Abstract
We have developed a procedure in which disulfide crosslinks are used to identify regions of proteins that undergo functionally important intramolecular motion. The approach was applied to the identification of disulfide bonds that stabilize the active state of the yeast α-mating pheromone receptor Ste2p, a member of the superfamily of G Protein Coupled Receptors. Cysteine residues were introduced at random positions in targeted regions of a starting allele of Ste2p that completely lacks cysteines. Libraries of mutated receptors were then screened for alleles that exhibit constitutive signaling. Two strongly activated alleles were recovered containing cysteine residues in transmembrane segments 5 and 6. Constitutive activity of these alleles was dependent on the presence of both introduced cysteines and was sensitive to reducing agent. Crosslinked peptides derived from the mutant receptors were detected by immunoblotting. Additional sites of crosslinking between transmembrane segments 5 and 6 that did not lead to constitutive activation were also identified. These results indicate that relative motion of the transmembrane segments 5 and 6 in the extracellular half of the membrane is sufficient to activate the receptor and that transmembrane segment 6, but not transmembrane segment 5, exhibits rotational mobility that is not associated with receptor activation.
G protein Coupled Receptors (GPCRs) comprise a large family of seven transmembrane segment (TM) receptors that recognize a wide array of extracellular, chemical and physical stimuli. As a consequence of their involvement in diverse signaling processes, GPCRs are widely targeted by pharmaceuticals. Upon activation, GPCRs undergo conformational changes leading to the nucleotide exchange and dissociation of their cognate hetero-trimeric G-proteins, triggering diverse downstream intracellular responses 1. Despite the availability of recent-solved crystal structures of activated forms of GPCRs, the molecular mechanisms of coupling between GPCR and G protein activation are not yet clear.
Previous approaches that have been used to monitor conformational changes in GPCRs associated with receptor activation include: 1) the determination of crystal structures of GPCRs under conditions thought to induce active states of receptors 2; 3; 4; 5; 6; 2) the use of site directed spin labeling coupled with electron paramagnetic resonance (EPR) to measure structural changes in rhodopsin 7; 8; 3) the use of fluorescence and infrared spectroscopies techniques to monitor conformational changes associated with activation 9; 10; 11; 4) mutagenic approaches for identifying amino acid substitutions capable of enhancing or preventing GPCR activation and signaling 12; 13; and 5) the use of disulfide cross-linking to detect changes in interacting residues upon activation 8; 14; 15. The major activation-associated conformational change detected by these studies is an outward motion of the cytoplasmic end of the sixth TM segment with respect to the rest of the TM helical bundle 2; 5; 6; 7; 9; 11. However it is not known to what extent this change is a requirement for activation as opposed to a consequence of changes initiated at other sites.
Disulfide cross-linking of introduced cysteine residues has previously been employed to study conformational changes in a variety of proteins, including TM channels and receptors 8; 16; 17; 18; 19; 20; 21; 22. One particularly informative type of crosslink in the context of GPCR activation would be a cysteine pair that locks a receptor in an active state, enhancing signaling activity in the absence of bound ligand. Identification of this type of crosslink could pinpoint a particular site of relative intramolecular motion that is sufficient to trap the conformational change associated with receptor activation, and is not just correlated with activation. In addition, such a disulfide locked constitutive mutant could be used as the basis for a strategy to maintain receptors in their active states for structural studies. Crosslinks that lead to increased levels of basal activity have been useful in understanding the structural basis of activation of bacterial chemotaxis receptors 16. Engineered disulfides have been identified that prevent rhodopsin from being activated 8, however, it is difficult to know whether such “lock-off” mutations directly block the normal activation pathway or simply force a receptor into an aberrant inactive state.
We have developed a strategy for screening directly for cysteine mutations that result in constitutive activation of GPCRs using a new approach for introducing multiple cysteine residues at random positions in a receptor, followed by application of a genetic selection to identify mutants from randomized libraries that exhibit constitutive activity. This procedure allows screening of much larger numbers of combinations of cysteine mutations than would be possible using site-directed approaches for engineering cysteine residues two-at-a-time into large numbers of positions in receptors. The feasibility of this approach was tested using the signaling system responsible for the response to mating pheromone of the yeast Saccharomyces cerevisiae. The yeast α-factor receptor (Ste2p) is a GPCR that, upon binding to α factor, a 13-residue peptide, triggers activation of a cytoplasmic heterotrimeric G protein in MATa haploid cells 23. We focused on Ste2p because of the usefulness of this receptor in serving as a model for understanding more general GPCR function, and because of the genetic tools available for manipulation and analysis of the yeast pheromone response pathway. Although Ste2p shows little sequence similarity to mammalian GPCRs, expression of some mammalian GPCRs in yeast is capable of activating the yeast mating pathway 24; 25 and Ste2p exhibits function when expressed in mammalian cells 26.
The combination of random cysteine mutagenesis with genetic screening has allowed us to identify two crosslink-dependent constitutively active mutants of Ste2p. The positions of the introduced cysteines suggest that relative motion of TM segments 5 and 6 plays an important role in activation of the receptor. In addition, we have identified a number of additional sites of crosslinking that do not lead to constitutive activation of receptors. The range of positions of these sites indicates that TM 6 is subject to rotational motion while the motion of TM 5 is more restricted.
Results
To identify regions of Ste2p that undergo conformational change upon activation we screened for disulfide crosslinks that can lock the receptor in an active state following the overall scheme depicted in Fig. 1. This procedure was performed starting with a Cys-less receptor (C59SC252S) in order eliminate possible involvement of endogenous cysteines in disulfide crosslinking. (Removal of the normal cysteine residues at positions 59 and 252 in Ste2p has previously been shown to have no effect on receptor function 27; 28). The use of a random screening approach avoided the need to individually create and test a large number of site-directed introductions of cysteine pairs or to make a priori assumptions about which parts of the receptor would be most likely to interact. While it is, in principle, possible to recover cysteine mutations using conventional random mutagenesis for generation of missense mutations, such procedures yield a low recovery of cysteines at any position, and, thus, a vanishingly small likelihood of obtaining two cysteine substitutions in a single allele. In order to specifically introduce cysteine residues at every position in the targeted regions of the receptor, we used a procedure commonly applied for conventional site-direct mutagenesis in which mutagenic oligonucleotides are hybridized to a single-stranded template derived from a STE2-encoding phagemid. However, instead of conducting the reaction with a single mutagenic oligonucleotide, we used a collection of synthetic oligonucleotides, each encoding a single cysteine substitution targeted to predicted TM segments TM5 and TM6 of Ste2p (see Fig. 2). These two segments were selected for initial screening because of evidence that they undergo activation-dependent changes in other GPCRs 2; 3; 5; 6, because they are adjacent in sequence and in the tertiary structures of other GPCRs 29; 30; 31, and because crosslinks that do not activate the receptor have previously been isolated between these segments 14; 15. In view of uncertainty about the actual topology of the predicted TM segments of Ste2p, we mutagenized more than thirty different residues in each segment. One oligonucleotide encoding a single cysteine substitution that was already known to cause high levels of constitutive activity of Ste2p (P258C 14; 32), was omitted from the collection. The primer:template ratio in the mutagenesis reactions was optimized to obtain the desired number of cysteine substitutions based on sequencing of random clones from the mutagenesis reaction, which also provided verification that the procedure yielded the desired diversity of clones.
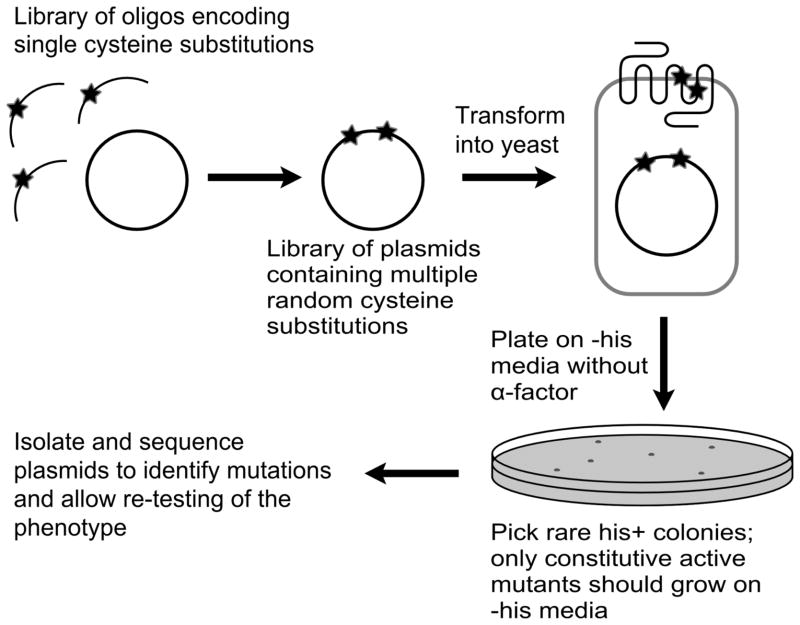
Figure 1.
Schematic view of the procedure for identifying disulfide-dependent constitutively activated receptors. First, random cysteine mutagenesis was performed using a mixture of oligonucleotides specifying each of the cysteine substitutions to be tested. Then the library of plasmids containing random cysteine substitutions was transformed into a yeast strain that allows for selection of constitutively active mutants. Finally, colonies that demonstrate constitutive behavior were isolated, sequenced to identify mutations, and transformed into a fresh yeast host for further testing.
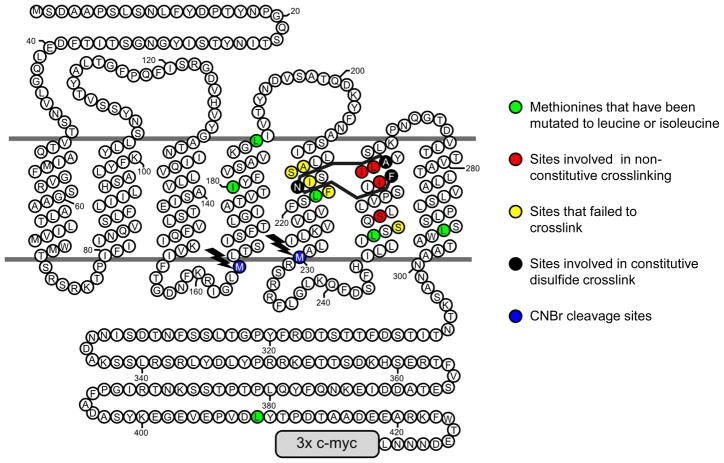
Figure 2.
Topological diagram of Ste2p showing positions of disulfides and modifications introduced to allow detection of crosslinks by immunoblotting of CNBr digests. Residues shown with green fills are naturally occurring methionines that were mutated to leucine or isoleucine to eliminate cleavage sites. Residues shown with blue fills (highlighted with lightning bolts) are remaining or introduced methionines where CNBr can cleave to create myc-tagged crosslinked or un-crosslinked fragments. The sites of disulfide bonded cysteines that result in constitutive activation are shown as residues with black fills with crosslinks indicated by solid black lines. Amino acids that, when mutated to cysteine, participate in non-constitutive crosslinks in combination with N216C are shown with red fills. Cysteine residues introduced at positions shown with yellow fills on TM5 did not form disulfide crosslinks with A265C or F262C substitutions on TM6. Cysteine introduced as a replacement for residue S252 in TM6, also shown with a yellow fill, did not form a disulfide crosslink (when mutated to cysteine) in conjunction with the N216C mutation on TM5.
Several independent libraries of cysteine-containing receptor mutants containing up to 10,000 clones each were subjected to screening by transformation into a yeast host lacking the normal chromosomal copy of the STE2 gene and containing chromosomally-encoded copies of FUS1-HIS3 and FUS1-lacZ reporter genes that are transcriptionally induced upon activation of the pheromone response pathway. When the largest tested library was plated on SD-ura-his plates to select for spontaneous induction of the FUS1-HIS3 reporter, approximately 200 colonies grew. To determine whether the observed phenotype was dependent on the STE2-encoding plasmid, we isolated plasmid from the his+ colonies and retransformed a fresh yeast host. A total of 33 clones that retained the his+ phenotype after re-transformation were subjected to DNA sequencing. Most of these exhibited only slightly enhanced constitutive activity based on α-factor-independent FUS1-lacZ expression compared with normal Ste2p receptors (see Supplementary Tables 1 and 2). In addition several clones that contained mutations at positions where a single substitution had previously been shown to result in constitutive activity were eliminated from further consideration. Since most of the recovered alleles contained more than two cysteine substitutions, site-directed mutagenesis was used to generate alleles containing pairs of introduced cysteines for further testing. The two strongest constitutive alleles that were recovered contained pairs of introduced cysteines were an allele containing the substitutions N216C and A265C, which exhibited approximately 6-fold enhancement of basal activity compared to normal receptors, and an allele containing the substitutions N216C and F262C, which exhibited 11-fold enhancement (see Fig. 3b). Alleles containing each of these substitutions were also isolated from a completely independent screen of a second cysteine-substitution mutagenesis reaction (see Supplementary Table 2).
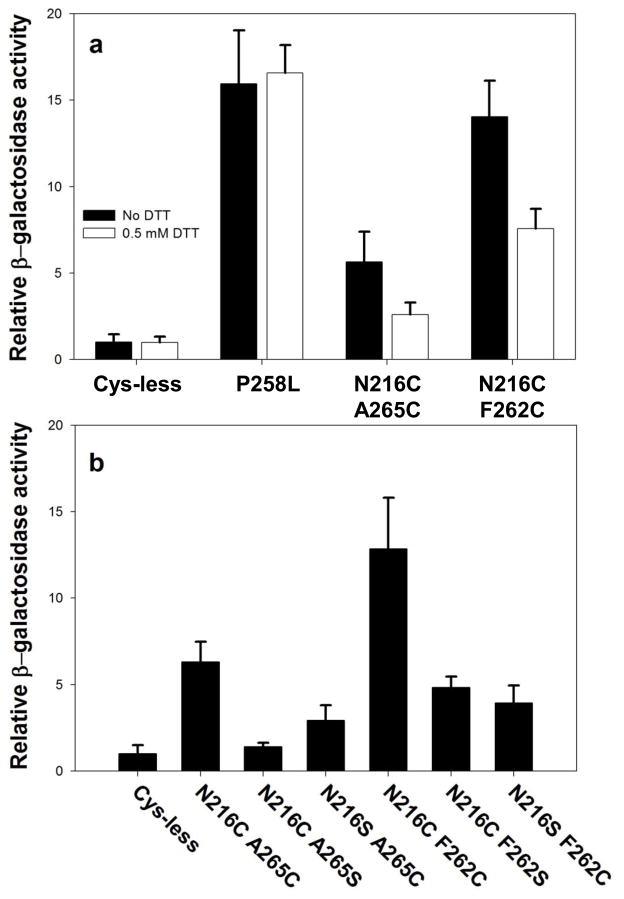
Figure 3.
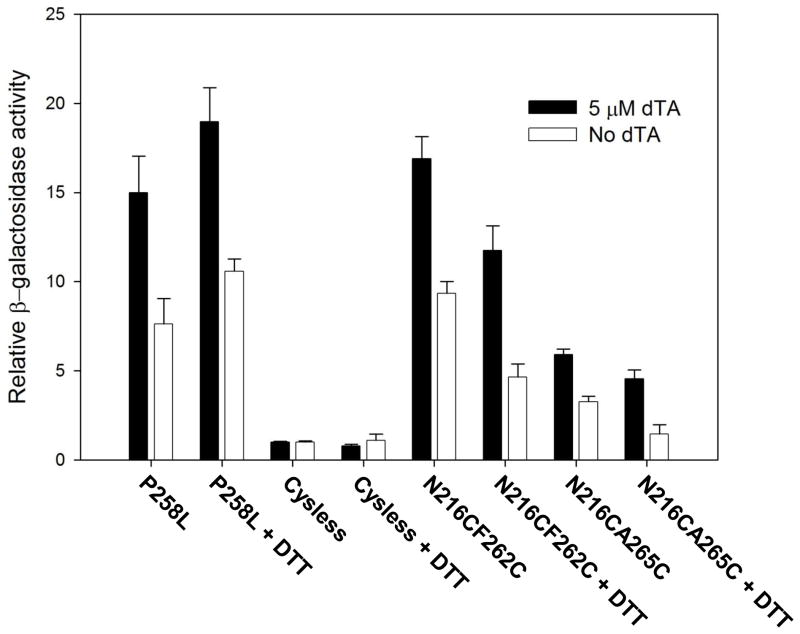
Constitutive levels of induction of expression of the FUS1 lacZ reporter. a) Effect of DTT on constitutive activity of various receptor alleles. Unfilled bars refer to β-galactosidase activity measured following treatment of intact cells treated with 0.5 mM DTT for 2 hours. Filled bars indicate activities of receptors expressed on cells that were not treated with DTT. The β-galactosidase activities of all mutants were normalized to the untreated normal Cys-less basal activity. b) Requirement for the presence of two cysteines to maintain full constitutive induction of FUS1-lacZ reporter. The graph shows the constitutive β-galactosidase activity induced by alleles containing two cysteine residues in comparison to alleles containing only one cysteine, Cys-less alleles, and the previously-characterized constitutive allele containing the P258L substitution.
We used three approaches to determine whether formation of a disulfide bond is responsible for the constitutive activity of the receptors containing the N216CA265C and N216CF262C substitutions:
Requirement for two cysteines. We individually mutated each of the two cysteine residues in each putative disulfide-forming allele to serine, an amino acid residue that lacks a sulfhydryl but maintains the closest possible steric similarity to cysteine. The constitutive activity of each of the cysteine-to-serine mutants was less than 40% of that of the double cysteine mutants (Fig. 3b), indicative of the importance of disulfide formation for the observed activity. Residual constitutive activity of some alleles containing only single cysteine residues is not unexpected, as a variety of mutations in this region of the receptor, including cysteine substitutions, have previously been shown to enhance basal levels of signaling 13; 14; 32; 33.
Dependence of constitutive signaling on cysteine oxidation state. Cells expressing receptor alleles containing the N216CA265C or N216CF262C cysteine pairs exhibited about a 50% decrease in constitutive activity after incubation with 0.5 mM DTT for 2 hours. In contrast, FUS1-lacZ induction by the previously-characterized strong constitutive receptor mutant containing the substitution P258L 13; 14; 32 was unaffected by similar treatment with DTT (Fig. 3a). There are several likely explanations for the fact that this DTT-treatment does not decrease constitutive activity of the double cysteine mutants to levels as low as those exhibited by the Cys-less control: 1) The conditions of treatment with DTT may have been inadequate to quantitatively reduce all disulfides, since reducing disulfides in the hydrophobic environment of a membrane protein interior can require lengthy treatments with high levels of reducing agents 20. The concentration of DTT that could be used for treating yeast cells was limited by secondary effects on the cells and signaling pathway – the use of concentrations higher than 0.5 mM resulted in inhibition of signaling by the P258L mutation, which contains no cysteine residues; 2) Cells in active culture may always contain a newly-synthesized population of receptors that have not been present on the cell surface for sufficient time to undergo reduction; and 3) Receptors containing single cysteine residues that do not participate in disulfides exhibit some constitutive activity.
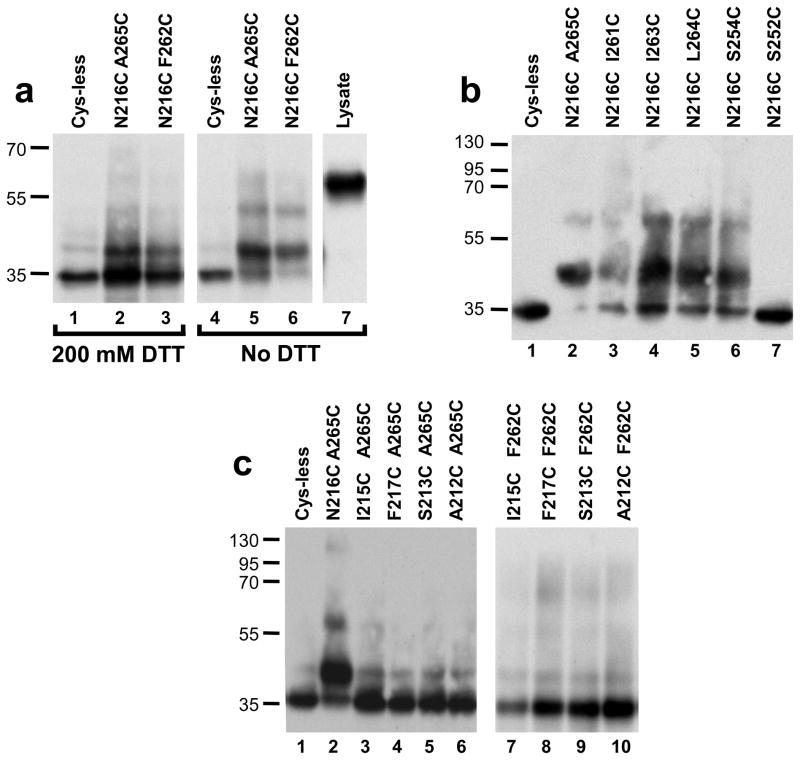
Detection of crosslinked cysteine-containing peptides derived from the receptor. To allow detection of crosslinked peptides in efficiently-cleaved receptors derived from minimally-processed samples, we used immunoblotting of cyanogen bromide (CNBr)-treated whole-cell lysates. To allow detection of the relevant peptides using the triple myc tag at the extreme C-terminal of the receptor, six methionine residues in the vicinity of the introduced cysteine residues and between the cysteines and the C-terminal were mutated to leucine or isoleucine (Fig. 2), removing potential CNBr cleavage sites. In addition, a methionine residue was introduced as a substitution for Ile230 to create a cleavage site between TM5 and TM6. These substitutions do not diminish basal or α-factor-dependent FUS1-lacZ induction Supplementary Fig. S1f) compared to normal Cys-less Ste2p. Furthermore, similar increases in basal activity of the N216CA265C and N216CF262C mutations relative to similar Cys-less receptor alleles were observed in the presence and absence of the methionine substitutions introduced to allow CNBR mapping (data not shown). The peptide analysis was conducted by lysing cells by agitation with zirconia beads, treating the lysate with CNBr, subjecting samples to SDS-PAGE under reducing and non-reducing conditions, and blotting with anti-myc antibodies. Cells and lysates were maintained at low pH during processing to prevent disulfide exchange. As shown in Fig. 4a, under both reducing and non-reducing conditions, the Cys-less receptor gives rise to a predominant immunoreactive band with an apparent mobility of 35 kDa, slightly greater than the predicted molecular mass of 28 kDa for the fragment extending from Arg231 to the C-terminal. Under non-reducing conditions the samples derived from cells expressing the constitutive N216CA265C and N216CF262C alleles exhibit a predominant band with an apparent molecular mass of 42kDa that comprises approximately 70%, of the total intensity of shifted plus non-shifted bands. Treatment of samples derived from these two alleles with reducing agent (200 mM DTT) just prior to SDS-PAGE results in a reduction in the intensity of the 42 kDa band to less than 20% of the total (for both alleles), accompanied by increased intensity of the band at 35 kDa. The 42 kDa band in cysteine-containing samples under non-reducing conditions corresponds to the size of the species expected for the product of crosslinking between the 8kDa CNBr fragment extending from Leu166 to Met230 and the C-terminal fragment beginning at Arg231 (see Fig. 2). The fact that only partial elimination of the crosslinked band is observed following DTT treatment may be explained by the difficulty of fully reducing disulfides in the hydrophobic interior of a membrane protein in a detergent micelle 20.
Figure 4.
Detection of disulfide bonds by immunoblotting of C-terminally c-myc tagged CNBr-cleaved alleles of Ste2p. a) Immunoblot of CNBr cleaved Ste2p alleles containing different cysteine pairs involving TM5 and TM6 that result in constitutive activity Where indicated, samples were incubated with 200 mM DTT at 37° C for 15 minutes prior to SDS polyacrylamide gel electrophoresis. b) Immunoblot of receptors containing cysteine pairs involving TM5 and TM6 that form disulfides but do not result in constitutive activation. c) Immunoblot of receptors containing cysteine pairs in TM5 and TM6 that do not form disulfides.
Cells expressing mutant receptors with the N216CA265C and N216CF262C substitutions maintained the ability to respond to α-factor (Supplementary Figs. S1a and S1b). The N216CA265C allele exhibited a ~3-fold decrease in EC50 and elevated maximal induction of FUS1-lacZ compared to Cys-less reference, whereas the response of the N216CF262C allele to α-factor was not distinguishable from that of the Cys-less reference (Table 1). Since peptide mapping experiments in Fig. 4a reveal a mixed population of crosslinked and non-crosslinked receptors in cells expressing these mutants, retention of α-factor responsiveness in cells expressing the constitutive alleles may either result from further activation of crosslinked constitutive receptors or from activation of the subset of receptors that do not contain crosslinks. (At normal expression levels, activation of only a small fraction of the population of receptors at the cell surface is sufficient for full responses to α-factor 34.)
Table 1.
Induction of the FUS1-lacZ reporter in response to α-factor for mutations containing intramolecular disulfides.
| Allele | EC50a | Maximal Activationb | Strain number |
|---|---|---|---|
| cys-less | 1 | 1 | A4061 |
| N216C/A265C | 0.3 ± 0.1 | 1.4 ± 0.2 | A4064 |
| N216C/F262C | 0.9 ± 0.2 | 1.1 ± 0.2 | A4082 |
| N216C/I261C | 1.2 ± 0.1 | 1.1 ± 0.1 | A4157 |
| N216C/I263C | 0.9 ± 0.1 | 1.0 ± 0.1 | A4158 |
| N216C/L264C | 1.1 ± 0.1 | 1.1 ± 0.1 | A4159 |
computed as the ratio EC50 (mutant)/EC50 (wild type) α-factor, where the absolute EC50 for activation of wild type receptors by α-factor was 33nM ± 5.
computed as [maximal FUS1-lacZ induction (mutant)]/[maximal FUS1-lacZ induction (wild type)] for α-factor.
As a test of the surface exposure of crosslinked receptors and their ability to be activated, we examined their interactions with the ligand [des-Trp1Ala3, Nle12]α-factor (dTA). This peptide acts as an antagonist or very weak agonist towards normal Ste2p, but as a strong agonist toward all previously-identified constitutively active alleles of Ste2p 13; 35. In accordance with this, both the N216CA265C and the N216CF262C alleles of Ste2p are effectively activated by dTA (Fig. 5). In addition, the dTA responsiveness of these alleles is decreased after pre incubation with 0.5 mM DTT while the Cys-less and P258L constitutive receptors are not (Fig. 5). The sensitivity of the dTA-responsive receptors to reducing agent indicates that it is the crosslinked population of receptors that is responsible for the dTA-sensitivity and, by extrapolation, for the constitutive activity of cells expressing the N216CA265C and the N216CF262C alleles. Furthermore, since α-factor and related peptides are not cell permeant, the observed responses to dTA must be reflect activation of crosslinked receptors that are present at the cell surface. As observed previously for other constitutively active alleles of STE2 13, low levels of cell surface expression of the cysteine-containing constitutvely active alleles precluded characterization of their ligand binding properties.
Figure 5.
DTT-treatment diminishes responses of disulfide-containing constitutive alleles to the α-factor analog dTA that activates constitutive receptors but behaves as an antagonist toward normal receptors. Where indicated, cells were treated with 0.5 mM DTT for 2 hours prior to the addition of dTA.
To examine the specificity of disulfide formation and crosslinking effects on receptor activation, we created additional alleles of Ste2p in which cysteine residues were introduced at positions adjacent to the sites of crosslinking identified from the genetic screen. Immunoblotting of CNBr fragments from the resulting alleles demonstrated a surprising promiscuity of disulfide formation between N216C on TM6 and cysteine substitutions S254C, I261C, I263C, and L264C on TM6 (Fig 4b). However, no crosslinking was observed between N216C and S252C further toward the intracellular surface of TM6. None of these additional sites of disulfide bonds resulted in more than a ~2-fold enhancement of constitutive activity of the mutant receptors compared to levels seen with receptors containing these mutations as single cysteine substitutions (Fig. 6). In striking contrast to the diversity of sites capable of forming disulfides on TM6, no crosslinking above background levels was detected between substitutions A212C, S213C, I215C, and F217C on TM5 and either F262C or A265C on TM6 (Fig. 4c).
Figure 6.
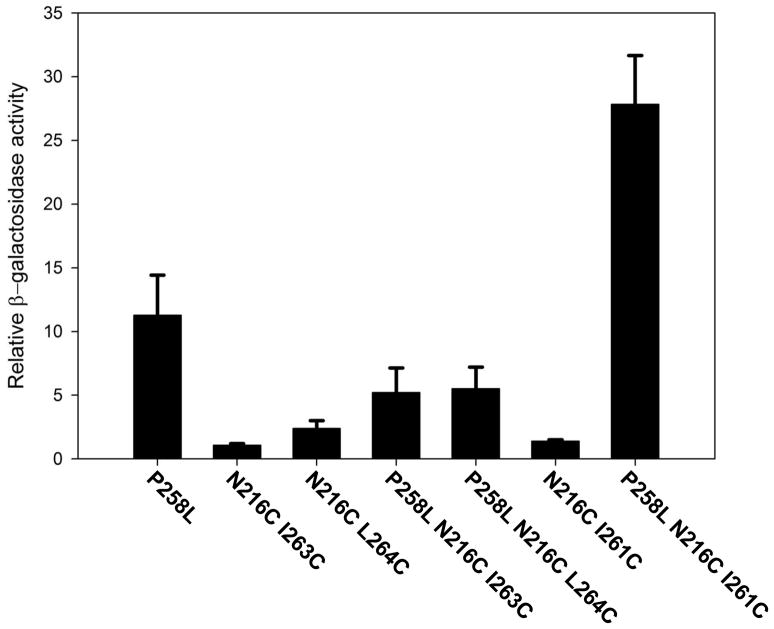
Levels of constitutive induction of FUS1−lacZ reporter by receptors containing the point mutation P258L in combination with cysteine pairs that do not, by themselves, result in constitutive activation. The β-galactosidase activities of all mutants were normalized to the untreated Cys-less untreated basal activity.
Cells expressing receptors containing each of the identified non-constitutive disulfide crosslinks between N216C and sites on TM6 retained α-factor responsiveness comparable to that of cells expressing Cys-less receptors (Supplementary Figs. S1c–e). This could either be because the crosslinking does not interfere with the conformational changes associated with receptor activation, or because disulfide formation is not quantitative. Based on peptide mapping, for each non constitutive disulfide crosslinked allele, at least 20% of the receptors in the cell are not in a crosslinked state (Fig. 4b). Since only a small fraction of the normal complement of receptors on the cell surface is sufficient to support responses to α-factor 34 the population of un-crosslinked receptors could be responsible for the observed cellular responses to α-factor.
To test the role of non-constitutive crosslinks in a context in which the number of receptors is more directly tied to signaling function, we created receptors that combine the constitutively-activating point mutation P258L with the cysteine pairs that form non-constitutive crosslinks. (The level of constitutive signaling by the P258L mutant has been shown to be dependent on expression levels 33). Whereas cells expressing receptors containing the P258L substitution in a Cys-less background exhibited a constitutive level of FUS1-lacZ induction of 11-fold over basal activity, the combination of P258L with the non-constitutive disulfide-inducing mutations N216CI263C and N216CL264C decreased this level by about 50% (Fig. 6), consistent with inability of the crosslinked allele to be able to adopt the activated state. However, surprisingly, the receptor allele containing the cysteine pair N216CI261C in conjunction with P258L exhibited more than 20-fold higher basal activity than the reference Cys-less allele, which is more than twice the activity of receptors containing P258L alone (Figure 6). Based on immunoblotting of whole cell extracts, the levels of expression of non-constitutive crosslinked mutants in combination with P258L were similar to those containing P258L alone (data not shown). Thus, differences in basal activity among the P258L-containing alleles do not appear to be due to expression levels. Based on, immunoblotting of CNBr digests, the extents of disulfide crosslinking of the alleles combining cysteine pairs with of these P258L-containing alleles were comparable to those for alleles containing the cysteine pairs alone (Supplementary Fig. S2).
Disulfide bond formation involving introduced cysteines could be intramolecular or intermolecular. To test for intermolecular crosslinking, lysates from cells expressing various combinations of cysteine residues were directly subjected to immunoblotting under non-reducing conditions without being subjected to CNBr treatment. Under the conditions used for immunoblotting (the presence of urea in the gel loading buffer, loading of low concentrations of receptors per lane), no oligomers could be detected, indicating that the disulfides are intramolecular (supplementary Fig. S3).
Discussion
Most proteins undergo some form of conformational change as part of their biological functions. The engineering of cysteine substitutions to create disulfide bonds that lock proteins in particular conformations provides a potentially widely applicable way of identifying changes in intramolecular protein contacts that drive functionally important conformational changes. However, using one-at-a-time site-directed introduction of cysteines to identify disulfides providing a conformational lock for proteins where the nature of the relevant conformational change is not yet known is a laborious and low-yield process. We describe here the combination of a new protocol for random introduction of cysteine residues into targeted regions of proteins in conjunction with the use of a genetic selection to identify alleles in which disulfide bonds alter the conformational equilibrium of a target protein in a specific pre-defined way. The approach was used to identify two disulfide bonds that lock the yeast α-factor receptor, a member of the GPCR superfamily, in an activated state. Identifying mutations that result in receptor activation is likely to be more informative than identifying mutations that prevent receptor activation, since loss of signaling function could occur if a receptor is locked in an aberrant conformation unrelated to either the active or inactive states.
The cysteine pairs that activate the α-factor receptor were recovered in two independent repetitions of the screen. The level of activation of the receptor by these mutations approaches that of the strongest previously-isolated mutation in this receptor. The effects of the cysteine pairs on signaling were observed in the context of the receptor’s normal environment in the yeast cell membrane in the presence of ambient oxygen. Formation of the observed disulfides does not depend on the addition of any additional oxidizing agents, which may, under some circumstances, promote promiscuous disulfide formation 36. High level constitutive signaling by the cysteine-containing mutants is dependent on the presence of both cysteine residues participating in the disulfides, and is diminished by treating receptor-expressing cells with a reducing agent. Based on the repeated isolation of the two constitutive alleles, it is likely that these two recovered pairs of cysteine substitutions provide the strongest constitutive activation of any two cysteines in the fifth and sixth TM segments of the receptor. Additional combinations of cysteine substitutions recovered from the screen that provided lower levels of constitutive activation (less than 5-fold) were not subjected to further analysis. The procedures we describe raises the possibility of searching for every disulfide bond in an entire GPCR that is capable of activating the receptor, screening 18,900 combinations of cysteine substitutions (allowing for 30 residues per TM segment to allow for uncertainties in the exact boundaries of the membrane inserted sequences), which could be subdivided into smaller libraries focusing on particular collections of TM segments. Testing this number of individual pairs of cysteine substitutions would likely be prohibitive using current technology.
The identification of disulfides involving TM5 and TM6 of Ste2p that enhance constitutive activity of the receptor suggests that the relative motion of these segments is sufficient for activating the signaling response. The involvement of TM5 and TM6 in activation is not surprising, given the previous structural, spectroscopic, and biochemical evidence for motions involving these portions of Ste2p and other GPCRs 2; 3; 4; 5; 6; 7; 8; 9; 11; 15; 21. However, our studies indicate that that imposing relative motion of these two helical segments is sufficient for activation, not simply a correlate or end result of a global change of conformation. Furthermore, the residues participating in the activating disulfide are predicted to reside closer to the external surface of the receptor than to its cytoplasmic surface (see model in Supplementary Fig. S4), despite the fact that only relatively small motions in the external portions of TM5 and TM6 are seen in comparing structures of inactive states with those of states that are thought to be activated. This suggests that a small relative motion of the extracellular half of these TM helices (which is the region where ligands normally bind to receptors) suffices to induce larger conformational changes at the cytoplasmic surface where activation of G proteins must occur.
The observation that the constitutively activating disulfide crosslinks do not activate signaling responses to the same extent as agonist binding (Supplementary Fig. S1) indicates that the crosslinked receptors are not trapped in a fully activated state. However, the presence of disulfides does not appear to prevent full activation, since cells expressing constitutive disulfide-containing receptors retain the ability to respond to α-factor. Such responsiveness appears to be an intrinsic property of crosslinked receptors at the cell surface, based the sensitivity to reducing agent of responses to the α-factor analog dTA (Fig. 5), which specifically activates constitutive receptors. The existence of differences between the disulfide-trapped and fully activated states is also suggested by the fact that neither the presence saturating concentrations of the normal agonist, α-factor (results not shown), nor the incorporation of the previously described activating mutation P258L into cysteine-containing receptors (Supplementary S. 2) results in detectable enhancement of the yield of crosslinked Ste2p-derived CNBr peptides.
We observed unexpected behavior of one pair of cysteines that does not induce constitutive signaling. The combination of substitutions N216C on TM5 and I261C on TM6 results in intramolecular crosslinking without inducing significant constitutive activation of Ste2p. However, when combined with the constitutively activating substitution P258L, the disulfide linking positions 216 and 261 leads to enhancement of constitutive activity compared to that of a receptor containing the P258L alone. This suggests that the activated state of a receptor containing the P258L substitution may be different from that of normal receptors, such that the disulfide linking positions 216 and 261 preferentially stabilizes the activated state of the P258L mutant, but not that of the receptors with the normal proline at position 258. This would be consistent with suggestions that receptors may occupy multiple active and inactive states 37.
The range of residues on TM6 that can crosslink to residue N216C on TM5 indicates that TM6 may undergo substantial rotational motion in the inactive state. The reduced constitutive activity observed when some of these combinations of cysteines were combined with a previously-described constitutive mutation suggests that these crosslinks between TM5 and a rotated configuration of TM6 restrict conversion to the receptor’s activated state. The observed rotational mobility does not appear to extend to all TM segments of the receptor, since we were only able to find one cysteine substitution (N216C) on TM5 that is capable of crosslinking to TM6. Disulfide linkages between TM segments that diminish transducing activation by rhodopsin have been identified previously 8; 38.
Random cysteine mutagenesis combined with screening for constitutive mutations proved to be an effective approach for identifying disulfide bonds that result in constitutive receptor activation in Ste2p. We anticipate that random cysteine mutagenesis followed by functional screening could be used to identify functionally important intramolecular motion of other receptors and non-receptor proteins. Since a variety of mammalian receptors can be expressed in yeast and coupled to the pheromone response pathway 24; 25, this method could be used in other GPCRs to identify general mechanisms of activation. The constitutively activating disulfide crosslinks that were detected in Ste2p demonstrate that in this receptor, which is a member of the class D family of GPCRs, TM5 and TM6 undergo relative motion upon activation and that such motion is capable of activating the receptor. Relative motion of these same helices is observed in structural comparisons of the activated and inactive states of members of the class A family of mammalian GPCRs. Thus, despite extensive diversity of sequences between different families of GPCRs, mechanisms of activation may be conserved.
Materials and Methods
Plasmids and yeast strains
A listing of plasmids and strains used in this study can be found in Supplementary Table 3. Two separate genetic screens for constitutively active disulfide-containing mutants were conducted using two different Ste2p-encoding starting plasmids: The first screen (Supplementary Table 1) used plasmid pMD354 28, encoding a form of Ste2p expressed from the native STE2 promoter in which the naturally-occurring cysteine residues in the protein were mutated to serines (C59S;C252S) and fused at its extreme C-terminal to three tandemly-repeated copies of a c-myc tag. pMD 354. To create plasmid pMD1736 with a distribution of methionine residues useful for peptide mapping using CNBr, pMD354 was mutagenized by site directed mutagenesis 39; 40 to contain the following substitutions: M409I, M294I, M218I, M189I. M180I I230M and M250L (also see Fig. 1). PMD 1736 was then used as a starting plasmid for the creation of site-directed mutants to be subjected to peptide mapping and for measurement of FUS1-lacZ induction. The second screen was conducted using plasmid pMD1210 (see supplemental text), encoding Ste2p fused to HA, His6, and ZZ IgG-binding affinity tags.
Yeast strain A2953 (see supplemental text) was used as a host in performing both the first and second screens for constitutively active receptors. Strain A575 13 (Mata ste2-Δ bar1-Δ far1cry1R ade2-1 his4-580 lys2oc trp1am tyr1oc SUP4-3ts leu2 ura3 FUS1::p[FUS1-lacZ TRP1]) was used for all characterization of the individual receptor alleles.
Random oligonucleotide directed cysteine mutagenesis
Random cysteine mutagenesis was performed using oligonucleotide-directed mutagenesis 39; 40. However, instead of using a single mutagenic oligonucleotide, as the technique is normally performed, we used a mixture of 61 mutagenic oligonucleotides encoding single cysteine substitutions at different positions in transmembrane segments TM5 (D201 to R231) and TM6 (S243 to T274 excluding P258). These oligonucleotides were designed so that the 5′ and 3′ regions flanking the mismatches all have similar annealing temperatures. The sequences of oligonucleotides described in this manuscript are available upon request. The 61 oligonucleotides was mixed in one reaction at a final concentration of 1 ng of total oligonucleotide mixture per 1 μg of single stranded DNA from phagemid template in a total volume of 50 μl. Transformation of the reaction products into E. coli (XL1-Blue) yielded approximately 20,000 transformants for each library. Clones picked at random from the bacterial transformations were subjected to DNA sequencing. The average number of substitutions per allele was 1.5 in the first library and 4.6 in the second library.
Genetic screening for constitutively active α-factor receptors
Plasmids isolated from pooled bacterial transformants were transformed 41 into Saccharomyces cerevisiae strain A2953 and plated on SD – ura medium to select for transformants based on the plasmid’s URA3 marker. In the first library a total of 12,000 transformants were initially initial selected on SD – ura, then replica plated on SD–ura–his media to test for induction of the FUS1-HIS3 reporter. (Cells were not directly plated on SD–ura–his plates in order to allow them to recover from transformation.) Colonies exhibiting growth on SD–ura–his media were sub-cloned on the same medium, then plasmids from the yeast colonies were isolated from cells that had been subjected to vortexing with zirconia/silica beads for 3 minutes. Plasmid isolation from the cell lysates was accomplished using the Qiagen® QIAprep spin miniprep kit according to the manufacturer’s directions. The resulting plasmids were transformed into E. coli, sequenced, and re-transformed into the yeast host. The screen for the second library was similar to the first except it contained 36,000 yeast transformants, and cells were replica plated on galactose-copntaining media SGa – ura–his to induce expression of the GAL1-controlled STE2. The plasmids were then isolated from E. coli, and subjected to DNA sequencing. Yeast that had been re- transformed with the isolated plasmids were tested for the ability to grow on SD-ura–his to confirm that the observed phenotype was plasmid-dependent.
Assays of α-factor receptor signaling function
Assays of FUS1-lacZ induction in response to pheromone were performed in triplicate from three independent isolates as previously described 39. Assays to determine levels of constitutive activity in absence of α-factor were incubated for 3–4 hours with FDG (fluorescein di-β-D-galactopyranoside) instead of the 30–45 minutes used to measure pheromone response. EC50s and maximal activation levels were calculated by nonlinear least squares fitting of β-galactosidase activities to a sigmoidal dose-response equation using the ligand binding module of Sigmaplot (SPSS Inc). Errors are reported as the standard error of the mean for assays done on three independent yeast transformants.
Peptide Mapping
Peptide mapping was performed essentially as described by Roth et al. 42. Cells were cultured to an optical density of 1.0 at OD600 in SD–ura media. 5 OD600 × ml of cells were centrifuged at 10,000 × g for 2 minutes and washed with an equal volume of de-ionized water and centrifuged again. 250 μl of zirconia/silica beads (BioSpec Products Inc.) and 300 μl of 20 mM sodium acetate pH 4.6 containing 1mM PMSF and 0.1 mM EDTA were added to the pellet. Cells were lysed in a cold room at 8° C by ten cycles of vortexing for 20 seconds, followed each time by cooling on ice for 2 minutes. 270 μL of 2.0 M CNBr (Sigma-Aldrich) in 70% formic acid were added to 30 μL of broken cells. The samples were then incubated at 37°C for 2 hours in the dark, then dried in a vacuum centrifuge for approximately 1.5 hours. 200 μl of distilled water were added and the sample was dried for an additional 1.5 hours to remove any residual CNBr. The pellet was then re-suspended in SDS gel-loading buffer (100 mM Tris pH 6.8, 0.1 mM EDTA, 9M urea, 5 % SDS, 0.02 mg/ml bromophenol blue) by pipetting and then vortexing for 5 seconds and then incubating at 37C ° for 15 minutes. DTT-treated samples were processed in the same way as non-DTT samples until the last 15-minute incubation, at which time DTT was added to tubes from a 2 M stock to a final concentration of 200mM. 30 μl of the sample was then loaded on a 15% SDS gel. A separation of at least three gel lanes was maintained between DTT-containing and non-DTT-containing samples, to prevent spurious reduction. All immunoblots were derived from gels maintained under non-reducing conditions unless otherwise noted. Immunoblotting was performed as described previously 28 except that 2.5 % non fat powdered milk was used as blocking reagent instead of newborn calf serum and the dilution of 1° anti-c-myc antibody (Roche Inc) was 1:5000. SuperSignal® West Dura Extended Duration Substrate (Thermo Scientific) was used for chemiluminescent detection of myc tagged receptors.
Intensities of bands were determined using the software Quantity One, using subtraction of a background level for each lane based on a region of the lane in which there were no visible bands and subtracted from intensity of each of its associated bands. The intensities of the experimental bands were calibrated against varying concentration of reference samples to insure that they were within the linear range of the detection system.
Supplementary Material
Acknowledgments
We thank Sara M. Connelly for technical assistance, Jeffrey Zuber for assistance in preparing figures, Dr. Patricia Hinkle for comments on this manuscript, and Dr. Fred Naider for providing the antagonist dTA. This study was supported by NIH grants GM084083 and GM059357 to M. Dumont.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–50. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 2.Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–80. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rasmussen SG, Devree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah ST, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Crystal structure of the beta(2) adrenergic receptor-Gs protein complex. Nature. 2011 doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SG, Choi HJ, Devree BT, Sunahara RK, Chae PS, Gellman SH, Dror RO, Shaw DE, Weis WI, Caffrey M, Gmeiner P, Kobilka BK. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469:236–40. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, Ernst OP. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 6.Xu F, Wu H, Katritch V, Han GW, Jacobson KA, Gao ZG, Cherezov V, Stevens RC. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011;332:322–7. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc Natl Acad Sci U S A. 2008;105:7439–44. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274:768–70. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 9.Ghanouni P, Steenhuis JJ, Farrens DL, Kobilka BK. Agonist-induced conformational changes in the G-protein-coupling domain of the beta 2 adrenergic receptor. Proc Natl Acad Sci U S A. 2001;98:5997–6002. doi: 10.1073/pnas.101126198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vilardaga JP, Bunemann M, Krasel C, Castro M, Lohse MJ. Measurement of the millisecond activation switch of G protein-coupled receptors in living cells. Nat Biotechnol. 2003;21:807–12. doi: 10.1038/nbt838. [DOI] [PubMed] [Google Scholar]
- 11.Ye S, Zaitseva E, Caltabiano G, Schertler GF, Sakmar TP, Deupi X, Vogel R. Tracking G-protein-coupled receptor activation using genetically encoded infrared probes. Nature. 2010;464:1386–9. doi: 10.1038/nature08948. [DOI] [PubMed] [Google Scholar]
- 12.Parnot C, Miserey-Lenkei S, Bardin S, Corvol P, Clauser E. Lessons from constitutively active mutants of G protein-coupled receptors. Trends Endocrinol Metab. 2002;13:336–43. doi: 10.1016/s1043-2760(02)00628-8. [DOI] [PubMed] [Google Scholar]
- 13.Sommers CM, Martin NP, Akal-Strader A, Becker JM, Naider F, Dumont ME. A limited spectrum of mutations causes constitutive activation of the yeast alpha-factor receptor. Biochemistry. 2000;39:6898–909. doi: 10.1021/bi992616a. [DOI] [PubMed] [Google Scholar]
- 14.Dube P, DeCostanzo A, Konopka JB. Interaction between transmembrane domains five and six of the alpha -factor receptor. J Biol Chem. 2000;275:26492–9. doi: 10.1074/jbc.M002767200. [DOI] [PubMed] [Google Scholar]
- 15.Lee YH, Naider F, Becker JM. Interacting residues in an activated state of a G protein-coupled receptor. J Biol Chem. 2006;281:2263–72. doi: 10.1074/jbc.M509987200. [DOI] [PubMed] [Google Scholar]
- 16.Chervitz SA, Falke JJ. Lock on/off disulfides identify the transmembrane signaling helix of the aspartate receptor. J Biol Chem. 1995;270:24043–53. doi: 10.1074/jbc.270.41.24043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeCaen PG, Yarov-Yarovoy V, Zhao Y, Scheuer T, Catterall WA. Disulfide locking a sodium channel voltage sensor reveals ion pair formation during activation. Proc Natl Acad Sci U S A. 2008;105:15142–7. doi: 10.1073/pnas.0806486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falke JJ, Koshland DE., Jr Global flexibility in a sensory receptor: a site-directed cross-linking approach. Science. 1987;237:1596–600. doi: 10.1126/science.2820061. [DOI] [PubMed] [Google Scholar]
- 19.Li JH, Hamdan FF, Kim SK, Jacobson KA, Zhang X, Han SJ, Wess J. Ligand-specific changes in M3 muscarinic acetylcholine receptor structure detected by a disulfide scanning strategy. Biochemistry. 2008;47:2776–88. doi: 10.1021/bi7019113. [DOI] [PubMed] [Google Scholar]
- 20.Lynch BA, Koshland DE., Jr Disulfide cross-linking studies of the transmembrane regions of the aspartate sensory receptor of Escherichia coli. Proc Natl Acad Sci U S A. 1991;88:10402–6. doi: 10.1073/pnas.88.23.10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Umanah GK, Huang LY, Maccarone JM, Naider FR, Becker JM. Changes in Conformation at the Cytoplasmic Ends of the Fifth and Sixth Transmembrane Helices of a Yeast G Protein-Coupled Receptor in Response to Ligand Binding. Biochemistry. 2011 doi: 10.1021/bi200254h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y, Madej MG, Guan L, Nie Y, Kaback HR. An early event in the transport mechanism of LacY: Interaction between helices V and I. J Biol Chem. 2011 doi: 10.1074/jbc.M111.268433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bardwell L. A walk-through of the yeast mating pheromone response pathway. Peptides. 2004;25:1465–76. doi: 10.1016/j.peptides.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 24.Dowell SJ, Brown AJ. Yeast assays for G protein-coupled receptors. Methods Mol Biol. 2009;552:213–29. doi: 10.1007/978-1-60327-317-6_15. [DOI] [PubMed] [Google Scholar]
- 25.King K, Dohlman HG, Thorner J, Caron MG, Lefkowitz RJ. Control of yeast mating signal transduction by a mammalian beta 2-adrenergic receptor and Gs alpha subunit. Science. 1990;250:121–3. doi: 10.1126/science.2171146. [DOI] [PubMed] [Google Scholar]
- 26.Yin D, Gavi S, Shumay E, Duell K, Konopka JB, Malbon CC, Wang HY. Successful expression of a functional yeast G-protein-coupled receptor (Ste2) in mammalian cells. Biochem Biophys Res Commun. 2005;329:281–7. doi: 10.1016/j.bbrc.2005.01.130. [DOI] [PubMed] [Google Scholar]
- 27.Akal-Strader A, Khare S, Xu D, Naider F, Becker JM. Residues in the first extracellular loop of a G protein-coupled receptor play a role in signal transduction. J Biol Chem. 2002;277:30581–90. doi: 10.1074/jbc.M204089200. [DOI] [PubMed] [Google Scholar]
- 28.Martin NP, Leavitt LM, Sommers CM, Dumont ME. Assembly of G protein-coupled receptors from fragments: identification of functional receptors with discontinuities in each of the loops connecting transmembrane segments. Biochemistry. 1999;38:682–95. doi: 10.1021/bi982062w. [DOI] [PubMed] [Google Scholar]
- 29.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–65. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–45. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 31.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–7. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 32.Stefan CJ, Overton MC, Blumer KJ. Mechanisms governing the activation and trafficking of yeast G protein-coupled receptors. Mol Biol Cell. 1998;9:885–99. doi: 10.1091/mbc.9.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konopka JB, Margarit SM, Dube P. Mutation of Pro-258 in transmembrane domain 6 constitutively activates the G protein-coupled alpha-factor receptor. Proc Natl Acad Sci U S A. 1996;93:6764–9. doi: 10.1073/pnas.93.13.6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah A, Marsh L. Role of Sst2 in modulating G protein-coupled receptor signaling. Biochem Biophys Res Commun. 1996;226:242–6. doi: 10.1006/bbrc.1996.1340. [DOI] [PubMed] [Google Scholar]
- 35.Mathew E, Bajaj A, Connelly SM, Sargsyan H, Ding FX, Hajduczok AG, Naider F, Dumont ME. Differential interactions of fluorescent agonists and antagonists with the yeast g protein coupled receptor ste2p. J Mol Biol. 2011;409:513–28. doi: 10.1016/j.jmb.2011.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Struthers M, Yu H, Kono M, Oprian DD. Tertiary interactions between the fifth and sixth transmembrane segments of rhodopsin. Biochemistry. 1999;38:6597–603. doi: 10.1021/bi9902384. [DOI] [PubMed] [Google Scholar]
- 37.Vaidehi N, Kenakin T. The role of conformational ensembles of seven transmembrane receptors in functional selectivity. Curr Opin Pharmacol. 2010;10:775–81. doi: 10.1016/j.coph.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Yu H, Oprian DD. Tertiary interactions between transmembrane segments 3 and 5 near the cytoplasmic side of rhodopsin. Biochemistry. 1999;38:12033–40. doi: 10.1021/bi9909492. [DOI] [PubMed] [Google Scholar]
- 39.Celic A, Connelly SM, Martin NP, Dumont ME. Intensive mutational analysis of G protein-coupled receptors in yeast. Methods Mol Biol. 2004;237:105–20. doi: 10.1385/1-59259-430-1:105. [DOI] [PubMed] [Google Scholar]
- 40.Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985;82:488–92. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen DC, Yang BC, Kuo TT. One-step transformation of yeast in stationary phase. Curr Genet. 1992;21:83–4. doi: 10.1007/BF00318659. [DOI] [PubMed] [Google Scholar]
- 42.Roth AF, Davis NG. Ubiquitination of the PEST-like endocytosis signal of the yeast a-factor receptor. J Biol Chem. 2000;275:8143–53. doi: 10.1074/jbc.275.11.8143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.