Abstract
Drought is one of the most severe environmental stresses affecting plant growth and limiting crop production. Although many genes involved in adaptation to drought stress have been disclosed, the relevant molecular mechanisms are far from understood. This study describes an Arabidopsis gene, ASPG1 (ASPARTIC PROTEASE IN GUARD CELL 1), that may function in drought avoidance through abscisic acid (ABA) signalling in guard cells. Overexpression of the ASPG1 gene enhanced ABA sensitivity in guard cells and reduced water loss in ectopically overexpressing ASPG1 (ASPG1-OE) transgenic plants. In ASPG1-OE plants, some downstream targets in ABA and/or drought-signalling pathways were altered at various levels, suggesting the involvement of ASPG1 in ABA-dependent drought avoidance in Arabidopsis. By analysing the activities of several antioxidases including superoxide dismutase and catalase in ASPG1-OE plants, the existence was demonstrated of an effective detoxification system for drought avoidance in these plants. Analysis of ProASPG1-GUS lines showed a predominant guard cell expression pattern in various aerial tissues. Moreover, the protease activity of ASPG1 was characterized in vitro, and two aspartic acid sites, D180 and D379, were found to be key residues for ASPG1 aspartic protease activity in response to ABA. In summary, these findings suggest that functional ASPG1 may be involved in ABA-dependent responsiveness and that overexpression of the ASPG1 gene can confer drought avoidance in Arabidopsis.
Keywords: ABA signalling, ASPG1, aspartic protease, drought avoidance, guard cell
Introduction
Abiotic stress conditions such as drought, salinity, and extreme temperatures all have a negative impact on the growth and productivity of plants (Boyer, 1982). For example, dehydration can lead to inhibition of physiological processes; thus, plants have to initiate adaptive mechanisms to survive (Luan, 2002; Kwak et al., 2008). Two pathways involved with drought stress adaptation have been characterized in Arabidopsis: (i) elevation of abscisic acid (ABA) levels can stimulate the activity of downstream targets; and (ii) an ABA-independent signal transduction pathway may direct counteraction against the dehydration (Yamaguchi-Shinozaki and Shinozaki, 2006). Numerous studies on ABA-regulated adaptation to drought stress have been reported (Schroeder et al., 2001; Zhu, 2002; Shinozaki and Yamaguchi-Shinozaki, 2006). The increase in ABA biosynthesis caused by dehydration indicates the importance of ABA signalling in adaptation to drought stress in plants (Guerrero and Mullet, 1986). ABA may trigger oscillations of cytosolic calcium in guard cells (McAinsh et al., 1990; Allan et al., 1994). Subsequently, the two types (S-type and R-type) of anion channels in the plasma membrane of guard cells can be activated (Schroeder and Hagiwara, 1989; Hedrich et al., 1990). Anion channels have been suggested to play a central role in stomatal closure. Genetic analysis has confirmed the regulatory role of SLAC1, the guard-cell S-type anion channel, in ABA-induced stomatal closure (Negi et al., 2008; Vahisalu et al., 2008). Under drought conditions, ABA triggers the production of reactive oxygen species (ROS) including hydrogen peroxide (H2O2) in guard cells, in turn promoting stomatal closure (Pei et al., 2000; Zhang et al., 2001; Kwak et al., 2003; Desikan et al., 2004; Kwak et al., 2008). ABA-induced elevation of cytosolic calcium can promote ROS production in guard cells through activation of NADPH oxidases (such as AtbohD and AtbohF), leading to stomatal closure (Kwak et al., 2003). OST1 (a member of the protein kinase SnRK2 family; Yoshida et al., 2002) acts upstream of ROS in guard cells in response to ABA (Mustilli et al., 2002). A recent study demonstrated that the OST1 can regulate AtbohF activity through phosphorylation (Sirichandra et al., 2009). In the primary ABA signal transduction pathway, the ABA receptor PYR/RCAR, protein phosphatase 2Cs (PP2Cs) ABI1, and protein kinase SnRK2 together act as essential components for initiating ABA signal transduction (Fujii et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009). Previous studies have also shown that H2O2 can inhibit the ABA-induced activities of two types of PP2Cs (ABI1 and ABI2) (Meinhard and Grill, 2001; Meinhard et al., 2002). Thus, ROS can function as the second messenger in mediating ABA signal transduction in guard cells (Kwak et al., 2008). Although ROS can act as a positive regulator in ABA signalling in guard cells, excessive accumulation of ROS during drought stress can be very toxic, killing the plant cells (Meinhard et al., 2002; Kwak et al., 2003). At least two regulatory mechanisms are required to balance the spatial–temporal dynamics of ROS production and scavenging: one to modulate low levels of ROS for signal transduction and another to detoxify excessive ROS in cells during stress (Dat et al., 2000; Mittler, 2002). Hence, antioxidase activity is extreme important to scavenge the excessive amount of ROS in order to defend against oxidative damage in plants (Dat et al., 2000; Mittler, 2002).
Although positive and negative regulators involved in ABA-dependent drought signal transduction have been reported, we are still far from understanding the molecular basis by how plants to adapt to drought stress. Extensive studies on characterization of the components involved in drought signalling in plants are essential. Aspartic proteases comprise a subfamily of proteolytic enzymes that have two highly conserved aspartates for catalysis of their peptide substrates (Szecsi, 1992). They are distributed among various organisms including viruses, bacteria, fungi, plants and animals (Davies, 1990; Rawlings and Barrett, 1995). The Arabidopsis genome contains at least 51 putative aspartic proteases, but their physiological and biochemical functions have remained elusive (Faro and Gal, 2005). A number of studies have shown important roles for aspartic proteases during Arabidopsis development (Xia et al., 2004; Ge et al., 2005). For example, PCS1 functions in cell fate determination during reproductive processes and embryonic development in Arabidopsis. The loss-of-function mutant pcs1 results in excessive cell death in gametogenesis and embryogenesis, whereas overexpression of PCS1 leads to male sterility by blocking anther dehiscence (Ge et al., 2005). CDR1 is involved in salicylic acid-mediated disease resistance. Overexpression of CDR1 produces dwarf plants and triggers resistance to virulent Pseudomonas syringae in Arabidopsis plants (Xia et al., 2004). The Oryza sativa aspartic protease S5 can be functional in hybrid sterility, which acts as a major regulator for the reproductive barrier in indica-japonica (Chen et al., 2008). Overall, it is clear that aspartic proteases are important for plant development. The involvement of aspartic proteases in abiotic stress, however, is poorly understood.
In this report, we characterized an Arabidopsis aspartic protease gene, named ASPG1 (ASPARTIC PROTEASE IN GUARD CELL 1), which demonstrated preferential expression in guard cells of various aerial tissues in Arabidopsis. The data demonstrated that overexpression of ASPG1 could confer drought avoidance via ABA-dependent signalling in Arabidopsis.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia (Col) was used in this study and all transgenic plants were generated in a Col background. Both mutant alleles, aspg1-1 (SALK_045354) and aspg1-2 (SAIL_667_E02), were from the Arabidopsis Biological Resource Center (ABRC; http://www.arabidopsis.org/abrc/). Seeds were sown on MS medium (Murashige and Skoog, 1962) containing 0.8% (w/v) agar and 1% (w/v) sucrose. The sown seeds were stratified for 3 days in the dark at 4 °C before being transferred to a growth chamber for germination. All seeds were grown and stored under the same conditions. All plants were grown under a 16 h light/8 h dark photoperiod and 70% humidity at 23 °C.
Plasmids constructions
In order to generate ectopically overexpressing ASPG1 (ASPG1-OE) transgenic plants, the pBA002-ASPG1 plasmid was constructed by cloning the full-length cDNA sequence (1503 bp) of the ASPG1 gene into the pBA002 binary vector at the XmaI/SpeI cloning sites (Kost et al., 1998). To make the plasmid ProASPG1-GUS containing the β-glucuronidase (GUS) gene, the ASPG1 promoter fragment (1945 bp) was cloned into the binary vector pBI101-GUS at the SbfI/SmaI cloning sites. To analyse ASPG1 cellular localization, the plasmid pCFP-ASPG1 was constructed by a cloning cDNA fragment (1503 bp) of the ASPG1 gene in the KpnI/SacI cloning sites of vector p35S-CFP, which was made by inserting a cyan fluorescent protein (CFP) fragment into vector p35S-MCS. To obtain the recombinant ASPG1 protein expressed in Escherichia coli, pET-30c-ASPG1 was made by inserting the 1503 bp ASPG1 coding sequences with additional cloning sites (EcoRV and NotI) at the C terminus of a His tag into the pET-30c vector (Novagen, Germany). Site-directed mutations of pET-30c-ASPG1D180N, pET-30c-ASPG1D379N and pET-30c-ASPG1D180N/D379N were constructed with a similar cloning strategy. For transient expression assays in mesophyll protoplasts, the 1503 bp cDNA fragments of ASPG1, ASPG1D180N, ASPG1D379N and ASPG1D180N/D379N were subcloned into p35S-MCS at the KpnI/SacI cloning sites. All primers used for plasmid construction are listed in Supplementary Table S1 available at JXB online.
Generation of ASPG1-OE transgenic plants
To generate transgenic ASPG1-OE plants, plasmid pBA002-ASPG1 was introduced into the GV3101 strain of Agrobacterium tumefaciens. Arabidopsis wild-type (Col) plants were transformed with the GV3101 Agrobacterium by floral dip-mediated infiltration (Clough and Bent, 1998). Transgenic plants were selected using BASTA (glufosinate ammonium; Sigma, USA) resistance. The homozygous T3 transgenic lines were used for further analyses.
RT-PCR analyses
Semi-quantitative RT-PCR was performed to analyse the expression level of the ASPG1 gene. Total RNA was isolated from 4-week-old seedlings using an RNAprep Pure Plant kit (Tiangen Biotech, China), following the manufacturer’s instructions. RNA samples were reverse-transcribed with a ReverTra Ace-α-® kit (Toyobo, Japan). The expression level of the ACTIN2 gene (AT3G18780) was used as a loading control. To assess gene expression levels, quantitative RT-PCR analysis was performed. The cDNA was amplified using a SYBR Green master mixture (Applied Biosystems, USA) with a Rotor-Gene 6000 (Corbett Research, Australia). The expression level of the β-ACTIN8 gene (AT1G49240) was used as a loading control. The primer sequences for semi-quantitative and quantitative RT-PCR analyses are listed in Supplementary Table S2 available at JXB online.
Preparation of recombinant ASPG1 protein and assessment of ASPG1 protease activity
Plasmids pET-30c-ASPG1, pET-30c-ASPG1D180N, pET-30c-ASPG1D379N and pET-30c-ASPG1D180N/D379N were transformed into the BL21(DE3) strain of E. coli. Bacteria harbouring expression plasmids were first incubated at 37°C until their exponential growth reached an optical density of 0.6 at 600 nm. Next, 0.5 mM isopropyl-β-D-thiogalactopyranoside (Sigma, USA) was added for 3 h at 20 °C to induce the recombinant protein expression. Purification of ASPG1 protein was performed using a TALON® Metal Affinity Resin column (Clontech, USA) following the manufacturer’s instructions. The purity of the protein was determined by 10% SDS-PAGE and immunoblot analyses. An anti-His polyclonal antibody (Proteintech Group, China) was used for immunoblotting. Protease activity was assessed using a Protease Fluorescent Detection kit (Sigma, USA) followed the manufacturer’s instructions.
Stomatal bioassay, water-loss quantification and ABA content measurement
A bioassay of stomatal apertures was carried out as described by Zhang et al. (2001). Arabidopsis leaves from 4-week-old plants were incubated in buffer containing 10 mM KCl, 50 μM CaCl2, and 10 mM MES/KOH (pH 6.15). To induce stomatal opening, the leaves were first incubated in the light for 3 h and then treated with ABA for 3 h at 23°C. ROS production was detected in guard cells of the leaf epidermal peels using dichlorofluorescein (DCF) (Pei et al., 2000). All images were taken with a TE-2000U inverted fluorescence microscope (Nikon, Tokyo, Japan) equipped with a cooled charged-coupled device camera (Cool SNAP HQ2; Roper Scientific, Houston, TX, USA). To detect the DCF fluorescent signal, a fluorescent filter set with excitation at 488 nm and emission at 525 nm was used. The sizes of the stomatal apertures were quantified using MetaMorph 7.5 software (Molecular Devices, USA). For water-loss quantification, detached rosette leaves from 4-week-old plants were placed in weighing dishes on the laboratory bench at room temperature. Fresh weight was measured throughout the time course of the experiment. Water loss was calculated as the percentage of initial fresh weight. The ABA content was measured using a method described previously (Chen et al., 2011).
Determination of H2O2 content and assessment of antioxidant enzymes
To examine the response to drought stress, 2-week-old seedlings of wild-type Col ASPG1-OE2 and ASPG1-OE14 plants were grown without water for 14 days before re-watering. H2O2 content and activity of antioxidant enzymes were measured after drought treatment. Leaves (0.25 g) from Col and ASPG1-OE plants were extracted in 2 ml extraction buffer containing 50 mM potassium phosphate, 1 mM EDTA, 1% polyvinylpyrrolidone and 1 mM phenylmethylsulfonyl fluoride (pH 7.8). The protein content was normalized using the method of Bradford (1976). H2O2 content was determined as described by Bernt and Bergmeyer (1974). Superoxide dismutase (SOD) activity was analysed with a 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt assay following the method of Ukeda et al. (2002). Catalase (CAT) activity was measured as described by Aebi (1983).
Protoplasts preparation and transient expression assay
Arabidopsis guard cell protoplasts (GCPs) were prepared from rosette leaves of 4-week-old plants using a method established by Pandey et al. (2002). The yield of GCP obtained was 5×106 GCP per 100 leaves with 97.5–99% purity. The method for mesophyll cell protoplast (MCP) preparation was as reported by Yoo et al. (2007), and transient expression assays were performed with MCPs following this method. Plasmid ProRD29A-LUC containing the luciferase (LUC) gene was used as an ABA-responsive reporter and plasmid ProUBQ10-GUS was co-transfected as an internal control, and the relative LUC:GUS activity was scored (Wang et al., 2011). In order to determine the cellular localization of ASPG1, a transient expression experiment was performed using a biolistic bombardment method (Sanford et al., 1993). Images were taken with a laser-scanning confocal imaging system (FV1000; Olympus, Japan). CFP fluorescence was acquired with excitation at 435 nm and emission at 475 nm; green fluorescent protein (GFP) fluorescence was acquired with excitation at 488 nm and emission at 509 nm.
Results
Correlation of ASPG1 expression levels and ABA sensitivity
In order to understand better how ABA regulates many important cellular processes, this study aimed to identify new components involved in ABA signal transduction pathways. A putative ABA-insensitive mutant was screened from a LexA-VP16-estragon receptor (XVE)-tagged T-DNA insertion mutant pool (Zhang et al., 2005) in a seed germination assay (Wang et al., 2011). The original screened putative mutant showed ABA-insensitive seed germination that was not dependent on induction of estradiol (see Supplementary Fig. S1A and B available at JXB online). In analysis of this original mutant, the novel gene locus AT3G18490 was identified encoding a putative aspartic protease by thermal asymmetric interlaced-PCR (Liu et al., 1995; Zuo et al., 2000). The XVE T-DNA insert was found at nt 1275 in the putative aspartic protease domain of gene AT3G18490 (see Supplementary Fig. S1C).
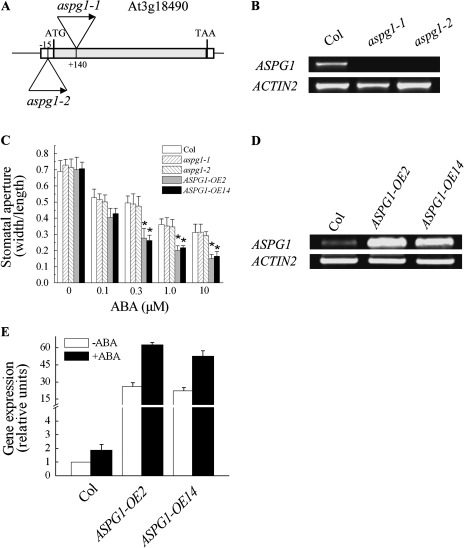
By searching public available microarray databases, it was found that AT3G18490 is expressed preferentially in guard cells in Arabidopsis (Leonhardt et al., 2004). This finding prompted further study of this gene. We named this gene ASPG1 (ASPARTIC PROTEASE IN GUARD CELL 1), and obtained the loss-of-function mutant alleles SALK_045354 (aspg1-1) and SAIL_667_E02 (aspg1-2) (Fig. 1A, B), from ABRC.
Fig. 1.
Responses to ABA of aspg1 mutant alleles and ASPG1-OE transgenic plants. (A) Schematic drawing (not to scale) showing the T-DNA insertion site in gene AT3G18490 revealed in the aspg1-1 (SAIL_667_E02) and aspg1-2 (SALK_045354) mutant alleles. (B) Semi-quantitative RT-PCR analysis of ASPG1 gene expression in wild type (Col) and in the aspg1-1 and aspg1-2 mutant alleles. ACTIN2 (AT3G18780) was used as the loading control. (C) Stomatal closure assay using ABA treatment. Values are means ±SE from three independent experiments (n=50). The epidermal peels of leaves from 4-week-old plants (Col, aspg1-1 and aspg1-2, ASPG1-OE2 and ASPG1-OE14) were first incubated in the light for 3 h to induce stomatal opening and then treated with ABA (0, 0.3, 1.0 and 10 μM) for 3 h. *, P<0.01 compared with wild-type Col in the same treatment. (D) ASPG1 expression was elevated in the ASPG1-OE2 and ASPG1-OE14 transgenic lines. ACTIN2 was used as a loading control. (E) Quantitative analysis of the expression level of ASPG1 in Col, ASPG1-OE2 and ASPG1-OE14 lines after ABA treatment for 3 h. Total RNA was extracted from 4-week-old plants. (–ABA, treated with 0.05% ethanol; +ABA, treated with 50 μM ABA). ASPG1 expression was analysed relative to the level of ABA-free treatment in Col plants, which was taken as 1. Results are shown as the mean ±SE of three independent experiments.
In scoring the stomatal closure with aspg1 mutant alleles, we failed to find a significant difference compared with wild-type Col (Fig. 1C). It was thus speculated that functional redundancy might exist in guard cells. A previous microarray analysis showed that a homologue of aspartic protease, AT3G20015 appeared to have a very low expression level in both guard cells and mesophyll cells (Leonhardt et al., 2004). Thus, an attempt was made to knock out/knock down the AT3G20015 gene (named ASPG2) in the aspg1-1 mutant background using an artificial microRNA (amiRNA) strategy (Schwab et al., 2006). The AT3G20015 gene was successfully knocked down in aspg1-1 plants as follows. Gene expression reduction was examined in aspg1-1 amiR-aspg2 lines by semi-quantitative and quantitative RT-PCR analyses, and 32 lines in which AT3G20015 gene expression was reduced were identified. The expression level of AT3G20015 gene in five lines was substantially reduced in comparison with that in wild-type Col. In line aspg1-1 amiR-aspg2 #3, the expression level of the AT3G20015 gene was reduced 4.3-fold; in line #5, it was reduced 6.7-fold; in line #11, it was reduced 4.2-fold; in line #14, it was reduced 5.3-fold; and in line #23, it was reduced 4.8-fold (see Supplementary Fig. S2A available at JXB online). Because line #5 (expression reduced by 6.7-fold) and line #14 (reduced by 5.3-fold) both showed a stronger reduction in AT3G20015 gene expression in comparison with Col, the behaviour of stomatal closure in these lines was analysed further (see Supplementary Fig. S2); however, neither line showed a differential response in the ABA-induced stomatal closure bioassay (see Supplementary Fig. S2B). Therefore, we turned to gain-of-function analysis by generating ASPG1-OE transgenic plants in Arabidopsis (Col background) (Fig. 1D). Overexpression of ASPG1 in these lines was found to significantly increase ABA sensitivity in guard cells (Fig. 1C). After 3 h of ABA treatment (0.1, 0.3, 1.0 and 10 μM), the ASPG1-OE2 and ASPG1-OE14 plants showed a significant hypersensitivity to ABA (Fig. 1C). These data suggested that ASPG1 might play a role in ABA-induced guard-cell movement.
To test whether ASPG1 expression was ABA inducible, the expression level of ASPG1 was analysed without or with ABA treatment. A 1.89-fold change in ASPG1 expression level was detected in ASPG1-OE and Col plants after treatment with 50 μM ABAfor 3 h (Fig. 1E), indicating that the ASPG1 gene is ABA inducible in Arabidopsis.
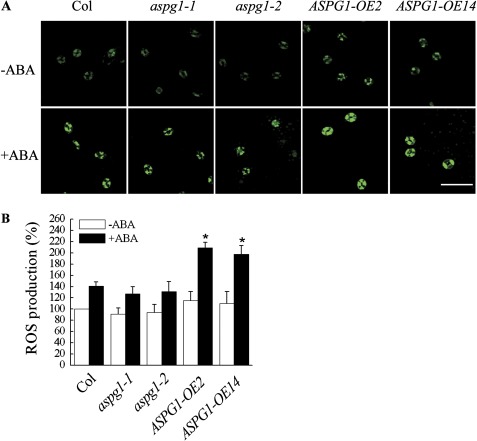
As a secondary messenger, ROS could modulate ABA-induced closure of guard cells (Pei et al., 2000; Zhang et al., 2001; Kwak et al., 2003; Desikan et al., 2004; Kwak et al., 2008). To investigate whether ABA-induced ASPG1 gene expression is associated with ROS production in guard cells, the ROS level was compared in Col, aspg1 and ASPG1-OE plants. An elevated ROS level was detected in the guard cells of ASPG1-OE2 and ASPG1-OE14 plants after 50 μM ABA treatment for 10 min (Fig. 2A). The ROS level increased by ∼80% (80.0–81.7%) in guard cells of ASPG1-OE plants; however, there was only a 41.0% increase in ROS levels in Col guard cells, and an ∼39% (39.4–39.6%) increase in ROS levels in the guard cells of aspg1 mutant allele plants (Fig. 2B). Together, these quantitative data on ROS levels implicated that ASPG1 may be able to trigger ROS production in guard cells in response to ABA.
Fig. 2.
Analysis of ROS production. (A) ROS production was detected using the fluorescent dye DCF. Epidermal peels were loaded with 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) for 10 min before adding 50 μM ABA. Bar, 50 μm. (B) Quantification of ROS levels in guard cells of Col, aspg1-1, aspg1-2, ASPG1-OE2 and ASPG1-OE14 plants after 50 μM ABA treatment. Results are shown as the mean ±SE of three independent experiments (n=50, *, P<0.01 compared with Col after ABA treatment). The fluorescent intensity of guard cells in Col before ABA treatment was taken as 100%.
The response to drought stress in ASPG1-OE plants
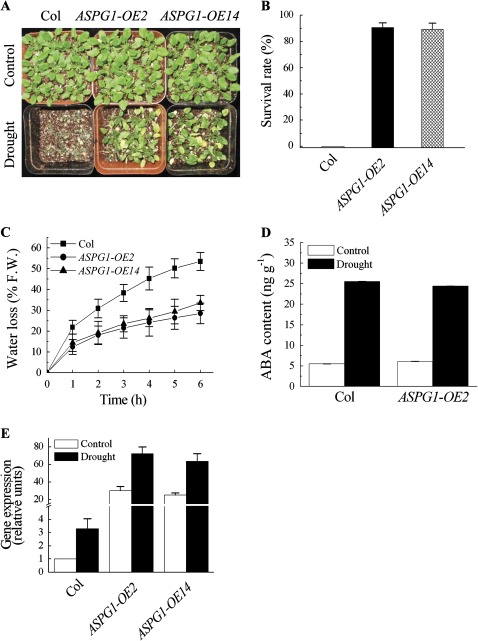
As ASPG1-OE plants showed a remarkable increase in ABA sensitivity in the stomatal closure assay (Fig. 1C), the response to drought stress was examined in ASPG1-OE plants. Two-week-old plants were first grown without water for 14 days and were then rewatered. The results showed that ASPG1-OE2 and ASPG1-OE14 plants could recover from wilting by rehydration in contrast to Col plants (Fig. 3A). After rewatering for 24 h, on average, 90% (89.3–90.7%) of ASPG1-OE plants recovered (Fig. 3B). Transpirational water loss of detached rosette leaves was then measured from 4-week-old Col and ASPG1-OE plants, which were left on the laboratory bench at room temperature with a humidity of ∼40–45%. Less water loss was detected in ASPG1-OE plants compared with Col plants (Fig. 3C). The endogenous ABA level was further analysed in 4-week-old Col and ASPG1-OE2 plants exposed to the same stress condition for 6 h with a humidity of ∼40–45% at 25 °C. The results showed that endogenous ABA content increased similarly in Col and ASPG1-OE plants (Fig. 3D). Taken together, these data suggested that the drought avoidance of ASPG1-OE plants was a consequence of ABA signalling, which might be modulated in guard cells (Figs 1C and 3C, D). To examine the expression level of ASPG1 under drought conditions, ASPG1 gene expression was characterized in Col and ASPG1-OE plants that were well watered (control) and in which water was withheld for 14 days (drought). ASPG1 gene expression was increased up to 3-fold in the drought-stressed Col plants, and a 2.5-fold change in ASPG1 expression was detected in drought-stressed ASPG1-OE lines (Fig. 3E). This further demonstrated that drought stress could also influence ASPG1 expression. To validate the correlation between drought avoidance and guard cell density, guard-cell density was compared in 4-week-old ASPG1-OE plants and Col plants. The data showed that guard-cell development was not affected in ASPG1-OE plants (see Supplementary Fig. S3 available at JXB online). In summary, these results suggested that increased expression levels of ASPG1 could confer drought avoidance of Arabidopsis plants by increasing ABA sensitivity in guard cells (Fig. 1C) accompanied by a reduction in transpirational water loss (Fig. 3C).
Fig. 3.
Response of ASPG1-OE transgenic plants to drought stress. (A) Two-week-old Col and ASPG1-OE plants were well watered (Control) or deprived of water for 14 days and then rewatered (Drought). The photos were taken on day 1 after rewatering. (B) Survival rates of Col and ASPG1-OE plants on day 1 after rewatering. Values are means ±SE from three independent experiments (n=50). (C) Transpirational water loss from detached leaves of 4-week-old Col and ASPG1-OE plants at the indicated time points. Water loss rates are indicated as the percentage of the initial fresh weight (% FW). Results are shown as the mean ±SD from four replicated samples with five leaves used for each sample. (D) Endogenous ABA levels of 4-week-old Col and ASPG1-OE2 plants without (Control) and with drought treatment for 6 h. Values are means ±SD of three independent experiments. (E) Expression levels of the ASPG1 gene in Col and ASPG1-OE plants under drought conditions as described in (A). The expression level of ASPG1 was analysed as relative to the level of the control treatment with Col plants, which was taken as 1. Results are shown as the mean ±SE of three independent experiments.
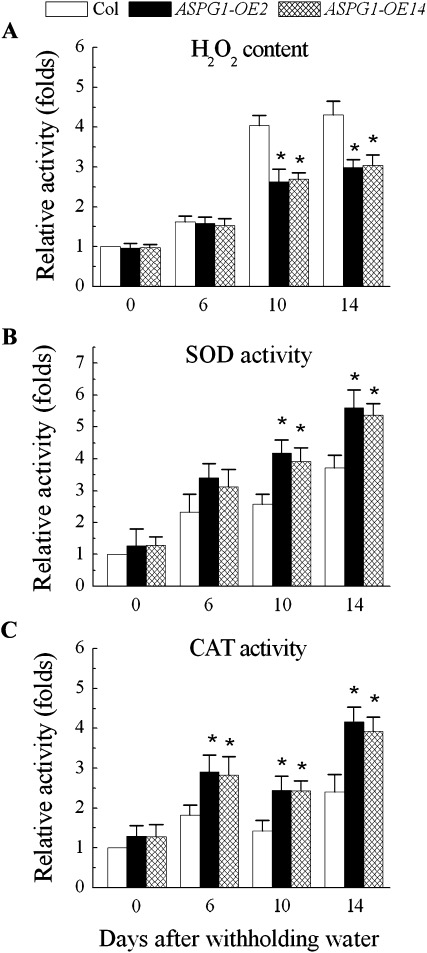
Drought stress is able to trigger ABA-induced ROS accumulation, and plants would thus need a ROS detoxification mechanism to achieve survival from drought stress (Smirnoff, 1993). To characterize whether ROS production accompanied the drought stress in ASPG1-OE plants, the level of H2O2 was analysed in 2-week-old ASPG1-OE plants under the conditions of drought (no water for 14 days). The H2O2 level decreased by 0.6- and 0.7-fold in ASPG1-OE plants on days and 14, respectively, after withholding water (Fig. 4A); thus, the reduction in H2O2 coincided with the mechanism of drought avoidance in ASPG1-OE plants (Fig. 3A, B). A previous study reported that ABA improves the drought adaptation of triploid bermudagrass by increases in the activities of the antioxidants SOD and CAT (Lu et al., 2009). To test the correlation between the reduction of H2O2 and the activity of antioxidases in ASPG1-OE plants, the enzymatic activities of SOD and CAT were measured under the same drought conditions. The results showed that the levels of the SOD activity increased significantly by 1.6- and 1.5-fold in ASPG1-OE plants on days 10 and 14, respectively, after withholding water (Fig. 4B), and the levels of CAT activity increased significantly by 1.4-, 1.7- and 1.6-fold on days 6, 10 and 14, respectively, after withholding water (Fig. 4C). Results from the measurement of ascorbate peroxidase and glutathione reductase activity showed no differential activities of these enzymes in Col and ASPG1-OE plants treated under the same drought conditions (data not shown). Nevertheless, these analyses on antioxidases demonstrated that ASPG1-OE plants were capable of scavenging excessive ROS to prevent oxidative damage to cells through SOD activation, which could convert to H2O2 at the first stage of defence; subsequently, CAT activation could detoxify H2O2 in the cells of ASPG1-OE plants, thus allowing ASPG1-OE plants to survive under drought conditions (Fig. 3A).
Fig. 4.
Determination of H2O2 levels and antioxidant enzymes activities. (A) Comparison of H2O2 levels in Col and ASPG1-OE plants. (B, C) Comparisons of SOD (B) and CAT (C) activity in ASPG1-OE and Col plants. Two-week-old plants had water withheld for the indicated number of days. The relative content of H2O2 and the activity of SOD and CAT were quantified as fold change compared with the Col control. Results are shown as the mean ±SD (n=6; *, P<0.01).
Transcriptional alterations of downstream targets in response to drought stress
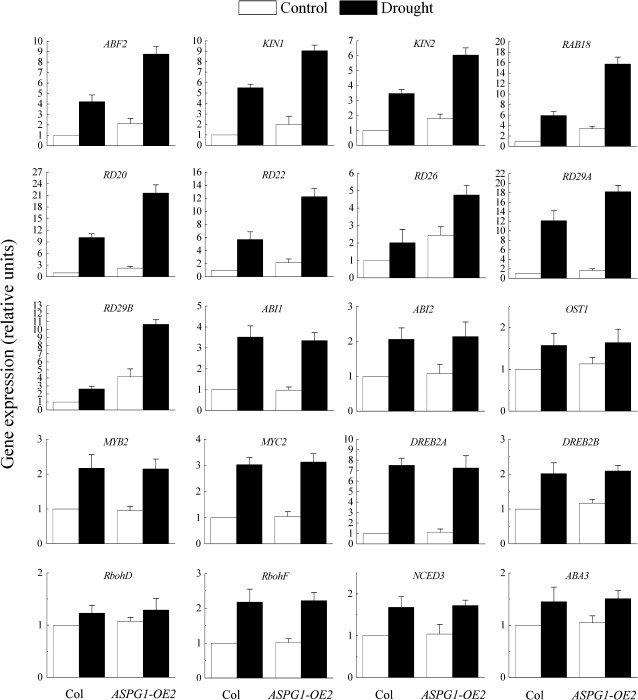
To understand better the scenario of drought avoidance in ASPG1-OE plants, some of the downstream targets of drought stress were analysed in 4-week-old Col and ASPG1-OE2 plants. Gene expression levels were analysed quantitatively after the plants had been subjected to drought stress for 6 h (Fig. 5). We analysed the expression of ABF2, a bZIP transcription factor that can bind to the ABA-responsive element (ABRE) (Kim et al., 2004; Riera et al., 2005; Shinozaki and Yamaguchi-Shinozaki, 2006). The level of ABF2 gene expression was 2.1-fold higher in ASPG1-OE2 plants than in Col plants after drought stress. Other drought- and/or ABA-inducible genes, such as KIN1, KIN2, RAB18, RD20, RD22, RD26, RD29A and RD29B (Riera et al., 2005; Shinozaki and Yamaguchi-Shinozaki, 2006) were also upregulated in ASPG1-OE2 plants in response to drought stress. The expression levels of these genes in ASPG1-OE2 plants were increased at least 2-fold compared with levels in Col plants (Fig. 5). In contrast, the expression levels of the genes for two transcription factors, MYB2 and MYC2, that are able to bind to the MYBR or MYCR element in drought- and ABA-inducible genes (Riera et al., 2005; Shinozaki and Yamaguchi-Shinozaki, 2006) were similar in ASPG1-OE2 and Col plants. Expression of the levels of primary ABA-responsive components such as ABI1, ABI2, OST1, RbohD and RbohF (Kwak et al., 2008) was also analysed. However, these genes displayed similar expression patterns in Col and ASPG1-OE2 plants (Fig. 5). Taken together, these data on gene expression suggested that ASPG1-OE plants are probably modulated by ABA-dependent signalling that involves bZIP transcription regulators (ABRE-binding factors or ABFs) such as ABF2. To clarify this notion, two AP2-type transcription factors, DREB2A and DREB2B, were analysed further and confirmed to be crucial for the ABA-independent drought response (Liu et al., 1998; Riera et al., 2005; Shinozaki and Yamaguchi-Shinozaki, 2006). Both DREB2A and DREB2B were maintained at similar levels in Col and ASPG1-OE2 plants after drought stress (Fig. 5), supporting the suggestion that the drought avoidance of ASPG1-OE plants was unlikely to involve an ABA-independent pathway. In addition, expression levels of ABA3 and NCED3 under drought treatment were determined to evaluate ABA biosynthesis. Both genes were induced at similar levels in Col and ASPG1-OE2 plants (Fig. 5). Thus, the gene expression patterns of ABA3 and NCED3 in Col and ASPG1-OE2 plants under conditions of drought stress reflected exactly the ABA levels in Col and ASPG1-OE2 plants (Fig. 3D). This finding indicated that the adaptation to drought stress in ASPG1-OE plants may be regulated by ABA signal transduction.
Fig. 5.
Expression levels of drought- and ABA-responsive genes in ASPG1-OE2 transgenic plants. The plants were subjected to drought conditions for 6 h; the control plants were not subjected to drought conditions. The expression levels of some drought- and ABA-responsive genes were analysed quantitatively by real-time RT-PCR with 4-week-old plants of Col and ASPG1-OE2. Gene expression levels were analysed relative to the expression levels in Col plants, which were taken as 1. Results are shown as the mean ±SE of three independent experiments.
Preferential guard-cell localization of ASPG1
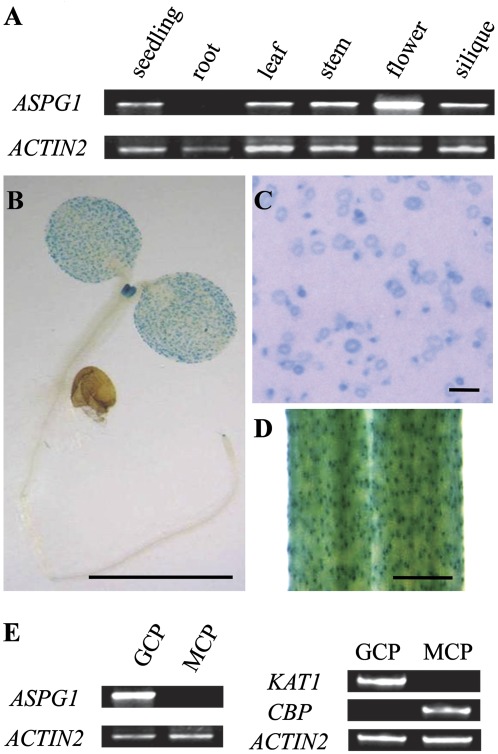
To understand the specificity and functionality of the ASPG1 gene, its expression pattern was characterized in various tissues. ASPG1 gene expression was detected in young seedlings, leaves, stems, flowers and siliques, but not in roots (Fig. 6A). Characterization of ProASPG1-GUS transgenic plants confirmed the tissue specificity of ASPG1 expression (Fig. 6B). ASPG1-expressed GUS was predominantly in guard cells (Fig. 6B–D). The unique guard-cell localization of ASPG1-GUS might imply its specific function for adaptation to drought stress in Arabidopsis. The expression specificity of the ASPG1 gene in guard cells was also quantified. ASPG1, remarkably, was found to be expressed in GCPs but not in MCPs (Fig. 6E). The purity of the GCP and MCP preparations was assessed by characterizing KAT1 expression (a marker gene for guard cells) and CBP expression (a marker gene for mesophyll cells) (Fig. 6E).
Fig. 6.
The predominant guard-cell expression pattern of ASPG1. (A) Semi-quantitative analysis of ASPG1 gene expression in various tissues. (B–D) The ProASPG1-GUS signal indicates the predominant guard-cell expression of ASPG1 in 10-day-old seedlings. Bars, 1 mm (B, D); 0.1 mm (C). (E) Semi-quantitative analysis of ASPG1 gene expression in GCPs and MCPs. Expression levels of the KAT1 (AT5G46240, leaf guard-cell marker) and CBP (AT4G33050, mesophyll cell marker) genes were analysed to determine the purity of GCPs and MCPs. ACTIN2 was used as a loading control.
Subcellular ER localization of ASPG1
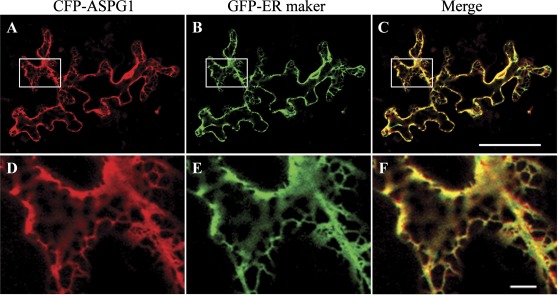
The subcellular localization of the CFP–ASPG1 fusion protein was analysed in leaf epidermal cells of Arabidopsis. Fluorescence analysis of the expression of CFP-ASPG1 revealed a meshwork-like structure that appeared to shown endoplasmic reticulum (ER) localization. The ER localization was further confirmed by co-expressing GFP–KDEL, a marker of the ER (ER-gb CD3-955) in plant cells (Nelson et al., 2007) (Fig. 7). The ER localization of the putative aspartic protease ASPG1 thus suggested its proteolytic characteristic under drought conditions.
Fig. 7.
ER localization of the CFP–ASPG1 fusion protein. (A–C) CFP–ASPG1 (red) and GFP–KDEL (ER-gb CD3-955, a well-known GFP–ER marker; green) were co-expressed in the leaves of 4-week-old Col plants following co-transformation of plasmids by biolistic bombardment. The overlay image (merge) shows co-localization of CFP–ASPG1 and GFP–KDEL (yellow). Bar, 60 μm. (D–F) Magnification of the areas outlined in (A)–(C). Bar, 5 μm.
Aspartic protease activity of ASPG1
ASPG1 is predicted to be an aspartic protease (PF00026) of 53 kDa belonging to the MEROPS peptidase family A1 (pepsin family) (http://arabidopsis.org/servlets/TairObject?id=40580&type=gene). ASPG1 does not have a signal peptide (http://smart.embl-heidelberg.de/smart/show_motifs.pl). Unlike other plant aspartic proteases, ASPG1 lacks a plant-specific sequence (Mutlu and Gal, 1999). By analysing the aligned sequences of ASPG1 and other aspartic proteases from different species, two putative aspartic acid residues (D180 and D379) were found to be conserved in the putative aspartic protease active sites of ASPG1 protein (see Supplementary Fig. S4 available at JXB online).
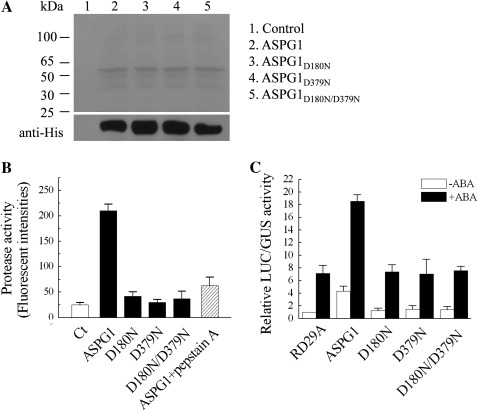
A previous study showed that the activity of aspartic proteases is pH dependent, and that pepstatin A is an inhibitor of aspartic proteases (Rawlings and Barrett, 1995). To determine the aspartic protease activity of ASPG1, the protease activity was assessed in vitro with a purified recombinant His–ASPG1 protein as well as the site-directed mutative proteins ASPG1D180N, ASPG1D379N and ASPG1D180N/D379N (Fig. 8A). The protease activity of His–ASPG1 was measured under different pH conditions (pH 2.0–8.0). Despite there being a broad spectrum of aspartic protease activities, His–ASPG1 displayed a higher activity at pH 2.0 and 6.0 in this assay (data not shown). The protease activity of His–ASPG1 was validated further at pH 6.0 and confirmed by a reduction of ∼70% activity when the protease inhibitor pepstatin A was added (Fig. 8B). In contrast, mutants His-ASPG1D180N, His-ASPG1D379N and His-ASPG1D180N/379N did not show any obvious protease activity (Fig. 8B). These results from aspartic protease analyses demonstrated that ASPG1 has protease activity in vitro and that the two active aspartic acid residues, D180 and D379, are vital for ASPG1 activity.
Fig. 8.
In vitro assay of ASPG1 protease activity. (A) Coomassie blue staining (top panel) and western blotting (bottom panel) showing the recombinant proteins of ASPG1 and mutants, which all were expressed in and purified from E. coli. The His tag was detected as a control of loading levels. (B) In vitro assay to analyse the protease activity of ASPG1. Fluorescent intensities denote the protease activity of ASPG1. Casein was used as the substrate and pepstatin A was used to inhibit the protease activity. Ct, casein; D180N, ASPG1D180N; D379N, ASPG1D379N; D180N/D379N, ASPG1D180N/D379N. Results are shown as means ±SD (n = 6). (C) Col MCPs were transfected with ProRD29A-LUC (RD29A), p35S-ASPG1 (ASPG1), p35S-ASPG1D180N (D180N), p35S-ASPG1D379N (D379N) or p35S-ASPG1D180N/D379N (D180N/D379N). The protoplasts were isolated from 4-week-old Col plants. ProRD29A-LUC was used as the ABA-responsive reporter and ProUBQ10-GUS was co-transfected as an internal control. After transfection, the protoplasts were incubated without ABA (–ABA) or with 50 μM ABA (+ABA) for 10 h in the dark at 23 °C. The relative LUC/GUS activity was quantified. Results are shown as mean ±SE of three independent experiments.
As ASPG1 might be involved in ABA-dependent drought signal transduction, further experiments were performed to investigate the correlation between ASPG1 and the ABA response. ABA responsiveness was analysed with the reporter RD29A–LUC using a transient expression assay. Plasmid ProRD29A-LUC with p35S-ASPG1, p35S-ASPG1D180N, p35S-ASPG1D379N or p35S-ASPG1D180N/D379N were co-expressed in Col mesophyll protoplasts. ProRD29A-LUC was used as an ABA-responsive reporter and ProUBQ10-GUS was co-transfected as an internal control to monitor the transformation efficiency (Wang et al., 2011). The relative LUC/GUS activity was quantified in the presence or absence of ABA. ProRD29A-LUC and ProUBQ10-GUS were transfected alone as negative controls. A significant increase in ProRD29A-LUC activity was obtained in the co-expressing plasmid p35S-ASPG1 with or without ABA (50 μM), but not when co-expressed with plasmid p35S-ASPG1D180N, p35S-ASPG1D379N or p35S-ASPG1D180N/D379N (Fig. 8C). Therefore, these results indicated that the aspartic protease activity of ASPG1 is required for the ABA-induced activity of ProRD29A-LUC.
Discussion
Genes encoding plant aspartic proteases have been identified from different plant species (Mutlu and Gal, 1999; Murakamia et al., 2000; Xia et al., 2004; Ge et al., 2005; Chen et al., 2008). Although studies have revealed the functions of aspartic proteases in various physiological processes during plant development including seed germination (Belozersky et al., 1989; Dunaevsky et al., 1989), leaf senescence (Kato et al., 2004), the immunity response (Xia et al., 2004), cell death (Ge et al., 2005) and reproduction (Chen et al., 2008), little is known about aspartic proteases involving in abiotic stress. The findings in this study have shown that the overexpression of ASPG1 can enhance ABA sensitivity in guard cells, in turn promoting adaptive drought avoidance in Arabidopsis.
ASPG1 confers drought avoidance through the ABA signal transduction pathway
Drought is one of the major abiotic stresses that can trigger severe damage to plants. It is imperative to search for genes that may be involved in plant adaptation to drought stress. In this study, it was found that an Arabidopsis aspartic protease, ASPG1, could play such a role. The findings demonstrated that overexpression of ASPG1 enhanced ABA sensitivity in guard cells (Fig. 1C) and ASPG1-OE plants showed an improvement in drought avoidance of plants (Fig. 3). In Arabidopsis, drought stress may initiate responses to either ABA-dependent or ABA-independent signal transduction (Shinozaki and Yamaguchi-Shinozaki, 2006). In this report, although the expression levels of upstream components of ABA-signalling such as ABI1, ABI2, OST1, RbohD and RbohF (Kwak et al., 2008) were all induced by drought stress, the expression patterns were not differentiated in Col and ASPG1-OE2 plants. Therefore, ASPG1 is probably functional downstream of these components (Fig. 5). It should be noted that expression of ABF2, a member of the ABF family (Jakoby et al., 2002), was significantly upregulated under drought conditions. ABFs are essential for ABA signal transduction (Fujii et al., 2009), and are able to bind to the ABRE of downstream drought- and/or ABA-responsive genes to initiate stomatal closure while responding to drought stress (Riera et al., 2005; Shinozaki and Yamaguchi-Shinozaki, 2006). In this study, the elevated expression level of ABF2 by drought stress in ASPG1-OE2 plants may suggest that ASPG1 involvement in drought adaptation may require ABF activity (at least that of ABF2); in turn, expression of downstream targets of ABA signalling could be stimulated under drought conditions (Fig. 5). This was confirmed by analysing ProRD29A-LUC activity (Fig. 8C). The RD29A promoter contains both an ABRE and a dehydration-responsive element (Yamaguchi-Shinozaki and Shinozaki, 2006). The data from the transient expression assay confirmed that ABA-induced activity of ProRD29A-LUC was significantly increased when co-expressed with plasmid p35S-ASPG1 (Fig. 8C). Thus, these results strongly suggested that ASPG1 may confer drought avoidance to plants via ABA signalling.
Drought stress causes endogenous ABA biosynthesis and in turn, ABA triggers ROS production to mediate downstream responses in guard cells (Pei et al., 2000; Zhang et al., 2001; Kwak et al., 2003; Desikan et al., 2004; Kwak et al., 2008). In this study, we observed that the ASPG1 gene was able to promote ROS (H2O2) production in guard cells after treatment with ABA (Fig. 2), indicating the involvement of ASPG1 in ROS-mediated ABA signal transduction. Accumulation of excessive ROS causes oxidative damage to cells and stimulates the activity of antioxidases (Mittler, 2002). Antioxidases SOD and CAT are two important enzymes that scavenge excessive ROS to prevent oxidative damage to cells. SOD could convert into H2O2 and CAT could then detoxify the H2O2 to defend cells against oxidative damage (Mittler, 2002). We determined significant activity of SOD and CAT in 2-week-old ASPG1-OE plants after withholding water for 10–14 days (Fig. 4B, C), suggesting that ASPG1-OE plants may be able to scavenge excessive ROS under severe dehydration conditions.
The possible function of ASPG1 aspartic protease in plant cells
By analysing public available microarray data (Leonhardt et al., 2004), it was noted that the ASPG1 gene showed higher expression levels in Arabidopsis guard cells but was almost absent in mesophyll cells. This predominant guard-cell expression trait of the ASPG1 gene was also observed by a histological assay in this study (Fig. 6). Coincidentally, ASPG1-OE plants were hypersensitive to ABA in stomatal closure assays (Fig. 1C). The appearance of drought avoidance in ASPG1-OE plants therefore may be relevant to the predominant guard-cell expression of ASPG1.
ASPG1 also showed proteolytic activity in vitro. Two aspartic residues D180 and D379 were identified within the highly conserved aspartic protease sites by a protease activity assay (Fig. 8A, B). The aspartic protease activity was found to be required for ASPG1 function in response to ABA (Fig. 8C). Nevertheless, the target proteins for ASPG1 remained unknown. Plant aspartic proteases have been demonstrated to play roles in pro-protein processing and protein degradation in vitro (Mutlu and Gal, 1999). A Brassica napus aspartic protease may process the polypeptide precursor of storage protein 2S albumins by cleaving within the N-terminal and internal pro-peptide linking the two subunits (D’Hondt et al., 1993). Several aspartic proteases conduct storage protein degradation during seed germination (Runeberg-Roos et al., 1994; Hiraiwa et al., 1997) and the degradation of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) in tobacco senescent leaves (Kato et al., 2004). The ER localization of CFP–ASPG1 in plant cells (Fig. 7) implies a possible role for ASPG1 in protein modification in the ER. Future analyses of the role of ASPG1 in protein processing and/or relevant protein modification in the ER would be interesting.
More than 50 putative aspartic proteases have been predicted in Arabidopsis (Faro and Gal, 2005). The functional redundancy of homologues of aspartic protease genes may complicate the scenario in guard cells when plants respond to drought stress, which may explain the insignificant phenotypes shown in the knockout aspg1 mutant alleles compared with the wild-type Col (Fig. 1C). In this study, we failed to determine which homologue(s) has a similar function in response to drought stress in Arabidopsis (see Supplementary Fig. S2). Further studies to clarify this homologue redundancy are imperative.
In summary, the findings of this study demonstrated ASPG1 function in ABA-dependent drought signalling. Under drought conditions, ABA induced ASPG1 gene expression. ASPG1 triggered stomatal closure to avoid water loss through the activation of antioxidases, thus preventing Arabidopsis plants from oxidative damage. As a consequence, the plant is able to adapt to drought stress.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Original screening with the XVE-tagged T-DNA insertion mutant lines. (A) Seeds of XVE T-DNA-tagged lines were screened on an MS plate containing 1% sucrose, 0.8% (w/v) agar, with or without 10 μM 17-β-estradiol (E) and 1.5 μM ABA. Photographs were taken to show the germination phenotypes on the day 7 after stratification. (B) Germination rates (%) were analysed on day 7 after stratification by scoring the number of open green cotyledons. Values are means ±SE from three independent experiments (n=100). (C) Schematic drawing (not to scale) showing the T-DNA insertion site in gene AT3G18490 revealed in the original mutant screen. The putative aspartic protease domain at nt 481–1494 is shaded black. Arrows denote the orientation of gene transcription.
Supplementary Fig. S2. Response to ABA of the amiRNA lines. (A) Analyses of the gene expression of ASPG2 (AT3G20015) in Col, aspg1-1 and aspg1-1 amiR-aspg2 lines (#3, #5, #11, #14 and #23). Expression levels were analysed relative to the level of ASPG2 in Col plants, which was taken as 1. Results are shown as mean ±SE of three independent experiments. (B) ABA-induced stomatal closure. Values are means ±SE from three independent experiments (n=50). The leaves from 4-week-old plants of Col, aspg1-1 and aspg1-1 amiR-aspg2 lines (#5 and #14) were first incubated in the light for 3 h to induce stomatal opening and then treated with ABA (0, 5, and 10 μM) for 3 h.
Supplementary Fig. S3. Overexpression of ASPG1 has no effect on the development of guard cells. (A) The epidermis of the abaxial surface of rosette leaves from Col and ASPG1-OE2 plants, Bar, 80 μm. (B) Number of stomata mm−2 in the epidermis of the abaxial surface of rosette leaves of Col, ASPG1-OE2, and ASPG1-OE14 lines were determined. Values are means ±SE from leaves of three individual plants of Col and ASPG1-OE lines. Three independent counts were performed on each leaf.
Supplementary Fig. S4. Two conserved putative aspartic activation sites in the ASPG1 protein. Alignments between predicted ASPG1 protein sequences containing two aspartic proteases in a number of organisms using ClustalX and GeneDoc3.2 tools. CDR1 and PCS1 are from Arabidopsis thaliana; CND41 is from Nicotiana tobacum. CNB-1 is from Brachypodium sylvaticum; S5 is from Oryza sativa; Q3UKT5 is from Mus musculus; Q9VLK3 is from Drosophila melanogaster; Q8WWD9 is from Homo sapiens. Arrows indicate the two catalytic aspartic acid residues in ASPG1.
Supplementary Table S1. Primer sequences used for plasmid constructions in this study.
Supplementary Table S2. Primer sequences used for semi-quantitative and quantitative RT-PCR experiments in this study.
Acknowledgments
We thank Dr Yu-Qi Feng (Department of Chemistry, Wuhan University) for his laboratory resources for carrying out the measurement of ABA content in this study; Dr Nam-Hai Chua (The Rockefeller University) for providing the pBA002 vector; Dr Benedikt Kost (University of Erlangen-Nuremberg) for sharing the p35S-MCS vector; Dr Nebenführ Andreas (University of Oklahama Health Sciences Center) for sharing the plasmid ER-gb CD3-955; and Dr Jianru Zou (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for generously providing XVE T-DNA-tagged mutant seeds and the pBI101-GUS vector. We thank all members of the Wu laboratory for their stimulating discussions and helpful comments on this manuscript. This work was supported by grants to Y.W. from the National Natural Science Foundation of China (90817013) and by the Chinese 111 Project (B06018).
Glossary
Abbreviations
- ABA
abscisic acid
- ABRC
Arabidopsis Biological Resource Center
- ABRE
abscisic acid-responsive element
- ABF
ABRE-binding factor
- amiRNA
artificial microRNA
- CAT
catalase
- CFP
cyan fluorescent protein
- DCF
dichlorofluorescein
- ER
endoplasmic reticulum
- GCP
guard cell protoplast
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- H2O2
hydrogen peroxide
- LUC
luciferase
- MCP
mesophyll cell protoplast
- ROS
reactive oxygen species
- SOD
superoxide dismutase
References
- Aebi HE. Catalase. In: Bergmeyer HU, editor. Methods of Enzymatic Analysis. Vol. 3. Weinheim: Verlag Chemie; 1983. pp. 273–286. [Google Scholar]
- Allan AC, Fricker MD, Ward JL, Beale MH, Trewavas AJ. Two transduction pathways mediate rapid effects of abscisic acid in Commelina guard cells. The Plant Cell. 1994;6:1319–1328. doi: 10.1105/tpc.6.9.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belozersky MA, Sarbakanova ST, Dunaevsky YE. Aspartic proteinase from wheat seeds: isolation, properties and action on gliadin. Planta. 1989;177:321–326. doi: 10.1007/BF00403589. [DOI] [PubMed] [Google Scholar]
- Bernt E, Bergmeyer HU. Inorganic peroxides. In: Bergmeyer HU, editor. Methods of enzymatic analysis. Vol. 14. New York: Academic Press; 1974. pp. 2246–2248. [Google Scholar]
- Boyer JS. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chen M, Huang Y, Liu J, Yuan B, Feng Y. Highly sensitive profiling assay of acidic plant hormones using a novel mass probe by capillary electrophoresis-time of flight-mass spectrometry. Journal of Chromatography B. 2011;879:938–944. doi: 10.1016/j.jchromb.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Chen JJ, Ding JH, Ouyang YD, et al. A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:11436–11441. doi: 10.1073/pnas.0804761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dat J, Vandenabeele S, Vranová E, Van Montagu M, Inzé D, van Breusegem F. Dual action of the active oxygen species during plant stress responses. Cell Molecular Life Sciences. 2000;57:779–795. doi: 10.1007/s000180050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DR. The structure and function of aspartic proteinases. Annual Review of Biophysics and Biophysical. Chemistry. 1990;19:189–215. doi: 10.1146/annurev.bb.19.060190.001201. [DOI] [PubMed] [Google Scholar]
- Desikan R, Cheung MK, Bright J, Henson D, Hancock JT, Neill SJ. ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. Journal of Experimental Botany. 2004;55:205–212. doi: 10.1093/jxb/erh033. [DOI] [PubMed] [Google Scholar]
- D’Hondt K, Bosch D, van Damme J, Goethals M, Vandekerckhove J, Krebbers E. An aspartic proteinase present in seeds cleaves Arabidopsis 2S albumin precursors in vitro. Journal of Biological Chemistry. 1993;268:20884–20891. [PubMed] [Google Scholar]
- Dunaevsky YE, Sarbakanova ST, Belozersky MA. Wheat seed carboxypeptidase and joint action on gliadin of proteases from dry and germinating seeds. Journal of Experimental Botany. 1989;40:1323–1329. [Google Scholar]
- Faro C, Gal S. Aspartic proteinase content of the Arabidopsis genome. Current Protein and Peptide Science. 2005;6:493–500. doi: 10.2174/138920305774933268. [DOI] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cuter SR, Sheen J, Rodriguez PL, Zhu JK. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XC, Dietrich C, Matsuno M, Li GJ, Berg H, Xia YJ. An Arabidopsis aspartic protease functions as an anti-cell-death component in reproduction and embryogenesis. EMBO Reports. 2005;6:282–288. doi: 10.1038/sj.embor.7400357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero F, Mullet JE. Increased abscisic acid biosynthesis during plant dehydration requires transcription. Plant Physiology. 1986;80:588–591. doi: 10.1104/pp.80.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrich R, Busch H, Raschke K. Ca2+ and nucleotide dependent regulation of voltage dependent anion channels in the plasma membrane of guard cells. EMBO Journal. 1990;9:3889–3892. doi: 10.1002/j.1460-2075.1990.tb07608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraiwa N, Kondo M, Nishimura M, Hara-Nishimura I. An aspartic endopeptidase is involved in the breakdown of propeptides of storage proteins in protein-storage vacuoles of plant. European Journal of Biochemistry. 1997;246:133–141. doi: 10.1111/j.1432-1033.1997.00133.x. [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. bZIP transcription factors in Arabidopsis. Trends in Plant Science. 2002;7:106–111. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- Kato Y, Murakami S, Yamamoto Y, Chatani H, Kondo Y, Nakano T, Yokota A, Sato F. The DNA-binding protease, CND41, and the degradation of ribulose-1,5-bisphosphate carboxylase/oxygenase in senescent leaves of tobacco. Planta. 2004;220:97–104. doi: 10.1007/s00425-004-1328-0. [DOI] [PubMed] [Google Scholar]
- Kim S, Kang JY, Cho DI, Park JH, Kim SY. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. The Plant Journal. 2004;40:75–87. doi: 10.1111/j.1365-313X.2004.02192.x. [DOI] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua NH. A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. The Plant Journal. 1998;16:393–401. doi: 10.1046/j.1365-313x.1998.00304.x. [DOI] [PubMed] [Google Scholar]
- Kwak JM, Mäser P, Schroeder JI. The Arabidopsis Book. Washington, DC: American Society of Plant Biologists; 2008. The clickable guard cell, version II: interactive model of guard cell signal transduction mechanisms and pathways. http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dang JL, Bloom RE, Bodde S, Jones JDG, Schroeder JI. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO Journal. 2003;22:2623–2633. doi: 10.1093/emboj/cdg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI. Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. The Plant Cell. 2004;16:596–615. doi: 10.1105/tpc.019000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. The Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. The Plant Journal. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Luan S. Signalling drought in guard cells. Plant, Cell and Environment. 2002;25:229–237. doi: 10.1046/j.1365-3040.2002.00758.x. [DOI] [PubMed] [Google Scholar]
- Lu SY, Su W, Li HH, Guo ZF. Abscisic acid improves drought tolerance of triploid bermudagrass and involves H2O2- and NO-induced antioxidant enzyme activities. Plant Physiology Biochemistry. 2009;47:132–138. doi: 10.1016/j.plaphy.2008.10.006. [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Brownlee C, Hetherington AM. Abscisic acid-induced elevation of guard cell cytosolic Ca2+ precedes stomatal closure. Nature. 1990;343:186–188. [Google Scholar]
- Meinhard M, Grill E. Hydrogen peroxide is a regulator of ABI1. a protein phosphatase 2C from Arabidopsis. FEBS Letters. 2001;508:443–446. doi: 10.1016/s0014-5793(01)03106-4. [DOI] [PubMed] [Google Scholar]
- Meinhard M, Rodriguez PL, Grill E. The sensitivity of AB12 to hydrogen peroxide links the abscisic acid-response regulator to redox signalling. Planta. 2002;214:775–782. doi: 10.1007/s00425-001-0675-3. [DOI] [PubMed] [Google Scholar]
- Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends in Plant Science. 2002;7:405–410. doi: 10.1016/s1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- Miyazono KI, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, Miyauchi Y, Takahashi M, Zhi YH, Fujita Y, Yoshida T, Kodaira KS, Yamaguchi-Shinozaki K, Tanokura M. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- Murakamia S, Kondob Y, Nakanoc T, Sato F. Protease activity of CND41, a chloroplast nucleoid DNA-binding protein, isolated from cultured tobacco cells. FEBS Letters. 2000;468:15–18. doi: 10.1016/s0014-5793(00)01186-8. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. The Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu A, Gal S. Plant aspartic proteinases: enzymes on the way to a function. Physiologia Plantarum. 1999;105:569–576. [Google Scholar]
- Negi J, Matsuda O, Nagasawa T, Oba Y, Takahashi H, Kawai-Yamada M, Uchimiya H, Hashimoto M, Iba K. CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature. 2008;452:483–486. doi: 10.1038/nature06720. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Wang XQ, Coursol SA, Assmann SM. Preparation and applications of Arabidopsis thaliana guard cell protoplasts. New Phytologist. 2002;153:517–526. doi: 10.1046/j.0028-646X.2001.00329.x. [DOI] [PubMed] [Google Scholar]
- Pei ZM, Murata Y, Benning G, Thomine S, Klüsener B, Allen GJ, Grill E, Schroeder JI. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature. 2000;406:731–734. doi: 10.1038/35021067. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ. Families of aspartic peptidases, and those of unknown catalytic mechanism. Methods in Enzymology. 1995;248:105–120. doi: 10.1016/0076-6879(95)48009-9. [DOI] [PubMed] [Google Scholar]
- Riera M, Valon C, Fenzi F, Giraudat J, Leung J. The genetics of adaptive responses to drought stress: abscisic acid-dependent and abscisic acid-independent signalling components. Physiologia Plantarum. 2005;123:111–119. [Google Scholar]
- Runeberg-Roos P, Kervinen P, Kovaleva J, Raikel NV, Gal S. The aspartic proteinase of barley is a vacuolar enzyme that processes probarley lectin in vitro. Plant Physiology. 1994;105:321–329. doi: 10.1104/pp.105.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JC, Smith FD, Rushell JA. Optimizing the biolistic process for different biological application. Methods in Enzymology. 1993;217:483–509. doi: 10.1016/0076-6879(93)17086-k. [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Hagiwara S. Cytosolic calcium regulates ion channels in the plasma membrane of Vicia faba guard cells. Nature. 1989;338:427–430. [Google Scholar]
- Schroeder JI, Kwak JM, Allen GJ. Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature. 2001;410:327–330. doi: 10.1038/35066500. [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski R, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. The Plant Cell. 2006;18:1121–1333. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Sirichandra C, Gu D, Hu HC, Davanture M, Lee S, Djaoui M, Valot B, Zivy M, Leung J, Merlot S, Kwak JM. Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Letters. 2009;583:2982–2986. doi: 10.1016/j.febslet.2009.08.033. [DOI] [PubMed] [Google Scholar]
- Smirnoff N. The role of active oxygen in the response of plants to water deficit and desiccation. New Phytologist. 1993;125:27–58. doi: 10.1111/j.1469-8137.1993.tb03863.x. [DOI] [PubMed] [Google Scholar]
- Szecsi PB. The aspartic protease. Scandinavian Journal of Clinical and Laboratory Investigation Supplementum. 1992;210:5–22. [PubMed] [Google Scholar]
- Ukeda H, Shimamura T, Tsubouchi M, Harada Y, Nakai Y, Sawamura M. Spectrophotometric assay of superoxide anion formed in Maillard reaction based on highly water-soluble tetrazolium salt. Analytical Science. 2002;18:1151–1154. doi: 10.2116/analsci.18.1151. [DOI] [PubMed] [Google Scholar]
- Vahisalu T, Kollist H, Wang YF, et al. SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature. 2008;452:487–491. doi: 10.1038/nature06608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YP, Li L, Ye TT, Zhao SJ, Liu Z, Feng YQ, Wu Y. Cytokinin antagonizes ABA suppression to seed germination of Arabidopsis by down-regulating ABI5 expression. The Plant Journal. 2011;68:249–261. doi: 10.1111/j.1365-313X.2011.04683.x. [DOI] [PubMed] [Google Scholar]
- Xia YJ, Suzuki H, Borevitz J, Blount J, Guo ZJ, Patel K, Dixon RA, Lamb C. An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO Journal. 2004;23:980–988. doi: 10.1038/sj.emboj.7600086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of plant Biology. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiology. 2002;43:1473–1483. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- Zhang J, Xu JX, Kong YZ, et al. Generation of chemical-inducible activation tagging T-DNA insertion lines of Arabidopsis thaliana. Acta Genetica Sinica. 2005;32:1082–1088. [PubMed] [Google Scholar]
- Zhang X, Zhang L, Dong F, Gao J, Galbraith DW, Song CP. Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiology. 2001;126:1438–1448. doi: 10.1104/pp.126.4.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. The Plant Journal. 2000;24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.