Abstract
Background
The basic Helix-Loop-Helix (bHLH) transcription factor Twist1 fulfills an essential function in neural crest cell formation, migration and survival and is associated with the craniosynostic Saethre-Chotzen syndrome in humans. However, its functions during mandibular development, when it may interact with other bHLH transcription factors like Hand2, are unknown since mice homozygous for the Twist1 null mutation die in early embryogenesis. To determine the role of Twist1 during mandibular development, we used the Hand2-Cre transgene to conditionally inactivate the gene in the neural crest cells populating the mandibular pharyngeal arch.
Results
The mutant mice exhibited a spectrum of craniofacial anomalies, including mandibular hypoplasia, altered middle ear development, and cleft palate. It appears that Twist1 is essential for the survival of the neural crest cells involved in the development of the mandibular ramal elements. Twist1 plays a role in molar development and cusp formation by participating in the reciprocal signaling needed for the formation of the enamel knot. This gene is also needed to control the ossification of the mandible, a redundant role shared with Hand2.
Conclusion
Twist1, along with Hand2, is essential for the proximo-distal patterning and development of the mandible and ossification.
Keywords: mandible, neural crest cells, Twist1, Hand2, palate, Meckel’s cartilage, mineralization, basic Helix-Loop-Helix transcription factor
Introduction
Mandibular development is characterized by the activity of complex regulatory pathways controlling the diverse cell types within the first pharyngeal arch to generate bones, cartilage, nerves and dentition (D'Souza et al., 2010). One of these cell types, the neural crest cells, migrates from the neural lip into the pharyngeal arches. The migration, patterning, proliferation and differentiation of these cells in the mandibular arch are tightly regulated by recursive signals from the surrounding ectoderm, mesoderm, endoderm and the crest cells themselves (Helms et al., 2005; Chai and Maxson, 2006; Depew and Simpson, 2006). While the neural crest cells are essential for the development of the craniofacial skeleton, the molecular pathways establishing the different territories within the mandibular arch are relatively unknown.
Axial territoriality is established early in the vertebrate embryo by the expression of homeobox genes from the four Hox clusters (Mallo et al., 2010). Since Hox genes are not expressed in the mandibular arch (Creuzet et al., 2002; Creuzet et al., 2005), the initial division of the mandibular arch neural crest cells is accomplished by the related homeobox genes of the distal-less/Dlx family (Depew et al., 2005). Dlx1 and Dlx2 regulate the development of the neural crest cells of the first pharyngeal arch giving rise to the upper jaw (Qiu et al., 1995; Qiu et al., 1997). Dlx5 and Dlx6 regulate the development of the distal mandibular arch and mandible proper. In mice homozygous for the Dlx5 and Dlx6 null mutations, the lower jaw is homeotically transformed into upper jaw-like structures (Beverdam et al., 2002; Depew et al., 2002). After the activation of the Dlx genes in the arch, the mandibular territory is further subdivided between the different basic Helix-Loop-Helix (bHLH) transcription factors, including Hand1, Hand2 and Twist1. The expression of these bHLH factors is established in unique areas of the mandibular arch in the mouse with overlapping boundaries by embryonic day (E) 10.5 (Ruest et al., 2004; Ruest and Clouthier, 2009). Hand1, which takes part in the development of the mandibular symphysis (Barbosa et al., 2007), is expressed the most distally, with the major part of its expression overlapping with Hand2 (Clouthier et al., 2000; Ruest et al., 2004). Their overlapping activity is essential for mandibular and incisor development (Barbosa et al., 2007). Hand2 is expressed medially in the mandibular arch (Clouthier et al., 2000; Ruest et al., 2004), plays an important role in the development and ossification of the mandibular dentary (Yanagisawa et al., 2003; Funato et al., 2009) and controls the development of the tongue (Barron et al., 2011). Its proximal expression coincides with that of Twist1, the most proximal factor (Ruest et al., 2004). The expression of Hand1 and Hand2 is regulated by endothelin signaling (Ruest et al., 2004; Ruest and Clouthier, 2009), with Dlx6 directly activating Hand2 expression (Charite et al., 2001). In turn, Hand2 partially represses the Dlx5 and Dlx6 expression (Barron et al., 2011). In the absence of endothelin activity, Twist1 expression extends more distally in the mandibular arch (Ruest et al., 2004), suggesting that the boundaries between the bHLH factors may be set by their mutual interactions.
The mammalian Twist1 gene associated with the Saethre-Chotzen syndrome (OMIM 101400) (Howard et al., 1997; Krebs et al., 1997) plays important roles during embryonic development. As in the Saethre-Chotzen syndrome, haploinsufficiency of Twist1 in the mouse results in craniosynostosis and polydactyly (Chen and Behringer, 1995; Bourgeois et al., 1998; Gorlin et al., 2001). However, mice deficient in Twist1 die by E11.5 due to neural tube, craniofacial and mesodermal defects (Chen and Behringer, 1995). Twist1 is essential for the development of mesodermal derivatives and the formation, migration and survival of the neural crest cells, limb development, cardiovascular development and neural tube closure (Chen and Behringer, 1995; Bourgeois et al., 1998; O'Rourke et al., 2002; Soo et al., 2002; Zuniga et al., 2002; Shelton and Yutzey, 2008; Vincentz et al., 2008). This gene likely fulfills other developmental functions such as palate closure (Rice et al., 2005; Yu et al., 2008) since it is involved in epithelial to mesenchymal transition (EMT) (Kang and Massague, 2004; Yang et al., 2004; Yu et al., 2009a), a function also associated with cancer and mesoderm formation. Based on its expression profile, Twist1 may also regulate tooth development (Rice et al., 2005). Twist1 activity involves its interaction with self or other bHLH factors and is influenced by its phosphorylation (Firulli et al., 2005; Connerney et al., 2006; Firulli et al., 2007; Connerney et al., 2008). Inactivation of the gene in forming neural crest cells produces severe craniofacial defects due to migratory and survival problems of these cells (Bildsoe et al., 2009), obscuring its function during mandibular formation.
To investigate the role of Twist1 during mandible development, we specifically inactivated it in the mandibular arch neural crest cells, which caused shortening of the mandible and impaired development of the ramus. However, these structural changes did not correlate with the alteration of the expression pattern of genes presumably regulated by Twist1. In the present study, we found evidence that Twist1 has unique functions that do not duplicate other bHLH factors; we also found functions shared with Hand2 during mandible development. In addition, our results showed that Twist1 is essential for the normal formation of the dentition, Meckel’s cartilage and the middle ear and to control mandibular ossification.
Results
Twist1 is essential for the development of proximal mandibular structures
Inactivation of the basic helix-loop-helix (bHLH) Twist1 gene in neural crest cells before or soon after their formation causes severe craniofacial defects (Bildsoe et al., 2009). These defects are largely due to the small number of cells detaching and migrating from the neural plate borber, consistent with Twist1 function in epithelial-mesenchymal transition (Kang and Massague, 2004; Yang et al., 2004) and its role in the migration toward and proper localization of these cells in the mandibular pharyngeal arch (Soo et al., 2002). The reduced number of cells and early inactivation of the gene masked the identification of Twist1 functions during mandibular development. To investigate its functions during lower jaw development, we bred the conditional Twist1 knockout mice [Twist1cko/cko (Chen et al., 2007)] with the Hand2-Cre line. In this line, the Cre recombinase is expressed specifically in the postmigratory neural crest cells populating the mandibular pharyngeal arch, starting at ~E9.5, continuing at E10.5 (Ruest et al., 2003). The activity of the enhancer driving the Cre transgene expression is decreased by E11.5, following the normal expression of the Hand2 gene (Charite et al., 2001; Ruest et al., 2003). Fate mapping analysis revealed that this Cre line allows deleting the Twist1 gene in the neural crest cells along the entire mandible/lower jaw complex, including cells involved in ramal development (Ruest et al., 2003; Ruest et al., 2004). Inactivation of the Twist1 gene was confirmed at E10.5 by whole mount in situ hybridization (Fig. 1A–D). In the mutant embryos, Twist1 expression was abrogated in the distal mandibular arch, consistent with the expression of the Hand2-Cre allele.
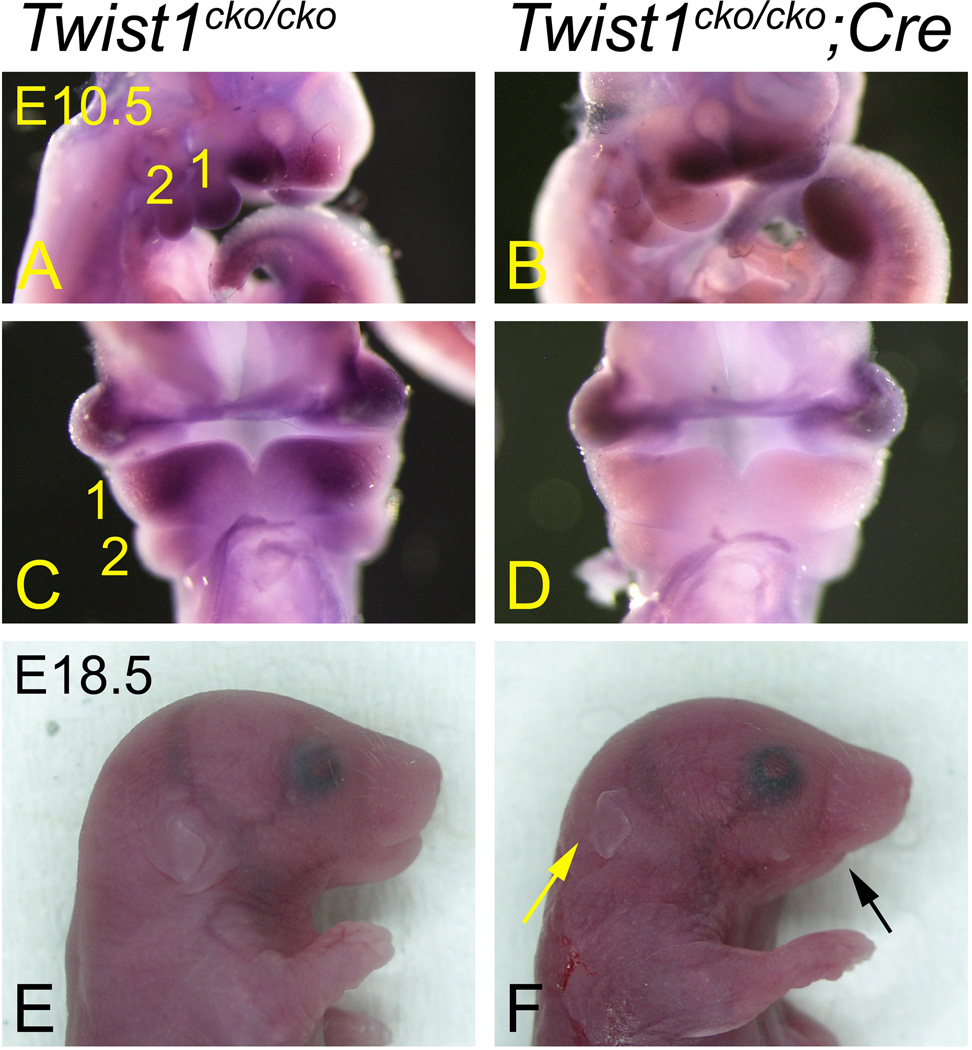
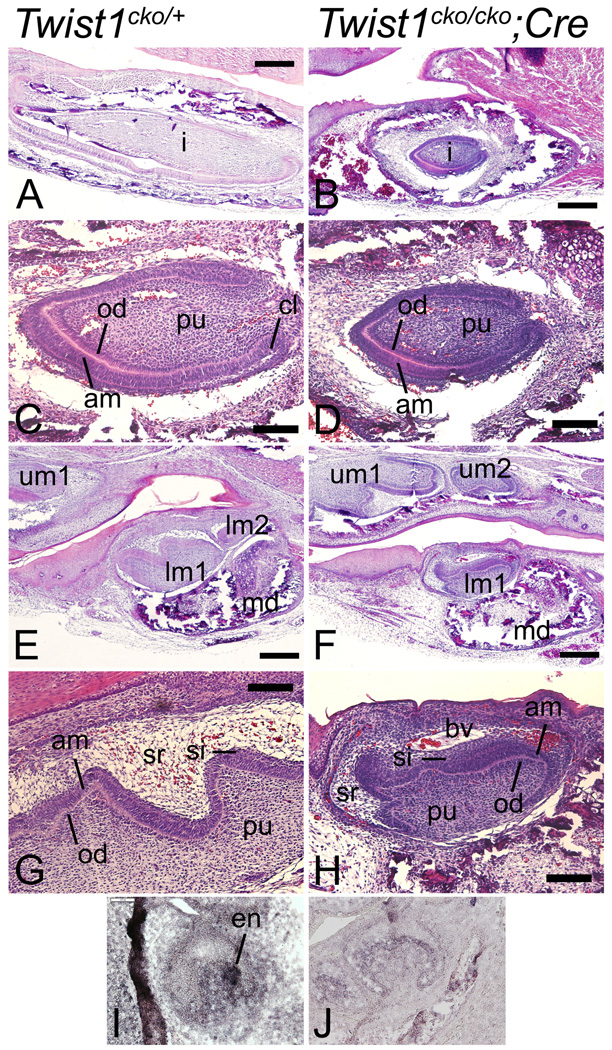
Figure 1. Twist1 is required for normal mandible development.
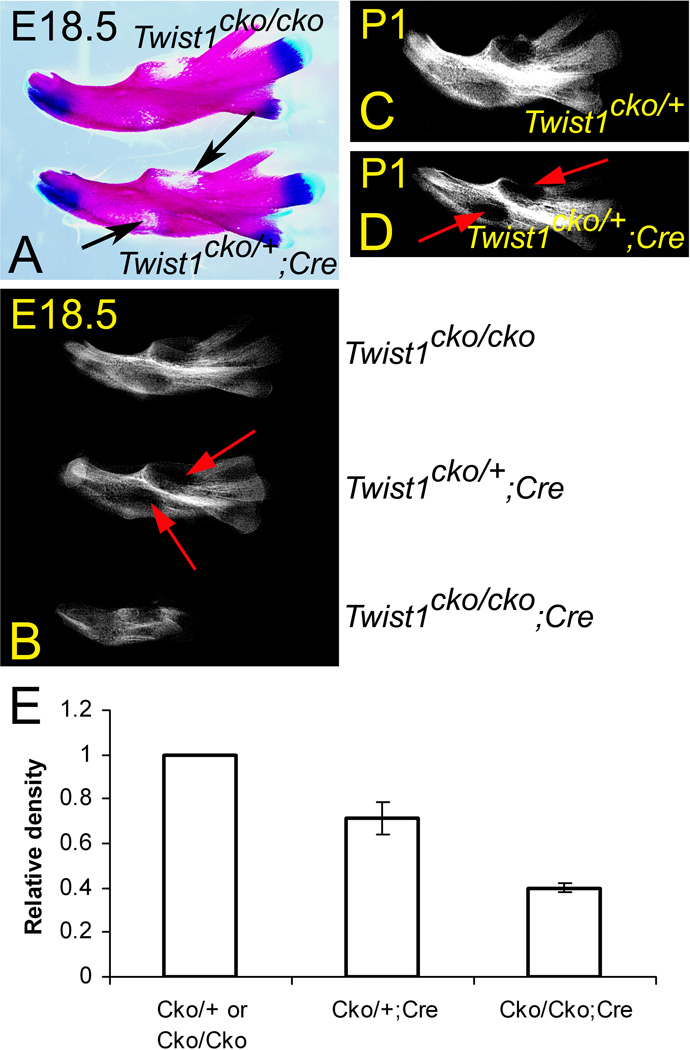
Lateral (A, B, E and F) and frontal (C and D) views of E10.5 (A–D) and E18.5 (E and F) Twist1cko/cko control (A, C and E) and Twist1cko/cko;Cre conditional mutant (B, D and F) embryos. A–D Whole mount in situ hybridization revealing that Twist1 expression is specifically lost in the mandibular pharyngeal arch (1) of mutant embryos, following inactivation with the Hand2-Cre transgene. E, F. In near-term conditional mutant embryos (F), the mandible is shorter (black arrow), and the external ear is displaced in a more posterior position (yellow arrow).
The Twist1cko/cko;Hand2-Cre mutant pups survived 1–2 days after birth, eventually dying of starvation from failure to feed as evidenced by the absence of milk in the stomach. Upon close examination of E18.5 embryos, a shorter lower jaw was noticeable (Fig. 1F), which likely prevented the pups from latching for feeding. The external ears were also located in a more posterior position. The skeleton preparations from the E18.5 Twist1cko/cko;Hand2-Cre embryos indicated that the mandible was shorter (Fig. 2). The dentary was fairly normal in shape and only slightly shorter than that in the wildtype littermates (Twist1cko/+;Hand2-Cre, Twist1cko/+ or Twist1cko/cko embryos) (Fig. 2 C–E). However, the incisors were not always visible, and the alveola for the molars was obstructed by a sheet of membranous bone. The ramal region was the most affected portion of the mandible and greatly reduced in size and lacking the typical processes associated with a mammalian mandible. The condylar process was clearly missing, along with the secondary cartilage of the condyle. The lack of a condyle prevented the formation of a proper temporomandibular articulation (Fig. 2E). The coronoid process, angular process and its associated secondary cartilage were absent as well.
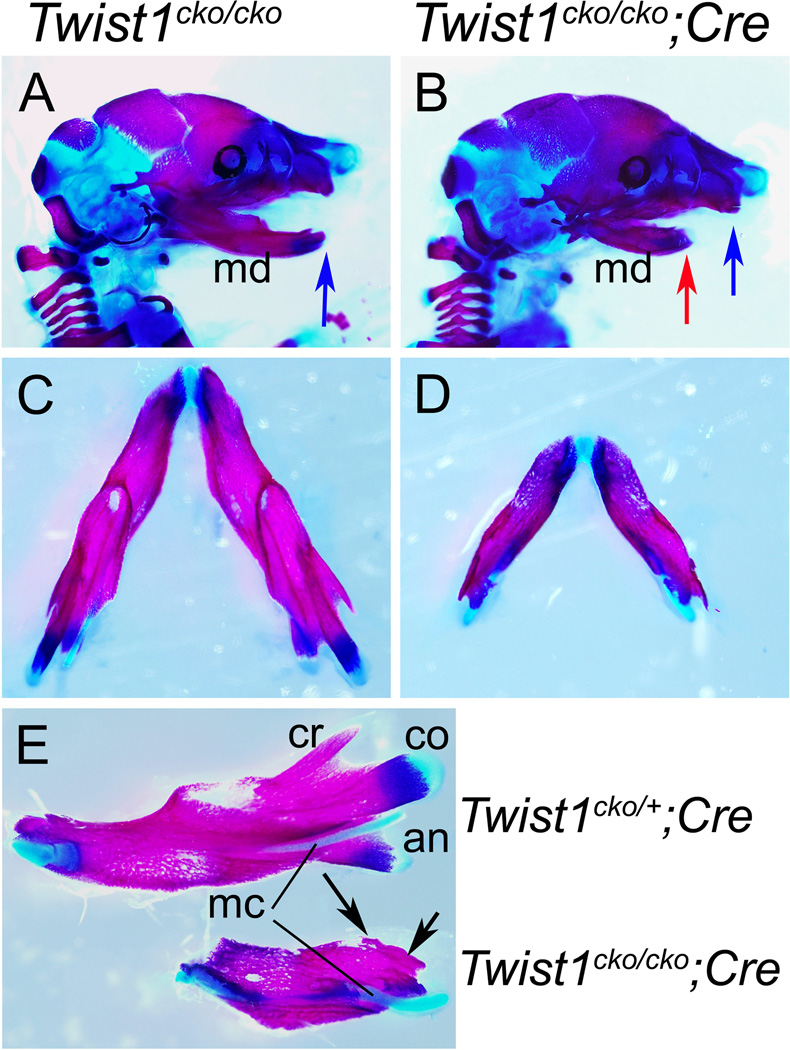
Figure 2. Twist1 is needed for the development of the proximal mandible.
Skeleton preparations with Alcian blue and Alizarin red revealing the cartilage and bone in blue and red, respectively, of E18.5 Twist1cko/cko or Twist1cko/+;Cre control (A, C and E) and Twist1cko/cko;Cre conditional mutant (B, D and E) embryos. A, B. In the mutant embryo (B), the mandible is hypoplastic (compare red arrow in B with blue arrow in A and B). The incisors are not visible. C–E. In the superior view of the mandible, the molar alveola is missing in the conditional mutant mandible (C). The ramal processes are also missing, which is more obvious in the lingual views of the mandibles (E), as the coronoid (cr), condylar (co) and angular (an) are missing in the mutant lower jaw (black arrows in E). The meckel’s cartilage (mc) was also shorter with a rounded end, and did not connect with the malleus in the middle ear.
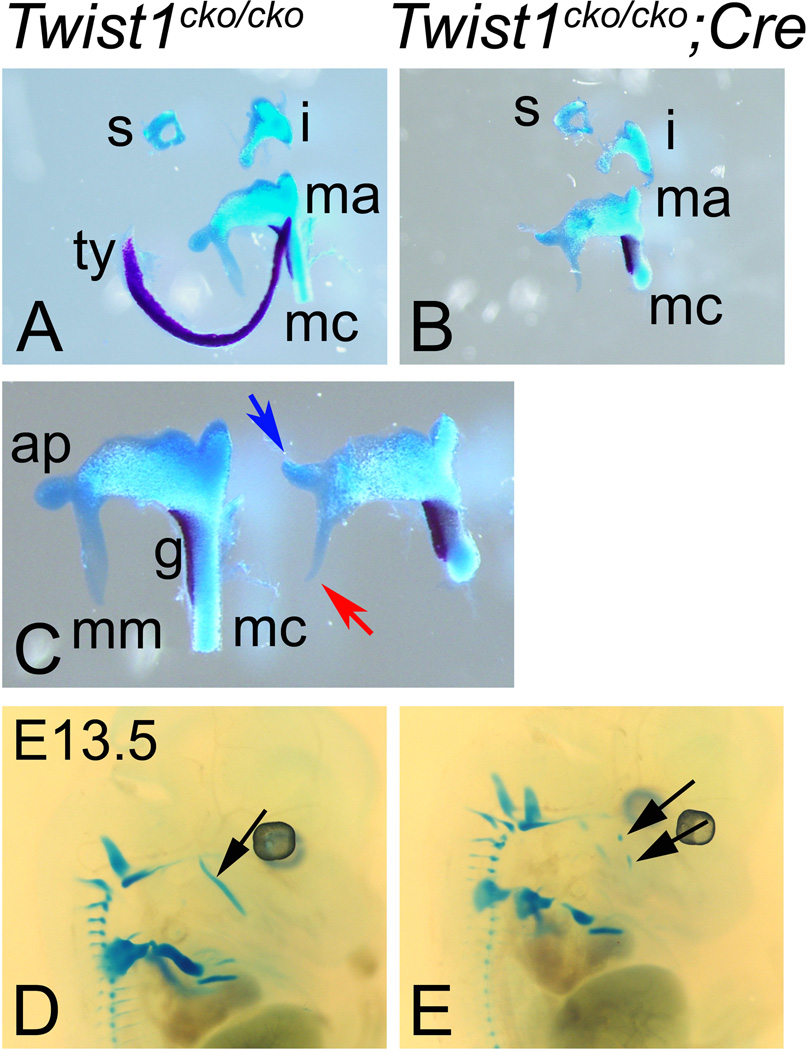
When the mandible was isolated in the mutant embryos, the Meckel’s cartilage did not appear as a continuous rod connecting the lower jaw with the malleus. Both disconnected ends of the cartilage were rounded, unlike the wild type cartilage, which had square ends after the required cutting during dissection (Fig. 3 A–C). To determine whether the cartilage grew into two segments or was severed during development, we stained the cartilage of E13.5 embryos with Alcian blue. In the wild type embryos, the cartilage appeared as two bilateral rods, one on each side of the forming lower jaw. In the Twist1cko/cko;Hand2-Cre embryos, the Meckel’s cartilage was clearly visible as two segments on each side (Fig. 3E), indicating that it had grown into two separate entities. The segmentation of the Mecklel’s cartilage suggests that the primary articulation of the lower jaw between the malleus and incus may have been altered. This alteration associated with the mostly missing ramal region may explain the lack of the condylar secondary cartilage that arises consequently to the embryonic jaw opening reflex (D'Souza et al., 2010). During the histological analysis, we also observed that the Meckel’s cartilage was slightly malformed. In the mutant embryos, the shape had changed from a circular rod into a “kidney bean” shape, with the concave surface toward the buccal area and the convex surface toward the lingual side of the oral cavity (Data not shown). At the symphysis in the mutant embryos, the normally reversed V-shaped cartilage was flattened (Data not shown).
Figure 3. Twist1 is needed for the development of the middle ear.
Alcian blue and Alizarin red skeleton staining revealing the cartilage and bone in blue and red, respectively, of E18.5 Twist1cko/cko control (A and C) and Twist1cko/cko;Cre conditional mutant (B and C) embryos. Lateral view of Alcian blue cartilage staining in E13.5 embryos (D and E). A–C. In the middle ear of the conditional mutant embryos (B and on the right in C), the tympanic ring (ty) is missing and the gonial (g) slightly shorter. The manubrium (mm and red arrow for the conditional mutant) and anterior process (ap and blue arrow) are malformed in the mutant embryos. D, E. The cartilage staining at E13.5 reveals that the meckel’s cartilage grew as two separated segments in the conditional mutants (black arrows in E) instead of the single cartilaginous rod (back arrow in D) connecting the lower jaw with the malleus. i, incus; mc, Meckel’s cartilage; s, stapes.
In the E18.5 mutant embryos, the middle ear defects reflected the expression of the Cre transgene (Ruest et al., 2003). The manubrium (handle) and anterior process of Folius (slender or gracillis process) of the malleus were smaller, but the collum (neck), capitulum (head) and its articular facet were normal (Fig. 3C). The gonial bone was shorter, and the tympanic annulus (ring) was missing (Fig. 3B, C). These defects suggest that Twist1 functions cell-autonomously, in agreement with a previous report (Soo et al., 2002).
Secondary cleft palate in the Twist1 conditional mutant embryos
Upon examination of the E18.5 skeleton preparations, a complete anterior-posterior cleft of the secondary palate was visible in the mutant embryos (Fig. 4A, B). While cleft palate was observed in the Twist1cko/cko;Wnt1-Cre embryos due to Twist1 function in neural crest cell migration, homing and development (Bildsoe et al., 2009), the phenotype was unexpected in our embryos since the Cre transgene is not expressed in the palate of Hand2-Cre mice (Ruest et al., 2003). Histological analysis of E18.5 embryos confirmed the presence of a complete cleft of the secondary palate, inferring the possibility of a submucosal cleft (Fig. 4D). In E18.5 mutant embryo sections, we noticed that the tongue was fairly normal in size, effectively creating a nasopharyngeal-like airway. Since mice are obligatory nose breathers, this substitute airway passage could explain why the conditional mutant pups were able to breathe and survive for 1–2 days after birth. With its malformed condyle and absent temporomandibular joint may have helped maintain a closed lower jaw and provide a functional airway.
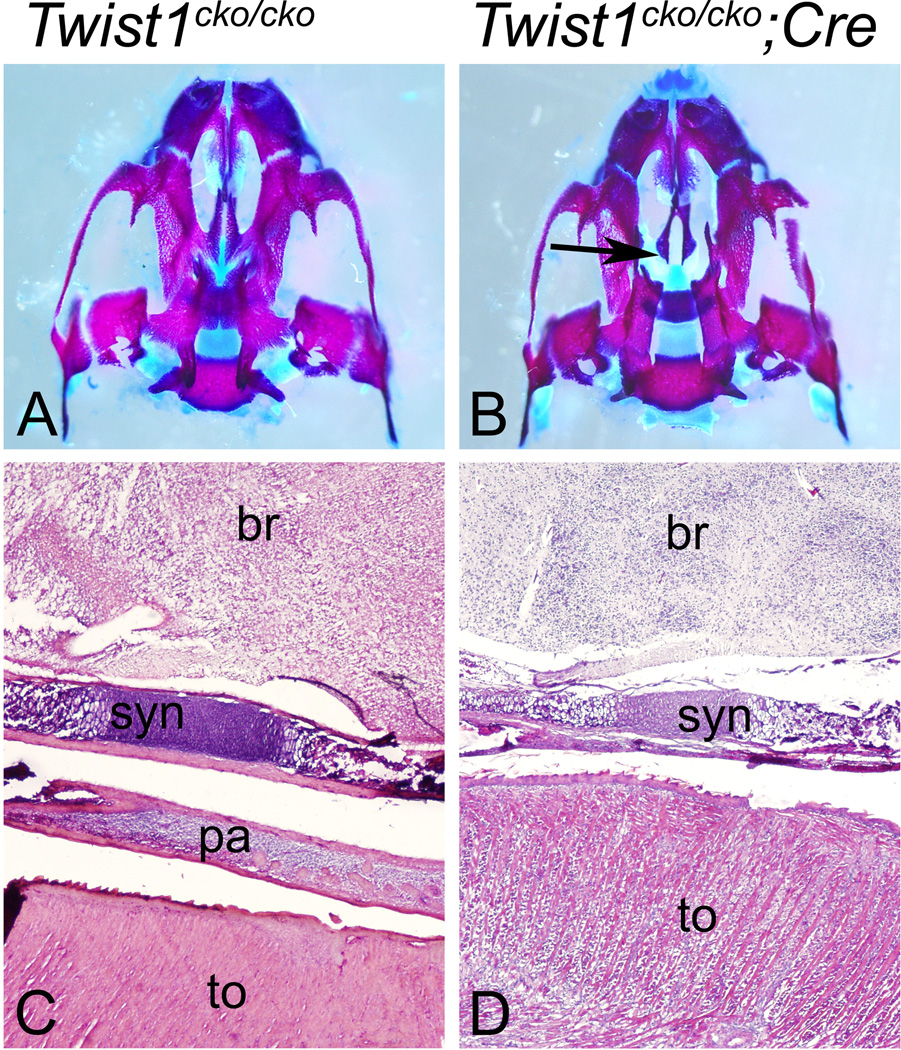
Figure 4. Inactivation of Twist1 in the mandible causes secondary defects of the posterior palate.
Ventral view of the upper jaw skeleton (A and B) and sagital hematoxylin and eosin-stained sections through the head of E18.5 Twist1cko/cko control (A and C) and Twist1cko/cko;Cre conditional mutant (B and C) embryos. A, B. In the conditional mutant embryos, the secondary palate was opened to the nasal cavity (black arrow in B), and the palatine bones were greatly reduced in size; their wings did not meet at the midline. C, D. The anterior-posterior (left to right) sagital sections confirmed the absence of a secondary (posterior) palate (pa) in the conditional mutant embryos (D) separating the tongue from the base of the skull below the brain (br). In these views, a cartilaginous synchondrose (syn) is visible.
Since the tongue in the Twist1cko/cko;Hand2-Cre embryos was relatively normal-sized despite the smaller lower jaw, we histologically analyzed the E13.5, 14.5 and 15.5 embryos to determine whether the cleft palate was a secondary defect caused by the tongue blocking the elevation of the palatal shelves (Yu et al., 2009a; Yu et al., 2009b; D'Souza et al., 2010). Results from our histological analysis confirmed this was the case (Data not shown). We concluded that the cleft palate was a secondary defect, resulting from the smaller lower jaw failing to drop downward and forward, which prevented the tongue from lowering, and then the tongue physically blocked the elevation and fusion of the palatal shelves. This series of events is similar to the clinical description of the Pierre-Robin sequence (OMIM: 261800)(Latham, 1966; Poswillo, 1967; Gorlin et al., 2001; Ricks et al., 2002).
Tooth development defects in the Twist1 conditional knockout embryos
The histological analysis of the E18.5 embryos revealed that the incisors were smaller in the Twist1 conditional mutant embryos (Fig. 5, B), which may explain why they were not always visible in the skeleton preparations located inside the incisor canal. The smaller size may be a simple adaptation to the smaller mandible. The cytodifferentiation of the mandibular incisors was comparable to that of the control wild type embryos, with evidence of dentin and enamel apposition (Fig. 5C, D). The cytodifferentiation of mandibular molar 1 was also normal in the mutant embryos (Fig. 5G, H) although this tooth was smaller than in the control embryos (Compare length between Figs. 5E and F), a likely adaptation to the smaller jaw. Cusp formation was altered, with the tooth surface appearing rounded and bulbous, a common characteristic of patients with Saethre-Chotzen syndrome (Gorlin et al., 2001). Molar agenesis has also been observed in some of these patients. Molars 2 and 3 were not observed in most of the Twist1cko/cko;Hand2-Cre embryos; a reduced-size secondary molar at the bud stage was sometimes seen. At this moment we cannot explain this phenotype and determine whether it is secondary to the smaller mandible or directly linked to Twist1 function in molar formation. Interestingly, the stellate reticulum cells failed to develop above the apical/occlusal surface of the first molar but the cells of the stratum intermedium that normally localize between the stellate reticulum and the inner enamel epithelium and ameloblasts were uncharacteristically numerous, producing a thickened layer inside the enamel organ (Fig. 5G, H). An ectopic blood vessel was commonly observed in the tooth organ of the mutant embryos. During the histological analysis of palate and tooth development, the presence of enlarged blood pooling was also observed in the proximal lower jaw of the conditional mutant embryos (Data not shown). Blood pooling has been observed in the Twist1 knockout embryos (Chen and Behringer, 1995), suggesting that our observed phenotype is a temporal continuity of the same defect.
Figure 5. The absence of Twist1 in neural crest cells causes molar defects.
Histological analysis of the dentition in E18.5 Twist1cko/+ (A, C, E and G) and Twist1cko/cko;Cre (B, D, F and H) conditional mutant embryos. A–D. In these sections through the incisor (i), the cytodifferentiation of the mutant incisor (B and D) appears normal since the apposition of enamel and dentin by the ameloblasts (am) and odontoblasts (od) is similar to that of the control incisors (A and C). However, the incisors in the mutant embryos are smaller than in the control embryos (compare the tooth length in A and B). E–H. In the first lower jaw molar (lm1) of the conditional mutant embryos, a similar reduction in size was observed (compare H and G). No second (lm2) or third molars were observed (F). Enamel and dentin apposition was observed in the molars (H), indicating the normal differentiation of the mesenchymal and inner enamel epithelium cells into odontoblasts and ameloblasts. However, the surface of the conditional mutant molar was unusually smooth with rounded cusps (H). The population of stellate reticulum (sr) cells was greatly reduced, apparently replaced by stratum intermedium (si) cells. An ectopic blood vessel (bv) is also visible above the molar. I, J. Expression analysis of the enamel knot marker Shh in the developing molar1 of E15.5–15.75 control (I) and mutant (J) embryos. Shh was only observed in the wild type control molar. Scale bar in A, B, E and F: 200 µm, and scale bar in C, D, G and H: 50µm. cl, cervical loop; pu, pulp; um1, upper molar 1; um2, upper molar 2.
To comprehend the phenotype, we analyzed tooth development in younger embryos. Histological analysis of the E13.5 embryos revealed that the dental laminas for mandibular molars 2 and 3 were induced (Data not shown) but then failed to progress further at E14.5 and E15.5. We also observed the presence of necrotic tissue in the molar tooth organ at the bud stage of the conditional mutant embryos at E13.5, which may explain the smaller tooth size (Data not shown). At E15.5, we observed in the first molar of the Twist1cko/cko;Hand2-Cre conditional mutant embryos that the stellate reticulum failed to develop normally and was replaced by what appears to be an increased population of stratum intermedium cells (Data not shown). However, the most striking change in molar development at that stage was the absence of an enamel knot. Analysis of Shh expression at that age or in slightly older embryos revealed the presence of a secondary enamel knot (Fig. 5I). In the mutant tooth organ, epithelial-derived cells did not express Shh, confirming the failed enamel knot formation (Fig 5J). This failure may explain the smoother molar surface and the absence of cusps at later stages of tooth development (Jernvall et al., 1994; Vaahtokari et al., 1996). Changes to the development of the enamel knots and the stellate reticulum are likely independent of the size reduction phenomenon mentioned above and were not observed in the upper molars (Data not shown).
Twist1 is needed for mandibular ossification
When we examined the mandible of E18.5 heterozygous Twist1cko/+;Hand2-Cre embryos, we noticed that the sub-molar region and diastema of the dentary appeared poorly ossified (Fig. 6A). The phenotype was confirmed by X-ray radiography in the lower jaws of E18.5 embryos and remained visible in the post-natal day 1 mice (Fig. 6B–D). On the digital X-ray images, we also observed poor mineralization of the molar alveolar ridge. This hypomineralization phenotype, visible in the heterozygous Twist1cko/+;Hand2-Cre embryos, was fully penetrant and can be used to accurately predict the genotype of the embryos in skeletal preparations. We measured the density of the dentary in the E18.5 embryos to confirm the phenotype. Our analyses revealed a significant (p<0.05, 1.20258E-05 and 2.21397E-06 respectively) reduction of approximately 28.8% in the heterozygous Twist1cko/+;Hand2-Cre embryos and about 60.1% in the mutant Twist1cko/cko;Hand2-Cre embryos (Fig.6E)
Figure 6. Twist1 is needed for mandibular ossification.
Intraoral (lingual) views of the mandible from E18.5 (A and B) and 1-day old (P1, C and D) control Twist1cko/cko or Twist1cko/+ and Twist1cko/+;Cre heterozygous conditional knockout embryos following Alcian blue/Alizarin red staining (A) or X-ray radiography (B–D). A, B. In the stained lower jaws, the alveolar margin for the molar teeth and lower portion of the dentary diastema appear hypomineralized in the heterozygous embryos (arrows in A and B). This hypocalcification is well evidenced in the reduced X-ray opacity (B). C, D. The hypomineralization of the mandible remained after birth (arrows in D), indicating that the phenotype does not result from delayed calcification of the intramembranous bone. E. Relative bone density of E18.5 control Twist1cko/cko or Twist1cko/+, heterozygous Twist1cko/+;Cre and mutant Twist1cko/cko;Cre dentaries. The differences between the controls and heterozygous or mutant mandibles are significant (p<0.05).
Twist1 is a known regulator of the obligatory ossification factor Runx2 by opposing its transcriptional activity in cranial sutures (Bialek et al., 2004). In the absence of Runx2, bones fail to mineralize (Komori et al., 1997; Otto et al., 1997). A reduction of Twist1 levels leads to the premature ossification and closure of the cranial sutures (Chen and Behringer, 1995; Bourgeois et al., 1998; Connerney et al., 2008), as evidenced by the clinical findings in Saethre-Chotzen patients (Howard et al., 1997; Krebs et al., 1997; Gorlin et al., 2001), contrary to the Fgf signaling controlling Runx2 activity (Aberg et al., 2004; Bialek et al., 2004; Connerney et al., 2008). However, it appears that this function is assumed by another bHLH in the mandible: Hand2 (Funato et al., 2009). In the absence of mandibular Hand2 expression, ossification occurs prematurely, rapidly depleting the pool of proliferating cells. In these embryos, the mandible appears porotic, resembling the phenotype of the heterozygous Twist1cko/+;Hand2-Cre embryos. Because of the known negative interaction between Runx2 and Twist1 (Bialek et al., 2004), we hypothesized that the same phenomenon occurs in the mandible of heterozygous embryos.
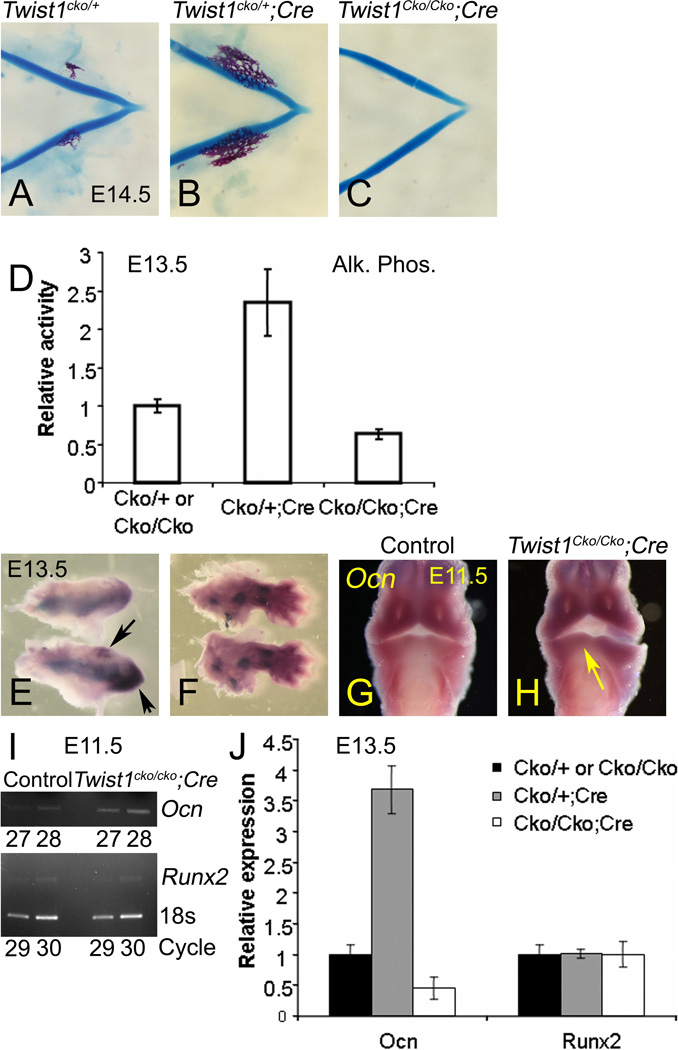
To test this hypothesis, we collected E14.5 embryos and stained their skeletons. At that age, mineral deposits stained with alizarin red are observed on the buccal side of the Meckel’s cartilage in the wild type control Twist1cko/+ and Twist1cko/cko embryos (Fig. 7A). In the lower jaw from heterozygous Twist1cko/+;Hand2-Cre embryos, ossification was more advanced (Fig.7B). Surprisingly, mineralization was not observed in the mutant lower jaw (Fig. 7C). Transformation of the Meckel’s cartilage at the symphysis was also observed in the mutant Twist1cko/cko;Hand2-Cre embryos. Before the symphysis, abnormal tapering of the cartilaginous rods was observed. At the mandibular symphysis, the anterior projection forming the mental protuberance was almost completely absent in the mutant embryos. We measured the alkaline phosphatase activity in the lower jaw to correlate the mineralization patterns. The activity was significantly higher (p=0.021) in the heterozygous embryos (Fig. 7D) and lower (p=0.004) in the mutant embryos. We also stained in whole mount the lower jaw of these control Twist1cko/+ and heterozygous Twist1cko/+;Hand2-Cre embryos for alkaline phosphatase activity and noticed enhanced activity in the distal mandible of the heterozygous embryos. We also observed the presence of cell clusters that appeared positive for the enzymatic activity (arrows in Fig. 7E, bottom). These clusters were not observed in the control embryos, in which the enzymatic activity remained confined along the Meckel’s cartilage (Fig. 7E, top). Similar activity was observed in the limbs of control (top) and heterozygous (bottom) (Fig. 7F), indicating that the increase in activity was specific to the mandible.
Figure 7. Twist1 controls the ossification of the mandible.
A–C. Mineralization in the lower jaw of E14.5 control Twist1cko/cko or Twist1cko/+ control (A), Twist1cko/+;Cre heterozygous (B) and Twist1cko/cko;Cre mutant (C) embryos. D. Standardized alkaline phosphatase activity in the lower jaws of E13.5 control, heterozygous or mutant embryos. E, F. Whole mount alkaline phosphatase activity in the lower jaws and limbs of E13.5 control (top) or heterozygous (bottom) embryos. In the lower jaws from heterozygous embryos (E), alkaline phosphatase staining activity is increased at the symphysis and in clusters of cells (black arrows). Limb buds (F) were used as staining controls. Differences were not observed in the developing limbs. G, H. Ocn gene expression analysis in E11.5 Twist1cko/cko;Cre conditional mutant embryos. I. Runx2 was normally expressed in the control and mutant embryos, as confirmed by the qRT-PCR results. Ocn expression was slightly increased in the mandibular arch of mutant embryos, as confirmed by the qRT-PCR results. J. Ocn and Runx2 expression analysis at E13.5 by qRT-PCR.
Interestingly, the delayed ossification in the mutant embryos did not correlate with the slight increase of the Runx2-dependent osteocalcin (Ocn) (Ducy et al., 1997) expression at E11.5 (Fig. 8H) in the distal mandibular arch. The increase was not accompanied by an increase in Runx2 expression at E11.5 (Fig. 7I). However, the increase in Ocn expression in the mutant embryos did not remain by E13.5. At that age, the inability to detect upregulation of Ocn expression correlated with the observed hypomineralization at E14.5 and lower alkaline phosphates activity at E13.5 (Fig. 7J). Correlation between Ocn and mineralization and alkaline phosphates activity was also clear in the heterozygous embryos (p=0.002). But these events occurred independently of Runx2 expression changes. Analysis of cell proliferation and death at E13.5, E14.5 and E15.5 did not reveal any significant differences (Data not shown). We concluded that the phenotype in the heterozygous embryos was likely due to the induction of premature ossification, which correlated with an increase of Runx2 transcriptional activity rather than an increase of Runx2 expression. These results indicate that Twist1, like Hand2, regulates Runx2 activity in the mandibular arch. However, more investigations are needed to determine whether the repression of Runx2 activity by both bHLH factors is concomitant and involves their mutual interactions or if these interactions with Runx2 occur at different periods of mandibular development.
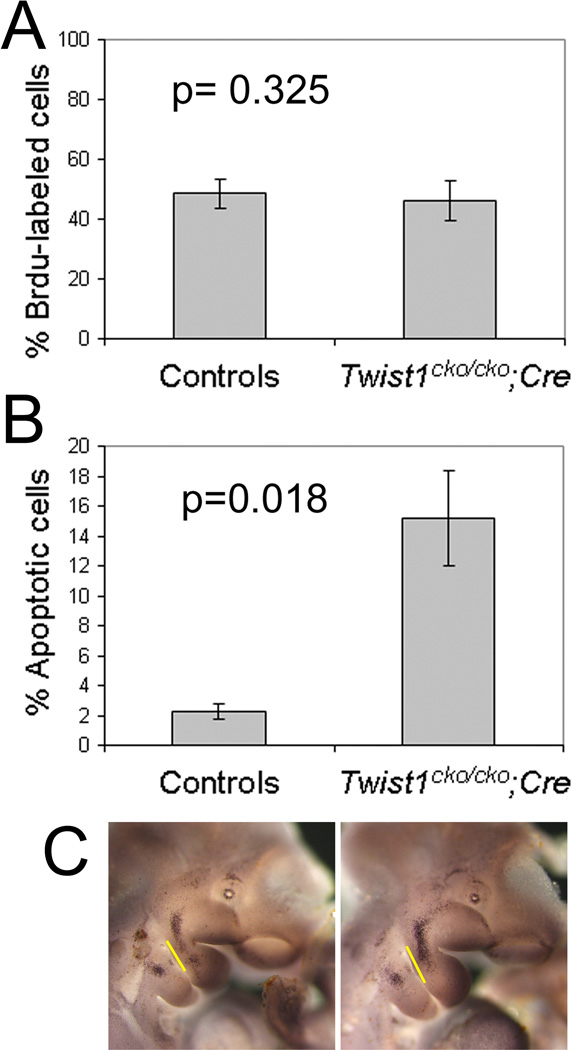
Figure 8. Twist1 is needed for the survival of the neural crest-derived mesenchyme.
Incidence, in percentage, of proliferating (A) and apoptotic (B) mesenchymal cells in the mandibular arch of control Twist1cko/cko or Twist1cko/+ and Twist1cko/cko;Cre conditional knockout embryos following Brdu immunodetection and TUNEL staining. The percentage of cells was calculated as the ratio of positively labeled nuclei over the total number of nuclei. A two-tailed t-test analysis was used to test the statistical significance. C. Whole mount TUNEL staining in E10.5 control (left) and conditional mutant (right) embryos. In the mutant embryo, more apoptotic cells are visible in the underlined proximal mandibular arch area.
Lack of Twist1in the mandibular arch affects neural crest cell survival but not proliferation
Since the lower jaw of the Twist1cko/cko;Hand2-Cre mutant embryos was smaller and the mandibular pharyngeal arch was slightly smaller at E10.5 (see Fig. 1), we examined whether the lack of Twist1 activity altered neural crest cell proliferation and survival at E10.5. Twist1 is a known factor regulating both cell proliferation and survival. The pregnant females were injected with Brdu before the embryos were collected, and a minimum of three sections from three control and mutant embryos were examined. The immunohistological detection of Brdu in the sections from the control and mutant embryos did not reveal any significant difference in the proliferation rate of the mandibular arch neural crest cells (p=0.325) (Fig. 8A and Data not shown).
The analysis of cell survival with the TUNEL method revealed a significant increase (p=0.018) in the ratio of apoptotic cell death in the conditional mutant embryos (15.18% ±3.21) compared to the control embryos (2.25% ± 0.54) (Fig.8B and Data not shown). The increase in cell death was more evident in the lateral portion of the mandibular arch (Fig. 8C and Data not shown), an area presumed to generate the mandibular ramus and associated processes (Ruest et al., 2003; Cerny et al., 2004; Lee et al., 2004). We also examined E11.5 embryos for evidence of increased apoptotic cell death in the mandibular arch. The phenotype was no longer apparent at that age (Data not shown). From these results, we concluded that the Twist1 function was essential for the survival of the neural crest cells populating the mandibular arch but not for their proliferation. This survival role is consistent with Twist1 function in migrating neural crest cells (Soo et al., 2002; Bildsoe et al., 2009) and in other cell populations, including the developing limb (Zuniga et al., 2002). However, this function appears to be limited in time, which was confirmed, as mentioned above, by our cell death analysis in E13.5, E14.5 and E15.5 lower jaws.
Gene expression changes in the Twist1cko/cko;Hand2-Cre mutant embryos
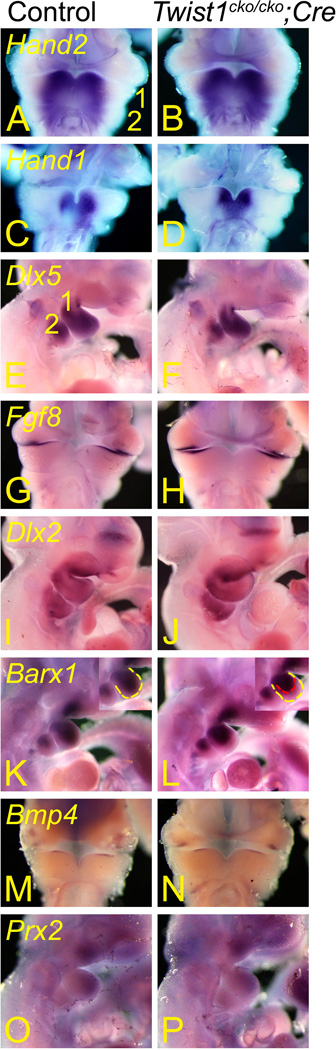
We examined different genes to determine the pathways affected by the loss of Twist1 function in the mandibular arch of E10.5 Twist1cko/cko;Hand2-Cre conditional mutant embryos. Twist1 is a bHLH transcription factor believed to interact with other bHLH factors (Barnes and Firulli, 2009), which would potentially be Hand1 and Hand2 in the mandibular arch (Ruest et al., 2004). Hand2 and Hand1 play a role in the development of the distal mandible (dentary) and incisors (Yanagisawa et al., 2003; Barbosa et al., 2007). The analysis of Hand2 and Hand1 expression did not reveal any changes in their respective expression patterns (Fig. 9A–D), which also correlated with the normal expression of Dlx5 and Dlx6 in the mandibular arch of the mutant embryos (Fig. 9E, F and Data not shown). Both Dlx5 and Dlx6 are involved in mandibular development, including the temporomandibular joint (Beverdam et al., 2002; Depew et al., 2002). These genes are regulated by endothelin signaling (Clouthier et al., 2000; Ruest et al., 2004), and Dlx6 directly regulates Hand2 expression (Charite et al., 2001). Endothelin signaling regulates Hand1 expression and represses Twist1 expression in the distal mandibular arch (Clouthier et al., 2000; Ruest et al., 2004; Ruest and Clouthier, 2009), potentially regulating the interacting bHLH partners for Twist1. The absence of changes in Dlx5 expression was unexpected since the earlier inactivation of the Twist1 conditional allele in forming neural crest cells with Wnt1-Cre and in the Twist1 knockout embryos showed decreased Dlx5 expression in the presumptive area where the mandibular arch produces the mandibular joint (Soo et al., 2002; Bildsoe et al., 2009).
Figure 9. Gene expression analysis in E10.5 Twist1cko/cko;Cre conditional mutant embryos.
Ventral and lateral views of E10.5 control Twist1cko/cko or Twist1cko/+ (A, C, E, G, I, K, M and O) and Twist1cko/cko;Cre conditional knockout (B, D, F, H, J, L, N and P) embryos following whole mount in situ hybridization analysis of Hand2 (A and B), Hand1 (C and D), Dlx5 (E and F), Fgf8 (G and H), Dlx2 (I and J), Barx1 (K and L), Bmp4 (M and N) and Prx2 (O and P).The expression pattern of these genes did not change in the mutant embryos with the exception of Barx1 in the distal mandibular arch (1). 2, pharyngeal arch 2.
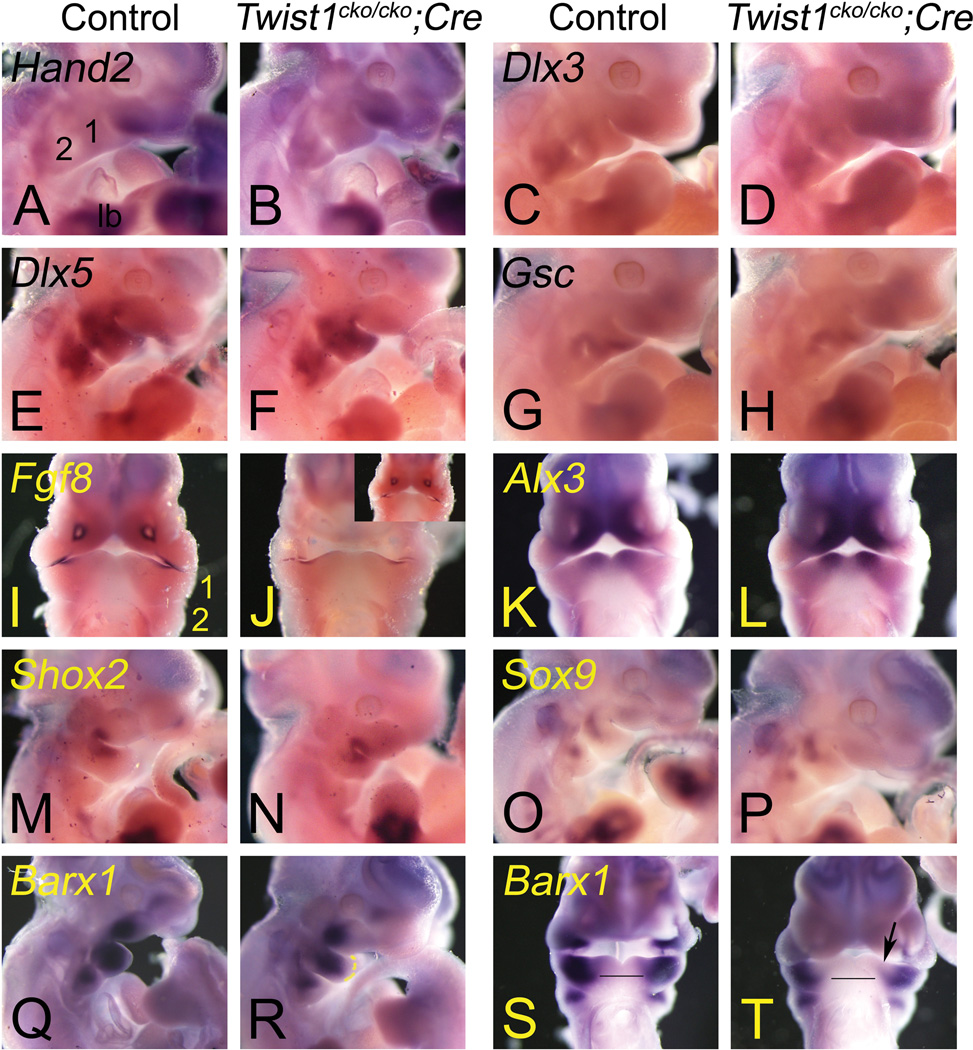
Twist1 and Fgf8 genetically interact during limb development but have opposing actions in cranial sutures (O'Rourke et al., 2002; Bialek et al., 2004; Firulli et al., 2005; Connerney et al., 2006; Connerney et al., 2008). Contrary to the loss of proximal Fgf8 expression in the Twist1−/− embryos (Soo et al., 2002), Fgf8 expression in our study was normal in the mutant embryos (Fig. 9G, H), suggesting that they did not genetically interact in the mandibular arch. The expression of the Fgf8-dependent gene Dlx2 (Thomas et al., 2000) in the arch mesenchyme was normal (Fig. 9I, J), confirming the absence of Twist1 effects on Fgf8 expression. Dlx2 expression was also normal in the pharyngeal arch ectoderm, indicating that Bmp4 signaling is likely normal in the conditional mutant embryos (Thomas et al., 2000). Dlx3 is another distal-less-related factor expressed in the mandibular arch in an area that may involve the development of the ramus (Ruest and Clouthier, 2009) and has recently been associated with the Tricho-dento-osseous syndrome (Choi et al., 2010); the expression of that gene was also normal (Data not shown). The same conclusion was also reached for the Aristaless-like factors Alx3 and Alx4 (Data not shown), two genes affected by the earlier inactivation of Twist1 in the neural crest cells or in the Twist1−/− embryos (Soo et al., 2002; Bildsoe et al., 2009).
Barx1 is a gene presumably involved in the development of the lower jaw articulation with the skull in zebrafish (Sperber and Dawid, 2008), but may also participate in molar development although mouse Barx1 mutant embryos appear to have gastric defects only (Kim et al., 2005; Mitsiadis and Drouin, 2008; MacKenzie et al., 2009). Surprisingly, its expression was not affected in the area presumed to produce the ramus but was unexpectedly reduced in the distal mandibular arch (Fig. 10K, L), the consequence of such a loss in that area are unknown. Proximal Barx1 expression is regulated by Fgf8 (Tucker et al., 1998) but the latter is normally expressed in the mutant embryos. Barx1 proximal expression is restricted by Bmp4 (Tucker et al., 1998; Mitsiadis and Drouin, 2008) but Bmp4 was normally expressed in our mutant embryos (Fig. 10M, N). Since both Fgf8 and Bmp4 are normally expressed, also confirmed by the normal Dlx2 expression, other mechanisms are also regulating Barx1 expression. We tested this hypothesis by examining Prx1 and Prx2 expression. When both genes are inactivated, Barx1 expression expands distally (Balic et al., 2009). However, both genes were normally expressed in the Twist1cko/cko;Hand2-Cre conditional mutant embryos (only Prx2 shown, Fig. 10O, P). These results suggest that yet another mechanism regulates Barx1 expression.
Figure 10. Gene expression analysis in E11.5 Twist1cko/cko;Cre conditional mutant embryos.
Ventral and lateral views of E11.5 control Twist1cko/cko or Twist1cko/+ (A, C, E, G, I, K, M, O, Q and S) and Twist1cko/cko;Cre conditional knockout (B, D, F, H, J, L, N, P, R, and T) embryos following whole mount in situ hybridization analysis of Hand2 (A and B), Dlx3 (C and D), Dlx5 (E and F), Gsc (G and H), Fgf8 (I and J), Alx3 (K and L), Shox2 (M and N), Sox9 (O and P) and Barx1 (Q–T).The expression pattern of these genes did not change in the mutant embryos with the exception of Barx1 in the distal mandibular arch (1). In the ventral view (S and T), the distance between the expression domains on each side of the lower jaw is wider in the mutant embryos (compare the control line in S and T). The expression is also reduced in the distal oral aspect of the mandibular arch (arrow in T). 2, pharyngeal arch 2.
We also examined the expression of goosecoid (gsc), a gene involved in mandibular development (Rivera-Perez et al., 1995; Yamada et al., 1995). Its expression appeared normal in the mandibular arch (Data not shown) albeit a previous report indicating the contrary (Soo et al., 2002). We came to the same conclusion for Runx2 (Data not shown), a factor physically interacting with Twist1 (Bialek et al., 2004) that was shown to be affected by the early loss of Twist1 expression in the neural crest cells (Bildsoe et al., 2009). Runx2 appeared to be mostly expressed in the maxillary prominence at E10.5. Its expression is weak at that age in the mandibular arch, but no differences were observed in the mandibular arch at E11.0 or E11.5 (Fig. 8I).
Since the inactivation of Twist1 in neural crest cells with Hand2-Cre occurs approximately one day later than with Wnt1-Cre, we examined whether the absence of gene expression changes at E10.5 was due to that delay. Our analyses of Hand2, Gsc, Dlx3, Dlx5, Fgf8 and Alx3 expression at E11.5 did not reveal any differences between the control and mutant embryos (Fig. 11A–L). Hand2 expression was greatly reduced at that age while robustly expressed in the limb buds, correlating with the eventual restricted expression of the gene in the distal mandible but not in the molar region or ramus at E13.5 (Ruest et al., 2003). This reduction restricts the pool and timing of likely dimerizing partners for Twist1 but suggests also that Hand2 can interact with Twist1 in the proximal mandible, where ossification defects were observed. When we examined Barx1 expression at the same age, the previously observed change in the distal mandibular arch of E10.5 mutant embryos remained apparent (Fig. 11Q–T). We also examined the expression of Shox2 and Sox9, two genes involved with the ramus/temporomandibular joint formation (Shibata et al., 2006; Gu et al., 2008; Hinton et al., 2009). The expression of these genes appeared normal in the E11.5 mutant embryos (Fig. 11M–P). Taken together with those of Barx1, Fgf8, Dlx2 and Dlx3 expression, these results suggest that other genetic pathways are in place to regulate early ramal and articulation development. However, during our gene expression analyses, we noticed changes in the expression of Hand2 and Fgf8 in the anterior limb buds of E10.5 embryos, correlating with the zeugopod and autopod defects we observed in the mutant embryos (Data not shown). Together, these results led us to conclude that the absence of changes at E10.5 was not because of a delay and that the mandibular phenotype does not involve genes appearing to be controlled by Twist1 in early neural crest cell development.
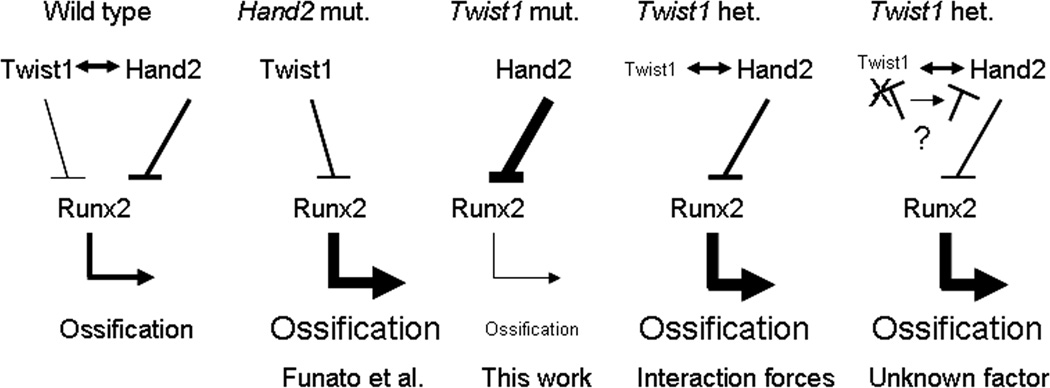
Figure 11. Model of ossification control by bHLH factors.
Thickness of line and bolding represent the strength of the interactions and effects on ossification. As such, based on Funato’s work, Hand2 is a better repressor than Twist1. In the wild type embryos, Hand2 and Twist1 are interacting, limiting the pool of factors repressing Runx2 activity. Cumulative repression is insufficient, allowing normal ossification. In the Hand2 mutant embryos (Funato et al.), cumulative repression on Runx2 is decreased, increasing ossification. In Twist1 mutants, more Hand2 molecules are free to repress Runx2 activity, decreasing ossification. In the heterozygous Twist1 embryos, two models are envisaged. In the interaction forces model (Twist1/Hand2 interactions favored), decreasing Twist1 dosage would only maintain its interaction with Hand2 but not its repression on Runx2, decreasing the total repression on Runx2 activity. In the alternative model with an unknown factor, decreasing amounts of Twist1 would transfer the repression exerted by this unknown factor unto Hand2, decreasing repression of Runx2. However, this alternative model cannot explain the decreased ossification in the Twist1 mutant embryos.
Discussion
In this project, we investigated the functions of the Twist1 gene during mandibular development. We specifically inactivated the gene in the neural crest cells populating the mandibular pharyngeal arch using the Hand2-Cre mouse line. The Cre expression in this line appears in the mandibular arch at E9.25–9.5 when the neural crest cells are still migrating into the arch and beginning to replicate (Ruest et al., 2003). This line allows inactivating Twist1 expression in all the neural crest cells forming the lower jaw by E10.5. This specific conditional inactivation of the gene in the mandibular arch produces a slightly different phenotype than when Twist1 is inactivated in forming or newly formed neural crest cells using Wnt1-Cre or other lines (Bildsoe et al., 2009). The ramus and its associated processes were greatly affected in our conditional mutant embryos. A previous study indicated that Twist1 likely functions cell autonomously (Soo et al., 2002), and the present study confirmed this aspect since the defects were restricted to locations where the Hand2-regulated Cre was expressed, with the exception of tooth development (discussed below). Overall, the hypoplasia and structural defects were less severe compared to those observed in the conditional mutants with Wnt1-Cre (Bildsoe et al., 2009) because more neural crest cells were present in the mandibular arch. Our results more likely reflect the functions of the Twist1 gene during mandible formation while defects caused by the earlier inactivation are influenced by other functions of the gene. These earlier functions include the important role Twist1 plays during neural crest cell formation, migration and survival (Soo et al., 2002).
The present study revealed Twist1 functions during lower jaw development, some of which were unknown. Twist1 role in neural crest cell survival was known, but we did not anticipate that this function was only restricted to a short period of time, as shown by our results. Analysis in E11.5, 13.5, 14.5 and 15.5 embryos did not reveal any significant differences. At E10.5, apoptotic cells were mostly restricted in the proximal area of the mandibular arch where Hand2-Cre fate mapping results indicate that these cells are involved in the development of the malleus, tympanic ring and ramus (Ruest et al., 2003; Ruest et al., 2004). These fate mapping results correlate with the observed mandibular phenotype and could explain why the malleus is malformed, tympanic ring is missing, ramus is practically absent and Meckel’s cartilage truncated. Twist1 was presumed to have a function in the formation of the Meckel’s cartilage but its roles in its full-length development and shape were unexpected. The transformation at the symphysis where Twist1 is not expressed and was not anticipated and supports the hypothesis of regional compartmentalization of the mandibular arch by bHLH factors during early lower jaw development (Ruest et al., 2004), with changes likely affecting the function of other bHLH factors.
Since Twist1 is expressed in the developing dentition, the gene is expected to play a role in tooth development (Rice et al., 2005). However, its function was unclear due to the early death of the knockout embryos (Chen and Behringer, 1995). Our study revealed that Twist1 participates in the reciprocal interactions between the dental ectoderm and dental mesenchyme, affecting the development of ectoderm-derived structures like the enamel knot and stellate reticulum cells, which influence cusp formation. However, dentin and enamel formation was normal. Future studies may investigate reciprocal signaling such as the expression of ectodermal Fgf4 and 9 and mesenchymal Fgf3 and 10.
Based on Bildsoe’s study, Twist1 function in ramal formation and dentary length was suspected but its role in the dentary ossification was unexpected since Runx2 activity is presumably controlled by Hand2 in that region (Funato et al., 2009), and earlier inactivation of Twist1 reduces the expression of Runx2 (Bildsoe et al., 2009). In our study, Runx2 expression was unaltered. This function will be discussed below.
Effect of timing on gene expression
The differences in the Twist1 inactivation timing between various studies had a tremendous effect on gene expression. Several genes whose expression in the mandibular arch was affected by the earlier conditional inactivation of Twist1 were normally expressed when Twist1 was inactivated only in the mandibular arch. Since a relatively similar mandibular phenotype was observed in our embryos, this surprising finding suggests that previously described gene expression changes (Soo et al., 2002; Bildsoe et al., 2009) are not related to Twist1 role in mandible development but rather to its regulatory role in neural crest cell formation, migration and homing to the mandibular arch, affecting the subsequent development of these cells. Further, there is no or little evidence that Twist1 directly binds the cis-regulatory elements of these previously described genes, though they may still be part of genetic pathways influenced by Twist1 in neural crest cells.
In the study by Bildsoe and colleagues (2009), the number of neural crest cells reaching and populating the mandibular arch was greatly reduced, as evidenced by its small size, corresponding to what was previously observed in the Twist1−/− embryos (Soo et al., 2002). The reduction in the critical mass caused by earlier inactivation apparently affected essential molecular pathways in the mandibular arch. In particular, the reciprocal signaling influences between the neural crest cells and the surrounding tissues (arch ectoderm, pharyngeal pouch endoderm and core paraxial mesoderm) may have been affected, masking the true function of Twist1 during mandibular development. This phenomenon is especially evident in our molar phenotype where ectoderm-derived structures were affected but not crest cell-derived structures, the only defect showing that Twist1 does not always function cell autonomously. In our study, we failed to identify early genes, particularly transcription factors that are clearly regulated by Twist1 and could characterize the observed phenotype. While Barx1 expression was altered, the effects of this distally restricted expression are difficult to judge since Barx1 mutant embryos do not appear to have a mandibular or tooth phenotype (Kim et al., 2005) and genes controlling its early expression boundaries (Fgf8, Bmp4 and Prx genes) were normally expressed. These normally expressed controlling genes suggest that another yet unidentified mechanism regulates Barx1 expression. Other changes observed were related to the control of ossification, mainly the absence of negative regulation of Runx2 by Twist1. These results emphasize the need to identify the genes and miRNA regulated by the transcriptional activity of Twist1 in the mandibular arch. However, we anticipate that Twist1 positive or negative transcriptional activity may depend on its interacting partners.
Twist1 interactions
Twist1 is mostly viewed as a transcriptional repressor (Spicer et al., 1996; Yin et al., 1997; Sharif et al., 2006; Vesuna et al., 2008). As a bHLH factor, Twist1 likely interacts with other bHLH factors (Barnes and Firulli, 2009; Franco et al., 2010). These interactions may be modified by the availability of interacting partners and the phosphorylated state of Twist1 itself and may change the E boxes bound by the dimer pairs. In the mandibular arch, Twist1 could form homodimers or heterodimers with class I bHLH (E2A gene products E12 and E47), class II bHLH factors Hand1, Hand2 or Twist2 (Dermo1), or the inhibitory class V bHLH Id proteins. Interactions between class II factors are important to regulate their functions and a shift in the balance between these factors can alter cell response (Firulli et al., 2000; Firulli et al., 2005; Barbosa et al., 2007; Firulli and Conway, 2008). Changing bHLH gene dosage is expected to alter these interactions.
In our study, we did not observe any changes in the expression of Hand1, Hand2 or E2A gene products. Although Twist1 and Twist2 are highly conserved proteins expressed in similar tissues and may share some in vitro functions, the consensus is that these factors have different functions and do not interact in vivo (Franco et al., 2010; Tukel et al., 2010), thus excluding Twist2 as the interacting partner for Twist1 in the mandibular arch. Hand1 interaction with Twist1 is likely quite limited, based on their respective expression pattern at E10.5 and the incisor phenotypes (this study and (Ruest et al., 2004; Barbosa et al., 2007)), leaving Hand2 as the likely interacting class II factor for Twist1. The expression of these two factors largely overlaps in the mandibular arch (Ruest et al., 2004); they have been shown to interact during limb development, although having opposing functions (Firulli et al., 2005; Firulli et al., 2007; Zhang et al., 2010). However, Twist1 interactions with similar genetic pathways could have opposing results, depending on the location and timing, such as Twist1 and Fgf8 during limb (O'Rourke et al., 2002; Zuniga et al., 2002), cranial sutures (Bialek et al., 2004; Connerney et al., 2008) and tooth (Thesleff, 2006) development. If they interact, Twist1 and Hand2 likely have redundant functions, and we can expect similar phenotypes when their respective genes are inactivated, toward which our study seems to point.
The length of the mandibular dentary (particularly its diastema) is reduced in the Hand2 branchial arch enhancer mutant pups (Yanagisawa et al., 2003), a phenotype observed in our conditional Twist1 mutant embryos. Further, the reduction in the size of the mandibles in both mutants did not involve the tongue, causing a secondary cleft palate. This chain of events is a clinical characterization of the Pierre-Robin sequence (Ricks et al., 2002). Interestingly, recent studies have indicated that HAND2 is a candidate gene for this condition (Huang et al., 2002; Kaalund et al., 2008; Rossi et al., 2009). If normal Hand2 functions involve its interaction with Twist1 during mandibular development, abrogating this interaction in our conditional mutants is expected to produce results similar to the inactivation of the Hand2 gene, thus creating the Pierre-Robin sequence-like phenotype of our embryos. The hypomineralization of the mandible in the Hand2 branchial arch enhancer mutants (Funato et al., 2009) and our heterozygous embryos are further evidence of the possible interaction between the two factors. These results also suggest that their interaction involves the regulation of Runx2 activity, a function associated with both factors (Bialek et al., 2004; Funato et al., 2009). Together, these similar results suggest that Twist1 and Hand2 interact and cooperate during mandibular development. Our preliminary results with the Dynamin 3 opposite strand (Dmn3os) promoter (Loebel et al., 2005) confirm their interaction (Data not shown). In these experiments, Hand2 did not repress Twist1 transcriptional activation of Dmn3os and likely compensate, in the mutant embryos, for the loss of Twist1 to maintain the expression of the microRNAs miR-199a and miR-214 encoded within Dmn3os (Lee et al., 2009). However, more investigations are needed to confirm Hand2-Twist1 interaction and identify other commonly regulated genes or miRs.
Control of mandibular ossification by bHLH factors
Our results indicate that Twist1 and Hand2 interact and these interactions likely control mandibular ossification. However, it appears that Hand2 is a better repressor of Runx2 activity (Funato et al., 2009). This complicates our interpretation of the results because both Hand2 and Twist1 are involved in the development of the area where premature ossification was observed and, Hand2 should compensate for the loss of Twist1. At this moment we cannot fully comprehend the phenotype and several mechanisms are likely involved. One of the possible mechanisms altered by gene dosage is the titration/balance of these two bHLH factors by their mutual interactions (Firulli et al., 2000; Barbosa et al., 2007; Barnes and Firulli, 2009). In Hand2 absence, since Twist1 repression of Runx2 activity is weaker (Funato et al., 2009), more Runx2 remains free thus, promoting ossification (Fig 11.). This gene dosage mechanism would predict that by liberating Hand2 from Twist1 interaction, more Hand2 would be available to repress Runx2 activity, consequently diminishing ossification as we observed in our mutant embryos. However, these models based on gene dosage cannot explain the observed phenotype in the heterozygous embryos. Evaluation of the interaction/dissociation forces between Twist1, Hand2 and Runx2 may help elucidate this issue. However, based on the observed phenotypes in Funato’s study, our present work and the better capacity for Hand2 to repress Runx2 activity, we can anticipate that the forces between Twist1 and Runx2 are weaker than between Twist1 and Hand2. Predictions based on this hypothesis suggest that the loss of one Twist1 allele would change the balance between the factors. In this case maintaining the interactions between Twist1 and Hand2 but freeing Runx2 from Twist1 and preventing Hand2 to interact with Runx2. Consequently, this balance shift would cause the premature ossification that we observed in the heterozygous embryos (Fig. 11). The same model could explain the absence of early ossification in our Twist1 mutant embryos and why the premature ossification defect was observed in the homozygous but not the heterozygous Hand2 pharyngeal arch enhancer mutant embryos (Funato et al., 2009). Alternatively, if another factor is preferably and negatively interacting with Twist1 but could also interact with Hand2, decreasing the amount of available Twist1 would transfer the repression toward Hand2, allowing Runx2 transcriptional activity (Fig. 11). However, this model could not explain the decreased ossification in the mutant embryos thus, the least favored model. More studies are needed to evaluate the interactions between Twist1 and Hand2 and their roles in mandibular development.
In conclusion, we identified in this study that Twist1 regulates the development of the ramal elements of the mandible, including the condyle, the length and ossification of the dentary. Its function in mandibular development involves the interaction with Hand2. We also identified the role of Twist1 in molar cusp formation and the genesis of posterior molars, consistent with its function in Saethre-Chotzen syndrome. However, more work is needed to fully comprehend its function in tooth development and determine whether interactions with other bHLH are needed to fully control mandibular development.
Experimental Procedures
Mice, breeding and genotyping
The Twist1 conditional mutant mice (Twist1cko/cko) and Hand2-Cre (formerly “dHAND-Cre”) transgenic mouse strains have been described previously (Ruest et al., 2003; Chen et al., 2007). Twist1cko/cko mice were bred with Hand2-Cre transgenic mice to generate Twist1cko/+;Hand2-Cre animals bred with Twist1cko/cko mice to generate Twist1cko/cko;Hand2-Cre embryos. Mice and embryos were genotyped by PCR amplification using genomic DNA obtained from tail biopsies or yolk sacs. The Twist1cko and Hand2-Cre genotyping was performed using primers previously described (Ruest et al., 2003; Chen et al., 2007; Bildsoe et al., 2009; Ruest and Clouthier, 2009). The protocol for the use of animals was approved by the Institutional Animal Care and Use Committee at Baylor College of Dentistry, and the animals were euthanized following NIH guidelines. A minimum of 3 specimens were examined or sectioned to ensure the repeatability of the results.
Skeleton staining
Skeleton staining was preformed as described earlier (Ruest et al., 2004; Ruest and Clouthier, 2009). Skeletons were analyzed and photographed using an Olympus SZX16 stereomicroscope fitted with a digital camera. Control n=16; Twist1cko/+;Hand2-Cre n=38 and mutants Twist1cko/cko;Hand2-Cre n=18.
Histology
Histological analysis was performed as previously described (Ruest et al., 2004; Ruest and Clouthier, 2009). Sections were stained with hematoxylin and eosin before mounting for examination. They were then examined and photographed with an Olympus BX51 microscope fitted with a DP72 digital camera. N=3 for each geneotype.
Cell proliferation and apoptosis analysis
Cell proliferation analysis was performed through the incorporation of bromodeoxyuridine (BrdU) as previously reported (Abe et al., 2007) with a Zymed’s BrdU Staining Kit. Pregnant mice were injected intraperitoneally with 200 mg/kg body weight of BrdU one hour before embryo collection. The percentage of proliferating cells was calculated as the ratio of positively labeled nuclei over the total nuclei. N=3 for each genotype.
The detection of apoptotic cells was performed using the Roche’s TUNEL assay In Situ Death Detection Kit (TMR Red) using sections consecutive to those used for the cell proliferation assay. The incidence of apoptosis was calculated as the ratio of positively labeled nuclei to the total number of nuclei. Whole mount detection of apoptotic cells in embryos was performed by the vital dye exclusion assay using Trypan blue (Trumpp et al., 1999). Freshly collected embryos were incubated in a 1:1 PBS and Trypan blue mix for 30 minutes. After rinsing in PBS, embryos were rapidly photographed as described above. TUNEL staining with HRP-conjugated streptavidin was also used.
In situ hybridization
Whole mount and sectional in situ hybridization analysis of gene expression was performed as previously indicated using digoxigenin (DIG)-labeled riboprobes (Ruest et al., 2004; Ruest and Clouthier, 2009). The following genes were analyzed: Twist1, Hand1, Hand2, Dlx2, Dlx3, Dlx5, Dlx6, Fgf8, Bmp4, Alx3, Alx4, Barx1, Runx2, Prx1, Prx2, Shh, Sox9, Shox2, Ocn and Gsc. The embryos or sections were photographed as above.
X-ray radiography
Digital X-ray radiographs of the mandibles and forelimbs were made using the Specimen Radiography System (Faxitron X-Ray) following the manufacturer’s recommendations. The specimens were dissected prior to radiographic imaging. Bone density was measured using the manufacturer’s software on images containing the control (n=8), heterozygous (n=8) and mutant (n=3) mandibles. The results were calculated as the ratio of the raw density of the experimental sample over the density of the control mandible on the same image.
Alkaline phosphatase activity
The lower jaw complex was dissected from E13.5 embryos and bisected in the middle with one half used for whole mount staining and the other for quantifying the enzymatic activity. For the whole mount staining, mandibles were fixed in 4% paraformaldehyde, rinsed twice in 0.2M sodium phosphate buffer pH7.3 (supplemented with 2mM magnesium chloride, 0.02% NP40 and 0.01% sodium deoxycholate) and then processed for staining as for whole mount in situ with BCIP and NBT. To measure the enzymatic activity, the samples were lysed in 200 µl of reaction buffer (100mM Tris pH9.5, 100mM sodium chloride, 20mM magnesium chloride and 1% triton). For the assay, 30 µl of the cleared lysate was added to 900 µl of the reaction buffer supplemented with 5mM p-nitrophenyl phosphate and incubated for 60 minutes at 37°C. Each sample was assayed in triplicate, and the reactions were stopped by the addition of 70 µl of 10N sodium hydroxide. The optical density was read at 405 nm, and the specific enzymatic activity was determined by dividing the resulting density readings by the protein concentration of each sample. Protein concentration was measured using the modified Bradford assay and compared to a bovine serum albumin curve. (controls n=13; heterozygotes n=8 and mutants n=4).
Real-time PCR
Osteocalcin (Ocn) and Runx2 expression were analyzed in dissected E11.5 and E13.5 mandibular tissues by qRT-PCR using a BioRad CFX96 apparatus. Micro-RNAs miR-199a and miR-214 were analyzed from E13.5 mandibular explants. Total RNA was obtained using the Trizol method (Mraz et al., 2009) and treated with DNase before the reverse transcription reaction. Amplification was measured using the Finnzymes’ DyNamo SYBR green qPCR reagents and compared to 18s rRNA (Ambion). Oligonucleotide primers were (5’ to 3’) for Ocn: CCGGGAGCAGTGTGAGCTTA and TAGATGCGTTTGTAGGCGGTC; for Runx2: GTGCGGTGCAAACTTTCTCC and CTACCAGCCTCACCATAC; for miR-199a (GI: 262205950): AAGCTTCAGGAGATCCTGCTC and CCTTGCCCAGTCTAACCAAT; for miR-214 (GI: 262205879): CTGGCTGGACAGAGTTGTCA and AGGCTGGGTTGTCATGTG. These primers produced a single product as confirmed by their respective single-peak dissociation curve and gel electrophoresis. Each sample was analyzed in triplicate and n=3 or more for each genotype.
Cloning of Dmn3os promoter and luciferase assays
The Dynamin 3 opposite strand (Dmn3os) promoter was cloned using the previously described primers (Lee et al., 2009) to which a SacI or HindIII site were added for cloning into the pGL4.26 luciferase vector (Promega). High fidelity Pfu Turbo DNA polymerase (Agilent) was used to generate the insert that was confirmed by restriction digestions and sequencing. For the luciferase assays, HEK-293E cells were transfected with the pDmn3os-luc vector along with an empty mammalian expression vector or with vectors encoding for Twist1 and Hand2. A con-transfected vector encoding for the lacZ gene product β-galactosidase was used for transfection control and standardization. Luciferase assays were performed following the manufacturer’s instructions (Promega). Each sample was analyzed in triplicate and n=4 for each gene.
Western blotting
Protein analyses were performed as previously described (Ruest et al., 2002) using proteins extracted from dissected E14.5 mandibles (n=3 for each genotype). The phospho-PTEN ser380 (9551), phospho-Akt ser473 (9271S) and Akt (4691S) antibodies were used following the vendor’s instructions (Cell Signaling).
Statistical analysis
A two-tailed Student’s t-test analysis was used to evaluate the statistical significance.
Data not shown
Data indicated as “Data not shown” were presented and reviewed by the reviewers.
Highlights.
Twist1 regulates the development of the mandibular ramus
Twist1 regulates molar development and cusp formation
Mandibular hypoplasia causes cleft palate following a Pierre-Robin-like sequence
Twist1 controls the ossification of the mandible with Hand2
Acknowledgements
This work was supported by NIH U24 DE16472 and a research development grant from the TAMHSC Office of the Vice-President for Research and Graduate Studies (LBR), as well as fellowships from NIH T32-DE018380 (YZ) and the Baylor Oral Health Foundation summer student research program (ELB and GRK). The authors thank Drs. Yiping Chen, Mina Mina and Jerry Feng for probes, Chunlin Qin for the qRT-PCR primers, Hiromi Yanagisawa and Bob Lu for the mammalian expression vectors, David Clouthier for the Hand2-Cre mice and Richard Behringer and Rena D’Souza for the Twist1 conditional knockout mice. We also thank Mr. Eric Jansen for his technical help and Ms. Jeanne Santa Cruz for her editorial help.
References
- Abe M, Ruest LB, Clouthier DE. Fate of cranial neural crest cells during craniofacial development in endothelin-A receptor-deficient mice. Int J Dev Biol. 2007;51:97–105. doi: 10.1387/ijdb.062237ma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberg T, Wang XP, Kim JH, Yamashiro T, Bei M, Rice R, Ryoo HM, Thesleff I. Runx2 mediates FGF signaling from epithelium to mesenchyme during tooth morphogenesis. Dev Biol. 2004;270:76–93. doi: 10.1016/j.ydbio.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Balic A, Adams D, Mina M. Prx1 and Prx2 cooperatively regulate the morphogenesis of the medial region of the mandibular process. Dev Dyn. 2009;238:2599–2613. doi: 10.1002/dvdy.22092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AC, Funato N, Chapman S, McKee MD, Richardson JA, Olson EN, Yanagisawa H. Hand transcription factors cooperatively regulate development of the distal midline mesenchyme. Dev Biol. 2007;310:154–168. doi: 10.1016/j.ydbio.2007.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes RM, Firulli AB. A twist of insight - the role of Twist-family bHLH factors in development. Int J Dev Biol. 2009;53:909–924. doi: 10.1387/ijdb.082747rb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron F, Woods C, Kuhn K, Bishop J, Howard MJ, Clouthier DE. Downregulation of Dlx5 and Dlx6 expression by Hand2 is essential for initiation of tongue morphogenesis. Development. 2011;138:2249–2259. doi: 10.1242/dev.056929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverdam A, Merlo GR, Paleari L, Mantero S, Genova F, Barbieri O, Janvier P, Levi G. Jaw transformation with gain of symmetry after Dlx5/Dlx6 inactivation: mirror of the past? Genesis. 2002;34:221–227. doi: 10.1002/gene.10156. [DOI] [PubMed] [Google Scholar]
- Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, Wu H, Yu K, Ornitz DM, Olson EN, Justice MJ, Karsenty G. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- Bildsoe H, Loebel DA, Jones VJ, Chen YT, Behringer RR, Tam PP. Requirement for Twist1 in frontonasal and skull vault development in the mouse embryo. Dev Biol. 2009;331:176–188. doi: 10.1016/j.ydbio.2009.04.034. [DOI] [PubMed] [Google Scholar]
- Bourgeois P, Bolcato-Bellemin AL, Danse JM, Bloch-Zupan A, Yoshiba K, Stoetzel C, Perrin-Schmitt F. The variable expressivity and incomplete penetrance of the twist-null heterozygous mouse phenotype resemble those of human Saethre-Chotzen syndrome. Hum Mol Genet. 1998;7:945–957. doi: 10.1093/hmg/7.6.945. [DOI] [PubMed] [Google Scholar]
- Cerny R, Lwigale P, Ericsson R, Meulemans D, Epperlein HH, Bronner-Fraser M. Developmental origins and evolution of jaws: new interpretation of "maxillary" and "mandibular". Dev Biol. 2004;276:225–236. doi: 10.1016/j.ydbio.2004.08.046. [DOI] [PubMed] [Google Scholar]
- Chai Y, Maxson RE., Jr Recent advances in craniofacial morphogenesis. Dev Dyn. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- Charite J, McFadden DG, Merlo G, Levi G, Clouthier DE, Yanagisawa M, Richardson JA, Olson EN. Role of Dlx6 in regulation of an endothelin-1-dependent, dHAND branchial arch enhancer. Genes Dev. 2001;15:3039–3049. doi: 10.1101/gad.931701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YT, Akinwunmi PO, Deng JM, Tam OH, Behringer RR. Generation of a Twist1 conditional null allele in the mouse. Genesis. 2007;45:588–592. doi: 10.1002/dvg.20332. [DOI] [PubMed] [Google Scholar]
- Chen ZF, Behringer RR. twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 1995;9:686–699. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- Choi SJ, Song IS, Feng JQ, Gao T, Haruyama N, Gautam P, Robey PG, Hart TC. Mutant DLX 3 disrupts odontoblast polarization and dentin formation. Dev Biol. 2010;344:682–692. doi: 10.1016/j.ydbio.2010.05.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier DE, Williams SC, Yanagisawa H, Wieduwilt M, Richardson JA, Yanagisawa M. Signaling pathways crucial for craniofacial development revealed by endothelin-A receptor-deficient mice. Dev Biol. 2000;217:10–24. doi: 10.1006/dbio.1999.9527. [DOI] [PubMed] [Google Scholar]
- Connerney J, Andreeva V, Leshem Y, Mercado MA, Dowell K, Yang X, Lindner V, Friesel RE, Spicer DB. Twist1 homodimers enhance FGF responsiveness of the cranial sutures and promote suture closure. Dev Biol. 2008;318:323–334. doi: 10.1016/j.ydbio.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connerney J, Andreeva V, Leshem Y, Muentener C, Mercado MA, Spicer DB. Twist1 dimer selection regulates cranial suture patterning and fusion. Dev Dyn. 2006;235:1345–1357. doi: 10.1002/dvdy.20717. [DOI] [PubMed] [Google Scholar]
- Creuzet S, Couly G, Le Douarin NM. Patterning the neural crest derivatives during development of the vertebrate head: insights from avian studies. J Anat. 2005;207:447–459. doi: 10.1111/j.1469-7580.2005.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creuzet S, Couly G, Vincent C, Le Douarin NM. Negative effect of Hox gene expression on the development of the neural crest-derived facial skeleton. Development. 2002;129:4301–4313. doi: 10.1242/dev.129.18.4301. [DOI] [PubMed] [Google Scholar]
- D'Souza RN, Ruest LB, Hinton RJ, Svoboda KKH. Development of the craniofacial complex. In: Bronner F, Farach-Carson MC, Roach HI, editors. Topics in Bone Biology: Bone and Development. London: Springer-Verlag; 2010. pp. 153–181. [Google Scholar]
- Depew MJ, Lufkin T, Rubenstein JL. Specification of jaw subdivisions by Dlx genes. Science. 2002;298:381–385. doi: 10.1126/science.1075703. [DOI] [PubMed] [Google Scholar]
- Depew MJ, Simpson CA. 21st century neontology and the comparative development of the vertebrate skull. Dev Dyn. 2006;235:1256–1291. doi: 10.1002/dvdy.20796. [DOI] [PubMed] [Google Scholar]
- Depew MJ, Simpson CA, Morasso M, Rubenstein JL. Reassessing the Dlx code: the genetic regulation of branchial arch skeletal pattern and development. J Anat. 2005;207:501–561. doi: 10.1111/j.1469-7580.2005.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- Firulli AB, Conway SJ. Phosphoregulation of Twist1 provides a mechanism of cell fate control. Current medicinal chemistry. 2008;15:2641–2647. doi: 10.2174/092986708785908987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli BA, Hadzic DB, McDaid JR, Firulli AB. The basic helix-loop-helix transcription factors dHAND and eHAND exhibit dimerization characteristics that suggest complex regulation of function. J Biol Chem. 2000;275:33567–33573. doi: 10.1074/jbc.M005888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli BA, Krawchuk D, Centonze VE, Vargesson N, Virshup DM, Conway SJ, Cserjesi P, Laufer E, Firulli AB. Altered Twist1 and Hand2 dimerization is associated with Saethre-Chotzen syndrome and limb abnormalities. Nat Genet. 2005;37:373–381. doi: 10.1038/ng1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli BA, Redick BA, Conway SJ, Firulli AB. Mutations within helix I of Twist1 result in distinct limb defects and variation of DNA binding affinities. J Biol Chem. 2007;282:27536–27546. doi: 10.1074/jbc.M702613200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco HL, Casasnovas J, Rodriguez-Medina JR, Cadilla CL. Redundant or separate entities?--roles of Twist1 and Twist2 as molecular switches during gene transcription. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato N, Chapman SL, McKee MD, Funato H, Morris JA, Shelton JM, Richardson JA, Yanagisawa H. Hand2 controls osteoblast differentiation in the branchial arch by inhibiting DNA binding of Runx2. Development. 2009;136:615–625. doi: 10.1242/dev.029355. [DOI] [PubMed] [Google Scholar]
- Gorlin RJ, Cohen MM, Hennekam RCM. Syndromes of the head and neck. 4th ed. Oxford [England]; New York: Oxford University Press; 2001. [Google Scholar]
- Gu S, Wei N, Yu L, Fei J, Chen Y. Shox2-deficiency leads to dysplasia and ankylosis of the temporomandibular joint in mice. Mech Dev. 2008;125:729–742. doi: 10.1016/j.mod.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms JA, Cordero D, Tapadia MD. New insights into craniofacial morphogenesis. Development. 2005;132:851–861. doi: 10.1242/dev.01705. [DOI] [PubMed] [Google Scholar]
- Hinton RJ, Serrano M, So S. Differential gene expression in the perichondrium and cartilage of the neonatal mouse temporomandibular joint. Orthod Craniofac Res. 2009;12:168–177. doi: 10.1111/j.1601-6343.2009.01450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard TD, Paznekas WA, Green ED, Chiang LC, Ma N, Ortiz de Luna RI, Garcia Delgado C, Gonzalez-Ramos M, Kline AD, Jabs EW. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nat Genet. 1997;15:36–41. doi: 10.1038/ng0197-36. [DOI] [PubMed] [Google Scholar]
- Huang T, Lin AE, Cox GF, Golden WL, Feldman GL, Ute M, Schrander-Stumpel C, Kamisago M, Vermeulen SJ. Cardiac phenotypes in chromosome 4q- syndrome with and without a deletion of the dHAND gene. Genet Med. 2002;4:464–467. doi: 10.1097/00125817-200211000-00011. [DOI] [PubMed] [Google Scholar]
- Jernvall J, Kettunen P, Karavanova I, Martin LB, Thesleff I. Evidence for the role of the enamel knot as a control center in mammalian tooth cusp formation: non-dividing cells express growth stimulating Fgf-4 gene. Int J Dev Biol. 1994;38:463–469. [PubMed] [Google Scholar]
- Kaalund SS, Moller RS, Teszas A, Miranda M, Kosztolanyi G, Ullmann R, Tommerup N, Tumer Z. Investigation of 4q-deletion in two unrelated patients using array CGH. Am J Med Genet A. 2008;146A:2431–2434. doi: 10.1002/ajmg.a.32458. [DOI] [PubMed] [Google Scholar]
- Kang Y, Massague J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Kim BM, Buchner G, Miletich I, Sharpe PT, Shivdasani RA. The stomach mesenchymal transcription factor Barx1 specifies gastric epithelial identity through inhibition of transient Wnt signaling. Dev Cell. 2005;8:611–622. doi: 10.1016/j.devcel.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Krebs I, Weis I, Hudler M, Rommens JM, Roth H, Scherer SW, Tsui LC, Fuchtbauer EM, Grzeschik KH, Tsuji K, Kunz J. Translocation breakpoint maps 5 kb 3' from TWIST in a patient affected with Saethre-Chotzen syndrome. Hum Mol Genet. 1997;6:1079–1086. doi: 10.1093/hmg/6.7.1079. [DOI] [PubMed] [Google Scholar]
- Latham RA. The pathogenesis of cleft palate associated with the Pierre Robin syndrome. An analysis of a seventeen-week human foetus. Br J Plast Surg. 1966;19:205–214. doi: 10.1016/s0007-1226(66)80044-9. [DOI] [PubMed] [Google Scholar]
- Lee SH, Bedard O, Buchtova M, Fu K, Richman JM. A new origin for the maxillary jaw. Dev Biol. 2004;276:207–224. doi: 10.1016/j.ydbio.2004.08.045. [DOI] [PubMed] [Google Scholar]
- Lee YB, Bantounas I, Lee DY, Phylactou L, Caldwell MA, Uney JB. Twist-1 regulates the miR-199a/214 cluster during development. Nucleic Acids Res. 2009;37:123–128. doi: 10.1093/nar/gkn920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebel DA, Tsoi B, Wong N, Tam PP. A conserved noncoding intronic transcript at the mouse Dnm3 locus. Genomics. 2005;85:782–789. doi: 10.1016/j.ygeno.2005.02.001. [DOI] [PubMed] [Google Scholar]
- MacKenzie B, Wolff R, Lowe N, Billington CJ, Jr, Peterson A, Schmidt B, Graf D, Mina M, Gopalakrishnan R, Petryk A. Twisted gastrulation limits apoptosis in the distal region of the mandibular arch in mice. Dev Biol. 2009;328:13–23. doi: 10.1016/j.ydbio.2008.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo M, Wellik DM, Deschamps J. Hox genes and regional patterning of the vertebrate body plan. Dev Biol. 2010;344:7–15. doi: 10.1016/j.ydbio.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsiadis TA, Drouin J. Deletion of the Pitx1 genomic locus affects mandibular tooth morphogenesis and expression of the Barx1 and Tbx1 genes. Dev Biol. 2008;313:887–896. doi: 10.1016/j.ydbio.2007.10.055. [DOI] [PubMed] [Google Scholar]
- Mraz M, Malinova K, Mayer J, Pospisilova S. MicroRNA isolation and stability in stored RNA samples. Biochem Biophys Res Commun. 2009;390:1–4. doi: 10.1016/j.bbrc.2009.09.061. [DOI] [PubMed] [Google Scholar]
- O'Rourke MP, Soo K, Behringer RR, Hui CC, Tam PP. Twist plays an essential role in FGF and SHH signal transduction during mouse limb development. Dev Biol. 2002;248:143–156. doi: 10.1006/dbio.2002.0730. [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Poswillo D. The Pierre Robin syndrome: etiology and early treatment. Trans Int Conf Oral Surg. 1967:425–429. [PubMed] [Google Scholar]
- Qiu M, Bulfone A, Ghattas I, Meneses JJ, Christensen L, Sharpe PT, Presley R, Pedersen RA, Rubenstein JL. Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev Biol. 1997;185:165–184. doi: 10.1006/dbio.1997.8556. [DOI] [PubMed] [Google Scholar]
- Qiu M, Bulfone A, Martinez S, Meneses JJ, Shimamura K, Pedersen RA, Rubenstein JL. Null mutation of Dlx-2 results in abnormal morphogenesis of proximal first and second branchial arch derivatives and abnormal differentiation in the forebrain. Genes Dev. 1995;9:2523–2538. doi: 10.1101/gad.9.20.2523. [DOI] [PubMed] [Google Scholar]
- Rice R, Thesleff I, Rice DP. Regulation of Twist, Snail, and Id1 is conserved between the developing murine palate and tooth. Dev Dyn. 2005;234:28–35. doi: 10.1002/dvdy.20501. [DOI] [PubMed] [Google Scholar]
- Ricks JE, Ryder VM, Bridgewater LC, Schaalje B, Seegmiller RE. Altered mandibular development precedes the time of palate closure in mice homozygous for disproportionate micromelia: an oral clefting model supporting the Pierre-Robin sequence. Teratology. 2002;65:116–120. doi: 10.1002/tera.10022. [DOI] [PubMed] [Google Scholar]
- Rivera-Perez JA, Mallo M, Gendron-Maguire M, Gridley T, Behringer RR. Goosecoid is not an essential component of the mouse gastrula organizer but is required for craniofacial and rib development. Development. 1995;121:3005–3012. doi: 10.1242/dev.121.9.3005. [DOI] [PubMed] [Google Scholar]
- Rossi MR, DiMaio MS, Xiang B, Lu K, Kaymakcalan H, Seashore M, Mahoney MJ, Li P. Clinical and genomic characterization of distal duplications and deletions of chromosome 4q: study of two cases and review of the literature. Am J Med Genet A. 2009;149A:2788–2794. doi: 10.1002/ajmg.a.33088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest LB, Clouthier DE. Elucidating timing and function of endothelin-A receptor signaling during craniofacial development using neural crest cell-specific gene deletion and receptor antagonism. Dev Biol. 2009;328:94–108. doi: 10.1016/j.ydbio.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest LB, Dager M, Yanagisawa H, Charite J, Hammer RE, Olson EN, Yanagisawa M, Clouthier DE. dHAND-Cre transgenic mice reveal specific potential functions of dHAND during craniofacial development. Dev Biol. 2003;257:263–277. doi: 10.1016/s0012-1606(03)00068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest LB, Marcotte R, Wang E. Peptide elongation factor eEF1A-2/S1 expression in cultured differentiated myotubes and its protective effect against caspase-3-mediated apoptosis. J Biol Chem. 2002;277:5418–5425. doi: 10.1074/jbc.M110685200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest LB, Xiang X, Lim KC, Levi G, Clouthier DE. Endothelin-A receptor-dependent and -independent signaling pathways in establishing mandibular identity. Development. 2004;131:4413–4423. doi: 10.1242/dev.01291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif MN, Sosic D, Rothlin CV, Kelly E, Lemke G, Olson EN, Ivashkiv LB. Twist mediates suppression of inflammation by type I IFNs and Axl. J Exp Med. 2006;203:1891–1901. doi: 10.1084/jem.20051725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton EL, Yutzey KE. Twist1 function in endocardial cushion cell proliferation, migration, and differentiation during heart valve development. Dev Biol. 2008;317:282–295. doi: 10.1016/j.ydbio.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Suda N, Suzuki S, Fukuoka H, Yamashita Y. An in situ hybridization study of Runx2, Osterix, and Sox9 at the onset of condylar cartilage formation in fetal mouse mandible. J Anat. 2006;208:169–177. doi: 10.1111/j.1469-7580.2006.00525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo K, O'Rourke MP, Khoo PL, Steiner KA, Wong N, Behringer RR, Tam PP. Twist function is required for the morphogenesis of the cephalic neural tube and the differentiation of the cranial neural crest cells in the mouse embryo. Dev Biol. 2002;247:251–270. doi: 10.1006/dbio.2002.0699. [DOI] [PubMed] [Google Scholar]
- Sperber SM, Dawid IB. barx1 is necessary for ectomesenchyme proliferation and osteochondroprogenitor condensation in the zebrafish pharyngeal arches. Dev Biol. 2008;321:101–110. doi: 10.1016/j.ydbio.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer DB, Rhee J, Cheung WL, Lassar AB. Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein Twist. Science. 1996;272:1476–1480. doi: 10.1126/science.272.5267.1476. [DOI] [PubMed] [Google Scholar]
- Thesleff I. The genetic basis of tooth development and dental defects. Am J Med Genet A. 2006;140:2530–2535. doi: 10.1002/ajmg.a.31360. [DOI] [PubMed] [Google Scholar]
- Thomas BL, Liu JK, Rubenstein JL, Sharpe PT. Independent regulation of Dlx2 expression in the epithelium and mesenchyme of the first branchial arch. Development. 2000;127:217–224. doi: 10.1242/dev.127.2.217. [DOI] [PubMed] [Google Scholar]
- Trumpp A, Depew MJ, Rubenstein JL, Bishop JM, Martin GR. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. 1999;13:3136–3148. doi: 10.1101/gad.13.23.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker AS, Matthews KL, Sharpe PT. Transformation of tooth type induced by inhibition of BMP signaling. Science. 1998;282:1136–1138. doi: 10.1126/science.282.5391.1136. [DOI] [PubMed] [Google Scholar]
- Tukel T, Sosic D, Al-Gazali LI, Erazo M, Casasnovas J, Franco HL, Richardson JA, Olson EN, Cadilla CL, Desnick RJ. Homozygous nonsense mutations in TWIST2 cause Setleis syndrome. Am J Hum Genet. 2010;87:289–296. doi: 10.1016/j.ajhg.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaahtokari A, Aberg T, Jernvall J, Keranen S, Thesleff I. The enamel knot as a signaling center in the developing mouse tooth. Mech Dev. 1996;54:39–43. doi: 10.1016/0925-4773(95)00459-9. [DOI] [PubMed] [Google Scholar]
- Vesuna F, van Diest P, Chen JH, Raman V. Twist is a transcriptional repressor of E-cadherin gene expression in breast cancer. Biochem Biophys Res Commun. 2008;367:235–241. doi: 10.1016/j.bbrc.2007.11.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz JW, Barnes RM, Rodgers R, Firulli BA, Conway SJ, Firulli AB. An absence of Twist1 results in aberrant cardiac neural crest morphogenesis. Dev Biol. 2008;320:131–139. doi: 10.1016/j.ydbio.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada G, Mansouri A, Torres M, Stuart ET, Blum M, Schultz M, De Robertis EM, Gruss P. Targeted mutation of the murine goosecoid gene results in craniofacial defects and neonatal death. Development. 1995;121:2917–2922. doi: 10.1242/dev.121.9.2917. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Clouthier DE, Richardson JA, Charite J, Olson EN. Targeted deletion of a branchial arch-specific enhancer reveals a role of dHAND in craniofacial development. Development. 2003;130:1069–1078. doi: 10.1242/dev.00337. [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Yin Z, Xu XL, Frasch M. Regulation of the twist target gene tinman by modular cis-regulatory elements during early mesoderm development. Development. 1997;124:4971–4982. doi: 10.1242/dev.124.24.4971. [DOI] [PubMed] [Google Scholar]
- Yu W, Kamara H, Svoboda KK. The role of twist during palate development. Dev Dyn. 2008;237:2716–2725. doi: 10.1002/dvdy.21627. [DOI] [PubMed] [Google Scholar]
- Yu W, Ruest LB, Svoboda KK. Regulation of epithelial-mesenchymal transition in palatal fusion. Exp Biol Med (Maywood) 2009a;234:483–491. doi: 10.3181/0812-MR-365. [DOI] [PubMed] [Google Scholar]
- Yu W, Serrano M, Miguel SS, Ruest LB, Svoboda KK. Cleft lip and palate genetics and application in early embryological development. Indian J Plast Surg. 2009b;42(Suppl):S35–S50. doi: 10.4103/0970-0358.57185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Sui P, Dong A, Hassell J, Cserjesi P, Chen YT, Behringer RR, Sun X. Preaxial polydactyly: interactions among ETV, TWIST1 and HAND2 control anterior-posterior patterning of the limb. Development. 2010;137:3417–3426. doi: 10.1242/dev.051789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga A, Quillet R, Perrin-Schmitt F, Zeller R. Mouse Twist is required for fibroblast growth factor-mediated epithelial-mesenchymal signalling and cell survival during limb morphogenesis. Mech Dev. 2002;114:51–59. doi: 10.1016/s0925-4773(02)00048-5. [DOI] [PubMed] [Google Scholar]