Abstract
Terpenoids are the largest class of plant secondary metabolites and have attracted widespread interest. Salvia miltiorrhiza, belonging to the largest and most widely distributed genus in the mint family, is a model medicinal plant with great economic and medicinal value. Diterpenoid tanshinones are the major lipophilic bioactive components in S. miltiorrhiza. Systematic analysis of genes involved in terpenoid biosynthesis has not been reported to date. Searching the recently available working draft of the S. miltiorrhiza genome, 40 terpenoid biosynthesis-related genes were identified, of which 27 are novel. These genes are members of 19 families, which encode all of the enzymes involved in the biosynthesis of the universal isoprene precursor isopentenyl diphosphate and its isomer dimethylallyl diphosphate, and two enzymes associated with the biosynthesis of labdane-related diterpenoids. Through a systematic analysis, it was found that 20 of the 40 genes could be involved in tanshinone biosynthesis. Using a comprehensive approach, the intron/exon structures and expression patterns of all identified genes and their responses to methyl jasmonate treatment were analysed. The conserved domains and phylogenetic relationships among the deduced S. miltiorrhiza proteins and their homologues isolated from other plant species were revealed. It was discovered that some of the key enzymes, such as 1-deoxy-D-xylulose 5-phosphate synthase, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, hydroxymethylglutaryl-CoA reductase, and geranylgeranyl diphosphate synthase, are encoded by multiple gene members with different expression patterns and subcellular localizations, and both homomeric and heteromeric geranyl diphosphate synthases exist in S. miltiorrhiza. The results suggest the complexity of terpenoid biosynthesis and the existence of metabolic channels for diverse terpenoids in S. miltiorrhiza and provide useful information for improving tanshinone production through genetic engineering.
Keywords: Salvia, Salvia miltiorrhiza, tanshinone, terpenoid biosynthesis
Introduction
Terpenoids, also known as isoprenoids or terpenes, are the largest class of plant secondary metabolites. They play important roles in plant growth, development, general metabolism, and defence against predators, pathogens, and competitors (Gershenzon and Dudareva, 2007). Many terpenoids have been used as pharmaceuticals, cosmetics, pesticides, and potential biofuels. In the last two decades, the molecular biochemistry and genomics of terpenoid biosynthesis have attracted widespread interest (Bohlmann and Keeling, 2008).
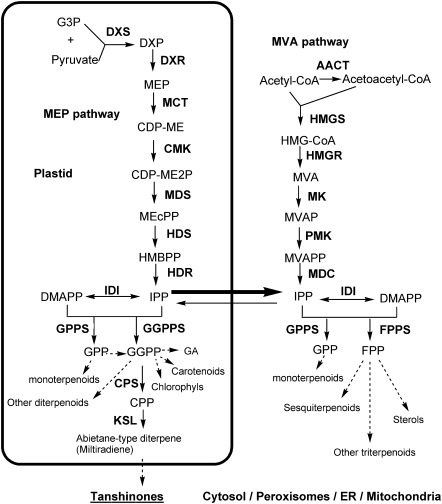
Generally, the biosynthetic pathway of plant terpenoids can be divided into three stages (Fig. 1). The first stage leads to the synthesis of the universal isoprene precursor ispentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP) through the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway and/or the mevalonate (MVA) pathway. In the second stage, the intermediate diphosphate precursors, including geranyl diphosphate (GPP), farnesyl diphosphate (FPP), and geranylgeranyl diphosphate (GGPP), are synthesized under the catalysis of isoprenyl diphosphate synthases (IDSs), including geranyl diphosphate synthase (GPPS), farnesyl diphosphate synthase (FPPS), and geranylgeranyl diphosphate synthase (GGPPS). The last stage involves the formation of diverse terpenoids under the catalysis of terpene synthases/cylases (TPSs), such as copalyl diphosphate synthase (CPS) and kaurene synthase (KS), and various terpenoid-modifying enzymes. Enzymes involved in terpenoid biosynthesis have different subcellular localizations. All MEP pathway enzymes are located in plastids, whereas the MVA pathway enzymes can be in the cytosol or peroxisomes (Reumann et al., 2007; Sapir-Mir et al., 2008; Simkin et al., 2011). The localizations of IDSs and TPSs are more diversified and often correlated with the subcellular location of terpenoid biosynthesis.
Fig. 1.
Proposed pathways of terpenoid biosynthesis in S. miltiorrhiza.
Because of the important role of terpenoids in plant development and the potential value of metabolic engineering of terpenoid biosynthesis pathways, identification and characterization of the genes encoding the enzymes involved in terpenoid biosynthesis have been carried out in various plant species, such as Arabidopsis, conifers, and Hevea brasiliensis (Lange and Ghassemian, 2003; Sando et al., 2008a,b; Zulak and Bohlmann, 2010). However, due to the complexity of terpenoid biosynthesis pathways, many enzyme-encoding genes are still not well defined. For example, the terpenoid-modifying enzymes involved in the last stage of terpenoid biosynthesis are largely unknown. In addition, 1-deoxy-D-xylulose 5-phosphate synthases (DXSs) involved in the MEP pathway, hydroxymethylglutaryl-CoA reductases (HMGRs) involved in the MVA pathway, and the IDSs involved in the second stage of terpenoid biosynthesis are encoded by small gene families with at least two members, many of which have not been identified, and the physiological functions of each member are not well known in most plant species. It is particularly true for those species without their whole genome sequence available.
Salvia, including ∼900 species, is the largest genus in the economically and medicinally important Labiatae family and is widely distributed throughout the world. Chemical constituents of Salvia plants have become a major focus in the related field. Considerable reports about the isolation, identification, structure modification and synthesis, and biology activities of diterpenoids in Salvia have been published. The results suggest that Salvia produce diverse diterpenoids, such as tanshinone IIA, salvicine, and neotanshinlactone, many of which have significant bioactivities (Honda et al., 1988; Wang et al., 2004; Munro et al., 2005).
Salvia miltiorrhiza Bunge (Danshen in Chinese) is a significant Salvia species with great economic and medicinal value. It is the first Chinese medicinal material entering the international market and has been widely used in traditional Chinese medicine (TCM) for treating dysmenorrhoea, amenorrhoea, and cardiovascular diseases (Cheng, 2006). The main lipophilic bioactive components of S. miltiorrhiza are diterpenoid tanshinones, including tanshinone I, tanshinone IIA, cryptotanshinone, and so forth. To date, >30 tanshinones and related diterpenoid quinines have been isolated and characterized (Li et al., 2009) and the biosynthesis of tanshinone has been shown to be stimulated by methyl jasmonate (MeJA) treatment in S. miltiorrhiza (Gao et al., 2009). These constituents are known to possess a variety of pharmacological effects, such as antibacterial, antioxidant, anti-inflammatory, and antineoplastic activities (Honda et al., 1988). Salvia miltiorrhiza has been developed to be a potential model medicinal plant because of its relatively small genome size (∼600 Mb), short life cycle, undemanding growth requirements, and significant medicinal value. A total of 13 genes involved in tanshinone biosynthesis have been cloned from S. miltiorrhiza (Table 1). Among them, only three, SmDXS1, SmHMGR1, and SmGGPPS1, have been characterized through genetic transformation (Cui et al., 2011). Many genes and gene family members involved in terpenoid biosynthesis are unknown.
Table 1.
Terpenoid biosynthesis-related genes in S. miltiorrhiza
| Name | Accession no. | Lena | pI | Mol. wt (kDa) | Locb | TMHc | Reference |
| SmDXS1 | EU670744 | 714 | 6.37 | 76.7 | C | 0 | |
| SmDXS2 | FJ643618 | 725 | 6.51 | 78.4 | C | 0 | |
| SmDXS3 | JN831116 | 713 | 7.01 | 76.5 | C | 0 | This study |
| SmDXS4 | JN831117 | 713 | 5.93 | 77.9 | C | 0 | This study |
| SmDXS5 | JN831118 | 703 | 6.83 | 75.5 | C | 0 | This study |
| SmDXR | FJ476255 | 474 | 5.99 | 51.6 | C | 0 | Wu et al. (2009) |
| SmMCT | JN831096 | 304 | 5.72 | 33.5 | C | 0 | This study |
| SmCMK | EF534309 | 396 | 6.41 | 43.4 | C | 0 | Wang et al. (2010) |
| SmMDS | JN831097 | 234 | 8.53 | 24.6 | C | 0 | This study |
| SmHDS | JN831098 | 742 | 6.02 | 82.4 | C | 0 | This study |
| SmHDR1 | JN831099 | 463 | 5.72 | 51.9 | C | 0 | This study |
| SmHDR2 | JN831100 | 462 | 5.86 | 52.1 | C | 0 | This study |
| SmAACT1 | EF635969 | 403 | 6.33 | 41.2 | _ | 0 | Cui et al. (2011) |
| SmAACT2 | JN831101 | 403 | 8.07 | 41.6 | _ | 0 | This study |
| SmHMGR1 | EU680958 | 565 | 7.1 | 60.5 | C | 2 | Liao et al. (2009) |
| SmHMGR2 | FJ747636 | 550 | 6.07 | 58.7 | _ | 2 | Dai et al. (2011) |
| SmHMGR3 | JN831102 | 562 | 5.74 | 60.5 | _ | 3 | This study |
| SmHMGR4 | JN831103 | 550 | 7.51 | 58.9 | _ | 2 | This study |
| SmHMGS | FJ785326 | 460 | 6.04 | 50.7 | _ | 0 | Cui et al. (2011) |
| SmMK | JN831104 | 387 | 5.61 | 40.8 | S | 0 | This study |
| SmPMK | JN831095 | 509 | 5.3 | 54.9 | _ | 0 | This study |
| SmMDC | JN831105 | 422 | 7.58 | 46.5 | _ | 0 | This study |
| SmIDI1 | EF635967 | 236 | 5.29 | 27.2 | _ | 0 | Cui et al. (2011) |
| SmIDI2 | JN831106 | 269 | 5.49 | 30.6 | C | 0 | This study |
| SmGPPS | JN831107 | 424 | 6.18 | 46.5 | M | 0 | This study |
| SmFPPS | EF635968 | 349 | 5.63 | 40.0 | _ | 0 | Cui et al. (2011) |
| SmGGPPS1 | FJ643617 | 364 | 5.9 | 39.0 | C | 0 | Unpublished |
| SmGGPPS2 | JN831112 | 346 | 6.52 | 37.4 | _ | 0 | This study |
| SmGGPPS3 | JN831113 | 379 | 8.35 | 41.3 | C | 0 | This study |
| SmGPPS.SSUI | JN831108 | 314 | 6.76 | 34.5 | C | 0 | This study |
| SmGPPS.SSUII.1 | JN831109 | 290 | 6.66 | 31.8 | C | 0 | This study |
| SmGPPS.SSUII.2 | JN831110 | 331 | 5.81 | 36.3 | C | 0 | This study |
| SmGPPS.LSU | JN831111 | 344 | 6.47 | 37.4 | M | 0 | This study |
| SmCPS1 | EU003997 | 793 | 6.04 | 90.5 | C | 0 | Cui et al. (2011) |
| SmCPS2 | JN831114 | 757 | 5.97 | 86.6 | M | 0 | This study |
| SmCPS3 | JN831115 | 701* | This study | ||||
| SmCPS4 | JN831120 | 670* | This study | ||||
| SmCPS5 | JN831121 | 445* | This study | ||||
| SmKSL1 | EF635966 | 595 | 5.7 | 68.4 | C | 0 | Cui et al. (2011) |
| SmKSL2 | JN831119 | 762* | This study |
Len represents the number of amino acid residue. * indicates that the predicted sequence is partial.
Loc represents the protein localization predicted by TargetP. ‘C’ stands for chloroplast, suggesting that the sequence contains a chloroplast transit peptide. ‘M’ stands for mitochondrial, suggesting the sequence contains a mitochondrial targeting peptide. ‘S’ stands for secretory pathway, showing that the sequence contains a signal peptide. ‘–’ indicates any other location.
TMH represents the number of predicted transmembrane helices.
Recently, the genome sequencing programme of S. miltiorrhiza has been initiated. A working draft of the genome has been obtained (Chen et al., unpublished data). The current assembly has ∼20× coverage and consists of 611 208 contigs representing ∼92% of the entire S. miltiorrhiza genome and 96% of the protein-coding genes. It allows a genome-wide identification and characterization of genes involved in terpenoid biosynthesis to be performed.
Materials and methods
Plant materials
Salvia miltiorrhiza Bunge (line 993) with whole genome sequences available was grown in a field nursery. Flowers, leaves, stems, root cortices, and root steles were collected from 2-year-old plants in June when the pharmacologically active components were rapidly accumulated (Xu et al., 2010). Plant tissues were stored in liquid nitrogen until use.
Plantlets used for MeJA treatment were prepared from stem segments with node and shoot tips of field-grown S. miltiorrhiza (line 993). Explants were surface-sterilized in a 0.1% HgCl2 solution for 5–10 min followed by washing four times in sterile water. The sterilized explants were cultivated on MS agar medium (Murashige and Skoog). Shoots were excised from the explants and rooted on 6,7-V agar medium (Chen et al., 1997) for ∼6 weeks under a 16/8 h light/dark photoperiod at 25 °C.
MeJA treatment
Plantlets with regenerated roots were transferred to 6,7-V liquid medium and cultivated for 2 d. MeJA in carrier solution containing 0.1% Tween-20 and 5% ethanol was added to the medium to obtain a final concentration of 200 μM. Plantlets were treated for 0 h and 24 h and then leaves and roots of similar sizes were collected separately. Plantlets treated with carrier solution were used as controls. Tissues from two individual plants were pooled. All samples were frozen and stored in liquid nitrogen until use.
Sequence retrieval and gene prediction
The current assembly of S. miltiorrhiza genome sequences (Chen et al., unpublished data) was searched for homologues of terpenoid biosynthesis-related proteins from various plant species using the BLASTx algorithm (Altschul et al., 1997). An e-value cut-off of 10−5 was applied to the homologue recognition. All retrieved sequences were used for gene prediction on the Genscan web server (http://genes.mit.edu/GENSCAN.html) (Burge and Karlin, 1998). The predicted gene models were further examined and corrected manually by comparison with related genes identified from other plant species.
Sequence feature analysis
Intron/exon structures were predicted using the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/chinese.php) (Guo et al., 2007). The theoretical isoelectric point and molecular weight were predicted using the Compute pI/MW tool on the ExPASy server (Bjellqvist et al., 1994) (http://web.expasy.org/compute_pi/). The localizations of deduced proteins were predicted on the TargetP 1.1 server (http://www.cbs.dtu.dk/services/TargetP/) (Emanuelsson et al., 2007). Transmembrane domains were analysed on the TRMHMM server v 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/) (Krogh et al., 2001). Conserved domains were searched against the Pfam protein families database locally using the Perl script ‘pfam_scan.pl’ (Finn et al., 2009) (ftp://ftp.sanger.ac.uk/pub/databases/Pfam/Tools/README). The conserved amino acids were analysed by protein alignment using tools such as ClustalW and then checked manually (Thompson et al., 1994; Hall, 1999).
Phylogenetic analysis
Phylogenetic relationships were analysed using MEGA version 4.0 (Tamura et al., 2007). The Poisson correction parameter and pairwise deletion of gaps were applied. The reliability of branching was assessed by the bootstrap re-sampling method using 1000 bootstrap replications. For each analysis, only nodes supported by bootstrap values >50% are shown.
RNA extraction
Total RNA was extracted from plant tissues using the plant total RNA extraction kit (BioTeke, China) and pre-treated with RNase-Free DNase (Promega, USA) to eliminate genomic DNA contamination. RNA integrity was analysed on a 1% argarose gel. RNA quantity was determined using a NanoDrop 2000C Spectrophotometer (Thermo Scientific, USA).
Quantitative real-time reverse transcription-PCR (qRT-PCR)
Total RNA was reverse-transcribed by Superscript III Reverse Transcriptase (Invitrogen, USA). The PCRs were performed according to the instructions of the SYBR premix Ex Taq™ kit (TaKaRa, China) and carried out in triplicate using the CFX96™ real-time PCR detection system (Bio-Rad, USA). Gene-specific primers were designed using primer designing tools such as Primer3 (http://frodo.wi.mit.edu/primer3/) (Rozen and Skaletsky, 2000). The primer sequences are listed in Supplementary Table S1 available at JXB online. The lengths of amplicons are between 100 bp and 250 bp. SmUBQ10 was chosen as an endogenous control in this study. The expression of the genes in plantlets treated with MeJA for 24 h was further normalized to their expression in plantlets treated with carrier solution for 24 h. Standard deviations were calculated from three PCR replicates. The specificity of amplification was assessed by dissociation curve analysis, and the relative abundance of genes was determined using the comparative Ct method as suggested by the CFX-manager software (Bio-Rad, USA).
Results and Discussion
Identification of 40 terpenoid biosynthesis-related genes
Using a systematic computational approach, 40 terpenoid biosynthesis-related genes were identified from the current S. miltiorrhiza genome assembly, of which 27 are novel (Table 1). These genes are members of 19 gene families, which encode all enzymes involved in the biosynthesis of the universal isoprene precursor IPP and its isomer DMAPP, and two associated with the biosynthesis of labdane-related diterpenoids (Fig. 1). The exon/intron structures of these genes and the features of deduced proteins were predicted using several web tools (Bjellqvist et al., 1994; Krogh et al., 2001; Emanuelsson et al., 2007; Guo et al., 2007; Finn et al., 2009). These genes have different exon/intron structures. The deduced proteins show different length, isoelectric point (pI), molecular weight, subcellular localization, and transmembrane helix number, and contain conserved domains and motifs (Table 1; Supplementary Fig. S1–S12 and Table S2 at JXB online). Phylogenetic analysis shows that these proteins are highly similar to those involved in the biosynthesis of terpenoids in various plants (Supplementary Figs S13–S26). However, tissue-specific expression patterns and different responses to MeJA treatment were found for members in a gene family, suggesting they probably play distinct roles in terpenoid biosynthesis (Supplementary Figs S27, S28). In the following paragraphs, the possible physiological functions of these genes are analysed and discussed in detail based on their sequence features, tissue-specific expression patterns, responses to MeJA treatment, and phylogenetic relationships to homologues in other plant species.
Characterization and expression analysis of genes involved in the MEP pathway
The MEP pathway is mainly present in eubacteria and plants, but it is absent in other eukaryotes, including fungi and animals (Lange et al., 2000). In plants, enzymes involved in this pathway usually operate in plastids to synthesize monoterpenes, diterpenes, carotenoids, and the phytol chain of chlorophyll. Among the 40 identified terpenoid biosynthesis-related genes, 12 encode enzymes involved in the MEP pathway. They include five DXS genes, two HDR (4-hydroxy-3-methylbut-2-enyl diphosphate reductase) genes, and one each of DXR (1-deoxy-D-xylulose 5-phosphate reductoisomerase), MCT (2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase), CMK [4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol kinase], MDS (2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase), and HDS (4-hydroxy-3-methylbut-2-enyl diphosphate synthase), suggesting that genes encoding all seven MEP pathway enzymes have been identified. Among the 12 genes, seven, namely SmDXS3, SmDXS4, SmDXS5, SmMCT, SmMDS, SmHDS, and SmHDR2, are reported for the first time. Protein subcellular localization prediction indicates that enzymes encoded by these genes are most possibly located in chloroplasts (Table 1). These findings are consistent with the previous reports for Arabidopsis MEP pathway enzymes (Hsieh et al., 2008).
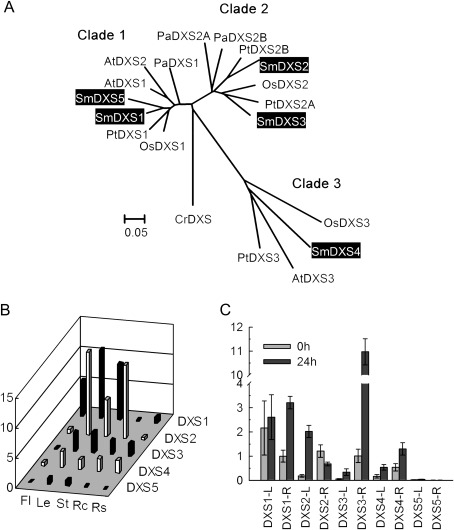
DXS is the first enzyme of the MEP pathway. It catalyses the transketolase-type condensation reaction of pyruvate and glyceraldehyde 3-phosphate to yield 1-deoxy-D-xylulose 5-phosphate (DXP) and plays a critical role in the biosynthesis of terpenoids (Chappell, 1995; Estevez et al., 2001). The present results suggest that DXS is encoded by a small gene family of five members in S. miltiorrhiza. The sequences of SmDXS1 and SmDXS2 have previously been submitted to GenBank, while SmDXS3, SmDXS4, and SmDXS5 are newly identified in this study. All five genes encode proteins with domains and motifs conserved among previously known DXSs. They include the consensus thiamine pyrophosphatase-binding motif and the pyridine-binding DRAG domain, suggesting that SmDXSs have the same type of biochemical activity (Supplementary Figs S2, S3 at JXB online).
Previous studies suggested that two different classes of DXS genes existed in plants. Genes in the DXS1 clade are probably involved in primary metabolism, such as the biosynthesis of carotenoids and the phytol chain of chlorophyll, and play housekeeping roles. Genes in the DXS2 clade are probably involved in secondary terpenoid biosynthesis (Walter et al., 2002; Phillips et al., 2007). Recent studies revealed the presence of the third, DXS3, clade. Genes in this clade were proposed to be involved in the biosynthesis of some products essential for plant survival and required at very low levels (Cordoba et al., 2011). Among the five SmDXS genes, SmDXS1 and SmDXS5 belong to the DXS1 clade, SmDXS2 and SmDXS3 are members of the DXS2 clade, while SmDXS4 is in the more divergent DXS3 clade (Fig. 2A). These results suggest the existence of members of all three DXS clades in S. miltiorrhiza and indicate the different roles of each SmDXS gene in terpenoid biosynthesis. Consistently, differential expression of SmDXS genes was observed. SmDXS1 is highly expressed in leaves, stems, and flowers. SmDXS2 is predominantly expressed in leaves, stems, and root cortices. SmDXS3 is expressed in all the tissues analysed except for flowers. The expression of SmDXS4 appears to be ubiquitious. SmDXS5 is mainly expressed in leaves and stems and its expression is low compared with that of the other four SmDXS genes (Fig. 2B). These expression patterns of SmDXS genes are in agreement with the proposed functions of DXS genes in each clade. In root cortices, which is the main location of major bioactive constituents, such as tanshinones, the level of SmDXS2 is the highest compared with other SmDXS genes, suggesting the importance of SmDXS2 in tanshinone biosynthesis in S. miltiorrhiza. Like SmDXS2, SmDXS3 is one of the DXS2 clade genes that are probably involved in secondary terpenoid biosynthesis. In addition to leaves, stems, and root cortices, where SmDXS2 is highly expressed, SmDXS3 is also expressed in root steles (Fig. 2B). These findings, together with the results showing that SmDXS3 is highly induced by MeJA in the roots of S. miltiorrhiza plantlets (Fig. 2C), indicate the involvement of SmDXS3 in the biosynthesis of defence-related terpenoids in roots. The present results are consistent with those from some other plant species. For instance, both of the Populus abies DXS genes (PaDXS2A and PaDXS2B) belong to the DXS2 clade. However, they are differentially expressed and only one is induced under MeJA treatment (Phillips et al., 2007).
Fig. 2.
Expression patterns of SmDXS genes and the phylogenetic relationship of their deduced proteins with various other plant species. (A) Phylogenetic relationship of plant DXSs. The rooted Neighbor–Joining tree was constructed using the MEGA program (version 4.0) with default parameters. CrDXS (Chlamydomonas reinhardtii, CAA07554) was used as an outgroup. Transit peptides of DXSs were trimmed for the analysis of sequence data. DXSs included are Arabidopsis thaliana AtDXS1 (At4g15560), AtDXS2 (At3g21500), AtDXS3 (At5g11380), Oryza sativa OsDXS1 (NP_001055524), OsDXS2 (NP_001059086), OsDXS3 (BAA83576), Populus trichocarpa PtDXS1 (XP_002312717), PtDXS2A (XP_002303416), PtDXS2B (XP_002331678), PtDXS3 (XP_002308644), Picea abies PaDXS1 (ABS50518), PaDXS2A (ABS50519), PaDXS2B (ABS50520), and five S. miltiorrhiza SmDXSs (highlighted). (B) Fold changes of SmDXS genes in flowers (Fl), leaves (Le), stems (St), root cortices (Rc), and root steles (Rs) of S. miltiorrhiza plants grown in soil. The expression level of SmDXS1 in root steles was arbitrarily set to 1. (C) Fold changes of SmDXS genes in leaves (L) and roots (R) of S. miltiorrhiza plantlets treated with MeJA for 0 h and 24 h. The level of SmDXS1 in roots of plantlets without treatment was arbitrarily set to 1.
DXR is the second enzyme of the MEP pathway. It is involved in an intramolecular rearrangement and reduction step to form MEP from DXP in the presence of NADPH. Searching the current assembly of the S. miltiorrhiza genome, only one DXR gene was found (SmDXR). It contains 12 exons and 11 introns (Supplementary Fig. S1 at JXB online). SmDXR was previously reported to be expressed constitutively and to play a significant role in the MEP pathway, and it was suggested to regulate the production and accumulation of tanshinones in S. miltiorrhiza (Wu et al., 2009). In this study, SmDXR shows a tissue-specific expression, with the highest level in leaves, followed by stems and flowers. The expression of SmDXR in roots is very low, which is consistent with the low level of tanshinones, indicating that DXR is a rate-limiting enzyme of tanshinone biosynthesis (Supplementary Figs S27, S28).
MCT catalyses the conversion of MEP to CDP-ME [2-phospho-4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol] in a CTP-dependent reaction. In plants, the MCT gene was first cloned from Arabidopsis and was showed to be a key enzyme in the MEP pathway (Rohdich et al., 2000). However, it has never been isolated from S. miltiorrhiza before. Using a computational approach, an SmMCT gene was identified from the current genome assembly. The deduced protein contains the conserved IspD motif and shows high identities with other plant MCTs (Supplementary Fig. S2 at JXB online). SmMCT is mainly expressed in leaves and can also be expressed in stems, roots, and flowers. SmMCT is not obviously induced by MeJA, indicating that it is a constitutive gene in S. miltiorrhiza (Supplementary Figs S27, S28).
CMK catalyses the phosphorylation reaction of the 2-hydroxy group of CDP-ME and converts it into CDP-ME2P. SmCMK has been reported recently (Wang et al., 2010). Analysis of gene expression shows that SmCMK is expressed in all tissues analysed, including leaves, stems, roots, and flowers (Supplementary Figs S27, S28 at JXB online), which is similar to the expression of SmMCT. These results are consistent with the functions of SmMCT and SmCMK in the biosynthesis of diverse terpenoids.
In the next two steps of the MEP pathway, CDP-ME2P is converted by MDS into a cyclic intermediate MEcPP (2-C-methyl-D-erythritol 2,4-cyclodiphosphate), which is then converted into HMBPP (4-hydroxy-3-methylbut-2-enyl diphosphate) by HDS. The genes encoding MDS and HDS have not been characterized in most plant species. From the S. miltiorrhiza genome, an SmMDS gene and an SmHDS gene were identified and their sequence features were analysed. SmMDS contains only three exons, which is the smallest exon number among all 12 MEP pathway genes. In contrast, SmHDS has the most complex intron/exon structure, containing 19 exons and 18 introns (Supplementary Fig. S1 at JXB online). The expression patterns of SmMDS and SmHDS are similar to that of other single gene family members of the MEP pathway, showing expression in all tissues analysed and exhibiting slight induction by MeJA.
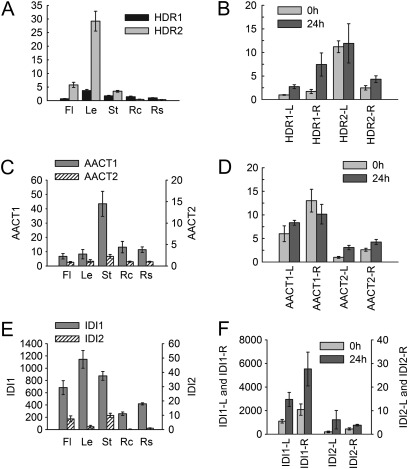
HMBPP produced under the catalysis of MDS can be further converted into the isoprene precursor IPP by HDR, an enzyme also playing a key role in the supply of plastidial terpenoid precursors (Botella-Pavia et al., 2004). In this study, two HDR genes were identified in the S. miltiorrhiza genome. The deduced amino acid sequences of SmHDR1 and SmHDR2 show 82.1% identity. The two SmHDR genes have similar exon/intron structures, indicating they are probably derived from gene duplication events (Supplementary Fig. S1 at JXB online). SmHDR1 is expressed in all tissues analysed and the expression can be induced by MeJA. In contrast, SmHDR2 exhibits more tissue-specific expression with very high levels in leaves, flowers, and stems. The expression of SmHDR2 appears not to be affected in S. miltiorrhiza plantlets under MeJA treatment (Fig. 4A, B). These results indicate that SmHDR1 is probably involved in the biosynthesis of secondary terpenoids, such as tanshinones, and also plays a significant role in defence responses in S. miltiorrhiza, while SmHDR2 is probably involved in primary metabolism and plays a housekeeping role.
Fig. 4.
Expression patterns of SmHDR, SmAACT, and SmIDI genes. (A, C, and E) Fold changes of SmHDR genes (A), SmAACT genes (C), and SmIDI genes (E) in flowers (Fl), leaves (Le), stems (St), root cortices (Rc), and root steles (Rs) of S. miltiorrhiza plants grown in soil. The expression level of SmHDR1 (A), SmAACT2 (B), and SmIDI2 (C) in root steles was arbitrarily set to 1. (B, D, and F) Fold changes of SmHDR genes (B), SmAACT genes (D), and SmIDI genes (F) in leaves (L) and roots (R) of S. miltiorrhiza plantlets treated with MeJA for 0 h and 24 h. The level of SmHDR1 (B), SmAACT2 (D), and SmIDI2 (F) in leaves of plantlets without treatment was arbitrarily set to 1.
Characterization and expression analysis of genes involved in the MVA pathway
The MVA pathway is an ancestral metabolic route existing in all three domains of life, such as eukaryotes and a few bacteria (Lombard and Moreira, 2011). It mainly operates in the cytoplasm and mitochondria, and predominantly synthesizes sterols, sesquiterpenes, and ubiquinones. A total of six enzymes are involved in this pathway (Fig. 1). From the S. miltiorrhiza genome, a total of 10 genes were identified for the six MVA pathway enzymes (Table 1). These include four (SmAACT1, SmHMGS, SmHMGR1, and SmHMGR2) reported previously and six newly identified (SmAACT2, SmHMGR3, SmHMGR4, SmMK, SmPMK, and SmMDC).
Acetyl-CoA C-acetyltransferase (AACT) catalyses the condensation of two acetyl-CoA molecules to form acetoacetyl-CoA. Two AACT genes (SmAACT1 and SmAACT2) were identified in the S. miltiorrhiza genome. They encode proteins with a conserved thiolase domain (Supplementary Fig. S2 at JXB online). SmAACT1 and SmAACT2 show 76.7% identity at the amino acid level and have the same number of exons and introns (Supplementary Fig. S1). The deduced SmAACT1 and SmAACT2 proteins are predicted to be cytoplasmic (Table 1), and both of them lack PTS1 peroxisomal targeting sequences in the C-terminus, suggest they could not localized in peroxisomes (Supplementary Fig. S4). Gene expression analysis shows that SmAACT1 and SmAACT2 are expressed in all of analysed tissues, with predominant expression in stems. However, the levels of SmAACT1 are much higher than those of SmAACT2, suggesting the importance of SmAACT1 in S. miltiorrhiza (Fig. 4C, D). SmAACT1 has previously been reported to be involved in tanshinone biosynthesis (Cui et al., 2011), whereas the exact functions of SmAACT2 need to be characterized further.
Hydroxymethylglutaryl-CoA synthase (HMGS) catalyses the condensation reaction of acetyl-CoA and acetoacetyl-CoA to produce 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA). An SmHMGS gene was identified from the current assembly of the S. miltiorrhiza genome. It contains four exons and three introns (Supplementary Fig. S1 at JXB online). SmHMGS is expressed in all tissues analysed and is not significantly induced by MeJA (Supplementary Figs S27, S28). SmHMGS was indicated to be involved in tanshinone biosynthesis (Cui et al., 2011; Zhang et al., 2011). However, its roles in the biosynthesis of other terpenoids remain to be identified.
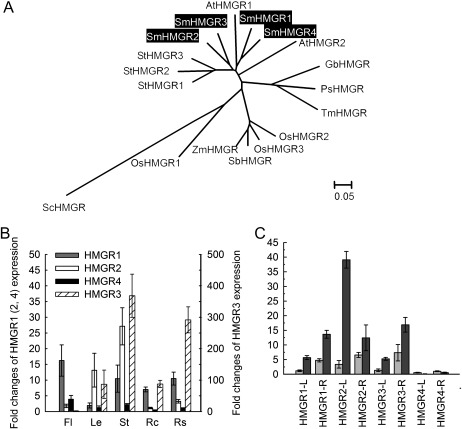
HMGR catalyses the conversion of HMG-CoA to MVA, which is the first committed step in the MVA pathway. Although HMGR is encoded by a single gene in higher animals, archaea, and eubacteria, it is usually encoded by multiple genes in plants. This implies that plant HMGR genes have arisen by gene duplication and subsequent sequence divergence (Friesen and Rodwell, 2004). From the S. miltiorrhiza genome, four HMGR genes (SmHMGR1, SmHMGR2, SmHMGR3, and SmHMGR4) were identified, two of which have been described in previous studies (Liao et al., 2009; Dai et al., 2011). The SmHMGR proteins show high sequence similarities in the 3′-terminal region encoding the C-terminal catalytic domain, whereas the 5′-terminal region is highly divergent. Consistent with many other plant HMGRs, all of the deduced SmHMGR proteins contain two potential N-linked glycosylation sites (N-X-S/T), two HMG-CoA-binding motifs (EMPVGYVQIP and TTEGCLA), and two NADPH-binding motifs (DAMGMNM and GTCGGG) in the conserved C-terminal catalytic domain. An additional N-linked glycosylation site (NST) associated with the production of elicited defensive compounds is present in SmHMGR1 and SmHMGR3 (Choi et al., 1992; Ha et al., 2003) (Supplementary Fig. S5 at JXB online). SmHMGR1 and SmHMGR4 have the same exon/intron structures and show very high sequence similarity, implying that they are probably duplicated genes within the S. miltiorrhiza genome. Similarly, SmHMGR2 and SmHMGR3 are probably duplicated genes because they also show a high sequence similarity and both of them contain only one exon (Supplementary Fig. S1). In contrast to DXSs, all of the plant HMGRs are derived from a common ancestor in evolution (Fig. 3A). Significant differential expression was observed for four SmHMGR genes. SmHMGR1 is highly expressed in flowers, followed by stems and root steles, and shows the lowest expression in leaves. SmHMGR2 is mainly expressed in stems and leaves. Compared with the other three SmHMGR genes, SmHMGR3 is highly expressed in all of the analysed tissues other than flowers. In contrast, SmHMGR4 is more flower specific and its expression levels in the other four tissues are the lowest among the four SmHMGR genes (Fig. 3B). SmHMGR1, SmHMGR2, and SmHMGR3 can be induced by MeJA, whereas the level of SmHMGR4 is more stable in plants treated with MeJA (Fig. 3C). These results are consistent with previous observations for SmHMGR1 and SmHMGR2, indicating that different HMGR isoforms are probably involved in the biosynthesis of different terpenoids (Liao et al., 2009; Dai et al., 2011). In root cortices, which is the main location of tanshinones, the expression of SmHMGR3 is the highest, followed by SmHMGR1. The expression of SmHMGR2 and SmHMGR4 in root cortices is very low (Fig. 3B). This suggests the importance of SmHMGR3 and SmHMGR1 in tanshinone biosynthesis. In addition, SmHMGR2 is also likely to be involved in tanshinone biosynthesis, although its expression in the cortex of roots is low. Consistently, it was shown that overexpression of SmHMGR2 resulted in the enhancement of tanshinone production in cultured hairy roots of S. miltiorrhiza (Dai et al., 2011).
Fig. 3.
Expression patterns of SmHMGR genes and the phylogenetic relationship of their deduced proteins with various other plant species. (A) Phylogenetic relationship of HMGRs in various plant species. The rooted Neighbor–Joining tree was constructed using the MEGA program (version 4.0) with default parameters. ScHMGR (Saccharomyces cerevisiae, AAA34676) was used as an outgroup. HMGRs included are Arabidopsis AtHMGR1 (CAA33139), AtHMGR2 (AAA67317), Solanum tuberosum StHMGR1 (AAA93498), StHMGR2 (AAB52551), StHMGR3 (AAB52552), rice OsHMGR1 (AAA21720), OsHMGR2 (AAD08820), OsHMGR3 (AF110382), Zea mays ZmHMGR (O24594), Sorghum bicolor SbHMGR (XP_002445887), Ginkgo biloba GbHMGR (AAU89123), Picea sitchensis PsHMGR (ACN40476), Taxus×media TmHMGR (AAQ82685), and four S. miltiorrhiza SmHMGRs (highlighted). (B) Fold changes of SmHMGR genes in flowers (Fl), leaves (Le), stems (St), root cortices (Rc), and root steles (Rs) of S. miltiorrhiza plants grown in soil. The expression level of SmHMGR4 in root steles was arbitrarily set to 1. (C) Fold changes of SmHMGRs in leaves (L) and roots (R) of S. miltiorrhiza plantlets treated with MeJA for 0 h and 24 h. The level of SmHMGR4 in roots of plantlets without treatment was arbitrarily set to 1.
Mevalonate kinase (MK), 5-phosphomevalonate kinase (PMK), and mevalonate pyrophosphate decarboxylase (MDC) proteins catalyse the last three steps of the MVA pathway. The plant MK gene was first cloned from Arabidopsis, and there is only one in the Arabidopsis genome (Riou et al., 1994; Lluch et al., 2000). Consistent with this, an MK gene was obtained from the current assembly of the S. miltiorrhiza genome. Arabidopsis MK is preferentially expressed in roots and inflorescences (Lluch et al., 2000), whereas SmMK exhibits the highest expression level in stems, followed by root cortices, root steles, leaves, and flowers, and is induced >2-fold in leaves and roots of plantlets under MeJA treatment (Supplementary Figs S27, S28 at JXB online), suggesting that MK may have distinct spatial and temporal expression patterns in different plant species. The information about plant PMK and MDC is very limited. Similar to MK, PMK and MDC are also encoded by a single gene and exhibit higher expression level in stems and roots than in leaves and flowers in S. miltiorrhiza. The levels of MK and MDC are induced to various degrees by MeJA (Supplementary Figs S27, S28). These results suggest the coordination of SmMK, SmPMK, and SmMDC in the biosynthesis of terpenoids. SmMK, SmPMK, and SmMDC contain the PTS2 peroxisomal targeting signal motif previously found in various MVA pathway enzymes in other plant species, such as Catharanthus roseus (Cr) MK, PMK, and MDC (Supplementary Figs S6–S8) (Simkin et al., 2011). However, the presence of PTS2 in SmMK, SmPMK, and SmMDC does not mean that all of them are peroxisomal enzymes. It has been shown that PTS2 is not necessarily sufficient to target the protein to the peroxisome. For instance, CrPMK and CrMDC are targeted to peroxisomes, whereas CrMK is cytosolic, although they all possess the PTS2 motif (Simkin et al., 2011). Thus, the subcellular localization of SmMK, SmPMK, and SmMDC remains to be determined.
Characterization and expression analysis of IDI genes
Isopentenyl diphosphate isomerase (IDI) catalyses the reversible conversion of IPP to DMAPP (Ramos-Valdivia et al., 1997). Two IDI genes (SmIDI1 and SmIDI2) exist in S. miltiorrhiza. SmIDI1 has been demonstrated to be a candidate gene involved in tanshinone biosynthesis in hairy roots of S. miltiorrhiza (Cui et al., 2011), whereas SmIDI2 is newly identified. SmIDI1 and SmIDI2 have different gene structures and show 65.6% identity at the nucleotide level and 71.5% at the amino acid level. SmIDI2 possesses a chloroplast localization transit peptide, whereas no such a peptide is predicted for SmIDI1 (Table 1). Consistently, TargetP prediction suggests that SmIDI1 is cytosolic, whereas SmIDI2 is chloroplastic. In addition, both SmIDI1 and SmIDI2 contain the PTS1 peroxisomal targeting signal motif (HKL) (Supplementary Fig. S9 at JXB online), suggesting that they are also possibly targeted to peroxisomes. Thus, the localization of SmIDIs is complex and need to be analysed further. Gene expression analysis reveals that the transcripts of SmIDI1 are more abundant than those of SmIDI2 in all tissues analysed and can be induced to ∼2.5 times in leaves and roots of plantlets treated with MeJA for 24 h, suggesting the importance of SmIDI1 in the biosynthesis of terpenoids (Fig. 4E, F).
Characterization and expression analysis of IDS genes
After IPP and DMAPP formation, the following steps utilize them to form prenyl diphosphates with various chain lengths under the catalysis of IDSs, which are also known as prenyltransferases (PTs). According to the chain length of products, IDSs can be classified into three subfamilies: short-, medium-, and long-chain IDS. Among them, the short-chain IDS subfamily is the most intensively studied. It consists mainly of GGPPS, FPPS, and GPPS. From the current assembly of the S. miltiorrhiza genome, nine short-chain IDS genes were identified, three GGPPS genes (SmGGPPS1, SmGGPPS2, and SmGGPPS3), one FPPS gene (SmFPPS), and five GPPS genes (SmGPPS, SmGPPS.LSU, SmGPPS.SSUI, SmGPPS.SSUII.1, and SmGPPS.SSUII.2) (Fig. 5, Table 1). SmGPPS and SmFPPS, consisting of 12 and 10 exons, respectively, are two IDS genes with complex gene structures. The structures of other SmIDS genes are much simpler. SmGGPPS3 probably contains two introns, SmGGPPS1, SmGPPS.SSUII.1, and SmGPPS.SSUII.2 contain one intron, while SmGGPPS2, SmGPPS.SSU, and SmGPPS.LSU are intron free (Supplementary Fig. S1 at JXB online). The proteins encoded by SmGPPS and SmGPPS.LSU are probably localized in mitochondria. SmGGPPS2 proteins appear to be localized in the cytosol, whereas other deduced IDS proteins are most probably localized in plastids (Table 1). Although all of the deduced IDS proteins contain the conserved polyprenyl_synt domains (Supplementary Fig. S2), the motifs of IDSs could be different. Like other plant GGPPSs and FPPSs, all three SmGPPSs and the SmFPPS contain the conserved FARM motif (the first aspartate-rich motif, DDX2–4D) and the SARM motif (the second aspartate-rich motif, DDXXD), which are important in prenyl-substrate binding (Wang and Ohnuma, 1999). The motifs of GPPSs in plants are more diverse. Interestingly, most of the small subunits of heterodimeric GPPSs discovered in plants contain two conserved CXXXC motifs (where ‘X’ can be a hydrophobic amino acid, such as alanine, leucine, isoleucine, valine, glycine, or serine) that are crucial for physical interaction between two GPPS subunits (Wang and Dixon, 2009), whereas the large subunit of hererodimeric GPPSs and the subunits of homodimeric GPPSs do not contain these motifs (Supplementary Figs S10–S12; Table S2).
Fig. 5.
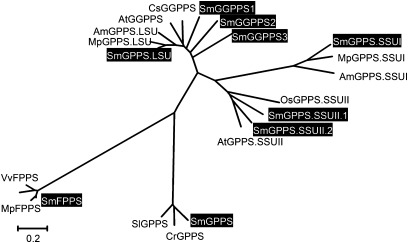
Phylogenetic analysis of homomeric and heteromeric GPPSs, GGPPSs, and FPPSs in S. miltiorrhiza and various other plant species. The unrooted Neighbor–Joining tree was constructed using the MEGA program (version 4.0) with default parameters. Proteins included are Mentha×piperita MpGPPS.LSU (AF182828), Antirrhinum majus AmGPPS.LSU (AAS82860), Croton sublyratus CsGGPPS (BAA86284), Arabidopsis AtGGPPS (AAM65107); MpGPPS.SSUI (AF182827), AmGPPS.SSUI (AAS82859), rice OsGPPS.SSUII (EAY87007), AtGPPS.SSUII (At4g38460), Solanum lycopersicum SlGPPS (ABB88703), Catharanthus roseus CrGPPS (ACC77966), Hevea brasiliensis HbFPPS (AAM98379), Vitis vinifera VvFPPS (AAX76910), MpFPPS (AF384040), and nine S. miltiorrhiza SmIDSs (highlighted).
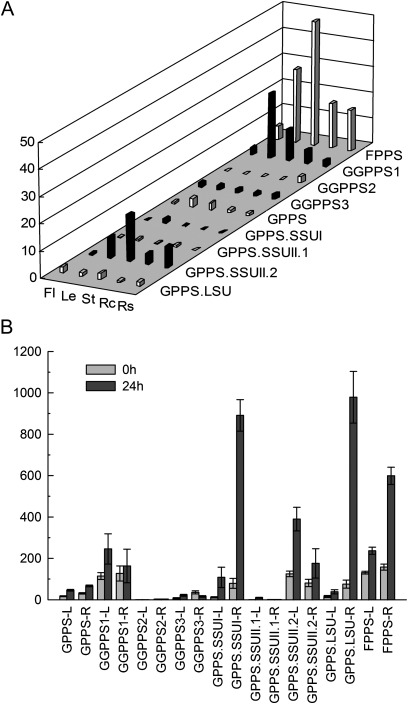
In plants and bacteria, GGPPS catalyses the condensation reactions of IPP and DMAPP to form GGPP, a precursor for the biosynthesis of a structurally diverse group of compounds that includes some specific diterpenoids, carotenoids, chlorophylls, and geranylgeranylated proteins. cDNAs encoding GGPPS have been isolated and characterized from diverse plant species (Okada et al., 2000; Engprasert et al., 2004; Liao et al., 2005). Phylogenetic analysis shows that SmGGPPS1, SmGGPPS2, and SmGGPPS3 proteins are similar to OsGGPPS and AtGGPPS proteins. SmGGPPS1 is expressed in all of the tissues analysed, with high levels in leaves, stems, and root cortices, and the expression level can be induced 2-fold in leaves of plantlets by MeJA (Fig. 6). These results are consistent with the previous report for SmGGPPS1 (Kai et al., 2010). The expression of SmGGPPS1 in Escherichia coli has been shown to accelerate the biosynthesis of carotenoids (Kai et al., 2010). Overexpression of SmGGPPS1 in transgenic S. miltiorrhiza hairy roots resulted in the enhancement of tanshinone production (Kai et al., 2011). Thus, SmGGPPS1 appears to play a significant role in the biosynthesis of diterpenoids, such as tanshinones that are synthesized mainly in the cortex of roots, and tetraterpenoids, such as carotenoids that are produced in chloroplasts and chromoplasts of plants. SmGGPPS2 and SmGGPPS3 are newly identified genes. SmGGPPS2 is predominantly expressed in the stele of roots, whereas SmGGPPS3 exhibits similar expression levels in all the tissues analysed (Fig. 6). The functions of SmGGPPS2 and SmGGPPS3 remain to be elucidated.
Fig. 6.
Expression patterns of SmIDS genes. (A) Tissue-specific expression patterns of SmIDS genes. Fold changes of SmIDS genes in flowers (Fl), leaves (Le), stems (St), root cortices (Rc), and root steles (Rs) of S. miltiorrhiza plants grown in soil. The relative abundance of genes is determined using a comparative Ct method, and the expression level of SmGPPS in root steles was arbitrarily set to 1. (B) The expression of SmIDS genes with or without MeJA treatment. Fold changes of SmIDI genes in leaves (L) and roots (R) of S. miltiorrhiza plantlets treated with MeJA for 0 h and 24 h. The level of SmGPPS.SSUII.1 in roots of plantlets without treatment was arbitrarily set to 1.
FPPS catalyses the sequential head-to-tail condensation of two molecules of IPP with one molecule of DMAPP to form the sesquiterpenoid precursor, FPP. This enzyme is a homodimer of subunits. The deduced SmFPPS protein shows high identities with known FPPSs isolated from various plants, such as Mentha×piperita and Vitis vinifera (Fig. 5). Compared with other short-chain SmIDSs, SmFPPS shows the highest expression level in all of the tissues analysed, including flowers, leaves, stems, root cortices, and root steles (Fig. 6A). The expression of SmFPPS can be induced to a higher level by MeJA, particularly in the roots of plantlets (Fig. 6B), indicating the involvement of SmFPPS in defence responses in S. miltiorrhiza. SmFPPS was considered to be a candidate gene associated with tanshinone biosynthesis because of the relationship between its expression level and the accumulation of tanshinones (Cui et al., 2011). However, the involvement of SmFPPS in tanshinone biosynthesis is very doubtful. No evidence has shown that FPP can serve as a precursor of tanshinones, a group of labdane-related diterpenoids. The increase in SmFPPS expression and the accumulation of tanshinones observed previously are probably two independent events caused by the increase in the IPP level.
GPPS is generally considered to be involved only in monoterpene biosynthesis in plastids, but recent studies show that GPPS is also required for the biosynthesis of some diterpenoids, such as gibberellins (van Schie et al., 2007). Both homodimeric and heterodimeric GPPSs have been discovered in plants. The homodimeric GPPS exists in both angiosperms and gymnosperms (Bouvier et al., 2000; Burke and Croteau, 2002; van Schie et al., 2007), whereas the heterodimeric GPPS has only been found in some angiosperm plant species, such as snapdragon (Antirrhinum majus), Clarkia breweri, and hop (Humulus lupulus) (Burke et al., 1999; Tholl et al., 2004; Wang and Dixon, 2009). The heterodimeric GPPS is composed of two types of subunits, a large subunit and a small subunit, known as LSU and SSU, respectively (Chang et al., 2010). LSU shows significant homology to homomeric IDSs, such as FPPS and GGPPS, while the homology between SSU and other IDSs is very low. Additionally, two types of SSUs (GPPS.SSUI and GPPS.SSUII) have been described (Wang and Dixon, 2009). In some plants, such as mint, two LSU/SSU heterodimers may form a tetramer (LSU/SSU)2 to catalyse the production of C10-GPP in vivo (Chang et al., 2010). Based on phylogenetic analysis, the five newly discovered SmGPPS genes can be classified into SmGPPS that encodes the homomeric GPPS subunit and SmGPPS.LSU, SmGPPS.SSUI, and SmGPPS.SSUII encoding heteromeric GPPS subunits (Fig. 5). SmGPPS.SSUII is represented by two genes, SmGPPS.SSUII.1 and SmGPPS.SSUII.2, whereas each of the other SmGPPS genes is represented by one gene. The discovery of both homomeric and heteromeric GPPSs suggests the complexity of terpenoid biosynthesis in S. miltiorrhiza.
SmGPPS.SSUI and SmGPPS.LSU show the highest homologies with mint MpGPPS.SSUI and MpGPPS.LSU, respectively, implying the existence of a (LSU/SSU)2 tetramer in S. miltiorrhiza (Fig. 5; Supplementary Fig. S10 at JXB online) (Chang et al., 2010). Consistent with the probable role of SmGPPS.SSUI in the production of some volatile monoterpenoids, SmGPPS.SSUI is predominantly expressed in leaves. The expression of SmGPPS.LSU is less tissue specific compared with that of SmGPPS.SSUI (Fig. 6A), suggesting that SmGPPS.LSU and SmGPPS.SSUI are regulated differently in S. miltiorrhiza. Both SmGPPS.LSU and SmGPPS.SSUI can be induced to very high levels in roots of plantlets, indicating the involvement of heteromeric SmGPPSs in plant defence responses (Fig. 6B). SmGPPS.SSUII.2 accumulates to higher levels than SmGPPS.SSUII.1 in all of the tissues analysed. SmGPPS.SSUII.1 is predominantly expressed in leaves and root cortices, whereas SmGPPS.SSUII.2 is highly expressed in stems, followed by leaves and root steles (Fig. 6A), indicating they may be involved in the biosynthesis of different monoterpenoids. The significance of the existence of two types of SSUs (SSUI and SSUII) in a plant species is currently unknown. The aerial parts of many Salvia species are covered with trichomes that can produce and accumulate essential oils (volatile oils), a group of monoterpenoid and sesquiterpenoid derivatives with antimicrobial, antioxidant, and antigerminative activities (Bozin et al., 2007; Yousefzadi et al., 2007; De Martino et al., 2010). The biosynthetic mechanism of these essential oils is still unknown. Identification and characterization of sesquiterpenoid and monoterpenoid biosynthesis-related SmFPPS and SmGPPS will definitely help in elucidating the mechanism of essential oil biosynthesis in plants.
Characterization and expression analysis of genes encoding CPS and KSL
CPS and KSL (kaurene synthase-like) are two important terpenoid synthases involved in the biosynthesis of labdane-related diterpenoids. In addition to SmCPS1 and SmKSL1 that were reported to be involved in tanshinone biosynthesis (Gao et al., 2009 Cui et al., 2011), a new full-length CPS (SmCPS2), three partial CPS genes (SmCPS3, SmCPS4, and SmCPS5), and one partial KSL (SmKSL2) were identified through the sequence homology-based search of the current assembly of the S. miltiorrhiza genome (Table 1). Full-length sequences of three SmCPS genes and a SmKSL could not be obtained, probably because the genes involved downstream of the terpenoid biosynthetic pathway are less conserved compared with those involved upstream, as shown in a recent report (Ramsay et al., 2009). It is also probably due to the incomplete S. miltiorrhiza genome sequence.
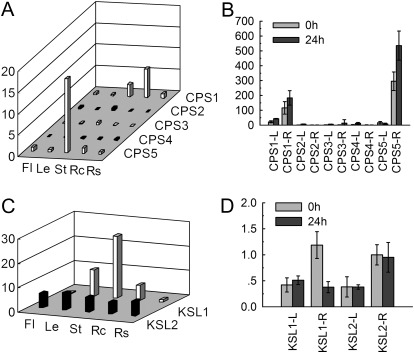
All of the deduced CPS and KSL proteins contain the terpene synthase domains (Supplementary Fig. S2 at JXB online). SmCPS1 and SmCPS5 exhibit higher expression levels than other SmCPS genes in all tissues analysed (Fig. 7). SmCPS1 is highly expressed in root cortices, followed by stems, root steles, flowers, and leaves. This finding is consistent with previous reports showing the involvement of SmCPS1 in tanshinone biosynthesis (Gao et al., 2009). SmCPS5 is predominantly expressed in stems and is also expressed in root cortices, flowers, leaves, and roots. The expression of SmCPS5 is induced under MeJA treatment in roots of S. miltiorrhiza plantlets. These results suggest that SmCPS5 is also important in terpenoid biosynthesis. Phylogenetic analysis shows that the SmKSL1 protein is highly similar to tobacco KS, whereas SmKSL2 is highly similar to Arabidopsis and Chinese chestnut KS (Supplementary Fig. S26), implying that the two SmKSLs may be functionally distinct. Consistent with this, SmKSL1 exhibits differential expression patterns, with the highest level in stems, followed by leaves, root cortices, root steles, and flowers, while the levels of SmKSL2 are similar in all of the tissues analysed (Fig. 7).
Fig. 7.
Expression patterns of SmCPS and SmKSL genes. (A and C) Fold changes of SmCPS genes (A) and SmKSL genes (C) in flowers (Fl), leaves (Le), stems (St), root cortices (Rc), and root steles (Rs) of S. miltiorrhiza plants grown in soil. The expression level of SmCPS1 (A) and SmKSL1 (C) in root steles was arbitrarily set to 1. (B and D) Fold changes of SmCPS genes (B) and SmKSL genes (D) in leaves (L) and roots (R) of S. miltiorrhiza plantlets treated with MeJA for 0 h and 24 h. The level of SmCPS2 (B) and SmKSL2 (D) in roots of plantlets without treatment was arbitrarily set to 1.
Genes probably involved in tanshinone biosynthesis
The biosynthesis of tanshinones is mainly via the MEP pathway, but also depends on cross-talk between the MEP and MVA pathways (Laule et al., 2003; Ge and Wu, 2005). In this study, all of the genes encoding enzymes of the two pathways were systemically characterized in S. miltiorrhiza. Among the seven MEP pathway enzymes, five are encoded by single genes (SmDXR, SmMCT, SmCMK, SmMDS, and SmHDS), whereas the other two, DXS that catalyses the first reaction and HDR that is involved in the last step of the MEP pathway, are encoded by multigene families with five and two members, respectively. Based on gene expression patterns and the results from phylogenetic analysis, it is proposed that SmDXS2 and SmHDR1 are probably involved in tanshinone biosynthesis.
Genes encoding the five MVA pathway enzymes in S. miltiorrhiza were identified and characterized. HMGS, MK, PMK, and MDC are encoded by single genes, while AACT and HMGR are encoded by small gene families with two and four members, respectively. Compared with SmAACT2, SmAACT1 exhibits higher homology with Arabidopsis AACT2, which has been suggested to play a role in terpenoid biosynthesis. Thus, SmAACT1 has a higher possibility to be associated with tanshinone biosynthesis than SmAACT2. Among the four SmHMGR genes, SmHMGR2 has previously been demonstrated to be involved in tanshinone biosynthesis through genetic transformation (Dai et al., 2011). In this study, it is shown that SmHMGR3 and SmHMGR1 are highly expressed in root cortices, the main location of tanshinones. Thus, SmHMGR1, SmHMGR2, and SmHMGR3 are probably involved in tanshinone biosynthesis.
Of the two IDI genes, SmIDI1 is probably involved in tanshinone biosynthesis because it shows a much higher expression level than SmIDI2 and has previously been demonstrated to be associated with tanshinone biosynthesis in hairy roots of S. miltiorrhiza (Cui et al., 2011). Among nine short-chain IDS genes, SmGGPPS1 is most probably involved in tanshinone biosynthesis. This gene belongs to the GGPPS subfamily and is highly expressed in the cortex of roots. Overexpression of SmGGPPS1 resulted in the enhancement of tanshinone production in transgenic S. miltiorrhiza hairy roots (Kai et al., 2011). Moreover, based on expression patterns, SmCPS1, SmCPS5, and SmKSL1 appear to be tanshinone biosynthesis-associated terpene synthases.
In summary, a total of 20 genes probably involved in tanshinone biosynthesis have been identified. SmDXS2, SmDXR, SmMCT, SmCMK, SmMDS, SmHDS, and SmHDR2, involved in the MEP pathway, seem to play the main role in supplying the isoprene precursor for tanshinone biosynthesis. SmAACT1, SmHMGS, SmHMGR1, SmHMGR2, SmHMGR3, SmMK, SmPMK, and SmMDC in the MVA pathway may indirectly affect the supply of IPP precursor. SmIDI1 and SmGGPPS1 play significant roles in the second stage of tanshinone biosynthesis, whereas SmKSL1, SmCPS1, and SmCPS5 are probably the terpene synthases involved in the third stage of tanshinone biosynthesis. Further characterization of the 40 genes using transgenics may help give a clear picture and add new insights into tanshinone biosynthesis in S. miltiorrhiza.
Conclusions
Based on the S. miltiorrhiza genome information, 40 terpenoid biosynthesis-related genes were obtained, of which 13 have been reported previously, while the other 27 are novel. These genes can be grouped into 19 families, which include 10 single- and nine multigene families. They encode all of the enzymes involved in the first and second stages of terpenoid biosynthesis and two associated with the third stage. The genomic DNA sequences for these genes were also identified. Using a comprehensive approach, the gene structures and gene expression patterns were analysed. The conserved domains and phylogenetic relationships among the deduced S. miltiorrhiza proteins and their homologues isolated from other plant species were analysed. Many unique features of terpenoid biosynthesis-related genes were revealed in S. miltiorrhiza. Some of the key enzymes, such as DXS, HDR, HMGR, and GGPPS, are encoded by multiple gene members with different expression patterns and subcellular localizations, suggesting the complexity of terpenoid biosynthesis in S. miltiorrhiza. The results support the view that specific groups of terpenoids can be synthesized by specific isoenzymes organized by metabolic channels within the pathway. On the other hand, an isoenzyme may be involved in the biosynthesis of various terpenoids by inserting into different metabolic units (Chappell, 1995; Lluch et al., 2000). Through a systematic analysis, a total of 20 genes were identified that could be involved in the biosynthesis of tanshinones, a group of diterpenoids with significant bioactivities. These results will provide a better understanding of terpenoid biosynthesis in S. miltiorrhiza and other plant species and provide the basis for improving tanshinone production through genetic engineering.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Exon and intron structures of 40 terpenoid biosynthesis-related genes.
Figure S2. Conserved domains of enzymes involved in terpenoid biosynthesis in S. miltiorrhiza.
Figure S3–26. Sequence alignment and phylogenetic analysis of deduced terpenoid biosynthesis-related proteins from S. miltiorrhiza and various other plants.
Figure S27. Expression patterns of 40 terpenoid biosynthesis-related genes in various tissues of S. miltiorrhiza plants.
Figure S28. Expression patterns of 40 terpenoid biosynthesis-related genes in S. miltiorrhiza plantlets treated with MeJA.
Table S1. Primers used for quantitative real-time RT-PCR.
Table S2. Conserved motifs of IDSs in various plant species.
Acknowledgments
We thank the sequencing group in our institute for kindly providing the S. miltiorrhiza genome sequence. We also thank Dr Chang Liu for his assistance in sequence analysis, and Dr Xu Zeng for aiding in phylogenetic tree construction. This work was supported by grants from the Beijing Natural Science Foundation (grant no. 5112026 to SL), the Major Scientific and Technological Special Project for Significant New Drugs Creation (grant no. 2012ZX09301002-001-030 to SL), and the Natural Science Foundation of China (grant no. 81102727 to YM).
Glossary
Abbreviations
- AACT
acetyl-CoA C-acetyltransferase
- CDP-ME
4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol
- CDP-ME2P
2-phospho-4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol
- CMK
4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol kinase
- CPS
copalyl diphosphate synthase
- DMAPP
dimethylallyl diphosphate
- DXP
1-deoxy-D-xylulose 5-phosphate
- DXR
1-deoxy-D-xylulose 5-phosphate reductoisomerase
- DXS
1-deoxy-D-xylulose 5-phosphate synthase
- FPP
farnesyl diphosphate
- FPPS
farnesyl diphosphate synthase
- G3P
glyceraldehyde 3-phosphate
- GA
gibberellin
- GGPP
geranylgeranyl diphosphate
- GGPPS
geranylgeranyl diphosphate synthase
- GPP
geranyl diphosphate
- GPPS
geranyl diphosphate synthase
- HDR
4-hydroxy-3-methylbut-2-enyl diphosphate reductase
- HDS
4-hydroxy-3-methylbut-2-enyl diphosphate synthase
- HMBPP
4-hydroxy-3-methylbut-2-enyl diphosphate
- HMG-CoA
3-Hydroxy-3-methylglutaryl-CoA
- HMGR
hydroxymethylglutaryl-CoA reductase
- HMGS
hydroxymethylglutaryl-CoA synthase
- IDI
isopentenyl diphosphate isomerase
- IDS
isoprenyl diphosphate synthase
- IPP
isopentenyl diphosphate
- KS
kaurene synthase
- KSL
kaurene synthase-like
- MCT
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase
- MDC
mevalonate pyrophosphate decarboxylase
- MDS
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
- MEcPP
2-C-methyl-D-erythritol 2,4-cyclodiphosphate
- MeJA
methyl jasmonate
- MEP
2-C-methyl-D-erythritol 4-phosphate
- MK
mevalonate kinase
- MVA
mevalonate
- MVAP
mevalonate-5-phosphate
- MVAPP
mevalonate-5-diphosphate
- PMK
5-phosphomevalonate kinase
- TPS
terpene synthase
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjellqvist B, Basse B, Olsen E, Celis JE. Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis. 1994;15:529–539. doi: 10.1002/elps.1150150171. [DOI] [PubMed] [Google Scholar]
- Bohlmann J, Keeling CI. Terpenoid biomaterials. The Plant Journal. 2008;54:656–669. doi: 10.1111/j.1365-313X.2008.03449.x. [DOI] [PubMed] [Google Scholar]
- Botella-Pavia P, Besumbes O, Phillips MA, Carretero-Paulet L, Boronat A, Rodriguez-Concepcion M. Regulation of carotenoid biosynthesis in plants: evidence for a key role of hydroxymethylbutenyl diphosphate reductase in controlling the supply of plastidial isoprenoid precursors. The Plant Journal. 2004;40:188–199. doi: 10.1111/j.1365-313X.2004.02198.x. [DOI] [PubMed] [Google Scholar]
- Bouvier F, Suire C, d’Harlingue A, Backhaus RA, Camara B. Molecular cloning of geranyl diphosphate synthase and compartmentation of monoterpene synthesis in plant cells. The Plant Journal. 2000;24:241–252. doi: 10.1046/j.1365-313x.2000.00875.x. [DOI] [PubMed] [Google Scholar]
- Bozin B, Mimica-Dukic N, Samojlik I, Jovin E. Antimicrobial and antioxidant properties of rosemary and sage (Rosmarinus officinalis L. and Salvia officinalis L., Lamiaceae) essential oils. Journal of Agriculture and Food Chemistry. 2007;55:7879–7885. doi: 10.1021/jf0715323. [DOI] [PubMed] [Google Scholar]
- Burge CB, Karlin S. Finding the genes in genomic DNA. Current Opinion in Structural Biology. 1998;8:346–354. doi: 10.1016/s0959-440x(98)80069-9. [DOI] [PubMed] [Google Scholar]
- Burke C, Croteau R. Geranyl diphosphate synthase from Abies grandis: cDNA isolation, functional expression, and characterization. Archives of Biochemistry and Biophysics. 2002;405:130–136. doi: 10.1016/s0003-9861(02)00335-1. [DOI] [PubMed] [Google Scholar]
- Burke CC, Wildung MR, Croteau R. Geranyl diphosphate synthase: cloning, expression, and characterization of this prenyltransferase as a heterodimer. Proceedings of the National Academy of Sciences, USA. 1999;96:13062–13067. doi: 10.1073/pnas.96.23.13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TH, Hsieh FL, Ko TP, Teng KH, Liang PH, Wang AH. Structure of a heterotetrameric geranyl pyrophosphate synthase from mint (Mentha piperita) reveals intersubunit regulation. The Plant Cell. 2010;22:454–467. doi: 10.1105/tpc.109.071738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J. The biochemistry and molecular biology of isoprenoid metabolism. Plant Physiology. 1995;107:1–6. doi: 10.1104/pp.107.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Yuan JP, Chen F, Zhang YL, Song JY. Tanshinone production in Ti-transformed Salvia miltiorrhiza cell suspension cultures. Journal of Biotechnology. 1997;58:147–156. doi: 10.1016/s0168-1656(97)00144-2. [DOI] [PubMed] [Google Scholar]
- Cheng TO. Danshen: a popular Chinese cardiac herbal drug. Journal of the American College of Cardiology. 2006;47:1498. doi: 10.1016/j.jacc.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Choi D, Ward BL, Bostock RM. Differential induction and suppression of potato 3-hydroxy-3-methylglutaryl coenzyme A reductase genes in response to Phytophthora infestans and to its elicitor arachidonic acid. The Plant Cell. 1992;4:1333–1344. doi: 10.1105/tpc.4.10.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordoba E, Porta H, Arroyo A, San Roman C, Medina L, Rodriguez-Concepcion M, Leon P. Functional characterization of the three genes encoding 1-deoxy-D-xylulose 5-phosphate synthase in maize. Journal of Experimental Botany. 2011;62:2023–2038. doi: 10.1093/jxb/erq393. [DOI] [PubMed] [Google Scholar]
- Cui G, Huang L, Tang X, Zhao J. Candidate genes involved in tanshinone biosynthesis in hairy roots of Salvia miltiorrhiza revealed by cDNA microarray. Molecular Biology Reports. 2011;38:2471–2478. doi: 10.1007/s11033-010-0383-9. [DOI] [PubMed] [Google Scholar]
- Dai Z, Cui G, Zhou SF, Zhang X, Huang L. Cloning and characterization of a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Salvia miltiorrhiza involved in diterpenoid tanshinone accumulation. Journal of Plant Physiology. 2011;168:148–157. doi: 10.1016/j.jplph.2010.06.008. [DOI] [PubMed] [Google Scholar]
- De Martino L, Roscigno G, Mancini E, De Falco E, De Feo V. Chemical composition and antigerminative activity of the essential oils from five Salvia species. Molecules. 2010;15:735–746. doi: 10.3390/molecules15020735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protocols. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Engprasert S, Taura F, Kawamukai M, Shoyama Y. Molecular cloning and functional expression of geranylgeranyl pyrophosphate synthase from Coleus forskohlii Briq. BMC Plant Biology. 2004;4:18. doi: 10.1186/1471-2229-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez JM, Cantero A, Reindl A, Reichler S, Leon P. 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. Journal of Biological Chemistry. 2001;276:22901–22909. doi: 10.1074/jbc.M100854200. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, et al. The Pfam protein families database. Nucleic Acids Research. 2009;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen JA, Rodwell VW. The 3-hydroxy-3-methylglutaryl coenzyme-A (HMG-CoA) reductases. Genome Biology. 2004;5:248. doi: 10.1186/gb-2004-5-11-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Hillwig ML, Huang L, Cui G, Wang X, Kong J, Yang B, Peters RJ. A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Organic Letters. 2009;11:5170–5173. doi: 10.1021/ol902051v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Wu JY. Tanshinone production and isoprenoid pathways in Salvia miltiorrhiza hairy roots induced by Ag+ and yeast elicitor. Plant Science. 2005;168:487–491. [Google Scholar]
- Gershenzon J, Dudareva N. The function of terpene natural products in the natural world. Nature Chemical Biology. 2007;3:408–414. doi: 10.1038/nchembio.2007.5. [DOI] [PubMed] [Google Scholar]
- Guo AY, Zhu QH, Chen X, Luo JC. GSDS: a gene structure display server. Yi Chuan. 2007;29:1023–1026. [PubMed] [Google Scholar]
- Ha SH, Kim JB, Hwang YS, Lee SW. Molecular characterization of three 3-hydroxy-3-methylglutaryl-CoA reductase genes including pathogen-induced Hmg2 from pepper (Capsicum annuum) Biochimica et Biophysica Acta. 2003;1625:253–260. doi: 10.1016/s0167-4781(02)00624-3. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Honda G, Koezuka Y, Tabata M. Isolation of an antidermatophytic substance from the root of Salvia miltiorrhiza. Chemical and Pharmaceutical Bulletin (Tokyo) 1988;36:408–411. doi: 10.1248/cpb.36.408. [DOI] [PubMed] [Google Scholar]
- Hsieh MH, Chang CY, Hsu SJ, Chen JJ. Chloroplast localization of methylerythritol 4-phosphate pathway enzymes and regulation of mitochondrial genes in ispD and ispE albino mutants in Arabidopsis. Plant Molecular Biology. 2008;66:663–673. doi: 10.1007/s11103-008-9297-5. [DOI] [PubMed] [Google Scholar]
- Kai G, Liao P, Zhang T, Zhou W, Wang G, Xu H, Liu Y, Zhang L. Characterization, expression profiling, and functional identification of a gene encoding geranylgeranyl diphosphate synthase from Salvia miltiorrhiza. Biotechnology and Bioprocess Engineering. 2010;15:236–245. [Google Scholar]
- Kai G, Xu H, Zhou C, Liao P, Xiao J, Luo X, You L, Zhang L. Metabolic engineering tanshinone biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. Metabolic Engineering. 2011;13:319–327. doi: 10.1016/j.ymben.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lange BM, Ghassemian M. Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Molecular Biology. 2003;51:925–948. doi: 10.1023/a:1023005504702. [DOI] [PubMed] [Google Scholar]
- Lange BM, Rujan T, Martin W, Croteau R. Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proceedings of the National Academy of Sciences, USA. 2000;97:13172–13177. doi: 10.1073/pnas.240454797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laule O, Furholz A, Chang HS, Zhu T, Wang X, Heifetz PB, Gruissem W, Lange M. Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 2003;100:6866–6871. doi: 10.1073/pnas.1031755100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YG, Song L, Liu M, Hu ZB, Wang ZT. Advancement in analysis of Salviae miltiorrhizae Radix et Rhizoma (Danshen) Journal of Chromatogry A. 2009;1216:1941–1953. doi: 10.1016/j.chroma.2008.12.032. [DOI] [PubMed] [Google Scholar]
- Liao P, Zhou W, Zhang L, Wang J, Yan X, Zhang Y, Zhang R, Li L, Zhou G, Kai G. Molecular cloning, characterization and expression analysis of a new gene encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase from Salvia miltiorrhiza. Acta Physiologiae Plantarum. 2009;31:565–572. [Google Scholar]
- Liao Z, Gong Y, Kai G, Zuo K, Chen M, Tan Q, Wei Y, Guo L, Tan F, Sun X, Tang K. An intron-free methyl jasmonate inducible geranylgeranyl diphosphate synthase gene from Taxus media and its functional identification in yeast. Molekuliarnaia Biologiia. 2005;39:14–20. [PubMed] [Google Scholar]
- Lluch MA, Masferrer A, Arro M, Boronat A, Ferrer A. Molecular cloning and expression analysis of the mevalonate kinase gene from Arabidopsis thaliana. Plant Molecular Biology. 2000;42:365–376. doi: 10.1023/a:1006325630792. [DOI] [PubMed] [Google Scholar]
- Lombard J, Moreira D. Origins and early evolution of the mevalonate pathway of isoprenoid biosynthesis in the three domains of life. Molecular Biology and Evolution. 2011;28:87–99. doi: 10.1093/molbev/msq177. [DOI] [PubMed] [Google Scholar]
- Munro TA, Rizzacasa MA, Roth BL, Toth BA, Yan F. Studies toward the pharmacophore of salvinorin A, a potent kappa opioid receptor agonist. Journal of Medicinal Chemistry. 2005;48:345–348. doi: 10.1021/jm049438q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Okada K, Saito T, Nakagawa T, Kawamukai M, Kamiya Y. Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiology. 2000;122:1045–1056. doi: 10.1104/pp.122.4.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MA, Walter MH, Ralph SG, et al. Functional identification and differential expression of 1-deoxy-D-xylulose 5-phosphate synthase in induced terpenoid resin formation of Norway spruce (Picea abies) Plant Molecular Biology. 2007;65:243–257. doi: 10.1007/s11103-007-9212-5. [DOI] [PubMed] [Google Scholar]
- Ramos-Valdivia AC, van der Heijden R, Verpoorte R. Isopentenyl diphosphate isomerase: a core enzyme in isoprenoid biosynthesis. A review of its biochemistry and function. Natural Product Reports. 1997;14:591–603. doi: 10.1039/np9971400591. [DOI] [PubMed] [Google Scholar]
- Ramsay H, Rieseberg LH, Ritland K. The correlation of evolutionary rate with pathway position in plant terpenoid biosynthesis. Molecular Biology and Evolution. 2009;26:1045–1053. doi: 10.1093/molbev/msp021. [DOI] [PubMed] [Google Scholar]
- Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, Antonicelli GE, Rasche N, Luder F, Weckwerth W, Jahn O. Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. The Plant Cell. 2007;19:3170–3193. doi: 10.1105/tpc.107.050989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou C, Tourte Y, Lacroute F, Karst F. Isolation and characterization of a cDNA encoding Arabidopsis thaliana mevalonate kinase by genetic complementation in yeast. Gene. 1994;148:293–297. doi: 10.1016/0378-1119(94)90701-3. [DOI] [PubMed] [Google Scholar]
- Rohdich F, Wungsintaweekul J, Eisenreich W, Richter G, Schuhr CA, Hecht S, Zenk MH, Bacher A. Biosynthesis of terpenoids: 4-diphosphocytidyl-2C-methyl-D-erythritol synthase of Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 2000;97:6451–6456. doi: 10.1073/pnas.97.12.6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods in Molecular Biology. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Sando T, Takaoka C, Mukai Y, Yamashita A, Hattori M, Ogasawara N, Fukusaki E, Kobayashi A. Cloning and characterization of mevalonate pathway genes in a natural rubber producing plant, Hevea brasiliensis. Bioscience, Biotechnology, and Biochemistry. 2008a;72:2049–2060. doi: 10.1271/bbb.80165. [DOI] [PubMed] [Google Scholar]
- Sando T, Takeno S, Watanabe N, Okumoto H, Kuzuyama T, Yamashita A, Hattori M, Ogasawara N, Fukusaki E, Kobayashi A. Cloning and characterization of the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway genes of a natural-rubber producing plant, Hevea brasiliensis. Bioscience, Biotechnology, and Biochemistry. 2008b;72:2903–2917. doi: 10.1271/bbb.80387. [DOI] [PubMed] [Google Scholar]
- Sapir-Mir M, Mett A, Belausov E, Tal-Meshulam S, Frydman A, Gidoni D, Eyal Y. Peroxisomal localization of Arabidopsis isopentenyl diphosphate isomerases suggests that part of the plant isoprenoid mevalonic acid pathway is compartmentalized to peroxisomes. Plant Physiology. 2008;148:1219–1228. doi: 10.1104/pp.108.127951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin AJ, Guirimand G, Papon N, Courdavault V, Thabet I, Ginis O, Bouzid S, Giglioli-Guivarc'h N, Clastre M. Peroxisomal localisation of the final steps of the mevalonic acid pathway in planta. Planta. 2011;234:903–914. doi: 10.1007/s00425-011-1444-6. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tholl D, Kish CM, Orlova I, Sherman D, Gershenzon J, Pichersky E, Dudareva N. Formation of monoterpenes in Antirrhinum majus and Clarkia breweri flowers involves heterodimeric geranyl diphosphate synthases. The Plant Cell. 2004;16:977–992. doi: 10.1105/tpc.020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schie CC, Ament K, Schmidt A, Lange T, Haring MA, Schuurink RC. Geranyl diphosphate synthase is required for biosynthesis of gibberellins. The Plant Journal. 2007;52:752–762. doi: 10.1111/j.1365-313X.2007.03273.x. [DOI] [PubMed] [Google Scholar]
- Walter MH, Hans J, Strack D. Two distantly related genes encoding 1-deoxy-D-xylulose 5-phosphate synthases: differential regulation in shoots and apocarotenoid-accumulating mycorrhizal roots. The Plant Journal. 2002;31:243–254. doi: 10.1046/j.1365-313x.2002.01352.x. [DOI] [PubMed] [Google Scholar]
- Wang G, Dixon RA. Heterodimeric geranyl (geranyl) diphosphate synthase from hop (Humulus lupulus) and the evolution of monoterpene biosynthesis. Proceedings of the National Academy of Sciences, USA. 2009;106:9914–9919. doi: 10.1073/pnas.0904069106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Ohnuma S. Chain-length determination mechanism of isoprenyl diphosphate synthases and implications for molecular evolution. Trends in Biochemical Science. 1999;24:445–451. doi: 10.1016/s0968-0004(99)01464-4. [DOI] [PubMed] [Google Scholar]
- Wang X, Bastow KF, Sun CM, Lin YL, Yu HJ, Don MJ, Wu TS, Nakamura S, Lee KH. Antitumor agents. 239. Isolation, structure elucidation, total synthesis, and anti-breast cancer activity of neo-tanshinlactone from Salvia miltiorrhiza. Journal of Medicinal Chemistry. 2004;47:5816–5819. doi: 10.1021/jm040112r. [DOI] [PubMed] [Google Scholar]
- Wang X, Cui G, Huang L, Gao W, Yuan Y. A full length cDNA of 4-(cytidine 5'-2 diphospho)-2-C-methyl-D-erythritol kinase cloning and analysis of introduced gene expression in Salvia miltiorrhiza. Acta Pharmaceutica Sinica. 2010;43:1251–1257. [PubMed] [Google Scholar]
- Wu SJ, Shi M, Wu JY. Cloning and characterization of the 1-deoxy-D-xylulose 5-phosphate reductoisomerase gene for diterpenoid tanshinone biosynthesis in Salvia miltiorrhiza (Chinese sage) hairy roots. Biotechnology and Applied Biochemistry. 2009;52:89–95. doi: 10.1042/BA20080004. [DOI] [PubMed] [Google Scholar]
- Xu C, Shu Z, Wang Y, Miao F, Zhou L. The accumulation rule of the main medicinal components in different organs of Salvia miltiorrhiza Bunge. and Salvia miltiorrhiza Bunge. f. alba. Lishizhen Medicine and Materia Medica Research. 2010;21:2129–2132. [Google Scholar]
- Yousefzadi M, Sonboli A, Karimic F, Ebrahimi SN, Asghari B, Zeinalia A. Antimicrobial activity of some Salvia species essential oils from Iran. Zeitschrift für Naturforschung C. 2007;62:514–518. doi: 10.1515/znc-2007-7-809. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yan X, Wang J, Li S, Liao P, Kai G. Molecular cloning and expression analysis of a new putative gene encoding 3-hydroxy-3-methylglutaryl-CoA synthase from Salvia miltiorrhiza. Acta Physiologiae Plantarum. 2011;33:953–961. [Google Scholar]
- Zulak KG, Bohlmann J. Terpenoid biosynthesis and specialized vascular cells of conifer defense. Journal of Integrative Plant Biology. 2010;52:86–97. doi: 10.1111/j.1744-7909.2010.00910.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.