Abstract
Native to South America, Alstroemeria flowers are known for their colourful tepals, and Alstroemeria hybrids are an important cut flower. However, in common with many commercial cut flowers, virtually all the commercial Alstroemeria hybrids are not scented. The cultivar ‘Sweet Laura’ is one of very few scented commercial Alstroemeria hybrids. Characterization of the volatile emission profile of these cut flowers revealed three major terpene compounds: (E)-caryophyllene, humulene (also known as α-caryophyllene), an ocimene-like compound, and several minor peaks, one of which was identified as myrcene. The profile is completely different from that of the parental scented species A. caryophyllaea. Volatile emission peaked at anthesis in both scented genotypes, coincident in cv. ‘Sweet Laura’ with the maximal expression of a putative terpene synthase gene AlstroTPS. This gene was preferentially expressed in floral tissues of both cv. ‘Sweet Laura’ and A. caryophyllaea. Characterization of the AlstroTPS gene structure from cv. ‘Sweet Laura’ placed it as a member of the class III terpene synthases, and the predicted 567 amino acid sequence placed it into the subfamily TPS-b. The conserved sequences R28(R)X8W and D321DXXD are the putative Mg2+-binding sites, and in vitro assay of AlstroTPS expressed in Escherichia coli revealed that the encoded enzyme possesses myrcene synthase activity, consistent with a role for AlstroTPS in scent production in Alstroemeria cv. ‘Sweet Laura’ flowers.
Keywords: Alstroemeria, caryophyllene, gene expression, humulene, myrcene, scent emission, terpene synthase, volatiles
Introduction
The main biological function of floral scent is as an attractant for pollinators. Scent forms an important component of the ‘pollination syndrome’ that includes other characters such as visual cues and nectar production, and has played a key role in floral evolution (Fenster et al., 2004; Galliot et al., 2006). Scent composition varies between species based on a combination of diverse volatile compounds, including terpenoids, benzenoid aromatics, and fatty acid derivatives (Pichersky et al., 2006). Scent compounds are produced de novo, and the wide range of volatile organic compounds (VOCs) and their relative amounts in the final bouquet determine a high specificity of floral scent for different pollinator species (Galliot et al., 2006). Scent compound profiles are species specific and can vary between closely related species or even between different varieties of the same species (e.g. in Petunia, Klahre et al., 2011), attracting different pollinators. Variations both in the total amount and specific composition of floral scent profiles have been detected during the floral life span, often with maximal scent production coinciding with reproductive organ maturity (Dudareva and Pichersky, 2000; Galliot et al., 2006). Thus scent emission normally ceases after pollination, accompanied by floral senescence. The peak of emission often also coincides with pollinator activity, for example scent emission from Antirrhinum (Dudareva et al., 2003) and petunia flowers (Hoballah et al., 2005; Verdonk et al., 2005) are both controlled by a circadian rhythm.
The most common components of floral bouquets are terpenoids and benzenoids (van Schie et al., 2006). Terpenoids are important components of floral scent in a wide range of species (Knudsen et al., 1993). They represent the largest and most diverse family of natural products, including >30 000 individual compounds, half of them synthesized by plants (Buckingham, 1998). All terpenoids are derived from the basic C5 units isopentenyl diphosphate (IDP) and dimethylallyl diphosphate (DMADP), which are synthesized both in the cytosol (Qureshi and Porter, 1981) and in the chloroplast (Rodríguez-Concepción and Boronat, 2002). During the second phase of terpene biosynthesis they give rise to geranyl diphosphate (GPP), farnesyl diphosphate (FPP), and geranylgeranyl diphosphate (GGPP) via elongation reactions (Koyama and Ogura, 1999; Liang et al., 2002). These are the precursors of monoterpenes, sesquiterpenes, and diterpenes, respectively, whose synthesis is catalysed by terpene synthase (TPS) enzymes, to generate the vast range of terpenoid hydrocarbons (Alleman, 2008). The large TPS family also includes enzymes able to catalyse the synthesis of multiple terpenoid structures (Steele et al., 1998; O’Maille et al., 2006; Degenhardt et al., 2009). TPS genes have been cloned from a variety of species and present two well-conserved motifs: an aspartate-rich region DDXXD that interacts with Mg2+ ions involved in positioning the substrate for catalysis (Little and Croteau, 2002; Seemann et al., 2002; Prosser et al., 2004; Christianson, 2006; Shishova et al., 2008), and an arginine-rich region R(R)X8W thought to be required for the cyclization of the GPP substrate (Williams et al., 1998). It was widely accepted that monoterpene and diterpene biosynthesis occurs only in plastids and sesquiterpene biosynthesis only in the cytoplasm. However, recent studies have shown a variable degree of cross-talk between compartments for isoprenoid synthesis (e.g. Bick and Lange, 2003; Lichtenthaler, 2007; Davidovich-Rikanati et al., 2008). Furthermore, some TPSs with a dual targeting signal sequence have been reported (Davidovich-Rikanati et al., 2008; Lee and Chappell, 2008). Thus, in principle, a TPS could utilize both GPP and FPP substrates to produce both/either kind(s) of monoterpene and/or sesquiterpene compounds based on its catalytic properties and access to substrate.
Analysis of TPS sequences suggests that they share a common evolutionary origin (Cseke et al., 1998). Seven gene subfamilies within the plant TPS gene family (Tpsa–Tpsg) have been identified based on their predicted amino acid sequences (Bohlmann et al., 1998), while an analysis of intron–exon structures groups TPS genes into three different classes: 12–14 introns (class I), 8–9 introns (class II), and six introns (class III) (Trapp and Croteau, 2001).
TPS genes associated with floral scent have been isolated from a number of model or economically important flower crops including Clarkia breweri (Dudareva et al., 1996), snapdragon (Dudareva et al., 2003; Nagegowda et al., 2008), lavender (Landmann et al., 2007), Arabidopsis (Tholl et al., 2004), and rose (Guterman et al., 2002). However, none to our knowledge is available from the ornamentally important Liliales. In several of these species, TPS gene expression is temporally regulated with flower opening, and TPS transcript levels increase just before terpenoid scent emission, suggesting that they are synthesized de novo.
Alstroemeria spp. originate in South America, with Chile and Brazil as the main centres of diversity (Bayer, 1987; Muñoz and Moreira, 2003). They are herbaceous, perennial, and rhizomatous plants with large colourful flowers, living in a wide range of habitats from rainforest to desert areas and from the Andes to the coast (Muñoz and Moreira, 2003). Alstroemeria has been a target for breeders who have developed many commercial cultivars. These are mainly sold as cut flowers but also as pot plants, and have been developed through interspecific hybridization (Burchi et al., 2000), selection of mutants, and polyploidization (Broertjes and Verboom, 1974). However, few native Alstroemeria species are scented, and use of a narrow genetic background in breeding programmes (Aros et al., 2006) including almost exclusively non-scented species resulted in the presence of only two scented cultivars in the European–American market. These are ‘Sweet Laura’, obtained though an interspecific cross between Chilean non-scented A. aurea and Brazilian scented A. caryophyllaea (Pounders et al., 2003; M. Bridgen, personal communication) and cv. ‘Ajax’, obtained from crossing A. caryophyllaea with six or seven other lines (R. Meijles, personal communication). Wild species of Alstroemeria have been described as naturally cross-pollinated by Hymenopteran insects; however, these studies have been performed only in the non-scented Chilean species A. aurea (Aizen and Basilio, 1998a, b), A. pallida (Cavieres et al., 1998), A. umbellata (Valdivia and Niemeyer, 2005), and A. ligtu (Botto-Mahan and Ojeda-Camacho, 2000). No information is available on pollinators of the Brazilian scented A. caryophyllaea.
In this work, the VOC emissions in scented genotypes of Alstroemeria related to the native A. caryophyllaea have been analysed in cut flowers, revealing that terpenes are the major scent components. This work focused on cut flowers due to their commercial relevance. Furthermore, an Alstroemeria TPS with myrcene synthase activity has been characterized. Its gene expression in scented Alstroemeria cv. ‘Sweet Laura’ tepals is well correlated with scent emission and it is likely to be responsible for the myrcene fraction of the Alstroemeria cv. ‘Sweet Laura’ fragrance.
Materials and methods
Plant materials and reagents
Flowers from four different genotypes of Alstroemeria were grown in a greenhouse (minimum temperature 14 °C) or obtained as cut flowers from commercial sources. The cv. ‘Rebecca’ is a non-scented cultivar belonging to Royal Van Zanten. Alstroemeria caryophyllaea is a scented species native to southern Brazil. The cv. ‘Sweet Laura’ is a scented cultivar derived from the A. aurea × A. caryophyllaea cross produced by Professor Mark Bridgen at Cornell University, USA. The cv. ‘Ajax’ is a scented cultivar bred by Könst with a pedigree including A. caryophyllaea as the only contributor of scent and another seven non-scented ancestors (R. Meijles, personal communication). Eight stages of floral development were defined according to Breeze et al. (2004): S0, coloured bud; S1, first tepal opening; S2, fully open flower; S3, three dehiscent anthers; S4, six dehiscent anthers; S5, open stigma; S6, loss of colour and wilting; and S7, abscission of wilted tepals. All reagents were from Sigma Aldrich (St Louis, MO, USA) unless stated otherwise.
Floral volatile analysis
To collect VOC emissions, flowers were enclosed in 300 ml flasks with 30 ml of distilled water under ambient temperature and light. Headspace VOCs were collected by solid-phase microextraction (SPME). Three SPME fibres were used: 100 μm, polydimethylsiloxane (PDMS) on 1 cm fused silica, optimized to extract low-boiling mostly non-polar compounds with a mol. wt of 30–225 Da (red fibre); 85 μm, polyacrylate on 1 cm fused silica optimized to extract low to medium boiling slightly polar compounds with a mol. wt of 80–300 Da (white fibre); and 50/30 μm divinylbenzene/carboxene/PDMS composite fibre on 2 cm fused silica for very volatile and low concentration compounds (grey fibre) (Sigma Aldrich). SPME fibres were exposed to the flowers in the flasks for 1 h (2.5 h and 17.5 h for the grey fibre). Fibres were injected manually and desorbed in the injection port of the gas chromatograph (GC) using a GC 8000 (Fisons, Ipswich, UK), as detailed here or a 6890 GC (Hewlett Packard, Palo Alto, CA, USA), as detailed below. The fibres were desorbed for 2 min at 220 °C in splitless mode. Samples were separated over a 30 m×0.25 mm ID×0.25 μm VF-23ms (FactorFour™, Varian) using the following temperature program: initial temperature 40 °C for 5 min, linear increase of 5 °C min−1 to 250 °C, followed by 5 min at 250 °C constant. Electron impact mass spectra were recorded in full scan mode from m/z 35 to 500 in an MD800 (Thermo-Finnigan, Manchester, UK) mass spectrometer (MS) coupled to the GC. Before each set of samples, the fibre was conditioned for 1 h at 200 °C in the injection port of the GC-MS and a fibre blank was recorded before sampling was started. A C8–C20 alkane standard solution was analysed regularly to provide retention time references for calculation of retention (Kovats) indices (RIs) and to monitor system performance. Data were analysed using Masslab v1.4 (Thermo-Finnigan). Signals were integrated using total ion count and quantities normalized to the largest signal in the chromatogram. Putative identification was achieved by comparing mass spectra with the NIST mass spectra library (v. 1.2) taking into account available information on Kovats indices. Further identification was achieved by comparing mass spectra with those of pure compounds tested on the same GC-MS machine using settings identical to those used for analysis of flowers, since co-injection is not possible when using SPME fibres. Analysis of samples on grey fibres and a further C8–C20 alkane standard was carried out on an Agilent GC-MS system (GC 6890, MSD 5973). Fibre samples were desorbed for 2 min at 240 °C in the injection port and standards were injected manually (0.1 μl, Hamilton) and separated on a 30 m×0.25 mm ID 0.25 μm HP-5MS (Agilent) using the following temperature program: initial temperature 35 °C for 2.5 min, linear increase of 3 °C min−1 to 140 °C and 16°C min−1 to 300 °C, followed by 1.5 min constant at 300 °C. Mass spectra were recorded from m/z 35 to 450 and data evaluated as before using Chemstation and the NIST mass spectra library. The following standard compounds were tested on both GC-MS machines as described above: myrcene, (–)-β-pinene, (+)-α-pinene, (Z)-ocimene (25% limonene), (+)-2-carene, (+)-3-carene, (+)-limonene, (E)-caryophyllene, and humulene.
All VOC collections were carried out during the morning. Further VOC collections aimed at identifying whether a circadian rhythm affects the floral scent emission were performed with cv. ‘Ajax’ and cv. ‘Sweet Laura’, performing at least two replicates in each case. Time points included morning (9:00–11:00 h GMT) and afternoon (17:00–18:00 h GMT) (collections were in July/August at 51°29′N 3°11′W).
Isolation of partial and full-length AlstroTPS cDNAs
A partial TPS cDNA clone of 310 bp (GenBank accession no. CF588473) was obtained from an Alstroemeria tepal S2-S0 subtracted library of cv. ‘Rebecca’ (Breeze et al., 2004). Using primers NCF327F (TAGAAAATGCAAGGAATTCGG) and NCF327R (TAGCCTTGCGGTCTTTGTTC) designed to this sequence. The homologous sequence was obtained from Alstroemeria cv. ‘Sweet Laura’ cDNA and used for 3′ and 5′ rapid amplification of cDNA ends (RACE) to obtain a full-length TPS clone from Alstroemeria cv. ‘Sweet Laura’, named AlstroTPS.
RNA was extracted essentially as described by Breeze et al. (2004), using ∼200 mg of plant material collected from at least three independent samples (flowers or leaves). DNA was removed using RQ1 DNase (Promega, Madison, WI, USA). For PCR amplification using the specific primers above, cDNA was synthesized from total RNA using oligo(dT) and M-MLV RT (Promega).
5′ and 3′-RACE-Ready cDNA was synthesized using the BD SMART™ RACE cDNA Amplification Kit (Clontech, Mountain View, CA, USA). The following primers: ALSTGSP1 (CACCTCTCTCTCCTTCTGCCTCTGAAGTTG), ALSTGSP2 (TGCATGAAACGGATGCTTCAGAGGTGATGG), ALSTGSP3 (CAGCTTCTCGAGCCTGGCAGCACATTCC), and ALSTGSP4 (GTTTGCAAATAGTTGTTGTTCCAGACGG) were used in two rounds of both 5′ and 3′ RACE. Thermal cycling was performed using a PTC-100 thermocycler (MJ Research Inc., Waltham, MA, USA) and the amplification was conducted with the following thermal profile: initial denaturation at 95 °C for 1 min; cycles of 1 min at 95 °C, 1 min at the appropriate primer Tm, 1 min at 72 °C; final extension at 72 °C for 15 min. The number of cycles was specific for each experiment.
RACE fragments were cloned into pGEM-EasyT (Promega), fully sequenced, and assembled using SEQMAN (DNASTAR, Lasergene, MA, WI) to provide a consensus sequence for the whole TPS cDNA. The consensus sequence was used to design primers to amplify 1620 bp of the open reading frame (ORF) (ALSTER-F: TAGAGGATCCACGCCGCTCGGCAAATTAT and ALSTER-R: CGATGCACGGTCGACTAGCAATATAGGTTCCAC), which also included BamHI and SalI, respectively, to facilitate cloning into the pET21b expression vector. Three independent clones were fully sequenced to check for PCR errors and the sequence was deposited in the EMBL database. Accession numbers for the sequences are FR822739 (1620 bp of the ORF) and FR822740 (N-terminal region). Signal peptide analysis was performed using ChloroP1.1 (Emanuelsson et al., 1999) and WoLF PSORT (Horton et al., 2007).
Real-time PCR
RNA was extracted as above and treated with DNase to eliminate contaminating genomic DNA. Removal of the DNA was tested by PCR of the treated RNA with 18S rRNA primers as described below. Forward and reverse primers were designed using Biotools (http://biotools.umassmed.edu/). For real-time quantitative reverse transcription-PCR (qRT-PCR). the primers selected for AlstroTPS were RTALSTERSF: ACTTTATGACGACTTGGGAACTTCA and RTALSTERSR: CCTTATAAACATGCGAGCCATCA, flanking a nucleotide sequence of 121 bp. The real-time qRT-PCR was conducted in a 6 μl volume containing 3 μl of SyBR Green Mix (Applied Biosystems, Warrington, UK), 5 pmol of each primer (Sigma Aldrich), and ∼20 ng of cDNA. Two biological replicates (i.e. cDNA synthesized from two separate RNA extractions) were tested, and triplicate reactions were carried out. As a negative control, a PCR containing water instead of cDNA was included for each set of PCRs. The 18S rRNA was used as a housekeeping gene amplified for data normalization, using two primers (Alstro18Sfor CGAACACTTACCACGACGACTCT; Alstro18Srev CGTTCAAAGACTCGATGGTTCAC) flanking a 120 bp DNA sequence. Amplifications were carried out using an ABI 7900 thermocycler (Applied Biosystems, Monza, Italy) with the following program: 50 °C for 2 min; 95 °C for 10 min; 45 cycles of 95 °C for 15 s and 58 °C for 45 s. Real-time PCR amplification was followed by a dissociation cycle (95 °C for 5 min; 60 °C for 1 min; 95 °C for 1 min) to validate the denaturation temperature of the amplicons. Calibration curves were calculated for the 18S rRNA and the target gene AlstroTPS using 10-fold serial cDNA dilutions. Relative transcript levels were calculated from the calibration curves and normalized against the 18S rRNA transcript level. Analysis of variance (ANOVA) on gene expression was performed using SPSS 17.0 for Windows, using Tukey’s HSD (honestly significant difference) test (P=0.05).
DNA extraction and analysis of gene structure and phylogeny
An unrooted phylogenetic tree based on amino acid alignments of AlstroTPS with 45 TPSs was constructed by the Neighbor–Joining method using MEGA 4.0.2 (Tamura et al., 2007) (Supplementary Table S1 available at JXB online).
Genomic DNA was extracted from young leaves of Alstroemeria cv. ‘Sweet Laura’, according to Dempster et al. (1999). Approximately 200 mg of plant material were placed into sterile 1.5 ml Eppendorf tubes and ground in liquid nitrogen with an Eppendorf pestle to a fine dust. Then, 700 μl of 2% cetyltrimethylammonium bromide (CTAB) buffer (100 mM TRIS-HCl, 1.4 M NaCl, 20 mM EDTA, 2% CTAB, 1% PVP, and 0.3% β-mercaptoethanol) were added and the samples were incubated for 30 min at 65 °C, mixing gently every 10 min. The DNA was extracted twice with 1 vol. (700 μl) of chloroform:isoamyl alcohol (24:1) and precipitated with 1 vol. (700 μl) of cold isopropanol. The pellet was washed with 200 μl of 70% ethanol by vortexing and then centrifuged at room temperature in a microcentrifuge. The air-dried pellet was resuspended in 50 μl of TE and treated with RNase (Invitrogen, Carlsbad, CA, USA).
Primers were designed using IDT Scitools, Oligo Analyzer 3.1 (http://eu.idtdna.com) to divide the AlstroTPS ORF sequence into segments that overlapped each other by ∼100 nucleotides (Supplementary Table S2 at JXB online). PCRs were set up as described above for cDNA using the following thermal profile in a PTC-100 machine: 95 °C for 1 min; 40 cycles of 95 °C for 1 min, 55 °C for 1 min, 72 °C for 1 min; final extension at 72 °C for 15 min. Genomic sequences obtained were assembled into a finished contig using the programs EDITSEQ and SEQMAN (DNASTAR), and aligned against the AlstroTPS ORF using BioEdit (v. 7.0.5.3) (Hall, 1999) to identify intron positions.
Expression of AlstroTPS in Escherichia coli and enzyme assays
A single colony of E. coli BL21 carrying the AlstroTPS 1620 bp ORF fragment in the pET-21 expression vector (see above) was inoculated into 100 ml of LB medium with antibiotic (100 μg ml−1 ampicillin). The culture was incubated overnight at 37 °C at 150 rpm. A 20 ml aliquot of overnight culture was used to inoculate 2 litres of fresh LB medium with antibiotic as above and the culture was again incubated at 37 °C at 150 rpm until it reached an OD600 of 0.6–0.8. Protein expression was induced by adding isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. After an additional 4 h incubation, the culture was centrifuged for 10 min at 4000 g and the pellet stored at –80 °C. To check for induction, samples from the cultures before and after induction were analysed by SDS–PAGE.
Pellets containing induced proteins were thawed on ice and resuspended in 100 ml of cell lysis buffer (20 mM TRIS, 5 mM EDTA, 5 mM β-mercaptoethanol, pH 8.0). The samples were sonicated and then centrifuged (4 °C, 30 000 g, 30 min). The protein was expressed as insoluble inclusion bodies, so the supernatant was discarded and the pellet resuspended in 50 ml of cell lysis buffer by stirring at 4 °C for 30 min. The pH of the solution containing the completely dissolved pellets was raised to 12 with 5 M NaOH. The solution was again stirred at 4 °C for 30 min and then the pH was adjusted to 8.0 with 1 M HCl, and β-mercaptoethanol was added to a final concentration of 5 mM. After 30 min of stirring at 4 °C, the solution was centrifuged (4 °C, 30 000 g, 30 min) and the supernatant dialysed overnight against 3 litres of dialysis buffer (10 mM TRIS-HCl, 5 mM β-mercaptoethanol, pH 7.0).
The protein solution was tested for TPS activity of recombinant AlstroTPS. The reaction mixture (∼500 μl) contained 400 μl of incubation buffer [25 mM HEPES, 15 mM MgCl2, and 5 mM dithiothreitol (DTT), pH 7.5], 100 μl of E. coli AlstroTPS extract, and 2 mM substrate (GPP or FPP). Two negative controls for each substrate were also performed using only buffer alone or with bovine serum albumin (BSA). A layer of 1 ml of pentane was added to the top of each reaction to trap the resulting organic compounds. The reaction was incubated for 24 h at 25 °C in sealed 1 ml vials. The olefin products were extracted three times with 1 ml of pentane and then passed through a short pad of silica gel (∼500 mg). The products were analysed with a GC-MS Hewlett Packard 6890 GC fitted with a J&W scientific DB-5MS column (30 m×0.25 mm internal diameter) and a Micromass GCT Premiere detecting in the range m/z 50–800 in EI+ mode, scanning once every second with a scan time of 0.9 s. Injections were performed in split mode (split ratio 5:1) using the following thermal profile: starting oven temperature 50 °C, temperature increase rate 4 °C min−1 up to 150 °C, and then at 20 °C min−1 for 5 min up to 250 °C final temperature. The identification of compounds was achieved by co-injection with synthetic standards (as above).
Results
Analysis of floral VOC emission profiles from Alstroemeria genotypes
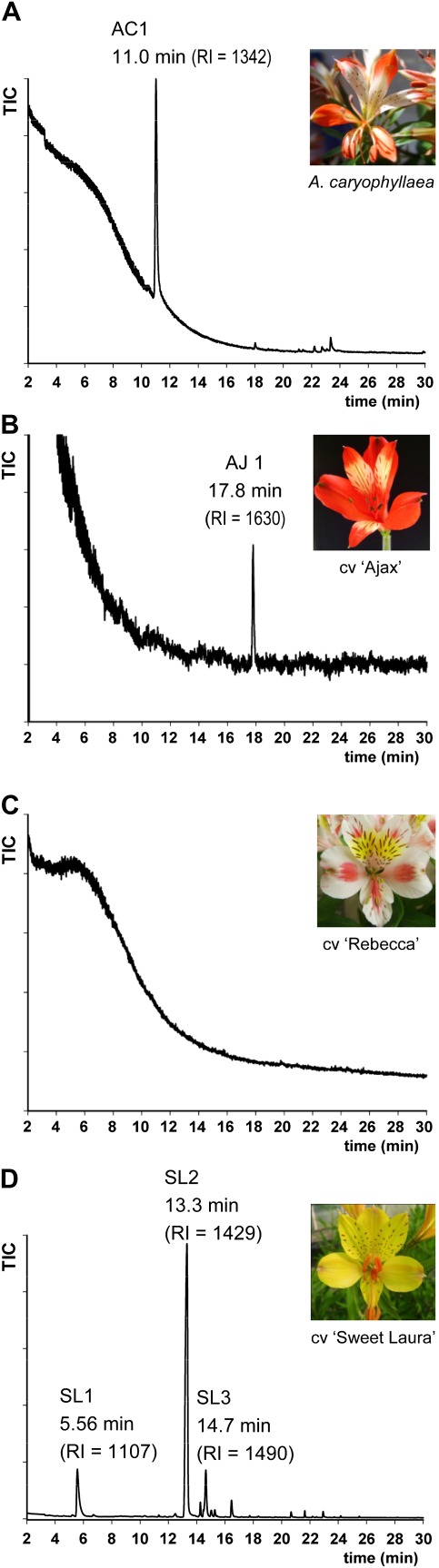
Preliminary investigations indicated that VOC emission in the scented genotypes was highest in open flowers, thus stage 4–5 flowers were selected for the detailed VOC analyses. At this stage all anthers are dehisced and the stigmatic lobes are separating to form the fully mature stigma. GC-MS analysis of the three scented accessions revealed genotype-specific VOC profiles. Only one volatile compound was detected from A. caryophyllaea (Fig. 1A, AC1), a sesquiterpene that could not be unequivocally identified from either NIST libraries or pure compounds tested. A single compound was also detected in cv. ‘Ajax’, which could be a terpene, but the concentration was very low and hence it was not possible to obtain a good mass spectrum from it (Fig. 1B, AJ1). However, its Kovats index (RI=1630) was different from those of terpenes detected in the other scented genotypes. No additional floral VOC compounds were detected in either of these two genotypes using two different SPME fibre types (100 μm PDMS or 60 μm polyacrylate). VOC analysis of the non-scented Alstroemeria cv. ‘Rebecca’ confirmed the lack of emission of VOCs at the detection level of the GC-MS (Fig. 1C).
Fig. 1.
GC trace of Alstroemeria floral VOC emission (stage 4–5). Peaks corresponding to terpene compounds are labelled. (A) Alstroemeria caryophyllaea, (B) cv. ‘Ajax’, (C) cv. ‘Rebecca’, (D) cv. Sweet Laura. (This figure is available in colour at JXB online.)
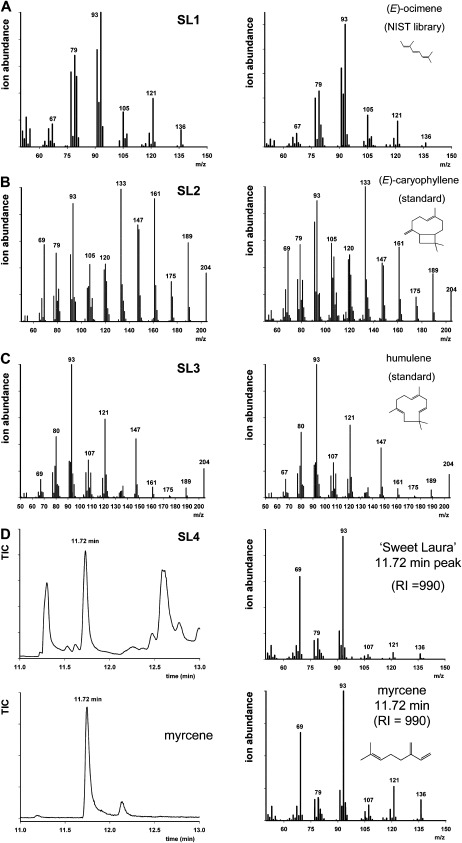
In contrast, the cv. ‘Sweet Laura’ chromatogram showed three major terpene peaks (Fig. 1D) as well as several minor peaks. The most abundant (SL2) was identified as the sesquiterpene (E)-caryophyllene, based on comparison of mass spectra and Kovats index with the pure compound (Fig. 2B). The second largest peak (SL1) was more problematic. The monoterpene (Z)-ocimene was suggested in an NIST library search to be a component of this fraction. (Z)-Ocimene could, however, be ruled out as a component of this peak as its RI (1107) was not consistent with the RI of the purified compound (1155) (Fig. 1D). (E)-Ocimene was identified from the NIST libraries as another possibility for this component (Fig. 2A), but the unavailability of a standard precluded an identification by comparison of the retention times and Kovats indices. A third major terpene peak (SL3) was identified as the sesquiterpene humulene (Fig, 2C) with an excellent match to the standard both in Kovats index and mass spectrum. To improve sensitivity of the detection, VOCs were re-analysed using a third SPME fibre (2 cm, DVB/CAR/PDMS) and a longer collection time with more flowers (2.5 h and 17.5 h). One of the minor terpene fractions (SL4) was identified as myrcene from its Kovats index and mass spectrum in comparison with the synthetic standard (Fig. 2D). Benzenoid aromatic compounds were also detected in the cv. ‘Sweet Laura’ floral headspace analysis but were not analysed further due to their low abundance.
Fig. 2.
GC-MS analysis of volatile emissions from Alstroemeria cv. ‘Sweet Laura’ flowers: (A–C) mass spectra for Alstroemeria cv. ‘Sweet Laura’ peaks SL1, SL2, and SL3, respectively, compared with the spectra from standards for SL2 and SL3, and the nearest match from the NIST libraries for SL1. (D) GC trace and mass spectra of Alstroemeria cv. ‘Sweet Laura’ minor peak SL4 and myrcene standard.
Since in many flowers VOC emission follows a circadian rhythm, emission levels of the main terpene VOCs from cv. ‘Sweet Laura’ and from cv. ‘Ajax’ were analysed at two time points (morning and afternoon). However, no clear pattern of emissions related to circadian rhythm was detected with either cultivar (data not shown).
Isolation and expression profile of an Alstroemeria TPS gene
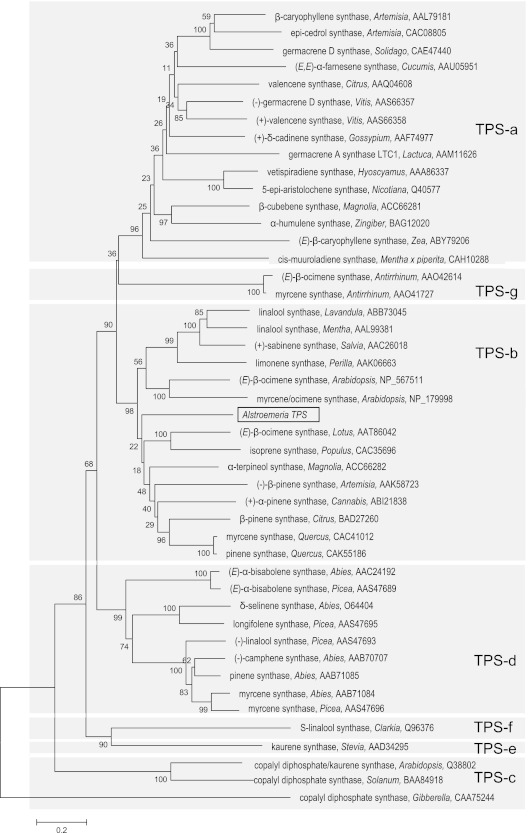
Given the prevalence of terpenoid compounds in scented Alstroemeria genotypes, four sequences related to TPS genes (which fell into two contigs) were selected for further study from expressed sequence tag (EST) collections derived from cv. ‘Rebecca’ tepals (Breeze et al., 2004). Amplification of the cognate sequence from cv. ‘Sweet Laura’ followed by successive rounds of 5' and 3' RACE yielded an ORF sequence of 1701 bp, similar in size to other TPS genes. Phylogenetic analysis of the predicted amino acid sequence compared with 45 TPS protein sequences available in the databases placed the Alstroemeria TPS (AlstroTPS) in the TPS-b family (Fig. 3). The AlstroTPS amino acid sequence was aligned with three closely homologous sequences: Magnolia grandiflora α-terpineol synthase (70% similarity, 50% identity), Quercus ilex myrcene synthase (67% similarity, 47% identity), and Arabidopsis thaliana myrcene/ocimene synthase (62% similarity, 40% identity). The AlstroTPS has the shortest sequence, with 567 amino acids compared with the M. grandiflora α-terpineol synthase (592 amino acids), the Q. ilex myrcene synthase (597 amino acids), and the A. thaliana myrcene/(E)-ocimene (591 amino acids) (Fig. 4). The conserved domain DDXXD is observed in all the sequences analysed and is located at D321 in the AlstroTPS. The arginine pair ‘RR’ of the R(R)X8W motif is also present in all the sequences but at different positions. ‘RR’ in the Alstroemeria TPS is located at R28, while in M. grandiflora, Q. ilex, and A. thaliana sequences it is located at amino acids 53, 56, and 45, respectively.
Fig. 3.
Phylogenetic tree of 46 plant TPS genes based on estimation of pair-wise distances at the amino acid level. The evolutionary history was inferred using the Neighbor–Joining method (Saitou and Nei, 1987). The optimal tree with the sum of branch length=16.56 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches (Felsenstein, 1985). The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the data set (Complete deletion option). There were a total of 398 positions in the final data set. Phylogenetic analyses were conducted in MEGA4 (Tamura et al., 2007). The six subfamilies proposed by Bohlmann et al. (1998) (TPS-a, -b, -c, -d, -e, and -f) and the additional subfamily TPS-g suggested by Dudareva et al. (2003) are shaded. AlstroTPS is shown boxed in black. Sequences were obtained from public databases and their details are shown in Supplementary Table S1 at JXB online.
Fig. 4.
Alignment of deduced amino acid sequences of AlstroTPS with three closely related sequences: Magnolia α-terpineol synthase, Quercus myrcene synthase, and Arabidopsis myrcene/ocimene synthase, TPS10 (protein accession nos ACC66282, CAC41012, and NP_179998, respectively). * indicates the conserved R(R)X8W motif; ** indicates the conserved DDXXD motif.
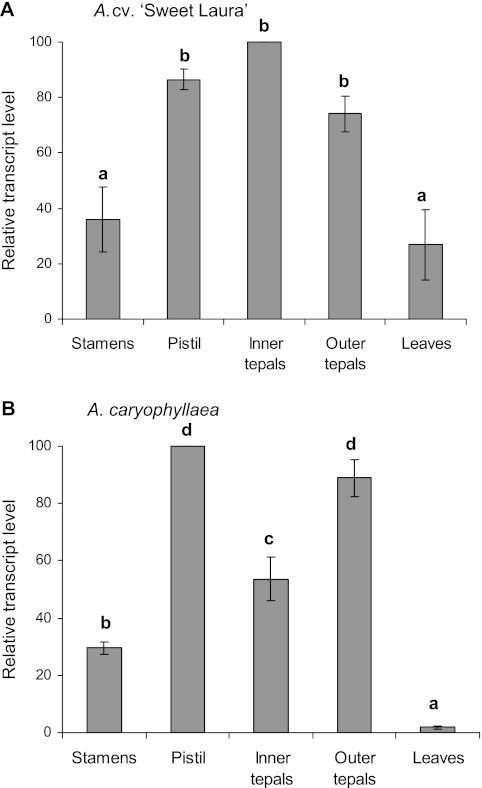
AlstroTPS was preferentially expressed in floral tissues of both cv. ‘Sweet Laura’ and A. caryophyllaea (Fig. 5). In cv. ‘Sweet Laura’ expression levels were maximal in tepals and pistils, intermediate in expression in stamens, and lowest in leaves (Fig. 5A). There was no significant difference in expression levels between inner and outer tepals. In A. caryophyllaea, the AlstroTPS expression level in leaves was similar to that in stamens; moreover, expression levels were significantly higher in outer tepals compared with inner tepals (Fig. 5B).
Fig. 5.
Spatial analysis of AlstroTPS gene expression in flower organs and leaves of (A) Alstroemeria cv. ‘Sweet Laura’ and (B) A. caryophyllaea, evaluated by semi-quantitative RT-PCR. Mean values (±SE, n=3) are expressed as a percentage of the maximum peak value and normalized against 18S rRNA.
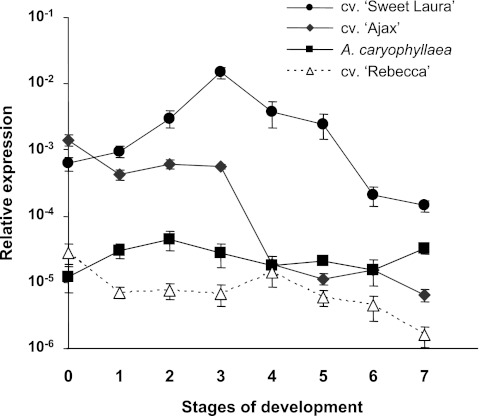
Analysis of AlstroTPS expression in tepals during flower development and senescence in the four genotypes revealed different patterns and levels of expression (Fig. 6). AlstroTPS expression was highest in cv. ‘Sweet Laura’ and lowest in the non-scented cv. ‘Rebecca’, with intermediate levels in cv. ‘Ajax’ and A. caryophyllaea. In all three scented genotypes, AlstroTPS expression peaked around stage 2–3, representing young to open flowers in which the first three anthers are reaching anthesis, and the stigma is not yet fully mature (stigmatic lobes as yet unseparated). This precedes the maximal scent production at stages 4–5 when the sexual organs are all fully mature. The peak in AlstroTPS expression was most notable in cv. ‘Sweet Laura’ where it was very marked. In cv. ‘Ajax’, TPS expression was high in early stages of tepal development and dropped dramatically after stage 3. In the non-scented cv. ‘Rebecca’, TPS expression was highest in young buds and gradually declined throughout development and senescence, except a minor peak at stage 4 (Fig. 6).
Fig. 6.
Developmental analysis of AlstroTPS gene expression in tepals of cvs. ‘Ajax’ ‘Sweet Laura’, ‘Rebecca’, and A. caryophyllaea, evaluated by quantitative real-time RT-PCR. Filled symbols, scented genotypes; open triangles, non-scented ‘Rebecca’. The eight developmental stages are described in the Materials and methods. Mean values (±SE, n=12) are given, following normalization with 18S rRNA transcript values (y-axis in log scale).
Intron–exon structure of AlstroTPS places it as an anomalous class III TPS
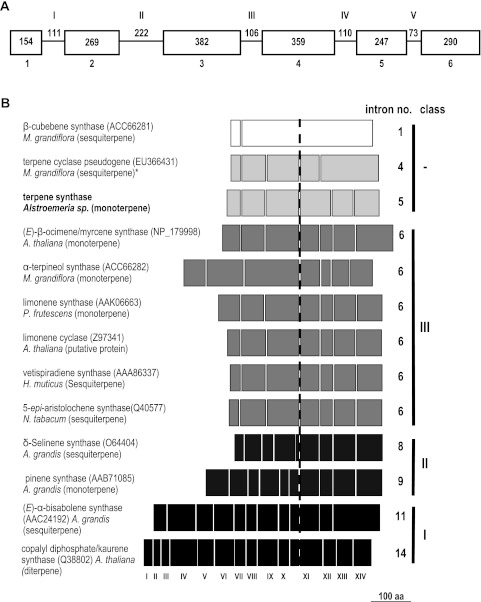
Using 12 PCR primers designed to the AlstroTPS ORF, a region of 2323 bp was amplified from cv. ‘Sweet Laura’ genomic DNA. Sequence alignment with the AlstroTPS cDNA revealed the presence of five introns (I–V, Fig. 7A). Comparison of the AlstroTPS intron–exon structure with that of other TPS genes revealed some unique features (Fig. 7B). According to the nomenclature based on A. thaliana copalyl disphosphate/kaurene synthase (Q38802; Sun and Kamiya, 1994), AlstroTPS has only five introns: III, VIII, XI, XIII, and XIV. While the intron position was conserved, introns of the TPS genes included in Fig. 7 showed a strong length polymorphism (Supplementary Table S3 at JXB online).
Fig. 7.
Intron–exon structure of (A) AlstroTPS alone and (B) AlstroTPS compared with other selected TPS genes. In (A), white boxes, exons (Arabic numerals); lines, introns (Roman numerals), both drawn to scale. Numbers represent the length (in bp) of each exon and intron. (B) Comparison of the AlstroTPS intron–exon structure with the structure of other TPS genes. Boxes show exons and are drawn to scale. Gaps between exons correspond to introns and are numbered at the bottom of the figure with Roman numerals. All the sequences have been aligned to intron XI (dashed line) of copalyl diphosphate/kaurene synthase (A. thaliana) as the following exon contains the highly conserved aspartate-rich domain (DDXXD), common to all plant TPSs. The number of introns and the TPS class (according to Trapp and Croteau, 2001) are shown on the right. Classes I, II, and III are highlighted in different colours, while ‘–’ identifies TPSs with unique intron–exon organization. An asterisk identifies the M. grandiflora sesquiterpene synthase, described as a member of a putative subclass III-a (Lee and Chappell, 2008). Sequences were obtained from public databases and their details are shown in Supplementary Tables S1 and S3 at JXB online.
Functional characterization of AlstroTPS as a myrcene synthase
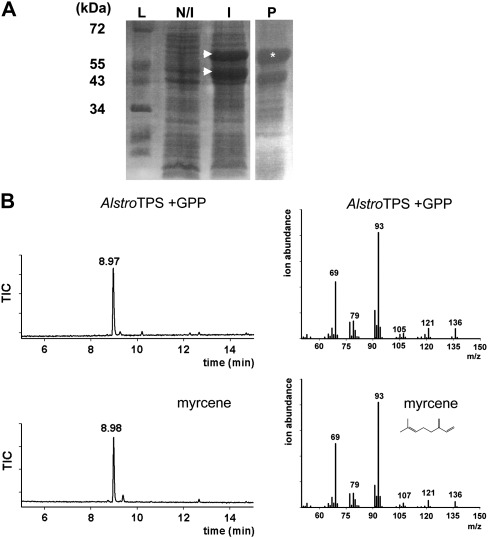
To determine whether AlstroTPS encoded a functionally active TPS, 1620 bp of its ORF, starting from the conserved arginine pair ‘RR’ of the R(R)X8W motif, was expressed in E. coli. Bacterial lysates were partially purified prior to enzymatic activity assays with GPP and FPP substrates. Two proteins of 63 kDa and 50 kDa were detected by SDS–PAGE (Fig. 8A). The upper band is consistent with the molecular weight of AlstroTPS (65.3 kDa) predicted from the amino acid sequence.
Fig. 8.
Detection of AlstroTPS protein and enzymatic assay of AlstroTPS. (A) Crude bacterial protein extracts showing induction of AlstroTPS on SDS–PAGE: (I induced, N/I non-induced), and partially purified AlstroTPS protein (P). Arrows indicate major bands after induction; an asterisk indicates a major protein band after partial purification (B) GC-MS mass spectra of the standard myrcene and the peak obtained from the AlstroTPS + GPP reaction.
Incubation of partially purified AlstroTPS protein with GPP resulted in a single product, whereas no peaks were evident following enzyme assays with FPP. The product from the reaction with the GPP substrate was identified by GC-MS as myrcene by comparing the mass spectra with those of a standard (Fig. 8B). Thus AlstroTPS is a myrcene synthase.
Discussion
Scent emission in Alstroemeria flowers
VOC analysis of scented Alstroemeria revealed very different profiles for the different genotypes. Alstroemeria caryophyllaea, the scented progenitor of both cvs ‘Sweet Laura’ and ‘Ajax’, produced only one sesquiterpene volatile compound which could not be unequivocally identified. In contrast the VOC profiles of cvs ‘Sweet Laura’ and ‘Ajax’ differed from that of the scented parent. Changes in scent profiles of hybrids have been observed in other species, for example in the orchid Ophrys, hybrids produce more and different compounds than their parents (Vereecken et al., 2010). This presumably reflects the complex pathways generating the volatile compounds and the polymorphic nature of the genes encoding the key enzymes regulating scent production. This polymorphism may be an important contributor to shifts in pollinator specificity during hybrid formation leading to reproductive isolation of new hybrids from their parents. Analysis of VOC patterns in hybrids is also a powerful method for investigating the genetic basis of VOC production and has been used successfully in Petunia to identify scent-associated quantitative trait loci (Stuurman et al., 2004) and genetic loci (Klahre et al., 2011).
The volatile profile of cv. ‘Sweet Laura’ was the most complex of the three scented genotypes analysed, the sesquiterpene (E)-caryophyllene being the most abundant compound. (E)-Caryophyllene is a common component of both floral scent and VOCs from green tissues (Knudsen et al., 1993), and has been identified as a VOC component in a wide range of taxonomically unrelated flowers such as Ophrys (Borg-Karlson et al., 1985), rose (Bu et al., 1987), and clover (Buttery et al., 1984). The VOC profile of cv. ‘Sweet Laura’ also contained two other major peaks, one of which was identified as humulene, and the other as an ocimene isomer. A minor fraction was identified as myrcene. All three of these compounds are also found in the volatile emissions of a range of taxonomically distinct flowers (http://www.pherobase.com).
Scent emission in cv. ‘Sweet Laura’ coincided with full anthesis of all six anthers and the final stages of maturation of the stigma; however, its regulation appeared not to be under circadian control, unlike Antirrhinum (Dudareva et al., 2003) or petunia (Verdonk et al., 2005). Further information on the behaviour of pollinators of native scented Alstroemeria genotypes would be needed to determine whether this constant production of volatiles reflects pollinator activity. Volatile emission can also be affected by growth conditions and temperature during volatile collection. In these experiments, a constant ambient temperature of 20 °C was maintained during volatile collection from cut flowers. In further work it would be interesting to test volatile profiles from flowers attached to the plant and under different environmental conditions.
Functional and phylogenetic characterization of an Alstroemeria TPS
The finding that mono- and sesquiterpenes were the major components of the Alstroemeria VOC profiles led to the investigation of the expression and function of TPS genes. The identified cv. ‘Sweet Laura’ TPS gene shared high homology with other TPS-encoding genes, and its expression profile and in vitro activity are consistent with a role for this gene in the production of a scent-related terpene.
AlstroTPS expression correlates well with floral VOC emission in scented genotypes. This correlation is both spatial and temporal, since AlstroTPS is predominantly expressed in floral organs of A. caryophyllaea and cv. ‘Sweet Laura’, with much lower expression in leaves. Furthermore, its transcript levels anticipate the VOC emission peak during development. This is in agreement with expression patterns of floral scent-related genes in other species, which are high in petals and/or pistils (Pichersky et al., 1994; D’Auria et al., 2002; Tholl et al., 2005). The AlstroTPS expression pattern in tepals also coincides with the beginning of reproductive organ maturity, falling off during tepal senescence stages when scent emission also declines. Expression of AlstroTPS in the non-scented cv. ‘Rebecca’ and in the scented cv. ‘Ajax’ was relatively high at early flower developmental stages. This is a similar pattern to that of a chloroplast TPS from non-scented Iris flowers (van Doorn et al., 2003), suggesting that early TPS expression in these genotypes could be associated with other roles not necessarily linked with the biosynthesis of scent volatiles. These could, for example, be related to the regulation of carotenoid (including red, orange, and yellow pigments) or plant hormone biosynthetic pathways (McGarvey and Croteau, 1995), both likely to be active in young flowers.
From the sequence alone it was not obvious whether AlstroTPS encoded a mono- or sesqui-TPS. The signal peptide is relatively short, which in other TPS genes has been associated with sesquiterpene synthases rather than monoterpene synthases, since in the latter the longer sequence encodes the plastid targeting peptide. However, bioinformatic analysis of the signal sequence predicted that the protein is plastid located. In addition, phylogenetic analysis placed AlstroTPS in the TPS-b subfamily, which includes only monoterpene synthases (Bohlmann et al., 1998). Although the 27 amino acid putative signal sequence for AlstroTPS is substantially shorter than the average of ∼49 residues for subfamily TPS-b, another two members of this subfamily have signal peptides of comparable length: the Arabidopsis (E)-β-ocimene synthase TPS03 (NP_567511) and the Lavandula angustifolia linalool synthase (ABB73045) with signal peptides of 25 and 26 amino acids, respectively. Phylogenetic analysis based on intron–exon structure placed AlstroTPS as an anomalous member of class III according to the classification of Trapp and Croteau (2001), together with both mono- and sesqui-TPS sequences. AlstroTPS lacks intron XII; however, analysis of exon size revealed that the length of AlstroTPS exon 4 corresponds to that of exon 4 plus exon 5 of class III TPS genes (Table 1), indicating that it might have resulted from the fusion of exons 4 and 5. Except for exon 1, the remaining exons of AlstroTPS are very similar in size to those of other class III TPS, consistent with its clustering in this class.
Table 1.
Comparison of the sizes of exons 4 and 5 of AlstroTPS with six other terpenoid synthases grouped in class III
| AlstroTPS | Myrcene/ocimene synthase (NP_179998) A. thaliana | α-Terpineol synthase (ACC66282) M. grandiflora | Limonene synthase (AAK06663) P. frutescens | Limonene cyclase (Z97341) A. thaliana | Vetispiradiene synthase (AAA86337) H. muticus | 5-Epi-aristolochene synthase (Q40577) N. tabacum | |
| Exon 4 | – | 73 | 73 | 73 | 73 | 73 | 73 |
| Exon 5 | 119 | 46 | 46 | 47 | 47 | 47 | 46 |
| Total | 119 | 119 | 119 | 120 | 120 | 120 | 119 |
Numbers indicate the number of amino acids for each exon and for the total (exon 4 + exon 5).
Since the major peaks of volatiles emitted from cv. ‘Sweet Laura’ included both mono- and sesquiterpenes, the enzyme activity of cv. ‘Sweet Laura’ recombinant TPS was tested to establish whether it could be responsible for one of the terpenoid compounds emitted. Substrate specificity and analysis of reaction products established that AlstroTPS encodes a monoterpene synthase with myrcene synthase activity. Myrcene was identified as a minor component of the cv. ‘Sweet Laura’ floral VOC profile, supporting a role for AlstroTPS as a functional myrcene synthase contributing to the complex volatile mixture emitted by this cultivar.
The isolation and characterization of scent emission profiles and TPS genes from Alstroemeria opens the way to a better understanding of the role of scent in the ecology and physiology, and the temporal and spatial regulation of VOC emission, in this species. It also provides opportunities for breeding and manipulation of scent related-enzymes in Alstroemeria hybrids to enhance the commercial value of this important cut flower.
Supplementary data
Supplementary data are available at JXB online
Table S1. Terpenoid synthases genes used for comparisons with AlstroTPS.
Table S2. Primers used for the amplification of genomic DNA comprising the AlstroTPS gene sequence showing their sequence (5′–3′), length (bp), and Tm (°C).
Table S3. Comparison of the size and position of the introns of AlstroTPS with other TPS genes.
Acknowledgments
This work was supported by the UK Biotechnology and Biological Sciences Research Council (BBSRC) through grant BB/G003572/1 (RKA, VG) and the School of Chemistry. We would like to thank Lyndon Tuck for his assistance with plant growth and propagation, Ronald Meijles from Könst for provision of material and useful discussions, Dr Rob Jenkins and Mike O’Reilly for invaluable technical assistance with the GC-MS, and Steve Hope for sequencing. DA was funded by a CONICYT Scholarship, Ministry of Education, Government of Chile.
References
- Aizen MA, Basilio A. Within and among flower sex-phase distribution in Alstroemeria aurea (Alstromeriaceae) Canadian Journal of Botany. 1998a;73:1986–1994. [Google Scholar]
- Aizen MA, Basilio A. Sex differential nectar secretion in protandrous Alstroemeria aurea (Alstroemeriaceae): is production altered by pollen removal and receipt? American Journal of Botany. 1998b;85:245–252. [PubMed] [Google Scholar]
- Allemann RK. Chemical wizardry? The generation of chemical diversity in terpenoid biosynthesis. Pure and Applied Chemistry. 2008;80:1773–1780. [Google Scholar]
- Arimura G, Ozawa R, Kugimiya S, Takabayashi J, Bohlmann J. Herbivore-induced defense response in a model legume. Two-spotted spider mites induce emission of (E)-β-ocimene and transcript accumulation of (E)-β-ocimene synthase in Lotus japonicus. Plant Physiology. 2004;135:1976–1983. doi: 10.1104/pp.104.042929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aros D, Meneses C, Infante R. Genetic diversity of wild species and cultivated varieties of Alstroemeria estimated through morphological descriptors and RAPD markers. Scientia Horticulturae. 2006;108:86–90. [Google Scholar]
- Bayer E. Mitteilungen der Botanischen. Staatsamml: Munchen; 1987. Die Gattung Alstroemeria in Chile; pp. 241–362. [Google Scholar]
- Bick JA, Lange BM. Metabolic cross talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: unidirectional transport of intermediates across the chloroplast envelope membrane. Archives of Biochemistry and Biophysics. 2003;415:146–154. doi: 10.1016/s0003-9861(03)00233-9. [DOI] [PubMed] [Google Scholar]
- Bohlmann J, Meyer-Gauen G, Croteau R. Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proceedings of the National Academy of Sciences, USA. 1998;95:4126–4133. doi: 10.1073/pnas.95.8.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg-Karlson A-K, Bergstriim G, Groth I. Chemical basis for the relationship between Ophrys orchids and their pollinators. I. Volatile compounds of Ophrys lutea and O. fusca as insect mimetic attractants/excitants. Chemica Scripta. 1985;25:283–294. [Google Scholar]
- Botto-Mahan C, Ojeda-Camacho M. The importance of floral damage for pollinator visitation in Alstroemeria ligtu L. Revista Chilena de Entomologia. 2000;26:73–76. [Google Scholar]
- Breeze E, Wagstaff C, Harrison E, Bramke I, Rogers H, Stead A, Thomas B, Buchanan-Wollaston V. Gene expression patterns to define stages of post-harvest senescence in Alstroemeria petals. Plant Biotechnology Journal. 2004;2:155–168. doi: 10.1111/j.1467-7652.2004.00059.x. [DOI] [PubMed] [Google Scholar]
- Broertjes C, Verboom H. Mutation breeding of Alstroemeria. Euphytica. 1974;23:39–44. [Google Scholar]
- Bu X, Huang A, Sun Y, Wu Z, Liu M. Essence constituents of Rosa chinensis flower. Acta Botanica Sinica. 1987;29:297–301. [Google Scholar]
- Buckingham J. Dictionary of Natural Products on CD-ROM. 1998. Version 6.1. London: Chapman & Hall. [Google Scholar]
- Burchi G, Mercuri A, Bianchini C, Bregliano R, Schiva T. New interspecific hybrids of Alstroemeria obtained through in vitro embryo-rescue. Acta Horticulturae. 2000;508:233–235. [Google Scholar]
- Buttery RG, Kamm JA, Ling LC. Volatile components of red clover leaves, flowers, and seed pods: possible insect attractants. Journal of Agricultural and Food Chemistry. 1984;32:254–256. [Google Scholar]
- Cavieres L, Peñaloza AP, Arroyo MTK. Efectos del tamaño floral y densidad de flores en la visita de insectos polinizadores en Alstroemeria pallida Graham (Amaryllidaceae) Gayana Botánica. 1998;55:1–10. [Google Scholar]
- Christianson DW. Structural biology and chemistry of the terpenoid cyclases. Chemical Reviews. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- Cseke L, Dudareva N, Pichersky E. Structure and evolution of linalool synthase. Molecular Biology and Evolution. 1998;15:1491–1498. doi: 10.1093/oxfordjournals.molbev.a025876. [DOI] [PubMed] [Google Scholar]
- D’Auria JC, Chen F, Pichersky E. Characterization of an acyltransferase capable of synthesizing benzylbenzoate and other volatile esters in flowers and damaged leaves of Clarkia breweri. Plant Physiology. 2002;130:466–476. doi: 10.1104/pp.006460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich-Rikanati R, Lewinsohn E, Bar E, Iijima Y, Pichersky E, Sitrit Y. Overexpression of the lemon basil α-zingiberene synthase gene increases both mono- and sesquiterpene contents in tomato fruit. The Plant Journal. 2008;56:228–238. doi: 10.1111/j.1365-313X.2008.03599.x. [DOI] [PubMed] [Google Scholar]
- Degenhardt J, Köllner TG, Gershenzon J. Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry. 2009;70:1621–1637. doi: 10.1016/j.phytochem.2009.07.030. [DOI] [PubMed] [Google Scholar]
- Dempster E, Pryor K, Francis D, Young J, Rogers H. Rapid DNA extraction from ferns for PCR based analysis. Biotechniques. 1999;27:66–68. doi: 10.2144/99271bm13. [DOI] [PubMed] [Google Scholar]
- Dudareva N, Cseke L, Blanc VM, Pichersky E. Evolution of floral scent in Clarkia: novel patterns of S-linalool synthase gene expression in the C. breweri flower. The Plant Cell. 1996;8:1137–1148. doi: 10.1105/tpc.8.7.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Martin D, Kish CM, Kolosova N, Gorenstein N, Faldt J, Miller B, Bohlmann J. (E)-beta-ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon: function and expression of three terpene synthase genes of a new terpene synthase subfamily. The Plant Cell. 2003;15:1227–1241. doi: 10.1105/tpc.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Pichersky E. Biochemical and molecular genetic aspects of floral scents. Plant Physiology. 2000;122:627–633. doi: 10.1104/pp.122.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Science. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fenster CB, Armbruster WS, Wilson P, Dudash MR, Thomson JD. Pollination syndromes and floral specialization. Annual Review of Ecology, Evolution and Systematics. 2004;35:375–403. [Google Scholar]
- Galliot C, Stuurman J, Kuhlemeier C. The genetic dissection of floral pollination syndromes. Current Opinion in Plant Biology. 2006;9:78–82. doi: 10.1016/j.pbi.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Guterman I, Shalit M, Menda N, et al. Rose scent: genomics approach to discovering novel floral fragrance-related genes. The Plant Cell. 2002;14:2325–2338. doi: 10.1105/tpc.005207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hoballah ME, Stuurman J, Turlings TCJ, Guerin PM, Connétable S, Kuhlemeier C. The composition and timing of flower odour emission by wild Petunia axillaris coincide with the antennal perception and nocturnal activity of the pollinator. Manduca sexta. Planta. 2005;222:141–150. doi: 10.1007/s00425-005-1506-8. [DOI] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Research. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U, Gurba A, Hermann K, Saxenhofer M, Bossolini E, Guerin PM, Kuhlemeier C. Pollinator choice in Petunia depends on two major genetic loci for floral scent production. Current Biology. 2011;21:730–739. doi: 10.1016/j.cub.2011.03.059. [DOI] [PubMed] [Google Scholar]
- Knudsen JT, Tollsten L, Bergstrom LG. Floral scents—a checklist of volatile compounds isolated by head-space techniques. Phytochemistry. 1993;33:253–280. [Google Scholar]
- Koyama T, Ogura K. Isopentenyl diphosphate isomerase and prenyltransferases. In: Cane DE, editor. Comprehensive natural products chemistry: isoprenoids including steroids and carotenoids. Vol. 2. Oxford: Pergamon; 1999. pp. 69–96. [Google Scholar]
- Landmann C, Fink B, Festner M, Dregus M, Engel K-H, Schwab W. Cloning and functional characterization of three terpene synthases from lavender (Lavandula angustifolia) Archives of Biochemistry and Biophysics. 2007;465:417–429. doi: 10.1016/j.abb.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Lee S, Chappell J. Biochemical and genomic characterization of terpene synthases in Magnolia grandiflora. Plant Physiology. 2008;147:1017–1033. doi: 10.1104/pp.108.115824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P-H, Ko T-P, Wang AH- J. Structure, mechanism and function of prenyltransferases. European Journal of Biochemistry. 2002;269:3339–3354. doi: 10.1046/j.1432-1033.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. Biosynthesis, accumulation and emission of carotenoids, α-tocopherol, plastoquinone, and isoprene in leaves under high photosynthetic irradiance. Photosynthesis Research. 2007;92:163–179. doi: 10.1007/s11120-007-9204-y. [DOI] [PubMed] [Google Scholar]
- Little DB, Croteau R. Alteration of product formation by directed mutagenesis and truncation of the multiple-product sesquiterpene synthases δ-selinene synthase and γ-humulene synthase. Archives of Biochemistry and Biophysics. 2002;402:120–135. doi: 10.1016/S0003-9861(02)00068-1. [DOI] [PubMed] [Google Scholar]
- McGarvey D J, Croteau R. Terpenoid metabolism. The Plant Cell. 1995;7:1015–1026. doi: 10.1105/tpc.7.7.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz M, Moreira A. Alstroemerias de Chile: diversidad, distribución y donservación. Santiago: Taller La Era; 2003. [Google Scholar]
- Nagegowda DA, Gutensohn M, Wilkerson CG, Dudareva N. Two nearly identical terpene synthases catalyze the formation of nerolidol and linalool in snapdragon flowers. The Plant Journal. 2008;55:224–239. doi: 10.1111/j.1365-313X.2008.03496.x. [DOI] [PubMed] [Google Scholar]
- O’Maille PE, Chappell J, Noel JP. Biosynthetic potential of sesquiterpene synthases: alternative products of tobacco 5-epi-aristolochene synthase. Archives of Biochemistry and Biophysics. 2006;448:73–82. doi: 10.1016/j.abb.2005.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E, Noel JP, Dudareva N. Biosynthesis of plant volatiles: nature’s diversity and ingenuity. Science. 2006;311:808–811. doi: 10.1126/science.1118510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E, Raguso RA, Lewinsohn E, Croteau R. Floral scent production in Clarkia (Onagraceae): I. Localization and developmental modulation of monoterpene emission and linalool synthase activity. Plant Physiology. 1994;106:1533–1540. doi: 10.1104/pp.106.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pounders C, Nyochembeng L, Brown E. Breeding Alstroemerias for the South. In: Plant Breeding & Evaluation. SNA Research Conference. 2003;48:482–484. [Google Scholar]
- Prosser I, Altug IG, Phillips AL, Konig WA, Bouwmeester HJ, Beale MH. Enantiospecific (+)- and (–)-germacrene D synthases, cloned from goldenrod, reveal a functionally active variant of the universal isoprenoid-biosynthesis aspartate-rich motif. Archives of Biochemistry and Biophysics. 2004;432:136–144. doi: 10.1016/j.abb.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Qureshi N, Porter JW. Conversion of acetyl-Coenzyme A to isopentenyl pyrophosphate. In: Porter JW, Spurgeon SL, editors. Biosynthesis of isoprenoid compounds. Vol. 1. New York: John Wiley & Sons; 1981. pp. 47–94. [Google Scholar]
- Rodríguez-Concepción M, Boronat A. Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiology. 2002;130:1079–1089. doi: 10.1104/pp.007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor–joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Seemann M, Zhai GZ, de Kraker JW, Paschall CM, Christianson DW, Cane DE. Pentalenene synthase. Analysis of active site residues by site-directed mutagenesis. Journal of the American Chemical Society. 2002;124:7681–7689. doi: 10.1021/ja026058q. [DOI] [PubMed] [Google Scholar]
- Shishova EY, Yu F, Miller DJ, Faraldos JA, Zhao Y, Coates RM, Allemann RK, Cane DE, Christianson DW. X-ray crystallographic studies of substrate binding to aristolochene synthase suggest a metal ion binding sequence for catalysis. Journal of Biological Chemistry. 2008;283:15431–15439. doi: 10.1074/jbc.M800659200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele CL, Crock J, Bohlmann J, Croteau R. Sesquiterpene synthases from grand fir (Abies grandis). Comparison of constitutive and wound-induced activities, and cDNA isolation, characterization and bacterial expression of δ-selinene synthase and γ-humulene synthase. Journal of Biological Chemistry. 1998;273:2078–2089. doi: 10.1074/jbc.273.4.2078. [DOI] [PubMed] [Google Scholar]
- Stuurman J, Hoballah ME, Broger L, Moore J, Basten C, Kuhlemeier C. Dissection of floral pollination syndromes in Petunia. Genetics. 2004;168:1585–1599. doi: 10.1534/genetics.104.031138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Kamiya Y. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. The Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tholl D, Chen F, Petri J, Gershenzon J, Pichersky E. Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers. The Plant Journal. 2005;42:757–771. doi: 10.1111/j.1365-313X.2005.02417.x. [DOI] [PubMed] [Google Scholar]
- Tholl D, Kish CM, Orlova I, Sherman D, Gershenzon J, Pichersky E, Dudareva N. Formation of monoterpenes in Antirrhinum majus and Clarkia breweri flowers involves heterodimeric geranyl diphosphate synthases. The Plant Cell. 2004;16:977–992. doi: 10.1105/tpc.020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp SC, Croteau RB. Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics. 2001;158:811–832. doi: 10.1093/genetics/158.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia CE, Niemeyer HM. Reduced maternal fecundity of the high Andean perennial herb Alstroemeria umbellata (Alstroemeriaceae) by aphid herbivory. New Zealand Journal of Ecology. 2005;292:321–324. [Google Scholar]
- van Doorn WG, Balk PA, van Houwelingen AM, et al. Gene expression during anthesis and senescence in Iris flowers. Plant Molecular Biology. 2003;53:845–863. doi: 10.1023/B:PLAN.0000023670.61059.1d. [DOI] [PubMed] [Google Scholar]
- van Schie CCN, Haring MA, Schuurink RC. Regulation of terpenoid and benzenoid production in flowers. Current Opinion in Plant Biology. 2006;9:203–208. doi: 10.1016/j.pbi.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Verdonk J C, Haring MA, van Tunen A J, Schuurink RC. ODORANT1 regulates fragrance biosynthesis in petunia flowers. The Plant Cell. 2005;17:1612–1624. doi: 10.1105/tpc.104.028837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vereecken NJ, Cozzolino S, Schiestl FP. Hybrid floral scent novelty drives pollinator shift in sexually deceptive orchids. BMC Evolutionary Biology. 2010;10:103–114. doi: 10.1186/1471-2148-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DC, McGarvey DJ, Katahira EJ, Croteau R. Truncation of limonene synthase preprotein provides a fully active ‘pseudomature’ form of this monoterpene cyclase and reveals the function of the amino-terminal arginine pair. Biochemistry. 1998;37:12213–12220. doi: 10.1021/bi980854k. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving genes and proteins. New York: Academic Press; 1965. pp. 97–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.