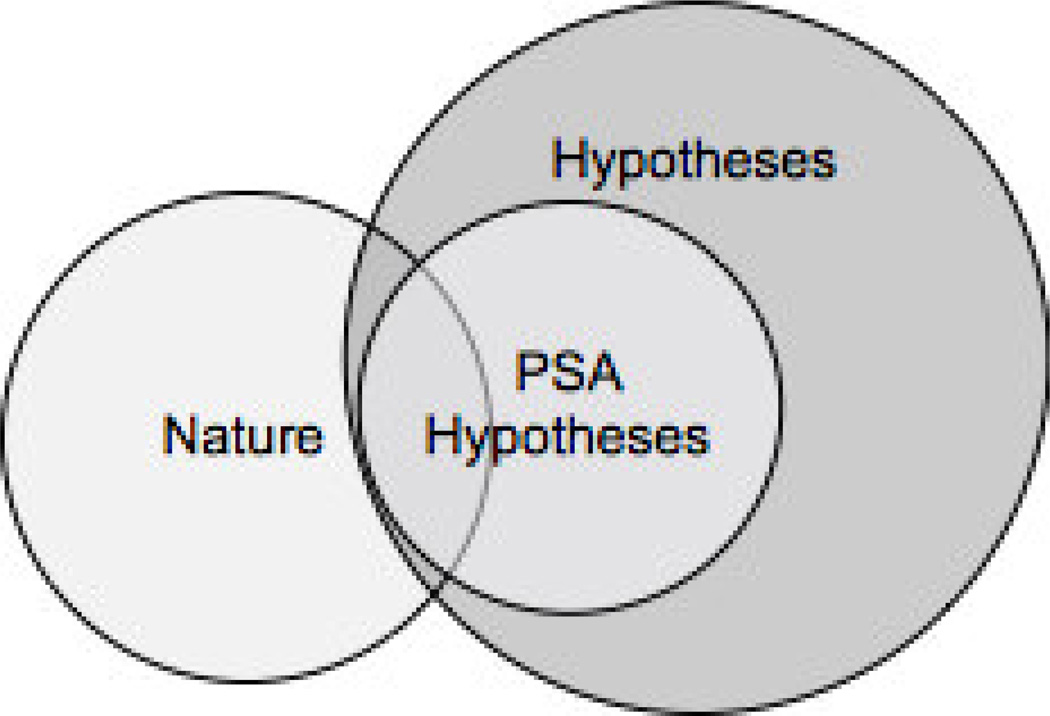
Abstract
Motivation
Although trauma is the leading cause of death for those below 45 years of age, there is a dearth of information about the temporal behavior of the underlying biological mechanisms in those who survive the initial trauma only to later suffer from syndromes such as multiple organ failure. Levels of serum cytokines potentially affect the clinical outcomes of trauma; understanding how cytokine levels modulate intra-cellular signaling pathways can yield insights into molecular mechanisms of disease progression and help to identify targeted therapies. However, developing such analyses is challenging since it necessitates the integration and interpretation of large amounts of heterogeneous, quantitative and qualitative data. Here we present the Pathway Semantics Algorithm (PSA), an algebraic process of node and edge analyses of evoked biological pathways over time for in silico discovery of biomedical hypotheses, using data from a prospective controlled clinical study of the role of cytokines in multiple organ failure (MOF) at a major US trauma center. A matrix algebra approach was used in both the PSA node and PSA edge analyses with different matrix configurations and computations based on the biomedical questions to be examined. In the edge analysis, a percentage measure of crosstalk called XTALK was also developed to assess cross-pathway interference.
Results
In the node/molecular analysis of the first 24 hours from trauma, PSA uncovered 7 molecules evoked computationally that differentiated outcomes of MOF or non-MOF (NMOF), of which 3 molecules had not been previously associated with any shock / trauma syndrome. In the edge/molecular interaction analysis, PSA examined four categories of functional molecular interaction relationships – activation, expression, inhibition, and transcription – and found that the interaction patterns and crosstalk changed over time and outcome. The PSA edge analysis suggests that a diagnosis, prognosis or therapy based on molecular interaction mechanisms may be most effective within a certain time period and for a specific functional relationship.
Keywords: Systems biology, signaling pathways, trauma, hypothesis generation, biomedical informatics
1. Introduction
In recent years, advances in technology have made it possible to measure a wide variety of molecules and molecular interactions in cell lines, bio-fluids and tissues. The increasing availability of these data has opened new avenues of biomedical research, and challenged the scientific community to uncover the meaning of molecular data in contexts ranging from cell signaling pathways to phenotype/genotype associations to personalized medicine [8]. Plausible and meaningful molecular hypotheses that support clinical diagnosis, prognosis and therapies must be derived from a deluge of quantitative and qualitative experimental data that are spread over a variety of experimental paradigms such as clinical outcome, time, cell cycle phase, or molecular localization.
Current approaches to collecting data about molecular patterns in disease include the use of high throughput measurement techniques such as mass spectrometry and microarray immunoassays. Mass spectrometry is the most common technique for “unbiased” discovery where all protein and peptide components of tissues and biofluids are identified within the capability of the equipment. Microarray immunoassays are more sensitive and specific; they measure the concentrations of pre-determined analytes using immunological reactions. Both assay methods have benefits and drawbacks for clinical usage [11].
A wide variety of analytical approaches, both qualitative and quantitative, are being explored to understand these data [18]. Text mining algorithms search published literature for information about molecular function and disease associations while graphical analysis uses algorithms from computer science to identify subgraph motifs in canonical pathway networks of molecular interactions found in diseases. Network-based graphical analysis using gene expression patterns has been shown to generate novel hypotheses about the classification of breast cancer metastasis, including the finding that some gene associations can only be detected using network rather than conventional analysis [20]. Statistical biomedical informatics methods, such as gene set enrichment analysis (GSEA), identify gene sets, based on gene expression data, that are correlated with phenotypic classes, and generate hypotheses for further exploration [22, 23]. Systems biology tools model in silico biological pathway systems using computational methods that parallel in vitro cell-line and in vivo animal models for hypothesis discovery and instantiation [24].
Although these approaches are useful, there are limitations for the study of disease progression over time. For example, the most significant molecular interactions associated with the disease may appear in a non-canonical pathway [25] that text mining and in silico modeling may overlook. Time-based models of biological pathways can be explored using ordinary differential equations (ODEs); however, they usually model a small group of canonical pathways within a single cell and are not easily computable at the organism level. For example, an ODE model of one NF-kappa B signaling pathway in one cell activated by one TNF-α signaling molecule uses 18 nonlinear differential equations, with 33 independent variables and 16 dependent variables in a simplified reaction kinetics model [26].
Studies of scientific discovery have demonstrated that most new findings arise from data-driven hypotheses generated from unexpected observations rather than from verification of pre-determined hypotheses based on theories [27]. In a bedside-to-bench approach, discovery is driven by patient data collected at the bedside. Mechanisms or therapies are confirmed later at the lab bench. Data-driven, evidence-based molecular patterns are a fundamental component of personalized medicine research. Notable diagnostic successes based on the molecular patterns found in patient data include the validation of 14-3-3 proteins found in cerebrospinal fluid (CSF) as diagnostic of transmissible spongiform encephalopathies [28] and the validation of a panel of 18 urinary molecules that discriminate antibody-associated vasculitis from other renal diseases [29].
Here we present the Pathway Semantics Algorithm (PSA) that converts the directed graphs of the most likely biological pathways evoked from patients’ molecular data into transformed matrices of various formats for algebraic analysis, with the goal of generating hypotheses addressing specific biomedical questions about the meaning, or “semantics”, of the pathways. The term hypothesis is used in its broadest sense as a potential explanation or conclusion that is to be tested by collecting and presenting evidence [30]. Generating hypotheses computationally based on scientific and plausible reasoning extends the domain of search beyond that which was originally observed or “known”, while reducing the size of the solution space. In the sample disease progression analysis given in Section 4, the pathway generation algorithm gave a potential solution space of more than 1,000 molecule/time points. Using PSA algebraic node analysis, the solution space was reduced to 7 molecules that differentiated outcomes at different times. The pathway graphs contain two major types of entities: nodes that correspond to specific bio-molecules and edges that correspond to the interactions between the molecules. PSA constructs the matrices to represent biomedical queries for comparative analyses of the pathways over stratifications such as time and/or outcome. Matrix construction is specific to the query because scientific discovery is strongly influenced by data representation [31]. The transformation of graphs to matrices enables the application of powerful computationally tractable techniques that scale well from matrix algebra to develop mathematical comparison methods, analyses, and metrics leading to useful insights into disease progression across time and clinical outcomes.
PSA was applied to patient data from a shock / trauma study of multiple organ failure, first analyzing the nodes of the likely biological pathways and then examining the edges. A matrix format called a Temporal Dependency Matrix(TDM) was instrumental in revealing novel patterns of molecules evoked from patient data over time in shock/trauma, where disease progression is rapid yet not clinically visible. The computational results predicted seven molecules, based on input from the original assays, associated with the biological mechanisms underlying multiple organ failure; only three had been recognized as associated with any shock / trauma syndrome. Next PSA examined the edges of the pathway graphs, corresponding to interactions between molecules including genes, RNAs, proteins, or chemicals. We applied matrix methods to investigate patterns of molecular interactions across time and across clinical outcomes in terms of four functional relationship categories: activation, expression, transcription and inhibition. Applying graph theory and linear algebra, we found that the interaction patterns of relationship sub-graphs changed rapidly within the first 24 hours of insult, and that these patterns differed across clinical outcomes of multiple organ failure (MOF) and non-multiple organ failure (non-MOF). In addition, we developed a numerical metric of crosstalk in molecular pathways called XTALK. In contrast to current practice that merely classifies a network in strictly binary fashion as having crosstalk or not, XTALK quantifies crosstalk among molecular interactions from 0% to 100%, thereby leading to a deeper, fine-grained understanding of crosstalk and its variation due to disease progression. Results obtained suggest that a diagnosis, prognosis or therapy based on molecular interaction mechanisms may be most effective within a certain time period and for a certain functional interaction relationship.
In the following sections, we first present background information and definitions relating to molecular interactions and mathematical notation, followed by a description of the Pathway Semantics Algorithm, its application to analysis of patterns of molecules and molecular interactions in the first 24 hours of trauma progression, the results and a discussion of their meaning, concluding with our plans for future work.
2. Background
At a sub-cellular level, molecular interactions can be analyzed using the rules of biochemistry when they are represented as sets of differential equations. However, due to computational complexity and lack of interaction parameter rate data, this approach is not suitable for larger comparative analyses. Instead, molecular interactions, such as protein-protein or gene-protein interactions, are commonly combined into biological pathway networks represented as graphs, where the node, or vertex, is the molecule and the edge is the interaction. This representation facilitates the use of qualitative and quantitative methods derived from graph theory and algebra because the same biological pathway network graph can be mapped to a matrix in different ways, allowing for a choice of mathematical methods appropriate to the biomedical question under study.
Biological pathway networks can be generated manually through direct observation of patient data, such as performed in morphoproteomic tissue analysis [32], and computationally through software, such as Ingenuity Pathway Analysis (IPA) [www.ingenuity.com][33] that uses a proprietary algorithm to evoke likely pathways generated from the measurements of molecular data in bio-fluids and tissues.
Comparative analyses of nodes in pathways can reveal key molecules that likely play significant roles in disease progression over all time, or only at certain times. Comparative analyses of edges, or links between the nodes, in pathways parallels research into “link communities” in social networks, where one person may be connected to several overlapping communities of home, work, and interests [34, 35]. In both social and biological networks, the edges are directional, showing the influence from one node (a person or molecule) upon another in a multi-directional cascade. Biological link communities also overlap; a molecule may participate in several different interaction categories simultaneously with the same target molecule, or inversely, several interactions may occur simultaneously with different molecules to achieve the same target function. This latter property has been defined as degeneracy – the ability of structurally different elements to perform the same function or yield the same output; in contrast, redundancy requires identical elements to perform the same function. [36, 37]. Degeneracy is a key property underlying the robustness of complex adaptive biological systems, such as the immune system [38–40].
We define crosstalk in biological pathways to consist of the redundant signaling messages sent over degenerate edges to achieve the same biological function. This is consistent with Bruni’s definition that crosstalk exists when edges are functionally compatible to, or dependent, on other edges [41]. Crosstalk relates to how pathways determine functional specificity, how ubiquitous messengers transmit specific information, and how similar messages crosslink within the system while undesired signals are minimized. Quantifying crosstalk in patient data-driven biological pathways can give insights into the relative robustness of different biological functions and suggest timing and approaches for therapies directed at pathway modulation. For simplicity, this study measured crosstalk in one molecular interaction function at a time in each pathway; cascades of “mixed-function” molecular interactions that overall would result in execution of the same target function were not considered.
2.1 Additional definitions
Notation and definitions used correspond to those used by Ingenuity Pathways Analysis (IPA) [42], the pathway generation software used in this study. The term node is used rather than vertex, and the term edge rather than arc.
A molecule is any gene, RNA, protein or chemical. A molecule is represented by a node on the directed graph of a biological pathway.
A relationship is a functional interaction from one molecule to another. The relationships used in this study are defined by Ingenuity Systems (Ingenuity Systems, personal communication) as follows:
Activation: includes activation events such as activation, activity, stimulation, reactivation, and specific activity.
Inhibition: includes inhibition events such as inhibition, desensitization, inactivation, repression and autoinhibition.
Expression: includes expression events such as expression, upregulation, downregulation, translation, production, microRNA targeting, and induction.
Transcription: includes transcription events such as transcription, germline transcription, transactivation, and transrepression.
A relationship is represented by an edge on the directed graph of a biological pathway.
A directed graph, in mathematical terminology, has specific properties that can be exploited computationally. IPA designates relationships as direct or indirect, in a different sense of the word “direct”. A direct relationship is a direct physical contact interaction between the two molecules; it includes chemical modifications, such as phosphorylations, if there is evidence that the two factors involved interact directly rather than through an intermediary. It is represented by a solid line edge. An indirect relationship is an interaction that does not require physical contact but is explicitly documented in the literature [42]. It is represented by a dotted line edge. A relationship graph is a directed graph whose edges are in the same relationship category. Molecules or edges are called invariant when they are the same in different stratifications. For example, edges are invariant over all time in one outcome if they do not change over all time periods for that outcome; alternatively, edges are invariant over outcome if they are the same in both outcomes in one time period or more as specified.
Let B(E,N) be a directed graph with E edges and N nodes that represents a biological pathway with relationship interactions as edges and molecules as nodes. Then A is a relationship sub-graph of B with A ⊆B when ∀E in A are in the same relationship category.
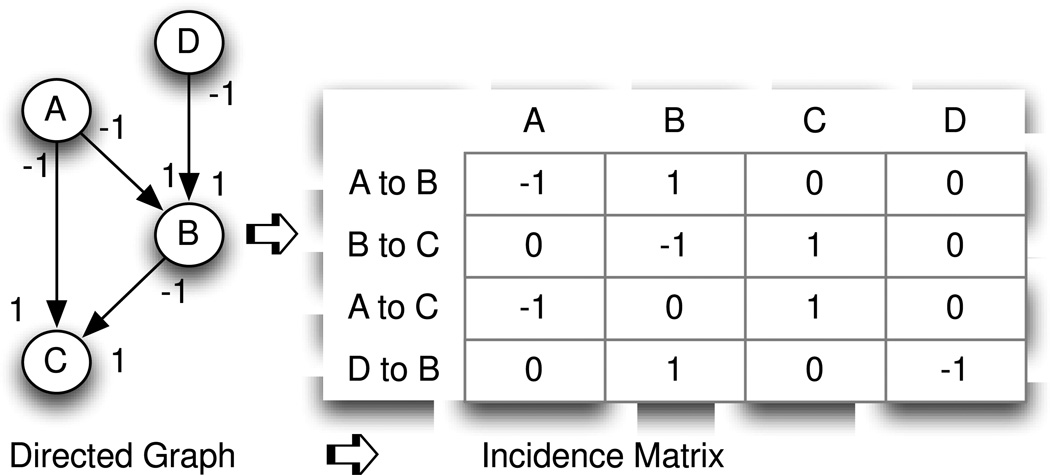
The incidence matrix M = [mij] of a directed graph B = B(E,N) is a E × N′ matrix, M(E,N′) where E = number of edges and N′ = number of nodes (with duplicate nodes for self-loops) such that mij = −1 if edge i leaves node j, +1 if edge i enters node j, 0 otherwise [43].
3. Pathway Semantics Algorithm
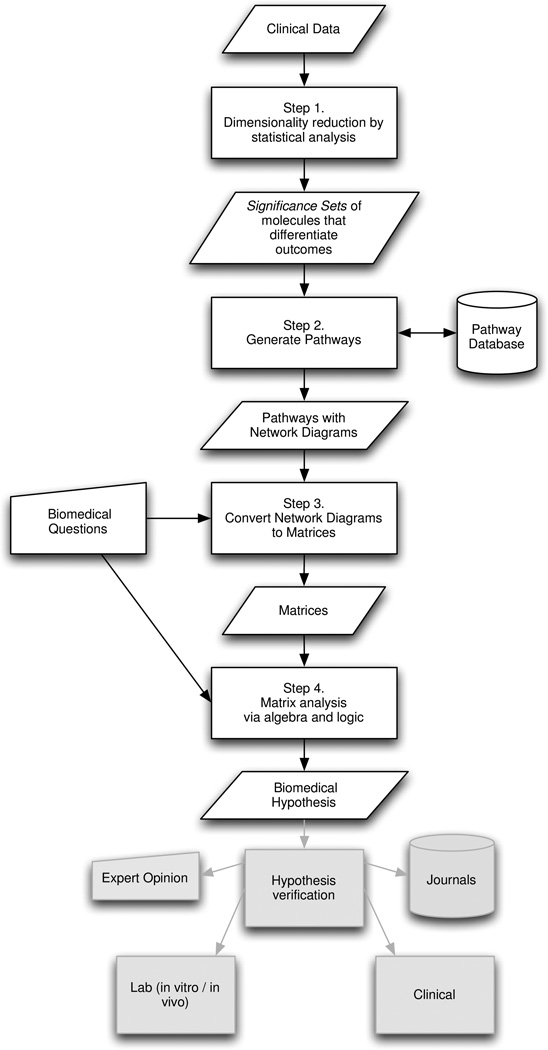
The Pathway Semantics Algorithm augments pathway generation and core analyses, such as those in IPA, through customized preprocessing of the measured molecular data and post-processing of the evoked pathways so that both input data and output matrices are tailored to the biological and clinical questions under study. The goal is to narrow down potential answers to those most likely and useful as clinical hypotheses (see Fig. 1).
Fig. 1.
The overarching goal of the Pathway Semantics Algorithm (PSA) is to efficiently generate clinically useful hypotheses about disease progression using matrix algebra to integrate quantitative and qualitative data.
PSA first processes the input data to generate biological pathways (Steps 1–2) and then maps the results to matrices constructed to answer the biomedical questions under study (Steps 3–4). If biological pathways are already available, for example, from morphoproteomic tissue analysis [44], only Steps 3 and 4 need be performed. See Fig. 2.
Fig. 2.
Pathway Semantics Algorithm (PSA) flow diagram
Step 1. Dimensionality Reduction
This process selects characteristic subsets of the measured molecules. The assayed molecules are assembled into Significance Sets of those molecules that statistically differentiate the disease states over the stratifications under study, such as outcome, time period of measurement, cell cycle phase observed, or a combination of stratifications. The statistical analysis is utilized as a feature extraction tool to identify significant molecules.
Step 2.Pathway Generation
The Significance Set for each stratification group plus the statistically observed average values (means or medians as appropriate) for each molecule in the group are input to a pathway generation algorithm that expands each set to include its likely neighboring molecules, based on published literature and pathway databases. A network diagram is then created of the biological pathways showing the interactions among the molecules for each stratification group.
Step 3. Convert Network Diagrams to Matrices
Matrix representations, suitable for the biomedical questions under study, are created from the network diagrams. For example, the molecules, or nodes, in the network diagrams can be mapped to node matrices (or vectors) of molecules over stratifications such as disease state and time. In a similar manner, molecular interactions, or edges, can be converted to edge matrices (or vectors) of molecular interactions over stratifications such as functional interaction types. In the simplest form, the resulting matrices have a 1 in a row/column cell if the row molecule (or molecular interaction) is present in the column stratification; 0 otherwise.
Step 4. Matrix Analysis
Algebra is used to compare the matrices to identify differential patterns of molecules and molecular interactions of biomedical significance over outcome, time and other stratifications. The specific calculations used depend on the biomedical questions represented by the matrices. For example, in PSA node analysis, matrices of node molecules over time and outcome can be added, subtracted, or logically compared through “ands” and “ors”. Similar calculations can be done with matrices of edge molecular interactions over time, outcome and functional relationship. In addition, when molecular interactions are represented as edges in an incidence matrix, matrix properties such as rank can be used to infer biological processes such as crosstalk.
Definition. The rank R of a matrix M is the maximal number of its linearly independent columns or rows [45]. Rank can be calculated using Gaussian elimination or singular value decomposition.
If rank R is greater than or equal to E, the number of edges (rows), then all the edges act independently. The percentage, or ratio, of independent edges = R/E, and the ratio of dependent edges is 1 – R/E.
We propose the biological interpretation that the maximum number of independent molecular interactions (edges) required for a molecular function is the same as the rank of the incidence matrix constructed from the functional relationship sub-graph, and that a measure of crosstalk for that function can be based on the percentage of dependent edges.
Definition. The XTALK ratio of a directed graph B = B(E,N) with incidence matrix M(E,N′) is defined as 1 – (rank (M(E,N′))/E).
If XTALK = 0%, then all edges act independently for a particular function. The XTALK measure includes normalization by the total number of edges in a graph to allow comparisons of crosstalk over time and outcome.
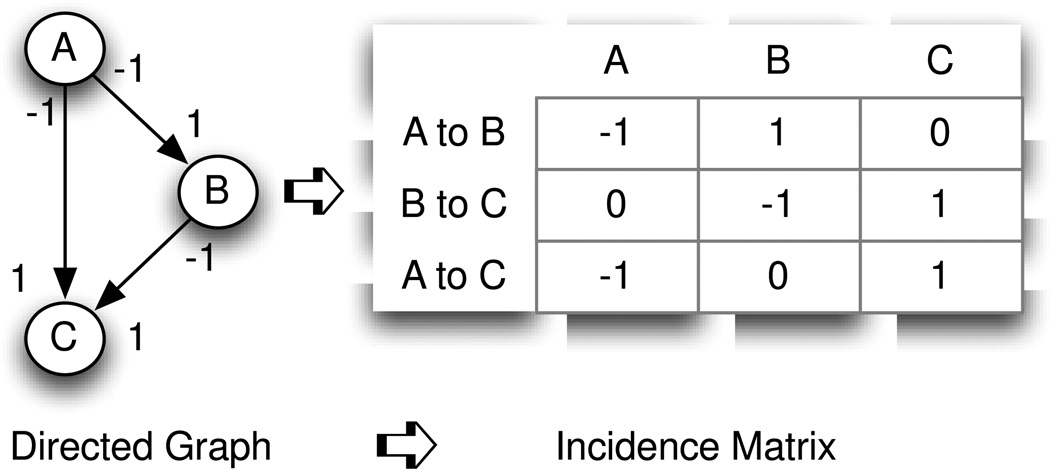
In Fig. 3, rank R = 2, number of edges = 3. XTALK = 1–(2/3) = 33%, suggesting there exists one-third crosstalk in the biological functional relationship represented by the graph.
Fig. 3.
The directed graph on the left with 3 nodes and 3 edges is a simplified representation of a common sub-graph found in a biological pathway network. To enable algebraic computation, the graph is mapped to the incidence matrix on the right. The column headers name the nodes of the graph (representing molecules in a pathway) and the rows represent the interaction pairs between two molecules, with the “from” node represented by a “−1” and the “to” node represented by a “1”. The calculated rank of this incidence matrix is 2, meaning that 2 edges are independent and 1 edge is dependent. It can be seen that the edge from A to C is a combination of edge A to B followed by edge B to C. The graph shows the property of degeneracy: the molecular interaction function – for example activation – can be achieved solely by molecule A acting on C or, instead, by the cascade of the molecule A acting on B followed by the molecule B acting upon C.
4. PSA gives novel hypotheses for shock/trauma progression
Trauma refers to serious bodily injury such as penetrating injuries from gunshots and stab wounds, blunt injuries such as those sustained during automotive accidents, and burns; trauma is the cause of 74% of all deaths for people ages 15–24 [46]. The term shock / trauma is used in this manuscript to refer to trauma that is associated with the clinical signs of shock, defined physiologically as oxygen consumption (VO2) inadequate to meet the oxygen demands of peripheral tissue. Disease progression in shock / trauma is rapid and deadly; patients who survive the initial trauma may suffer morbidity from potentially preventable syndromes such as multiple organ failure (MOF) [47, 48]. MOF is unique in that the organs that fail are not necessarily injured from the trauma and that late MOF may arise days to weeks after the initial incident. The pathophysiology underlying MOF is still not well understood [49, 50]. Patterns of signaling molecules called cytokines [51] have been associated with patient outcomes in trauma and critical care for some time [52–56], and analysis of the biological pathways evoked from cytokines may offer insights into disease progression. Cytokines are small proteins released by stimulated macrophages, monocytes, T cells, and other cells; they bind to specific receptors to induce a wide variety of local and systemic responses particularly within the innate and adaptive immune systems [51].
4.1 Data
PSA was applied to data from the Jastrow MOF study [54] that associated certain cytokine patterns within the first 24 hours from trauma with the outcome of multiple organ failure before other symptoms were visible [54]. In contrast, traditional predictors of MOF were not significantly different between MOF and non-MOF outcomes. The PSA goal was uncover patterns of evoked molecules and molecular interactions associated with shock / trauma progression that would lead to clinical hypotheses.
De-identified patient data from the Jastrow study [54] were extracted from the UTHSC-H Trauma Research Database with the approval of the Committee for the Protection of Human Subjects (Institutional Review Board / IRB) of the UTHSC-H (HSC-SHIS-09-0237). The data included serum cytokine measurements, collection times, and MOF outcomes for 48 patients from an IRB approved prospective observational trauma study conducted in the shock / trauma Intensive Care Unit (STICU) at Memorial Hermann Hospital, a Level I trauma center in Houston, Texas from January through December 2005. The 48 patients had a mean age of 39 ± 3 years, 67% were male, 88% of the insult was blunt mechanism, and the mean Injury Severity Score was 25 ± 2. MOF developed in 11 (23%) of the patients. Twenty-seven cytokines were measured every 4 hours from the start of the resuscitation protocol and were later assayed by the Bio-Plex Human Cytokine 27-Plex Panel. The measurement times were adjusted to time from insult, and grouped into 4 hour time periods starting at hour 2 from insult and ending at the study limit of hour 24. Twenty-seven cytokines were measured by Bio-Plex immunoassay. All were used for the PSA-Node analysis; eleven were used for the PSA-Edge analysis (see Table 1).
Table 1.
Cytokines were assayed using the Bio-Plex Human Cytokine 27-Plex Panel
| Cytokine | Gene Name | UNIPROT ID | PSA (Node, Edge) |
|---|---|---|---|
| Eotaxin | CCL11 | P51671 | N, E |
| FGF Basic | FGF2 | P09038 | N |
| G-CSF | CSF3 | P09919 | N, E |
| GM-CSF | CSF2 | P04141 | N, E |
| IFN-γ | IFNG | P01579 | N, E |
| IL-1β | IL1B | P01584 | N, E |
| IL-1ra | IL1RN | P18510 | N, E |
| IL-2 | IL2 | P60568 | N, E |
| IL-4 | IL4 | P05112 | N |
| IL-5 | IL5 | P05113 | N |
| IL-6 | IL6 | P05231 | N, E |
| IL-7 | IL7 | P13232 | N |
| IL-8 | IL8 | P10145 | N, E |
| IL-9 | IL9 | P15248 | N |
| IL-10 | IL10 | P22301 | N, E |
| IL12 (p70) | IL12A/B | P29459/P29460 | N |
| IL-13 | IL13 | P35225 | N |
| IL-15 | IL15 | P40933 | N |
| IL-17 | IL17A | Q16552 | N |
| IP-10 | CXCL10 | P02778 | N |
| MCP-1 | CCL2 | P13500 | N |
| MIP-1α | CCL3 | P10147 | N |
| MIP-1β | CCL4 | P13236 | N |
| PDGF-BB | PDGFB | P01127 | N |
| RANTES | CCL5 | P13501 | N |
| TNF-α | TNF | P01375 | N, E |
| VEGF | VEGFA | P15692 | N |
4.2 Data Pre-processing
The cytokine data were partitioned for analysis purposes into 6 groups by time periods: hours 2–6, 6–10, 10–14, 14–18, 18–22 and 22–24. The four-hour time period was chosen because that was the scheduled time between clinical measurements. The clinical data were pre-processed before Step 1 (see Fig. 2) as follows:
In order to preserve biological relationships over time, the measurement times were adjusted to biologically relevant start times, so that the biological activities “lined up” for analysis. Here, measurement times were adjusted to time from insult, since it was hypothesized that cytokine pattern activities would start changing at that time.
In order to preserve rankings among data, “low” and “high” nominal measurement data were replaced with calculated ordinal data instead of treating that data as missing values. Only true missing values were retained. Low measurements were replaced by 50% of the minimum value of the data over all stratifications and “high” by 150% of the maximum value of the data over all stratifications. These quantitative values were used only for ranked analysis. For example, [5, 2, 7, low, 9] -> [5, 2, 7, 1, 9]. All five data points would be retained and the rank order would be the same.
Because the measured molecules were signaling molecules, the number of molecules available to trigger biological pathways was considered more important than their total mass. The cytokine data were converted from pg/mL units to SI units before input to the software that generated the most likely biological pathways based on relative concentrations of molecules.
The data were grouped over stratifications to facilitate discrete analysis. This preserved the original data without making the continuity assumption that the concentrations of the cytokine molecules varied smoothly between measurement times.
For clarity and simplicity, the mathematical representation used was limited to vectors over time in the form of two-dimensional matrices.
Additional details on data preparation can be found in the Supplement 1, Section 1.
4.3 PSA-Node and PSA-Edge: Steps 1 and 2
In Steps 1 and 2, the Pathway Semantics Algorithm (PSA) reduces the dimensionality of the pre-processed input data to generate targeted biological pathways. The description that follows is for the PSA-Node analysis based on 27 cytokines.
Step 1: Dimensionality Reduction
Notation: I = number of time periods; A = number of significant molecules in a time period. Significance Sets Si=1,I of molecules ci=1,I;a=1,A that statistically differentiated the K outcomes qk=1,K over time periods xi=1,I were created based on the non-parametric Mann–Whitney–Wilcoxon (MWW) test (p<.05) executed in each of 6 time periods within the first 24 hours from insult. Outcomes were q1 = MOF (multiple organ failure) or q2 = NMOF (non-multiple organ failure). Time periods from insult were xi=1,6 = 2–6, 6–10, 10–14, 14–18, 18–22 and 22–24. The Significance Sets S1, S2 and S6 contained the names of 10 of the 27 measured cytokines; S3 and S5 contained 14 cytokines; and S4 had 15 cytokines. The names of the cytokines differed in each Si. For example, S1 contained: c1,1= Eotaxin; c1,2= G-CSF; c1,3= GM-CSF; c1,4= IFN-γ; c1,5= IL-1ra; c1,6= IL-6; c1,7= IL-8; c1,8= IP-10; c1,9= MCP-1 and c1,10= MIP-1 (See Table 2).
Table 2.
Dimensionality reduction was achieved by selecting for further analysis only the group of cytokine molecules identified as statistically significant outcome differentiators in each time period. Si contains the names of the molecules in the Significance Set in time period xi. X represents the median values vi,a,k for each outcome. Note that the significant molecules in Si differ by time period, reflecting the dynamic nature of the cytokine signaling patterns in shock / trauma progression.
| Cytokine | S1 | S2 | S3 | S4 | S5 | S6 |
|---|---|---|---|---|---|---|
| Eotaxin | X | X | X | X | X | X |
| G-CSF | X | X | X | X | X | X |
| GM-CSF | X | X | X | X | X | X |
| IFN-γ | X | X | X | X | X | |
| IL-1ra | X | X | X | X | X | X |
| IL-5 | X | X | ||||
| IL-6 | X | X | X | X | X | |
| IL-7 | X | X | X | |||
| IL-8 | X | X | X | X | X | X |
| IL-9 | X | |||||
| IL-10 | X | X | X | X | ||
| IL-13 | X | X | ||||
| IP-10 | X | X | X | X | X | X |
| MCP-1 | X | X | X | X | X | X |
| MIP-1β | X | X | X | X | X | |
| RANTES | X | |||||
| TNF-α | X | X | X |
Step 2: Pathway Generation
Ingenuity Pathways Analysis (IPA) was used to find the likely biological pathway networks associated with the levels of the measured molecules. IPA provides a literature and pathway database search along with a pathway generation algorithm that utilizes weighted lists of molecules (Ingenuity® Systems, www.ingenuity.com). The algorithm breaks “ties” about which neighbors to add to an evoked network based on the relative weightings of the input molecules [33]. Because the analytes were signaling molecules, the relative numbers of molecular signals, rather than the relative weights of the molecules, generate more representative biological pathways [57]. Therefore, two additional data modifications were performed. First, the units for the median values vi,a,k were converted from concentrations in pg/mL to v′i,a,k, the number of molecules per liter (pmol/L) based on the mass of the cytokine in kDa as reported in UniProt. Second, certain cytokines must be present in multiples or have multiple receptors to send signals. Therefore the v′i,a,k were further adjusted to v″i,a,k by how many molecules were required for one signal. The adjusted calculation details are given in Supplement 1, Section 2.
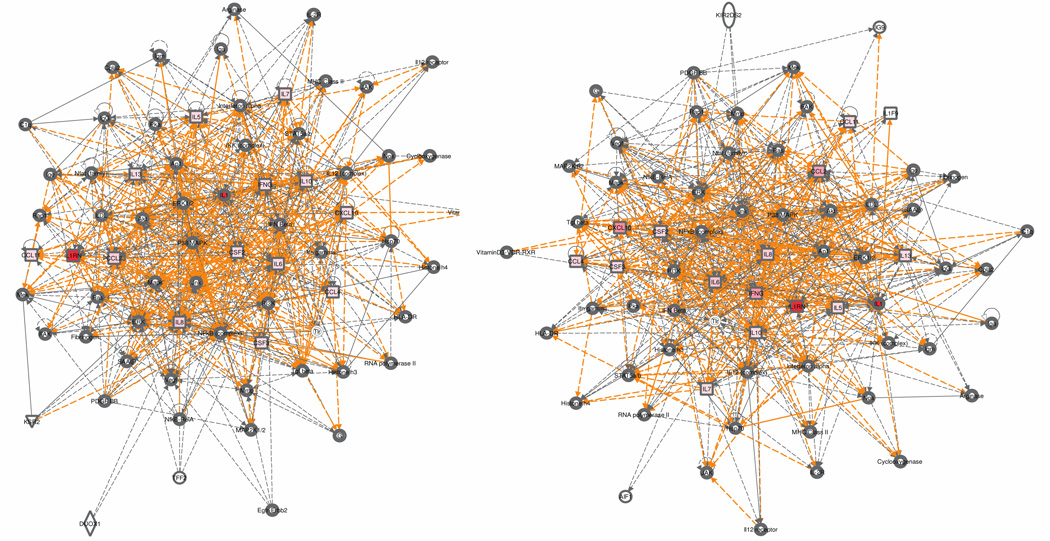
An IPA data template was prepared for each Si with the assayed molecule weightings v″i,a,k (intensities) for both outcomes qk in time period xi and the molecule’s “Gene/Protein ID”. The molecule was identified by its UniProt Knowledgebase (UniProtKB) Accession Number, based on the best match for human (subunit A or chain A). Each v″i,a,k was entered as an “Observation/Expression k”, with k=1 for MOF and k=2 for non-MOF. The 6 datasets generated 12 time-stamped network groups with one to three 35-molecule networks in each group (the default 35-molecule limit is adjustable.) Each group was exported as a text list of molecules (network nodes) and as a graphic image of molecular interactions (network edges) (See Fig. 4).
Fig. 4.
It is very difficult to discern differences between graphs by visual inspection (above); when converted to matrices, the graphs can be compared computationally. Shown are the networks for multiple organ failure (left) and non-multiple organ failure (right) based on patient cytokine data at hours 10–14 from trauma. Both networks were evoked from the same set of molecules, S3, with different median concentrations v″3,a,k for each outcome qk=1, 2. See Supplement 1, Section 3 for all 12 graphs that generated the 193 unique subject molecules (best viewed online). Graphs © 2000 –2011 Ingenuity Systems, Inc. All rights reserved. Used with permission.
4.4 PSA-Node Steps 3 and 4
For the PSA-Node analysis, two biomedical questions were addressed: first, were there molecular patterns in the evoked pathways that were time-shifted differently in outcomes of MOF vs. non-MOF, and secondly, were there molecules that were primarily associated with only one outcome over time?
Step 3. Convert Network Diagrams to Matrices
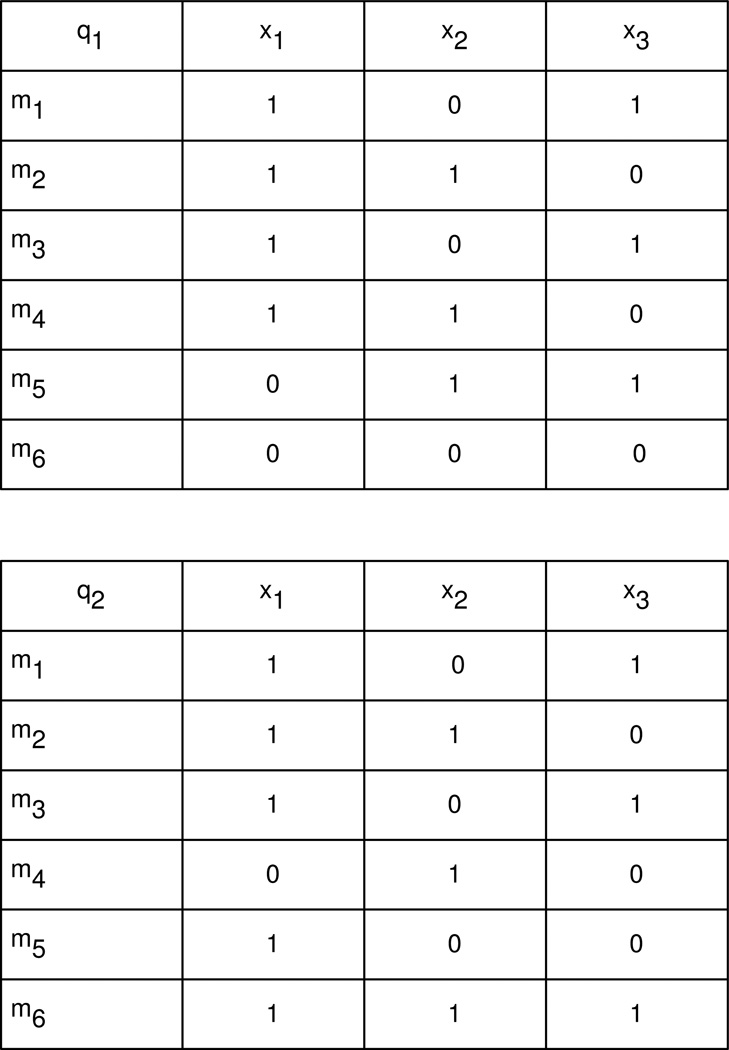
Given the analysis focus on time, the questions were embedded in a matrix format called a Temporal Dependency Matrix (TDM), using the 12 pathway network graphs (6 for MOF and 6 for non-MOF) generated in Step 2. A general example of the TDM format is shown in Fig. 5.
Fig. 5.
Temporal Dependency Matrices (TDM) example.
In Fig. 5, TDMq1 (above) and TDMq2 (below) show 6 molecules mr, r=1…6 over 3 time periods xi, i=1…3 in 2 outcomes qk, k=1,2. To identify molecular patterns by outcome and over time, a summary list mr was compiled of the names of the molecules present in any of the biological networks evoked from the assayed molecules. Then a temporal dependency matrix (TDM) matrix was constructed for each outcome qk, with the molecule names mr as the first column and the time periods xi as the headers across the remaining columns. If the molecule was present in the time period in the outcome, a 1 was placed in the row r, column i cell zkri of the TDM for outcome k; otherwise 0. The rationale behind this process was to facilitate computational comparisons over time and outcome using matrix algebra and logic.
For the trauma application, a summary list Tr of the 193 molecule names mr that were evoked in any outcome at any time were entered into both columns 1 of two temporal dependency matrices TDMMOF(mr, xi) and TDMNMOF(mr, xi). The subscript r ranged from 1 to 193 (number of molecules) and the subscript i ranged from 1 to 6 (number of time periods). For clarity of notation, the TDMs were subscripted by “MOF” for k=1 and “NMOF” for k=2. The headers for the six columns 2 –7 were set as the time periods xi and a 1 or 0 was placed in matrix/row/column cells zkri denoting the presence or absence of the molecule as depicted in the example matrices in Fig. 5. Matrix algebra was then used to compare the TDMs over disease state stratifications to elucidate disease progression and explore the given biomedical questions.
Step 4. Matrix Analysis
Node Analysis 1
Identify molecules mr that appear at least once in both outcomes in the same time period xi and at least once in either outcome in a different time period.
Background
Danger-associated molecular patterns (DAMP) in the systemic inflammatory response syndrome (SIRS) and sepsis induce the production of pro and anti-inflammatory mediators by pattern-recognition receptors (PRR). A dysfunctional acute inflammatory response may lead to MOF [58, 59].
Biomedical questions
In this study, are there molecules that are “time-shifted” in different outcomes? Is a molecular interaction continuing past its “normal” innate response?
Hypothesis
If the identified molecules appear in both outcomes at different times, then additional research may show how to modulate those molecules to minimize negative outcomes.
Notation
To simplify notation in the following TDMs, the k subscript is deleted. It is assumed = 1 in the row / column cells in ZMOF; k = 2 is indicated by “′” in ŹNMOF. Z″ is the summation of both TDMs, and k = 0 is indicated by “″” in its row / column cells.
Let Z″ = ZMOF + Z′NMOF
The cells z″ri of the resulting matrix Z″ had a 2 if the molecule mr was present in both outcomes in time period xi, a 1 if it was present in one outcome or the other, and 0 if it was not present in either. A molecule mr was selected if there was at least one 2 and one 1 in its row. Using these criteria, four molecules were identified that appeared at least once in both outcomes in the same time period and at least once in either outcome in a different time period: CIITA, HIRA, IG9, and KSR2.
Node Analysis 2
Identify molecules that appeared only in one outcome or the other in more than one time period.
Background
Cytokine patterns are associated with different trauma outcomes [50, 54].
Biomedical question
Are there molecules in the pathways triggered by the measured cytokines that are associated only with one outcome in at least 2 of the 6 time periods under study?
Hypothesis
Molecules that meet these criteria may reveal underlying mechanisms that have not yet been associated with specific clinical outcomes.
Notation
To simplify notation, the k subscript is deleted. It is assumed = 1 in the row / column cells zri (MOF) and k=2 in the row / column cells z′ri (NMOF). I = 6, the number of time periods.
Let MOF_SELECT (mr) = 1,
| (1) |
NMOF_SELECT (mr) was also calculated using equation (1) exchanging zri and z′ri. Based on these criteria, four molecules were identified as appearing only in MOF: Egfr-Erbb2, IFI6, MRAS and NOD1; no molecules appeared solely in non-MOF.
4.5 PSA-Node Results
The matrix analysis in Step 4 identified eight molecules from the 193 molecules evoked by the assayed cytokines whose patterns at different times differentiate outcomes. Literature searches were performed on each molecule to ascertain associations with multiple organ failure or other shock / trauma syndromes. Although IG9 [60] was generated by Ingenuity Pathway Analysis (IPA), no other published references to the named molecule were found. The investigator confirmed that research on IG9 had ceased and requested that it be deleted from the findings. (T. M. Calderon, personal communication). See Table 3.
Table 3.
Evoked differential molecular patterns of multiple organ failure based on algebraic comparisons. M: appears in MOF, N: appears in non-MOF, M*: appears only in MOF. The header row shows the time in hours from trauma. Bold italics not previously associated with trauma.
Based on a PubMed search for the molecule name and the MeSH term “shock,” which includes syndromes other than MOF, only three of the seven molecules listed in Table 3 have been previously been associated with shock / trauma: CIITA, EGFR and NOD1 (see Supplement 1, Section 4). All three maintain intestinal epithelial cell homeostasis during immune and inflammatory responses and appear in MOF pathways in this study. This is consistent with previous findings that pathophysiology of the gut (epithelium, mucosal immune system, and the commensal bacteria) contributes to critical illness [61] and to multiple organ failure [62].
Although four molecules - HIRA, IFI6, KSR2, and MRAS - have not yet been associated with shock / trauma, their biological functions seem to be consistent with trauma progression. MRAS appears in hours 2–10 solely in MOF; it is implicated in the regulation of integrin-mediated leukocyte adhesion in inflammatory and immune responses [15]. IFI6 appears in hours 14– 22 solely in MOF; it regulates apoptosis, suggesting that programmed cell death is essential to MOF [9]. HIRA is observed in non-MOF in the first hours, and later in MOF. It promotes nucleosome assembly [7]. This may indicate either the activation of gene transcription or silencing, with different timings associated with different outcomes. Likewise, KSR2 is associated with both outcomes early on, but appears solely in MOF in hours 22–24. It regulates insulin sensitivity [10] and, through inhibition of MAP3K8, decreases pro-inflammatory mediators [13],[14]. Hence, the presence of KSR2 may reflect the up-regulation of pathways in an attempt to modulate the inflammatory response after injury. This may be an underlying mechanism related to the fact that insulin resistance and hyperglycemia are common in non-diabetic critically ill patients [12]. See Table 4 for a summary list of the seven molecules that differentiated outcomes over time.
Table 4.
Summary list of molecules that differentiated outcomes over time. Bold italics not previously associated with trauma.
| Molecule | Known Functions | Citations |
|---|---|---|
| CIITA | CIITA is up-regulated by PPARγ in vascular smooth muscle cells, which enhances IFNγ-mediated transcription and rescues the TGFβ antagonism. | [1] |
| CIITA directly inhibits viral replication and spreading. CIITA triggers antigen presentation to CD4+ T cells leading to an adaptive immune response. |
[2] | |
| Enteral glutamine decreases infectious complications in trauma by protecting the gut. Glutamine administered to the post-ischemic gut has been correlated with transcriptional activation of PPARγ. There is also smooth muscle in the gut; therefore CIITA may be up-regulated due to the PPARγ activated by the administration of enteral glutamine, which has been shown to be safe during active shock resuscitation. |
[3], [4] | |
| EGFR | Transactivation of EGFR and ErbB2 protects intestinal epithelial cells from TNF-induced apoptosis. | [5] |
| EGF is a potential therapeutic agent for the treatment of sepsis. | [6] | |
| HIRA | HIRA promotes replication-independent nucleosome assembly. | [7] |
| IFI6 | IFI6 is believed to play a critical role in the regulation of apoptosis, or programmed cell death and is a marker for interferon beta (IFNB) activity. | [9] |
| KSR2 | KSR2 regulates insulin sensitivity and glucose. Hyperglycemia associated with insulin resistance is common in critically ill patients. |
[10], [12] |
| KSR2 inhibits MAP3K8 (Cot, Tpl2) kinase activity and signaling. Inhibition of MAP3K8 in primary human cell types can decrease the production of TNF alpha and other pro-inflammatory mediators such as MAP3K3-mediated IL-8 (EG 283455) during inflammatory events. | [13], [14] | |
| MRAS | MRAS is involved with adhesion signaling, inducing lymphocyte function-associated antigen 1 (LFA-1)-mediated cell aggregation. | [15] |
| NOD1 | Activation of NOD1 has been shown to induce septic shock and multiple organ injury. | [16] |
| NOD1 protects the intestine from inflammation-induced tumorigenesis. | [17] | |
| NOD1 is involved in the direct killing of Helicobacter pylori bacteria in the stomach and duodenum by epithelial cells. | [19] | |
| Commensal bacteria promote immune homeostasis via the innate immune receptor NOD1. | [21] |
4.6 PSA-Edge Steps 3 and 4
The PSA Edge analysis addressed two biomedical questions in the trauma study: did the types of molecular interactions change over time, and did the crosstalk within the interaction categories also change over time? As a demonstration of edge analysis, PSA Steps 1 and 2 were re-run using eleven of the 27 cytokines chosen by the clinicians as those most likely related to multiple organ failure (see Table 1). The number of cytokines was limited due to edge export restrictions of the pathway generation software (Ingenuity Pathway Analysis) and the fact that, as a result, all edges had to be manually transcribed visually from the generated pathway graphs. IPA generated 12 combined network graphs of the most likely biological pathways evoked from the assay results of the 11 cytokines during 6 time periods and 2 outcomes. There were a total of 132 different molecules evoked in silico across all 24 hours.
The PSA Edge analysis evaluated three of the six time periods in the study: hours 6 – 10, 10 – 14, and 22 – 24 hours from trauma; two outcomes: multiple organ failure (MOF) and non-multiple organ failure (non-MOF); and four relationship categories of molecular interactions: activation, expression (including metabolism and synthesis for chemicals), inhibition and transcription, for a total of 24 relationship sub-graphs. Both direct and indirect interactions were used in the edge analysis. See Fig. 6 for the highlighted expression relationship sub-graph for MOF at hours 6 – 10; all are shown in Supplement 2.
Fig. 6.
Biological Pathways Graph: Hours 6 – 10, MOF. The “expression” interactions are highlighted. Graphs © 2000 –2011 Ingenuity Systems, Inc. All rights reserved. Used with permission.
Step 3. Convert Network Diagrams to Matrices
Four relationship sub-graphs were extracted from each of the 6 evoked network pathway graphs for both outcomes over the three chosen time periods. The 24 sub-graphs were identified by interactively highlighting the edges for each of the 4 interaction categories of activation, expression, inhibition and transcription. The sub-graphs were represented as cyclic digraphs (directed graphs with cycles). Each directed edge, or arc, of a sub-graph was a one-way interaction relationship from one molecule to another. The sub-graphs could also contain loops, or cycles because feedback, feed forward, and self-loops occurred in molecular interactions. This necessitated the use of incidence matrices for computation and limited graph metrics to those for cyclic digraphs. 1,264 graph edges were manually logged by visual inspection into a FileMaker database (www.filemaker.com). Each edge record was identified by its outcome, time period, “FROM” molecule, “TO” molecule, and molecular interaction relationship category.
Using custom software, the edge records for each relationship sub-graph for each time and outcome were converted to an incidence matrix, called an Edge-Molecule (EM) matrix, where each row represented a from-to edge, and each column represented a molecule, with doubles for self-loops. A -1 was placed in the from molecule column, a +1 in the to column and 0 otherwise. All 132 unique molecules evoked in Steps 1 and 2 were placed in the column header row. 12 molecules had self-loop feedback and required duplicate columns: CCL11, CCNA1, Cyclin A, Cyclin E, IL6, TNF, IFNG, IL1, IL10, Hsp70, RARB, and MYBL2. The final number of molecule name columns in each EM matrix was 144, with the number of row edges (molecule – molecule interactions) changing according to the interaction type and the time period. Fig. 7 shows a portion of the EM matrix for the Fig. 6 graph.
Fig. 7.
Portion of EM Matrix for Hours 6 – 10, MOF (full graph in Fig. 6). In this incidence matrix representation, the “from” molecule is mapped to −1, the “to” molecule to 1, and 0 otherwise.
Step 4. Matrix Analysis
The 24 EM matrices were exported for mathematical analysis into MATLAB (www.mathworks.com).
Edge Analysis 1
A descriptive analysis was performed to count the number of edges in each relationship in each outcome over time and to identify edges that were unchanged over time and outcome.
Edge Analysis 2
The crosstalk for each relationship, time period, and outcome was calculated as the measure XTALK using linear algebra as shown in Section 3. Relationship sub-graphs were then analyzed using XTALK to uncover which functional relationships had the most or the least crosstalk in different outcomes and how crosstalk changed over stratifications such as time.
4.7 PSA-Edge Results
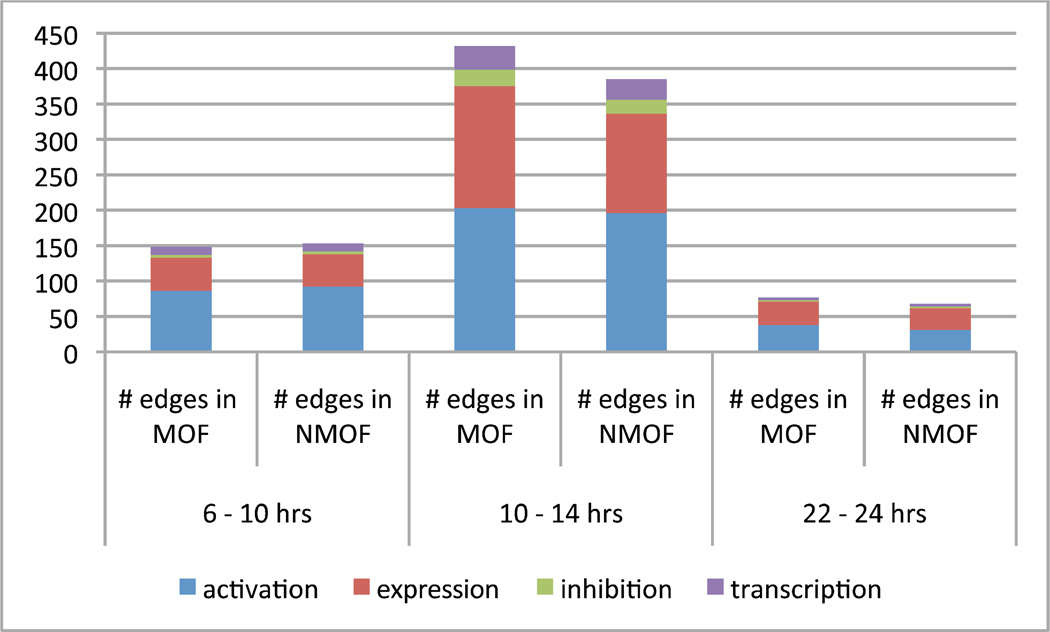
Dominant functions
Based on the edge count, the most interactions per time period were in the activation function category, except in hours 22 – 24 for non-MOF when activation interactions were fewer than expression interactions. Inhibition and transcription interactions were most active in hours 10 – 14. See Fig. 8.
Fig. 8.
Counting edge interactions over time, outcome, and functional relationship category show the most activity in hours 10 – 14 from trauma.
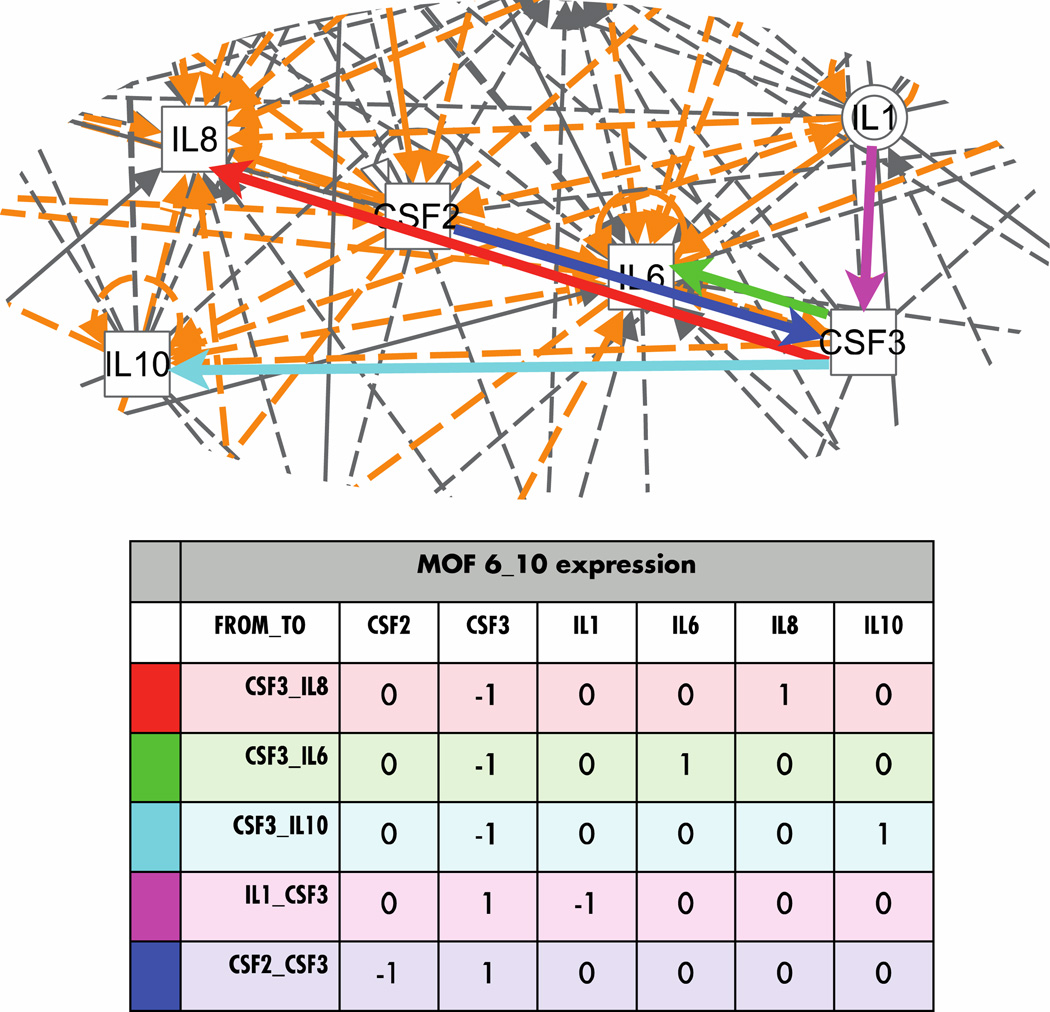
Invariant interactions across all time
Only two molecular interactions were present in both MOF and non-MOF over all time periods; both affected transcription: PDGF BB→ CSF2 (GM-CSF) and IL1 (IL-1β)→ IL8. PDGF BB is a platelet-derived growth factor homodimer that causes mitosis in cells of mesenchymal origin; here it affects the transcription of CSF2, which encodes a cytokine that controls the production, differentiation, and function of granulocytes and macrophages. IL1 is a cytokine produced by activated macrophages that mediates the inflammatory response, in this case by increasing transcription of IL8, a chemokine that functions as a neutrophil polymorphonuclear cell (PMN) chemoattractant. It is also a potent angiogenic factor.
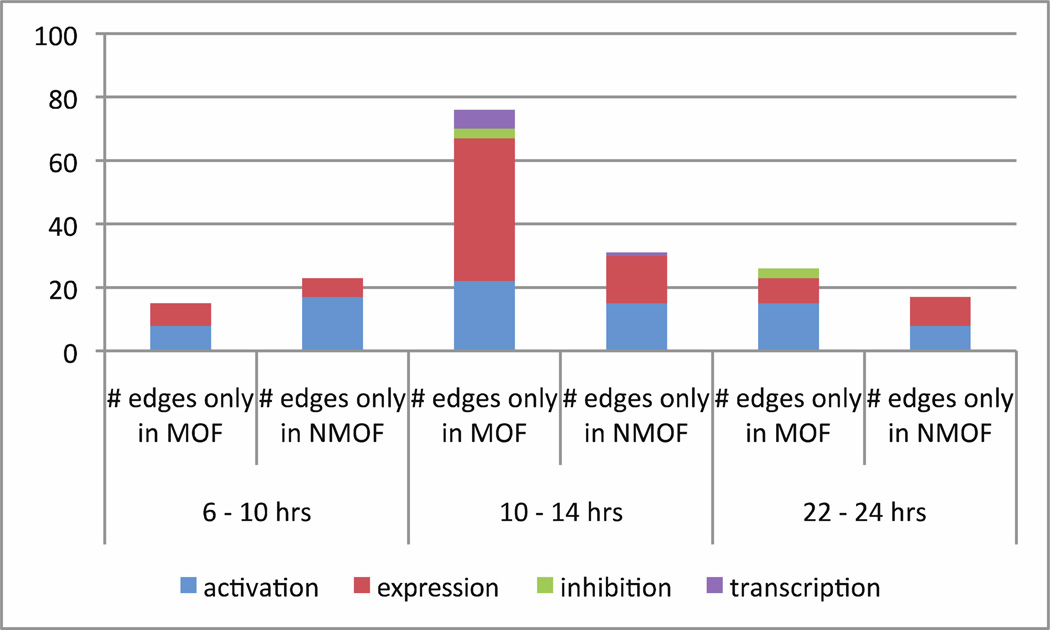
Unique interactions in each time period
Although the majority of molecular interactions were similar in each time period over both outcomes, distinct differences were revealed by a count of the edges unique to MOF or non-MOF. See Fig. 9. In hours 6 to 10 from trauma, there were twice as many unique activation interactions in non-MOF than MOF; whereas by hours 10 – 14, MOF surpassed non-MOF with a greater number of unique interactions in all categories. In hours 22 – 24, MOF had twice as many unique activation edges than non-MOF, although both had the same number of unique expression edges. There were few unique inhibition or transcription interactions. Overall, there were more interactions that appeared solely in MOF than in non-MOF. Another point of interest is that IL6 was involved in ~50% of the unique expression interactions in both outcomes in the first 6 – 10 hours, while IFNG became dominant in hours 10 – 14.
Fig. 9.
Counting unique edge interactions by outcome, over time and functional relationship category. These are in addition to the invariant interactions in each time period that are in both outcomes.
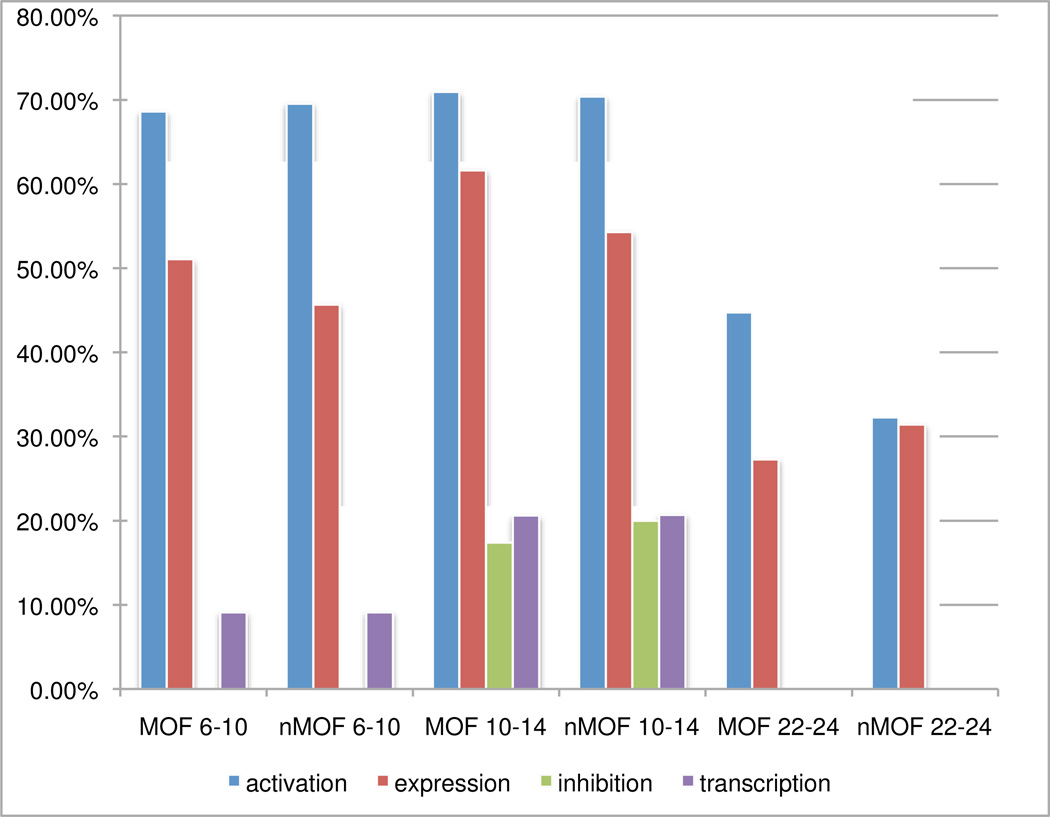
Crosstalk
XTALK, a measure of crosstalk based on the dependency between the functional edges as calculated by matrix rank, ranged from 0% to a high of 71%, and changed over time. (See Fig. 10). Activation crosstalk was calculated at ~69% in hours 6 – 10, staying steady to 71% at hours 10 – 14, and decreasing in hours 22 – 24 to 45% in MOF and 32% in non-MOF. In hours 6 – 10, expression edge crosstalk was 51% in MOF and 46% in non-MOF. This increased in hours 10 – 14 with MOF rising to 62% and non-MOF to 54%. Crosstalk then decreased in hours 22 – 24 to 27% in MOF and 31% in non-MOF. There was no crosstalk in inhibition interactions in hours 6 – 10 and 22 – 24; however, crosstalk increased to 17% in MOF and 20% in non-MOF in hours 10 – 14. 9% transcription crosstalk was calculated in both outcomes in hours 6 – 10, rising to ~21% in hours 10 – 14, then decreasing to 0% by hours 22 – 24.
Fig. 10.
Percentages of crosstalk in functional relationships across time and outcome based on the XTALK measure.
Activation
In hours 6 – 10, there were twice as many unique activation edges in non-MOF compared to MOF; however the reverse was the case in the later time periods. This may imply that in non-MOF, a large number of favorable molecular interactions were underway early on, so fewer unique activations were needed as the pathways approached a favorable outcome of non-MOF. The percentage of activation crosstalk was about the same in hours 6 – 10 and 10 – 14 in both outcomes, decreasing only in hours 22 – 24.
Expression
By hours 10 – 14, MOF had more than three times the number of unique expression edges than non-MOF, implying a higher energy consumption in MOF metabolism than in non-MOF at this time. The percentage of expression crosstalk was slightly lower in non-MOF than MOF in the first 2 time periods, changing to slightly higher by the end.
Inhibition
Unique inhibition interactions appeared solely in MOF in the last 2 time periods. Crosstalk appeared in both outcomes only during hours 10 – 14; it was slightly higher in non-MOF. Again, this suggests an attempt to damp down molecular interactions in both outcomes starting in hours 10 – 14 that was continued in hours 22 – 24 by additional unique inhibitory interactions in MOF.
Transcription
Unique transcription interactions appeared in both outcomes in hours 10 – 14, with the majority in MOF. Crosstalk in transcription interactions increased initially, and disappeared in both outcomes by hours 22 – 24 when only 2 transcription interactions occurred in each outcome.
5. Discussion
Today it is generally accepted that there is a need to develop computational, data-driven algorithms to exploit the vast quantity of molecular information available in knowledge bases in order to advance systems biology and to improve patient care [63–67]. Due to several successes [68–70], in silico hypotheses generators are no longer denigrated as “fishing expeditions” [71].
The Pathway Semantics Algorithm (PSA) presented in this manuscript is an initial in silico data integration and analysis step towards formulating hypotheses about disease progression for personalized diagnosis, prognosis, and therapies that can be validated in the laboratory and in the clinic. PSA is based on a novel, flexible approach that uses graph theory and numerical algebra to computationally compare non-canonical biological pathways evoked from patient data over time. The use of matrix representation and algebra, as used in the Pathway Semantics Algorithm (PSA), offers a way to computationally integrate qualitative and quantitative approaches for improved hypothesis generation about disease progression. PSA identifies molecular patterns in biological pathways derived from patient data, an important benefit that supports personalized medicine. PSA preprocesses the molecular concentration data, tailoring it to the biological and clinical questions under study, before submitting it to a network generation algorithm (in this case, IPA). PSA then algebraically post-processes the evoked pathway networks to reveal changing molecular patterns not easily observed in the static text and graphical formats output by IPA. This algebraic post-processing changes the data representation. It is important because the data representation space is one of the four inter-related problem spaces in scientific discovery, along with the hypothesis space, the experiment space, and the experimental paradigm. Changes in data representation uncover regularities and invariants, facilitate categorization, and suggest alternative search strategies key to scientific discovery [31]. PSA differs from graphical analysis since it does not start with predetermined graphs of canonical pathways. Instead, PSA is data-driven; the algorithmis initialized with clinical data from patients upon which biological pathway networks are constructed based on most likely interactions even if they are not part of canonical pathways. As a result, PSA supports personalized medicine. Although both Gene Set Enrichment Analysis (GSEA) [22, 23] and PSA generate hypotheses correlated with phenotype, their inputs, methods and goals are substantially different. The goal of GSEA is to provide a more robust way to compare independently derived gene expression data sets (possibly obtained with different platforms) and obtain more consistent results than single gene analysis. In contrast, the goal of PSA is to efficiently generate clinically useful hypotheses about disease progression over time using matrix algebra. PSA frames quantitative and qualitative data in matrix representation to answer biomedical questions and the PSA matrix node analysis can be applied to the gene sets evoked from GSEA for further hypothesis generation. Insights can be gained, not only into expression of genes as in GSEA, but also to changes in activation, inhibition, transcription and other activities of molecular interactions over time. Finally, PSA uses mathematical algorithms for matrix representation and computation that are readily available and can be implemented in a wide variety of software.
PSA was applied to a prospective observational study of shock / trauma, a research area where patient data is sparse and difficult to obtain even at a Level I trauma center; randomized controlled trials are not an option. By using patients’ molecular cytokine data to evoke non-canonical biological pathways from the Ingenuity Pathway Knowledge Base, PSA expanded the existing information to include the most likely molecules and molecular interactions evoked by the patients’ cytokines. With the expanded information set, and its representation as pathway graphs, PSA was able to use computational tools and algorithms from graph theory and numerical algebra to compare patterns of molecules and molecular interactions over different stratifications. In particular, PSA was able to analyze patterns over time – an absolute necessity for clinicians who treat disease as it unfolds [72]. This feature shows the potential of PSA to support temporal reasoning in medical decision-making and support systems.
5.1 Overall response to insult
Applied to the trauma study, PSA Node analysis identified and qualified 7 molecules in patterns across time of the progression of multiple organ failure; of these, only 3 had been previously associated with any shock / trauma syndrome. A literature search confirmed that the molecules’ biological functions were consistent with the current understanding of MOF. PSA also highlighted the dynamic nature of trauma response, indicating that molecular patterns are specific to certain time periods from insult. PSA uncovered novel molecular patterns in shock / trauma using an unbiased data-driven approach that integrated what was known about the patient and what was known about molecular interactions. The appearance of these patterns made sense within the disease context, and suggested hypothetical answers to the biomedical questions about which molecules differentiated patient outcomes. All 7 of these molecules were in the evoked biological pathways over time and were not measured directly. Instead, they were inferred from published literature documenting molecular interactions.
The results from the PSA Edge analyses suggest that molecular interaction activity – and the nature of that activity – changed dramatically within the first 24 hours of trauma. In both outcomes, the number of interactions peaked during hours 10 – 14 from insult, lessening to about half of the initial activity by hours 22 – 24; this may be due to the effects of interventions during the first 24 hours combined with the innate systemic response. There were core sets of molecular interactions that were invariant over outcomes in each time period plus unique interactions only in one outcome or the other. This suggests a primary molecular response to the injury that was modulated by the unique interactions edges towards favorable or unfavorable outcomes. MOF had fewer unique interactions early in response, but by hours 10 – 14, MOF had almost three times as many unique edges as non-MOF – perhaps an excessive number.
5.2 Changes in the gene regulation process
Multiple organ failure has been characterized as an adaptive, multilevel time-based stress response with marked changes in gene expression [73–75]. We believe that ours is the first study to quantify the changing aspects of gene expression in MOF over time. By examining edge interactions in silico, changes in functional relationships and their crosstalk over time and outcome were revealed.
Molecules must be activated before they can be transcribed and then expressed, and inhibition can halt any step in the gene regulation process. It is known that cells respond quickly to stress by altering their metabolism; they can induce apoptosis or cell-cycle arrest and alter nuclear pathways for DNA repair [76]. Activation interactions dominated the initial response in both outcomes through hours 10 –14, showing the immediate cellular response to stress. Expression was higher in MOF, suggesting a higher metabolic load on the system. Inhibition and transcription interactions were a small proportion of the overall count.
5.3 Variations in crosstalk over time and outcome
For demonstration purposes, we performed a simple analysis that did not include interaction cascades of different functions in order to focus on a “black box” of four dominant functions. Even with this limitation, differences were observed across time and outcome. This is important because it suggests that a diagnosis, prognosis or therapy based on molecular data might only be valid within a certain time period and for a certain functional relationship, due to the degeneracy in the biological network. For example, because there appear to be few inhibition relationships and little or no inhibition crosstalk in initial trauma, it may be worth exploring increasing inhibition interactions early on in order to limit the excessive unique expression interactions in MOF in hours 10 – 14. Crosstalk decreased over time in the first 24 hours from trauma, suggesting that therapies should consider time from insult as well as which interaction functions they are targeting in order to be effective. This also suggests that trauma therapies may have to be administered in a particular sequence, similar to certain cancer therapies.
5.4 PSA considerations and limitations
The quality of the PSA analysis results depends on the quality of the patient data, the clinical study protocol, the assay method, the choice of statistical analysis, and the accuracy of the biological pathway networks generated by the Ingenuity Pathway Algorithm from its knowledge base.
Validation. The Pathway Semantics Algorithm uses generally accepted methods of statistics and matrix algebra, along with a widely used commercial algorithm and knowledge base for pathway generation. Therefore, the overall Pathway Semantics Algorithm and its resulting hypotheses have at least face validity. This has been confirmed in the previous sections though correlation of the results with published literature and expert opinion as is the usual practice [77].
Because PSA was illustrated based on cytokine time series data from a completed trauma patient study, it was not possible to re-test the patients for empirical validation of the hypotheses generated. Subsequent to the trauma research, PSA was applied to a study of cytokine time series data of a mouse model of inflammatory immune response in hemophilia. Molecular patterns predicted by PSA to occur at specific times were later validated in the mouse model, as documented in the author’s dissertation. [78].
Evaluation. PSA’s extensive use of matrix algebra for analysis minimizes computational complexity while allowing computationally tractable scaling over large numbers of molecules, molecular interactions, outcomes, time periods and other stratifications. In addition, the matrix algebra reduces the size of the solution space, that is, the set of hypotheses generated from the evoked pathways in response to specific biomedical questions. For example, in the trauma PSA node analysis, the 193 molecules in the pathways evoked by the assayed cytokines over 6 time periods resulted in a potential solution space of 1,158 molecules/times. Algebra reduced that to 7 molecules that differentiated outcomes at different times. Finally, the XTALK measure derived from PSA can be shown to be robust under small changes. Expanding the Fig. 3 three-node graph to 4 nodes only modifies XTALK from 33% to 25% as shown in Fig. 11.
Fig. 11.
The directed graph on the left with 4 nodes and 4 edges is the same as Fig. 3 with one added node D and one edge D to B. The calculated rank of the incidence matrix is 3. XTALK for this variation is 1 – (rank/edges) = 1 – (3/4) = 25%. This compares to Fig. 3, where XTALK was 33%.
General Applicability. It is well understood that intracellular signaling processes play an important role in disease progression [79–81]. The Pathway Semantics Algorithm is designed to be generally applicable to the development of hypotheses regarding the roles of signaling molecules, such as cytokines, in disease progression, independent of data set, disease, disease state, or specific method of pathway generation. In addition, as mentioned previously, PSA has been empirically validated in a mouse model of immune response in hemophilia also based on cytokine time series data and published in the author’s dissertation. The authors believe that validation with independent data for a different species and a different disease over a different time progression shows that PSA is a general method; it was not “optimized” for a specific data set, domain, or context.
Following are some key application considerations:
Quality of the patient data and the assay method. In the MOF application, 8.5% of the data were missing. Only one assay method was used, and, its working ranges and limits of detection (LOD) varied depending on the cytokine being assayed. (See Supplement 1, Section 1).
Quantity of the patient data. Only 11 of the 48 patients had outcomes of multiple organ failure; however, there were several thousand cytokine measurements taken on a regular time basis. Because the time periods were based on time from trauma, the number of measurements differed in each time period, with the fewest being in the first time period 2–6 due to patient travel time and the time of protocol entry. In comparison, this sample contained more cytokine data than found in the Trauma Related Database (TRDB) of the multi-center, multi-year Inflammation and the Host Response to Injury Large Scale Collaborative Program. As of 2008, the TRDB contained only 80 trauma subjects with cytokine data sampled irregularly (www.gluegrant.org).
Dimensionality reduction through Significance Sets. Dimensionality reduction, or limiting the number of variables under consideration, was performed to reduce false positives, noise and redundancy in the input data and to reduce the computational burden in subsequent steps. The trade-off was loss of pattern information.
Choice of statistical analysis used to identify Significance Sets. In this exploratory analysis, we identified six time-based Significance Sets using the Mann–Whitney–Wilcoxon (MWW) test on two independent samples (MOF or non-MOF) over 27 observed molecules in each time period. MWW was selected because more sophisticated techniques rely on normality, a condition not satisfied in these data sets. In this exploratory analysis, we chose to identify six Significance Sets rather than one Significance Set from a repeated measures test in order to yield more a detailed understanding of disease progression. With our focus on inclusiveness for hypothesis generation, we tolerated the 5% false positive rate in the Significance Sets and the assumption of independence of the observed molecules. However, if enough data are available, multivariate methods such as MANOVA could be applied to account for correlations among the observations. Note that the statistical analysis is being used to judge the significance of a variable (e.g. a cytokine in a time period), not the significance of a value (e.g. an observation of a patient’s cytokine in a time period.) Given a larger sample size with a normal distribution, exploratory factor analysis methods could be used to identify the Significance Sets.
Linearity assumption. Using matrix rank as a basis for the XTALK measure implies that the edges are related in a linear manner - that is, each edge can be represented as a combination of nodes with coefficients of −1, 0, or 1. This can be considered to be a linear approximation to a non-linear function, computed by taking the first term in the representative Taylor series.
Quality of the biological pathway knowledge base and the algorithm used to evoke biological pathways based on assay measurements. PSA used the commercial product Ingenuity Pathway Analysis. IPA is well accepted in the biological sciences community as seen in several hundred references in PubMed. We chose to use IPA because it is capable of using concentration data to generate pathway networks, and has the flexibility to generate biological networks of any size incorporating the closest interaction neighbors to the input data. To minimize the effects of noise in the data, median values were used as input to IPA. The default size of 35 nodes per network was used in this study, with 1 to 3 networks generated for each outcome in each time period. Each network group was combined before matrix analysis, resulting in up to 105 nodes connected by direct and indirect molecular interaction edges per time period per outcome.
Incomplete pathway data. Some functional relationships may be more highly represented in the Ingenuity Pathways Knowledge Base than others due to the type of experiments performed in the published research, rather than the reality of the true proportion of those relationships in nature. This was addressed in the crosstalk calculation by normalizing XTALK by the number of edges in each relationship sub-graph, to facilitate comparison across stratifications.
Changing nature of the biological pathway knowledge base. IPA generated the biological networks evoked in this study during 2008–2009. Since that time, there have been extensive, continuous updates to the IPA knowledge base. It is not possible to access older versions of the knowledge base (Ingenuity Systems, personal communication) nor is it possible to export interaction data in other than graphical formats, resulting in extensive manual transcription before computation can be done. Therefore, this manuscript is intended as a demonstration of the algorithm, and the actual biomedical results may differ somewhat based on current research. Our assumption is that the evoked biological networks will primarily be the same, with the difference that new discoveries may bring new “closest neighbors” into the graph, pushing out existing molecules past the default 35 node limit per graph. This can be addressed by generating new graphs with larger node limits. In addition, the relationships between molecules may be augmented with new relationships or reclassified to related relationships. However, as with published research, older information about relationships is rarely deleted.
Biological scope of the generated network. If the biological scope is limited to certain species or disease states, the generated network will reflect only current knowledge with the result that potential molecular interactions in other species and disease states may be overlooked. Since the goal of applying PSA to MOF was to uncover hypotheses about potential molecular patterns underlying trauma, it was preferable to run the IPA network generation algorithm without constraints, with the understanding that some of the molecular patterns identified may need to be verified in human shock / trauma progression.
Categories of interaction relationships. Ingenuity Pathway Analysis broadly defines the categories of functional interaction relationships. Each edge on an IPA generated graph is annotated with a single letter, such as E for expression, followed by a number in parentheses, which gives the number of references for that interaction. An “E” annotated edge means that the “from” molecule affects the expression of the “to” molecule. As noted in IPA’s definitions in Section 2.1, the result may be up or down-regulation or another modifier; that information is available in IPA by examining each listed reference online. A more detailed analysis could be performed by changing the categories to include the most common modifiers identified in the references for each interaction category on each edge. At the time of this study, that information was not readily exportable from IPA.
Utility of the molecular patterns. The identified molecules may be difficult to assay clinically due to their primary presence in tissue rather than biofluids, low concentrations, or lack of existing assays. However, the molecular patterns may be useful for in vitro and in vivo verification of the underlying biological mechanisms that may present more clinically useful information.
Resource requirements to implement PSA. Published data for time-based analysis of biofluids and tissues in disease progression may not be readily available although access to biological pathway algorithms and data ranges from free open source to commercial products. This presents opportunities for research studies to collect more data in areas such as trauma and critical care where rapid changes are seen and rapid response to changing patient condition is required.
6. Conclusions
The Pathway Semantics Algorithm identified different patterns of molecules and molecular interactions over time, outcomes, and functional relationships in biological networks that would not be easily found through direct assays, literature or database searches. By framing biomedical questions within a variety of matrix representations, PSA had the flexibility to analyze combined quantitative and qualitative data over a wide range of stratifications and generate hypotheses addressing those specific biomedical questions.
The algorithm was illustrated with an application to disease progression in trauma; the results show promise for further clinical investigation. The seven evoked molecules that differentiated outcomes of MOF and non-MOF in specific time periods suggest novel hypotheses for underlying mechanisms of shock / trauma progression. The differences in the number of edges, the number of unique edges, and XTALK showed the utility of evaluating a molecular interaction not just as a connection between two molecules, but as a directed interaction from one molecule to another that may carry out one or many specific functions [82]. The crosstalk measure XTALK provided a novel perspective on the changing functional interaction relationships in disease progression; the results supported the existence of the property of degeneracy in biological networks. Next steps in this work include exploring the biological significance of other matrix-based numerical algebra methods, analysis of other diseases of clinical interest, and laboratory validation of results. Substantial progress has been made in this regard. PSA was applied and empirically validated in a mouse model of hemophilia; the results are being prepared for separate publication at the request of the co-authors.
Supplementary Material
Acknowledgements
We acknowledge the participation of the Memorial Hermann Hospital STICU, Houston in the prospective study that collected the data used in this study.
Funding: This work was supported by the National Institutes of Health [T15-LM07093-16 to M.F.M.; GM-38529, GM-08792, CReFF UCRC #M01RR002558 to D.W.M.] and by the Harvey S Rosenberg, Endowed Chair in Pathology for the Morphoproteomics Initiative, UT Medical School at Houston to M.F.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary information: Supplementary data are available at the Journal of Biomedical Informatics online.
Conflict of Interest: None declared.
References
- 1.Kong X, Fang M, Fang F, Li P, Xu Y. PPAR-gamma enhances IFN-gamma-mediated transcription and rescues the TGf-beta antagonism by stimulating CIITA in vascular smooth muscle cells. Journal of Molecular and Cellular Cardiology. 2009;46:748–757. doi: 10.1016/j.yjmcc.2009.01.011. [PubMed: 19358337] [DOI] [PubMed] [Google Scholar]
- 2.Tosi G, Bozzo L, Accolla RS. The dual function of the MHC class II transactivator CIITA against HTLV retroviruses. Front Biosci. 2009;14:4149–4156. doi: 10.2741/3519. [PubMed: 19273341] [DOI] [PubMed] [Google Scholar]
- 3.Santora R, Kozar RA. Molecular Mechanisms of Pharmaconutrients. J Surg Res. 2009 doi: 10.1016/j.jss.2009.06.024. [PubMed: 20080249] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McQuiggan M, Kozar R, Sailors RM, Ahn C, McKinley B, Moore F. Enteral glutamine during active shock resuscitation is safe and enhances tolerance of enteral feeding. JPEN J Parenter Enteral Nutr. 2008;32:28–35. doi: 10.1177/014860710803200128. [PubMed: 18165444] [DOI] [PubMed] [Google Scholar]
- 5.Yamaoka T, Yan F, Cao H, Hobbs SS, Dise RS, Tong W, Polk DB. Transactivation of EGF receptor and ErbB2 protects intestinal epithelial cells from TNF-induced apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:11772–11777. doi: 10.1073/pnas.0801463105. [PubMed: 18701712] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark JA, Clark AT, Hotchkiss RS, Buchman TG, Coopersmith CM. Epidermal growth factor treatment decreases mortality and is associated with improved gut integrity in sepsis. Shock. 2008;30:36–42. doi: 10.1097/shk.0b013e31815D0820. [PubMed: 18004230] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eitoku M, Sato L, Senda T, Horikoshi M. Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell Mol Life Sci. 2008;65:414–444. doi: 10.1007/s00018-007-7305-6. [PubMed: 17955179] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weng G, Bhalla US, Iyengar R. Complexity in biological signaling systems. Science. 1999;284:92–96. doi: 10.1126/science.284.5411.92. [PubMed: 10102825] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serrano-Fernandez P, Moller S, Goertsches R, Fiedler H, Koczan D, Thiesen HJ, Zettl UK. Time course transcriptomics of IFNB1b drug therapy in multiple sclerosis. Autoimmunity. 2009 doi: 10.3109/08916930903219040. [PubMed: 19883335] [DOI] [PubMed] [Google Scholar]
- 10.Costanzo-Garvey DL, Pfluger PT, Dougherty MK, Stock JL, Boehm M, Chaika O, Fernandez MR, Fisher K, Kortum RL, Hong EG, Jun JY, Ko HJ, Schreiner A, Volle DJ, Treece T, Swift AL, Winer M, Chen D, Wu M, Leon LR, Shaw AS, McNeish J, Kim JK, Morrison DK, Tsch••p MH, Lewis RE. KSR2 Is an Essential Regulator of AMP Kinase, Energy Expenditure,, and Insulin Sensitivity. Cell Metabolism. 2009;10:366–378. doi: 10.1016/j.cmet.2009.09.010. [PubMed: 19883615] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoofnagle AN, Wener MH. The fundamental flaws of immunoassays and potential solutions using tandem mass spectrometry. J Immunol Methods. 2009;347:3–11. doi: 10.1016/j.jim.2009.06.003. [PubMed: 19538965] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Den Berghe G, Wouters P, Weekers F, Verwaest C, Bruyninckx F, Schetz M, Vlasselaers D, Ferdinande P, Lauwers P, Bouillon R. Intensive insulin therapy in the critically ill patients. New England Journal of Medicine. 2001;345:1359–1367. doi: 10.1056/NEJMoa011300. [PubMed: 11794168] [DOI] [PubMed] [Google Scholar]
- 13.Channavajhala PL, Wu L, Cuozzo JW, Hall JP, Liu W, Lin LL, Zhang Y. Identification of a novel human kinase supporter of Ras (hKSR-2) that functions as a negative regulator of Cot (Tpl2) signaling. J Biol Chem. 2003;278:47089–47097. doi: 10.1074/jbc.M306002200. [PubMed: 12975377] [DOI] [PubMed] [Google Scholar]
- 14.Hall JP, Kurdi Y, Hsu S, Cuozzo J, Liu J, Telliez JB, Seidl KJ, Winkler A, Hu Y, Green N, Askew GR, Tam S, Clark JD, Lin LL. Pharmacologic inhibition of tpl2 blocks inflammatory responses in primary human monocytes, synoviocytes, and blood. J Biol Chem. 2007;282:33295–33304. doi: 10.1074/jbc.M703694200. [PubMed: 17848581] [DOI] [PubMed] [Google Scholar]
- 15.Yoshikawa Y, Satoh T, Tamura T, Wei P, Bilasy SE, Edamatsu H, Aiba A, Katagiri K, Kinashi T, Nakao K, Kataoka T. The M-Ras-RA-GEF-2-Rap1 pathway mediates tumor necrosis factor-alpha dependent regulation of integrin activation in splenocytes. Mol Biol Cell. 2007;18:2949–2959. doi: 10.1091/mbc.E07-03-0250. [PubMed: 17538012] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cartwright N, Murch O, McMaster SK, Paul-Clark MJ, van Heel DA, Ryffel B, Quesniaux VF, Evans TW, Thiemermann C, Mitchell JA. Selective NOD1 agonists cause shock and organ injury/dysfunction in vivo. Am J Respir Crit Care Med. 2007;175:595–603. doi: 10.1164/rccm.200608-1103OC. [PubMed: 17234906] [DOI] [PubMed] [Google Scholar]
- 17.Chen GY, Shaw MH, Redondo G, Nunez G. Innate immune receptor nod1 protects the intestine from inflammation-induced tumorigenesis. Cancer Research. 2008;68:10060–10067. doi: 10.1158/0008-5472.CAN-08-2061. [PubMed: 19074871] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGuire MF, Iyengar MS, Mercer DW. Computational Approaches for Translational Clinical Research in Disease Progression. J Investig Med. 2011;59:893–903. doi: 10.231/JIM.0b013e318224d8cc. [PubMed: 21712727] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grubman A, Kaparakis M, Viala J, Allison C, Badea L, Karrar A, Boneca IG, Le Bourhis L, Reeve S, Smith IA, Hartland EL, Philpott DJ, Ferrero RL. The innate immune molecule, NOD1, regulates direct killing of Helicobacter pylori by antimicrobial peptides. Cell Microbiol. 2009 doi: 10.1111/j.1462-5822.2009.01421.x. [PubMed: 20039881] [DOI] [PubMed] [Google Scholar]
- 20.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [PubMed: 17940530] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen GY, Nunez G. Gut Immunity: A NOD to the Commensals. Current Biology. 2009:19. doi: 10.1016/j.cub.2008.12.027. [PubMed: 19243695] [DOI] [PubMed] [Google Scholar]
- 22.Subramanian A, Tamayo P, Mootha V, Mukherjee S, Ebert B, Gillette M, Paulovich A, Pomeroy S, Golub T, Lander E, Mesirov J. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [PubMed: 16199517] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamayo P, Steinhardt G, Liberzon A, Mesirov JP. Gene Set Enrichment Analysis Made Right. Statistical Methods in Medical Research. 2011 doi: 10.1177/0962280212460441. (submitted); arXiv: 1110.4128v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGuire MF, Iyengar MS. Software Tools for Biological Pathway Modeling. In: Iyengar MS, editor. Symbolic Systems Biology: Theory and Methods. Sudbury: Jones & Bartlett Publishers; 2010. pp. 175–195. [Google Scholar]
- 25.Li WX. Canonical and non-canonical JAK-STAT signaling. Trends Cell Biol. 2008;18:545–551. doi: 10.1016/j.tcb.2008.08.008. [PubMed: 18848449] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho KH, Shin SY, Lee HW, Wolkenhauer O. Investigations into the analysis and modeling of the TNF alpha-mediated NF-kappa B-signaling pathway. Genome Res. 2003;13:2413–2422. doi: 10.1101/gr.1195703. [PubMed: 14559780] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klahr D, Simon HA. Studies of scientific discovery: Complementary approaches and convergent findings. Psychological Bulletin. 1999;125:524–543. [PubMed: NA] [Google Scholar]
- 28.Hsich G, Kenney K, Gibbs CJ, Lee KH, Harrington MG. The 14-3-3 brain protein in cerebrospinal fluid as a marker for transmissible spongiform encephalopathies. N Engl J Med. 1996;335:924–930. doi: 10.1056/NEJM199609263351303. [PubMed: 8782499] [DOI] [PubMed] [Google Scholar]
- 29.Haubitz M, Good DM, Woywodt A, Haller H, Rupprecht H, Theodorescu D, Dakna M, Coon JJ, Mischak H. Identification and validation of urinary biomarkers for differential diagnosis and evaluation of therapeutic intervention in anti-neutrophil cytoplasmic antibody-associated vasculitis. Mol Cell Proteomics. 2009;8:2296–2307. doi: 10.1074/mcp.M800529-MCP200. [PubMed: 19564150] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heuer RJ. Psychology of Intelligence Analysis History Staff, Center for the Study of Intelligence, Central Intelligence Agency. 1999 [Google Scholar]
- 31.Schunn CD, Klahr D. A 4-space model of scientific discovery; Proceedings of the 17th Annual Conference of the Cognitive Science Society; 1995. [Google Scholar]
- 32.Brown RE. Morphogenomics and morphoproteomics: a role for anatomic pathology in personalized medicine. Arch Pathol Lab Med. 2009;133:568–579. doi: 10.5858/133.4.568. [PubMed: 19391654] [DOI] [PubMed] [Google Scholar]
- 33.Ingenuity S. IPA Network Generation Algorithm Whitepaper © 2005 Ingenuity Systems Proprietary and Confidential. Ingenuity Systems. 2005:26. [Google Scholar]
- 34.Evans TS, Lambiotte R. Line graphs, link partitions, and overlapping communities. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;80 doi: 10.1103/PhysRevE.80.016105. 016105. [PubMed: 19658772] [DOI] [PubMed] [Google Scholar]
- 35.Ahn YY, Bagrow JP, Lehmann S. Link communities reveal multiscale complexity in networks. Nature. 2010;466:761–764. doi: 10.1038/nature09182. [PubMed: 20562860] [DOI] [PubMed] [Google Scholar]
- 36.Tononi G, Sporns O, Edelman GM. Measures of degeneracy and redundancy in biological networks. Proc Natl Acad Sci U S A. 1999;96:3257–3262. doi: 10.1073/pnas.96.6.3257. [PubMed: 10077671] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edelman GM, Gally JA. Degeneracy and complexity in biological systems. Proc Natl Acad Sci U S A. 2001;98:13763–13768. doi: 10.1073/pnas.231499798. [PubMed: 11698650] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macia J, Sole RV. Distributed robustness in cellular networks: insights from synthetic evolved circuits. J R Soc Interface. 2009;6:393–400. doi: 10.1098/rsif.2008.0236. [PubMed: 18796402] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitacre JM. Degeneracy: a link between evolvability, robustness and complexity in biological systems. Theor Biol Med Model. 2010;7:6. doi: 10.1186/1742-4682-7-6. [PubMed: 20167097] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tieri P, Grignolio A, Zaikin A, Mishto M, Remondini D, Castellani GC, Franceschi C. Network, degeneracy and bow tie integrating paradigms and architectures to grasp the complexity of the immune system. Theor Biol Med Model. 2010;7:32. doi: 10.1186/1742-4682-7-32. [PubMed: 20701759] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bruni LE. Cellular Semiotics And Signal Transduction. In: Barbieri M, editor. Introduction to Biosemiotics: The New Biological Synthesis. Berlin: Springer; 2007. pp. 365–408. [Google Scholar]
- 42.Ingenuity. Pathways Knowledge. [Accessed April 13];2010 Available from: http://www.ingenuity.com/products/pathways_knowledge.html. [Google Scholar]
- 43.Bondy A, Murty USR. Graph Theory (Graduate Texts in Mathematics) Springer; 2008. [Google Scholar]
- 44.Brown RE. Morphoproteomics: exposing protein circuitries in tumors to identify potential therapeutic targets in cancer patients. Expert Rev Proteomics. 2005;2:337–348. doi: 10.1586/14789450.2.3.337. [PubMed: 16000081] [DOI] [PubMed] [Google Scholar]
- 45.Birkhoff G, MacLane S. A Survey of Modern Algebra. Vol. 6. New York: A K Peters/CRC Press; 1998. Feb, p. 1953. [Google Scholar]
- 46.Heron M, Hoyert D, Xu J, Scott C, Tejada-Vera B. National vital statistics reports. no 16. vol 56. Hyattsville, MD: National Center for Health Statistics; 2008. Deaths: Preliminary data for 2006. [Google Scholar]
- 47.Stewart RM. Injury prevention: why so important? J Trauma. 2007;62:S47–S48. doi: 10.1097/TA.0b013e31806546ab. [PubMed: 17556969] [DOI] [PubMed] [Google Scholar]
- 48.Watson GA, Sperry JL, Rosengart MR, Minei JP, Harbrecht BG, Moore EE, Cuschieri J, Maier RV, Billiar TR, Peitzman AB. Inflammation, Host Response to Injury I. Fresh frozen plasma is independently associated with a higher risk of multiple organ failure and acute respiratory distress syndrome. J Trauma. 2009;67:221–227. doi: 10.1097/TA.0b013e3181ad5957. discussion 228-30. [PubMed: 19667872] [DOI] [PubMed] [Google Scholar]
- 49.Deitch EA. Multiple organ failure. Pathophysiology and potential future therapy. Ann Surg. 1992;216:117–134. doi: 10.1097/00000658-199208000-00002. [PubMed: 1503516] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maier B, Lefering R, Lehnert M, Laurer HL, Steudel WI, Neugebauer EA, Marzi I. Early versus late onset of multiple organ failure is associated with differing patterns of plasma cytokine biomarker expression and outcome after severe trauma. Shock. 2007;28:668–674. [PubMed: 18092384] [PubMed] [Google Scholar]
- 51.Janeway C, Travers P, Walport M, Shlomchik M. Immunobiology. 6th Edition. Garland Science; 2004. [Google Scholar]
- 52.Vodovotz Y. Translational systems biology of inflammation and healing. Wound Repair Regen. 2010;18:3–7. doi: 10.1111/j.1524-475X.2009.00566.x. [PubMed: 20082674] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hranjec T, Swenson BR, Dossett LA, Metzger R, Flohr TR, Popovsky KA, Bonatti HJ, May AK, Sawyer RG. Diagnosis-dependent relationships between cytokine levels and survival in patients admitted for surgical critical care. J Am Coll Surg. 2010;210:833–844. 845–846. doi: 10.1016/j.jamcollsurg.2009.12.042. [PubMed: 20421061] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jastrow KM, 3rd, Gonzalez EA, McGuire MF, Suliburk JW, Kozar RA, Iyengar S, Motschall DA, McKinley BA, Moore FA, Mercer DW. Early cytokine production risk stratifies trauma patients for multiple organ failure. J Am Coll Surg. 2009;209:320–321. doi: 10.1016/j.jamcollsurg.2009.05.002. [PubMed: 19717036] [DOI] [PubMed] [Google Scholar]
- 55.Visser T, Pillay J, Koenderman L, Leenen LP. Postinjury immune monitoring: can multiple organ failure be predicted? Curr Opin Crit Care. 2008;14:666–672. doi: 10.1097/MCC.0b013e3283196522. [PubMed: 19005307] [DOI] [PubMed] [Google Scholar]
- 56.Roumen RM, Redl H, Schlag G, Zilow G, Sandtner W, Koller W, Hendriks T, Goris RJ. Inflammatory mediators in relation to the development of multiple organ failure in patients after severe blunt trauma. Crit Care Med. 1995;23:474–480. doi: 10.1097/00003246-199503000-00010. [PubMed: 7874897] [DOI] [PubMed] [Google Scholar]
- 57.McGuire MF, Iyengar MS, Mercer DW. Measurement units may impact results of pathway analysis. Journal of Critical Care. 2007;22:342–343. [PubMed: NA] [Google Scholar]
- 58.Castellheim A, Brekke OL, Espevik T, Harboe M, Mollnes TE. Innate immune responses to danger signals in systemic inflammatory response syndrome and sepsis. Scand J Immunol. 2009;69:479–491. doi: 10.1111/j.1365-3083.2009.02255.x. [PubMed: 19439008] [DOI] [PubMed] [Google Scholar]
- 59.Bianchi ME. DAMPs, PAMPs and alarmins: All we need to know about danger. Journal of Leukocyte Biology. 2007;81:1–5. doi: 10.1189/jlb.0306164. [PubMed: 17032697] [DOI] [PubMed] [Google Scholar]
- 60.Calderon TM, Gertz SD, Sarembock IJ, Berliner JA, Fallon JT, Taubman MB, Berman JW. Induction of IG9 monocyte adhesion molecule expression in smooth muscle and endothelial cells after balloon arterial injury in cholesterol-fed rabbits. Arterioscler Thromb Vasc Biol. 2000;20:1293–1300. doi: 10.1161/01.atv.20.5.1293. [PubMed: 10807745] [DOI] [PubMed] [Google Scholar]
- 61.Clark JA, Coopersmith CM. Intestinal crosstalk: A new paradigm for understanding the gut as the “motor” of critical illness. Shock. 2007;28:384–393. doi: 10.1097/shk.0b013e31805569df. [PubMed: 17577136] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hassoun HT, Weisbrodt NW, Mercer DW, Kozar RA, Moody FG, Moore FA. Inducible nitric oxide synthase mediates gut ischemia/reperfusion-induced ileus only after severe insults. J Surg Res. 2001;97:150–154. doi: 10.1006/jsre.2001.6140. [PubMed: 11341791] [DOI] [PubMed] [Google Scholar]
- 63.Li W, Xu M, Zhou XJ. Unraveling complex temporal associations in cellular systems across multiple time-series microarray datasets. Journal of Biomedical Informatics. 2010;43:550–559. doi: 10.1016/j.jbi.2009.12.006. [PubMed: 20083231] [DOI] [PubMed] [Google Scholar]
- 64.Veliz-Cuba A, Jarrah AS, Laubenbacher R. Polynomial algebra of discrete models in systems biology. Bioinformatics. 2010;26:1637–1643. doi: 10.1093/bioinformatics/btq240. [PubMed: 20448137] [DOI] [PubMed] [Google Scholar]
- 65.Tipney HJ, Leach SM, Feng W, Spritz R, Williams T, Hunter L. Leveraging existing biological knowledge in the identification of candidate genes for facial dysmorphology. BMC Bioinformatics. 2009;10(Suppl 2):S12. doi: 10.1186/1471-2105-10-S2-S12. [PubMed: 19208187] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruths DA, Nakhleh L, Iyengar MS, Reddy SA, Ram PT. Hypothesis generation in signaling networks. J Comput Biol. 2006;13:1546–1557. doi: 10.1089/cmb.2006.13.1546. [PubMed: 17147477] [DOI] [PubMed] [Google Scholar]
- 67.Aristotelis T. Pattern discovery for hypothesis generation in biology. New York University; 2006. p. 167. [Google Scholar]
- 68.Sam L, Liu Y, Li J, Friedman C, Lussier YA. Discovery of protein interaction networks shared by diseases. Pac Symp Biocomput. 2007:76–87. [PubMed: 17992746] [PMC free article] [PubMed] [Google Scholar]
- 69.Sachs K, Gentles AJ, Youland R, Itani S, Irish J, Nolan GP, Plevritis SK. Characterization of patient specific signaling via augmentation of Bayesian networks with disease and patient state nodes. Conf Proc IEEE Eng Med Biol Soc. 2009;2009:6624–6627. doi: 10.1109/IEMBS.2009.5332563. [PubMed: 19963681] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucl. Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [PubMed: 19880382] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brent R, Lok L. Cell biology. A fishing buddy for hypothesis generators. Science. 2005;308:504–506. doi: 10.1126/science.1110535. [PubMed: 15845840] [DOI] [PubMed] [Google Scholar]
- 72.Shahar Y. Dimensions of time in illness: an objective view. Ann Intern Med. 2000;132:45–53. doi: 10.7326/0003-4819-132-1-200001040-00008. [PubMed: 10627251] [DOI] [PubMed] [Google Scholar]
- 73.Cobb JP, Buchman TG, Karl IE, Hotchkiss RS. Molecular biology of multiple organ dysfunction syndrome: injury, adaptation, and apoptosis. Surg Infect (Larchmt) 2000;1:207–213. doi: 10.1089/109629600750018132. discussion 214-5. [PubMed: 12594891] [DOI] [PubMed] [Google Scholar]
- 74.Adib-Conquy M, Cavaillon JM. Compensatory anti-inflammatory response syndrome. Thromb Haemost. 2009;101:36–47. [PubMed: 19132187] [PubMed] [Google Scholar]
- 75.Warren HS, Elson CM, Hayden DL, Schoenfeld DA, Cobb JP, Maier RV, Moldawer LL, Moore EE, Harbrecht BG, Pelak K, Cuschieri J, Herndon DN, Jeschke MG, Finnerty CC, Brownstein BH, Hennessy L, Mason PH, Tompkins RG. Inflammation, Host Response to Injury Large Scale Collaborative Research P. A genomic score prognostic of outcome in trauma patients. Mol Med. 2009;15:220–227. doi: 10.2119/molmed.2009.00027. [PubMed: 19593405] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [PubMed: 20965417] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klahr D, Dunbar K. Dual space search during scientific reasoning. Cognitive Science. 1988;12:1–48. [PubMed: NA] [Google Scholar]
- 78.McGuire MF. Pathway Semantics: An Algebraic Data Driven Algorithm to Generate Hypotheses About Molecular Patterns Underlying Disease Progression. School of Biomedical Informatics Houston: University of Texas Health Science Center at Houston. 2011:173. [Google Scholar]
- 79.Monnier M, Boehm F. Prufung der Leistungsfahigkeit des optischen Systems durch kombinierte Elektroretinographie und Elektroencephalographie beim Menschen. Helv Physiol Pharmacol Acta. 1947;5 C rend, 33.[PubMed: 18932909] [PubMed] [Google Scholar]
- 80.Reddy SA. Signaling pathways in pancreatic cancer. Cancer J. 2001;7:274–286. [PubMed: 11561604] [PubMed] [Google Scholar]
- 81.Lee E, Chuang HY, Kim JW, Ideker T, Lee D. Inferring pathway activity toward precise disease classification. PLoS Comput Biol. 2008;4 doi: 10.1371/journal.pcbi.1000217. e1000217. [PubMed: 18989396] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu Y, Zhang X, Yu J, Ouyang Q. Identification of a topological characteristic responsible for the biological robustness of regulatory networks. PLoS Comput Biol. 2009;5 doi: 10.1371/journal.pcbi.1000442. e1000442. [PubMed: 19629157] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.