Abstract
Biomolecular assays are continually being developed that use progressively smaller amounts of material, often precluding the use of conventional cuvette-based instruments for nucleic acid quantitation for those that can perform microvolume quantitation.
The NanoDrop microvolume sample retention system (Thermo Scientific NanoDrop Products) functions by combining fiber optic technology and natural surface tension properties to capture and retain minute amounts of sample independent of traditional containment apparatus such as cuvettes or capillaries. Furthermore, the system employs shorter path lengths, which result in a broad range of nucleic acid concentration measurements, essentially eliminating the need to perform dilutions. Reducing the volume of sample required for spectroscopic analysis also facilitates the inclusion of additional quality control steps throughout many molecular workflows, increasing efficiency and ultimately leading to greater confidence in downstream results.
The need for high-sensitivity fluorescent analysis of limited mass has also emerged with recent experimental advances. Using the same microvolume sample retention technology, fluorescent measurements may be performed with 2 μL of material, allowing fluorescent assays volume requirements to be significantly reduced. Such microreactions of 10 μL or less are now possible using a dedicated microvolume fluorospectrometer.
Two microvolume nucleic acid quantitation protocols will be demonstrated that use integrated sample retention systems as practical alternatives to traditional cuvette-based protocols. First, a direct A260 absorbance method using a microvolume spectrophotometer is described. This is followed by a demonstration of a fluorescence-based method that enables reduced-volume fluorescence reactions with a microvolume fluorospectrometer. These novel techniques enable the assessment of nucleic acid concentrations ranging from 1 pg/ μL to 15,000 ng/ μL with minimal consumption of sample.
Keywords: Basic Protocols, Issue 45, NanoDrop, Microvolume Quantitation, DNA Quantitation, Nucleic Acid Quantitation, DNA Quantification, RNA Quantification, Microvolume Spectrophotometer, Microvolume Fluorometer, DNA A260, Fluorescence PicoGreen
Protocol
1. Microvolume Nucleic Acid Quantification Using a NanoDrop 2000c Spectrophotometer
To begin, clean the upper and lower optical surfaces of the microvolume spectrophotometer sample retention system by pipetting 2 to 3 μL of clean deionized water onto the lower optical surface.
Close the lever arm, ensuring that the upper pedestal comes in contact with the deionized water. Lift the lever arm and wipe off both optical surfaces with a clean, dry, lint-free lab wipe.
Open the NanoDrop software and select the Nucleic Acid application. Use a small-volume, calibrated pipettor to perform a blank measurement by dispensing 1 μL of buffer onto the lower optical surface. Lower the lever arm and select "Blank" in the Nucleic Acid application.
Once the blank measurement is complete, clean both optical surfaces with a clean, dry, lint-free lab wipe.
- Choose the appropriate constant for the sample that is to be measured.
Sample Type Select Option Constant Used to Calculate Concentration dsDNA DNA-50 50 ssDNA DNA-33 33 RNA RNA-40 40 Oligo Custom 15-150 Dispense 1 μL of nucleic acid sample onto the lower optical pedestal and close the lever arm. Because the measurement is volume independent, the sample only needs to bridge the gap between the two optical surfaces for a measurement to be made.
Select "Measure" in the application software. The software will automatically calculate the nucleic acid concentration and purity ratios. Following sample measurement, review the spectral output.
The software will automatically calculate the nucleic acid concentration and purity ratios. Following sample measurement, review the spectral image to assess sample quality.
A typical nucleic acid sample will have a very characteristic profile.
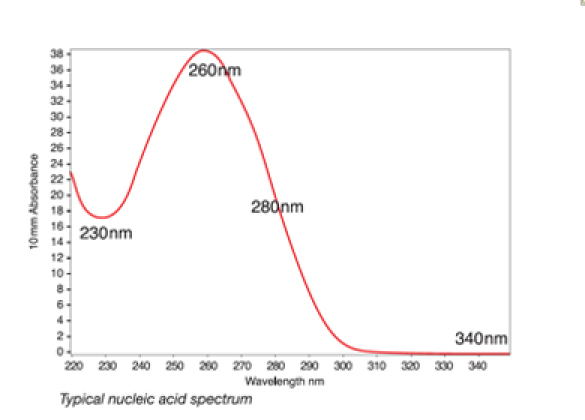 Figure 1. A typical nucleic acid sample will have a very characteristic profile.
Figure 1. A typical nucleic acid sample will have a very characteristic profile.Common sources of contaminants associated with specific nucleic acid isolation techniques include Phenol/Trizol and column extraction. In the case of Phenol/Trizol extraction, residual reagent contamination may be indicated by abnormal spectra between 220 to 240 nm as well as by shifts in the 260 to 280 nm region. Conversely, residual guanidine from column extraction may contribute to a peak near 230 nm and a shift in the trough from 230 nm to approximately 240 nm.
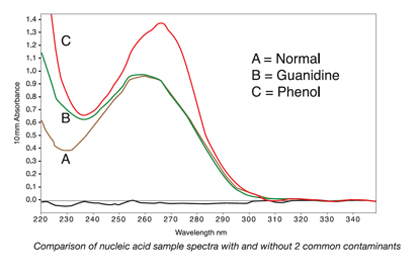 Figure 2. The shifts in the peaks and troughs of samples B and C as compared to sample A illustrate how contaminants can affect the spectra of nucleic acid samples.
Figure 2. The shifts in the peaks and troughs of samples B and C as compared to sample A illustrate how contaminants can affect the spectra of nucleic acid samples.To accurately assess sample quality, 260/280 or 260/230 ratios should be analyzed in combination with overall spectral quality. Pure nucleic acids typically yield a 260/280 ratio of ~1.8 and a 260/280 ratio of ~2.0 for DNA and RNA, respectively. This ratio is dependent on the pH and ionic strength of the buffer used to make the blank and sample measurements. Acidic solutions will under-represent the ratio by 0.2-0.3, while a basic solution will over-represent the ratio by 0.2-0.3. Significantly different purity ratios may indicate the presence of protein, phenol or other contaminants that absorb strongly at or near 280 nm. The 260/230 purity ratio is a second measure of DNA purity with values for a "pure" nucleic acid commonly in the range of 1.8-2.2. Purity ratios that are significantly lower than the expected values may indicate the isolation technique used may require further optimization.
NanoDrop spectrophotometers can also be used to quantify proteins using direct absorbance or by protein colorimetric assays. To ensure proper column formation and reproducibility, 2-μl samples are advised for protein samples. Refer to the following JoVE video: Microvolume Protein Concentration Determination Using the NanoDrop Spectrophotometer http://www.jove.com/index/Details.stp?ID=1610
2. High-Sensitivity Microvolume Nucleic Acid Quantitation Using the NanoDrop 3300 Fluorospectrometer
To demonstrate high-sensitivity microvolume nucleic acid quantitation using the NanoDrop 3300, a PicoGreen fluorescence kit is used. To begin, equilibrate the kit standards and all nucleic acid samples to room temperature. Fluorescent samples are light sensitive and should be kept in amber or aluminum wrapped tubes.
Prepare enough 1x TE buffer for all standards and samples to be measured as well as for the PicoGreen working solution that will be needed. Prepare 1xTE buffer by diluting the provided 20x TE buffer with nuclease-free water per the manufacturer's protocol. Reaction volumes can range from 200 μL to as little as 10 ul. In this example, 10 μL reaction volumes will be prepared.
Prepare serially diluted dsDNA standards at 2x the final desired concentration in nuclease-free vials or tubes. Transfer 5 μL of each of the diluted dsDNA standards into an individual labeled nuclease-free amber or foil-covered tube.
Aliquot 5 μL of each dsDNA sample of interest into appropriately labeled nuclease-free amber or foil-covered tubes.
Prepare PicoGreen working solution according to the manufacturer's protocol. Transfer an equal volume of the PicoGreen working solution to each tube containing either a standard or sample of interest (5 μL in this example).
Combine equal volumes of 1x TE buffer and PicoGreen working solution to prepare the negative control, which serves as a reference solution.
Mix the contents of each reaction tube thoroughly by gentle pipetting and incubate the reactions at room temperature for 5 minutes. All standards and samples should be equilibrated to the same temperature before measurement as fluorescence signal is affected by temperature.
To prepare the instrument and perform measurements, first clean the lower and upper surfaces of the sample retention system. Pipet 2 to 3 μL of deionized water onto the lower optical surface, close then open the lever arm, and blot the upper and lower pedestals with a clean, lint-free, laboratory wipe. Ensure the surfaces are free of lint before proceeding as lint can exhibit fluorescence in the presence of an excitation source and can interfere with the measurement.
Open the instrument software, select the "Nucleic Acids" application, and select the "PicoGreen-dsDNA" option.
To blank the instrument, add 2 μL of 1x TE to the lower pedestal, close the arm, and select "Blank" in the instrument software field. Once blank is complete, blot off the blank solution from both surfaces of the sample retention system.
Mix the reference solution by gentle pipetting. Using low retention tips, pipette 2 μL onto the lower pedestal and close the arm.
Select "Reference" in the measurement type, and select the desired units. Click "Measure" to initiate the measurement cycle.
When the measurement cycle is complete, open the arm and thoroughly blot off the sample retention system surfaces. Measure up to five replicates of the reference, using a fresh aliquot for each replicate.
Repeat the process for the additional standards to build a standard curve. Up to seven standards may be used. The software is designed to store up to five replicates for each standard. Replicates of the standards are averaged to generate a standard curve from which the sample concentrations are automatically determined.
Once the standard curve is complete, select the "Samples" tab, enter the respective sample ID information and measure each unknown sample.
Discussion
The quantitation protocols presented here are based on the most widely accepted microvolume systems. Such systems provide practical alternatives for traditional nucleic acid quantitation methodology, which rely on greater sample volumes and the use of containment apparatus.
NanoDrop microvolume technology employs a sample retention system that relies on the surface tension properties of the sample being measured to form a liquid column. It is essential that the sample makes contact with the upper and lower optical measurement surfaces for proper column formation. If at any point the measurement results seem inaccurate or not reproducible, it is most likely the result of sample heterogeneity or liquid column breakage.
Detergents and isopropyl alcohol are not recommended cleaning agents as they may uncondition the pedestal measurement surfaces. An unconditioned pedestal occurs when the hydrophobic surface properties of the pedestal have been compromised, resulting in a flattening of the sample droplet. Unconditioned pedestals can lead to breakage of the liquid column during measurement.
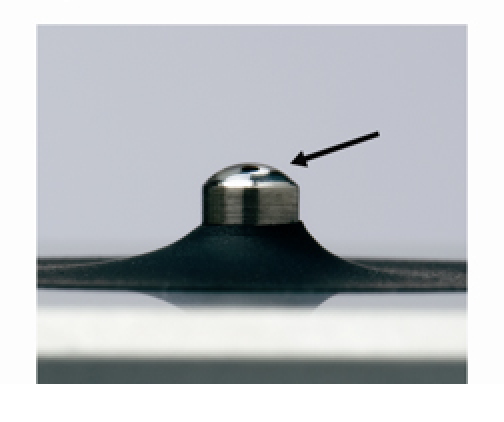 Figure 3. Unconditioned surface is characterized by a flattening of a water droplet.
Figure 3. Unconditioned surface is characterized by a flattening of a water droplet.
Uncondtioned pedestal surfaces may be reconditioned using NanoDrop Pedestal Reconditioning Compound (PR-1, Thermo Scientific NanoDrop Products). A thin layer of reconditioning compound is applied to the upper and lower pedestal surfaces, allowed to dry for ~30 seconds, then rubbed off using a clean, dry, lint-free lab wipe. A successfully reconditioned state is characterized by a water droplet beading up when applied to the bottom pedestal surface.
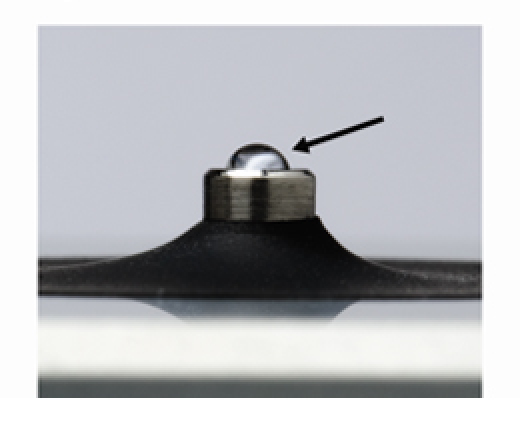 Figure 4. A reconditioned surface is characterized by a beading up of a water droplet.
Figure 4. A reconditioned surface is characterized by a beading up of a water droplet.
Microvolume quantitation systems greatly reduce sample consumption and dramatically increase the concentration range when compared to more traditional quantification systems. Although microvolume quantitation has often become an enabling technology for circumstances involving limited cell mass such as needle biopsies and laser-capture microdissection, the efficiency and ease-of-use of this methodology has made it a widely accepted alternative to traditional nucleic acid quantitation methods even when sample is plentiful.
Disclosures
Philippe Desjardins and Debra Conklin are employed by Thermo Scientific NanoDrop Products, Inc that produces the instruments used in this article.
Acknowledgments
Richard W. Beringer and David L. Ash, Applications Scientists, Thermo Scientific NanoDrop products, for technical assistance and filming.
References
- Wilfinger WW, Mackey K, Chomczynski P. Effect of pH and Ionic Strength on the Spectrophotometric Assessment of Nucleic Acid Purity. BioTechniques. 1997;22:474–481. doi: 10.2144/97223st01. [DOI] [PubMed] [Google Scholar]
- Voolstra C, Jungnickel A, Borrmann L, Kirchner R, Huber A. Implen Applications Note. Munich, Germany: 2006. Spectrophotometric Quantification of Nucleic Acids: LabelGuard enables photometric quantification of submicroliter samples using a standard photometer. [Google Scholar]
- Jr Ingle, D J, Crouch SR. Spectrochemical analysis. Englewood Cliffs, NJ: Prentice Hall; 1988. pp. XV–590. [Google Scholar]
- Van Lancker M, Gheyssens LC. A comparison of four frequently used assays for quantitative determination of DNA. Anal. Lett. 1986;19:615–623. [Google Scholar]


