Abstract
Gibberellin (GA) signalling during pumpkin male flower development is highly regulated, including biosynthetic, perception, and transduction pathways. GA 20-oxidases, 3-oxidases, and 2-oxidases catalyse the final part of GA synthesis. Additionally, 7-oxidase initiates this part of the pathway in some cucurbits including Cucurbita maxima L. (pumpkin). Expression patterns for these GA-oxidase-encoding genes were examined by competitive reverse transcription-PCR (RT-PCR) and endogenous GA levels were determined during pumpkin male flower development. In young flowers, GA20ox3 transcript levels are high in stamens, followed by high levels of the GA precursor GA9. Later, just before flower opening, transcript levels for GA3ox3 and GA3ox4 increase in the hypanthium and stamens, respectively. In the stamen, following GA3ox4 expression, bioactive GA4 levels rise dramatically. Accordingly, catabolic GA2ox2 and GA2ox3 transcript levels are low in developing flowers, and increase in mature flowers. Putative GA receptor GID1b and DELLA repressor GAIPb transcript levels do not change in developing flowers, but increase sharply in mature flowers. Emasculation arrests floral development completely and leads to abscission of premature flowers. Application of GA4 (but not of its precursors GA12-aldehyde or GA9) restores normal growth of emasculated flowers. These results indicate that de novo GA4 synthesis in the stamen is under control of GA20ox3 and GA3ox4 genes just before the rapid flower growth phase. Stamen-derived bioactive GA is essential and sufficient for male flower development, including the petal and the pedicel growth.
Keywords: Cucurbita maxima, GA-oxidases, gibberellin, flower development
Introduction
Bioactive gibberellins (GAs) regulate distinct developmental processes during the entire life cycle of a higher plant, including growth and flowering (Davies, 2004; Fleet and Sun, 2005; Mutasa-Göttgens and Hedden, 2009). Reduced GA signalling (including GA synthesis, perception, and signal transduction pathways) typically leads to plants with dwarf phenotype, delayed flowering, and reduced fertility. In some species (e.g. Arabidopsis) enhanced GA signalling typically results in tall phenotype, and in earlier flowering (Radi et al., 2006). Consequently, fine-tuning of GA signalling is necessary for normal plant growth and development.
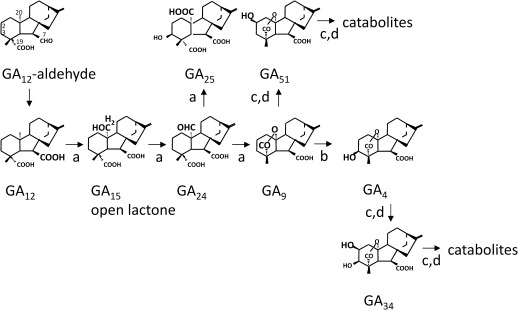
GAs are formed via complex biosynthetic pathways, the final part of which is catalysed by 2-oxoglutarate-dependent dioxygenases, referred to as GA-oxidases (Fig. 1; for a review, see Yamaguchi, 2008). Typically, this final part starts with GA12, which is formed from GA12-aldehyde by membrane-bound cytochrome P450 monooxygenases. In pumpkin, however, an additional soluble 2-oxoglutarate-dependent dioxygenase, GA 7-oxidase, has been found to catalyse this step (Lange et al., 1993, 1994b; Lange, 1997). In all higher plants the following steps of the pathway are catalysed by 2-oxoglutarate-dependent dioxygenases. GA 20-oxidases (GA20oxs) oxidize GA12 to GA9 via GA15 and GA24. Then, GA 3-oxidases (GA3oxs) synthesize the plant hormone GA4 from GA9. Finally, GA 2-oxidases (GA2oxs) inactivate the plant hormone GA4 but also the precursor GA9 by forming GA34 and GA51, respectively (Fig. 1). Additionally, some GA2oxs form catabolites from GA34 and GA51 (Fig. 1). In pumpkin plants, GA4 is the main bioactive GA (Pimenta Lange and Lange, 2006).
Fig. 1.
Gibberellin biosynthesis in pumpkin plants. This simplified pathway only shows the final part for the synthesis of the non-13-hydroxylated GAs. Reactions are catalysed by recombinant GA20ox4 (a), GA3ox4 (b), GA2ox2 (c), and GA2ox3 (d).
Small multigene families code for these GA-oxidases that ensure dynamic GA signalling during plant development in a tissue- and organ-specific manner (Pimenta Lange and Lange, 2006; Yamaguchi, 2008; Mutasa-Göttgens and Hedden, 2009). Several factors regulate GA-oxidase gene expression, including photoperiod (Zeevaart and Gage, 1993; Xu et al., 1997), feedback and feedforward regulatory mechanisms (Phillips et al., 1995; Xu et al., 1995; Martin et al., 1996; Rebers et al., 1999; Thomas et al., 1999; Itoh et al., 2001; Lange et al., 2005), and ‘cross-talk’ with other plant hormones (van Huizen et al., 1997; Ross et al., 2000). More recently, Griffiths et al. (2006) found that all three Arabidopsis GID1 GA receptor genes are also subject to feedback regulation mechanisms. Therefore, the late steps of GA biosynthesis and perception are important targets for GA regulation of plant development.
GAs regulate flower development and are essential for male and female fertility. In GA-deficient Arabidopsis and tomato mutants, stamen growth is arrested. No viable pollen develops in severe GA-deficient mutants, and sepals, petals, and pistils are underdeveloped, leading in some cases even to premature abortion of the flower (Koornneef and van der Veen, 1980; Nester and Zeevaart, 1988; Goto and Pharis, 1999). Application of bioactive GAs or even of the GA precursor GA9 restores normal flower development, and Arabidopsis stamens require higher GA concentrations than do the other floral organs (Goto and Pharis, 1999). Stamens offer a rich source for GAs, as has been demonstrated, for example, for rice (Hirano et al., 2008). Moreover, for a long time it has been known that in Glechoma hederacea, stamen-derived GAs stimulate corolla growth (Plack, 1958). More recently, Griffiths et al. (2006) found in Arabidopsis triple GID1 receptor mutants not only the stamen and petal development arrested and the pistil length reduced, but also a striking reduction in elongation of the pedicel. Furthermore, Hu et al. (2008) identified two potential sites for bioactive GA synthesis in Arabidopsis flowers, stamens and/or flower receptacles, and suggest that GAs are transported from these organs to promote petal growth.
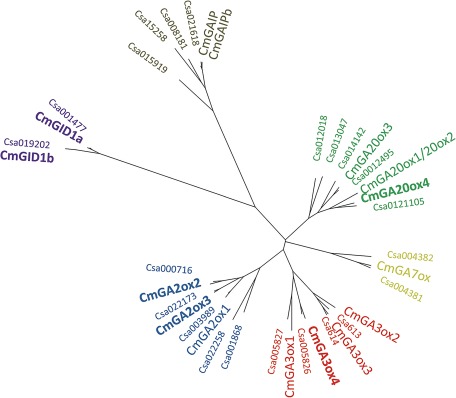
In this study, the function of GAs was examined for male flower development of monoecious pumpkin plant. Cucurbits have contributed fundamentally to our knowledge of GA signalling (Graebe, 1987; Haywood et al., 2005; Pimenta Lange and Lange, 2006). Several GA-related genes have been identified in pumpkin (Haywood et al., 2005; Lange et al., 2005). Recently, the genomic sequence of cucumber (Cucumis sativus) became available (Huang et al., 2009). Phylogenetic analysis reveals high sequence homology between GA-related genes from pumpkin and the respective genes derived from the cucumber genome (Fig. 2). It also suggests the existence of additional GA-related genes in pumpkin. Cucumber genome information was utilized to isolate four new pumpkin GA-oxidase cDNAs (GA20ox4, GA3ox4, GA2ox2, and GA2ox3). In addition, two putative pumpkin GA receptor cDNAs (GID1a and GID1b) were cloned for this study. To identify sites and key regulators of GA signalling during pumpkin male flower development, profiles of endogenous GAs and of transcript levels of GA-related genes were analysed in floral organs at specific developmental stages. Moreover, emasculation experiments demonstrate the role of GA at specific floral sites in male flower development.
Fig. 2.
Phylogenetic relationship of pumpkin (Cm) and cucumber (Csa) GA-related proteins (Huang et al., 2009) using the program Geneious Pro 5.4. For this study newly isolated genes from pumpkin are in bold. CmGAIP and CmGAIPb were isolated originally by Haywood et al. (2005). For the other CmGA-oxidases see Lange et al. (2005). For Cucumis sativus: Csa613 (ACHR01000613; ORF, bp 29 702–30 198 and 30 355–30 982), Csa614 (ACHR01000614; ORF, bp 8355–8848 and 8929–9559); for other Csa genes see the Cucurbit Genomics Database (www.icugi.org/cgi-bin/ICuGI/genome/cuke.cgi).
Materials and methods
Plant material
Cucurbita maxima L. cv. ‘Riesenmelone’, seeds (from Samenhaus Knieke, Braunschweig, Germany) were imbibed for 2 h and sown in soil (100 ml pots) moistened with 50 ml of water. After 2 weeks, the plants were transferred to 2500 ml pots and watered every day. Growth, sampling, and total RNA extraction conditions were essentially the same as described by Lange et al. (2005) for seedlings and adult plants, with the exception that extra DNase I treatment and the respective inactivation was done according to the manufacturer’s protocol (Fermentas, Thermo Fisher Scientific Inc., Waltham, MA, USA).
Emasculation experiments
Emasculation experiments were performed with C. maxima male flower buds at development stage II (Fig. 3, 1.5–2.0 cm flower heads). A small longitudinal incision was made with a scalpel through the sepals, and stamens were removed with tweezers. Through the same incision GA12-aldehyde (10−4 M), GA9 (10−4 M), or GA4 (10−4 M) were added in 5 μl of 0.5% Tween-20 in 1% methanol in addition to a solvent-only mock treatment at the site where stamens were removed. GA12-aldehyde and GA9 were gifts from Professor J.E. Graebe, Göttingen, and GA4 was a gift from Professor L. Mander, Canberra, Australia. The identity of all unlabelled GAs was verified and the purity was >95% as determined by full scan gas chromatography–mass spectrometry (GC-MS).
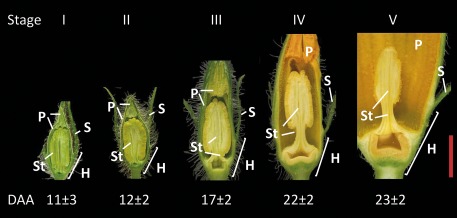
Fig. 3.
Stages of flower development. Values for day after appearance (DAA) of flower buds shown are ±SEM (n = 12). Tissues are as indicated: hypanthium (H), sepals (S), petals (P), and stamens (St). Bar=1 cm.
Cloning of GA20ox4, GA3ox4, GA2ox2, GA2ox3, GID1a, GID1b, GAIP, and GAIPb
A PCR-based strategy was used to clone GA20ox4, GA3ox4, GA2ox2, and GA2ox3 from root tips of 7-day-old pumpkin seedlings. Amino acid sequences of known GA20oxs, GA3oxs, and GA2oxs from C. maxima and C. sativus were aligned (ClustalW2, http://www.ebi.ac.uk/Tools/msa/clustalw2/) and consensus peptide sequences identified. Total RNA (0.5–1 μg) from 7-day-old pumpkin root tips was used in a 10 μl reverse transcription reaction (RevertAid®H Minus-MuL V RT, Fermentas; Thermo Fisher Scientific Inc.) using an oligo(dT)18 primer to produce cDNA molecules according to the manufacturer’s recommendations. The cDNA molecules obtained were used as templates (1/10 reaction volume) in PCRs with the respective degenerate primers designed according to consensus peptide sequences (Supplementary Table S2 available at JXB online).
All the PCRs (10 μl) contained 1 μl of cDNA, 20 pmol of each degenerate primer, 1.5 mM MgCl2, 0.2 mM dNTPS, 2% dimethylsulphoxide (DMSO), 0.3 U of Taq DNA polymerase (Fermentas, Thermo Fisher Scientific Inc.), and the respective PCR buffer. The annealing temperatures were 50 °C for the GA20ox and GA2oxs and 55 °C for the GA3ox. After re-amplification the expected PCR bands of 430 bp (GA20ox), 290 bp (GA3ox), and 420 bp (GA2oxs) were purified by agarose gel electrophoresis using the GeneJetTM Gel Extraction-Kit and ligated into pJet1.2 (CloneJET™ PCR Cloning-Kit, Fermentas, Thermo Fisher Scientific Inc.). Transformations were done using GM2163 competent cells. Positive clones were checked by PCR using M13 primers and sequenced. Fragments of one new GA20ox (GA20ox4), one GA3ox (GA3ox4), and two GA2oxs (GA2ox2 and GA2ox3) were obtained. The corresponding full-length sequences were obtained by RLM-RACE (FirstChoice® RLM-RACE, Ambion-Applied Biosystems, Foster City, CA, USA) following the manufacturer’s recommendations and specific gene RACE (rapid amplification of cDNA ends) primers (Supplementary Table S2 at JXB online). The four new GA20ox4, GA3ox4, GA2ox2 and GA2ox3 open reading frames (ORFs) were amplified by PCR using as template cDNA from 7-day-old pumpkin root tips (as described above with the exception that random hexamer primers were used). PCR conditions were as described above using the respective gene primers with restriction sites (Supplementary Table S2) and annealing temperatures of 50 °C for GA20ox4 and GA3ox4, 52 °C for GA2ox2, and 55 °C for GA2ox3. The PCR products obtained for the ORFs of the new GA20ox4, GA3ox4, GA2ox2, and GA2ox3 were 1160, 1050, 1000, and 1020 bp, respectively. After digestion with the respective restriction endonucleases (Supplementary Table S2), the PCR products were ligated into pBluescript II SK(+) (Stratagene, La Jolla, CA, USA) at the corresponding restriction sites. Transformations were carried out using XL1-Blue competent cells. Positive clones were checked by PCR using M13 primers and sequenced.
For GID1a and GID1b, a slightly different cloning strategy was used. First, amino acid sequences of known GID1 homologues from Oryza sativa and Arabidopsis were aligned and consensus peptide sequences identified. Degenerate primers were designed (Supplementary Table S2) and used to amplify two 270 bp fragments by RT-PCR as described above with the exception that total RNA from pumpkin 3-day-old shoot (GID1a) and 7-day-old root (GID1b) was used as template to produce the cDNA molecules. The PCR fragments were ligated into pCR®2.1-TOPO (Invitrogen, Carlsbad, CA, USA) and transformations was carried out using XLI-Blue competent cells. Positive clones were checked by PCR using M13 primers and sequenced. The 5' and 3' sequences of GID1a and GID1b were amplified by PCR using a pumpkin 7-day-old root tip cDNA library (Lange et al., 2005) as template together with M13 primers. The same cDNA library was used as template to amplify the ORF of GID1a (1041 bp) and GID1b (1029 bp) by PCR as described above, with the respective gene-specific primers containing restriction sites (Supplementary Table S2) at annealing temperatures of 56 °C and 54 °C, respectively.
GAIP (accession no. AY326306) and GAIPb (AY326307) sequence data were obtained from the GenBank database. Accordingly, gene-specific primers were designed (Supplementary Table S2 at JXB online) and used to amplify full-length sequences by RT-PCR as described above with the difference that the cDNA molecules were obtained from pumpkin 7-day-old shoot (GAIP) and pumpkin 7-day-old root (GAIPb) and that the PCR annealing temperature was 60 °C and 55 °C, respectively. After restriction digestion with endonucleases (Supplementary Table S2), the full-length PCR fragments were ligated into pJET1.2 (Fermentas, Thermo Fisher Scientific Inc.) and transformed into XLI-Blue cells.
Heterologous expression of recombinant GA-oxidases, standard enzyme assay, and analysis of incubation products
First the ORFs of GA20ox4, GA3ox4, GA2ox2, and GA2ox3 were amplified by PCR using primers for the respective genes containing restriction sites (Supplementary Table S2) that enable ligation in-frame into pUC18. Heterologous expression of the recombinant new GA-oxidases and respective enzyme assays were performed as described previously (Lange et al., 2005).
Quantitative analysis of endogenous GAs
Quantitative analysis of endogenous GA levels was done as described previously (Lange et al., 2005).
Competitive RT-PCR
Methods to produce standard RNA and quantitative RT-PCR were essentially as described elsewhere (Lange et al., 1997, 2005). Detailed information on the production of the mRNA standards and on primer sequences are provided as Supplementary Materials and methods at JXB online.
Reproducibility
All experiments have been repeated at least once, giving very similar results.
Results
Cloning and expression of GA20ox4, GA3ox4, GA2ox2, and GA2ox3, and cloning of putative GID1a and GID1b
Phylogenetic analysis of the cucumber genome suggests the presence of additional GA-related genes that had not been identified in pumpkin so far (Fig. 2). In earlier work, Lange et al. (2005) found that pumpkin seedlings offer a convenient source for the isolation of GA-related genes. For this study, total RNA was used, derived from root tips of 7-day-old developing pumpkin seedlings, that was reverse transcribed. cDNA molecules were amplified by a PCR-based method using suitable degenerate oligonucleotide primers. Four new GA-oxidase cDNA molecules were isolated, including one GA20ox cDNA (designated GA20ox4), one GA3ox cDNA (GA3ox4), and two GA2ox cDNAs (GA2ox2 and GA2ox3). Similarly, two putative GA receptor-encoding cDNAs (GID1a and GID1b) were cloned from reverse-transcribed total RNA of 3-day-old shoot and 7-day-old root, respectively. The new GA-oxidases share high sequence homology to known sequences of other GA-oxidases (Fig. 2), including motifs typical for 2-oxoglutarate-dependent dioxygenases (Xu et al., 1995; Prescot and John, 1996). The putative pumpkin GID1 receptors share essential regions for GA binding and DELLA interaction with known GID1 receptor molecules (Ueguchi-Tanaka et al., 2007; Ueguchi-Tanaka and Matsuoka, 2010).
The catalytic properties of recombinant GA20ox4, GA3ox4, GA2ox2, and GA2ox3 proteins were examined by expression of the respective predicted ORF in the pUC18 vector in Escherichia coli BL-21 cells (Table 1). Cell lysates of recombinant GA20ox4 converted [14C]GA12 to [14C]GA15, [14C]GA24, and [14C]GA9 (Table 1, Fig. 1). Incubations with [14C]GA24 gave two products, a minor one, [14C]GA25, and a major one, [14C]GA9. Recombinant GA3ox4 oxidized precursor [14C]GA9 to the hormone [14C]GA4. Both recombinant 2-oxidases converted [14C]GA9 and [14C]GA4 to [14C]GA51 and [14C]GA34, respectively. Additionally, the 2-oxidases formed [14C]GA51 catabolite and [14C]GA34 catabolite from [14C]GA51 and [14C]GA34, respectively (Table 1, Fig. 1, Supplementary Table S1 at JXB online). Other minor products from [14C]GA4 and [14C]GA34 incubations were not identified conclusively. However, the resulting mass spectra show similarities to spectra of [14C]GA34 catabolites (see Supplementary Table S1). Recombinant GA20ox4, GA3ox4, and GA2ox3 also oxidize 13-hydroxylated substrates [14C]GA53, [14C]GA20, and [14C]GA1, to form [14C]GA44, [14C]GA1, and [14C]GA8, respectively, but product formation was less efficient (see Supplementary Table S1).
Table 1.
Mass spectra of products of the methyl ester trimethylsilylether derivatives from incubations of [14C]GAs with cell lysates from Escherichia coli expressing recombinant pumpkin GA20ox4, GA3ox4, GA2ox2, and GA2ox3
| Enzyme | Substrate | Compound formed | Mass spectrum m/z (relative intensity) |
| GA20ox4 | [1-,7-,12-,18-14C]GA12 | [1-,7-,12-,18-14C]GA15 | M+ 352(5), 344(3), 320(11), 312(6), 306(8), 298(4), 290(34), 284(20), 245(100), 239(52), 201(47), 195(22) |
| [1-,7-,12-,18-14C]GA24 | M+ 382(0), 350(11), 342(6), 322(39), 314(30), 293(28), 285(16), 232(86), 231(100), 226(49), 225(52) | ||
| [1-,7-,12-,18–14C]GA9 | M+ 338(6), 330(3), 306(75), 298(45), 276(100), 270(54), 251(60), 243(42), 233(66), 232(78), 227(38), 226(48), 189(48), 183(36) | ||
| [17-14C]GA24 | [17-14C]GA9 | M+ 332(4), 300(87), 288(11), 272(100), 229(74), 228(88), 213(32), 185(47) | |
| [17-14C]GA25 | M+ 406(0), 374(12), 346(1), 314(66), 286(100), 271(10), 255(14), 227(77) | ||
| GA3ox4 | [17-14C]GA9 | [17-14C]GA4 | M+ 420(11), 388(18), 360(11), 330(27), 291(63), 286(100), 263(35), 235(62) |
| GA2ox2 | [17-14C]GA4 | [17-14C]GA34 | M+ 508(100), 418(8), 389(12), 374(13), 359(14), 358(17), 329(15), 315(25), 290(31), 263(13), 231(59), 225(64), 203(71) |
| [17-14C]GA34 catabolite (as ketone) | M+ 448(28), 373(62), 341(15), 329(100), 313(77), 299(38), 260(51), 239(69), 201(74) | ||
| [17-14C]GA9 | [17-14C]GA51 | M+ 420(1), 388(15), 330(15), 298(19), 286(67), 270(75), 243(29), 229(45), 227(100), 226(55), 225(28), 143(44) | |
| [17-14C]GA51 catabolite (as enol) | M+ 432(100), 417(3), 373(15), 357(17),313(79), 283(14), 269(26) | ||
| GA2ox3 | [17-14C]GA4 | [17-14C]GA34 | M+ 508(100), 418(9), 389(11), 374(11), 359(13), 358(12), 329(10), 315(27), 290(33), 263(27), 231(72), 225(79), 203(80) |
| [17-14C]GA9 | [17-14C]GA51 | M+ 420(1), 388(14), 330(14), 298(18), 286(67), 270(75), 243(28), 229(44), 227(100), 226(56), 225(30), 143(44) | |
| [17-14C]GA51 catabolite (as enol) | M+ 432(100), 417(2), 373(10), 357(12), 313(82), 283(14), 269(20) |
Development of pumpkin male flowers
In the monoecious pumpkin plant, first male flowers appear, followed by female flowers. Under the growth conditions chosen, male flower buds appear at day 17±2 (mean ±SD, n=19). Initially, growth rates of individual flower buds are slow and vary considerably. Therefore, from the day of appearance (DAA) until fully opened, flower development was divided into five stages, focused on the stamen and petal development (Fig. 3). Flower buds reach stage I at ∼11 DAA. At this stage, buds contain immature greenish floral organs, and stamens stick to the petals. At stage II the stamen tissue turns yellowish and is disconnected from the petal tissue. From stage II onwards, flowers develop more uniformly and were therefore chosen for emasculation experiments (see below). At stage III, the rapid growth phase starts and flowers begin to mature. The petals start turning yellowish, and elongation of the filament and the pedicel has just begun. At stage IV, flowers are just ready to unfold, and the filaments and the pedicels are fully developed. Finally, at stage V, flowers are mature with a fully open corolla.
Profiling endogenous GAs in developing male pumpkin flowers
It has been known for a long time that GAs are involved in floral developmental processes (Plack, 1958; Koornneef and van der Veen, 1980), and that anthers are a rich source of GAs (Hirano et al., 2008). To investigate sites and timing of GA accumulation during pumpkin male flower development, endogenous GAs were quantified from different floral parts of male flowers at five developmental stages by combined GC-MS (Table 2). For the first four developmental stages, the hypanthium, the sepal, and the petal tissues were analysed together, and the stamen tissues were analysed separately. Stage V mature flowers, however, were dissected into the hypanthium, the sepal, the petal, and the stamen tissues. Sepals of stage V flowers were used for analysing transcript levels only (see below).
Table 2.
Endogenous GA levels (ng g−1 fresh weight) in different floral organs during pumpkin male flower development
| Stages of floral organ development | GA12 | GA15 | GA24 | GA9 | GA4 | GA34 | GA51 |
| Stamen | |||||||
| I | 1.1 | 1.5 | 0.3 | 3.6 | 1.0 | ND | 1.9 |
| II | 1.2 | 3.1 | 0.9 | 11.0 | 2.4 | ND | 2.8 |
| III | 3.0 | 3.3 | 1.7 | 25.6 | 25.6 | ND | 1.5 |
| IV | 5.8 | 2.1 | 1.2 | 12.2 | 56.1 | 0.2 | 0.3 |
| V | 2.1 | 1.9 | 0.3 | 3.8 | 16.2 | 0.2 | 0.2 |
| Hypanthium, sepal, and petal | |||||||
| I | 1.2 | 0.5 | 0.3 | 0.7 | 3.4 | ND | 0.5 |
| II | 0.7 | 0.7 | 1.3 | 0.7 | 3.2 | ND | 0.2 |
| III | 1.1 | 0.6 | 0.2 | 0.9 | 6.5 | ND | 0.1 |
| IV | 1.5 | 1.0 | 1.5 | 0.4 | 20.7 | ND | 0.4 |
| Hypanthium | |||||||
| V | 0.8 | 0.3 | 0.3 | 1.4 | 3.5 | 0.1 | 0.2 |
| Petal | |||||||
| V | 0.1 | 0.1 | 0.5 | 0.2 | 3.8 | ND | 0.1 |
Stages of floral organ development were as described in Fig. 3.
ND, not detected (endogenous GA not detected, internal standard recovered).
GA12, GA15, and GA24 are present mostly in stamen tissues and throughout flower development (Table 2). However, GA9 and bioactive GA4 levels in the stamen undergo dramatic changes during flower development. GA9 levels increase at stage II, reach a maximum at stage III, and decrease again a few days later at stage IV. Bioactive GA4 levels are low at stages I and II, increase later at stage III, and reach a maximum at stage IV, just before the flower unfolds (Table 2, Fig. 3). A couple of days later, at developmental stage V when the corolla is fully open, endogenous GA4 together with GA precursor levels decrease again (Fig. 3). Interestingly, while GA precursors are found mainly in the stamen, considerable levels of bioactive GA4 are also detected in the other floral organs. Biologically inactive GA34 levels were low in all tissues (Table 2), but some inactive GA51 was present at early developmental stages in the stamen. Endogenous GAs of the 13-hydroxylated pathway, including GA1, GA8, and GA20, were not detectable during male flower development (data not shown).
The presented results show that stamens are a rich source of GA precursors and of bioactive GAs in pumpkin male flowers just before and during the rapid growth phase. In addition, considerable levels of bioactive GA4, found in the hypanthium, sepals, and/or petals, did not exclude the hypothesis at this point that sites other than stamens might be important for GA biosynthesis in flowers as proposed by Hu et al. (2008).
Transcript profiles of GA-oxidase and putative GA receptor and GA signal transduction genes during pumpkin male flowers development
To address further the question of potential sites of GA signalling, transcript levels for the known pumpkin GA-related genes were examined at all developmental stages with the exception of stage II (Fig. 4, Supplementary Fig. S1 at JXB online). Transcripts were quantified by competitive RT-PCR as described previously (Lange et al., 2005). New internal standard RNA molecules were designed for the newly identified pumpkin GA-oxidase genes, and for putative GID1 receptor and DELLA protein sequences (Haywood et al., 2005).
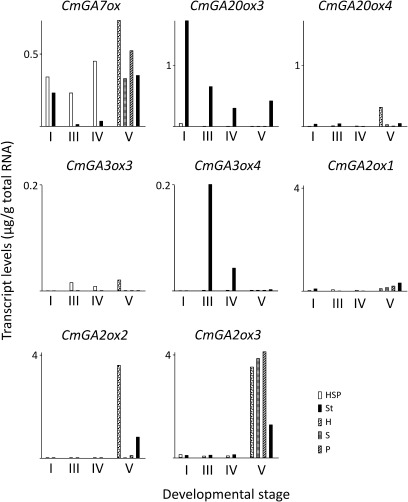
Fig. 4.
Transcript levels of gibberellin oxidase genes in different floral organs during male flower development. Developmental stages and floral organs used for analysis by competitive RT-PCR were as described in Fig. 3. For gene abbreviations, see Fig. 2. HSP, H, S, and P were analysed together.
One GA 7-oxidase (GA7ox) has been isolated from pumpkin (Lange et al., 1997). Transcripts encoding GA7ox were present in all floral organs, but levels were low in the stamens from flowers at developmental stages III and IV, where, in general, endogenous GA levels are highest (Fig. 4). Thus, the transcript profile of GA7ox does not mirror well endogenous GA levels found in the stamen. Transcript levels of genes coding for the four known pumpkin GA20oxs were analysed (Lange et al., 2005; this study). Two of them (GA20ox1 and GA20ox2) are not expressed in male flowers (data not shown). Transcript profiles of GA20ox3 project endogenous GA15, GA24, and, particularly, GA9 levels in stamens (compare Fig. 4 and Table 2). In stamens, GA20ox3 transcript levels are high in immature flowers and decrease as the flower develops. GA20ox4, however, is expressed mainly in the hypanthium of mature flowers (stage V, Fig. 4). Four GA3oxs were identified in pumpkin (Lange et al., 2005; this study). GA3ox1 and GA3ox2 are not expressed in male flowers (data not shown). GA3ox3 and GA3ox4 are not expressed in young flower buds (stage I, Fig. 4). GA3ox3 is not expressed in stamens but is expressed at low levels in other floral organs, including the hypanthium. GA3ox4 is the only GA3ox expressed in stamens with a sharp increase at stage III. It was not detected in other floral organs and its transcript profile predicts the obtained endogenous bioactive GA4 levels observed in stamens (compare Fig. 4 and Table 2). Transcript levels of all three known pumpkin GA2oxs are low in the flower until developmental stage IV (Lange et al. 2005; Fig. 4). Nevertheless, high levels of GA2ox2 (particularly in the hypanthium and stamen) and of GA2ox3 (in all floral organs) are found in developed flowers (stage V, Fig. 4).
Two putative GID1 GA receptor cDNAs were isolated from pumpkin seedlings (designated GID1a and GID1b; Uegushi-Tanaka et al., 2005; this study). In pumpkin, two DELLA protein-encoding genes (GAIP and GAIPb) were identified previously (Haywood et al., 2005). Transcripts for these four genes were quantified in the same pumpkin male flower tissues as above. In all floral organs, expression of GID1a is low (see Supplementary Fig. S1 at JXB online). Transcript levels of GID1b are also low in male flowers until developmental stage IV, when endogenous bioactive GA4 levels are particularly high, but increase sharply when GA4 levels fall and the flower is open. The changes of transcript levels encoding DELLA proteins were not as dramatic as of GID1b, but GAIPb levels also peak at late stage V of flower development (see Supplementary Fig. S1).
In summary, transcript profiles obtained for GA-oxidases strongly suggest the stamen as a main site of de novo GA synthesis during flower development. GA3ox4 transcript levels peak in the stamen just before bioactive GA4 levels rise in this tissue, and before the filament, petal, and pedicel growth accelerates. However, in other floral organs, bioactive GA4 is also present together with transcripts that encode GA-oxidases, including GA3ox3; although at much lower levels. These results question the idea that the stamen is the (only) source for bioactive GAs that regulate growth of other floral organs. To address this problem further, emasculation experiments were performed.
Effect of stamen removal and GA application on flower development
GA biosynthesis is particularly active in pumpkin flowers during the rapid growth phase from developmental stage III onwards (see above). To examine further the site of GA synthesis and action, pumpkin male flower buds of stage II were chosen, when stamens can be separated easily from petals. At this stage, in the stamen, bioactive GA4 levels are still low and precursor GA9 levels are just about to rise (Table 2). The flower buds were either kept intact (Fig. 5A), or they were emasculated (Fig. 5B). Emasculated buds received a single treatment, either with water or with an aqueous solution of bioactive GA4 at the site, where stamens were removed. Eleven days after the treatment, flowers were photographed (Fig. 5C–E). At this developmental stage, intact male flowers are just before opening (Fig. 5C). Development of emasculated flowers treated with water is arrested completely, and the flowers die a few days later after stamens are removed (Fig. 5D). However, emasculated flowers treated with GA4 reach normal size, but development is hastened. It is noteworthy, that growth not only of floral organs, but also of the pedicel is dependent on the stamen (Fig. 5E).
Fig. 5.
Emasculation arrests flower growth that is rescued by bioactive GA4 application. Male flower buds of developmental stage II (Fig. 3) were either left intact (A) or were emasculated (B). Eleven days after the treatment flowers are illustrated that developed from intact buds (C) or from emasculated buds treated either with water (D) or with an aqueous solution of GA4 as described in the Materials and methods (E). Bar=1 cm.
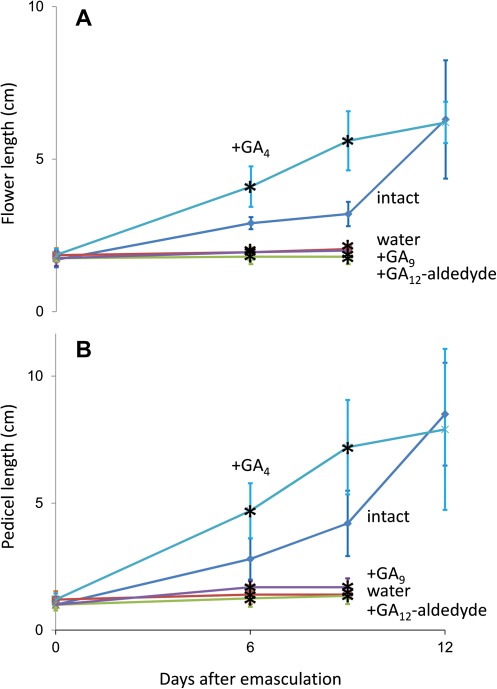
Flower development can be normalized in Arabidopsis GA-deficient mutants by application of bioactive GAs, including GA4, and, less efficiently, by application of the precursor GA9 (Goto and Pharis, 1999). In these experiments, GA9 was probably not active per se but was first oxidized at the 3β-position to form GA4 (Fig. 1). Because GA9 is also present in pumpkin stamens (Table 2), one could speculate that this precursor moves first to the hypanthium where it is oxidized by GA3ox3 to form bioactive GA4. From the hypanthium, bioactive GA4 could then reach the petals. Therefore, in addition to GA4, the effects of precursors GA12-aldehyde and GA9 on growth of emasculated flowers and their pedicel were also examined (Fig. 6). Initially, growth of emasculated flowers treated with GA4 is hastened compared with intact flowers, but both reach the same size 12 d after treatment (Fig. 6A). Similar growth patterns were observed for the respective pedicels (Fig. 6B). However, treatment of emasculated flower buds with water, GA12-aldehyde, or GA9 failed to restore the flower growth or the pedicel growth, and the flowers are aborted 6–9 d after the treatment (Fig. 6).
Fig. 6.
Effect of the gibberellin precursors GA12-aldehyde and GA9, and of bioactive GA4 on growth of emasculated pumpkin flowers. Male flower buds of developmental stage II (Fig. 3) were either kept intact or emasculated. The latter ones were treated either with water, or with an aqueous solution of GA12-aldehyde, GA9, or GA4 as described in the Materials and methods. The length of the flower (A) and of the pedicel (B) was measured at day 6, day 9, and day 12 after the treatments. Values shown are ±SEM (n=10). Asterisks indicate values that are statistically different from those of intact flowers (Student’s t test, P < 0.05).
These experiments demonstrate that bioactive GA4, but not its biosynthetic precursors, moves from the stamen to the other floral organs and that this hormone is a sufficient substitute for the stamen during pumpkin male flower development.
Discussion
GAs regulate flower development. The first evidence came from studies on GA-deficient mutants, in which flower development is arrested but can be rescued by applied GAs (Koornneef and van der Veen, 1980; Nester and Zeevaart, 1988; Goto and Pharis, 1999). There is evidence that anthers produce high levels of bioactive GAs (Hirano et al., 2008). Recent investigations suggest that bioactive GAs are made in stamens and/or flower receptacles and these GAs are transported to petals to promote their growth (Hu et al., 2008).
Male flowers of monoecious pumpkin that have organs of sufficient size enabling detailed investigation of GA biosynthesis and distribution were examined. The major site of GA accumulation in pumpkin male flowers is the stamen, where precursor GA9 levels rise first, followed by a sharp increase in bioactive GA4 levels. Thus, GAs accumulate in the stamen, just before the rapid flower growth phase starts, and might move to other floral organs to support their growth, including the petal and the pedicel. However, endogenous GA4 levels detected in floral organs other than stamens do not exclude the presence of additional sites for GA production.
To clarify where and when GA biosynthesis takes place, transcripts coding for the known GA-oxidases were quantified during pumpkin male flower development. The GA7ox transcript profile does not match the respective endogenous GA levels. Moreover, the expression of additional GA7ox in pumpkin flowers cannot be excluded. A second putative GA7ox gene was identified recently in the genome of the closely related species cucumber (Huang et al., 2009). Also, as in other plant species, the 7-oxidation step may be catalysed by the P450 monooxygenase, kaurenoic acid oxidase (Yamaguchi, 2008).
Four GA20ox cDNAs have been identified so far from pumpkin. Two of them, GA20ox1 and GA20ox2, are expressed in developing pumpkin seeds but not in flowers (Lange et al., 1994a; Frisse et al., 2003; data not shown). GA20ox3 and GA20ox4 show typical catalytic properties as they produce mainly the hormone precursor GA9 (Fig. 1; Lange et al., 2005; this study) and their genes are expressed during pumpkin male flower development. GA20ox4 transcript levels are low, apart from mature flowers at developmental stage V, making this gene unlikely to have a major impact in GA biosynthesis during flower development. In contrast, GA20ox3 transcript levels correlate well with endogenous GA9 levels during flower development, indicating that this is the principal GA20ox gene operating in male flowers. However, it is possible that in pumpkin, as in cucumber, additional GA20oxs are present (Huang et al., 2009).
The pumpkin GA3ox gene family appears to be complete. For all four pumpkin GA3oxs, closely related putative GA3ox-encoding genes are present in the cucumber genome (Fig. 2). Two of the pumpkin GA3ox genes (GA3ox1 and GA3ox2) are expressed in developing seeds but not in the male flower (Frisse et al., 2003; data not shown). Transcripts for the other two (GA3ox3 and GA3ox4) are found in seedlings and flowers (Lange et al., 2005; this study). The catalytic properties of recombinant GA3ox3 and GA3ox4 are similar: both oxidize the precursor GA9 at the 3β-position to form bioactive GA4 (Lange et al., 2005; this study). During male flower development, GA3ox3 transcript levels are relatively low and found mainly in the hypanthium. Transcripts of GA3ox4 are found in the stamen only, and its expression pattern correlates well with endogenous GA4 levels in this organ. Moreover, its expression pattern correlates well with growth of the entire male flower and the pedicel. Thus, GA3ox4 appears to be a key gene for bioactive GA4 synthesis in the stamen and for the regulation of entire male flower development.
Three GA2ox-encoding genes are known from pumpkin, and closely related genes have been identified in the cucumber genome. However, the cucumber genome contains two additional putative GA2ox genes (Huang et al., 2009). Enzymatic characterization revealed that all three pumpkin GA2oxs convert bioactive GAs to inactive GAs and further to catabolites. Similar catalytic properties have been described previously for recombinant GA2oxs from Arabidopsis (Thomas et al., 1999). However, inactive GA51 and GA34 levels are particularly low in mature pumpkin flowers of developmental stage V, where GA2ox2 and GA2ox3 expression is high. Catabolite levels were not included in the present analysis and, thus, endogenous GA51 and GA34 levels might not reflect overall GA 2-oxidation of the floral organ. However, a strong decrease in bioactive GA4 (and GA9) levels in pumpkins flowers at developmental stage V suggests the presence of catabolic GA2-ox(s), which is typical for mature tissues (Ross et al., 2003). Moreover, particularly high expression levels of GA2ox2 in the hypanthium might indicate that movement of bioactive GAs from the stamen to the petal and the pedicel is arrested in mature flowers due to inactivation and catabolism of bioactive GA4.
Elements of the GA signal perception and transduction pathways have been identified as important regulators of flower development (Cheng et al., 2004; Fleet and Sun, 2005; Griffiths et al., 2006). It is likely that the pumpkin GID1 family consist of two members as the cucumber GID1 family does (Fig. 2). Transcript levels of one of the two GA receptor genes, GID1b, increase sharply in mature flowers of developmental stage V. Moreover, during the rapid flower growth phase GID1b levels are low when bioactive GA4 levels are particularly high. This might indicate feedback regulatory processes acting on pumpkin GID1b gene expression as described previously for the three Arabidopsis GID1 genes (Griffiths et al., 2006). Senescence processes take place soon after the flower opens. An increase in DELLA repressor as found for GAIPb transcript levels in the mature pumpkin male flowers might be important to initiate this process in addition to terminating the petal, stamen, and anther development as has been shown for Arabidopsis flowers (Cheng et al., 2004). Other hormones (e.g. ethylene) are known to be involved in up-regulating DELLA repressors (Archard et al., 2003). In contrast, high levels of GID1b receptor in the open flower might help to delay pumpkin male flower senescence processes. However, the relatively low and constant transcript levels of GID1a/GID1b and of GAIP/GAIPb during the rapid growth phase question their function as key regulators for pumpkin male flower development.
The importance of the stamen as a source of GA that promotes male flower growth was examined using emasculated flowers. Genetic approaches are not suitable for pumpkin, because mutants are not available and this species is recalcitrant to genetic manipulation (data not shown). However, for a long time emasculation experiments have been used successfully to study the role of the stamen for flower development in Glechoma hederacea and in Petunia hybrida (Plack, 1958; Weiss and Halevy, 1989). In Arabidopsis, Griffiths et al. (2006) observed in triple GID1 GA receptor mutants not only a severe effect on floral organ growth and development, but also a dramatic reduction in elongation of the pedicel. In pumpkin male flowers, removal of the stamen completely arrests the petal and the pedicel growth and results in early abscission of the male flower bud. It is noteworthy that emasculated flowers were always surrounded by intact flowers, indicating that there is no movement of the hormonal signal in sufficient quantities from intact to emasculated flowers. Abortion of emasculated flowers was prevented by exogenous application of bioactive GA4 only (but not by application of its biosynthetic GA precursors). Recently, Hu et al. (2008) suggested that bioactive GAs produced in stamens and/or flower receptacles of Arabidopsis are transported to the petals to promote their growth. In the present study, the possibility of long-distance transport of the hormonal signal was discussed. The data presented here, however, indicate that long-distance transport of the GA signal, if present, is not sufficient to rescue development of emasculated pumpkin male flowers.
This study extends our knowledge of flower development. First, the stamen is the essential site of GA synthesis. Other sites cannot replace the stamen. Secondly, GA20ox3 and GA3ox4 are key regulators of GA biosynthesis in the stamen. Thirdly, short-distance movement of bioactive GA (but not of its biosynthetic precursors) from the stamen to the other floral organs and the pedicel is essential and sufficient for flower development. Thus, the stamen is the site that regulates, via bioactive GA, the male flower and the pedicel growth in pumpkin. However, this opens up the question of how female flowers regulate growth and development, since regulatory mechanisms/organs other than those in male flowers are mandatory.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: CmGA20ox4 (FN808418); CmGA3ox4 (FN808419); CmGA2ox2 (FN808420); CmGA2ox3 (FN808421); CmGID1a (AM745266); and CmGID1b (AM745267).
Supplementary data
Supplementary data are available at JXB online.
Supplementary Materials and methods
Figure S1. Transcript levels of putative elements of the gibberellin signalling pathway in different floral organs during male flower development.
Table S1. Mass spectra of additional products of the methyl ester trimethylsilylether derivatives from incubations of [14C]GAs with cell lysates from Escherichia coli expressing recombinant pumpkin GA20ox4, GA3ox4, GA2ox2, and GA2ox3.
Table S2. List of primers and their respective use.
Acknowledgments
We thank Professor Dr Peter Hedden, Rothamsted Research, Harpenden, Herts AL5 2JQ, UK, for help with interpreting mass spectra of GA catabolites, and Anja Liebrandt, Institut für Pflanzenbiologie der Technischen Universität Braunschweig, for technical assistance. This work was supported by a grant from the DFG (La880/7-1).
References
- Achard P, Vriezen WH, van der Straeten D, Harberd NP. Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. The Plant Cell. 2003;15:2816–2825. doi: 10.1105/tpc.015685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Qin L, Lee S, Fu X, Richards DE, Cao D, Luo D, Harberd NP, Peng J. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development. 2004;131:1055–1064. doi: 10.1242/dev.00992. [DOI] [PubMed] [Google Scholar]
- Davies PJ. Plant hormones: biosynthesis, signal transduction, action! Dordrecht, The Netherlands: Kluwer Academic Publishers; 2004. [Google Scholar]
- Fleet CM, Sun TP. A DELLAcate balance: the role of gibberellin in plant morphogenesis. Current Opinion in Plant Biology. 2005;8:77–85. doi: 10.1016/j.pbi.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Frisse A, Pimenta MJ, Lange T. Expression studies of gibberellin oxidases in developing pumpkin seeds. Plant Physiology. 2003;131:1220–1227. doi: 10.1104/pp.015206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto N, Pharis RP. Role of gibberellins in the development of floral organs of the gibberellin-deficient mutant, ga1-1, of Arabidopsis thaliana. Canadian Journal of Botany. 1999;77:944–954. [Google Scholar]
- Graebe JE. Gibberellin biosynthesis and control. Annual Review of Plant Biology. 1987;38:419–465. [Google Scholar]
- Griffiths J, Murase K, Rieu I, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. The Plant Cell. 2006;18:3399–3414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood V, Yu TS, Huang NC, Lucas WJ. Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. The Plant Journal. 2005;42:49–68. doi: 10.1111/j.1365-313X.2005.02351.x. [DOI] [PubMed] [Google Scholar]
- Hirano K, Aya K, Hobo T, Sakakibara H, Kojima M, Shim RA, Hasegawa Y, Ueguchi-Tanaka M, Matsuoka M. Comprehensive transcriptome analysis of phytohormone biosynthesis and signaling genes in microspore/pollen and tapetum of rice. Plant and Cell Physiology. 2008;49:1429–1450. doi: 10.1093/pcp/pcn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Mitchum MG, Barnaby N, et al. Potential sites of bioactive gibberellin production during reproductive growth in Arabidopsis. The Plant Cell. 2008;20:320–336. doi: 10.1105/tpc.107.057752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Li R, Zhang Z, et al. The genome of the cucumber, Cucumis sativus L. Nature Genetics. 2009;41:1275–1281. doi: 10.1038/ng.475. [DOI] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sentoku N, Kitano H, Matsuoka M, Kobayashi M. Cloning and functional analysis of two gibberellin 3β-hydroxylase genes that are differently expressed during the growth of rice. Proceedings of the National Academy of Sciences, USA. 2001;98:8909–8914. doi: 10.1073/pnas.141239398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh. Theoretical and Applied Genetics. 1980;58:257–263. doi: 10.1007/BF00265176. [DOI] [PubMed] [Google Scholar]
- Lange T. Cloning gibberellin dioxygenase genes from pumpkin endosperm by heterologous expression of enzyme activities in Escherichia coli. Proceedings of the National Academy of Sciences, USA. 1997;94:6553–6558. doi: 10.1073/pnas.94.12.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T, Hedden P, Graebe JE. Biosynthesis of 12α- and 13-hydroxylated gibberellins in a cell-free system from Cucurbita maxima endosperm and the identification of new endogenous gibberellins. Planta. 1993;189:340–349. doi: 10.1007/BF00194430. [DOI] [PubMed] [Google Scholar]
- Lange T, Hedden P, Graebe JE. Expression cloning of a gibberellin 20-oxidase, a multifunctional enzyme involved in gibberellin biosynthesis. Proceedings of the National Academy of Sciences, USA. 1994a;91:8552–8556. doi: 10.1073/pnas.91.18.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T, Kappler J, Fischer A, Frisse A, Padeffke T, Schmidtke S, Pimenta Lange MJ. Gibberellin biosynthesis in developing pumpkin seedlings. Plant Physiology. 2005;139:213–223. doi: 10.1104/pp.105.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T, Robatzek S, Frisse A. Cloning and expression of a gibberellin 2β,3β-hydroxylase cDNA from pumpkin endosperm. The Plant Cell. 1997;9:1459–1467. doi: 10.1105/tpc.9.8.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T, Schweimer A, Ward DA, Hedden P, Graebe JE. Separation and characterisation of three 2-oxoglutarate-dependent dioxygenases from Cucurbita maxima L. endosperm involved in gibberellin biosynthesis. Planta. 1994b;195:98–107. [Google Scholar]
- Martin DN, Proebsting WM, Parks TD, Dougherty WG, Lange T, Lewis MJ, Gaskin P, Hedden P. Feedback regulation of gibberellin metabolism and gene expression in Pisum sativum L. Planta. 1996;200:159–166. doi: 10.1007/BF00208304. [DOI] [PubMed] [Google Scholar]
- Mutasa-Göttgens E, Hedden P. Gibberellin as a factor in floral regulatory networks. Journal of Experimental Botany. 2009;60:1979–1989. doi: 10.1093/jxb/erp040. [DOI] [PubMed] [Google Scholar]
- Nester JE, Zeevaart JAD. Flower development in normal tomato and a gibberellin-deficient (ga-2) mutant. American Journal of Botany. 1988;75:45–55. [Google Scholar]
- Phillips AL, Ward DA, Uknes S, Appleford NEJ, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P. Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiology. 1995;108:1049–1057. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimenta Lange MJ, Lange T. Gibberellin biosynthesis and the regulation of plant development. Plant Biology. 2006;8:281–290. doi: 10.1055/s-2006-923882. [DOI] [PubMed] [Google Scholar]
- Plack A. Effect of gibberellic acid on corolla size. Nature. 1958;182:610. [Google Scholar]
- Prescot AG, John P. Dioxygenases: molecular structure and role in plant metabolism. Annual Review of Plant Biology. 1996;47:245–271. doi: 10.1146/annurev.arplant.47.1.245. [DOI] [PubMed] [Google Scholar]
- Radi A, Lange T, Niki T, Koshioka M, Pimenta Lange MJ. Ectopic expression of pumpkin gibberellin oxidases alters gibberellin biosynthesis and development of transgenic Arabidopsis plants. Plant Physiology. 2006;140:528–536. doi: 10.1104/pp.105.073668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebers M, Kaneta T, Kawaide H, Yamaguchi S, Yang YY, Imai R, Sekimoto H, Kamiya Y. Regulation of gibberellin biosynthesis genes during flower and early fruit development of tomato. The Plant Journal. 1999;17:241–250. doi: 10.1046/j.1365-313x.1999.00366.x. [DOI] [PubMed] [Google Scholar]
- Ross JJ, Davidson SE, Wolbang CM, Bayly-Stark E, Smith JJ, Reid JB. Developmental regulation of the gibberellin pathway in pea shoots. Functional Plant Biology. 2003;30:83–89. doi: 10.1071/FP02108. [DOI] [PubMed] [Google Scholar]
- Ross JJ, O’Neill DP, Smith JJ, Huub L, Kerckhoffs J, Elliott RC. Evidence that auxin promotes gibberellin A1 biosynthesis in pea. The Plant Journal. 2000;21:547–552. doi: 10.1046/j.1365-313x.2000.00702.x. [DOI] [PubMed] [Google Scholar]
- Thomas SG, Phillips AL, Hedden P. Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proceedings of the National Academy of Sciences, USA. 1999;96:4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;437:693–698. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Matsuoka M. The perception of gibberellins: clues from receptor structure. Current Opinion in Plant Biology. 2010;13:503–508. doi: 10.1016/j.pbi.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Nakajima M, Katoh E, et al. Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. The Plant Cell. 2007;19:2140–2155. doi: 10.1105/tpc.106.043729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Huizen R, Ozga JA, Reinecke DM. Seed and hormonal regulation of gibberellin 20-oxidase expression in pea pericarp. Plant Physiology. 1997;115:123–128. doi: 10.1104/pp.115.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D, Halevy AH. Stamens and gibberellin in the regulation of corolla pigmentation and growth in Petunia hybrida. Planta. 1989;179:89–96. doi: 10.1007/BF00395775. [DOI] [PubMed] [Google Scholar]
- Xu YL, Gage DA, Zeevaart JAD. Gibberellins and stem growth in Arabidopsis thaliana. Effects of photoperiod on expression of the GA4 and GA5 loci. Plant Physiology. 1997;114:1471–1476. doi: 10.1104/pp.114.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YL, Li L, Wu KQ, Peeters AJM, Gage DA, Zeevaart JAD. The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proceedings of the National Academy of Sciences, USA. 1995;92:6640–6644. doi: 10.1073/pnas.92.14.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. Gibberellin metabolism and its regulation. Annual Review of Plant Biology. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- Zeevaart JAD, Gage DA. ent-Kaurene biosynthesis is enhanced by long photoperiods in the long-day plants Spinacia oleracea L. and Agrostemma githago L. Plant Physiology. 1993;101:25–29. doi: 10.1104/pp.101.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.