Summary
Purpose
Mapping seizure susceptibility loci in mice provides a framework for identifying potentially novel candidate genes for human epilepsy. Using C57BL/6J × A/J chromosome substitution strains (CSS), we previously identified a locus on mouse chromosome 10 (Ch10) conferring susceptibility to pilocarpine, a muscarinic cholinergic agonist that models human temporal lobe epilepsy by inducing initial limbic seizures and status epilepticus (status), followed by hippocampal cell loss and delayed-onset chronic spontaneous limbic seizures. Herein we report further genetic mapping of pilocarpine quantitative trait loci (QTLs) on Ch10.
Methods
Seventy-nine Ch10 F2 mice were used to map QTLs for duration of partial status epilepticus and the highest stage reached in response to pilocarpine. Based on those results we created interval-specific congenic lines to confirm and extend the results, using sequential rounds of breeding selectively by genotype to isolate segments of A/J Ch10 genome on a B6 background.
Key Findings
Analysis of Ch10 F2 genotypes and seizure susceptibility phenotypes identified significant, overlapping QTLs for duration of partial status and severity of pilocarpine-induced seizures on distal Ch10. Interval-specific Ch10 congenics containing the susceptibility locus on distal Ch10 also demonstrated susceptibility to pilocarpine-induced seizures, confirming results from the F2 mapping population and strongly supporting the presence of a QTL between rs13480781 (117.6 Mb) and rs13480832 (127.7 Mb).
Significance
QTL mapping can identify loci that make a quantitative contribution to a trait, and eventually identify the causative DNA-sequence polymorphisms. We have mapped a locus on mouse Ch10 for pilocarpine-induced limbic seizures. Novel candidate genes identified in mice can be investigated in functional studies and tested for their role in human epilepsy.
Keywords: Epilepsy, Genetics, Mouse, Quantitative trait locus, Pilocarpine
The importance of genetic factors in epilepsy is well accepted, but the specific genes responsible have been identified for only a few rare Mendelian epilepsy syndromes in humans (Winawer & Shinnar, 2005; Baulac & Baulac, 2010). The difficulty in finding genes results in part from the fact that most epilepsy is “complex,” with multiple genetic, nongenetic, and possibly interacting causes, and temporal lobe epilepsy (TLE) exemplifies this problem (Mathern et al., 1996; Cendes et al., 1998; Semah et al., 1998; Engel, 2001; Fuerst et al., 2001; Kobayashi et al., 2001; Stephen et al., 2001; Ottman, 2005; Santos et al., 2002; Vadlamudi et al., 2003; Scharfman & Pedley, 2006). Epilepsy is also heterogeneous because of the variety of underlying biologic processes that may give rise to seizure susceptibility and act as potential therapeutic targets.
Mapping seizure susceptibility loci in mice provides a framework for identifying potentially novel candidate genes for complex human disorders (Frankel, 2009). Prolific breeding and short gestation expedite genetic linkage methods (Palmer & Phillips, 2002; Phillips et al., 2002), and the availability of genetic maps and sequence data for multiple inbred strains also facilitate genetic research in mice. Conservation of genes from mouse to human genomes allows genetic discoveries in the mouse to be translated for study in human populations.
Inbred mouse strains differ in susceptibility to develop seizures and seizure-induced hippocampal damage in response to many different convulsant stimuli, and these differences have been shown to be heritable ((Neumann & Collins, 1991, 1992; Ferraro et al., 1995, 1997; Schauwecker & Steward, 1997; Ferraro et al., 1998; Gershenfeld et al., 1999; Hain et al., 2000; Schauwecker, 2002; McKhann et al., 2003; Frankel, 2009). The A/J strain has been shown to be more susceptible than B6 to most (though not all) types of convulsant stimuli (Kosobud & Crabbe, 1990; Mathis et al., 1994; Gershenfeld et al., 1999; Golden et al., 2001; Ferraro et al., 2002), and has recently been described to have sleep-related spontaneous seizures (Strohl et al., 2007). Genetic variation among strains is presumed to be a result of functional polymorphisms in genes that affect the threshold for neuroexcitability in response to exogenous stimuli (Frankel et al., 2001). Identifying these genes can provide insight into epilepsy pathophysiology and lead to improved and novel therapeutic strategies.
Quantitative trait loci (QTL) are genomic loci that make a quantitative contribution to a trait, such as seizure susceptibility. QTL mapping can identify these loci, and eventually the causative DNA-sequence polymorphisms. This powerful strategy is designed specifically to investigate oligogenic models, in which several genes of small to moderate effect act to influence a trait. Chromosome substitution strains (CSS) are strains in which a single chromosome from one inbred strain (donor) has been transferred onto a second strain (host) by repeated backcrossing, and provide an invaluable resource for mapping mouse QTLs. Chromosome substitution strain analysis provides quick and efficient localization at the chromosomal level without the need for cross-breeding or genotyping (Nadeau et al., 2000). Using C57BL/6J (B6, host) × A/J (donor) CSS, we recently identified contributions to susceptibility to pilocarpine-induced seizures on mouse chromosomes 10 and 18 (Winawer et al., 2007).
Pilocarpine, a muscarinic cholinergic agonist, induces initial severe continuous limbic seizures (status epilepticus) in rodents, followed by cell loss similar to that observed in TLE, and delayed onset of chronic spontaneous limbic seizures (Turski et al., 1989; Turski, 2000; Leite et al., 2002; Borges et al., 2003). Prior to our report (Winawer et al., 2007), limbic seizures had not been studied in the A/J strain, and susceptibility to pilocarpine-induced seizures had not been studied with QTL mapping. Herein we report fine mapping of a pilocarpine seizure susceptibility locus on mouse chromosome 10 (Ch10), using F2 mouse populations and interval-specific congenic strains. (See Methods for breeding scheme details.)
Methods
Animals
A/J, B6, and CSS breeder pairs were purchased from the Jackson Laboratories (Bar Harbor, ME, U.S.A.), and colonies of each strain, F1s, F2s, and congenics were bred in our laboratory. Mice were housed in a temperature- and humidity-controlled environment, with a 12-h light/dark schedule and food and water available ad libitum. Only male animals were tested for seizure susceptibility, to avoid estrus cycle effects on seizure susceptibility, and animals were tested at between 10 and 11 weeks of age. All methods were approved by the Institutional Animal Care and Use Committee of Columbia University and met the guidelines of the National Institutes of Health (NIH).
Breeding scheme
To investigate the mode of inheritance of the Ch10 susceptibility locus, we bred the CSS10 males—chromosome substitution strain mice containing Ch10 of A/J on a B6 background—with B6 females. We also tested 10F1 animals—the first generation offspring from a cross of CSS10 females with CSS10 males—for seizure susceptibility. F1 animals appeared to have intermediate seizure susceptibility between that of B6 and CSS10 animals, suggesting an additive mode of inheritance. We, therefore, bred CSS10 males with B6 females to produce a Ch10 × B6 F1 generation, and intercrossed these animals to produce the CSS-10 × B6F2 mapping population (n = 79). The genders of the CSS10 and B6 animals in the first cross were specified in order to limit potential variability from imprinting.
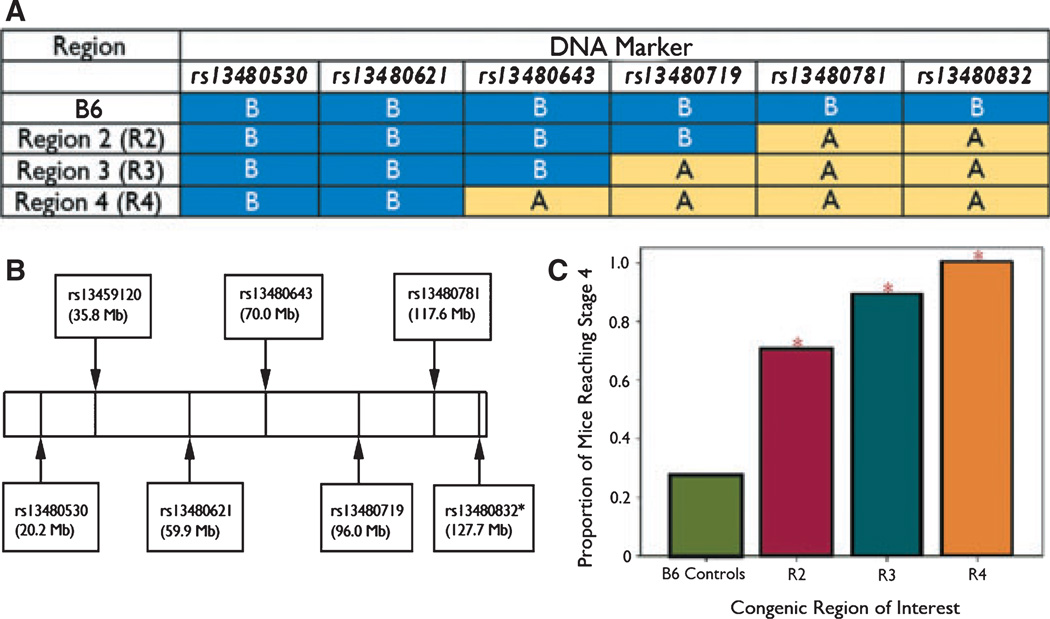
We created Ch10 interval-specific congenic strains by breeding successive rounds of littermates selectively by genotype in order to isolate subregions of A/J Ch10 on a B6 background. We began this process by using genotyped CSS-10 × B6F2 mice, selecting mice heterozygous for an A/J Ch10 segment of interest, and breeding them with B6 animals. Heterozygous offspring from these crosses were then brother–sister mated to produce a first round of congenics. These mice were genotyped, and subsequent generations were bred selectively by genotype to isolate the region of interest. Heterozygote mating was used to perpetuate congenic lines, and to create B6 homozygous littermate controls for comparison. Fig. 1A illustrates the panel of congenic strains used in the analyses presented here, and Fig. 1B the marker positions in Mb.
Figure 1.
(A) Marker genotypes across the Ch10 region of interest for C57BL/6J and interval-specific congenic strains. (B) Markers genotyped and relative positions on Ch10. *Used in congenic strains only. (C) Seizure susceptibility in Ch10 interval-specific congenic strains. *Statistically significant difference from B6 control.
Epilepsia © ILAE
Pilocarpine administration and seizure scoring
Animals were weighed immediately prior to seizure testing, and pilocarpine dose was calculated based on this weight. Animals were injected with intraperitoneal (i.p.) pilocarpine hydrochloride (Sigma-Aldrich, St. Louis, MO, U.S.A.). A dose of 250 mg/kg was used for the CSS-10 × B6F2 mapping population, and a dose of 300 mg/kg was used for all congenic studies. We increased the dose from 250–300 mg/kg to accentuate differences between congenic and control phenotypes and facilitate fine mapping. All seizure susceptibility testing was performed between the hours of 12:00 and 5:00 p.m. to limit any effects of diurnal variation on results (the light cycle begins at 7 a.m. and ends at 7 p.m. in our facility). To limit peripheral side-effects, 30 min before pilocarpine injection, all mice were given atropine methyl nitrate (5 mg/kg, i.p., TCI America, Portland, OR, U.S.A.), a competitive muscarinic acetylcholine receptor antagonist that does not cross the blood–brain barrier. CSS-10 × B6F2 animals were observed continuously for 3 h. In the course of seizure susceptibility testing, we found that over observation of >550 animals, 99% progressed to their highest stage by 2.5 h. We, therefore, subsequently shortened our observation period from 3 to 2.5 h for all congenic animals. The time of onset and offset for each seizure stage was recorded in minutes from time of pilocarpine injection. From these data we extracted the highest stage reached, the proportion of animals having generalized seizures (reaching stage 4 or higher), duration of stages, and latency to reach each stage (see Table 1 for seizure staging). We used a seizure staging system adapted from established rodent seizure scales (Racine, 1972), as described previously (Winawer et al., 2007), with several modifications.
Table 1.
Limbic seizure stages in pilocarpine-treated mice
| Stage 1 | Immobility/lying low |
| Stage 2 | Partial (limbic) seizures: twitching/tremor/shaking of tail/head/body/or limbs, not continuous, forelimb and/or tail extension, rigid posture, repetitive movements, head bobbing |
| Stage 3 | Partial status epilepticus: continuous tremor/clonic seizures of body and tail while retaining posture |
| Stage 3.1 | Partial status epilepticus lasting 10 min or more |
| Stage 4 | Generalized seizures: rearing/hyperexcitability/running/falling, tonic extension/clonic seizuresa with loss of posture |
| Stage 4.5 | Stage 4 seizures, recurrent, but not continuous for 3 or more minutes |
| Stage 4.75 | Stage 4 seizures, recurrent and continuous for 3 or more minutes |
| Stage 5 | Generalized status epilepticus (continuous stage 4 seizures) resulting in death |
Tonic seizures are characterized by whole body stiffening and extension and clonic seizures by repetitive rhythmic jerking. These two phenomena are typically seen together in whole body convulsive seizures.
Since our last publication, we have further refined seizure scoring, adding intermediate levels between stages 3 and 4 and between 4 and 5 (Table 1). The intermediate seizure stages we have added are reliably identified by our observation protocol, and were developed after observation of more than a thousand mice. More detailed phenotyping allows better discrimination among animals with different degrees of seizure susceptibility.
We mapped two variables in the CSS-10 × B6F2 population: time spent in stage 3, which corresponds to the duration of continuous partial seizures, or partial status epilepticus, and highest stage reached, which corresponds to the most severe seizure stage attained. Use of these two variables captures both latency to and severity of pilocarpine-induced seizures, two essential measures of susceptibility.
After mapping both phenotypes with confidence to the same region on distal Ch10, we used a dichotomous version of highest stage reached, whether an animal reached stage 4 or higher (had generalized seizures) for fine mapping in congenic strains. Stage 4 is reliably identified in mice, and occurred more frequently in our congenic animals because of the higher dose used for testing (300 mg/kg) compared with CSS-10 × B112 animals (250 mg/kg). We also performed analyses with the original phenotypes, time in stage 3 and highest stage reached, for comparison.
Genotyping
We used the Qiagen DNAeasy kit (Qiagen Sample and Assay Technologies, Germantown, MD, U.S.A.) to extract DNA from mouse tail and spleen. F2 mice were genotyped at six polymorphic single-nucleotide polymorphism (SNP) markers that were evenly spaced across chromosome 10 (rs13480530, rs13459120, rs13480621, rs13480643, rs13480719, and rs13480781.) For congenic line construction, in order to further saturate the region of interest, we used rs13480832 as well (Fig. 1). Genotyping was performed at the University of Chicago using the ABI TaqMan Assay (Applied Biosciences, Foster City, CA, U.S.A.) and a Step One PCR (polymerase chain reaction) machine (Applied Biosciences) in accordance with the manufacturer’s instructions.
Statistical methods
Analysis of the CSS-10 × B6F2 population
We used the R/QTL software (Version 1.14-2; http://www.rqtl.org/) (Broman et al., 2003), which runs in the R software environment (Version 2.9.2; http://www.r-project.org/) to map susceptibility loci in the F2 population. R/qtl is essentially a linkage program used for complex trait analysis, and calculates a logarithm of the odds (LOD) score, providing evidence for linkage of a trait of interest to a chromosomal region delineated by genetic markers. To map the variable time spent in stage 3 (partial status epilepticus), we used the “normal” model and the “EM algorithm” method with the age and weight of the mice as covariates. For highest stage reached, we used the nonparametric “np” model and the “EM algorithm” method (nonparametric models do not accommodate covariates). We used 1,000 permutations of these data using the “perm” argument in the scanone function in R/qtl to estimate significance (Churchill & Doerge, 1994). Confidence intervals were calculated using a Bayesian method as implemented in R/qtl. The effect of genotype at the most associated marker was estimated using “effect plot” function in R/qtl. We empirically estimated the distance between markers and additionally estimated genotype at 1 cM intervals between each marker using R/qtl.
Analysis of interval-specific congenic strains
In order to fine map the QTL contributing to seizure susceptibility on distal Ch10, analyses were carried out for which attaining stage 4 or higher was the outcome variable, and only animals homozygous at all four loci—rs13480643, rs13480719, rs13480781, and rs13480832—were included. Three sets of analyses were carried out. In the first set, interval-specific congenic strains defined by their genotype profile at the four loci were compared separately from the control animals using a chi-square statistic. In the second, a chi-square statistic was used to compare animals with the AJ genotype at an SNP to animals with the B6 genotype, separately for each of the loci. In the third set, a multiple logistic regression analysis of reaching stage 4 or higher was carried out to narrow the susceptibility region further, including genotypes at the top markers as predictors.
Results
QTL analysis
CSS-10 × B6F2 animals
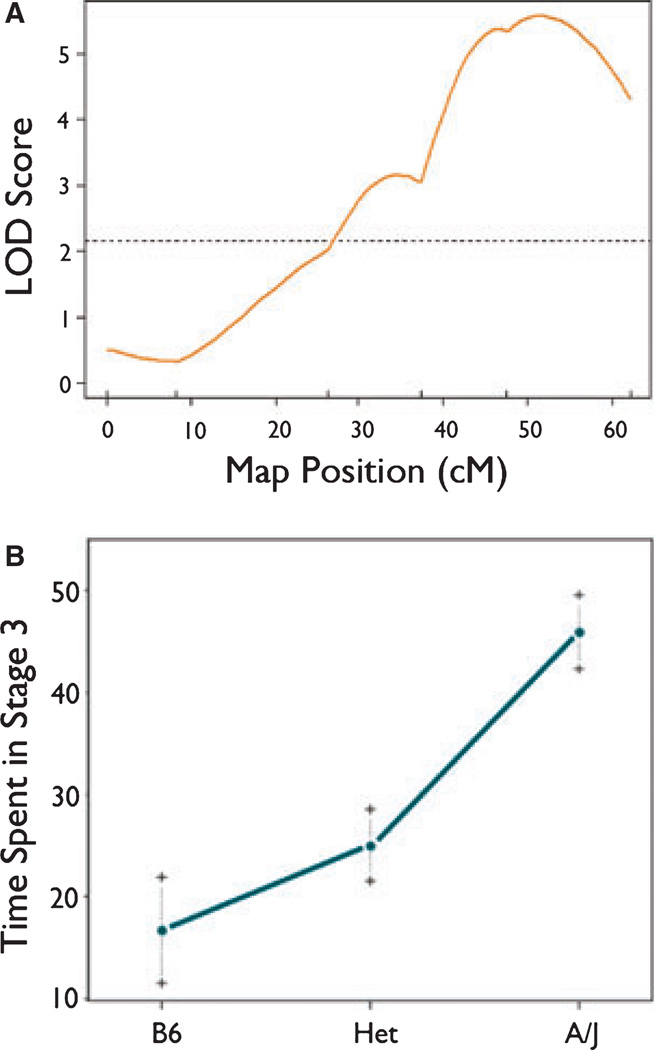
R/qtl analysis of the F2 mice identified significant QTLs for the variables time in stage 3 and highest stage reached. For time in stage 3 (Fig. 2A), there was a highly significant QTL in the distal portion of chromosome 10 that was nearest to marker rs13480719. The Bayes 95% confidence interval for this QTL spanned from 43–60 cM, with a maximum LOD score of 5.34 at rs13480719. The QTL for time in stage 3 accounted for approximately 27% of the variance for this trait in the F2 population. The effect of this QTL appeared to be approximately additive (Fig. 2B). In other words, there is a relatively linear effect of gene dose on phenotype, with the heterozygote phenotype being intermediate to the two homozygotes (B6/B6 and A/A) at the top locus. Results were similar if the covariates were not included in the model, but we included them a priori to be consistent with other analyses.
Figure 2.
(A) rQTL-generated LOD curve for time in stage 3 (partial status) on mouse Ch10. Marker positions are represented by vertical tick marks above the x axis. The dotted horizontal line is the line for statistical significance (p < 0.05). (B) Effect of genotype on time in stage 3 (partial status) at marker rs1340719. On the x axis, B6 refers to homozygosity for the B6 allele at the locus, Het refers to heterozygosity at the locus, and A/J refers to homozygosity for the A/J allele at the locus.
Epilepsia © ILAE
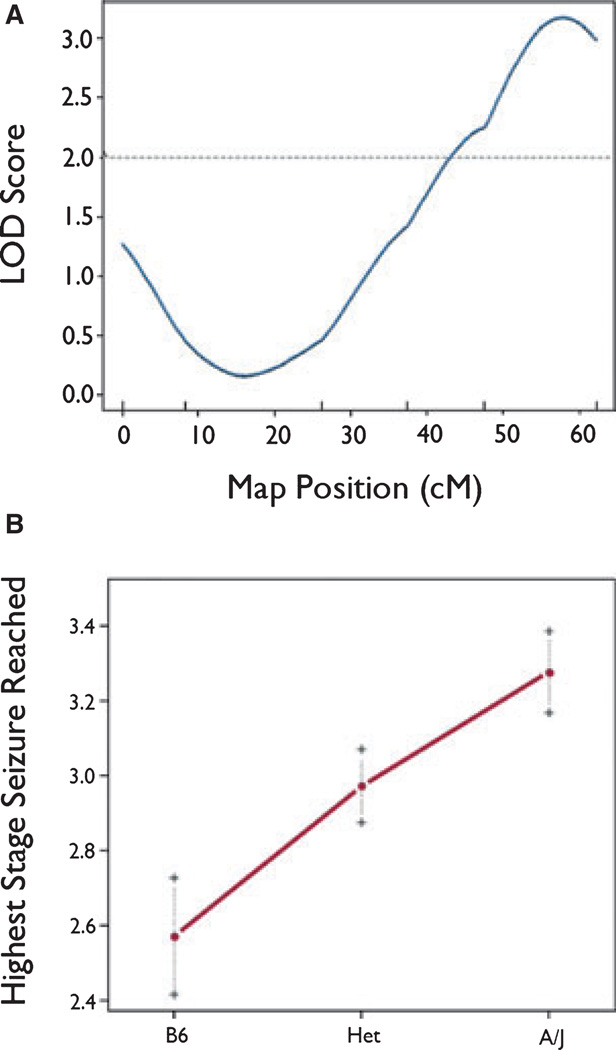
For highest stage reached (Fig. 3A) there was a highly significant QTL that was also in the distal portion of chromosome 10 and was nearest to marker rs13480781, with a maximum LOD score of 2.99 at marker rs13480781. The QTL for highest stage reached accounted for approximately 16% of the variance for this trait in the F2 population. The Bayes 95% confidence interval for this QTL spanned from 45 cM until the last marker in our map (rs13480781). The effect of this QTL also appeared to be approximately additive (Fig. 3B). Although not formally tested, we hypothesize that loci for time in stage 3 and highest stage reached reflect the pleiotropic action of a single QTL. Although these methods are robust to deviations from normality in the sample, we repeated the rQTL analyses using nonparametric (np) methods to account for nonnormality. All results were quantitatively similar (not shown).
Figure 3.
(A) rQTL-generated LOD curve for highest stage reached on mouse Ch10. Marker positions are represented by vertical tick marks above the x axis. The dotted horizontal line is the line for statistical significance (p < 0.05). (B) Effect of genotype on highest seizure stage reached at marker rs13480871. On the x axis, B6 refers to homozygosity for the B6 allele at the locus, Het refers to heterozygosity at the locus, and A/J refers to homozygosity for the A/J allele at the locus.
Epilepsia © ILAE
Chromosome 10 interval-specific congenic strain
As described above, we performed analyses in which reaching stage 4 or higher was the outcome variable. The first set of analyses, which compared each interval-specific congenic strain (ISCS) with the B6 control animals, resulted in statistically significant evidence for differences between strain and control for congenic strains R2, R3, and R4 (Fig. 1A–C; Tables 2 and 3). These results were replicated for the variables of time in stage 3 and highest stage reached (data not shown). The second set of analyses, which examined the effect of genotype at each marker locus separately, resulted in statistically significant evidence for association at rs13480781 and rs13480832. Finally, logistic regression analyses were performed to identify the interval most likely to harbor the susceptibility locus (data not shown). This third analysis examined the effects of multiple loci simultaneously. Regression analyses demonstrated statistically significant evidence of an association with rs13480781 after adjusting for rs13480832, and statistically significant evidence for an association with rs13480832 after adjusting for rs13480781. The association with rs13480719 did not persist after adjusting for rs13480781. This method is comparable to one recently described (Shao et al., 2010). These results correspond to the presence of a trait locus between rs13480781 and rs13480832. We repeated all analyses using logistic regression and adjusting for covariates of mouse age and weight at testing. Results were quantitatively similar (data not shown).
Table 2.
Comparison of individual interval-specific congenic strains with B6 controls
This table reports chi-square statistics and p-values for the test of independence between congenic status and the indicator of reaching stage 4 in analyses restricted to controls compared with individual congenics.
Values marked with * reflect a significant difference from B6 controls.
Table 3.
Effect of genotype at each marker locus in interval-specific congenic strains
| Observation | Value | p-value | Locus |
|---|---|---|---|
| 1 | 0.0506 | 0.8221 | rs13480643 |
| 2 | 1.0347 | 0.3090 | rs13480719 |
| 3 | 16.7317 | <0.0001* | rs13480781 |
| 4 | 17.1236 | <0.0001* | rs13480832 |
Chi-square statistics and p-values for the test of independence and genotype in a simultaneous analysis of all congenic strains at each locus.
Values marked with * represent significant effects.
Discussion
The purpose of this study is to explore the genetics of seizure susceptibility by identifying the loci contributing to susceptibility in a mouse model. This has potentially powerful implications for understanding the genetic contributions to human epilepsy. Using mouse CSS, Ch10 F2 populations, and ISCS, we have identified and fine mapped a locus on distal mouse chromosome 10 (Ch10) that contributes to limbic seizure susceptibility. Mapping of pilocarpine-induced seizures has not been reported previously, and there have been few prior reports of seizure susceptibility candidate loci on mouse Ch10 or syntenic human regions. Gershenfeld et al. reported an association of D10Mit180 (117.59 Mb) with seizure susceptibility in a B6 × A/J cross, in response to β-CCM, a γ-aminobutyric acid (GABA)ergic agent that produces generalized (not limbic) seizures (Gershenfeld et al., 1999). Subsequent studies of a congenic have suggested that this QTL may play a role in both anxiety-related endophenotypes and seizure susceptibility (Zhang et al., 2005).
We have also found evidence for the presence of a locus underlying both seizure susceptibility and anxiety endophenotypes on A/J Ch10 (Ponder et al., 2007; Winawer et al., 2007). In an independent dataset we have confirmed the location of this QTL, using a fear-conditioning paradigm (Palmer A, unpublished results). The possibility that a single allele pleiotropically influences both traits should be investigated further, especially in light of the known comorbidity between psychiatric disorders and epilepsy (Winawer & Hesdorffer, 2010). The region on distal Ch10 may contain one or more loci that play a role in epilepsy and psychiatric comorbidity.
D10Mit180 is located at approximately 117.59 Mb on Ch10, near marker rs13480781, which is located at 117.56 Mb (Fig. 1B). Marker rs13480719 maps to a homologous region on human CH12q. This may overlap a locus on chromosome 12q22–23, which has been found to be linked to familial TLE with febrile seizures in a single extended pedigree (Claes et al., 2004). To our knowledge, no other potential epilepsy genes have been mapped to this region.
We have mapped two seizure susceptibility phenotypes to the same region of mouse Ch10. The first, time in stage 3, is a measure of latency, or duration of partial status. The second, highest stage reached, is a measure of seizure severity. The colocalization of the latency and severity aspects of susceptibility suggests that this QTL has a broad effect on seizure susceptibility across different parameters. Defects in some molecular or biochemical pathways may act to prolong seizures once initiated, whereas others may affect the mechanism of propagation from partial to generalized seizures. Identification of genes that raise risk for one or both mechanisms can give insight into the biology of seizure susceptibility and development of targeted therapies.
These results may be limited, in that they map seizure susceptibility in response to an acute stimulus, not epilepsy. However, identification of genes influencing seizure susceptibility is fundamental to understanding human epilepsy (Noebels, 2003). For example, mutagenesis in a mouse model identified a single mutation on mouse Ch2, Szt1, conferring susceptibility to electroconvulsive threshold (ECT) minimal clonic seizures (Yang et al., 2003). Two of the three known deleted genes—Kcnq2 and Chrna4—are known to be mutated in human epilepsy families (Singh et al., 1998; Steinlein, 2000), and Kcnq2 haploinsufficiency was shown to determine the Szt1 susceptibility phenotype. These results demonstrate a direct correspondence between mouse seizure susceptibility genes and human epilepsy genes. Furthermore, screening compounds for seizure susceptibility in rodents has led to the development of many effective anticonvulsant agents (Frankel, 2009). Even if specific genes identified do not raise risk for epilepsy per se, the information obtained will still be valuable for elucidating the pathophysiology of seizures and therapeutic targets.
The identification of mouse QTLs and quantitative trait genes (QTGs) can provide a framework for identifying novel homologous candidate genes for human epilepsy. This has been demonstrated by the identification of a maximal electroshock seizure threshold (MEST) locus on mouse chromosome 1 (Ch1), fine mapping, selection of a potassium channel candidate gene, Kcnj10, and subsequent discovery of genetic association in a human epilepsy population (Ferraro et al., 2001; Buono et al., 2004; Ferraro et al., 2004). Mutations in Kcnj10 have also been reported in families with epilepsy, sensorineural deafness, and renal tubulopathy, supporting the gene’s role in cellular homeostasis in humans (Bockenhauer et al., 2009; Sala-Rabanal et al., 2010). Improving prior probability by the identification of biologically probable candidates can help minimize the false-positive results that plague human genetic association studies (Cardon & Bell, 2001; Emahazion et al., 2001) We have previously demonstrated success translating from mouse QTL to human genetic association with the identification of CSNK1E, a candidate gene for methamphetamine sensitivity in both mice and humans (Veenstra-Vander-Weele et al., 2006). QTL mapping is a powerful technique with the potential to identify novel loci and genes involved in epilepsy and other human complex disorders.
Analysis of chromosome substitution strains, Ch10 F2 populations, and ISCS created to date provides strong evidence for a seizure susceptibility locus on distal mouse Ch10 between markers rs13480781 and rs13480832. Further congenic strain creation is underway, and will help improve the resolution of our mapping and narrow the list of potential candidate genes.
Acknowledgments
Supported by grants NS061991 and MH079103. Technical laboratory support was provided by Shazeda Khan.
Footnotes
Disclosure
None of the authors has any conflict of interest to disclose. We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
References
- Baulac S, Baulac M. Advances on the genetics of mendelian idiopathic epilepsies. Clin Lab Med. 2010;30:911–929. doi: 10.1016/j.cll.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Bockenhauer D, Feather S, Stanescu HC, Bandulik S, Zdebik AA, Reichold M, Tobin J, Lieberer E, Sterner C, Landoure G, Arora R, Sirimanna T, Thompson D, Cross JH, van’t Hoff W, Al Masri O, Tullus K, Yeung S, Anikster Y, Klootwijk E, Hubank M, Dillon MJ, Heitzmann D, Arcos-Burgos M, Knepper MA, Dobbie A, Gahl WA, Warth R, Sheridan E, Kleta R. Epilepsy, ataxia, sensorineural deafness, tubulopathy, and KCNJ10 mutations. N Engl J Med. 2009;360:1960–1970. doi: 10.1056/NEJMoa0810276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges K, Gearing M, McDermott DL, Smith AB, Almonte AG, Wainer BH, Dingledine R. Neuronal and glial pathological changes during epileptogenesis in the mouse pilocarpine model. Exp Neurol. 2003;182:21–34. doi: 10.1016/s0014-4886(03)00086-4. [DOI] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Buono RJ, Lohoff FW, Sander T, Sperling MR, O’Connor MJ, Dlugos DJ, Ryan SG, Golden GT, Zhao H, Scattergood TM, Berrettini WH, Ferraro TN. Association between variation in the human KCNJ10 potassium ion channel gene and seizure susceptibility. Epilepsy Res. 2004;58:175–183. doi: 10.1016/j.eplepsyres.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet. 2001;2:91–99. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- Cendes F, Lopes-Cendes I, Andermann E, Andermann F. Familial temporal lobe epilepsy: a clinically heterogeneous syndrome. Neurology. 1998;50:554–557. doi: 10.1212/wnl.50.2.554. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138(3):963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claes L, Audenaert D, Deprez L, Van Paesschen W, Depondt C, Goossens D, Del-Favero J, Van Broeckhoven C, De Jonghe P. Novel locus on chromosome 12q22-q23.3 responsible for familial temporal lobe epilepsy associated with febrile seizures. J Med Genet. 2004;41:710–714. doi: 10.1136/jmg.2004.019257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emahazion T, Feuk L, Jobs M, Sawyer SL, Fredman D, St Clair D, Prince JA, Brookes AJ. SNP association studies in Alzheimer’s disease highlight problems for complex disease analysis. Trends Genet. 2001;17:407–413. doi: 10.1016/s0168-9525(01)02342-3. [DOI] [PubMed] [Google Scholar]
- Engel J., Jr A proposed diagnostic scheme for people with epileptic seizures and with epilepsy: report of the ILAE Task Force on Classification and Terminology. Epilepsia. 2001;42:796–803. doi: 10.1046/j.1528-1157.2001.10401.x. [DOI] [PubMed] [Google Scholar]
- Fehr C, Shirley RL, Belknap JK, Crabbe JC, Buck KJ. Congenic mapping of alcohol and pentobarbital withdrawal liability loci to a < 1 centimorgan interval of murine chromosome 4: identification of Mpdz as a candidate gene. J Neurosci. 2002;22:3730–3738. doi: 10.1523/JNEUROSCI.22-09-03730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Smith GG, Berrettini WH. Differential susceptibility to seizures induced by systemic kainic acid treatment in mature DBA/2J and C57BL/6J mice. Epilepsia. 1995;36:301–307. doi: 10.1111/j.1528-1157.1995.tb00999.x. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Smith GG, Schork NJ, St Jean P, Ballas C, Choi H, Berrettini WH. Mapping murine loci for seizure response to kainic acid. Mamm Genome. 1997;8:200–208. doi: 10.1007/s003359900389. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Snyder R, Laibinis M, Smith GG, Buono RJ, Berrettini WH. Genetic influences on electrical seizure threshold. Brain Res. 1998;813:207–210. doi: 10.1016/s0006-8993(98)01013-0. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Smith GG, Longman RL, Snyder RL, DeMuth D, Szpilzak I, Mulholland N, Eng E, Lohoff FW, Buono RJ, Berrettini WH. Quantitative genetic study of maximal electroshock seizure threshold in mice: evidence for a major seizure susceptibility locus on distal chromosome 1. Genomics. 2001;75:35–42. doi: 10.1006/geno.2001.6577. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Smith GG, DeMuth D, Buono RJ, Berrettini WH. Mouse strain variation in maximal electroshock seizure threshold. Brain Res. 2002;936:82–86. doi: 10.1016/s0006-8993(02)02565-9. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Smith GG, Martin JF, Lohoff FW, Gieringer TA, Zamboni D, Schwebel CL, Press DM, Kratzer SO, Zhao H, Berrettini WH, Buono RJ. Fine mapping of a seizure susceptibility locus on mouse chromosome 1: nomination of Kcnj10 as a causative gene. Mamm Genome. 2004;15:239–251. doi: 10.1007/s00335-003-2270-3. [DOI] [PubMed] [Google Scholar]
- Frankel WN. Genetics of complex neurological disease: challenges and opportunities for modeling epilepsy in mice and rats. Trends Genet. 2009;25:361–367. doi: 10.1016/j.tig.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel WN, Taylor L, Beyer B, Tempel BL, White HS. Electroconvulsive thresholds of inbred mouse strains. Genomics. 2001;74:306–312. doi: 10.1006/geno.2001.6564. [DOI] [PubMed] [Google Scholar]
- Fuerst D, Shah J, Kupsky WJ, Johnson R, Shah A, Hayman-Abello B, Ergh T, Poore Q, Canady A, Watson C. Volumetric MRI, pathological, and neuropsychological progression in hippocampal sclerosis. Neurology. 2001;57:184–188. doi: 10.1212/wnl.57.2.184. [DOI] [PubMed] [Google Scholar]
- Gershenfeld HK, Neumann PE, Li X, St Jean PL, Paul SM. Mapping quantitative trait loci for seizure response to a GABAA receptor inverse agonist in mice. J Neurosci. 1999;19:3731–3738. doi: 10.1523/JNEUROSCI.19-10-03731.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden GT, Ferraro TN, Smith GG, Snyder RL, Jones NL, Berrettini WH. Acute cocaine-induced seizures: differential sensitivity of six inbred mouse strains. Neuropsychopharmacology. 2001;24:291–299. doi: 10.1016/S0893-133X(00)00204-9. [DOI] [PubMed] [Google Scholar]
- Hain HS, Crabbe JC, Bergeson SE, Belknap JK. Cocaine-induced seizure thresholds: quantitative trait loci detection and mapping in two populations derived from the C57BL/6 and DBA/2 mouse strains. J Pharmacol Exp Ther. 2000;293:180–187. [PubMed] [Google Scholar]
- Kobayashi E, Lopes-Cendes I, Guerreiro CA, Sousa SC, Guerreiro MM, Cendes F. Seizure outcome and hippocampal atrophy in familial mesial temporal lobe epilepsy. Neurology. 2001;56:166–172. doi: 10.1212/wnl.56.2.166. [DOI] [PubMed] [Google Scholar]
- Kosobud AE, Crabbe JC. Genetic correlations among inbred strain sensitivities to convulsions induced by 9 convulsant drugs. Brain Res. 1990;526:8–16. doi: 10.1016/0006-8993(90)90243-5. [DOI] [PubMed] [Google Scholar]
- Leite JP, Garcia-Cairasco N, Cavalheiro EA. New insights from the use of pilocarpine and kainate models. Epilepsy Res. 2002;50:93–103. doi: 10.1016/s0920-1211(02)00072-4. [DOI] [PubMed] [Google Scholar]
- Mathern GW, Babb TL, Leite JP, Pretorius K, Yeoman KM, Kuhlman PA. The pathogenic and progressive features of chronic human hippocampal epilepsy. Epilepsy Res. 1996;26:151–161. doi: 10.1016/s0920-1211(96)00052-6. [DOI] [PubMed] [Google Scholar]
- Mathis C, Paul SM, Crawley JN. Characterization of benzodiazepine-sensitive behaviors in the A/J and C57BL/6J inbred strains of mice. Behav Genet. 1994;24:171–180. doi: 10.1007/BF01067821. [DOI] [PubMed] [Google Scholar]
- McKhann GM, Wenzel HJ, Robbins CA, Sosunov AA, Schwartzkroin PA. Mouse strain differences in kainic acid sensitivity, seizure behavior, mortality, and hippocampal pathology. Neuroscience. 2003;122:551–561. doi: 10.1016/s0306-4522(03)00562-1. [DOI] [PubMed] [Google Scholar]
- Nadeau JH, Singer JB, Matin A, Lander ES. Analysing complex genetic traits with chromosome substitution strains. Nat Genet. 2000;24:221–225. doi: 10.1038/73427. [DOI] [PubMed] [Google Scholar]
- Neumann PE, Collins RL. Genetic dissection of susceptibility to audiogenic seizures in inbred mice. Proc Natl Acad Sci USA. 1991;88:5408–5412. doi: 10.1073/pnas.88.12.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann PE, Collins RL. Confirmation of the influence of a chromosome 7 locus on susceptibility to audiogenic seizures. Mamm Genome. 1992;3:250–253. doi: 10.1007/BF00292152. [DOI] [PubMed] [Google Scholar]
- Noebels JL. The biology of epilepsy genes. Annu Rev Neurosci. 2003;26:599–625. doi: 10.1146/annurev.neuro.26.010302.081210. [DOI] [PubMed] [Google Scholar]
- Ottman R. Analysis of genetically complex epilepsies. Epilepsia. 2005;46(Suppl. 10):7–14. doi: 10.1111/j.1528-1167.2005.00350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AA, Phillips TJP. Quantitative trait locus (QTL) mapping in mice. In: Liu Y, Lovinger DM, editors. Methods in alcohol-related neuroscience research. Boca Raton, FL: CRC Press; 2002. pp. 1–30. [Google Scholar]
- Phillips TJ, Belknap JK, Hitzemann RJ, Buck KJ, Cunningham CL, Crabbe JC. Harnessing the mouse to unravel the genetics of human disease. Genes Brain Behav. 2002;1:14–26. doi: 10.1046/j.1601-1848.2001.00011.x. [DOI] [PubMed] [Google Scholar]
- Ponder CA, Munoz M, Gilliam TC, Palmer AA. Genetic architecture of fear conditioning in chromosome substitution strains: relationship to measures of innate (unlearned) anxiety-like behavior. Mamm Genome. 2007;18:221–228. doi: 10.1007/s00335-007-9013-9. [DOI] [PubMed] [Google Scholar]
- Racine RJ. Modification of seizure activity by electrical stimulation. II. Motor seizure. Electroencephalogr Clin Neurophysiol. 1972;32:281–294. doi: 10.1016/0013-4694(72)90177-0. [DOI] [PubMed] [Google Scholar]
- Sala-Rabanal M, Kucheryavykh LY, Skatchkov SN, Eaton MJ, Nichols CG. Molecular mechanisms of EAST/SeSAME syndrome mutations in Kir4.1 (KCNJ10) J Biol Chem. 2010;285:36040–36048. doi: 10.1074/jbc.M110.163170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos NF, Sousa SC, Kobayashi E, Torres FR, Sardinha JA, Cendes F, Lopes-Cendes I. Clinical and genetic heterogeneity in familial temporal lobe epilepsy. Epilepsia. 2002;43(Suppl. 5):136. doi: 10.1046/j.1528-1157.43.s.5.23.x. [DOI] [PubMed] [Google Scholar]
- Scharfman H, Pedley T. Temporal lobe epilepsy. In: Gilman S, editor. The neurobiology of disease. New York: Academic Press; 2006. pp. 349–370. [Google Scholar]
- Schauwecker PE. Modulation of cell death by mouse genotype: differential vulnerability to excitatory amino acid-induced lesions. Exp Neurol. 2002;178:219–235. doi: 10.1006/exnr.2002.8038. [DOI] [PubMed] [Google Scholar]
- Schauwecker PE, Steward O. Genetic determinants of susceptibility to excitotoxic cell death: implications for gene targeting approaches. Proc Natl Acad Sci USA. 1997;94:4103–4108. doi: 10.1073/pnas.94.8.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semah F, Picot MC, Adam C, Broglin D, Arzimanoglou A, Bazin B, Cavalcanti D, Baulac M. Is the underlying cause of epilepsy a major prognostic factor for recurrence? Neurology. 1998;51:1256–1262. doi: 10.1212/wnl.51.5.1256. [DOI] [PubMed] [Google Scholar]
- Shao H, Sinasac DS, Burrage LC, Hodges CA, Supelak PJ, Palmert MR, Moreno C, Cowley AW, Jr, Jacob HJ, Nadeau JH. Analyzing complex traits with congenic strains. Mamm Genome. 2010;2010:21. doi: 10.1007/s00335-010-9267-5. Epub 2010 Jun 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NA, Charlier C, Stauffer D, DuPont BR, Leach RJ, Melis R, Ronen GM, Bjerre I, Quattlebaum T, Murphy JV, McHarg ML, Gagnon D, Rosales TO, Peiffer A, Anderson VE, Leppert M. A novel potassium channel gene, KCNQ2, is mutated in an inherited epilepsy of newborns. Nat Genet. 1998;18:25–29. doi: 10.1038/ng0198-25. [DOI] [PubMed] [Google Scholar]
- Steinlein OK. Neuronal nicotinic receptors in human epilepsy. Eur J Pharmacol. 2000;393:243–247. doi: 10.1016/s0014-2999(00)00065-0. [DOI] [PubMed] [Google Scholar]
- Stephen LJ, Kwan P, Brodie MJ. Does the cause of localisation-related epilepsy influence the response to antiepileptic drug treatment? Epilepsia. 2001;42:357–362. doi: 10.1046/j.1528-1157.2001.29000.x. [DOI] [PubMed] [Google Scholar]
- Strohl KP, Gallaugher L, Lynn A, Friedman L, Hill A, Singer JB, Lander ES, Nadeau J. Sleep-related epilepsy in the A/J mouse. Sleep. 2007;30:169–176. doi: 10.1093/sleep/30.2.169. [DOI] [PubMed] [Google Scholar]
- Turski WA. Pilocarpine-induced seizures in rodents – 17 years on. Pol J Pharmacol. 2000;52:63–65. [PubMed] [Google Scholar]
- Turski L, Ikonomidou C, Turski WA, Bortolotto ZA, Cavalheiro EA. Review: cholinergic mechanisms and epileptogenesis. The seizures induced by pilocarpine: a novel experimental model of intractable epilepsy. Synapse. 1989;3:154–171. doi: 10.1002/syn.890030207. [DOI] [PubMed] [Google Scholar]
- Vadlamudi L, Scheffer IE, Berkovic SF. Genetics of temporal lobe epilepsy. J Neurol Neurosurg Psychiatry. 2003;74:1359–1361. doi: 10.1136/jnnp.74.10.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra-VanderWeele J, Qaadir A, Palmer AA, Cook EH, Jr, de Wit H. Association between the casein kinase 1 epsilon gene region and subjective response to D-amphetamine. Neuropsychopharmacology. 2006;31:1056–1063. doi: 10.1038/sj.npp.1300936. [DOI] [PubMed] [Google Scholar]
- Winawer MR, Hesdorffer DC. Migraine, epilepsy, and psychiatric comorbidity: partners in crime. Neurology. 2010;74:1166–1168. doi: 10.1212/WNL.0b013e3181d90065. [DOI] [PubMed] [Google Scholar]
- Winawer MR, Shinnar S. Genetic epidemiology of epilepsy or what do we tell families? Epilepsia. 2005;46(Suppl. 10):24–30. doi: 10.1111/j.1528-1167.2005.00354.x. [DOI] [PubMed] [Google Scholar]
- Winawer MR, Kuperman R, Niethammer M, Sherman S, Rabinowitz D, Guell IP, Ponder CA, Palmer AA. Use of chromosome substitution strains to identify seizure susceptibility loci in mice. Mamm Genome. 2007;18:23–31. doi: 10.1007/s00335-006-0087-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Beyer BJ, Otto JF, O’Brien TP, Letts VA, White HS, Frankel WN. Spontaneous deletion of epilepsy gene orthologs in a mutant mouse with a low electroconvulsive threshold. Hum Mol Genet. 2003;12:975–984. doi: 10.1093/hmg/ddg118. [DOI] [PubMed] [Google Scholar]
- Zhang S, Lou Y, Amstein TM, Anyango M, Mohibullah N, Osoti A, Stancliffe D, King R, Iraqi F, Gershenfeld HK. Fine mapping of a major locus on chromosome 10 for exploratory and fear-like behavior in mice. Mamm Genome. 2005;16:306–318. doi: 10.1007/s00335-004-2427-8. [DOI] [PubMed] [Google Scholar]