Abstract
HYL1 is an important regulator of microRNA (miRNA) biogenesis. A loss-of-function mutation of HYL1 causes the reduced accumulation of some miRNAs but fails to display the miRNA-deficient phenotypes of these miRNAs. In Arabidopsis, miR156 mediates phase transition through repression of SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE (SPL) genes. However, it remains unknown whether, and if so how, HYL1 enables phase transition through miR156. This study showed that a loss-of-function mutation of the HYL1 gene caused defects in the timing of the juvenile phase. In the primary leaves of hyl1-2 mutants, abaxial trichomes were generated prematurely, the leaf blades elongated, and the blade base angles enlarged, as is observed for adult leaves. In hyl1-2 p35S::miR156a and hyl1-2 spl9-4 spl15-1 plants, increased accumulation of miR156a and repressed expression of the SPL genes were concomitant with a complete or partial rescue of the hyl1-2 phenotype in phase defects. In contrast, overexpression of the SPL9 gene in hyl1-2 mutants led to total disappearance of the juvenile phase. Moreover, HYL1 prevented the premature accumulation of adult-related transcripts in the primary leaves. Taken together, these results suggest that HYL1 controls the expression levels of miR156-targeted SPL genes and enables plants to undergo the juvenile phase, an important and critical step during plant development to ensure maximum growth and productivity.
Keywords: HYL1, juvenile phase, miR156, SPL9, vegetative growth
Introduction
Plants undergo several developmental transitions, including the transition from an embryonic to post-embryonic mode of growth, the juvenile-to-adult vegetative transition, and the vegetative-to-reproductive transition. The juvenile phase is an important and critical step during plant development to ensure maximum growth and productivity. The juvenile-to-adult-phase transition (vegetative change) usually involves changes in a variety of species-specific traits, including leaf shape, presence of trichomes and thorns, production of phytochemicals, leaf retention, internode length, and disease and pest resistance (Willmann and Poethig, 2011). The timing of the transition from the juvenile to the adult phase is significantly influenced by environmental cues, such as day length, light intensity, ambient temperature, and the plant hormone gibberellic acid (Willmann and Poethig, 2005). However, this transition is also controlled genetically. Recently, genetic analyses of developmental maturation in plants have advanced significantly, and some of the genetic elements involved in phase transition have been identified.
One of these identified genetic elements is the microRNA (miRNA) miR156, which is highly expressed early in shoot development and decreases with time. Overexpression of miR156 prolongs the juvenile-phase length and delays flowering (Wu and Poethig, 2006; Chuck et al., 2007; Wang et al., 2009). miR156 targets the SQUAMOSA PROMOTER BINDING-LIKE (SPL) transcription factors, which influence developmental transitions. In Arabidopsis thaliana, there are 16 members of the SPL family of transcription factors. Ten of the miR156-targeted SPL genes are grouped into different clades: SPL3/SPL4/SPL5, SPL2/SPL10/SPL11, SPL6/SPL13 and SPL9/SPL15 (Guo et al., 2008).The first clade consists of SPL3, SPL4, and SPL5, which encode small proteins that contain a SQUAMOSA promoter DNA-binding protein domain. Constitutive overexpression of SPL3, SPL4, or SPL5 accelerates the juvenile-to-adult-phase change and advances flowering time (Wu and Poethig, 2006), whereas overexpression of SPL9 and its closely related paralogue SPL15 not only shortens the juvenile phase and produces an early flowering phenotype but also affects changes in the cell size and cell number typical of adult leaves (Usami et al., 2009). In addition, the gain-of-function phenotype of SPL10 or its closely related paralogues SPL2 and SPL11 also affects leaf initiation and developmental transitions (Shikata et al., 2009). Although it is well known that the juvenile-to-adult transition depends on the accumulation of miR156 (Chuck et al., 2007; Wang et al., 2008; Wu et al., 2009), the upstream elements that determine the temporal and spatial expression patterns of miR156 remain unclear.
The processing of miRNAs from longer primary transcripts (pri-miRNAs) requires the activity of several proteins, including DICER-LIKE1 (DCL1), the double-stranded RNA-binding protein HYPONASTIC LEAVES1 (HYL1), and the zinc-finger protein SERRATE (SE) (Hiraguri et al., 2005; Kurihara et al., 2006; Lobbes et al., 2006; Dong et al., 2008; Yang et al., 2010; Machida et al., 2011). All three proteins are found in nuclear processing centres called D-bodies (Fang and Spector, 2007; Fujioka et al., 2007; Song et al., 2007). Mutants deficient in miRNA biogenesis exhibit increased pri-miRNA levels and reduced amounts of mature miRNAs, and suffer from a wide range of morphological defects (Han et al., 2004; Vazquez et al., 2004; Lobbes et al., 2006; Yang et al., 2006). The loss-of-function mutants hyl1 and hyl1-2, which are disrupted by Ds and T-DNA insertions, respectively, display a series of mutant phenotypes such as shorter stature, narrower hyponastic leaves, delayed flowering, and reduced fertility (Lu and Fedoroff, 2000; Vazquez et al., 2004).
Changes in the transition from the juvenile to the adult phase are reported in loss-of-function mutants of SE (Clarke et al., 1999). Similarly, a mutation in a recessive allele of ARGONAUTE1 (AGO1), which encodes the enzyme responsible for cleavage of miRNA transcripts in Arabidopsis, significantly affects leaf initiation in plants (Wang et al., 2008). However, it remains unknown whether the juvenile-to-adult-phase transition is affected in hyl1 mutants. Many miRNAs act by silencing their target genes, and deregulation of these miRNAs usually produces morphological changes. However, deregulation of some miRNAs that occur in hyl1 mutants does not lead to obvious phenotypes (Liu et al., 2011). The morphological interaction between HYL1-regulated miRNA biogenesis and miRNA-mediated gene silencing is complicated. To answer whether, and if so how, HYL1 controls the miR156-regulated phase transition, we conducted a genetic and molecular analysis of HYL1, as well as miR156 and its targets. The results indicated that HYL1 functions as an upstream regulator of miR156 in maintenance of the juvenile phase.
Materials and methods
Plant material and growth conditions
The wild-type and mutant A. thaliana plants used in this study were of the Columbia ecotype (Col-0), unless otherwise indicated. The plasmids pSPL9::rSPL9 and pSPL10::rSPL10 were gifts from Dr R. Scott Poethig (University of Pennsylvania, PA, USA), and plasmid p35S::rSPL3 was a gift from Dr Detlef Weigel (The Salk Institute for Biological Sciences, CA, USA).
Seeds were sown and grown at 22 °C under long (16 h light/8 h dark) and short (8 h light/16 h dark) day conditions. hyl1-2 mutant T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center (OH, USA). The spl9-4 spl15-1 mutant was a gift from Dr Scott Poethig (Wu et al., 2009). Seeds were surface sterilized in 70% ethanol for 1 min, followed by 0.1% HgCl2 for 10 min, and then washed four times in sterile distilled water and plated in molten 0.1% agar in water on top of solid 1% sugar MS0 medium. The plates were sealed with Parafilm, incubated at 4 °C in the dark for 2–3 d, and then moved to a growth chamber at 22 °C with 8 h light (short day).
For phenotypic observations, the seeds were sown in pots with peat soil and grown in a growth chamber under the same conditions. The blade width, blade length, and petiole length were measured. For leaf-shape analysis, fully expanded leaves were removed, attached to A4 paper with double-sided tape, flattened, and measured using vernier calipers. The abaxial trichomes were scored 60 d after planting, using a stereomicroscope.
Transgenic plants
Fragments corresponding to precursor of miR156a were inserted into pCAMBIA3301 binary vectors and placed under the control of the cauliflower mosaic virus 35S promoter. The binary constructs were delivered into Agrobacterium tumefaciens strain GV3101 (pMP90RK) using a freeze–thaw method (Weigel and Glazebrook, 2006). The Arabidopsis plants were transformed using a flower-dip method (Clough and Bent, 1998). For selection of transgenic plants, the seeds were sterilized and germinated on agar medium containing 50 mg l−1 kanamycin and 10 mg l−1 phosphinothricin. Seedlings conferring resistance to the herbicide Basta or to kanamycin were transplanted in a greenhouse and grown at 22 °C under an 8 h light regime. The transgenic plants were selfed for at least three generations, and the seeds from each plant were harvested separately for subsequent observations.
Small-RNA deep sequencing
All hyl1 plants were grown under a 16 h light/8 h dark photoperiod at 22 °C for 3 weeks. RNA samples from the aboveground parts of the seedlings were prepared using the Illumina Alternative v1.5 Protocol, and small-RNA deep sequencing was performed using an Illumina GAII sequencer and the mirVana™ miRNA Isolation Kit (Ambion).
Real-time PCR
Total RNA was extracted with TRIzol (Invitrogen) and treated with DNase I (Takara) to remove DNA contamination. Approximately 4 μg RNA was used for reverse transcription with oligo(dT) primers. Real-time PCR was performed using specific pairs of primers (see Supplementary Table S3, available in JXB online. The comparative threshold cycle (Ct) method was used to determine relative transcript levels in the real-time PCR (MyiQ2 Two-color Real-time PCR Detection System; Bio-Rad). Three biological replicates and three technical replicates were performed.
In situ hybridization
HYL1 full-length coding sequence was PCR amplified and cloned into pBluescript SK. Digoxigenin-labelled sense and antisense probes were synthesized with T7 or T3 RNA polymerase (Takara). For the miR156a probe, locked nucleic acid-modified probes of miR156a were synthesized and labelled with digoxigenin at the 3′ end by TaKaRa and used for in situ hybridization. Shoot apices from 20-d-old short-day-grown wild-type plants were prepared following pre-treatment and hybridization methods described previously (Liu et al., 2011).
Northern blotting
A volume containing 30–50 μg RNA was separated on 19% polyacrylamide denaturing gels. The RNA was transferred to a Hybond membrane (Amersham Biosciences, GE Healthcare) for 2 h at 200 mA. After cross-linking by UV irradiation for 3 min, the Hybond membrane was hybridized with biotin-labelled DNA probes complementary to the predicted miRNA sequences at 42 °C overnight. The membrane was washed at 42 °C twice with 2× SSC and 0.1% SDS, followed by two higher stringency washes of 0.1× SSC and 0.1% SDS at 42 °C. Subsequently, the membrane was incubated with a stabilized streptavidin–horseradish peroxidase conjugate (Thermo Scientific) in nucleic acid detection blocking buffer and then washed five times with 1× washing buffer. After washing with substrate equilibration buffer and adding stable peroxide solution and enhancer solution, the blots were imaged using an FLA-5000 Phosphorimager (FujiFilm). To confirm uniform loading, the blots were also probed with a DNA probe complementary to the U6 gene as a control. The DNA probes used for small-RNA northern blotting were synthesized and biotin-labelled using a 3′-end DNA labelling method (Invitrogen).
Microarray analysis
The hyl1-2 and Nossen ecotype seedlings were grouped randomly and grown under identical conditions for 3 weeks. Each group contained five seedlings and was designated as one biological replicate. Six biological replicates for each plant line were prepared, and three were used for array experiments.
GeneChip array analysis was performed using Affymetrix ATH1 microarrays (ATH1 121501). Differentially expressed genes were identified from expression data acquired from six independent microarray hybridizations. Three replicates of hyl1 RNA and wild-type reference RNA were used to calculate the expression values for each gene. For analysis of the expression profile, the fold changes in the normalized signals derived from three hyl1 replicates and three wild-type replicates were calculated. Only the fold changes of genes that met a significance criterion of P <0.05 with a fold change of >1.5 are presented.
Results
HYL1 maintains the juvenile phase of primary leaves
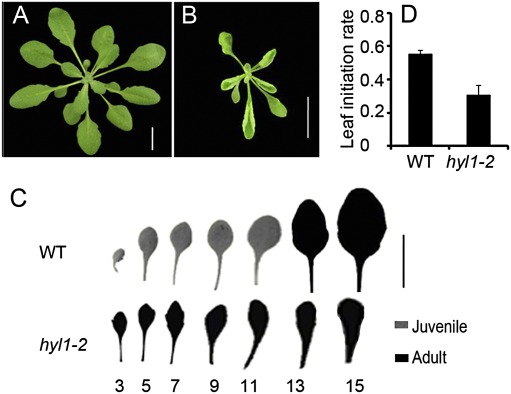
hyl1 mutants are deficient in miRNA processing and exhibit pleiotropic abnormalities that include a shorter stature and hyponastic leaves (Lu and Fedoroff, 2000). hyl1-2 is a hypomorphic allele of hyl1, and both share the same mutant phenotypes. Under short-day conditions, the vegetative period of hyl1-2 mutants was shortened compared with the wild type, and all of the rosette leaves curved upward, making it difficult to observe transition from the juvenile to the adult phase (Fig. 1A, B). To examine whether mutation of the HYL1 gene altered vegetative phase transition, we carried out phenotyping of the hyl1-2 plants under short-day conditions. In wild-type plants, abaxial trichomes were not observed on the first nine leaves but became evident on the 13th leaves and progressively numerous on the 15th leaves and thereafter (Fig. 2A). According to the timing of abaxial trichomes, the first nine leaves were juvenile and the 13th and subsequent leaves were adult (Fig. 1C). Among these leaves, the first two leaves (primary leaves) and third leaves were representative of the early juvenile phase, and the 13th to 15th leaves were representative of the adult phase, and thus we paid particular attention to the first three leaves and the 13th to 15th leaves. Compared with wild-type plants, the juvenile phase of the hyl1-2 plants was defective. Abaxial trichomes were observed on most of the primary leaves and on all of the third leaves of hyl1-2 plants (Fig. 2A). Precisely, the abaxial trichomes on hyl1-2 leaves were produced earlier than those in wild-type plants by 12 plastochrons. These observations demonstrated that the primary leaves of hyl1-2 plants had acquired adult characteristics. In addition, the leaf shape index (length-to-width ratio) of the third leaf of the hyl1-2 mutants resembled that of the 13th wild-type leaves. Due to their increased length, the hyl1-2 blades appeared narrow and elongated. Moreover, the leaf initiation rate of the mutant plants was lower than that of wild-type plants (Fig. 1D). Taken together, these results showed that most of the primary leaves in hyl1-2 plants were transformed from juvenile to adult leaves.
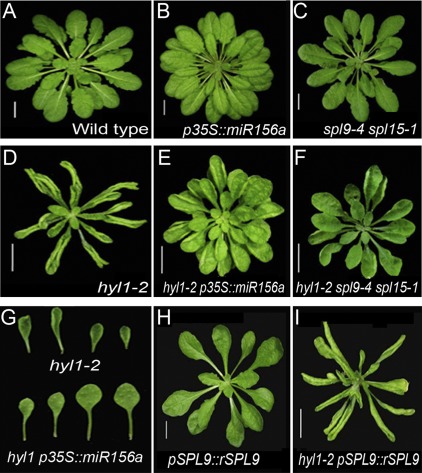
Fig. 1.
Early phase transition of hyl1-2 mutants. (A, B) Wild-type (A; WT) and hyl1-2 (B) seedlings 30 d after germination (DAG). (C) Fully expanded leaves showing the leaf shape. Black leaves represent the adult phase, while grey leaves represent the juvenile phase; the numbers beneath are the leaf order counting from the primary leaves (the first two rosette leaves). Bar, 1 cm. (D) Leaf initiation rate (calculated by the formula: number of leaves initiated per plant/DAG) of hyl1-2 30-d-old plants. The number of plants observed was >30. Results are shown as means ±SD. (This figure is available in colour at JXB online.)
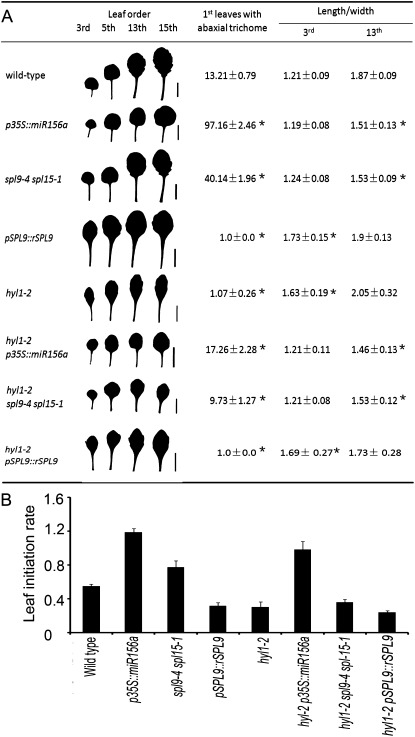
Fig. 2.
Parameters of hyl1-2 and transgenic plants, and triple mutants. (A) Leaf shape, abaxial trichomes, and leaf length-to-width ratios of hyl1-2 and related plants. Bars, 1 cm. (B) Leaf initiation rate of hyl1-2, hyl1-2 p35S::miR156a, and hyl1-2 pSPL9::rSPL9 plants (n=30). Asterisks indicate significant differences from the wild type (P <0.01) and results are shown as means ±SD.
HYL1 determines the temporal and spatial accumulation of miR156 in the juvenile phase
HYL1 is one of the major regulators of miRNAs in plants. To verify the function of HYL1 in the production of miR156 and other related miRNAs, we examined the abundance of small RNAs in hyl1 and wild-type seedlings (Nossen ecotype) by small-RNA deep sequencing. This high-throughput sequencing identified a total of 5.3 million and 6.2 million small RNA sequences from samples from hyl1 and wild-type seedlings, respectively. We normalized each dataset of small RNAs to transcripts per 5 million and observed that the hyl1 and wild-type datasets contained 61 and 77 miRNAs, respectively, that are conserved in Arabidopsis. The expression of most of these miRNAs was downregulated in the hyl1 seedlings compared with the wild-type (>1.5-fold), and among them, miR156a–f were the most downregulated (>10-fold) (Table 1). As miR156 and miR157 target SPL genes, we focused on differences in the expression of members of the miR156 family. The mature sequences of miR156g and miR156h presented several mismatches with miR156a, and these miRNAs were regulated differentially by HYL1. The expression of miR156g was downregulated 3.5-fold in the hyl1 seedlings. In contrast to miR156a and miR156g, expression of miR156h was not downregulated in the hyl1 seedlings. Nevertheless, miR156h was less abundant than miR156a in both the wild-type and hyl1 seedlings. As expected, miR157a–c and miR157f were downregulated in the hyl1 seedlings. Differential abundance of miR156 members might result in different silencing of the target genes. However, the total accumulation of miR156 in hyl1 seedlings was much lower than in the wild type.
Table 1.
Down- and upregulation of miRNAs in hyl1 seedlings relative to the wild type
| miRNA | Sequence | Reads |
hyl1/WT | ||
| WT | hyl1 | ||||
| Ten miRNAs most deregulated | |||||
| 1 | miR156a–f | TGACAGAAGAGAGTGAGCAC | 129 754 | 12 776 | 0.098 |
| 2 | miR158a | TCCCAAATGTAGACAAAGCA | 91 804 | 30 001 | 0.327 |
| 3 | miR167 | TGAAGCTGCCAGCATGATCTA | 29 246 | 24 066 | 0.403 |
| 4 | miR166a–g | TCGGACCAGGCTTCATTCCCC | 5 736 | 2 312 | 0.403 |
| 5 | miR157d | TGACAGAAGATAGAGAGCAC | 3 733 | 2 237 | 0.599 |
| 6 | miR843a | TTTAGGTCGAGCTTCATTGGA | 3 474 | 1 451 | 0.418 |
| 7 | miR161a.2 | TTGAAAGTGACTACATCGGGG | 2 587 | 552 | 0.213 |
| 8 | miR168 | TCGCTTGGTGCAGGTCGGGAA | 2 360 | 6 312 | 2.675 |
| 9 | miR396b | GCTCAAGAAAGCTGTGGGAAA | 2 291 | 761 | 0.332 |
| 10 | miR172a–b | AGAATCTTGATGATGCTGCAT | 2 123 | 2 514 | 1.184 |
| Different types of miR156 and miR157 | |||||
| 1 | miR156a–f | TGACAGAAGAGAGTGAGCAC | 129 754 | 12 776 | 0.098 |
| 2 | miR156g | CGACAGAAGAGAGTGAGCAC | 145 | 42 | 0.290 |
| 3 | miR156h | TGACAGAAGAAAGAGAGCAC | 10 | 11 | 1.100 |
| 4 | miR157a–c | TTGACAGAAGATAGAGAGCAC | 76 378 | 57 268 | 0.750 |
| 5 | miR157d | TGACAGAAGATAGAGAGCAC | 3 733 | 22.37 | 0.599 |
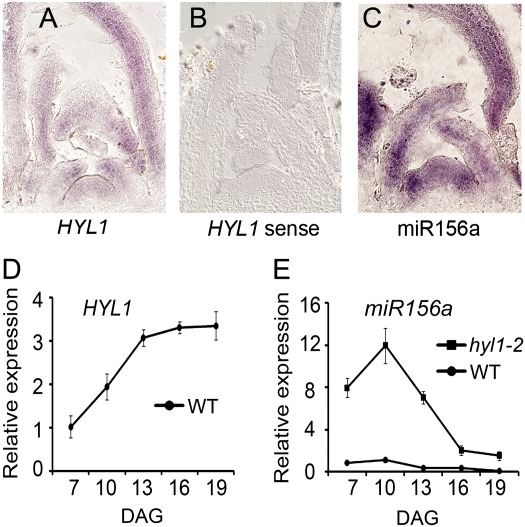
The vegetative-phase transition depends on the accumulation of miR156 and temporal changes in the expression of miR156-targeted SPL genes (Wu et al., 2009). If HYL1 controls the expression of miR156, the spatial and temporal patterns of miR156 should be correlated with those of HYL1. To address this point, we performed in situ hybridization and quantitative real-time PCR analyses of the HYL1 gene. The HYL1 gene was expressed preferentially in the shoot apical meristem, leaf primordial, and growing leaves (Fig. 3A, B). Real-time PCR revealed that HYL1 expression increased rapidly in 7–13-d-old seedlings and was maintained at relatively high levels from d 13 to 19 (Fig. 3D). In situ hybridization showed that miR156a accumulated in the same regions as HYL1 expression, except that stronger accumulation was observed in the shoot apical meristem (Fig. 3C). To confirm whether the accumulation of miR156a was upregulated during the vegetative phase, we harvested the shoot apices of hyl1-2 plants, and used small-RNA northern blotting to detect the accumulation of miR156a at various stages (10-, 20-, 30-, 40-, and 50-d-old seedlings). In wild-type plants, the levels of miR156a accumulation were highest at 10 d of age and subsequently declined over time. Similarly, the accumulation of miR156a in hyl1-2 seedlings decreased from 10 to 20 d. However, the accumulation of miR156a in hyl1-2 seedlings was much lower than that of the wild type at d 10, and almost undetectable from d 20 (Fig. 4A).
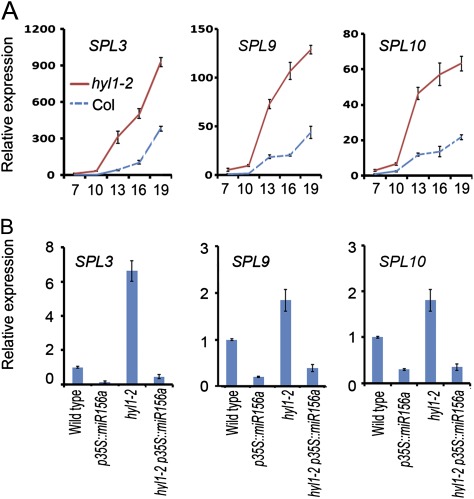
Fig. 3.
Expression pattern of HYL1 and pri-miR156a. (A–C) RNA in situ hybridization of HYL1 with antisense (A) and sense (B) probes and miR156a (C), showing spatial expression of HYL1 and miR156a in the shoot tips of Arabidopsis seedlings at 3 weeks grown under short-day conditions. (D) Temporal expression pattern of HYL1. (E) Temporal expression pattern of pri-miR156a. Three biological replicates were performed. DAG, Days after germination; WT, wild type. Results are shown as means ±SD.
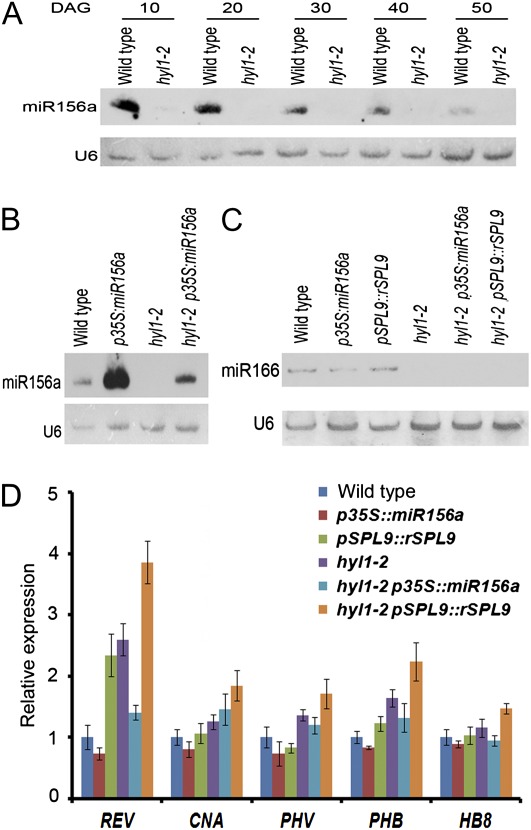
Fig. 4.
Expression of miR156, miR165/6 and HD-ZIP III. (A, B) Northern hybridization showing the accumulation of miR156a in shoot apices of hyl1-2 plants at different stages (10–50 d after germination (DAG)) (A), and of hyl1-2 p35S::miR156a plants at 20 DAG (B). (C) Northern hybridization showing the accumulation of miR165/6 in shoot apices of hyl1-2 p35S::miR156a and hyl1-2 pSPL9::rSPL9 plants at 10 DAG. U6 was used as a control. (D) Expression levels of HD-ZIP III genes in hyl1-2 p35S::miR156a and hyl1-2 pSPL9::rSPL9 plants at 10 DAG. Three biological replicates were performed.
It was possible that the reduced accumulation of miR156a in hyl1-2 mutants was dependent on the amount of pri-miR156a. To exclude this possibility, we analysed the amount of pri-miR156a in shoot apices of both wild-type and hyl1-2 plants using reverse transcription followed by real-time PCR. During the vegetative phase, the level of pri-miR156a in hyl1-2 seedlings was greater than that of the wild type, in contrast to the accumulation of miR156a (Fig. 3E). This was consistent with a previous report (Kurihara et al., 2009). Like the wild type, the transcript level degreased progressively from d 10 to 19 in hyl1-2 seedlings. In the primary leaves of hyl1-2 mutants, reduced accumulation of miR156a and increased levels of pri-miR156a were coincident with adult characteristics, indicating that HYL1 might control the accumulation of miR156 by post-transcriptional regulation rather than by transcriptional regulation.
If HYL1 promotes juvenile epidermal identity, increased accumulation of miR156 should rescue the epidermal defects seen in hyl1-2 mutants. To define the genetic interaction between HYL1 and miR156, we created hyl1-2 transgenic plants that accumulated miR156 under the control of a constitutive cauliflower mosaic virus 35S promoter (Fig. 5A, B, D and E). Wild-type plants transformed with p35S::miR156a presented a prolonged juvenile epidermal phenotype. Introduction of p35S::miR156a into hyl1-2 plants completely rescued the epidermal defects (Fig. 2A). Compared with hyl1-2 plants, hyl1-2 p35S::miR156a exhibited delayed production of abaxial trichomes by 16.2 plastochrons, which was almost what was seen in wild-type plants. Consistent with the rescue of the epidermal defects, the leaf initiation rate increased 1.5-fold relative to that of the wild type (Fig. 2B), and the juvenile leaf blades were rounded, similar to the wild type (Fig. 5G). To address whether there was overproduction of miR156a in hyl1-2 p35S::miR156a plants, we examined the accumulation of miR156a in 10-d-old seedlings by northern blotting. The accumulation of miR156a was enhanced in hyl1-2 p35S::miR156a seedlings relative to hyl1-2 and wild-type plants but did not overaccumulate as in the p35S::miR156a seedlings (Fig 4B). This indicated that the regulation of miR156a biogenesis by HYL1 was dose dependent. In this case, the abundance of miR156a in hyl1-2 p35S::miR156a seedlings was high enough and over the threshold required for the juvenile phase. We suggest that HYL1 maintains the juvenile state of primary leaves through temporal and spatial regulation of miR156.
Fig. 5.
Genetic interaction between HYL1, miR156a and SPL genes. (A–F) 60-d-old rosette seedlings of the wild-type (A), p35S::miR156a (B), spl9-4 spl15-1 (C), hyl1-2 (D), hyl1-2 p35S:: miR156a (E) and hyl1-2 spl9-4 spl15-1 (F) plants, grown under short-day conditions. (G) Individual leaves of hyl1-2 and hyl1-2 p35S::miR156a plants. (H, I) Rosette seedlings (60-d-old) of pSPL9::rSPL9 (H) and hyl1-2 pSPL9::rSPL9 (I), grown under short-day conditions. (This figure is available in colour at JXB online.)
HYL1 controls the expression levels of miR156-targeted SPL genes
To define the expression levels of miR156-targeted genes in hyl1 mutants, we applied ATH1 Affymetrix arrays to detect the global expression of miRNA-targeted genes. Among 22 810 genes, 657 genes were upregulated and 1247 genes were downregulated >1.5-fold in the hyl1 seedlings. Among the 657 upregulated genes, there were 35 target genes for miRNAs and/or trans-acting small interfering RNAs, which accounted for 3.6% of the upregulated genes (Table 2). Among the ten members of the miR156-targeted SPL genes, SPL3, SPL4, SPL5, SPL9, and SPL10 were upregulated 2.9-, 6.2-, 7.4-, 2.0-, and 2.2-fold, respectively.
Table 2.
miRNA-targeted genes upregulated >1.5-fold in 3-week-old hyl1 seedlings relative to the wild type
| Accession no.a | Gene | miRNAs | Fold change |
| AT1G72830 | HAP2C | miR169a, -b, -c, -h, -i, -j, -k, -l, -m, -n | 12.197156 |
| AT5G06510 | CBF-B/NF-YA | miR169b, -c, -d, -e, -f, -g, -h, -i, -j, -k, -l, -m, -n | 9.1556805 |
| AT3G15270 | SPL5 | miR156a, -b, -c, -d, -e, -f, -g; miR157d | 7.4200788 |
| AT3G05690 | HAP2B | miR169b, -c, -d, -e, -f, -g, -h, -i, -j, -k, -l, -m, -n | 7.0425992 |
| AT1G53160 | SPL4 | miR156a, -b, -c, -d, -e, -f, -g, -h; miR157a, -b, -c, -d | 6.1503794 |
| AT1g52070 | JR/MBP | miR846a | 5.352703 |
| AT1G63150 | PPR | miR161.1a, miR161.2a | 4.8119594 |
| AT3G57230 | AGL16 | miR824a | 4.7594344 |
| AT2G02850 | ARPN | miR408a | 4.6092139 |
| AT1G01040 | DCL1 | miR162a, -b | 4.462162 |
| AT1g52060 | JR/MBP | miR846a | 4.0534713 |
| AT1G17590 | rCBF-B/NF-YA | miR169a, -b, -c, -h, -i, -j, -k, -l, -m, -n | 3.8572252 |
| AT1G31280 | AGO2 | miR403a | 3.7480951 |
| AT1G63130 | PPR | miR161.1a; miR400a | 3.0100167 |
| AT1G30490 | PHV | miR165a, -b; miR166a, -b, -c, -d, -e, -f, -g | 3.0088157 |
| AT3G11440 | ATMYB65 | miR159c | 2.9420524 |
| AT2G33810 | SPL3 | miR156a, -b, -c, -d, -e, -f, -g | 2.9262585 |
| AT1G63080 | PPR | miR161.1a; miR400a | 2.8489617 |
| AT5G06100 | MYB33 | miR159c | 2.6038222 |
| AT3G09220 | LAC7 | miR857a | 2.5730044 |
| AT1G12290 | CC-NBS-LRR | miR472a | 2.5651874 |
| AT3G15030 | TCP4 | miR319a, -b, -c | 2.4187059 |
| AT1G54160 | CBF-B/NF-YA | miR169a, -b, -c, -h, -i, -j, -k, -l, -m, -n | 2.3524761 |
| AT1G27370 | SPL10 | miR156a, -b, -c, -d, -e, -f, -g, -h; miR157a, -b, -c, -d | 2.2149103 |
| AT3G20910 | CBF-B/NF-YA | miR169a, -b, -c, -d, -e, -f, -g | 2.0370432 |
| AT3G60630 | SCARECROW | miR170a; miR171a, -b, -c | 2.0144381 |
| AT2G42200 | SPL9 | miR156a, -b, -c, -d, -e, -f, -g, -h; miR157a, -b, -c, -d | 1.9632046 |
| AT1G56010 | NAC1 | miR164a, -b, -c | 1.9609002 |
| AT1G52150 | ATHB-15 | miR165a, -b; miR166a, -b, -c, -d, -e, -f, -g | 1.9513054 |
| AT1G77850 | ARF17 | miR160a, -b, -c | 1.8860873 |
| AT5G60690 | REV | miR165a, -b; miR166a, -b, -c, -d, -e, -f, -g | 1.7754512 |
| AT1G53230 | TCP3 | miR319a, -b, -c | 1.7221409 |
| AT4G30080 | ARF16 | miR160a, -b, -c | 1.6402834 |
| AT5G43780 | APS4 | miR395a, -b, -c, -d, -e, -f | 1.5858944 |
| AT1G76810 | eIF-2 | miR771a | 1.5199657 |
The Arabidopsis Information Resource database: http://www.arabidopsis.org/.
We chose SPL3, SPL9, and SPL10 as representative members to investigate whether their expression patterns were disrupted in hyl1-2 seedlings. In wild-type seedlings, the expression of SPL3, SPL9, and SPL10 increased over time from d 7 to 19 (Fig. 6A). In hyl1-2 seedlings, SPL3 expression was much higher than in the wild type and increased progressively with time, showing an inverse relationship to miR156. Similar expression profiles were observed for SPL9 and SPL10, although the differences between the expression pattern of the miRNAs and its target genes were not the same as for SPL3. To address whether the miR156-targeted genes were downregulated in hyl1-2 p35S::miR156a plants, we examined the expression levels of SPL3, SPL9, and SPL10 in 10-d-old seedlings by real-time PCR. Expression of the three SPL genes decreased in hyl1-2 p35S::miR156a seedlings relative to hyl1-2 and wild-type plants (Fig. 6B). We wondered whether HYL1 controlled the other miR156-targeted SPL genes. Real-time PCR revealed that the expression levels of SPL2, SPL4, SPL5, SPL6, SPL11, SPL13, and SPL15 were elevated in hyl1-2 to different extents, but expression levels of each in hyl1-2 35S::miR156a plants were reduced, except for SPL4, whose expression level was slightly higher than that of the wild type (see Supplementary Fig. S1, available in JXB online), suggesting that all of the miR156-targeted SPL genes were under the control of HYL1. The primary leaves of hyl1-2 p35S::miR156a seedlings became juvenile (Fig. 2A), apparently because the expression levels of the SPL genes in hyl1-2 p35S::miR156a seedlings were below the threshold required for the adult phase. We suggest that HYL1 controls the juvenile state of primary leaves through temporal regulation of miR156-targeted genes.
Fig. 6.
Expression of SPL3, SPL9, and SPL10 genes in hyl1-2 and transgenic plants. (A) Progressive expression levels of miR156-targeted SPL3, SPL9, and SPL10 genes in wild-type and hyl1-2 seedlings from 7 to 19 d after germination (DAG; x-axis) Total RNA was extracted from the shoot apex at different stages and analysed by real-time RT-PCR with three biological replicates. Expression was normalized relative to that of the wild type at 7 DAG. (B) Expression of SPL3, SPL9, and SPL10 genes in wild-type, hyl1-2, p35S::miR156a, and hyl1-2 p35S::miR156a 10-d-old seedlings. Three biological replicates were performed. Results are shown as means ±SD. (This figure is available in colour at JXB online.)
Single loss-of-function mutants of SPL3 and SPL10 show no obvious vegetative phenotype. In contrast, spl9-4 showed a slight juvenile epidermal phenotype, while an spl9-4 spl15-1 double mutant exhibited a delayed transition from the juvenile to the adult phase (Wu and Poethig, 2006; Wang et al., 2008; Wu et al., 2009). To address genetic interactions between the miR156-targeted SPL genes and HYL1, we constructed hyl1-2 spl9-4 spl15-1 triple mutants by crossing hyl1-2 plants with spl9-4 spl15-1 plants (Fig. 5C, D, F). The triple mutants significantly delayed abaxial trichome production compared with hyl1-2 single mutants but advanced abaxial trichome production compared with wild-type plants (Fig. 2A). In other words, the hyl1-2 phenotype was partially rescued by the spl9-4 spl15-1 alleles. Although the leaves of hyl1-2 spl9-4 spl15-1 plants were as round as the wild-type leaves, their initiation rates were not significantly increased (Fig. 2B).
The p35S::rSPL3 (miR156-resistant version of SPL3) plants showed early induction of flowering, but there was no significant effect on leaf shape. However, overexpression of SPL9 and SPL10 accelerated the expression of all adult leaf traits (Wang et al., 2008; Wu et al., 2009). We generated plants transgenic for p35S::rSPL3, pSPL9::rSPL9 and pSPL10::rSPL10, among which the rSPL9 plants were more similar to the hyl1-2 plants in the shape of the leaf and petiole (Fig. 2A). To define the genetic interaction between SPL genes and HYL1, we introduced rSPL9 into hyl1-2 mutants (Fig. 5H, I). In hyl1-2 pSPL9::rSPL9 plants, all of the primary leaves generated abaxial trichomes, and the number of abaxial trichomes present on the first two leaves was increased compared with that of the hyl1-2 mutants (Fig. 2A); the leaf initiation rate was also higher than that of hyl1-2 or pSPL9::rSPL9 plants (Fig. 2B), suggesting that the adult characteristics of the first leaves were enhanced.
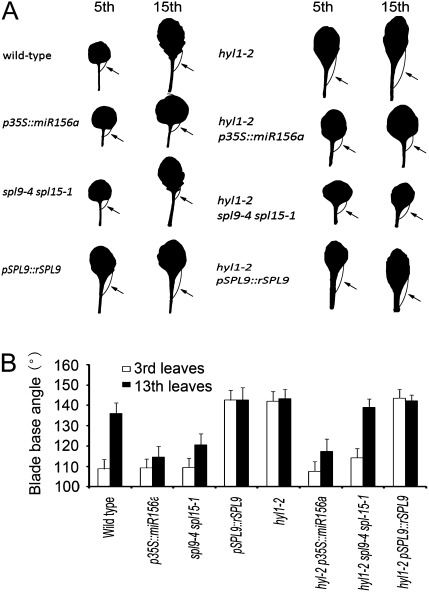
Blade base angle is a new morphological marker for the juvenile-to-adult-phase transition
During further phenotyping of hyl1-2 and wild-type leaves, we observed that the blade base angle (BBA; the angle formed by a common endpoint in the base that is shared by the petiole and blade) of wild-type leaves varied among developmental stages. In primary leaves, the BBA was nearly 90° (Fig. 7A), consistent with what is generally observed for round leaves. The BBA increased from the third to the tenth leaves, consistent with gradual elongation of the leaves. The BBA for the 13th leaves was 135°, whereas that of the third leaves was 107°, demonstrating a distinction between the juvenile and adult phases. Therefore, we designated BBA as a new quantitative marker for the juvenile or adult phase. In hyl1-2 mutants, the BBA values for the third and 13th leaves were almost the same (142°), and both were greater than those of wild-type plants, indicating that the third leaves of hyl1-2 plants displayed the same characteristics as the adult leaves of both hyl1-2 and wild-type plants (Fig. 7B). In hyl1-2 p35S::miR156a plants, the BBA of the third leaves was the same as that of the wild type, and the BBA of the 13th leaves was 110°, which was much lower than that of hyl1-2 and wild-type adult leaves. These results implied that, when p35S::miR156a recovers juvenile characteristics of the third leaves from the hyl1-2 phenotype, it enables the 13th adult leaves to acquire juvenile characteristics.
Fig. 7.
Blade base angles (BBA) of 50-d-old seedling of hyl1-2 and related mutants grown under short-day conditions. (A) Circular arcs in the fifth and 15th leaves indicate the BBA (arrows). (B) Comparison of BBA among hyl1-2 and related mutants. The number of leaves measured was >30. Results are shown as means ±SD.
In hyl1-2 spl9-4 spl15-1 plants, the BBA of the third leaves was lower than that of hyl1-2 but higher than that of the wild-type (Fig. 7B), while the BBA of the 13th leaves was lower than that of hyl1-2 and the wild-type but higher than that of spl9-4 spl15-1 plants. These results indicated that the adult characteristic of both the third and 13th leaves was compromised in hyl1-2 spl9-4 spl15-1 plants.
The transgenic p35S::miR156a plants exhibited a much stronger rescue of the hyl1-2 mutant phenotype than was seen in spl9-4 spl15-1 plants. The limited restoration of spl9-4 spl15-1 alleles to the hyl1-2 phenotype indicated that HYL1 regulates other SPL genes besides SPL9 and SPL15.
In all of these cases, high BBA values (>140°) for the leaves were correlated with the presence of abaxial trichomes and an elongated blade, while low BBA values (<120°) were correlated with an absence of abaxial trichomes and round blades. In summary, high BBA, the presence of abaxial trichomes on primary leaves, and a high length-to-width ratio of blades are three useful morphological markers to distinguish between adult and juvenile leaves.
HYL1 controls crosstalk between miR156 and miR165/166
hyl1-2 plants are characterized by leaf incurvature, which is attributable to adaxial–abaxial polarity defects due to reduced accumulation of miR165/166 (Yu et al., 2005). In Arabidopsis, five of the class III homeodomain-leucine zipper (HD-ZIP III) genes are targeted by miR165/166. Of these, PHABULOSA (PHB), PHAVOLUTA (PHV), and REVOLUTA (REV) act redundantly to promote the adaxial cell fates of the leaf primordium (McConnell and Barton, 1998; McConnell et al., 2001; Emery et al., 2003), while ATHB8 (HB-8) and CORONA (CNA) encode functions that are both antagonistic to those of REV within certain tissues and overlap those of REV in other tissues (Emery et al., 2003). In hyl1-2 p35S::miR156a and hyl1-2 spl9-4 spl15-1 plants, the complete rescue of the hyl1-2 phenotype in terms of early phase transition was concurrent with a reduction in leaf incurvature (Fig 5B–F), revealing that increased accumulation of miR156 or loss of function of the SPL genes contributed to the partial recovery from leaf incurvature. Our previous report confirmed that deficiency in the miR165/166 pathways was the primary cause for leaf incurvature of hyl1 mutants (Liu et al., 2011). To address the relationship between miR156 and miR165/166 or miR165/166-targeted genes, the accumulation of miR165/166 and miR165/166-targeted genes in hyl1-2 p35S::miR156a and hyl1-2 pSPL9::rSPL9 plants was analysed. The amount of miR165/166 decreased in plants transgenic for p35S::miR156a, whereas in those transgenic for pSPL9::rSPL9 it increased (Fig. 4C). To test whether SPL9 regulated miR165/166-targeted genes, we examined the expression level of HD-ZIP III genes using real-time PCR. Transcripts of REV were elevated about 2-fold in pSPL9::rSPL9 plants and decreased slightly in p35S::miR156a plants (Fig. 4D). In hyl1-2 plants, the expression of REV, PHB, and PHV increased in hyl1-2 pSPL9::rSPL9 plants but decreased in hyl1-2 p35S::miR156a plants (Fig. 4D). These results suggested that overaccumulation of miR156a or reduced expression of SPL genes downregulated the expression of some HD-ZIP III genes. Loss-of-function mutations of REV, PHB, and PHV rescue the incurvature phenotype of hyl1 leaves (Liu et al., 2011), which is why the extent of leaf incurvature decreased in hyl1-2 p35S::miR156a plants.
HYL1 prevents the premature transcription of adult-related genes in primary leaves
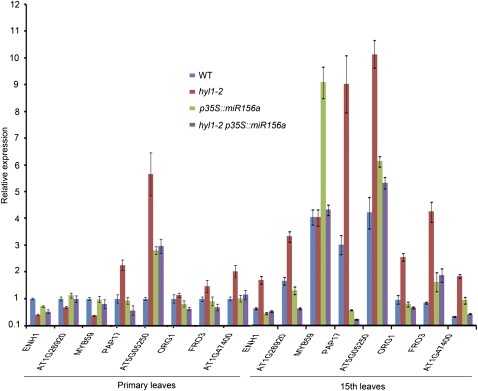
In hyl1-2 mutants, the adult characteristics of the primary leaves are consistent with the up- and downregulation of many genes, including miR156-mediated targets. These genes might be responsible for differentiation of the abaxial trichomes, proliferation of the proximo-distal axis, and cell division at the blade base. To examine the premature gene transcripts of adult-related genes, we searched for miR156-regulated genes among the differentially expressed transcripts in hyl1-2 mutants. These genes could be responsible for the premature gene expression observed in hyl1-2 primary leaves and might be partially required for functioning of the SPL genes. We compared microarray libraries from hyl1 with those from hyl1-2 p35S::miR156a and selected ten genes that were most differentially expressed between hyl1 and wild-type seedlings, and between hyl1-2 p35S::miR156a and hyl1 seedlings. We chose the three genes (ENH1, At1g26920, and MYB59) that were downregulated in hyl1 seedlings relative to the wild type and upregulated in hyl1-2 p35S::miR156a seedlings relative to hyl1 seedlings, and seven genes (PAP17, At5g05250, ORG1, FRO3, At1g47400, SPL3, and SPL5) that were upregulated in hyl1 seedlings relative to the wild type and downregulated in hyl1-2 p35S::miR156a seedlings relative to hyl1 seedlings (see Supplementary Table S1, available in JXB online).
To define the genes regulated by HYL1 through miR156-mediated SPL genes, we examined the premature expression of the differentially expressed genes by real-time PCR. In the primary leaves of 10-d-old seedlings (juvenile phase), the three downregulated genes exhibited decreased transcript levels in hyl1-2 plants relative to wild-type plants and increased transcription levels in hyl1-2 p35S::miR156a plants relative to hyl1-2 plants (Fig. 8). Four of the five upregulated genes (the exception was ORG1) showed increased transcript levels in hyl1-2 plants relative to the wild type and decreased transcript levels for hyl1-2 p35S::miR156a plants relative to hyl1-2 plants, suggesting that these genes are adult-stimulative. Surprisingly, the ENH1, At1g26920, and MYB59 genes all showed increased transcript levels in the 15th leaves of 50-d-old hyl1-2 plants relative to their primary leaves. Hence, we concluded that these genes are regulated differently in primary and adult leaves of hyl1-2 plants. Most of the adult-stimulative genes showed increased transcript levels in the 15th hyl1-2 leaves relative to hyl1-2 primary leaves, or in the 15th wild-type leaves relative to primary wild-type leaves. Therefore, they may play an important role in expression of adult traits in both wild-type and hyl1-2 leaves. These results suggest that HYL1 mediates the expression of adult-related genes that regulate adult-phase development of primary leaves.
Fig. 8.
Premature gene expression in the primary leaves and 15th leaves of hyl1-2, hyl1-2 p35S::miR156a, and the wild-type (WT). Three biological replicates were performed. Results are shown as means ±SD.
As SE, another important regulator of miRNA biogenesis, is involved in the vegetative-phase transition, we wondered whether hyl1 and se-1 mutants would exhibit similar premature gene expression patterns. To address this question, we examined the ten genes expressed most differentially between se-1 plants and the wild type, and between se-1 p35S::miR156f (miR156f is the same as miR156a) and se-1 seedlings using data described previously (Wang et al., 2008). Among these, six genes, including SPL3, were the most upregulated in se-1 seedlings relative to the wild type and downregulated in se-1 p35S::miR156f relative to se-1 seedlings. We observed that SPL5 was not downregulated in se-1 p35S::miR156f. A comparison of the genes most differentially expressed between se-1 and se-1 p35S::miR156f revealed that all of the most downregulated genes in hyl1 p35S::miR156a seedlings (see Supplementary Table S2, available in JXB online) were also downregulated in se-1 p35S::miR156f seedlings. In contrast, two of the most upregulated genes in se-1 seedlings were also upregulated in hyl1 seedlings. The premature expression of some SPL-regulated genes was clearly shared in hyl1 and se-1 seedlings. However, many of these genes were not common in hyl1 and se-1 seedlings. We wondered whether these genes were upregulated in hyl1-2 plants relative to wild-type primary leaves, and whether they were preferentially expressed in the 15th leaves of hyl1-2 mutants compared with the primary leaves. Compared with the control gene sets, the genes showing at least a 2-fold increase in their transcript levels in hyl1-2 primary leaves were significantly enriched in the 15th leaves relative to the primary leaves.
Discussion
HYL1 supplies abundant miR156 for the juvenilization of primary leaves
The periods of vegetative growth of higher plants vary with species, genotypes, and environmental conditions (Wang et al., 2011). In tomato and cucumber, vegetative growth and reproductive growth are intertwined throughout life, and pure vegetative growth is very short. In contrast, many Brassica crops such as Chinese cabbage and cabbage undergo vegetative growth in natural conditions for long period (>5 months). For example, the vegetative growth of Chinese cabbage is divided into four stages: seedling, rosette, folding, and heading (He et al., 2000), each of which is characterized by timing of abaxial trichomes, blade shape, and petiole length. A seedling stage is necessary for the formation of nutrient-rich heads. In Arabidopsis, the juvenile phase (equivalent to the seedling stage of Chinese cabbage) can be subdivided into the early juvenile phase and late juvenile phase. Normally, miR156a accumulates throughout vegetative growth, but it peaks during the early juvenile phase and decreases progressively during the subsequent vegetative phases with time. HYL1 controls the relative high accumulation of miR156a in primary leaves that is essential for the juvenile state, as it is preferentially expressed in the same regions where miR156a accumulates. In hyl1-2 mutants, a lack of HYL1 results in reduced accumulation of miR156 and in turn enhanced expression of miR156-targeted SPL genes compared with the wild type. During vegetative growth, reduced accumulation of miR156a causes an adult-like phenotype in the primary leaves and early transition of the adult phase in the first nine leaves. In this case, juvenile-to-adult-phase transition is determined by temporal expression of pri-miR156 and HYL1 genes. HYL1 must supply an abundance of miR156 for the juvenilization of primary leaves.
In hyl1-2 p35S::miR156a or hyl1-2 spl9-4 spl15-1 plants, the hyl1-2 phenotype in terms of abaxial trichome placement on primary leaves, elongation of the blade, and the increase in the BBA were completely or partially rescued. In hyl1-2 pSPL9::rSPL9 plants, the hyl1-2 phenotype was altered by the increased early appearance of abaxial trichomes, enhanced elongation of the blade, and an increased BBA. These observations revealed that HYL1 determines the timing of the juvenile-to-adult-phase transition through an miR156-mediated pathway.
HYL1 and SE might control different SPL-regulated genes
The genes that mediate the effects of the miR156-targeted SPL genes during leaf development are unknown. These SPL genes might exhibit similar functions to adaxial–abaxial identity genes, cell division genes, and auxin response factors that regulate the differentiation of abaxial trichomes, elongation of the blade and increases in the BBA. How HYL1 organizes these genes through miRNA-mediated SPL genes is not clear. In other aspects, the gain-of-function mutation of iaa18 or axr3-1, two members of the Aux/IAA transcriptional repressor family, has pleiotropic phenotypes, including long hypocotyls and up-curled leaves (Leyser et al., 1996; Cline et al., 2001; Uehara et al., 2008; Ploense et al., 2009). These phenotypes are similar to those of the hyl1-2 plants. It would be interesting to explore whether these genes were involved in HYL1-regulated developmental processes.
In Arabidopsis, the presence of trichomes on the abaxial surface of the leaf is often used as a morphological marker for the adult phase. The number of serrations, trichomes, and hydathodes, the size of the petiole and leaf blade, the length-to-width ratio of the leaf blade, and vascular complexity are also used to investigate vegetative-phase transition (Telfer et al., 1997; Tsukaya et al., 2000; Usami et al., 2009). In this study, we used the BBA as a new marker for analysis of phase transition. In general, differentiation of morphological markers, such as serrations, trichomes, hydathodes, petioles, blades, the length-to-width ratio of the leaf blade, and the BBA, is established by patterns of cell division that are often highly variable.
HYL1 defines the timing of abaxial trichome appearance, leaf shape, and BBA. However, the marginal serrations of the hyl1-2 leaves are not obvious, and the number of hydathodes was not changed compared with wild-type leaves. These results indicate that HYL1 regulates trichome patterns, the length-to-width ratio of the leaf blade, and the BBA, rather than serrations and hydathodes. In contrast, SE determines the timing of abaxial trichome, serration, and hydathode appearance. In theory, the SPL-regulated genes regulate leaf shape and BBA, while a different set of SPL-regulated genes determines serration and hydathodes.
SPL3 and SPL5 belong to the same class of the miR156-mediated SPL gene family. Both genes are downregulated in hyl1-2 p35S::miR156a relative to hyl1-2 seedlings, while SPL3 alone is downregulated in se-1 p35S::miR156f relative to se-1 seedlings. It remains unclear whether these subtle differences alter the premature expression of SPL-regulated genes. Nevertheless, not all of the genes that are most differentially expressed between hyl1-2 p35S::miR156a and se-1 p35S::miR156f are the same. This suggests that the premature gene expression in the primary leaves of hyl1-2 is not the same as for se-1. We suggest that HYL1 and SE might regulate different SPL-regulated genes and control different morphological traits.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table S1. The ten genes that were most deregulated (upregulated and downregulated) in hyl1 seedlings relative to the wild type and in hyl1-2 p35S::miR156a (hyl1 156) relative to hyl1 seedlings.
Supplementary Table S2. The ten genes that were most deregulated (upregulated and downregulated) in se-1 seedlings relative to the wild type and se-1 p35S::miR156a relative to se-1 seedlings. The data were derived from GEO GSE16061.
Supplementary Table S3. Oligonucleotide primer sequences used in this study.
Supplementary Fig. S1. Expression of SPL2, SPL4, SPL5, SPL6, SPL11, SPL1I3, and SPL15 genes in hyl1-2 and transgenic plants. Expression was normalized relative to that of the wild type and are shown as means ±SD.
Acknowledgments
We are grateful to Dr R. Scott Poethig for providing spl9-4 spl15-1 seeds and plasmids pSPL9::rSPL9 and pSPL10::rSPL10 and to Dr Detlef Weigel for providing plasmid p35S::rSPL3. This work was supported by grants from the Natural Science Foundation of China (grant no. 30730053) and National Basic Research Program of China (grant no. 2012CB113903).
Glossary
Abbreviations
- BBA
blade base angle
- miRNA
microRNA
- pri-miRNA
primary transcript miRNA
References
- Chuck G, Cigan AM, Saeteurn K, Hake S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nature Genetics. 2007;39:544–549. doi: 10.1038/ng2001. [DOI] [PubMed] [Google Scholar]
- Clarke JH, Tack DT, Findlay K, Van Montagu M, Van Lijsebettens M. The SERRATE locus controls the formation of the early juvenile leaves and phase length in Arabidopsis. The Plant Journal. 1999;20:493–501. doi: 10.1046/j.1365-313x.1999.00623.x. [DOI] [PubMed] [Google Scholar]
- Cline MG, Chatfield SP, Leyser O. NAA restores apical dominance in the axr3-1 mutant of Arabidopsis thaliana. Annals of Botany. 2001;87:61–65. [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dong Z, Han MH, Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proceedings of the National Academy of Sciences USA. 2008;105:9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. Radial patterning of Arabidopsis shoots by class IIIHD-ZIP and KANADI genes. Current Biology. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Fang YD, Spector DL. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Current Biology. 2007;17:818–823. doi: 10.1016/j.cub.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y, Utsumi M, Ohba Y, Watanabe Y. Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant and Cell Physiology. 2007;48:1243–1253. doi: 10.1093/pcp/pcm099. [DOI] [PubMed] [Google Scholar]
- Guo AY, Zhu QH, Gu X, Ge S, Yang J, Luo J. Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene. 2008;418:1–8. doi: 10.1016/j.gene.2008.03.016. [DOI] [PubMed] [Google Scholar]
- Han MH, Goud S, Song L, Fedoroff N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proceedings of the National Academy of Sciences USA. 2004;101:1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YK, Xue WX, Sun YD, Yu XH, Liu PL. Leafy head formation of the progenies of transgenic plants of Chinese cabbage with exogenous auxin genes. Cell Research. 2000;10:151–160. doi: 10.1038/sj.cr.7290044. [DOI] [PubMed] [Google Scholar]
- Hiraguri A, Itoh R, Kondo N, Nomura Y, Aizawa D, Murai Y, Koiwa H, Seki M, Shinozaki K, Fukuhara T. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Molecular Biology. 2005;57:173–188. doi: 10.1007/s11103-004-6853-5. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Kaminuma E, Matsui A, et al. Transcriptome analyses revealed diverse expression changes in ago1 and hyl1 Arabidopsis mutants. Plant and Cell Physiology. 2009;50:1715–1720. doi: 10.1093/pcp/pcp109. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Takashi Y, Watanabe Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006;12:206–212. doi: 10.1261/rna.2146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser HM O, Pickett FB, Dharmasiri S, Estelle M. Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. The Plant Journal. 1996;10:403–413. doi: 10.1046/j.1365-313x.1996.10030403.x. [DOI] [PubMed] [Google Scholar]
- Liu Z, Jia L, Wang H, He Y. HYL1 regulates the balance between adaxial and abaxial identity for leaf flattening via miRNA-mediated pathways. Journal of Experimental Botany. 2011;62:4367–4381. doi: 10.1093/jxb/err167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J. SERRATE: a new player on the plant microRNA scene. EMBO Reports. 2006;7:1052–1058. doi: 10.1038/sj.embor.7400806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Fedoroff N. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell. 2000;12:2351–2365. doi: 10.1105/tpc.12.12.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida S, Chen HY, Yuan YA. Molecular insights into miRNA processing by Arabidopsis thaliana SERRATE. Nucleic Acids Research. 2011;39:7828–7836. doi: 10.1093/nar/gkr428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JR, Barton MK. Leaf polarity and meristem formation in Arabidopsis. Development. 1998;125:2935–2942. doi: 10.1242/dev.125.15.2935. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- Ploense SE, Wu MF, Nagpal P, Reed JW. A gain-of-function mutation in IAA18 alters Arabidopsis embryonic apical patterning. Development. 2009;136:1509–1517. doi: 10.1242/dev.025932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata M, Koyama T, Mitsuda N, Ohme-Takagi M. Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant and Cell Physiology. 2009;50:2133–2145. doi: 10.1093/pcp/pcp148. [DOI] [PubMed] [Google Scholar]
- Song L, Han MH, Lesicka J, Fedoroff N. Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proceedings of the National Academy of Sciences USA. 2007;104:5437–5442. doi: 10.1073/pnas.0701061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A, Bollman KM, Poethig RS. Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development. 1997;124:645–654. doi: 10.1242/dev.124.3.645. [DOI] [PubMed] [Google Scholar]
- Tsukaya H, Shoda K, Kim GT, Uchimiya H. Heteroblasty in Arabidopsis thaliana (L.) Heynh. Planta. 2000;210:536–542. doi: 10.1007/s004250050042. [DOI] [PubMed] [Google Scholar]
- Uehara T, Okushima Y, Mimura T, Tasaka M, Fukaki H. Domain II mutations in CRANE/IAA18 suppress lateral root formation and affect shoot development in Arabidopsis thaliana. Plant and Cell Physiology. 2008;49:1025–1038. doi: 10.1093/pcp/pcn079. [DOI] [PubMed] [Google Scholar]
- Usami T, Horiguchi G, Yano S, Tsukaya H. The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development. 2009;136:955–64. doi: 10.1242/dev.028613. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Gasciolli V, Crete P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Current Biology. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Wang JW, Park MY, Wang LJ, Koo Y, Chen XY, Weigel D, Poethig RS. miRNA control of vegetative phase change in trees. PLoS Genet. 2011 doi: 10.1371/journal.pgen.1002012. 7, e1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Schwab R, Czech B, Mica E, Weigel D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell. 2008;20:1231–43. doi: 10.1105/tpc.108.058180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. In planta transformation of Arabidopsis. Cold Spring Harbor Protocols. 2006 doi: 10.1101/pdb.prot4668. doi:10.1101/pdb.prot4668. [DOI] [PubMed] [Google Scholar]
- Willmann MR, Poethig RS. Time to grow up: the temporal role of smallRNAs in plants. Current Opinion in Plant Biology. 2005;8:548–52. doi: 10.1016/j.pbi.2005.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann MR, Poethig RS. The effect of the floral repressor FLC on the timing and progression of vegetative phase change in Arabidopsis. Development. 2011;138:677–85. doi: 10.1242/dev.057448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138:750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu ZQ, Lu F, Dong AW, Huang H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant Journal. 2006;47:841–850. doi: 10.1111/j.1365-313X.2006.02835.x. [DOI] [PubMed] [Google Scholar]
- Yang SW, Chen HY, Yang J, Machida S, Chua NH, Yuan YA. Structure of Arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing. Structure. 2010;18:594–605. doi: 10.1016/j.str.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Yu X, Shen R, He Y. HYL1 gene maintains venation and polarity of leaves. Planta. 2005;221:231–242. doi: 10.1007/s00425-004-1439-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.