Abstract
The plant-specific WRKY transcription factor (TF) family with 74 members in Arabidopsis thaliana appears to be involved in the regulation of various physiological processes including plant defence and senescence. WRKY53 and WRKY70 were previously implicated as positive and negative regulators of senescence, respectively. Here the putative function of other WRKY group III proteins in Arabidopsis leaf senescence has been explored and the results suggest the involvement of two additional WRKY TFs, WRKY 54 and WRKY30, in this process. The structurally related WRKY54 and WRKY70 exhibit a similar expression pattern during leaf development and appear to have co-operative and partly redundant functions in senescence, as revealed by single and double mutant studies. These two negative senescence regulators and the positive regulator WRKY53 were shown by yeast two-hydrid analysis to interact independently with WRKY30. WRKY30 was expressed during developmental leaf senescence and consequently it is hypothesized that the corresponding protein could participate in a senescence regulatory network with the other WRKYs. Expression in wild-type and salicylic acid-deficient mutants suggests a common but not exclusive role for SA in induction of WRKY30, 53, 54, and 70 during senescence. WRKY30 and WRKY53 but not WRKY54 and WRKY70 are also responsive to additional signals such as reactive oxygen species. The results suggest that WRKY53, WRKY54, and WRKY70 may participate in a regulatory network that integrates internal and environmental cues to modulate the onset and the progression of leaf senescence, possibly through an interaction with WRKY30.
Keywords: Arabidopsis thaliana, ROS, SA, senescence, WRKY transcription factors
Introduction
Leaf senescence is the latest stage of leaf development that involves a slow and fine-tuned programmed cell death for recycling and re-use of valuable resources. Senescence is an active degenerative process under genetic control that begins with chloroplast dismantling followed by catabolism of macromolecules such as chlorophyll, proteins, lipids, and RNA (Hortensteiner and Feller, 2002; Buchanan-Wollaston et al., 2003; Lim et al., 2003, 2007; Guo et al., 2004; Lin and Wu, 2004; Guo and Gan, 2005; Hopkins et al., 2007). General catabolism converts cellular materials into easily exportable nutrients. These remobilized nutrients from senescing leaves are transported to reproductive and developing structures. Leaf senescence is therefore of pivotal importance for plant overall development.
Leaf senescence occurs in an age-dependent manner (Hensel et al., 1993; Nooden and Penney, 2001) influenced by various endogenous factors including developmental cues and reproductive growth (Gan and Amasino, 1995; Pic et al., 2002; Riefler et al., 2006). In this context, cytokinin, a phytohormone implicated in cell proliferation control during leaf development, acts as a negative regulator of senescence. Cytokinin amounts decrease during leaf development, resulting in avoidance of premature senescence in young leaves but allowing it in mature leaves (Singh et al., 1992; Gan and Amasino, 1995; Hwang and Sheen, 2001). In addition, alterations in sugar metabolism and accumulation of reactive oxygen species (ROS) in old leaves have been suggested as possible mechanisms through which age induces senescence (Munne-Bosch and Alegre, 2002; Moore et al., 2003; Guo and Gan, 2005; Pourtau et al., 2006; Wingler et al., 2006; Wingler and Roitsch, 2008). On the other hand, leaf senescence can also be triggered and modulated by various environmental factors, including photoperiod, light intensity, nutrient availability, as well as abiotic and biotic stress (Butt et al., 1998; Weaver et al., 1998; Miller et al., 1999; Quirino et al., 2000; Weaver and Amasino, 2001; Pic et al., 2002; Buchanan-Wollaston et al., 2003; Lim et al., 2003; Navabpour et al., 2003; Lin and Wu, 2004; Guo and Gan, 2005; Xiong et al., 2005). Consequently, perception of external factors and subsequent signals required for plant stress responses seem to be also shared by senescence regulation including stress-related hormones and the mitogen-activated protein kinase (MAP kinase) cascade (Guo and Gan, 2005; Zhou et al., 2009). Application of hormones and studies with hormonal signalling mutants have implicated abscisic acid (ABA), salicylic acid (SA), jasmonic acid (JA), and ethylene as positive modulators of leaf senescence and/or as inducers of senescence-associated genes (SAGs; Zacarias and Reid, 1990; Grbic and Bleecker, 1995; Park et al., 1998; Weaver et al., 1998; Morris et al., 2000; He et al., 2002; Guo and Gan, 2005; Jing et al., 2005). However, many of these hormones are considered as enhancers rather than triggering factors for leaf senescence. Consequently, it appears that the onset and progression of senescence are controlled by integration of complex signalling pathways mediated by both developmental and environmental factors.
Transcriptome studies using expressed sequence tag (EST) libraries and Arabidopsis thaliana genomic arrays have revealed thousands of genes that are up- or down-regulated during developmental leaf senescence and respectively called SAGs and senescence down-regulated genes (SDGs) (Gepstein et al., 2003; Guo et al., 2004; Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006; Balazadeh et al., 2008). This massive reprogramming of gene expression during senescence is mediated by a complex transcriptional regulatory network with >100 transcription factors (TFs) identified within SAG genes. The largest groups of senescence-related TFs include members of the NAC, WRKY, MYB, C2-H2 zinc-finger, bZIP, and AP2/EREBP families. Among these TFs, very few have been functionally related to senescence but they are likely to participate in coordinating the initiation and progression of leaf senescence.
The WRKY TF family with 74 members in Arabidopsis is specific to plants and appears to be involved in the regulation of various physiological processes including plant defence and senescence (Eulgem et al., 2000; Pandey and Somssich, 2009; Rushton et al., 2010). The 60 amino acid DNA-binding domain of WRKY proteins is highly conserved and contains a zinc-finger motif. WRKY TFs are classified into three groups depending on the number of WRKY domains and zinc-finger motifs. WRKY TFs are the second largest TF family to be induced during senescence, whereas the biological function in senescence of individual WRKY factors is not so far known. Indeed, to date, only WRKY group III TF members WRKY53 and WRKY70 have been functionally characterized as leaf senescence regulators (Miao et al., 2004; Ulker et al., 2007). Functional redundancy exists among the WRKY TFs due to the large number of members in the family and may explain the difficulties in identifying the specific contribution of individual WRKY factors (Xu et al., 2006). One example is WRKY6 that has been shown to be up-regulated during the progression of leaf senescence (Robatzek and Somssich, 2001). It is considered as a senescence regulator because of its binding to promoters of target genes known to be important for senescence such as SEN1 and SIRK. However, probably due to functional redundancy, wrky6 mutants do not show an altered phenotype during leaf senescence (Robatzek and Somssich, 2002).
The first WRKY TF demonstrated as a senescence regulator is WRKY53. Plants where expression of WRKY53 is altered present senescence-associated phenotypes that indicate a function as a positive senescence regulator for this protein (Miao et al., 2004). Moreover, WRKY53 is induced at an early stage of leaf senescence, before expression of several SAG genes, indicating a crucial function for the onset of senescence (Hinderhofer and Zentgraf, 2001). Following identification of WRKY53 as a senescence regulator, studies have focused on elucidating downstream target genes, cellular interactors, and signalling pathways (Zentgraf et al., 2010). Factors that regulate WRKY53 expression and DNA binding of the corresponding protein in senescence include hydrogen peroxide (Miao et al., 2004), other WRKY TFs (Miao et al., 2004), and the MAP kinase MEKK1 (Miao et al., 2007). In contrast, the premature senescence phenotype of wrky70 mutants suggests that WRKY70 could act as a negative regulator of senescence, with gradually increasing expression during leaf development to reach a maximum at the beginning of senescence (Ulker et al., 2007). WRKY70 is also known to be crucial in plant defence against pathogens, controlling the cross-talk of SA and JA signalling in plant defence (Li et al., 2004, 2006). This dual function in both senescence and plant defence, also observed for WRKY53 and WRKY6 (Robatzek and Somssich, 2001; Murray et al., 2007), was explained by conserved perception of external factors and subsequent signal transduction needed in both physiological processes.
Here the putative function of WRKY group III proteins in Arabidopsis leaf senescence has been explored. WRKY54 and WRKY70 exhibit a similar expression pattern during leaf development and appear to have a redundant function in senescence as revealed by single and double mutant studies. These two negative senescence regulators, WRKY54 and WRKY70, and the positive regulator of senescence WRKY53 were shown by yeast two-hydrid assay to interact independently with the so far uncharacterized WRKY30. Although micro RNA (miRNA) lines silenced for WRKY30 did not present a senescence phenotype, real-time quantitative PCR (RT-qPCR) measurement showed that WRKY30 was expressed during developmental leaf senescence. Finally, RT-qPCR analysis of WRKY expression in wild-type and SA-deficient mutants suggests a common but not exclusive role for SA in induction of WRKY30, 53, 54, and 70 during senescence. Additional signals such as ROS are needed for induction of WRKY30 and WRKY53. This work highlights the possibility of integration of internal and environmental factors at the transcription level to modulate the onset and the progression of leaf senescence.
Materials and methods
Plant growth conditions
Arabidopsis thaliana were germinated and grown on soil in a climatic chamber at 22 °C with 70/90% relative humidity and under a light/dark cycle of 12/12 h. For experiments on seedlings, seeds were surface sterilized and grown on MS medium plates (Duchefa). They were exposed for 2 weeks to 22 °C under a light/dark cycle of 16/8 h.
Plant material and transgenic lines
Each A. thaliana line used is in the Columbia (Col-0) ecotype. The sid2.1 mutant was kindly provided by J.P. Metraux (University of Fribourg, Switzerland). T-DNA mutant lines for wrky54 (SALK_111964) and wrky70 (SALK_025198) were obtained from the NASC. Homozygous T-DNA insertion lines were identified using PCR with gene-specific primers and T-DNA left border primers. Single mutants were crossed to obtain the double mutant wrky54/wrky70. To produce the miRNA-WRKY30 line, the MIR319a precursor (Schwab et al., 2006) included in the pRS300 vector was modified by directed PCR mutagenesis (S. Ossowski, J. Fitz, R. Schwab, M. Riester, and D. Weigel, personal communication) and cloned under the 35S promoter of the pCP60 binary vector (Kariola et al., 2006). The new amiRNA targets specifically WRKY30 with the following sequence: TTAGTTGATACTAGTTCCTAG. Transformation of Arabidopsis was performed by floral dip with the Agrobacterium GV3101 strain as described previously (Clough and Bent, 1998). Transgenic plants were selected by seed germination on MS (Murashige and Skoog) medium with kanamycin (50 μg ml−1).
Developmental senescence
For developmental leaf senescence studies, plants were kept under the growth conditions described above. Individual leaves of a plant have different ages and are not synchronized in their development; therefore, senescence was followed specifically in rosette leaves 5 and 6. Each RNA extraction was performed on a mix of eight leaves picked from four plants.
Chemical treatments
SA application was performed on 4-week-old plants grown in soil. Whole plants were sprayed with 5 mM SA; water was used as a control. Hydrogen peroxide (H2O2) treatment was performed on 2-week-old seedlings grown in vitro. Seedlings were submerged in half-strength MS liquid medium with or without 10 mM H2O2. Ozone exposure was performed on 3-week-old plants grown in soil and consisted of a single ozone pulse of 250 nl l−1 (ppb). Times of measurement refer to hours after the start of exposure. Uncontaminated air was used with plant controls.
Quantitative RT-PCR
Total RNA from Arabidopsis leaves or seedlings was prepared by TRIS-SDS/phenol/chloroform extraction and consecutive NaAc/ethanol and LiCl precipitations. RNA samples were treated with DNase using a TURBO DNase kit (Ambion), and first-strand cDNAs were synthesized using superscript III reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. qPCR was performed on an equal amount of cDNAs with Sybr green I master (Roche) and specific primers (see Supplementary Table S1 available at JXB online). Triplicate measurements were carried out to determine the mRNA abundance of each gene in each sample. The qPCR was performed in 384-well plates using the LightCycler 480 system (Roche). Reaction mixtures were denaturated at 95 °C for 10 min followed by 45 amplification cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 1 min. Melt curve analysis was performed on the end products of PCR, to determine the specificity of reactions. Relative quantification of gene expression was calculated according to the ΔΔCt method. Amplification of transcript from the At4g26410 gene served as a reference (Czechowski et al., 2005). Each expression profile measurement was performed at least twice with independent experimental replicates.
RNA gel blot analysis
Total RNA samples (10 μg) prepared in 1× MOPS/50% formamide/10% formaldehyde were denatured and separated by electrophoresis on a denaturing formaldehyde agarose gel. The gel was transferred by capillary elution to a positively charged nylon membrane (Amersham Biosciences). The membrane was hybridized with PCR-labelled gene-specific digoxigenin (DIG) probes (Roche). DNA probes were amplified from the cDNA of WRKY30. Membrane pre-hybridization and hybridizations were performed with Dig-Easy Hyb buffer (Roche) at 50 °C. The membrane was washed twice in 2× SSC/0.1% SDS at room temperature and in 0.1× SSC/0.1% SDS at 50 °C. After membrane blocking, immunodetection was done with an alkaline phosphatase-conjugated anti-DIG antibody and was visualized with the chemiluminescent substrate CSPD according to the instructions of the manufacturer (Roche).
Measurement of chlorophyll content
Chlorophyll was extracted from two calibrated leaf discs in 80% acetone, overnight at 4 °C. Total chlorophyll content was determined according to Porra (2002) by measuring absorbance at 646.6 nm and 663.6 nm.
Yeast two-hybrid analysis
Protein interaction between the WRKY III TF family was examined in yeast using the DUALhunter kit, which takes advantage of a split-ubiquitin system, according to the manufacturer’s protocol (Dualsystems Biotech). The full-length sequences of all WRKY III TFs were amplified from cDNA of SA-treated Arabidopsis leaves by PCR using Pfu DNA polymerase (Promega). SfiI restriction sites were introduced with each WRKY-specific primer. PCR products were subcloned into pGEM-T easy vector (Promega). The derived plasmids were digested with SfiI (Fermentas) and generated fragments were cloned in-frame into pDHB1 (Bait vector) and pPR3-N (prey vector). All of the constructs were confirmed by sequencing. LargeT was used as bait control and Alg5 fused to NubG or NubI was used as the negative and positive prey control, respectively. For the interaction screen, each bait construct was co-transformed with each prey construct in the NMY51 yeast strain, plated on minimum medium, and grown at 30 °C for 5 d. Construct expression in yeast was tested by western blot. In this system, protein interaction leads to expression of the lacZ, HIS3, and ADE2 reporter genes. Two SD media were used: without Leu and Trp to select transformed yeast and without Leu, Trp, His, and Ade for protein interaction. Pellet X-gal assay was used to confirm reporter gene induction: liquid-grown yeast were pelleted and lysed by three cold/heat treatments before adding 0.5% agar mix containing phosphate-buffered saline (PBS), 500 μg ml−1 X-gal, and 0.05% β-mercaptoethanol. A blue colour was observed after 30 min at 37 °C.
Results
The WRKY group III TF family in Arabidopsis
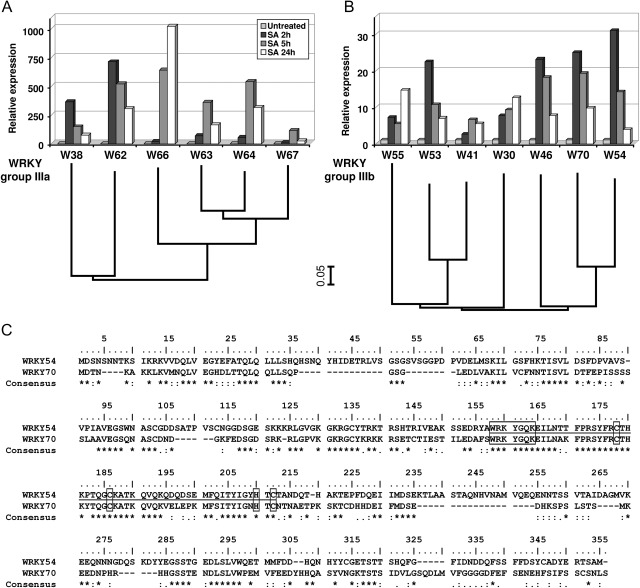
The WRKY TF family contains 74 members in Arabidopsis, with 13 members included in group III and presented in Fig. 1. Based on the highly conserved WRKY domain and structural organization of the genes, the monophyletic WRKY group III has originated from a common ancestral gene that diverged from the other WRKY groups by a slightly modified zinc-finger motif C2-HC within the WRKY domain. Outside the DNA-binding WRKY domain, WRKY group III TFs do not share extensive sequence similarities, indicating divergence in the potential activation and protein–protein interaction domains. However, despite this diversity, more related proteins can be readily identified within subgroups IIIa and IIIb (Fig. 1A, B).
Fig. 1.
Arabidopsis WRKY group III transcription factor family. RT-qPCR time course study of WRKY group IIIa (A) and IIIb (B) gene expression in wild-type leaves treated with 5 mM salicylic acid (SA). Phylogenetic relationships between these WRKY group III transcription factors are indicated below the expression data. Protein alignment was carried out with ClustalX and the trees were constructed by Neighbor–Joining distance analysis. Line lengths indicate the relative distances between nodes. (C) Protein sequence alignment of WRKY54 and WRKY70. The WRKY domain is underlined, with the consensus motif WRKYGQK and the zinc-finger motif C2-HC in boxes. Symbols on the consensus lines represent amino acid positions: ‘*’ fully conserved, ‘:’ one of the strong amino acids group is conserved, and ’.’ one of the weak amino acid groups is conserved.
Previous studies established that nearly all WRKY III TFs were responsive to SA (Kalde et al., 2003), which indicates a putative function for the whole family in defence signalling as already shown for WRKY70, WRKY41, WRKY62, and WRKY38 (Li et al., 2004, 2006; Higashi et al., 2008; Kim et al., 2008). To obtain a more detailed view of the induction profile of WRKY III TF genes in defence, their expression was characterized by RT-qPCR in response to SA (Fig. 1A, B). Differences and redundancies in WRKY expression parameters were evident. First, the fold induction of WRKY group IIIa genes is considerably higher than those of group IIIb. This difference can be partly explained by a difference in the basal expression levels between these two subgroups. While WRKY group IIIa genes are not expressed in non-stressed leaves, WRKY group IIIb genes could share a function in plant development in addition to plant defence, as demonstrated for WRKY53 and WRKY70 in senescence. Secondly, the related WRKY66, WRKY63, WRKY64, and WRKY67 reach maximal induction 5–24 h after treatment and may have a function in secondary signalling for late defence responses. In contrast, WRKY42, WRKY36, and several WRKY group IIIb genes are rapidly induced, with maximal expression 2 h after SA application, and could participate in early defence signalling. Finally, the related WRKY54 and WRKY70 present an identical expression pattern similar to that of WRKY46 and WRKY53.
These results suggest possible functions in distinct defence signalling pathways for some of these factors but also confirm a recent duplication of genes that may still have redundant functions such as WRKY54 and WRKY70. Protein sequence alignment of WRKY54 and WRKY70 (Fig. 1C) revealed that the WRKY domain is highly conserved, with both common WRKYGQK and zinc-finger motifs. The whole WRKY domain shares 80% similarity between these two proteins that decreases to 35% outside of the WRKY domain, but is still fairly extensive compared with other WRKY III TFs with only 6–12% similarity. In addition, these two WRKY genes also present very similar expression profiles in response to a number of biotic and abiotic stress factors tested (unpublished data).
Interaction network of WRKY group III TFs
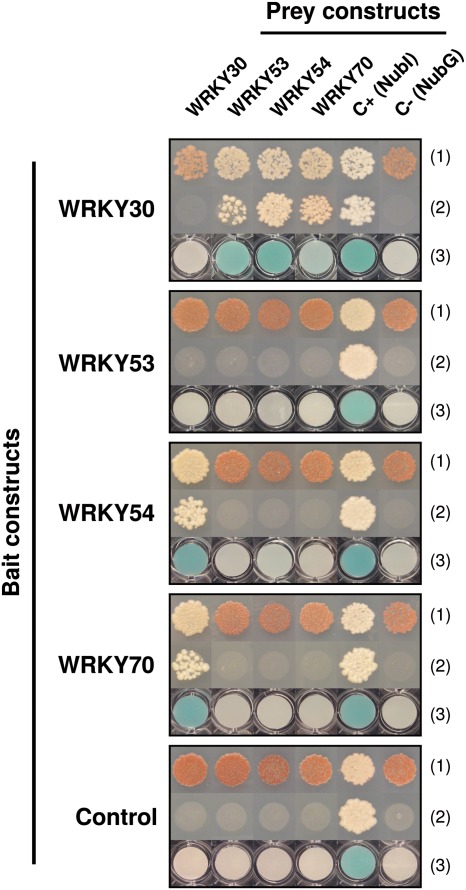
The WRKY III TF family members appear to control different aspects of the defence response and related physiological processes such as senescence (Rushton et al., 2010). It was postulated that some of these TFs may interact to participate in specific regulatory networks, based on their distinct expression patterns induced by specific stress conditions (Berri et al., 2009). To explore specific protein–protein interactions between WRKY III TFs, yeast two-hybrid analysis was employed. In the GAL4 yeast two-hybrid system, auto-activation of reporter genes was found for many WRKYs due to their activation domain. To maintain full-length WRKY cDNAs but avoid auto-activation, WRKY interactions were screened with a split-ubiquitin yeast two-hybrid system. All 13 WRKY III TFs were cloned in both bait and prey vectors. The baits and preys were co-transformed two by two into the NMY51 yeast strain and transformants plated on selective medium to visualize the pair-wise interactions between the WRKYIII proteins.
The most prominent interactions were observed between WRKY30, WRKY53, WRKY54, and WRKY70 (Fig. 2). With WRKY30 as bait, reporter genes were activated in yeast co-transformed with WRKY53, WRKY54, or WRKY70 as a prey. When WRKY54 or WRKY70 were used as bait, the observed interaction with WRKY30 was confirmed in both cases. However, when using WRKY53 as bait, no interaction was found with any of the tested preys even with WRKY30, most probably due to an inaccessible interaction domain of WRKY53 in the bait fusion protein. The data indicate that WRKY30 interacts independently with WRKY54, WRKY70, and WRKY53. No homodimer formation was detected between any of these WRKYs. The four WRKYs are apparently able to form heterodimers that could have the potential to disturb or regulate their binding activity, and to target specificity or activation efficiency in planta.
Fig. 2.
Identification of WRKY group III transcription factor interactions with yeast two-hybrid analysis. A split-ubiquitin system was used to screen interactions. Yeast strain NMY51 was co-transformed with various bait and prey constructs as indicated and plated on SD medium without Leu and Trp (line 1: all transformed yeast grown with red/white colonies depending on protein interactions) and without Leu, Trp, His, and Ade (line 2: transformed yeast grown depending on protein interactions). Each transformed yeast line was used to perform X-gal assays on the pellet (line 3). The largeT gene was used as bait control. Vectors carrying NubI or NubG were used as a prey control for negative and positive interactions, respectively.
Expression of WRKY30, 53, 54, and 70 during Arabidopsis leaf development
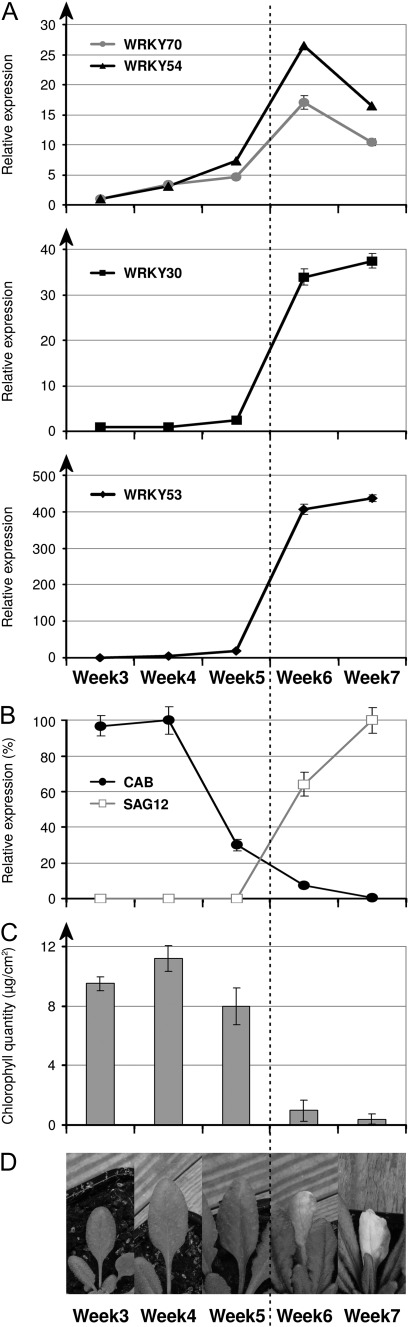
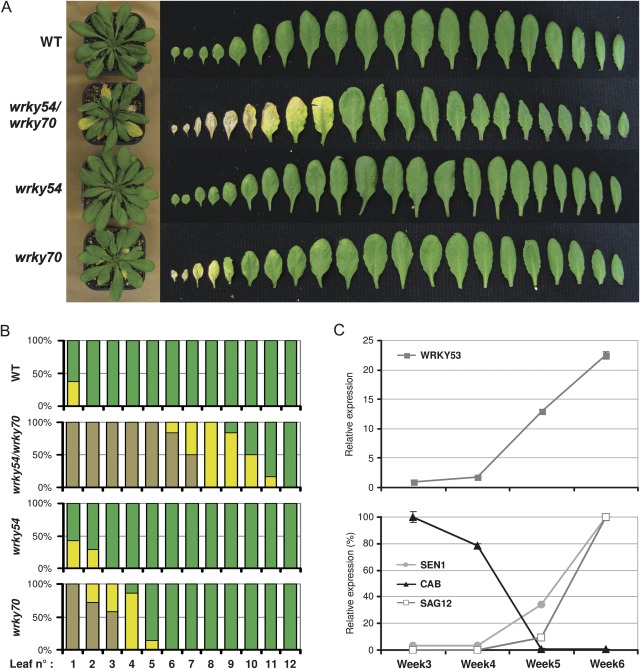
WRKY53 and WRKY70 have been shown to participate in regulation of leaf senescence (Hinderhofer and Zentgraf, 2001; Miao et al., 2004; Ulker et al., 2007). Consequently it was hypothesized that the WRKY partners detected by yeast two-hybrid analysis (WRKY30, 53, 54, and 70) could all have a function in this physiological process. The expression profiles of WRKY30 and WRKY54 were compared with those of WRKY53 and WRKY70 during developmental leaf senescence in Arabidopsis by RT-qPCR. Establishment of senescence in soil-grown plants was followed for leaves 5 and 6 by three cellular parameters: chlorophyll catabolism, change in expression of the photosynthesis-related CAB gene (chlorophyll a/b-binding protein), and change in expression of the senescence-related gene SAG12 (Lohman et al., 1994). Leaf phenotype, expression of senescence marker genes, and chlorophyll content indicated that the senescence process of leaves 5 and 6 was readily detectable in 6-week-old plants (Fig. 3B–D). In accordance with the results of Hinderhofer and Zentgraf (2001), induction of WRKY53 was correlated with senescence establishment (Fig. 3A). Interestingly, WRKY30 presented a similar induction profile to WRKY53, with a high level of expression maintained throughout the senescence process (Fig. 3A). In contrast WRKY54 and WRKY70 showed a somewhat different expression profile compared with WRKY30 and WRKY53, with a slow increase of transcripts during leaf growth and a strong but transient induction at the onset of senescence (Fig. 3A). The prominent up-regulation of WRKY30 and WRKY54 during leaf senescence, together with the ability of WRKY30 to form heterodimers with WRKY53 and WRKY70 in yeast, could suggest possible functions during leaf senescence for these four WRKYs in a TF network.
Fig. 3.
Time course of WRKY30 and WRKY54 expression during developmental leaf senescence. (A) WRKY expression was measured by RT-qPCR on RNA isolated from wild-type leaves 5 and 6 of different developmental stages. RNA samples were collected each week, from 3-week-old plants. (B) Expression of the senescence-related genes CAB and SAG12 was measured by RT-qPCR from the same samples to monitor progress of senescence. (C) Chlorophyll content in wild-type leaves 4 and 5 at each senescence stage. (D) Picture of leaf number 5 at each time point of collection.
Effect of WRKY54 and WRKY30 on leaf senescence
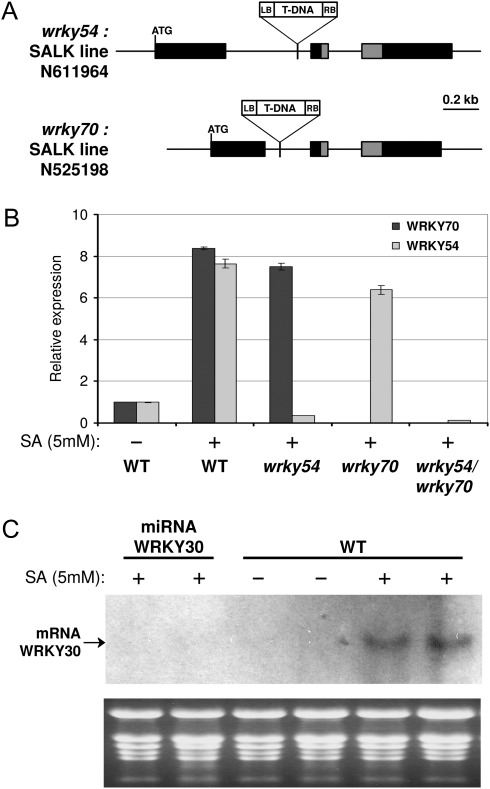
To address the role in planta of WRKY54 and WRKY30 in senescence, plants down-regulated for the corresponding genes were utilized. The wrky54 (SALK_111964) insertion mutant with T-DNA located within the first intron was used (Fig. 4A). The location of the T-DNA insertion and isolation of the homozygous knock-out line were performed with the help of PCR using allele-specific primers. Due to similarities between WRKY54 and WRKY70, the wrky54 mutant was subsequently crossed with wrky70 (SALK_025198) (Li et al., 2006) to obtain homozygous wrky54/wrky70 double mutants. RT-qPCR analysis of wrky54 and wrky70 single mutants shows the absence of WRKY54 and WRKY70 transcripts, respectively, even after SA treatment (Fig. 4B). Similarly, in the wrky54/wrky70 double mutant, neither of the transcripts could be detected even when induced by SA.
Fig. 4.
Characterization of WRKY transgenic lines. (A) Schematic representation of WRKY54 and WRKY70 gene structure indicating the location of T-DNA insertions. Exons are shown as dark boxes. The grey part indicates the region encoding the WRKY domain. (B) RT-qPCR analysis of WRKY54 and WRKY70 transcript levels in wrky54/wrky70 single and double mutants sprayed with 5 mM SA, compared with wild-type plants. Measurements were done 5 h after treatment. (C) RNA gel blot analysis of the WRKY30 transcript level in two independent miRNA-WRKY30 lines sprayed with 5 mM SA compared with wild-type plants. Measurments were done 5 h after treatment. EtBr (ethidium bromide) staining of the gel was used as loading control.
Leaf development of wrky54 and wrky70 single mutants was compared with that of the wrky54/wrky70 double mutant and wild-type plants grown under standard conditions in growth chambers. Figure 5B gives an overall view of the status of each leaf in a pool of plants of each genotype at 5.5 weeks post-germination. Representative plants of each population were used to visualize the developmental phenotype (Fig. 5A). Even the oldest wild-type leaves were green and healthy without any visual senescence symptoms. In contrast, the wrky54/wrky70 double mutant exhibits clearly premature senescence, with leaves 1–5 completely dried out and brown, and leaves 6–9 showing total to partial yellowing, suggesting chlorophyll degradation and indicating an ongoing senescence process. Leaf number 10 is the oldest leaf without any visible senescence symptoms. In comparison, the wrky70 mutant showed a somewhat enhanced senescence phenotype but less drastic than that of the double mutant, while no clear visual symptoms of premature senescence were evident in the wrky54 mutant when compared with the wild type. To confirm that the premature senescence phenotype was indeed caused by the wrky54/wrky70 double mutant and not by unlinked additional mutations, the co-segregation of the early senescence phenotype with the homozygosity for T-DNA insertions was characterized in both WRKY54 and WRKY70. This was achieved by screening both the senescence phenotype and the WRKY54 WRKY70 genotype in the F2 progeny from a cross between homozygous wrky54 and wrky70 single mutants. Of 104 F2 progeny genotyped, six homozygous double mutants were detected, all showing the premature senescence phenotype (data not shown). The much more precocious senescence phenotype of the wrky54/wrky70 double mutant compared with those in single mutants (Fig. 5A) suggests that WRKY70 and WRKY54 co-operate to contain development of senescence. These results also indicate that WRKY54 and WRKY70 present partly redundant functions as negative regulators of senescence.
Fig. 5.
Early senescence phenotype of the wrky54/wrky70 double mutant compared with single mutants and wild-type plants. (A) Phenotype of rosette leaves in 5.5-week-old plants: whole plants and excised leaves are arranged according to their age from older to younger. (B) Distribution of leaf senescence stages in 5.5-week-old plants. Leaves were classified into three groups according to their colour: brown/dry, yellow, and green. Seven plants of each line were used. (C) RT-qPCR analysis of expression of senescence-related genes (WRKY53, CAB, SEN1, and SAG12) during developmental leaf senescence in the wrky54/wrky70 double mutant.
To confirm that the observed leaf phenotype of the wrky54/wrky70 double mutant is due to a normal senescence-related cell death process, expression of senescence-related genes was measured during development of leaves 5 and 6. As observed for wild-type leaves (Fig. 3), senescence in the double mutant was accompanied by decreased expression of CAB and increased expression of SAG12 and SEN1 (Oh et al., 1996) (Fig. 5C). In accordance with the premature senescence symptoms observed visually, this altered expression of senescence-associated marker genes was also premature in the wrky54/wrky70 double mutant. Interestingly, while WRKY53 was also expressed at the onset of senescence in wrky54/wrky70, the expression level was 16-fold less than in the wild type. This suggests that the absence of the negatives regulators (WRKY54 and WRKY70) could allow a reduced amount of the positive regulator to be sufficient for induction of premature senescence.
As no T-DNA insertion mutants were available for WRKY30, miRNA-silenced lines were generated. Arabidopsis were transformed with the miRNA precursor miR319a carrying a specific sequence of AtWRKY30 driven by the 35S promoter to induce RNA silencing of WRKY30 transcripts. Homozygous lines for the construct were obtained from two independent transformants. SA-induced accumulation of WRKY30 transcripts observed in wild-type plants by northern blot hybridization was undetectable in these miRNA-silenced lines (Fig. 4C). Unfortunatly, no significant differences in senescence phenotype were observed for miRNA-WRKY30-silenced plants when compared with the wild type (data not shown).
WRKY54 and WRKY30 signalling pathway in senescence
SA is known to be a key signalling compound to trigger the plant defence response in the case of pathogen infection (Lu, 2009; Vlot et al., 2009). The SA-mediated pathway has also been shown to control gene expression during developmental senescence (Morris et al., 2000; Yoshimoto et al., 2009). SA inducibility of WRKY group III TFs prompted the investigation of whether induction of WRKY30, WRKY53, WRKY54, and WRKY70 during the senescence process was SA dependent. Induction of these WRKY genes was measured by RT-qPCR during developmental senescence at 3 and 6 weeks after seed germination in leaves 5 and 6 of the SA-deficient mutant sid2 (Nawrath and Metraux, 1999) and wild-type plants (Table 1). Transcript accumulation of each WRKY gene studied was clearly reduced in the sid2 background when compared with the wild type. Levels of induction in the sid2 mutant represent 25–55% of the corresponding wild-type values. These data suggest that the expression of WRKY30, 53, 54, and 70 during the senescence process is partially SA dependent.
Table 1.
Expression of WRKY III genes during senescence in an SA-deficient mutant sid2 compared with the wild type
| Genes | Fold induction between plants of 3- and 6-weeks old |
|
| Wild-type | sid2 | |
| WRKY54 | 9.6±1.8 | 5.3±1.1 |
| WRKY70 | 17.2±4.3 | 4.2±0.9 |
| WRKY30 | 65±5.8 | 26.1±3.6 |
| WRKY53 | 340±37.8 | 122±30.6 |
WRKY expression was measured by RT-qPCR on RNA isolated from leaves 4 and 5.
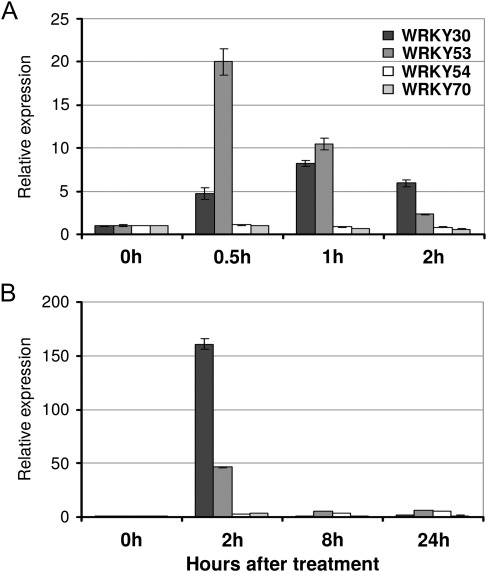
ROS are key components in senescence and cell death. Some regulators of senescence such as WRKY53 are known to be induced by H2O2 (Miao et al., 2007). To elucidate the participation of ROS in regulation of WRKY30 and WRKY54, expression of these genes was measured by RT-qPCR under two different oxidative stress treatments: exposure to H2O2 and ozone (Fig. 6). WRKY53 and WRKY30 were rapidly and transiently induced by both treatments. An increased expression level was observed after 30 min for WRKY53 and 1 h for WRKY30 under H2O2 treatment, and both were induced after 2 h of ozone exposure. Moreover, WRKY53 was much more responsive to H2O2 than WRKY30 and inversely to ozone. In contrast, WRKY54 and WRKY70 were induced neither by H2O2 nor by ozone.
Fig. 6.
Expression of WRKY30, WRKY53, WRKY54, and WRKY70 under oxidative stress. WRKY expression was measured by RT-qPCR. (A) RNA samples were isolated from 2-week-old wild-type seedlings submerged in liquid MS medium with 10 mM H2O2. (B) RNA samples were extracted from 3-week-old wild-type plants treated with 250 ppb ozone.
Discussion
Leaf senescence is basically governed by leaf age and global plant developmental stage, but onset and progression of senescence are also modulated by environmental factors (Buchanan-Wollaston et al., 2003). Integration of internal and external factors is therefore a critical point in senescence regulation that may implicate a complex regulation network. This is supported by the extensive transcriptome reprogramming during senescence, including induction of >100 TFs (Gepstein et al., 2003; Guo et al., 2004; Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006; Balazadeh et al., 2008). However, very little is known of the function of these TFs in senescence and of the integration of multiple signalling pathways. Of the WRKY TF family, WRKY53 and WRKY70 have been implicated in senescence regulation in addition to their function in plant defence (Li et al., 2004; Miao et al., 2004; Murray et al., 2007; Ulker et al., 2007). The present data demonstrate a functional overlap of WRKY54 and WRKY70 as negative senescence regulators. Both WRKY54 and WRKY70 appear to take part in the senescence regulatory network with positive senescence regulator WRKY53, possibly through an interaction with WRKY30.
WRKY70 was previously demonstrated to regulate both plant defence and leaf senescence in Arabidopsis, leading to an early senescence phenotype in wrky70 mutants (Ulker et al., 2007) and to enhanced resistance/susceptibility phenotypes to several pathogens in wrky70 overexpressor and mutant lines (Li et al., 2004; AbuQamar et al., 2006; Li et al., 2006). Within WRKY group III, WRKY54 is the closest homologue to WRKY70; moreover, the expression patterns of the corresponding genes in response to hormonal treatments or to various abiotic and biotic stresses were highly similar, suggesting a conserved function (Figs 1, 3, 6, 7, and unpublished data). This hypothesis was already investigated for plant defence (Wang et al., 2006). Unfortunately, no redundant function could be established on the basis of the resistance profiles of single and double mutants, but the wrky54/wrky70 double mutant showed a significant up-regulation of the SA biosynthesis gene ICS1 (isochorismate synthase) and consequently a high level of free SA compared with wrky70. Based on this observation, the authors suggested that WRKY70 and WRKY54 act as negative regulators of SA synthesis, but no further co-function was established for plant defence (Wang et al., 2006). Here the possible redundancy of WRKY70 and WRKY54 in plant senescence was investigated. The wrky70 mutant showed an early developmental senescence phenotype whereas the wrky54 mutant did not exhibit significant alterations in senescence (Fig. 5). However, the double mutant wrky54/wrky70 presents a drastically enhanced senescence phenotype clearly enhanced over that of wrky70, suggesting functional redundancy and possible co-operation of these two factors as negative regulators of senescence in leaves. Consistent with the large number of homologous members in the WRKY TF family, this kind of functional redundancy has already been demonstrated for several factors (Robatzek and Somssich, 2002; Journot-Catalino et al., 2006; Pandey and Somssich, 2009). Thus, WRKY54 and WRKY70 appear to have a common function in senescence regulation, although differences in factor efficiency were evident from the distinct senescence phenotypes of the single mutants. Similar observations were previously reported, for example for redundant WRKY11 and WRKY17 TFs in plant resistance against Pseudomonas syringae infection. In that study, a difference in compensation of single mutants was noted and was linked to a partially redundant function. Indeed, target screen and transcriptome analysis showed only a partial overlap in downstream components. This could also be the case for WRKY54 and WRKY70, although, a difference in expression level and efficacy between these two factors could not be excluded. Indeed, they share a highly conserved DNA-binding domain with 80% homology (Fig. 1), decreasing to <65% with other WRKYs that may indicate conserved targets. However, outside of the binding domain including the activation domain, important divergences exist between WRKY54 and WRKY70 that could explain the differences in factor efficacy. Taken together, the present results argue for a partly redundant function of WRKY54 with WRKY70 in senescence regulation, but it seems that WRKY54 is not sufficient to replace WRKY70 fully in senescence.
An extensive screen using a yeast split-ubiquitin two-hydrid system was employed to gain deeper insight into the possible interaction network of WRKY III TFs in plant gene regulation, and it was demonstrated that WRKY54, WRKY70, and WRKY53 interact independently with WRKY30 (Fig. 2). Homodimer and heterodimer formation between members of WRKY group IIa have been demonstrated, generated by leucine zipper motifs in the N-terminus of the proteins (Xu et al., 2006). This kind of motif is not found in proteins of the WRKY III family; moreover, no conserved motif can be identified outside of the WRKY domain. WRKY30 has never been functionally characterized and the interactions detected in yeast with the other WRKYs implicated in senescence suggest that WRKY30 might also have a senescence-associated function. Further support for its role in senescence comes from expression studies showing that WRKY30 was strongly induced during developmental leaf senescence (Fig. 3). Unfortunately, silencing of the WRKY30 gene by miRNA did not seem to affect the leaf senescence phenotype (unpublished data). However, the possibility that the absence of phenotype could be due to a low level expression of WRKY30 in the silenced line sufficient for its physiological function cannot be excluded.
Temporal expression patterns of WRKY30, WRKY54, WRKY70, and WRKY53 during leaf development reveal two distinct profiles in accordance with putative functions in leaf senescence. As previously demonstrated, WRKY53 is a positive regulator of senescence important for the onset of the process and is induced at the early stage of senescence (Fig. 3; Hinderhofer and Zentgraf, 2001). Interestingly, the WRKY30 expression profile was almost identical to that of WRKY53. In contrast, the negative senescence regulators WRKY54 and WRKY70 exhibit identical expression profiles in accordance with their suggested functional redundancy. Their expression slowly increases during leaf development, reaching a maximum at an early stage of senescence to decrease finally until the end of cell death (Fig. 3). These expression profiles suggest three different phases for the action of WRKY senescence regulators in leaf development: expression of negative regulators during leaf development prior to senescence, co-induction of both positive and negative factors at the onset of senescence, and finally predominance of positive regulators during the progression of senescence. Activation of critical physiological processes in plants that generate major changes are rigorously controlled and induced in accordance with the fitness of the whole plant (Heil and Baldwin, 2002). In leaf senescence, premature onset has to be prevented and progression has to be controlled to allow effective nutrient recycling before the final stages of cell death. The combination of WRKY54 and WRKY70 as negative senescence regulators with the positive regulator WRKY53 would permit such fast and fine-tuned control of senescence. Furthermore, the ability of WRKY30 to interact in yeast with characterized WRKY senescence regulators in addition to its expression during senescence suggests the presence of a WRKY interaction network in planta that could integrate both positive and negative signals at the TF level to fine-tune balanced leaf development. In this respect, variation in the expression ratio between WRKY54/WRKY70 and WRKY53 caused by internal factors or environmental conditions would affect heterodimer formation with displacement or preferential WRKY30 binding, and thereby alter the outcome of the leaf senescence programme. Such heterodimer formation would allow adjustment of their activities by modification of binding efficiency and activation properties, as has been demonstrated for the rice proteins OsWRKY51 and OsWRKY71. OsWRKY51 interaction will enhance OsWRKY71 binding of the Amy32b promoter, whereas OSWRKY51 does not bind to that promoter alone (Xie et al., 2006). It would be of interest to examine in depth WRKY30 function as a senescence regulator and its role in the cross-talk between positive and negative induction pathways to confirm these hypotheses and identify underlying molecular mechanisms.
WRKY70 expression in plant defence was shown to be mediated by SA (Li et al., 2004, 2006). Accumulation of WRKY70 transcripts in defence is strongly reduced in mutants defective in SA signlaling, pad4 and npr1, and absent in NahG plants (Li et al., 2004; Ulker et al., 2007). In the senescence context, WRKY70 induction was reduced but not completely suppressed in the SA-deficient mutant sid2 (Table 1). This result, also observed for WRKY30, WRKY53, and WRKY54, indicates that these four WRKY genes are dependent on the presence of SA for maximal expression in leaf senescence, but also suggests additional signalling pathways. These results are in accordance with previous work on several SAGs that were identified as partially SA dependent such as LSC460 (cytosolic glutamine synthetase) (Morris et al., 2000; Yoshimoto et al., 2009). ROS also appear important in senescence, either causing oxidative damage or as signal molecules (Finkel, 2003; Foyer and Noctor, 2005; Pitzschke et al., 2006; Moller et al., 2007). H2O2 was an element that regulates WRKY53 expression (Miao et al., 2004). Similarly, WRKY30 was induced by H2O2 treatment (Fig. 6); moreover, both WRKY53 and WRKY30 were highly induced by ozone exposure. Interestingly, paraquat treatment did not induce WRKY53 or WRKY30 (unpublished data). The chemical nature of ROS and their subcellular site of production could be critical for the biological activities of ROS signals (Laloi et al., 2006). It seems that some ROS are crucial inducers for WRKY53 and WRKY30 but not for the negative regulators WRKY54 and WRKY70. In addition, MAP kinases must be implicated in this signalling process, as has already been shown for WRKY53 with MEKK1 (Miao et al., 2007; Zhou et al., 2009; Zentgraf et al., 2010).
Taking together previous studies and the current findings, a crucial function for WRKY group III TFs in regulation of developmental leaf senescence has been demonstrated. WRKY53, WRKY54, and WRKY70 appear to participate in a regulatory network that integrates, at the TF level, both positive and negative signalling pathways for senescence, possibly through an interaction with WRKY30. WRKY proteins have a high binding affinity for the cognate W-box DNA element that is also over-represented within the WRKY TF promoters themselves (Eulgem et al., 2000; Ciolkowski et al., 2008). Consequently, WRKY TFs are subject to autoregulation and cross-regulation. Interestingly, transcriptome studies of WRKY53 and WRKY70 overexpressor lines by microarrays showed increased expression of WRKY70 and WRKY53, respectively (Li et al., 2004; Miao et al., 2004). Thus another level of complexity in senescence regulation by WRKYs exists with transcriptional cross-modulation. Finally, new TFs are regularly found to participate in senescence regulation such as NAC TFs and RAV TFs (Guo and Gan, 2006; Woo et al., 2010; Balazadeh et al., 2011), showing that the WRKY network does not work alone but takes part in a highly complex web of TFs. To gain more insight into this WRKY senescence regulatory network at the molecular level, further investigations will have to be carried out to identify the influence of WRKY30 on the activity of other WRKYs. These kinds of studies could be performed by yeast one-hybrid analysis and provide the next step in gaining more knowledge of the function of this network.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Target genes and primers used for pPCR.
Acknowledgments
The work was supported by the European Research Area in Plant Genomics (ERAPGFP/06.023a) and the Academy of Finland Center of Excellence programme. We thank Dr Sims-Huopaniemi for the critical reading of the manuscript and Professor Weigel for providing the pRS300 vector (MIR319a precursor).
Glossary
Abbreviations
- ABA
abscisic acid
- JA
jasmonate
- SA
salicylic acid
- TFs
transcription factors
References
- AbuQamar S, Chen X, Dhawan R, Bluhm B, Salmeron J, Lam S, Dietrich RA, Mengiste T. Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. The Plant Journal. 2006;48:28–44. doi: 10.1111/j.1365-313X.2006.02849.x. [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Kwasniewski M, Caldana C, Mehrnia M, Zanor MI, Xue GP, Mueller-Roeber B. ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Molecular Plant. 2011;4:346–360. doi: 10.1093/mp/ssq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S, Riaño-Pachón DM, Mueller-Roeber B. Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biology (Stuttgart) 2008;1(10 Suppl):63–75. doi: 10.1111/j.1438-8677.2008.00088.x. [DOI] [PubMed] [Google Scholar]
- Berri S, Abbruscato P, Faivre-Rampant O, et al. Characterization of WRKY co-regulatory networks in rice and Arabidopsis. BMC Plant Biology. 2009;9:120. doi: 10.1186/1471-2229-9-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Earl S, Harrison E, Mathas E, Navabpour S, Page T, Pink D. The molecular analysis of leaf senescence: a genomics approach. Plant Biotechnology Journal. 2003;1:3–22. doi: 10.1046/j.1467-7652.2003.00004.x. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. The Plant Journal. 2005;42:567–585. doi: 10.1111/j.1365-313X.2005.02399.x. [DOI] [PubMed] [Google Scholar]
- Butt A, Mousley C, Morris K, Beynon J, Can C, Holub E, Greenberg JT, Buchanan-Wollaston V. Differential expression of a senescence-enhanced metallothionein gene in Arabidopsis in response to isolates of Peronospora parasitica and Pseudomonas syringae. The Plant Journal. 1998;16:209–221. doi: 10.1046/j.1365-313x.1998.00286.x. [DOI] [PubMed] [Google Scholar]
- Ciolkowski I, Wanke D, Birkenbihl RP, Somssich IE. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Molecular Biology. 2008;68:81–92. doi: 10.1007/s11103-008-9353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends in Plant Science. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- Finkel T. Oxidant signals and oxidative stress. Current Opinion in Cell Biology. 2003;15:247–254. doi: 10.1016/s0955-0674(03)00002-4. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. The Plant Cell. 2005;17:1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S, Amasino RM. Inhibition of leaf senescence by autoregulated production of cytokinin. Science. 1995;270:1986–1988. doi: 10.1126/science.270.5244.1986. [DOI] [PubMed] [Google Scholar]
- Gepstein S, Sabehi G, Carp MJ, Hajouj T, Nesher MF, Yariv I, Dor C, Bassani M. Large-scale identification of leaf senescence-associated genes. The Plant Journal. 2003;36:629–642. doi: 10.1046/j.1365-313x.2003.01908.x. [DOI] [PubMed] [Google Scholar]
- Grbic V, Bleecker AB. Ethylene regulates the timing of leaf senescence in Arabidopsis. The Plant Journal. 1995;8:595–601. [Google Scholar]
- Guo Y, Cai Z, Gan S. Transcriptome of Arabidopsis leaf senescence. Plant, Cell and Environment. 2004;27:521–548. [Google Scholar]
- Guo Y, Gan S. Leaf senescence: signals, execution, and regulation. Current Topics in Developmental Biology. 2005;71:83–112. doi: 10.1016/S0070-2153(05)71003-6. [DOI] [PubMed] [Google Scholar]
- Guo Y, Gan S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. The Plant Journal. 2006;46:601–612. doi: 10.1111/j.1365-313X.2006.02723.x. [DOI] [PubMed] [Google Scholar]
- He Y, Fukushige H, Hildebrand DF, Gan S. Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiology. 2002;128:876–884. doi: 10.1104/pp.010843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M, Baldwin IT. Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends in Plant Science. 2002;7:61–67. doi: 10.1016/s1360-1385(01)02186-0. [DOI] [PubMed] [Google Scholar]
- Hensel LL, Grbic V, Baumgarten DA, Bleecker AB. Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in arabidopsis. The Plant Cell. 1993;5:553–564. doi: 10.1105/tpc.5.5.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi K, Ishiga Y, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y. Modulation of defense signal transduction by flagellin-induced WRKY41 transcription factor in Arabidopsis thaliana. Molecular Genetics and Genomics. 2008;279:303–312. doi: 10.1007/s00438-007-0315-0. [DOI] [PubMed] [Google Scholar]
- Hinderhofer K, Zentgraf U. Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta. 2001;213:469–473. doi: 10.1007/s004250000512. [DOI] [PubMed] [Google Scholar]
- Hopkins M, Taylor C, Liu Z, Ma F, McNamara L, Wang TW, Thompson JE. Regulation and execution of molecular disassembly and catabolism during senescence. New Phytologist. 2007;175:201–214. doi: 10.1111/j.1469-8137.2007.02118.x. [DOI] [PubMed] [Google Scholar]
- Hortensteiner S, Feller U. Nitrogen metabolism and remobilization during senescence. Journal of Experimental Botany. 2002;53:927–937. doi: 10.1093/jexbot/53.370.927. [DOI] [PubMed] [Google Scholar]
- Hwang I, Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413:383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- Jing HC, Schippers JH, Hille J, Dijkwel PP. Ethylene-induced leaf senescence depends on age-related changes and OLD genes in Arabidopsis. Journal of Experimental Botany. 2005;56:2915–2923. doi: 10.1093/jxb/eri287. [DOI] [PubMed] [Google Scholar]
- Journot-Catalino N, Somssich IE, Roby D, Kroj T. The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. The Plant Cell. 2006;18:3289–3302. doi: 10.1105/tpc.106.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KC, Lai Z, Fan B, Chen Z. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. The Plant Cell. 2008;20:2357–2371. doi: 10.1105/tpc.107.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi C, Przybyla D, Apel K. A genetic approach towards elucidating the biological activity of different reactive oxygen species in Arabidopsis thaliana. Journal of Experimental Botany. 2006;57:1719–1724. doi: 10.1093/jxb/erj183. [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Kariola T, Palva ET. WRKY70 modulates the selection of signaling pathways in plant defense. The Plant Journal. 2006;46:477–491. doi: 10.1111/j.1365-313X.2006.02712.x. [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET. The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. The Plant Cell. 2004;16:319–331. doi: 10.1105/tpc.016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. Leaf senescence. Annual Review of Plant Biology. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- Lim PO, Woo HR, Nam HG. Molecular genetics of leaf senescence in Arabidopsis. Trends in Plant Science. 2003;8:272–278. doi: 10.1016/S1360-1385(03)00103-1. [DOI] [PubMed] [Google Scholar]
- Lin JF, Wu SH. Molecular events in senescing Arabidopsis leaves. The Plant Journal. 2004;39:612–628. doi: 10.1111/j.1365-313X.2004.02160.x. [DOI] [PubMed] [Google Scholar]
- Lohman KN, Gan S, John MC, M AR. Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiologia Plantarum. 1994;92:322–327. [Google Scholar]
- Lu H. Dissection of salicylic acid-mediated defense signaling networks. Plant Signaling and Behavior. 2009;4:713–716. doi: 10.4161/psb.4.8.9173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Laun TM, Smykowski A, Zentgraf U. Arabidopsis MEKK1 can take a short cut: it can directly interact with senescence-related WRKY53 transcription factor on the protein level and can bind to its promoter. Plant Molecular Biology. 2007;65:63–76. doi: 10.1007/s11103-007-9198-z. [DOI] [PubMed] [Google Scholar]
- Miao Y, Laun T, Zimmermann P, Zentgraf U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Molecular Biology. 2004;55:853–867. doi: 10.1007/s11103-004-2142-6. [DOI] [PubMed] [Google Scholar]
- Miller JD, Arteca RN, Pell EJ. Senescence-associated gene expression during ozone-induced leaf senescence in Arabidopsis. Plant Physiology. 1999;120:1015–1024. doi: 10.1104/pp.120.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller IM, Jensen PE, Hansson A. Oxidative modifications to cellular components in plants. Annual Review of Plant Biology. 2007;58:459–481. doi: 10.1146/annurev.arplant.58.032806.103946. [DOI] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science. 2003;300:332–336. doi: 10.1126/science.1080585. [DOI] [PubMed] [Google Scholar]
- Morris K, MacKerness SA, Page T, John CF, Murphy AM, Carr JP, Buchanan-Wollaston V. Salicylic acid has a role in regulating gene expression during leaf senescence. The Plant Journal. 2000;23:677–685. doi: 10.1046/j.1365-313x.2000.00836.x. [DOI] [PubMed] [Google Scholar]
- Munne-Bosch S, Alegre L. Plant aging increases oxidative stress in chloroplasts. Planta. 2002;214:608–614. doi: 10.1007/s004250100646. [DOI] [PubMed] [Google Scholar]
- Murray SL, Ingle RA, Petersen LN, Denby KJ. Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Molecular Plant-Microbe Interactions. 2007;20:1431–1438. doi: 10.1094/MPMI-20-11-1431. [DOI] [PubMed] [Google Scholar]
- Navabpour S, Morris K, Allen R, Harrison E, S AH-M, Buchanan-Wollaston V. Expression of senescence-enhanced genes in response to oxidative stress. Journal of Experimental Botany. 2003;54:2285–2292. doi: 10.1093/jxb/erg267. [DOI] [PubMed] [Google Scholar]
- Nawrath C, Metraux JP. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. The Plant Cell. 1999;11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nooden LD, Penney JP. Correlative controls of senescence and plant death in Arabidopsis thaliana (Brassicaceae) Journal of Experimental Botany. 2001;52:2151–2159. doi: 10.1093/jexbot/52.364.2151. [DOI] [PubMed] [Google Scholar]
- Oh SA, Lee SY, Chung IK, Lee CH, Nam HG. A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Molecular Biology. 1996;30:739–754. doi: 10.1007/BF00019008. [DOI] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE. The role of WRKY transcription factors in plant immunity. Plant Physiology. 2009;150:1648–1655. doi: 10.1104/pp.109.138990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Oh SA, Kim YH, Woo HR, Nam HG. Differential expression of senescence-associated mRNAs during leaf senescence induced by different senescence-inducing factors in Arabidopsis. Plant Molecular Biology. 1998;37:445–454. doi: 10.1023/a:1005958300951. [DOI] [PubMed] [Google Scholar]
- Pic E, de La Serve BT, Tardieu F, Turc O. Leaf senescence induced by mild water deficit follows the same sequence of macroscopic, biochemical, and molecular events as monocarpic senescence in pea. Plant Physiology. 2002;128:236–246. [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A, Forzani C, Hirt H. Reactive oxygen species signaling in plants. Antioxidants and Redox Signaling. 2006;8:1757–1764. doi: 10.1089/ars.2006.8.1757. [DOI] [PubMed] [Google Scholar]
- Porra RJ. The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynthesis Research. 2002;73:149–156. doi: 10.1023/A:1020470224740. [DOI] [PubMed] [Google Scholar]
- Pourtau N, Jennings R, Pelzer E, Pallas J, Wingler A. Effect of sugar-induced senescence on gene expression and implications for the regulation of senescence in Arabidopsis. Planta. 2006;224:556–568. doi: 10.1007/s00425-006-0243-y. [DOI] [PubMed] [Google Scholar]
- Quirino BF, Noh YS, Himelblau E, Amasino RM. Molecular aspects of leaf senescence. Trends in Plant Science. 2000;5:278–282. doi: 10.1016/s1360-1385(00)01655-1. [DOI] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmulling T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. The Plant Cell. 2006;18:40–54. doi: 10.1105/tpc.105.037796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S, Somssich IE. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. The Plant Journal. 2001;28:123–133. doi: 10.1046/j.1365-313x.2001.01131.x. [DOI] [PubMed] [Google Scholar]
- Robatzek S, Somssich IE. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes and Development. 2002;16:1139–1149. doi: 10.1101/gad.222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends in Plant Science. 2010;15:247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. The Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Evensen KB, Kao TH. Ethylene synthesis and floral senescence following compatible and incompatible pollinations in Petunia inflata. Plant Physiology. 1992;99:38–45. doi: 10.1104/pp.99.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulker B, Shahid Mukhtar M, Somssich IE. The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta. 2007;226:125–137. doi: 10.1007/s00425-006-0474-y. [DOI] [PubMed] [Google Scholar]
- van der Graaff E, Schwacke R, Schneider A, Desimone M, Flugge UI, Kunze R. Transcription analysis of arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiology. 2006;141:776–792. doi: 10.1104/pp.106.079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. Salicylic acid, a multifaceted hormone to combat disease. Annual Review of Phytopathology. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathogens. 2006;2:e123. doi: 10.1371/journal.ppat.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Amasino RM. Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiology. 2001;127:876–886. [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM. A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Molecular Biology. 1998;37:455–469. doi: 10.1023/a:1005934428906. [DOI] [PubMed] [Google Scholar]
- Wingler A, Purdy S, MacLean JA, Pourtau N. The role of sugars in integrating environmental signals during the regulation of leaf senescence. Journal of Experimental Botany. 2006;57:391–399. doi: 10.1093/jxb/eri279. [DOI] [PubMed] [Google Scholar]
- Wingler A, Roitsch T. Metabolic regulation of leaf senescence: interactions of sugar signalling with biotic and abiotic stress responses. Plant Biology (Stuttgart) 2008;(10 suppl 1):50–62. doi: 10.1111/j.1438-8677.2008.00086.x. [DOI] [PubMed] [Google Scholar]
- Woo HR, Kim JH, Kim J, Lee U, Song IJ, Lee HY, Nam HG, Lim PO. The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. Journal of Experimental Botany. 2010;61:3947–3957. doi: 10.1093/jxb/erq206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Zhang ZL, Zou X, Yang G, Komatsu S, Shen QJ. Interactions of two abscisic-acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. The Plant Journal. 2006;46:231–242. doi: 10.1111/j.1365-313X.2006.02694.x. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Bassham DC. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. The Plant Journal. 2005;42:535–546. doi: 10.1111/j.1365-313X.2005.02397.x. [DOI] [PubMed] [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. The Plant Cell. 2006;18:1310–1326. doi: 10.1105/tpc.105.037523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. The Plant Cell. 2009;21:2914–2927. doi: 10.1105/tpc.109.068635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacarias L, Reid MS. Role of growth-regulators in the senescence of Arabidopsis thaliana leaves. Physiologia Plantarum. 1990;80:549–553. [Google Scholar]
- Zentgraf U, Laun T, Miao Y. The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. European Journal of Cell Biology. 2010;89:133–137. doi: 10.1016/j.ejcb.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Zhou C, Cai Z, Guo Y, Gan S. An arabidopsis mitogen-activated protein kinase cascade, MKK9–MPK6, plays a role in leaf senescence. Plant Physiology. 2009;150:167–177. doi: 10.1104/pp.108.133439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.