Abstract
Conjugative plasmid transfer is key to the ability of bacteria to rapidly adapt to new environments, but there is no agreement on a single quantitative measure of the rate of plasmid transfer. Some studies derive estimates of transfer rates from mass-action differential equation models of plasmid population biology. The often-used ‘endpoint method’ is such an example. Others report measures of plasmid transfer efficiency that simply represent ratios of plasmid-bearing and plasmid-free cell densities and do not correspond to parameters in any mathematical model. Unfortunately, these quantities do not measure the same thing – sometimes differing by orders of magnitude – and their use is often clouded by a lack of specificity. Moreover, they do not distinguish between bulk transfer rates that are only relevant in well-mixed populations and the ‘intrinsic’ rates between individual cells. This leads to problems for surface-associated populations, which are not well-mixed but spatially structured. We used simulations of a spatially explicit mathematical model to evaluate the effectiveness of these various plasmid transfer efficiency measures when they are applied to surface-associated populations. The simulation results, supported by some experimental findings, showed that these measures can be affected by initial cell densities, donor-to-recipient ratios and initial cell cluster size, and are therefore flawed as universal measures of plasmid transfer efficiency. The simulations also allowed us to formulate some guiding principles on when these estimates are appropriate for spatially structured populations and how to interpret the results. While we focus on plasmid transfer, the general lessons of this study should apply to any measures of horizontal spread (e.g., infection rates in epidemiology) that are based on simple mass-action models (e.g., SIR models in epidemiology) but applied to spatial settings.
Keywords: Plasmid, Conjugation rate, Spatial, Transfer efficiency, Horizontal
1. Introduction
Plasmids are extra-chromosomal genetic elements that are common in bacteria and code for traits such as virulence factors and resistance to antibiotics and heavy metals. There are two modes of plasmid replication: by propagation along with the bacterial host via cell division and by infectious transfer to new host cells (also called horizontal or conjugative transfer or ‘conjugation’). The infectious transfer of plasmids provides a form of bacterial sex when plasmid DNA is incorporated into the chromosomal DNA of the host cell. In addition to this role in chromosome evolution, plasmids also lead directly (without integration into the chromosome) to rapid adaptation of bacteria to changes in environmental conditions, owing to their ability to transfer among diverse hosts in a wide range of bacterial species (Davies, 1994; Clewell et al., 1995; Scott, 2002).
Just as infection rates are essential for understanding disease dynamics, conjugation rates play a central role in the population dynamics of bacterial plasmids. Their estimation is confounded by the fact that plasmid transmission occurs vertically as well as horizontally, and the fact that conjugation requires contact between plasmid-bearing and plasmid-free cells. This latter feature becomes especially relevant in surface-associated bacterial communities because spatial structure imposes limits on the amount of cell–cell contact. Conjugation rates on surfaces are inherently different from those in traditional liquid-based measurements due to a lack of spatial homogeneity, restrictions on cell movement, and the fact that some plasmids transfer more efficiently on solid surfaces than in liquid cultures (Lawley et al., 2004). Therefore, methods for estimation of plasmid transfer rates on surfaces are needed. Those most commonly used are typically more appropriate for liquid cultures and their effectiveness in spatial populations has not been systematically evaluated.
The ‘endpoint method’ has been widely used to estimate plasmid conjugation rates since it was first proposed (Simonsen et al., 1990). It avoids some of the shortcomings of other measures of plasmid ‘transfer efficiency’ by providing a fairly robust estimate of conjugation rate that is insensitive to factors such as initial cell density, initial donor-to-recipient ratio, and sampling time—at least in well-mixed environments such as liquid cultures. This estimate is easy to obtain, requiring the total cell density at time 0 and densities of recipients, donors, and transconjugants at a single sampling time—the endpoint. Here, ‘recipients’ (R) are plasmid-free cells; ‘donors’ (D) are the original plasmid-bearing cells, and can either have the same genetic background as the recipient or be a completely different strain, species, or genus; ‘transconjugants’ (T) are plasmid-bearing cells that arise either from a recipient cell that receives a copy of the plasmid through conjugation or from cell division of a transconjugant.
The endpoint estimate, γep, of the conjugation rate is given by
| (1) |
where ψmax is the estimated maximum per capita growth rate based on total cell densities during exponential phase; R = R(t1), D = D(t1), T = T(t1) are the densities of recipients, donors, and transconjugants, respectively, at a given sampling time t1; N = N(t1) is the total cell density (N=R+D+T) at the sampling time and N0 is the total cell density at time 0 (Simonsen et al., 1990).
To understand the endpoint estimate and its potential limitations, it is helpful to consider the underlying model assumptions upon which its derivation was based. Start with a simple mass-action differential equation for the time-dependent densities of recipients, transconjugants, and donors, and the nutrient concentration (C) in a batch culture
| (2) |
where ψ (h−1) and γ (ml cell−1 h−1) denote growth and conjugation rates (with subscripts corresponding to particular strains), and e (μg) is the resource conversion rate specifying the amount of resource needed to produce a new cell. The following assumptions were in force for the derivation of the endpoint estimate by Simonsen et al. (1990): (a) Identical growth rates for donors, recipients, and transconjugants, with nutrient dependence of the Monod form ψ(C) = ψmaxC/(C+K). (b) Identical conjugation rates for transconjugants and donors, including the same Monod form γ(C) = γmaxC/(C +K) as reproduction. It is the maximum conjugation rate, γmax, that the endpoint method estimates. The assumptions regarding nutrient dependence, including identical half-saturation constants for growth and conjugation, were essential in the derivation.
These stringent simplifying assumptions allowed the derivation of an easily calculated estimate of conjugation rate. Moreover, Simonsen et al. (1990) demonstrated via simulations of more complicated differential equations, that their estimate was actually quite robust to changes in some of the simplifying assumptions. For example, they showed that differences in growth rates for R, D, and T had little effect except when plasmid-bearing cells have a growth advantage.
Although the endpoint estimate was derived with a well-mixed liquid culture in mind, it has also been applied to surface-associated bacterial populations (MacDonald et al., 1992; Normander et al., 1998; Licht et al., 1999; Lilley and Bailey, 2002). An application of the endpoint method to spatially structured populations amounts to providing an estimate of an ‘effective conjugation rate’ for an ‘ideal liquid culture’ that would produce the observed global densities of donors, recipients, and transconjugants. There are several potential problems with such an approach.
First of all, one must keep in mind that plasmid transfer is fundamentally different on surfaces than in liquids due to the means by which the requisite contact between donors and recipients is attained. This is reflected in differences of many orders of magnitude between the ‘intrinsic’ conjugation rates for neighboring cells on a surface and the averaged, or ‘bulk,’ conjugation rates usually described for well-mixed populations (Achtman, 1975; Freter et al., 1983; Simonsen et al., 1990; Normander et al., 1998; Licht et al., 1999; Lilley and Bailey, 2002; Krone et al., 2007; Zhong et al., 2010). In contrast to liquid cultures in which bacterial populations are well mixed and plasmid transmission occurs homogeneously throughout the population, surface-attached bacterial populations grow as micro-colonies and plasmid transfer occurs at spatial scales that are much smaller than that of the whole population (Normander et al., 1998; Licht et al., 1999; Häagensen et al., 2002).
Furthermore, even if one can make some sense of a bulk conjugation rate for a spatial population, such a quantity is likely to be time dependent and sensitive to initial cell distributions. This is due to the natural clustering that occurs in spatially distributed, sessile populations. Simonsen (1990), for example, showed that the extent of plasmid transfer on surfaces was highly dependent on the initial cell density, while in liquids this dependence was very slight. Thus, the general appropriateness of applying the endpoint method to surface bacterial populations needs to be investigated.
Other measures commonly used to report plasmid transfer efficiency typically involve ratios of various combinations of T, D, and R. These include T/R, T/D, T/RD, and T/N (Curtiss et al., 1969; Bale et al., 1987; Pinedo and Smets, 2005; Sorensen et al., 2005). For example, if one begins with a surplus of recipients, say D/R=0.1 or 0.01, then the measurement T/D after 20 min will indicate the fraction of donors that were able to undergo a single round of conjugation before growth dynamics started influencing the densities. Invasion-when-rare experiments might begin with D/R=10−6 and measure T/N after 24 h to give an indication of the extent to which the plasmid was able to spread in the population. Here, of course, one cannot decouple the roles of conjugation and growth. As we discuss below, and was pointed out in Simonsen et al. (1990), this leads to some of these measures of plasmid transfer being highly sensitive to sampling time. Thus, while these measures give some indication of the efficiency of transfer, they do not directly report the rate or frequencies of the conjugative transfer events and are significantly confounded by growth of T, R, and D during the course of an experiment (often several hours, or even days for studies in microcosms or the field) (Simonsen et al., 1990; Hill and Top, 1998; van Elsas and Bailey, 2002).
Our main goal was to determine how well the endpoint estimate and other measures of plasmid transfer efficiency perform when applied to surface-attached bacterial communities. We used an individual-based stochastic spatial model to simulate surface-associated plasmid population dynamics across a variety of conditions, but with a fixed (and known) conjugation rate. This model, which captures the essential features of plasmid population dynamics on surfaces (cf. Krone et al., 2007; Fox et al., 2008), was used to generate simulated time-dependent densities of donors, recipients, and transconjugants. These densities were used to derive the corresponding bulk conjugation rates (γ, for the endpoint method) and other measures of plasmid transfer efficiency (T/R, T/D, T/RD, and T/N). The sensitivity of these estimates to conditions such as initial cell density, initial donor-to-recipient ratio, and sampling time was then evaluated. Note that, since the actual intrinsic conjugation rate has the same value in every simulation, any variations in the estimates of plasmid transfer rate are reflective of problems in applying these estimates to spatially distributed populations.
2. Materials and methods
2.1. Mathematical model
The model used here is an individual-based lattice model known as an interacting particle system (IPS). It is similar to the one developed in Krone et al. (2007), and is built on a two-dimensional square lattice. Each site in the lattice can be either vacant (V) or occupied by a single bacterial cell with type donor (D), recipient (R), or transconjugant (T). An occupied site is assumed to reproduce at a rate ψ (with subscripts used later to distinguish growth rates of different cell types), and deposit its (single) offspring onto a randomly chosen neighboring site that is vacant. If there are no neighboring vacant sites, the reproduction event is suppressed providing a kind of local carrying capacity that maintains the 2-dimensional structure of the model. A plasmid-bearing cell (D or T) can transfer a copy of its plasmid at a rate γ0 to a nearby recipient that is randomly chosen. We assume identical conjugation rates for donors and transconjugants and only consider permanently derepressed plasmids. Furthermore, to keep the model simple, we ignore cell death and segregative plasmid loss.
When a cell reproduces or conjugates, it initiates a change to one of the neighboring sites, causing a local change in the current lattice configuration. A ‘local neighborhood’ specifies which sites are neighbors for the purposes of determining the locations of daughter cells after cell division or the locations of potential recipient cells for conjugation by a plasmid-bearing cell. In our simulations, we took the local neighborhood of a given site to be the 8 adjacent sites surrounding that site.
There is empirical evidence (Lilley et al., 1996; Kroer et al., 1998; Normander et al., 1998; Hausner and Weurtz, 1999) that – at least for certain plasmid–bacteria combinations – the dependence of conjugation rate on nutrient concentration is not of the first-order Monod form (see Section 1) used in Simonsen et al. (1990), but rather exhibiting more of a threshold dependence with little or no conjugation for low levels of nutrient concentration and a nearly constant maximum conjugation rate for intermediate and high levels. (However, see Turner, 2004 for an unusual example of a plasmid whose conjugation rate appears to decrease at high glucose concentrations.) Thus, a more appropriate dependence is likely to be given by a ‘second-order Monod’ relation: γ(C) = γmaxC2/(C2+K2). In fact, Krone et al. (2007) found this threshold-like character of conjugation rate essential in fitting an IPS model to the aforementioned dependence of transconjugant density on initial cell density (Simonsen, 1990). Such threshold-type dependence is common in theoretical biology, arising for example as a Holling type III functional response in ecology and in the Emax model (with Hill exponent 2) in pharmacodynamics.
Rates of conjugation and reproduction in our model were assumed to be functions of the local ‘nutrient availability’ C, here equated with the fraction of vacant sites in the 7 × 7 ‘nutrient neighborhood’ centered at the ‘focal’ site x (see below)
| (3) |
and
| (4) |
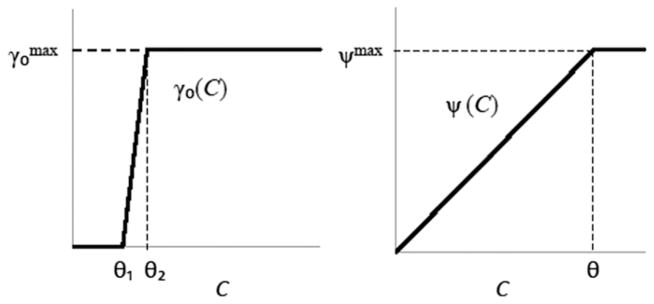
These piecewise linear ‘sigmoidal’ functions (Krone et al., 2007) are sketched in Fig. 1. They provide a simple way of generating approximations to the first and second order Monod functions described above and in Section 1, and are more appropriate in a discrete setting with only a limited number of nutrient ‘units’ available at a given site. (A Monod function reaches its maximum value asymptotically as the nutrient level tends to infinity.)
Fig. 1.
Conjugation rate γ0 (left) and growth rate ψ (right) as sigmoidal functions of resource concentration C.
This modeling approach, controlling growth via a (local) carrying capacity instead of explicitly modeling nutrient availability, is analogous to that of logistic growth dynamics in differential equation models (Brauer and Castillo-Chavez, 2001) where one assumes that per capita growth rates are proportional to the fraction of carrying capacity remaining to be filled. Indeed, the diffusion and consumption of nutrients and other growth-limiting quantities in spatially distributed populations can be quite complex and can vary significantly depending on species and experimental conditions. Not wanting to restrict our model to a specific case or to impose detailed assumptions that could influence behavior, we have chosen this more generic approach with fewer parameters. Our goal in the current study was to use as simple a model as possible in order to not bias the results by inclusion of too many specifics (which cannot all be measured in the lab) or unduly restrict applicability.
Note that the ‘intrinsic conjugation rate’ γ0(C) in Eq. (3) gives the rate of plasmid transfer from a specific plasmid-bearing cell to a neighboring recipient cell. This individual-based intrinsic conjugation rate differs fundamentally from the bulk conjugation rates of mass-action models (see Section 1 and Zhong et al., 2010).
2.2. Simulations
We used our spatially explicit IPS model to simulate the dynamics of bacterial growth and plasmid transfer on surfaces. Our simulations were carried out on a 1000 × 1000 lattice with wraparound boundaries to eliminate edge effects. With cell dimensions on the order of 1 μm, such a lattice can be thought of as corresponding to an area of approximately 1 mm2; thus a fully occupied lattice corresponds to 106 cells/mm2 or 108 cells/cm2. Simulations involve a series of (computer) time steps at each of which a ‘focal’ site is chosen at random in the lattice (with replacement); according to the rates of various events, it is determined whether a change will be initiated from that site and, if so, what that change will be.
The simulations began with donors and recipients being seeded on the lattice, with various levels of cell density, donor-to-recipient ratio, and spatial clustering. We set as a standard initial condition 106 cells/cm2 for initial cell density, 1:1 for donor-to-recipient ratio, and random cell placement. Other initial settings were deviations from this standard one: (i) donor-to-recipient ratio ranging from 1000:1 to 1:1000, (ii) initial cell density from 103 to 107 cells/cm2, and (iii) cluster size (described below) from 5 × 5 to 500 × 500.
The simulations were run until the lattice had 95% of its sites occupied by cells, an indication that the population density was reaching the carrying capacity of the lattice. These final cell densities are referred to as ‘stationary phase’ densities below. In addition to computing the endpoint estimate, γep, and the various other plasmid transfer efficiency measures, T/R, T/D, T/RD, and T/N, at stationary phase, we also tracked the endpoint estimates over time to examine the effect of sampling time.
We synchronized the simulation time steps with actual time using 30 min as the doubling time of the recipient cells during exponential growth. In simulations, it took about 7 × 106 time steps for a recipient population (started with low density) to double. By matching the two time scales (30 min=7 × 106 time steps), we were able to plot the simulated data on a scale of hours. For example, with an initial cell density of 106 cells/cm2, the 95% full stationary phase corresponded to a time of 9 h.
Model parameters used in the simulations were as follows: thresholds θ=0.8, θ1=0.2, θ2=0.3 for growth and conjugation rates, respectively; maximum growth rates ; maximum intrinsic conjugation rate . The choice of is similar to the intrinsic conjugation rates that provided the best fit to empirical data in our previous work (Krone et al., 2007; Fox et al., 2008; Zhong et al., 2010) using similar IPS models. Since conjugative DNA transfer happens on the order of minutes once contact has been made, for cell growth and conjugation to occur on similar time scales—matching what is seen in the lab, these intrinsic rates cannot be too different in the models.
2.3. Scaling for comparison of liquid- and IPS-based densities
Bulk conjugation rates, including estimates via the end-point method, are presented in units of ml cell−1 h−1. When comparing estimates of bulk conjugation rate for surface cultures or spatial simulations to these liquid-based quantities, the densities must be scaled so that one is comparing quantities in similar units. To come up with a scaling that compares lattice-based densities with liquid densities, we equate the densities at carrying capacity in the two settings. Suppose we use an L × L lattice and have at most 1 cell per site. This gives a carrying capacity of L2 cells/lattice (full grid). Suppose the corresponding carrying capacity in liquid is N̄ cells/ml. Equating these densities, we have L2 cells=lattice = N̄ cells=ml, or
| (5) |
In our simulations, we used a 1000 × 1000 lattice. So, based on an assumed liquid carrying capacity of N̄ = 109 cells=ml, lattice-based cell densities were multiplied by 103 to obtain an equivalent liquid density. This scaling up by a factor of 1000 all the densities appearing in the endpoint estimate cancels everywhere except in the denominator, N−N0. This meant that our IPS-generated end-point estimates of conjugation rate had to be divided by 1000. This, indeed, produced bulk conjugation rates that are of similar orders to those typically measured (Freter et al., 1983; Simonsen et al., 1990; Licht et al., 1999) in liquid cultures (see Section 3).
2.4. Statistical analysis
Statistical analysis, using log-transformed data, was based on two-sided t-tests and the Pearson correlation test, performed in Matlab (version 7.4).
2.5. Strains and media
E. coli K12 MG1655 (ATCC 47076) was used as the bacterial plasmid host. To be able to distinguish isogenic donor and recipient strains by selective plating, two antibiotic resistant mutants of this E. coli strain were used. As a donor we used a rifampicin (Rif) resistant mutant, designated K12Rif, while a nalidixic acid (Nal) resistant strain, K12Nal, was used as recipient (Fox et al., 2008). The plasmid was the 64.5 kb broad-host-range IncP-1β plasmid pB10, which confers resistance to four antibiotics (tetracycline, amoxicillin, streptomycin, and sulfonamides) and mercury chloride, transfers to various hosts at high rates (Dröge et al., 2000), and has been completely sequenced (Schlüter et al., 2003). A marked derivative of pB10 (pB10::rfp) has been constructed previously by insertion of the dsRed gene cassette (rfp) to visually distinguish between plasmid-bearing (red fluorescent) and plasmid-free (white) cells (De Gelder et al., 2005). Even though the red fluorescence feature was not used in this experiment, we used this marked plasmid since it is consistent with what we have used in other experiments carried out within the research project. Plasmid pB10::rfp was transferred to K12Rif in plate matings as described previously (Lejeune et al., 1983), with selection on LB amended with Rif (100 mg/l) and tetracycline (Tc) (10 mg/l).
2.6. Filter mating procedures
Plasmid transfer experiments were performed on 0.45 μm pore, 25 mm diameter filters (Whatman International Ltd., Maid-stone, England) on top of LB agar plates, essentially as previously described (Top et al., 1992). The donor strain was K12Rif (pB10::rfp), and the recipient strain K12Nal. Liquid cultures of donor and recipient strains were grown overnight at 30 °C in LB broth (with Tc 10 μg/ml for the donor), then centrifuged, rinsed and resuspended in an equal volume of LB broth. The OD600 of the donor and recipient suspensions were read to determine if the two strains had equal cell densities. The suspensions were also diluted and enumerated to determine the viable cell count at time 0. These suspensions were further diluted to obtain the desired initial densities (approximately 103–107 cfu/ml) of parental strains. Five microliter of first the recipient suspension was placed in the center of a sterile filter on top of an LB agar plate and allowed to dry; then the donor suspension was placed in the center of the same sterile filter on top of the LB agar plate. The droplet of cells dried in less than 1 min, thus limiting the amount of time cells spent in a liquid environment. Triplicate filters were prepared for each initial density. The plates were incubated for 30 h at 30 °C. To count the donor, recipient, and transconjugant populations, the filters were resuspended in 2 ml saline, and the respective population densities were enumerated by serial dilution in saline and plating appropriate dilutions on three different selective media using a spiral plater (Spiral Biotech, Inc.). The colony counts were determined using a QCount automatic colony counter (Spiral Biotech, Inc.). Colony forming units of donors were enumerated by plating on LB containing Rif (50 mg/l) and Tc (10 mg/l), recipients on LB with Nal (30 mg/l), and transconjugants on LB with Nal (30 mg/l) and Tc (10 mg/l). Appropriate controls of donor and recipient cultures placed separately on filters were always included, and no significant numbers of resistant colonies were observed on the transconjugant medium when plating these undiluted control suspensions. To determine the population densities per cm2, the diameter of the circular droplet after it had dried was measured and used to calculate the surface area covered by cells.
3. Results
When estimating the rate at which some event takes place in a population, it is important to understand how this estimate – or the rate itself – depends on various conditions. The endpoint estimate of plasmid transfer rate is known to be robust to changes in some experimental conditions for well-mixed liquid cultures. Whether or not this robustness holds when the estimate is applied to spatial populations has not been previously tested, even though the endpoint estimate has been applied to spatial data. Indeed, it is not obvious what such a ‘liquid-appropriate’ estimate would even mean in a spatial context. To address this, we measured in a spatial model how sensitive the endpoint estimate was to initial conditions in the parental populations, i.e., ratio and densities of donors and recipients, as well as their spatial configuration. The spatial simulations were run with prescribed parameters – including a fixed intrinsic conjugation rate – to produce time-dependent densities of donors, recipients, and transconjugants under the different scenarios described above. The simulated cell densities, under each ‘culture’ condition, were then used to compute the endpoint estimate of the (fictitious) ‘bulk’ conjugation rate γ. Other transfer efficiency measures were similarly computed. In this way, we were able to determine how various ‘experimental’ conditions in a spatially structured population might affect these estimates of plasmid transfer efficiency.
3.1. Effect of donor-to-recipient ratio
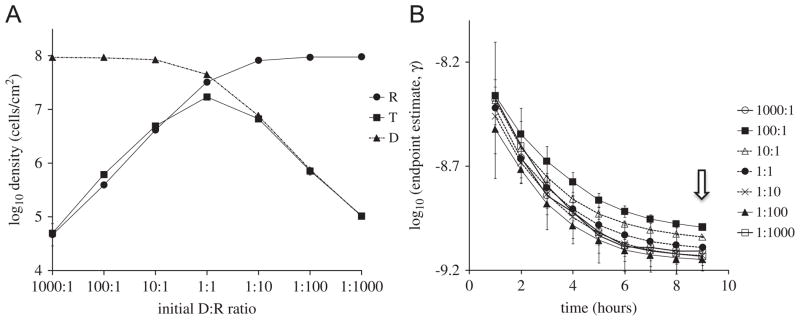
To assess the effect of initial donor-to-recipient ratios (D:R) on the endpoint estimate, γep, cells were randomly placed on the lattice at various D:R ratios, with the initial total density being fixed at 106 cells/cm2 (i.e., 104 cells/lattice). Here and in the treatments below, densities of R, D, and T were simulated at various time points and used to calculate endpoint estimates. The stationary phase densities of R, D, and T are shown in Fig. 2A. Although roughly half of the pairwise comparisons of the endpoint estimates for the data at 9 h resulted in significant differences (p<0.05), this appears to be mainly due to the small variances in the simulations at stationary phase. In fact, the endpoint estimates for the 9 h data were all within a factor of less than 1.5 as the initial densities were varied (see Fig. 2B or Table 1). The dependence of the endpoint estimates on sampling time is discussed below.
Fig. 2.
Simulated effect of initial donor-to-recipient ratio. (A) T, D, R densities sampled at stationary phase for different initial D:R ratios. (B) Endpoint estimates of conjugation rate as a function of sampling time for different initial D:R ratios, with the initial total cell density being set at 106 cells/cm2. The small scale on the y-axis is used to help distinguish between the curves. The arrow indicates the sampling time used in (A).
Table 1.
Comparison of plasmid transfer efficiency measures at stationary phase.
| Density (cells/cm2) | γep | T/R | T/D | T/(RD) | T/N |
|---|---|---|---|---|---|
| (a) Initial density | |||||
| 1 × 107 | 2.6 × 10−9 | 4.2 × 100 | 8.3 × 10−1 | 8.9 × 10−9 | 4.1 × 10−1 |
| 1 × 106 | 8.1 × 10−10 | 5.4 × 10−1 | 3.9 × 10−1 | 1.2 × 10−9 | 1.8 × 10−1 |
| 1 × 105 | 2.5 × 10−10 | 1.2 × 10−1 | 1.3 × 10−1 | 2.8 × 10−10 | 5.9 × 10−2 |
| 1 × 104 | 8.7 × 10–11 | 3.4 × 10−2 | 5.0 × 10−2 | 9.0 × 10–11 | 2.0 × 10−2 |
| 1 × 103 | 3.2 × 10–11 | 1.3 × 10−2 | 1.8 × 10−2 | 3.2 × 10–11 | 7.4 × 10−3 |
| Ratio | γep | T/R | T/D | T/(RD) | T/N |
|---|---|---|---|---|---|
| (b) Donor: recipient ratio | |||||
| 1000:1 | 8.0 × 10−10 | 9.9 × 10−1 | 5.3 × 10−4 | 1.1 × 10−9 | 5.3 × 10−4 |
| 100:1 | 1.0 × 10−9 | 1.5 × 100 | 6.5 × 10−3 | 1.7 × 10−9 | 6.5 × 10−3 |
| 10:1 | 9.1 × 10−10 | 1.2 × 100 | 5.8 × 10−2 | 1.4 × 10−9 | 5.2 × 10−2 |
| 1:1 | 8.1 × 10−10 | 5.3 × 10−1 | 3.8 × 10−1 | 1.2 × 10−9 | 1.8 × 10−1 |
| 1:10 | 7.4 × 10−10 | 8.1 × 10−2 | 8.5 × 10−1 | 1.1 × 10−9 | 6.9 × 10−2 |
| 1:100 | 7.1 × 10−10 | 7.6 × 10−3 | 9.4 × 10−1 | 1.0 × 10−9 | 7.5 × 10−3 |
| 1:1000 | 7.4 × 10−10 | 1.1 × 10−3 | 1.0 × 100 | 1.1 × 10−9 | 1.1 × 10−3 |
| Sub-square size (sites) | γep | T/R | T/D | T/(RD) | T/N |
|---|---|---|---|---|---|
| (c) Cluster size | |||||
| 500 × 500 | 2.6 × 10−11 | 1.2 × 10−2 | 1.2 × 10−2 | 2.6 × 10−11 | 6.1 × 10−3 |
| 125 × 125 | 1.1 × 10−10 | 5.2 × 10−2 | 5.1 × 10−2 | 1.1 × 10−10 | 2.5 × 10−2 |
| 50 × 50 | 2.6 × 10−10 | 1.3 × 10−1 | 1.2 × 10−1 | 2.9 × 10−10 | 6.0 × 10−2 |
| 20 × 20 | 5.4 × 10−10 | 3.2 × 10−1 | 2.6 × 10−1 | 7.0 × 10−10 | 1.3 × 10−1 |
| 5 × 5 | 8.1 × 10−10 | 5.5 × 10−1 | 3.9 × 10−1 | 1.2 × 10−9 | 1.8 × 10−1 |
This lack of sensitivity of the endpoint estimate to initial D:R ratio in the spatial model is similar to what has been observed in liquid cultures and corresponding models of well-mixed populations (Simonsen et al., 1990). For the spatial simulations, this can be explained from the final densities used to calculate the endpoint estimates (Fig. 2A). Namely, the quantities ψmax and N−N0 in Eq. (1) were nearly the same across all simulations. Thus, only the term TN/RD has the potential to significantly affect estimates of γ. A closer examination of Fig. 2A shows that TN/RD is approximately one for all the different initial D:R ratios. When the donor was initially dominant (D:R ratios of 1000:1, 100:1 and 10:1), D was close to N and T was close to R, thus TN/RD≈1; when recipients dominated initially, again TN/RD≈1; when D:R=1:1, T, D, R, N are of similar magnitude, thus TN/RD≈1.
3.2. Effect of initial density
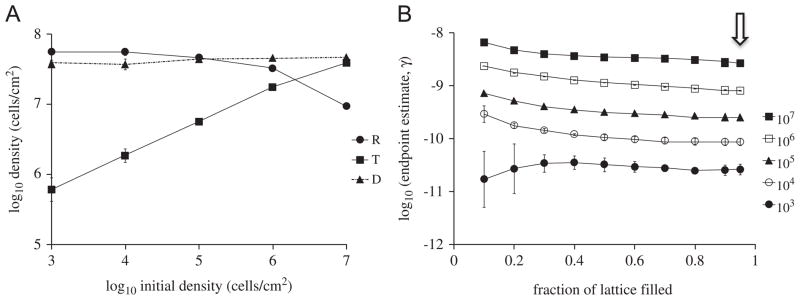
To determine the effect of initial cell densities on the endpoint estimate, donors and recipients (1:1 ratio) were randomly placed on the lattice with different initial densities. Densities of R, D, and T were simulated (stationary phase densities shown in Fig. 3A). The resulting values of γep (Fig. 3B) increased by two orders of magnitude as the initial density increased and depended very little on the sampling threshold, measured as the fraction of the lattice that is filled at the time of sampling. Thus, the initial cell density significantly affects (p=0.0002) the endpoint estimate of conjugation rate in spatial simulations. This is very different from the situation with well-mixed populations and it illustrates one of the pitfalls of applying the endpoint method to spatial populations. The above can be explained by observing (Fig. 3A) that the stationary phase transconjugant density was strongly dependent on initial density, while the donor and recipient densities were insensitive to initial density. This dependence of transconjugant density on initial cell density is, in fact, one of the signatures of plasmid spread on surfaces and is consistent with empirical results of Simonsen (1990) with the IncF plasmid R1drd19, as well as our own data (Fig. 4) with an IncP-1 plasmid. Note that the data in Fig. 3B are more easily interpretable, and make more sense, when presented as a function of sampling threshold –rather than elapsed time – since these simulations were started with different initial densities and hence take different times to reach the same density.
Fig. 3.
(A) Simulated T, D, R densities sampled at stationary phase for different initial densities. (B) Endpoint estimates of conjugation rate for different initial densities, as a function of sampling criterion (sampling time). The arrow indicates the sampling time used in (A).
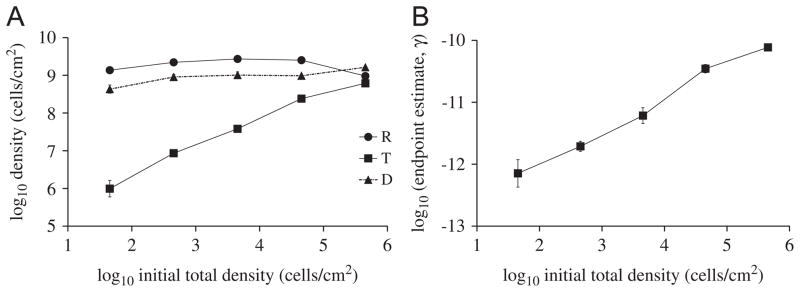
Fig. 4.
Stationary phase densities for the experiment with plasmid pB10::rfp in E. coli cells on filters on top of LB agar. (A) Effect of initial donor and recipient cell densities (in 1:1 ratio) on stationary densities of T, D, R. Population densities were measured after 30 h of incubation. ■: Recipient; ●: donor; ▲: transconjugants. Data represent the means of triplicate mating experiments, with error bars showing standard deviations. (B) Endpoint estimates of conjugation rate as a function of initial cell density.
This pronounced effect of spatial structure on the dynamics of conjugative plasmid transfer was demonstrated experimentally for the F-like plasmids R1 and its derepressed derivative (Simonsen, 1990). We evaluated whether this effect is also present for a very efficiently transferring broad-host-range plasmid of the IncP-1 group, pB10::rfp. IncP-1 plasmids behave very differently than F-like plasmids, presumably because their rigid pili allow more efficient transfer on surfaces than in liquid, while F plasmids with their flexible pili transfer just as efficiently in liquids as on surfaces (Bradley et al., 1980). Fig. 4A shows the experimentally derived stationary phase densities, including the characteristic rise in transconjugant density with increasing initial cell density. As shown in Fig. 4B, this leads to values of γep that increased as the initial cell density increased, very much similar to the predicted outcomes shown in Fig. 3. The simulated results are thus validated by two sets of experimental data obtained with two very different plasmids—a narrow-host-range IncF-II plasmid and a broad-host-range IncP-1 plasmid.
3.3. Effect of spatial clumping
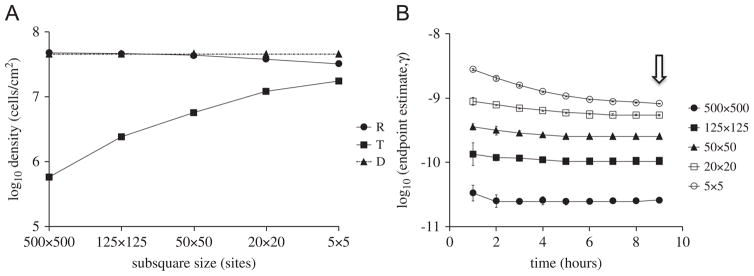
The effect of initial cell density on the endpoint estimate of bulk conjugation rate is due to the fact that conjugation only occurs at the interface between plasmid-bearing and plasmid-free colonies. When cell densities are initially low, conjugation is delayed until reproduction causes microcolonies to come into contact, and the extent of the resulting interfaces between donor and recipient colonies governs the amount of conjugation that can take place (Simonsen, 1990; Krone et al., 2007; Fox et al., 2008). We sought to further explore this effect by artificially controlling cluster size in a measurable way. Thus, we initialized our simulations with donors and recipients (1:1 ratio, total density 106 cells/cm2) placed on the lattice in a ‘checkerboard’ pattern, with alternating sub-squares containing exclusively donor or recipient cells. The sub-square size (or cluster size) was varied to artificially generate different degrees of spatial clumping: the larger the size of the clusters, the more the clumping and the smaller the region of donor-recipient interface. Stationary phase cell densities are shown Fig. 5A. The γep estimates were calculated at various time points after initial placement of D and R on the lattice (Fig. 5B). Transconjugant yields and γep estimates varied by approximately 1.5 to 2 orders of magnitude over the range of cluster sizes. As with the above results on the effects of initial cell density, the effects of initial spatial clumping on estimated γep were significant (p<0.0001).
Fig. 5.
(A) Simulated T, D, R densities sampled at stationary phase for different initial cluster sizes. (B) Endpoint estimates of conjugation rate as a function of sampling time for different initial cluster sizes. Donors and recipients were initiated in 1:1 ratio with a total density of 106 cells/cm2. The arrow indicates the sampling time used in (A).
3.4. Effect of sampling time
The endpoint estimate was tracked over time for all the simulations (Figs. 2, 3 and 5). As explained above, Fig. 3 had elapsed time measured not in hours, but in the progression of the lattice space being filled by cells. While the data indicated some correlation of γep with sampling time (p <0.05 for each case), the scale of the change in γep over time was relatively small. That is, the difference between the γep calculated during exponential phase and at stationary phase is within a factor of 5 (or 0.62 in log scale). Most of that variation can be accounted for at the earliest sampling times when cell densities were lowest. Thus, the values of γep for the spatial simulations were affected more by sampling time than what has been observed in liquid cultures (Simonsen et al., 1990), but this dependence was not nearly as strong as the dependence on initial density and clumping.
3.5. Other measures of transfer efficiency
Other transfer efficiency measures T/D, T/R, T/RD, and T/N were also evaluated (see Table 1) using the same stationary phase cell densities upon which the endpoint estimates were based. All of these alternative measures except T/RD showed variation across the spectrum of initial conditions (rows), at least as large as those of the endpoint estimate. The values for these measures differed from each other by orders of magnitude within each treatment and tended to be strongly dependent on sampling time. These are consistent with similar issues in well-mixed populations (Simonsen et al., 1990). The measure T/RD showed behavior similar to the endpoint estimate. This is not unexpected since (N−N0)−1 ln(1+(TN=DR)) ≈ T=DR when (TN)/(DR) is small and N−N0≈N.
4. Discussion
In surface-associated populations, spatial structure leads to heterogeneities in nutrient access, interactions between different cell types, etc. These have the potential to affect the performance of estimates that were designed with well-mixed populations in mind. Concerns about the limitations of measures of plasmid transfer efficiency and the need for predictive mathematical studies of plasmid transfer in surface-associated microbial populations were emphasized in Sorensen et al. (2005). Our simulation study seeks to address some of these issues.
All of the measures of plasmid transfer efficiency we investigated with our spatial simulations were sensitive to changes in initial cell density and clustering. This alone makes their application to surface-associated populations problematic unless care is taken to control for the natural clustering that is a hallmark of spatial growth. The endpoint method was the least sensitive to other culture conditions, and this is consistent with its performance in well-mixed populations. It also has the advantage of being interpretable as a bulk conjugation rate. The quantity T/RD approximates the endpoint estimate if the conditions mentioned earlier are satisfied, and so it has similar properties. The other transfer efficiency measures (T/D, T/R, and T/N), while easy to calculate, are very difficult to interpret in a consistent manner and by no means represent a ‘rate’ of transfer, but merely a proportion of transconjugants. These summary statistics can differ by orders of magnitude and are not useable as conjugation rates in mathematical models. They are sensitive to sampling time and initial conditions. At the very least, one should always specify clearly which measures of plasmid transfer efficiency are being used; they are not equivalent.
The endpoint estimate, applied to spatial cultures, can be thought of as providing an ‘effective conjugation rate’ that treats the population as if it were an idealized, perfectly mixed community with densities changing according to a mass-action differential equation. This is similar to the use of ‘effective population sizes’ in population genetics, where one tries to compare a population having some kind of structure or fluctuating population size with an ideal Wright–Fisher model of constant size. In both settings, the extent to which the idealized equations reflect empirical data (or predictions from more realistic models) depends on how well the effects of any heterogeneities average out or homogenize over the length and time scales being used.
It was suggested in Simonsen et al. (1990) that the endpoint estimate might also be applied to surface cultures provided the donor and recipient cells initially form a well-mixed confluent layer and are in 1:1 ratio. This suggestion was based on their observation that such a surface mating (using E. coli K12 hosts and the permanently derepressed plasmid R1drd19) had temporal dynamics that could be fit to a mass-action differential equation provided the incubation time was short (between 2 and 6 h). It is in restricted cases like this, for which one can think of an equivalent mass-action model, that the endpoint estimate has some merit in a spatial setting—provided it is interpreted appropriately. (Our results indicate that the 1:1 donor-to-recipient ratio is not necessary in the above suggestion from Simonsen et al. (1990), but having a well-mixed confluent layer is crucial.) In general, while some studies (e.g., Knudsen et al., 1988; Clewlow et al., 1990; Top et al., 1992) have used mass-action models to successfully simulate aspects of plasmid transfer in spatially structured communities such as soils and filter matings on agar plates, one must use care to ensure that the conditions that are implicit in these settings are not ignored when interpreting results.
Using a spatially explicit mathematical model to simulate plasmid population dynamics on surfaces, we saw that the endpoint estimate of the (effective bulk) conjugation rate was fairly stable to changes in sampling time and initial donor-to-recipient ratio, but changed by up to two orders of magnitude when initial cell densities or the amount of spatial clustering were varied. Cell density correlates positively with bulk conjugation rate estimates, while clustering correlates negatively. Both effects indicate that the degree of donor–recipient mixing is the variable that most affects bulk conjugation in surface-grown cultures. This strong effect of mixing on the level of plasmid transfer in surface-associated populations was also demonstrated experimentally in Fox et al. (2008).
We conclude with some recommendations for quantifying plasmid transfer efficiency. The best way to understand plasmid transfer in surface-associated populations is to estimate the intrinsic conjugation rate and use a spatially explicit mathematical model to determine the effects of various experimental conditions. The intrinsic conjugation rate can be estimated indirectly for a given model by matching the experimental spatial and temporal dynamics of donors, recipients, and transconjugants under a variety of initial configurations (cf. Krone et al., 2007). In principle, one can get a more direct estimate by spreading a confluent layer of donors and recipients – mixed with recipients in surplus – on a plate and measuring the time at which transconjugants start appearing. This, however, can be made difficult by issues such as physiological stresses that the cells experience when being transferred from a liquid culture to a plate environment (Cuny et al., 2007), spurious mating that occurs on selection plates used to count transconjugants, and other detection problems such as delays or inconsistent levels of fluorescent protein expression encountered when using fluorescently labeled plasmids.
If, instead, one uses the bulk estimates we have been discussing to attempt to quantify plasmid transfer in experimental cultures, the endpoint method provides the least problematic estimate for both liquid and spatial cultures. Furthermore, we suggest the following:
When applying the endpoint method to well-mixed (liquid) populations, take into account the restrictions outlined in Section 1 (and in Simonsen et al., 1990). For example, Simonsen et al. (1990) point out that their assumption of identical first-order Monod dependence on nutrient concentration for conjugation and growth rates means that one must take care when applying the endpoint method that both growth and conjugation are equally affected by nutrient availability during the incubation period. In particular, the incubation period should be short enough to avoid significant decreases in nutrient concentration. Remember also that the endpoint method is designed to provide an estimate of the maximum conjugation rate.
-
When applying the endpoint method to spatial populations,
Keep in mind that the endpoint estimate only provides an ‘effective conjugation rate’ by regarding the data as if arising from a well-mixed population. Applying it to spatially heterogeneous data will provide numbers, but the relevance and interpretation of those numbers requires caution. The endpoint estimate should not be confused with the actual ‘intrinsic’ conjugation rate that indicates the rate of plasmid DNA transfer between nearby (immobile) donor and recipient cells (cf. Zhong et al., 2010).
To minimize the effects of clustering in a spatial endpoint estimate, it is best to begin with donors and recipients well mixed on the surface with a high enough density so that the cells are in contact. Sampling (to estimate the densities required in the endpoint method) should be carried out before clustering and nutrient consumption lead to spatial heterogeneities and/or non-parallel changes in growth and conjugation rates. Thus applied, the endpoint estimate will provide a consistent upper bound on the effective bulk conjugation rate, keeping in mind that reduced densities and increased spatial clustering may decrease bulk conjugation in natural populations. Ultimately, the direction and magnitude of the effects of limited mixing will define the level of difficulty in (i) comparing structured populations to well-mixed ones and (ii) using bulk-type estimates to draw conclusions about structured populations.
When applying the transfer efficiency measures T/D, T/R, T/RD, and T/N to spatial populations, T/RD will be very close in behavior to the endpoint estimate, for reasons that are explained at the end of Results. In our simulations, all of these quantities had sensitivity to initial cell density and clustering of donors and recipients similar to that of the endpoint estimate and hence the afore-mentioned issues related to limited mixing apply. Unlike the endpoint estimate and T/RD, the measures T/D, T/R, and T/N are highly sensitive to donor-to-recipient ratios (cf. Table 1 where these measures differed by 3–4 orders of magnitude over the range of ratios) and to sampling time (data not shown, but see Simonsen et al. (1990) for similar effects in well-mixed populations). For these reasons, their use should always be accompanied by carefully measured indications of sampling times and donor-to-recipient ratios, and the results should be interpreted in light of these conditions.
Acknowledgments
This work benefitted from stimulating discussions with our colleagues Jarosław E. Król and Linda Rogers. This work was supported by NIH Grant R01 GM73821 and by NIH Grant P20 RR016454 from the INBRE Program of the National Center for Research Resources. We thank two anonymous reviewers for their comments on the manuscript.
References
- Achtman M. Mating aggregates in Escherichia coli conjugation. J Bacteriol. 1975;123:505–515. doi: 10.1128/jb.123.2.505-515.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale MJ, Fry JC, Day MJ. Plasmid transfer between strains of Pseudomonas aeruginosa on membrane filters attached to river stones. J Gen Microbiol. 1987;133:3099–3107. doi: 10.1099/00221287-133-11-3099. [DOI] [PubMed] [Google Scholar]
- Bradley DE, Taylor DE, Cohen DR. Specification of surface mating systems among conjugative drug resistance plasmids in Escherichia coli K-12. J Bacteriol. 1980;143:1466–1470. doi: 10.1128/jb.143.3.1466-1470.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer F, Castillo-Chavez C. Mathematical Models in Population Biology and Epidemiology. Springer; New York: 2001. [Google Scholar]
- Clewell DB, Flannagan SE, Jaworski DD. Unconstrained bacterial promiscuity: the Tn916–Tn1545 family of conjugative transposons. Trends Micro-biol. 1995;3:229–236. doi: 10.1016/s0966-842x(00)88930-1. [DOI] [PubMed] [Google Scholar]
- Clewlow LJ, Cresswell N, Wellington EMH. Mathematical model of plasmid transfer between strains of streptomycetes in soil microcosms. Appl Environ Microbiol. 1990;56:3139–3145. doi: 10.1128/aem.56.10.3139-3145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuny C, Lesbats M, Dukan S. Induction of a global stress response during the first step of Escherichia coli plate growth. Appl Environ Microbiol. 2007;73:885–889. doi: 10.1128/AEM.01874-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtiss R, III, Caro LG, Allison DP, Stallions DR. Early stages of conjugation in E. coli. J Bacteriol. 1969;100:1091–1104. doi: 10.1128/jb.100.2.1091-1104.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science. 1994;264:375–382. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- De Gelder L, Vandecasteele FP, Brown CJ, Forney LJ, Top EM. Plasmid donor affects host range of promiscuous IncP-1β plasmid pB10 in an activated-sludge microbial community. Appl Environ Microbiol. 2005;71:5309–5317. doi: 10.1128/AEM.71.9.5309-5317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dröge M, Pühler A, Selbitschka W. Phenotypic and molecular characterization of conjugative antibiotic resistance plasmids isolated from bacterial communities of activated sludge. Mol Gen Genet. 2000;263:471–482. doi: 10.1007/s004380051191. [DOI] [PubMed] [Google Scholar]
- Fox RE, Zhong X, Krone SM, Top EM. Spatial structure and nutrients promote invasion of IncP-1 plasmids in bacterial populations. ISME J. 2008;2:1024–1039. doi: 10.1038/ismej.2008.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freter R, Freter RR, Brickner H. Experimental and mathematical models of Escherichia coli plasmid transfer in vitro and in vivo. Infect Immun. 1983;39:60–84. doi: 10.1128/iai.39.1.60-84.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häagensen JAJ, Hansen SK, Johansen T, Molin S. In situ detection of horizontal transfer of mobile genetic elements. FEMS Microbiol Ecol. 2002;42:261–268. doi: 10.1111/j.1574-6941.2002.tb01016.x. [DOI] [PubMed] [Google Scholar]
- Hausner M, Weurtz S. High rates of conjugation in bacterial biofilms as determined by in situ analysis. Appl Environ Microbiol. 1999;65:3710–3713. doi: 10.1128/aem.65.8.3710-3713.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill KE, Top EM. Gene transfer in soil systems using microcosms. FEMS Microbiol Ecol. 1998;25:319–329. [Google Scholar]
- Knudsen GR, Walter MV, Porteous LA, Prince VJ, Armstrong JL, Seidler RJ. Predictive model of conjugative plasmid transfer in the rhizosphere and phyllosphere. Appl Environ Microbiol. 1988;54:343–347. doi: 10.1128/aem.54.2.343-347.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroer N, Barkay T, Sorensen S, Weber D. Effect of root exudates and bacterial metabolic activity on conjugal gene transfer in the rhizosphere of a marsh plant. FEMS Microbiol Ecol. 1998;25:375–384. [Google Scholar]
- Krone SM, Lu R, Fox R, Suzuki H, Top EM. Modelling the spatial dynamics of plasmid transfer and persistence. Microbiology. 2007;153:2803–2816. doi: 10.1099/mic.0.2006/004531-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Wilkins BM, Frost L. Bacterial conjugation in Gram-negative bacteria. In: Phillips G, Funnell BE, editors. Plasmid Biology. ASM Press; Washington, D.C: 2004. pp. 203–226. [Google Scholar]
- Lejeune P, Mergeay M, Vangijsegem F, Faelen M, Gerits J, Toussaint A. Chromosome transfer and R-prime plasmid formation mediated by plasmid pULB113 (RP4-Mini-Mu) in Alcaligenes eutrophus CH34 and Pseudomonas fluorescens 6.2. J Bacteriol. 1983;155:1015–1026. doi: 10.1128/jb.155.3.1015-1026.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht TR, Christensen BB, Krogfelt KA, Molin S. Plasmid transfer in the animal intestine and other dynamic bacterial populations: the role of community structure and environment. Microbiology. 1999;145:2615–2622. doi: 10.1099/00221287-145-9-2615. [DOI] [PubMed] [Google Scholar]
- Lilley AK, Bailey MJ, Day MJ, Fry JC. Diversity of mercury resistance plasmids obtained by exogenous isolation from the bacteria of sugar beet in three successive seasons. FEMS Microbiol Evol. 1996;20:211–228. [Google Scholar]
- Lilley AK, Bailey MJ. The transfer dynamics of Pseudomonas sp plasmid pQBR11 in biofilms. FEMS Microbiol Ecol. 2002;42:243–250. doi: 10.1111/j.1574-6941.2002.tb01014.x. [DOI] [PubMed] [Google Scholar]
- MacDonald JA, Smets BF, Rittmann BE. The effects of energy availability on the conjugative-transfer kinetics of plasmid RP4. Water Res. 1992;26:461–468. [Google Scholar]
- Normander B, Christensen BB, Molin S, Kroer N. Effect of bacterial distribution and activity on conjugal gene transfer on the phylloplane of the bush bean (Phaseolus vulgaris) Appl Environ Microbiol. 1998;64:1902–1909. doi: 10.1128/aem.64.5.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinedo CA, Smets BF. Conjugal TOL transfer from Pseudomonas putida to Pseudomonas aeruginosa: effects of restriction proficiency, toxicant exposure, cell density ratios, and conjugation detection method on observed transfer efficiencies. Appl Environ Microbiol. 2005;71:51–57. doi: 10.1128/AEM.71.1.51-57.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter A, Heuer H, Szczepanowski R, Forney LJ, Thomas CM, Pühler A, Top EM. The 64 508 bp IncP-1β antibiotic multiresistance plasmid pB10 isolated from a waste-water treatment plant provides evidence for recombination between members of different branches of the IncP-1β group. Microbiology. 2003;149:3139–3153. doi: 10.1099/mic.0.26570-0. [DOI] [PubMed] [Google Scholar]
- Scott KP. The role of conjugative transposons in spreading antibiotic resistance between bacteria that inhabit the gastrointestinal tract. Cell Mol Life Sci. 2002;59:2071–2082. doi: 10.1007/s000180200007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen L. Dynamics of plasmid transfer on surfaces. J Gen Microbiol. 1990;136:1001–1007. doi: 10.1099/00221287-136-6-1001. [DOI] [PubMed] [Google Scholar]
- Simonsen L, Gordon DM, Stewart FM, Levin BR. Estimating the rate of plasmid transfer: an endpoint method. J Gen Microbiol. 1990;136:2319–2325. doi: 10.1099/00221287-136-11-2319. [DOI] [PubMed] [Google Scholar]
- Sorensen SJ, Bailey M, Hansen LH, Kroer N, Wuertz S. Studying plasmid horizontal transfer in situ: a critical review. Nat Rev Micro. 2005;3:700–710. doi: 10.1038/nrmicro1232. [DOI] [PubMed] [Google Scholar]
- Top E, Vanrolleghem P, Mergeay M, Verstraete W. Determination of the mechanism of retrotransfer by mechanistic mathematical modeling. J Bacteriol. 1992;174:5953–5960. doi: 10.1128/jb.174.18.5953-5960.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner PE. Phenotypic plasticity in bacterial plasmids. Genetics. 2004;167:9–20. doi: 10.1534/genetics.167.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Elsas JD, Bailey MJ. The ecology of transfer of mobile genetic elements. FEMS Microbiol Ecol. 2002;42:187–197. doi: 10.1111/j.1574-6941.2002.tb01008.x. [DOI] [PubMed] [Google Scholar]
- Zhong X, Król J, Top EM, Krone SM. Accounting for mating pair formation in plasmid population dynamics. J Theor Biol. 2010;262:711–719. doi: 10.1016/j.jtbi.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]