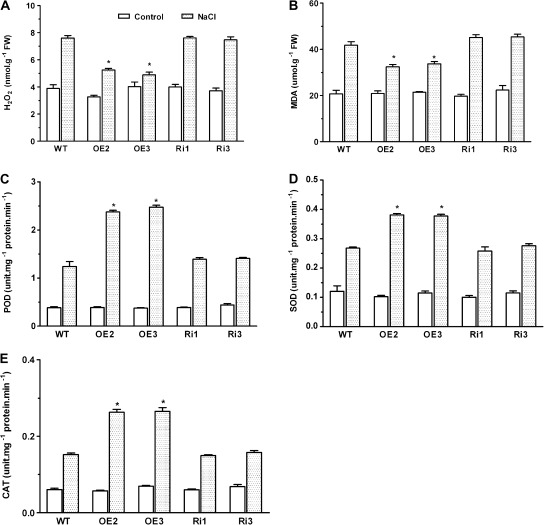
OsMYB2-overexpressing plants accumulated less H2O2 and MDA under salt stress
The effects of NaCl on H2O2 and MDA contents in wild-type and transgenic rice were investigated and no significant differences in H2O2 and MDA contents were found in the absence of NaCl in the incubation medium (Fig.8A, B). There were marked increases in H2O2 and MDA contents in both wild-type and transgenic plants upon exposure to 200 mM NaCl. However, the salt stress-induced increases in H2O2 and MDA contents were less in the OsMYB2-overexpressing plants than those in the wild-type and RNAi plants. These results indicate that overexpression of OsMYB2 confers greater tolerance of the oxidative stress associated with salt stress.
Fig. 8.
Effect of salt stress on contents of oxidants (A and B) and antioxidant enzymes (C–D) in wild-type and transgenic rice plants. (A and B) Plants were exposed to 200 mM NaCl for 2 days before determination of H2O2 (A) and malondialdehyde (MDA) (B). (C–E) were exposed to 200 mM NaCl for 24 hours and before determination of peroxidase (POD; C), superoxide dismutase (SOD; D), and catalase (CAT; E). Data are mean±SE of three replicates. Asterisks indicate statistically significant differences (P < 0.05) between wild-type (WT) and transgenic lines (OE and Ri).
The lower content of H2O2 in the OsMYB2-overexpressing plants under salt stress prompted this study to test whether the difference in H2O2 accumulation between wild-type and the OsMYB2-overexpressing lines resulted from differences in the activities of the major antioxidant enzymes. Under normal conditions, activities of POD, SOD, and CAT were comparable among wild-type, OsMYB2-overexpressing, and RNAi plants (Fig.8C-E). There were marked increases in activities of these enzymes for the rice seedlings upon exposure to salt stress. However, the salt stress-induced increases in activities of POD, SOD, and CAT were higher in the OsMYB2-overexpressing plants than the wild-type and RNAi plants (Fig. 8C-E). In contrast to OsMYB2-overexpressing plants, activities of these enzymes in the OsMYB2-RNAi lines did not differ from the wild-type plants under salt-stressed conditions (Fig.8C-E). These results imply that overexpression of OsMYB2 confers a more efficient antioxidant system to counteract oxidative stress under saline conditions.
Comparison of expression profiles between wild-type and OsMYB2-overexpressing rice plants
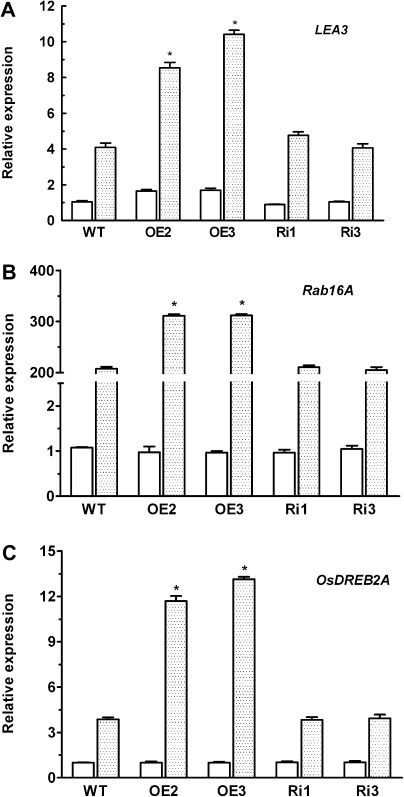
Up-regulation of several genes such as OsLEA3, OsRab16A, and OsDREB2A can contribute to enhanced tolerance of plants to salt stress (Zhang et al., 2009). To test whether these genes are also involved in the enhanced tolerance of OsMYB2-overexpressing plants to salt stress, real-time RT-PCR was used to study the effect of salt stress on the expression levels of these genes in wild-type and transgenic plants. When treated with 200 mM NaCl, a significant up-regulation of the salt-responsive genes was observed in both wild-type and transgenic plants, with higher expression levels in the OsMYB2-overexpressing plants than in the wild-type and RNAi plants (Fig. 9). No difference in expression of these genes between wild-type and RNAi plants was found in response to salt stress (Fig. 9)
Fig. 9.
Expression levels of some salt-responsive genes in wild-type and transgenic plants. (A) OsLEA3; (B) OsRab16A; (C) OsDREB2A. Total RNA was extracted from the 14-d-old rice seedlings grown under control (open columns) and salt stress (200 mM NaCl, filled columns) conditions for 24 hours. The transcript levels were measured by real-time reverse-transcription PCR. Actin was used as an internal control. Data are mean±SE of three replicates. Asterisks indicate statistically significant differences (P < 0.05) between wild-type (WT) and transgenic lines (OE and Ri).
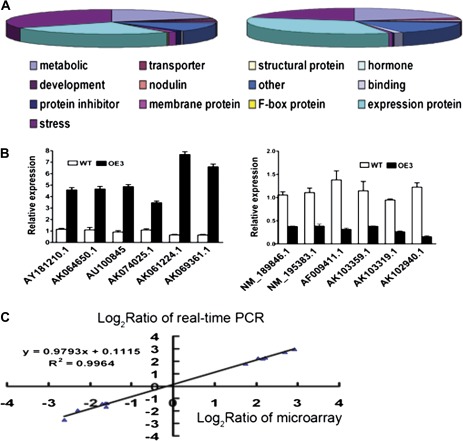
To identify the downstream genes targeted by OsMYB2, whole-genome expression profiling was performed using the Affymetrix Rice GeneChip array, which contains 55,515 probe sets. Relative change was determined by normalizing the data for the transgenic line (OE3) relative to wild-type plants. Total RNA was isolated from 14-d-old seedlings of the transgenic line and the wild type grown under normal conditions. A total of 519 genes was up-regulated (greater than 3-fold changes) and 423 genes were down-regulated (greater than 3-fold changes) in the overexpression line compared with wild-type plants (Supplementary Tables S2 a0nd S3). The detected genes were mainly involved in stress response (Fig. 10A). For example, genes encoding stress-related functional proteins, such as dehydrin proteins, LEA proteins, and stress-related regulatory factors such as transcription factor, protein kinase, and phosphatase, were greatly affected by overexpression of OsMYB2. The microarray data was further validated by real-time RT-PCR. Twelve genes representing different expression profiles were analysed, of which all exhibited expression patterns comparable to that obtained from the microarray data (Fig. 10B). A high degree of concordance was observed between the results generated by the two methods (Fig. 10C).
Fig. 10.
Global analysis of gene expression in OsMYB2-overexpressing rice (transgenic line OE3). (A) Predicted functions of the proteins encoded by up-regulated genes (left) and down-regulated genes (right). (B) Relative expression in the wild type and the transgenic line of six up-regulated genes (left) and six down-regulated genes (right) selected from the microarray data and confirmed by real-time reverse-transcription (RT) PCR. (C) Correlation between data obtained from microarray and RT-PCR data. Data are mean±SE of three replicates.
Discussion
In the rice genome, there are 183 MYB-encoding genes with diverse roles in developmental processes and defence responses (Chen et al., 2006). There is emerging evidence to support that many MYB proteins are involved in response and an adaptation to abiotic stress. For instance, the involvement of MYB proteins in tolerance of rice to cold stress has been demonstrated by overexpression of OsMYB4 (Vannini et al., 2004), OsMYB3R-2 (Dai et al., 2007; Ma et al., 2009), and OsMYBS3 (Su et al., 2010) in Arabidopsis and rice, which conferred tolerance to cold stress at the seedling stage. The present study isolated a novel R2R3-type MYB gene, OsMYB2, and demonstrated that overexpression of OsMYB2 greatly conferred tolerance of rice to salt, cold, and dehydration stress. This study also found that overexpression of OsMYB2 led to greater accumulation of soluble sugars and proline due to up-regulation of genes responsible for proline synthesis and transport, and less accumulation of H2O2 and MDA under salt stress. These metabolic changes due to overexpression of OsMYB2 would allow plants for effective osmo-regulation and less oxidative damage under salt stress, thus conferring tolerance to salt stress. In addition to these metabolic changes, overexpression of OsMYB2 also led to changes in expression of numerous genes involved in stress response as revealed by the microarray data (Fig. 10). These results also showed that the transcripts of salt-responsive genes such as OsLEA3, OsRAB16A, and OsDREB2A were higher in the OsMYB2-overexpressing plants than in wild-type plants under salt stress (Fig. 9), suggesting that the improved tolerance of transgenic plants overexpressing OsMYB2 may result from direct regulation of these stress-responsive genes by OsMYB2. Identification of targets of OsMYB2 and unravelling their signalling network may shed some light on the molecular mechanism underlying the OsMYB2-dependent tolerance to salt stress.
This study constructed a phylogenetic tree with a total of 31 MYB proteins and clustered them into six groups following the protocols used by Zhang et al. (2011). It has been suggested that the clusters C12, C20, and C25 are involved in stress response (Zhang et al., 2011). Cluster C6 was composed of four members with three MYB repeats, such as OsMYB3R-2. Four MYB-related proteins were grouped in Cr28. OsMYB2, together with AtMYB2, was clustered in C12, which has been implicated in stamen development (Zhang et al., 2011). AtMYB2, which encodes a R2R3-type MYB transcription factor, has been shown to regulate salt- and dehydration-responsive genes (Urao et al., 1993; Abe et al., 2003). Yoo et al. (2005) demonstrated that a specific calmodulin isoform mediates salt-induced Ca2+ signalling through activation of AtMYB2, conferring tolerance of Arabidopsis to salt stress. These results suggest that the cluster C12 may also participate in the abiotic stress signalling in addition to stamen development.
The expression of OsMYB2 was induced rapidly by cold such that its expression peaked after exposure to cold for 5 h and the expression declined thereafter (Fig. 2B). In contrast, cold-induced up-regulation of OsMYB3R-2 is sustained up to 72 h of exposure to cold (Dai et al., 2007). In addition, the present study found that the expression of OsMYB2 was also up-regulated by salt and dehydration stress (Fig. 2A, B). These findings are in contrast to OsMYB4 and OsMYBS3, which are induced by cold stress exclusively (Vannini et al., 2004; Su et al., 2010). Therefore, OsMYB2 is a novel R2R3-type MYB transcription factor in terms of its activation by abiotic stresses compared with the identified MYB proteins in rice.
To evaluate the role of OsMYB2 in tolerance to abiotic stress, this study generated transgenic plants with overexpression and underexpression of OsMYB2 by Agrobacterium-mediated transformation (Fig. 3). One important finding is that OsMYB2-overexpressing plants exhibited a higher survival rate than wild-type or RNAi plants grown in both hydroponic solution and soil when exposed to salt, cold, and dehydration stress (Figs. 4, 5). As far as is known, this is the first report demonstrating that a rice MYB protein confers tolerance to multiple abiotic stresses.
To elucidate the mechanisms responsible for the enhanced tolerance of OsMYB2-overexpressing plants to salt stress, several experiments were conducted to monitor changes in physiological processes associated with plant tolerance to salt stress. Accumulation of compatible solutes such as soluble sugars (Garg et al., 2002; Gupta and Kaur, 2005) and free proline (Liu and Zhu, 1997; Armengaud et al., 2004) is a common phenomenon in response to abiotic stress. The accumulated soluble sugars and free proline act as osmolytes to facilitate osmo-regulation, thus protecting plants from dehydration resulting from salt stress by reducing water potential of plant cells. In addition, proline can also function as a molecular chaperone to stabilize the structure of proteins as well as play a role in regulation of the antioxidant system (Hare et al., 1999; Szekely et al., 2008). These compatible solutes have been reported to play a predominant role in transgenic plants with enhanced tolerant to abiotic stress (Xiang et al., 2007; Xu et al., 2008). This study found greater accumulation of soluble sugars and free proline in the OsMYB2-overexpressing plants, which may partially account for the higher tolerance of OsMYB2-overexpressing plants to salt stress (Fig. 7B). In this context, similar increases in soluble sugar and proline levels have been demonstrated in transgenic plants with overexpressing OsMYB4 and OsMYB3R-2 under conditions of cold stress (Vannini et al., 2004; Pasquali et al., 2008; Ma et al., 2009).
Overexpression of OsMYB4 confers tolerance to chilling and freezing by up-regulating the cold-regulated (COR) gene and increasing the proline level in Arabidopsis (Vannini et al., 2004) and enhances tolerance to cold and drought by accumulating compatible solutes in transgenic apple (Malus pumila) (Pasquali et al., 2008). A recent study also demonstrated that overexpression of OsMYB3R-2 in rice leads to an increase in proline content and enhances the tolerance to cold (Ma et al., 2009). Thus, one important mechanism underlying the enhanced tolerance of overexpression of rice MYB genes may be accounted for by the enhanced accumulation of compatible solutes.
To further elucidate the mechanism underlying the greater accumulation of proline in the OsMYB2-overexpressing plants under salt stress, this study examined the changes in proline synthase and proline transporter genes in OsMYB2-overexpressing, RNAi, and wild-type plants in response to salt stress at the transcriptional level. Up-regulation of proline synthase and proline transporter genes has been reported in OsCIPK03- and OsCIPK12-overexpressing plants, leading to greater amounts of proline in these transgenic rice than in the wild type under conditions of cold and dehydration stress (Xiang et al., 2007). The present results demonstrate that expression of proline synthase and proline transporter genes was higher in OsMYB2-overexpressing plants than in wild-type plants under salt stress, suggesting that the higher proline contents in OsMYB2-overexpressing plants are likely to result from the greater up-regulation of these genes under salt stress (Fig. 7C-F). However, no significant difference in soluble sugars and free proline contents was found between the OsMYB2-overexpressing and wild-type plants under normal growth conditions, despite the fact that the constitutive promoter was used to drive the gene. This pattern is similar to that of other genes involved in abiotic stresses, such as OsZFP252, OsCIPK03, and OsCIPK12 (Xiang et al., 2007; Xu et al., 2008). One possible explanation is that other stress-responsive regulators are required to activate OsMYB2-dependent, stress-responsive genes under stressed conditions.
In addition to the genes encoding proline synthase and transporters, this study also found that overexpression of OsMYB2 led to higher expression of stress-related genes such as OsLEA3, OsRab16A, and OsDREB2A (Fig. 9). Both OsLEA3 and OsRab16A belong to the late embryogenesis abundant genes, which have been implicated in response to many abiotic stress (RoyChoudhury et al., 2007; Xiao et al., 2007; Xiang et al., 2008). OsRab16A encodes a group-2 late embryogenesis abundant protein and is significantly up-regulated by salt stress and ABA (Mundy and Chua, 1988) and transgenic tobacco plants overexpressing OsRab16A confer enhanced tolerance to salt stress (RoyChoudhury et al., 2007). OsLEA3 belongs to group 3 of the LEA gene family and is induced by dehydration, salt stress, and ABA (Xiao et al., 2007); for instance, under field conditions, overexpression of OsLEA3 in rice plants can significantly enhance the dehydration tolerance (Xiao et al., 2007). OsDREB2A is a stress-responsive, DRE/CRT gene that is involved in dehydration and salt stress (Dubouzet et al., 2003) and constitutive expression of Arabidopsis DREB2A and maize ZmDREB2A confers enhanced tolerance of transgenic Arabidopsis plants to drought stress (Sakuma et al., 2006; Qin et al., 2007). The present observations that overexpression of OsMYB2 led to a greater up-regulation of OsLEA3, OsRab16A, and OsDREB2A and enhanced tolerance to abiotic stress suggest that OsMYB2 may regulate the expression of LEA genes through the OsDREB2A-dependent signalling pathway. Moreover, this study also found by analysing the microarray data that overexpression of OsMYB2 altered the expression levels of a large number of genes and that many of the up- or down-regulated genes in the OsMYB2-overexpressing plants are predicted to be involved in stress tolerance. These results indicate that OsMYB2 play an important role in stress tolerance in rice by regulating a number of downstream genes that are closely associated with plant tolerance to abiotic stress.
The elevated concentrations of ROS can damage cellular structures and macromolecules, leading to cell death (Mittler, 2002). Malondialdehyde is widely recognized as a marker for lipid peroxidation (RoyChoudhury et al., 2007). The present study found that the contents of H2O2 and MDA in OsMYB2-overexpressing plants were markedly lower than in the wild-type and OsMYB2-RNAi plants under conditions of salt stress (Fig. 8A, B). The less accumulation of H2O2 under salt stress may result from an enhanced capacity for scavenging ROS, and the finding that POD, SOD, and CAT activities were higher in the OsMYB2-overexpressing plants than in the wild-type and OsMYB2-RNAi plants (Fig. 8, C-E) is in line with this proposition. The greater tolerance of OsMYB2-overexpressing plants to salt stress found in this study may be accounted for, at least in part, by mitigating oxidative damage due to suppression of ROS production.
Expression of OsMYB2 was up-regulated by exogenous ABA, and transgenic plants overexpressing OsMYB2 were more sensitive to exogenous ABA than the wild type in terms of seed germination and seedling growth (Fig. 6). Those findings are in contrast to other reported rice MYB genes in the literature. For example, expression of OsMYB4 is insensitive to ABA (Vannini et al., 2004). Overexpression of OsMYB3R-2 in Arabidopsis renders it less sensitive to ABA (Dai et al., 2007). Expression of OsMYBS3 in leaves is suppressed by exogenous application of ABA, while its expression in roots is not responsive to ABA (Su et al., 2010). The mechanism by which ABA regulates OsMYB2 in response of rice plants to salt stress warrants further investigation. These results suggest that MYB proteins play diverse roles in the ABA-dependent signalling transduction pathways.
Another interesting observation in this study is that the wild-type and RNAi plants showed no differences in terms of their response to the abiotic stress examined and metabolic changes induced by salt stress. This result may be because some other MYB proteins complement the function of OsMYB2. A similar observation has been reported in OsZFP252-knockdown lines (Xu et al., 2008). For example, overexpression of OsZFP252 in rice enhances accumulation of free proline and soluble sugars and improves the expression of stress-responsive genes, thus conferring OsZFP252-overexpressing rice with more tolerance to salt and drought stress than OsZFP252-antisense and wild-type rice (Xu et al., 2008). However, no difference in tolerance to salt and drought stress was observed between OsZFP252-knockdown lines and wild-type plants. Alternatively, it is possibile that the residual levels of OsMYB2 in the underexpressing RNAi plants may be sufficient for OsMYB2 functioning.
In summary, this study identified a transcription factor OsMYB2 that functions as positive regulator to mediate tolerance of rice seedlings to salt, cold, and dehydration stress. Overexpression of OsMYB2 led to greater accumulation of compatible osmolytes, such as soluble sugars, free proline, and LEA proteins in rice, and suppressed the accumulation of MDA and H2O2 under conditions of salt stress. The up-regulation of OsMYB2 may allow rice plants to effectively osmo-regulate their water potential by accumulating compatible solutes and minimize oxidative damage to plants under abiotic stress. More importantly, overexpression of OsMYB2 in rice seedlings did not affect their phenotypes under control conditions. Therefore, OsMYB2 provides a promising tool for improving the tolerance of rice to abiotic stress in general and to salt stress in particular.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Structure of the OsMYB2 protein and a phylogenetic tree of MYB proteins.
Supplementary Fig. S2. Distribution of stress-related cis-elements in the OsMYB2 promoter region.
Supplementary Fig. S3. Southern blot analysis of independent transgenic rice lines.
Supplementary Table S1. Primers used in real-time RT-PCR to verify the expression pattern of differentially expressed genes from the microarray experiment.
Supplementary Table S2. Genes up-regulated by overexpression of OsMYB2.
Supplementary Table S3. Genes down-regulated by overexpression of OsMYB2.
Acknowledgments
This work was supported by the National Science Foundation of China (30788003 and 30870188) and the Chinese Academy of Sciences (KSCX1-YW-03).
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. The Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. Journal of Biological Chemistry. 2006;281:37636–37645. doi: 10.1074/jbc.M605895200. [DOI] [PubMed] [Google Scholar]
- Alexieva V, Sergiev I, Mapelli S, Karanov E. The effect of drought and ultraviolet radiation on growth and stress markers in pea and wheat. Plant, Cell and Environment. 2001;24:1337–1344. [Google Scholar]
- Armengaud P, Thiery L, Buhot N, Grenier-De March G, Savoure A. Transcriptional regulation of proline biosynthesis in Medicago truncatula reveals developmental and environmental specific features. Physiologia Plantarum. 2004;120:442–450. doi: 10.1111/j.0031-9317.2004.00251.x. [DOI] [PubMed] [Google Scholar]
- Bailey RW. Reaction of pentoses with anthrone. Biochemical Journal. 1958;68:669–672. doi: 10.1042/bj0680669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels DSR. Drought and salt tolerance in plants. Critical Reviews in Plant Science. 2005;24:23–58. [Google Scholar]
- Bates LS, Waldren RP, Teare ID. Rapid determination of free proline for water-stress studies. Plant and Soil. 1973;39:205–207. [Google Scholar]
- Chen Y, Yang X, He K, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Molecular Biology. 2006;60:107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Current Biology. 2005;15:1196–1200. doi: 10.1016/j.cub.2005.05.048. [DOI] [PubMed] [Google Scholar]
- Dai X, Xu Y, Ma Q, Xu W, Wang T, Xue Y, Chong K. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiology. 2007;143:1739–1751. doi: 10.1104/pp.106.094532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denekamp M, Smeekens SC. Integration of wounding and osmotic stress signals determines the expression of the AtMYB102 transcription factor gene. Plant Physiology. 2003;132:1415–1423. doi: 10.1104/pp.102.019273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. The Plant Journal. 2003;33:751–763. doi: 10.1046/j.1365-313x.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- Garg AK, Kim JK, Owens TG, Ranwala AP, Choi YD, Kochian LV, Wu RJ. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proceedings of National Acaderemy of Sciences USA. 2002;99:15898–15903. doi: 10.1073/pnas.252637799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Chen H, Jiang JF, Zhao Y, Xu ML, Xu YY, Tan KH, Xu ZH, Chong K. Overexpression of OsRAA1 causes pleiotropic phenotypes in transgenic rice plants, including altered leaf, flower, and root development and root response to gravity. Plant Physiology. 2004;135:1502–1513. doi: 10.1104/pp.104.041996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta AK, Kaur N. Sugar signalling and gene expression in relation to carbohydrate metabolism under abiotic stresses in plants. Journal of Bioscience. 2005;30:761–776. doi: 10.1007/BF02703574. [DOI] [PubMed] [Google Scholar]
- Hare PD, Cress WA, van Staden J. Proline synthesis and degradation: a model system for elucidating stress-related signal transduction. Journal of Experimental Botany. 1999;50:413–434. [Google Scholar]
- Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Molecular Biology. 2008;67:169–181. doi: 10.1007/s11103-008-9309-5. [DOI] [PubMed] [Google Scholar]
- Ito M, Araki S, Matsunaga S, Itoh T, Nishihama R, Machida Y, Doonan JH, Watanabe A. G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. The Plant Cell. 2001;13:1891–1905. doi: 10.1105/TPC.010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, Nahm BH, Choi YD, Cheong JJ. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiology. 2008;146:623–635. doi: 10.1104/pp.107.110981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, et al. Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. The Plant Journal. 1998;16:263–276. doi: 10.1046/j.1365-313x.1998.00278.x. [DOI] [PubMed] [Google Scholar]
- Liang YK, Dubos C, Dodd IC, Holroyd GH, Hetherington AM, Campbell MM. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Current Biology. 2005;15:1201–1206. doi: 10.1016/j.cub.2005.06.041. [DOI] [PubMed] [Google Scholar]
- Lippold F, Sanchez DH, Musialak M, Schlereth A, Scheible WR, Hincha DK, Udvardi MK. AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis. Plant Physiology. 2009;149:1761–1772. doi: 10.1104/pp.108.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu JK. Proline accumulation and salt-stress-induced gene expression in a salt-hypersensitive mutant of Arabidopsis. Plant Physiology. 1997;114:591–596. doi: 10.1104/pp.114.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma HS, Liang D, Shuai P, Xia XL, Yin WL. The salt- and drought-inducible poplar GRAS protein SCL7 confers salt and drought tolerance in Arabidopsis thaliana. Journal of Experimental Botany. 2010;61:4011–4019. doi: 10.1093/jxb/erq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Dai X, Xu Y, Guo J, Liu Y, Chen N, Xiao J, Zhang D, Xu Z, Zhang X, Chong K. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiology. 2009;150:244–256. doi: 10.1104/pp.108.133454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao BH, Han XG, Zhang WH. The ameliorative effect of silicon on soybean seedlings grown in potassium-deficient medium. Annals of Botany. 2010;105:967–973. doi: 10.1093/aob/mcq063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends in Plant Sciences. 2002;7:405–410. doi: 10.1016/s1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- Mundy J, Chua NH. Abscisic acid and water-stress induce the expression of a novel rice gene. The EMBO Journal. 1988;7:2279–2286. doi: 10.1002/j.1460-2075.1988.tb03070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. The Plant Journal. 2007;51:617–630. doi: 10.1111/j.1365-313X.2007.03168.x. [DOI] [PubMed] [Google Scholar]
- Pasquali G, Biricolti S, Locatelli F, Baldoni E, Mattana M. Osmyb4 expression improves adaptive responses to drought and cold stress in transgenic apples. Plant Cell Reports. 2008;27:1677–1686. doi: 10.1007/s00299-008-0587-9. [DOI] [PubMed] [Google Scholar]
- Qin F, Kakimoto M, Sakuma Y, Maruyama K, Osakabe Y, Tran LS, Shinozaki K, Yamaguchi-Shinozaki K. Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. The Plant Journal. 2007;50:54–69. doi: 10.1111/j.1365-313X.2007.03034.x. [DOI] [PubMed] [Google Scholar]
- RoyChoudhury A, Roy C, Sengupta DN. Transgenic tobacco plants overexpressing the heterologous lea gene Rab16A from rice during high salt and water deficit display enhanced tolerance to salinity stress. Plant Cell Reports. 2007;26:1839–1859. doi: 10.1007/s00299-007-0371-2. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. The Plant Cell. 2006;18:1292–1309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SY, Chen Y, Chen J, Dai XY, Zhang WH. Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta. 2011;234:331–345. doi: 10.1007/s00425-011-1403-2. [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Current Opinion in Plant Biology. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- Su CF, Wang YC, Hsieh TH, Lu CA, Tseng TH, Yu SM. A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiology. 2010;153:145–158. doi: 10.1104/pp.110.153015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekely G, Abraham E, Cseplo A, et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. The Plant Journal. 2008;53:11–28. doi: 10.1111/j.1365-313X.2007.03318.x. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Mittler R. Reactive oxygen species and temperature stresses: a delicate balance between signaling and destruction. Physiologia Plantarum. 2006;126:45–51. [Google Scholar]
- Takasaki H, Maruyama K, Kidokoro S, Ito Y, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K, Nakashima K. The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Molecular and Genetic Genomics. 2010;284:173–183. doi: 10.1007/s00438-010-0557-0. [DOI] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. The Plant Cell. 1993;15:1529–1539. doi: 10.1105/tpc.5.11.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini C, Locatelli F, Bracale M, Magnani E, Marsoni M, Osnato M, Mattana M, Baldoni E, Coraggio I. Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. The Plant Journal. 2004;37:115–127. doi: 10.1046/j.1365-313x.2003.01938.x. [DOI] [PubMed] [Google Scholar]
- Wang Z, Xu Y, Jiang R, Xu Z, Chong K. A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.) Plant Molecular Biology Reports. 2004;22:409–417. [Google Scholar]
- Xiang Y, Huang Y, Xiong L. Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiology. 2007;144:1416–1428. doi: 10.1104/pp.107.101295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Tang N, Du H, Ye HY, Xiong LZ. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiology. 2008;148:1938–1952. doi: 10.1104/pp.108.128199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Huang Y, Tang N, Xiong L. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theorectical and Applied Genetics. 2007;115:35–46. doi: 10.1007/s00122-007-0538-9. [DOI] [PubMed] [Google Scholar]
- Xin Z, Browse J. Eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proceedings for National Academy of Sciences USA. 1998;95:7799–7804. doi: 10.1073/pnas.95.13.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu DQ, Huang J, Guo SQ, Yang X, Bao YM, Tang HJ, Zhang HS. Overexpression of a TFIIIA-type zinc finger protein gene ZFP252 enhances drought and salt tolerance in rice (Oryza sativa L.) FEBS Letters. 2008;582:1037–1043. doi: 10.1016/j.febslet.2008.02.052. [DOI] [PubMed] [Google Scholar]
- Xu ML, Jiang JF, Ge L, Xu YY, Chen H, Zhao Y, Bi YR, Wen JQ, Chong K. FPF1 transgene leads to altered flowering time and root development in rice. Plant Cell Reports. 2005;24:79–85. doi: 10.1007/s00299-004-0906-8. [DOI] [PubMed] [Google Scholar]
- Yoo JH, Park CY, Kim JC, et al. Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis. Journal of Biological Chemistry. 2005;280:3697–3706. doi: 10.1074/jbc.M408237200. [DOI] [PubMed] [Google Scholar]
- Zhang L, Tian LH, Zhao JF, Song Y, Zhang CJ, Guo Y. Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiology. 2009;149:916–928. doi: 10.1104/pp.108.131144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhao G, Jia J, Liu X, Kong X. Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. Journal of Experimental Botany. 2011 doi: 10.1093/jxb/err264. doi:10.1093/jxb/err264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MG, Tian QY, Zhang WH. Nitric oxide synthase-dependent nitric oxide production is associated with salt tolerance in Arabidopsis. Plant Physiology. 2007;144:206–217. doi: 10.1104/pp.107.096842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.