Abstract
We have performed a computational comparative analysis of six small non-coding RNA (sRNA) families in α-proteobacteria. Members of these families were first identified in the intergenic regions of the nitrogen-fixing endosymbiont S. meliloti by a combined bioinformatics screen followed by experimental verification. Consensus secondary structures inferred from covariance models for each sRNA family evidenced in some cases conserved motifs putatively relevant to the function of trans-encoded base-pairing sRNAs i.e., Hfq-binding signatures and exposed anti Shine-Dalgarno sequences. Two particular family models, namely αr15 and αr35, shared own sub-structural modules with the Rfam model suhB (RF00519) and the uncharacterized sRNA family αr35b, respectively. A third sRNA family, termed αr45, has homology to the cis-acting regulatory element speF (RF00518). However, new experimental data further confirmed that the S. meliloti αr45 representative is an Hfq-binding sRNA processed from or expressed independently of speF, thus refining the Rfam speF model annotation. All the six families have members in phylogenetically related plant-interacting bacteria and animal pathogens of the order of the Rhizobiales, some occurring with high levels of paralogy in individual genomes. In silico and experimental evidences predict differential regulation of paralogous sRNAs in S. meliloti 1021. The distribution patterns of these sRNA families suggest major contributions of vertical inheritance and extensive ancestral duplication events to the evolution of sRNAs in plant-interacting bacteria.
Keywords: Sinorhizobium meliloti, Brucella, Hfq, rhizobia, riboregulation, RNome, speF, suhB, symbiotic nitrogen fixation
Post-genomic research has rendered bacterial small non-coding RNAs (sRNAs) as major players in the post-transcriptional regulation of gene expression underlying a wide range of important cellular processes, e.g., general responses to abiotic stimuli, cell division, quorum sensing or virulence.1 However, very little is known about the role of riboregulation in the control of symbiotic and pathogenic plant-microbe interactions.
The α-subdivision of the proteobacteria includes Gram-negative microorganisms with diverse life styles, frequently involving long-term mutualistic or pathogenic interactions with higher eukaryotes.2 Sinorhizobium meliloti is an environmentally and agronomically relevant α-proteobacterium belonging to the order of the Rhizobiales. It is well recognized as a genetically tractable model microorganism for the investigation and exploitation of the nitrogen-fixing endosymbiosis with legume plants. The outcome of these interactions is the formation in the cognate legume (i.e., Medicago species for S. meliloti) of the so-called root nodules which finally host invading bacteria in their differentiated nitrogen-fixing competent form of bacteroids.3 The S. meliloti genome has a multipartite architecture consisting of a single chromosome (3.65 Mbp) and two large plasmids termed pSymA (1.35 Mbp) and pSymB (1.68 Mbp). Megaplasmid pSymA harbors the clusters of genes specifying symbiotic functions, among others, but is dispensable for bacterial free-living growth whereas pSymB exhibits chromosome-like features e.g., it accommodates the essential tRNA-Arg encoding gene.4 This composite arrangement is common to the genomes of many bacterial species of the order of the Rhizobiales in which second chromosomes have been proposed to evolve from an early-acquired ancestral plasmid.5 Similarly to S. meliloti, many α-proteobacteria interacting with plants usually host a variable number of accessory extrachromosomal replicons besides the ancestral set of primary chromosome and megaplasmid. These non-essential plasmids most likely have a mosaic origin and contribute to the adaptive flexibility demanded by the transition of bacteria from a free-living to an intracellular state. At the regulatory level, these adaptations require the coordinated expression of complex gene networks in which sRNAs are also expected to participate. Several recent computational comparative genomics and deep-sequencing approaches have identified more than thousand non-coding RNA elements in the S. meliloti genome.6-9 Nearly two hundred of these molecules have been cataloged as putative trans-encoded sRNAs.9 This is an abundant class of bacterial riboregulators, which mostly target mRNAs via discontinuous nucleotide stretches of sequence complementarity to control the translation and/or stability of the message, many of them in an Hfq-dependent manner.1,10
Rhizobial RNomics has been pioneered by a genome-wide comparative genomics screen conducted in our laboratory, which, in combination with the experimental verification of predictions, identified eight sRNA genes in the intergenic regions (IGRs) of the reference strain S. meliloti 1021.6 Northern hybridization experiments and RACE mapping revealed that these sRNAs are differentially expressed from independent transcription units in free-living and endosymbiotic bacteria, thus supporting their putative role as riboregulators in S. meliloti. In this work we have combined new experimental data with an extensive in silico structural comparative analysis to further characterize these sRNAs and assess their conservation across α-proteobacteria.
Results
Generation of Smr sRNA family models
The starting point of this study was the set of eight non-coding transcripts identified previously in our laboratory on the basis of structure conservation and experimental verification. These sRNAs were initially termed Smr7C, Smr9C, Smr14C, Smr15C, Smr16C Smr22C, Smr35B and Smr45C for S. meliloti RNA, where the suffix indicates their respective positions in the output table of candidates along with the genomic location of each locus on pSymA (A), pSymB (B) or chromosome (C) (Table 1). TAP-based 5′-RACE experiments mapped the transcription start sites (TSS) of each sRNA to defined positions in the S. meliloti genome. Their 3′-ends were assumed to map to the last residue of the consecutive stretches of Us of Rho-independent terminators predicted for most of the transcripts, except for Smr22C and Smr45C which 3′-ends have been inferred from published experimental data6,9 (Table 1). Recent RNA-Seq based characterization of the small RNA fraction (50–350 nt) of the closely related strain S. meliloti 2011 mapped the full-length Smr transcripts in the S. meliloti 1021 genome to essentially the same positions reported earlier.9
Table 1. Query S. meliloti sRNA sequences.
| Name | Alternative namesa | 5′-endb,c | 3′-endb | Length (nt) |
|---|---|---|---|---|
| Smr7C |
Sra03/Sm13/SmelC023 |
201,679 |
201,828 |
150 |
| Smr9C |
Sra32/Sm10/SmelC289 |
1398,425 |
1398,277 |
149 |
| Smr14C |
Sm7/SmelC397 |
1,667,613 |
1,667,491 |
123 |
| Smr15C |
Sra41/Sm3/SmelC411 |
1,698,731 |
1,698,617 |
115 |
| Smr16C |
Sra41/Sm3′/SmelC412 |
1,698,937 |
1,698,817 |
121 |
| Smr35B |
SmB6/SmelC053 |
577,730 |
577,868 |
139 |
| Smr45C |
SmelC706 |
3,105,445 |
3,105,298d |
148 |
| Smr22C | Sra56/Sm1/SmelC667/6S | 2,972,251 | 2,972,091c | 161 |
b Coordinates according to the S. meliloti 1021 genome database at http://iant.toulouse.inra.fr/bacteria/annotation/cgi/rhime.cgi or www.rhizogate.de;
c RACE-based mapping_ENREF_32;6
d Deep-sequencing data9
The nucleotide sequences of the full-length Smr transcripts were first used to query the Rfam database v. 10.0 (www.sanger.ac.uk/Software/Rfam).11 This search revealed full homology of Smr22C to the well characterized 6S RNA family and therefore, this sRNA was not further considered in this study. Of the remaining 7 RNAs, Smr15C/16C and Smr45C exhibited partial structural homology to the suhB (RF00519) and speF (RF00518) RNA families respectively, whereas the remaining query transcripts did not match any Rfam entry. These seven sRNA sequences, likely representing previously unknown bacterial sRNA families, were next BLASTed with default parameters against all available bacterial genomes (1,615 sequences at 20th April 2011; www.ncbi.nlm.nih.gov). The genomic regions exhibiting significant degree of homology to the query sequences (78–89% similarity) were collected to generate initial alignments for each RNA that were manually curated to construct an Infernal Model (covariance model; CM) for each sRNA. As expected from their primary nucleotide sequence similarity, this analysis merged the tandemly-encoded Smr15C and Smr16C transcripts into the same RNA family and they were renamed accordingly as Smr15C1 and Smr15C2, respectively. The six RNA families resulting from this study have homologies limited to species of the order of the Rhizobiales within the α-subgroup of proteobacteria. Consistent with the naming scheme of the query sRNAs, their family models have been referred to as αrn for α-proteobacteria RNA, where the suffix identifies the family according to the query sequence. Stockholm formatted alignments for each family is provided at en.wikipedia.org/wiki/Small_non_coding_RNAs_in_the_endosymbiotic_diazotroph_%CE%B1-proteobacterium_Sinorhizobium_meliloti.
Structural features of the αr sRNA families
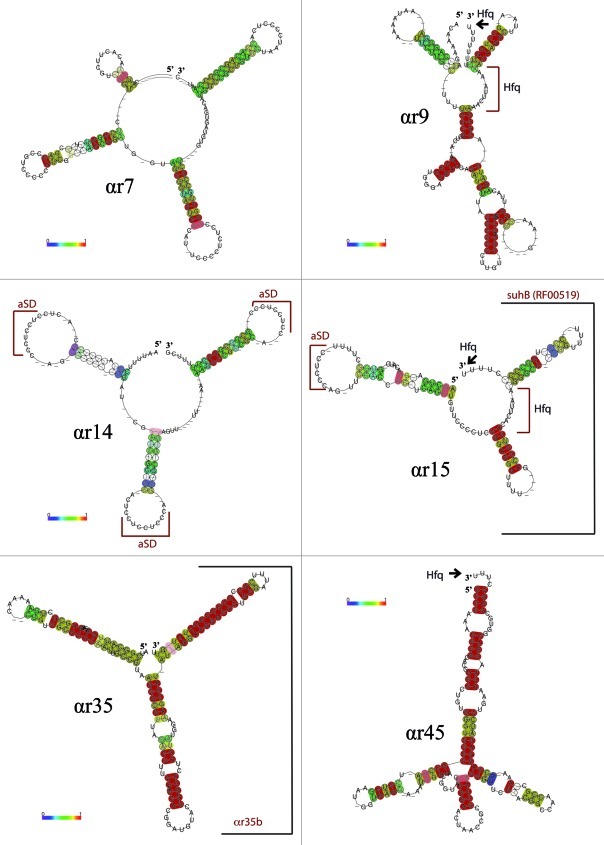
The inferred consensus secondary structures for each αr family model are shown in Figure 1.12 All six RNA families presented the typical sRNA arrangement in sub-structural domains with three to five main hairpin loops generally interrupted by internal stem-loops and/or single stranded sequence stretches. These structures are supported by a variable degree of nucleotide covariance that was particularly high in the three stem-loops of the αr7, αr14 and αr15 family members and the 5′ domains of αr9 and αr35 families. In most cases, the 3′ domain consists of a GC rich hairpin followed by tails of uridine residues, thus matching the main structural feature of the Rho-independent terminators of transcription. The exception was αr45 which last hairpin is supported by a strong conservation of the primary nucleotide sequences but does not resemble a bona fide Rho-independent terminator.
Figure 1. Consensus secondary structures of the αr sRNA families. Color code for base pairs is that of the Vienna RNA web suite.12 Putative Hfq-binding sites (Hfq) i.e., free 3′-hydroxyl end of an oligo-U stretch or internal single-stranded A/U-rich regions (Hfq) are marked with an arrowhead and a bracket, respectively. Ultraconserved anti Shine-Dalgarno sequences are indicated as (aSD), and identified sub-structural modules, suhB (RF00519) and αr35b, are marked with brackets.
A remarkable and complex structural situation was found in the αr15 and αr35 families. Members of the αr15 family showed partial homology to the Rfam model RF00519 known as suhB. In all cases this structural homology to the full-length suhB transcripts was restricted to the second hairpin and the Rho-independent terminator. SuhB-like genes have been computationally predicted to occur in multiple copies in a wide range of α-proteobacterial genomes and some meta-genomes.13
Similarly, αr35 sRNAs have three well-defined hairpin loops. The second and third structural motifs are maintained by extensive primary nucleotide sequence conservation and define a sequence stretch with wider occurrence in the genomes of the Rhizobiales (40 sequences) outside the full-length αr35 sRNAs (not shown). Therefore, suhB and this newly identified αr35 sub-structural domain (αr35b) likely represent widely distributed variants of the αr15 and αr35 sRNA families with a highly variable or even missing 5′ stem loops characteristic of the later transcripts.
The αr sRNA families mostly include putative trans-encoded transcripts, which are expected to influence translation of target mRNAs through short base-pairing interactions that usually occlude the ribosome-binding site (RBS). Interestingly, the loop anti Shine-Dalgarno sequence “CUCCUCCC” was found to be conserved in all the three hairpin loops of the αr14 family members as well as in the 5′ hairpin loop of αr15 sRNAs. Nonetheless, paired nucleotide stretches could also bind mRNA sequences if they are released and exposed to the target with the aid of proteins. The RNA chaperone Hfq has been shown to fulfill this function in most of the sRNA-mRNA target interactions documented to date. Internal single-stranded A/U-rich regions as well as a free 3′-hydroxyl end of an oligo-U stretch (e.g., of Rho-independent terminators) have been proposed as preferential sRNA interaction sites for Hfq.14-16 Both Hfq-binding signatures coexist in the αr9 and αr15 sRNAs, whereas exposed 3′-end poly-U tails of different lengths are also evident in αr45 transcripts. However, the terminal uridines of the Rho-independent terminators predicted for αr7, αr14 and αr35 family members are mostly base-paired to upstream sequences and hence could not be easily available for Hfq binding. In good correlation with these observations, the S. meliloti Smr9C (αr9), Smr15C1, Smr15C2 (both αr15) and Smr45C (αr45) sRNAs have been detected in the sub-population of transcripts co-inmunoprecipitated with a chromosomally-encoded epitope-tagged Hfq protein in lysates of free-living bacteria.17
Smr45C and speF are likely expressed as independent RNA elements in S. meliloti
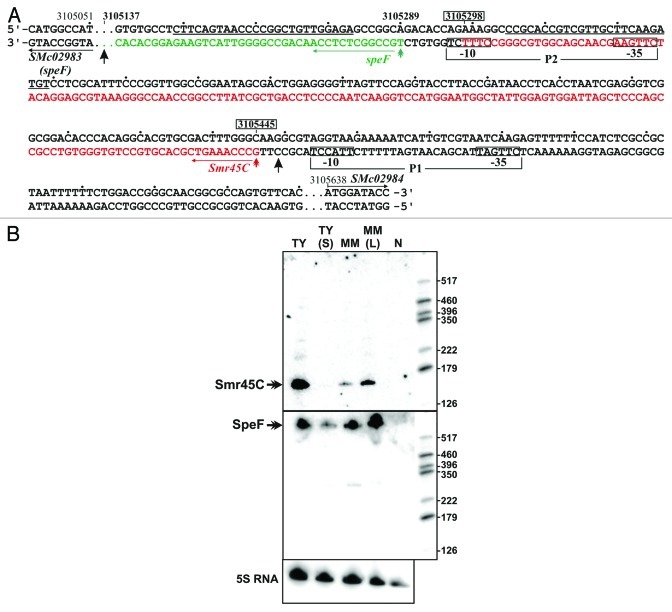
The αr45 RNA family partially matched the Rfam model speF (FR00518), a family of cis-acting RNA elements likely involved in the regulation of polyamine biosynthesis that have been identified in several α-proteobacterial species.13 Consistent with its proposed role, speF RNAs are mostly leader sequences of orthologs of ornithine decarboxylase-encoding genes.13 The S. meliloti speF structural homolog has been predicted to map between positions 3,105,448 and 3,105,137 in the chromosome of the reference strain 1021, upstream the SMc02983 gene which encodes a putative ornithine/arginine decarboxylase (Fig. 2) (rfam.sanger.ac.uk/genome/266834#tabview=tab1).13 Therefore, the 148 nt-long sequence of Smr45C, deduced from experimental mapping,6,9 would entirely match the 5′ region of speF (Fig. 2A). To solve this apparent inconsistency in the annotation of S. meliloti speF, the transcriptional output of this genomic region was further investigated. A closer inspection of the SMc02983/SMc02984 IGR identified two nucleotide sequence stretches that met the consensus CTTGAC-N17-CTATAT of σ70-dependent promoters in S. meliloti and other α-proteobacteria.18 One of these transcription signatures (P1) had been previously identified as the putative promoter of Smr45C and is located immediately upstream the TSS determined for this sRNA, whereas the second one (P2) overlaps the 3′ region of the Smr45C coding sequence (Fig. 2A). Transcription initiation from the P2 promoter is predicted to occur at the T residue at 3,105,289 nt position in the S. meliloti genome (Fig. 2A). Confirming previously reported data, a probe complementary to the 3′ region of Smr45C detected a unique RNA species of the expected size accumulating differentially in free-living microorganisms but not expressed in endosymbiotic bacteria (Fig. 2B). In contrast, a 25-mer oligonucleotide probe targeting a sequence 16 nt downstream the Smr45C 3′-end hybridized to a major RNA molecule visible at top of the gel with an expression profile very similar to that of Smr45C (Fig. 2B). The RNA species detected by this oligonucleotide most likely corresponds to the SMc02983 mRNA with a speF leader starting downstream the position previously predicted in silico. This RNA molecule could be originated either by processing of a larger undetectable and hence unstable RNA species transcribed from P1 or, most likely, by transcription from the newly identified promoter P2, independently of Smr45C in the biological conditions tested. In agreement with this observation, a S. meliloti map of TSS generated by RNA-Seq of total RNA revealed transcripts with 5′-ends at 3,105,292 and 3,105,166 nt positions in this region of the S. meliloti chromosome (A. Becker and J.P. Schlüter, personal communication). Altogether, these new experimental evidences further support classification of Smr45C as a Hfq-binding sRNA, likely unrelated to the speF RNA element.
Figure 2. Transcription of the speF and Smr45C RNAs. (A) Nucleotide sequence (both DNA strands) of the SMc02983-SMc02984 IGR expressing the speF and Smr45C RNA elements. Numbering indicates coordinates in the S. meliloti 1021 genome. The -35 and -10 hexamers of the predicted σ70-dependent promoters (P1 and P2) are boxed. Black arrowheads indicate the predicted start and end of the speF RNA as annotated in the Rfam database. Nucleotide positions of 5′ and 3′ ends previously determined for the Smr45C sRNA are boxed and a double arrowhead in red indicates its TSS. A double arrowhead in green indicates the predicted TSS for speF from the P2 promoter. The proposed speF and Smr45C coding sequences are in green and red letters, respectively. (B) Northern analysis of speF and Smr45C RNAs. Sequences of the 25-mer oligonucleotides used to probe the membranes are underlined in (A). RNA samples were: TY, log TY cultures; TY(S), stationary phase TY cultures; MM, log Minimal Medium cultures; MM(L), luteolin-induced log MM cultures; N, mature alfalfa nodules. Molecular weight markers are shown to the right of the panels. 5S RNA was also probed as RNA loading control.
Distribution of the αr sRNA families in the Rhizobiales
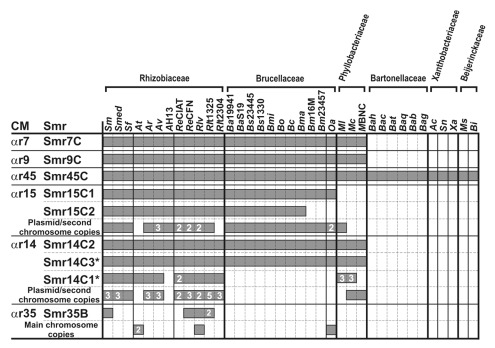
The occurrence of the αr sRNA families in sequenced bacterial species of the Rhizobiales was further assessed using the Infernal models (CMs) generated in this work. The results of this comparative analysis are summarized in Figure 3. With the only exception of Smr35B (αr35), which is encoded in the chromosome-like replicon pSymB, all our query sRNA genes are chromosomally located in S. meliloti 1021. Overall, structure-based clustering of the homologs identified with each of the CMs essentially correlates with the phylogeny of the order (en.wikipedia.org/wiki/Small_non_coding_RNAs_in_the_endosymbiotic_diazotroph_%CE%B1-proteobacterium_Sinorhizobium_meliloti). The dominant distribution pattern is represented by αr7, αr9 and αr14 CMs that identified members in the three taxonomic families of the order that include the bacterial species most closely related to S. meliloti i.e., Rhizobiaceae, Brucellaceae and Phyllobacteriaceae. The αr15 family was also found to be widely distributed in the Rhizobiales but lacks chromosomally-encoded relatives in Mesorhizobium species (family Phyllobacteriaceae). The widest distribution corresponded to αr45 which occurrence extended to species of other three taxonomic families with larger phylogenetic distances to S. meliloti i.e., Bartonellaceae, Xanthobacteriaceae and Beijerinckaceae. αr7, αr9 and αr45 members are all encoded by single-copy genes with well-defined promoter regions on the main bacterial chromosomes. Further, with a very few exceptions, complete microsynteny, i.e., conservation of upstream and downstream genes, was observed for representatives of all these three sRNA families in genomes of bacterial species from the same taxonomic family whereas only one of the two flanking genes appears variable across the Rhizobiales (en.wikipedia.org/wiki/%CE%B1r7_RNA; en.wikipedia.org/wiki/%CE%B1r9_RNA; en.wikipedia.org/wiki/%CE%B1r45_RNA). Thus, the current distribution pattern of the αr7, αr9 and αr45 sRNA families in bacteria is likely the result of the vertical inheritance of their respective sRNA genes located in the ancestral chromosome of the Rhizobiales.
Figure 3. Conservation of the S. meliloti Smr sRNAs in the Rhizobiales. CMs generated in this work along with the name of the query S. meliloti sRNA sequences are listed to the left. The newly predicted chromosomal copies of the Smr14 gene are indicated with an asterisk. All bacterial species with representatives of the αr RNA families are indicated on top of the panel grouped by taxonomic families i.e., Rhizobiaceae, Brucellaceae, Phyllobacteriaceae, Bartonellaceae, Xanthobacteriaceae and Beijerinckaceae, as follows; Sm, S. meliloti 1021; Smed, S. medicae WSM419; Sf, S. fredii NGR234; At, Agrobacterium tumefaciens C58; Ar, A. radiobacter K84; Av, A. vitis S4; AH13, A. sp H13–3; ReCIAT, Rhizobium etli CIAT652; ReCFN, R. etli CFN42; Rlv, R. leguminosarum bv. viceae 3841; Rlt1325, R. leguminosarum bv. trifolii WSM1325; Rlt2304, R. leguminosarum bv. trifolii WSM2304; Ba19941, Brucella abortus bv. One 9–941; BaS19, B. abortus S19; Bs23445, B. suis ATCC23445; Bs1330, B. suis 1330; Bmi, B. microti CCM4915; Bo, B. ovis ATCC25840; Bc, B. canis ATCC 23365; Bma, B. melitensis bv. abortus 2308; Bm16M, B. melitensis bv. 1 16M; Bm23457, B. melitensis ATCC23457; Oa, Ochrobactrum anthropi ATCC49188; Ml, Mesorhizobium loti MAFF303099; Mc, M. ciceri bv. biserrulae WSM1271; MBNC, M. sp BNC1; Bah, Bartonella henselae Houston-1; Bac, B. clarridgeiae 73; Bat, B. tribocorum CIP105476; Baq, B. quintana Toulouse; Bab, B. bacilliformis KC583; Bag, B. grahamii as4aup; Ac, Azorhizobium caulinodans ORS571; Sn, Starkeya novella DSM506; Xa, Xanthobacter autotrophicus Py2; Ms, Methylocella silvestris BL2, Bi, Beijerinckia indica subsp indica ATCC9039. Grey bars indicate distribution of each sRNA family in these bacterial species. If more than one, the number of chromosomal and extrachromosomal copies of each sRNA gene is also indicated.
In contrast, sRNA genes of the αr15 and αr14 families exist in highly variable copy numbers in the individual genomes; many of them located on extrachromosomal replicons i.e., large accessory plasmids in Rhizobiaceae/Phyllobacteriaceae representatives and the second chromosome in Brucella species. αr15 members occur in two chromosomal copies in 19 genomes of bacteria belonging to the Rhizobiaceae and Brucellaceae families. These two genes are clustered in the same IGR in genomes from Rhizobiaceae whereas in Brucella species map to distant positions on chromosome I. The second chromosomal αr15 loci were missed by our search in the genomes of B. melitensis bv. abortus 2308, B. melitensis bv.1 16M and Ochrobactrum anthropi ATCC49188. With the exceptions of A. tumefaciens C58 and R. leguminosarum bv. trifolii 2304, at least a third αr15 gene is located in extrachromosomal replicons of the host genomes. The αr14 RNA family showed an even more complex distribution pattern in the Rhizobiales. Two tandem copies of the S. meliloti Smr14C2 (formerly Smr14C) and Smr14C3 homologous genes were also identified in Sinorhizobium and Mesorhizobium species whereas in O. anthropi ATCC49188, Agrobacterium and Brucella species the second chromosomal gene predicted by the αr14 CM does not occur in such a syntenic context. A variable number of additional αr14 copies (up to six more in the genome of R. leguminosarum bv. trifolii WSM1325) were identified in the main chromosome and accessory plasmids of most of the bacterial species belonging to the Rhizobiaceae and Phyllobacteriaceae families. The αr15 and αr14 family members are mostly encoded in IGRs with a few exceptions of genes predicted within or antisense to annotated ORFs. However, these ORFs are frequently small, putatively coding for hypothetical proteins and/or absent from syntenic positions in bacterial genomes, thus representing probable mis-annotations as protein coding regions (en.wikipedia.org/wiki/%CE%B1r14_RNA#Genomic_Context; en.wikipedia.org/wiki/%CE%B1r15_RNA#Genomic_Context). In general, tandemly-arranged αr15 and αr14 genes occur in complete or partial microsynteny with the flanking genes in genomes of Rhizobiaceae and Phyllobacteriaceae as do their homologs on the main chromosome of O. anthropi ATCC49188 and Brucella species. However, microsynteny is much more fragmented or even absent for many of the remaining chromosomal and plasmidic copies of the αr14 and αr15 loci. Altogether, these observations suggest that αr14 and αr15 constitute families of paralogous sRNA gene copies in the Rhizobiales probably emanated from duplication events of their respective ancestral chromosomal genes over evolutionary time scales. Nonetheless, horizontal transfer events could certainly contribute to the current distribution patterns of some αr14 and αr15 gene copies, particularly of those occurring without signs of microsynteny in the accessory plasmids of plant-interacting bacteria. Noteworthy, some of the αr15 loci were flanked by insertion sequences or transposase-encoding genes, among other genetic elements involved in mobility events and genomic rearrangements (en.wikipedia.org/wiki/%CE%B1r15_RNA#Genomic_Context).
Finally, the αr35 family exhibits a more restricted and dispersed representation, not only at the species but also at the strain levels. Only seven candidates were identified by the αr35 Infernal models in addition to the S. meliloti Smr35B sRNA. Three of these predicted Smr35B homologs are encoded on the chromosomes of A. tumefaciens C58, O. anthropi ATCC49188, and R. leguminosarum bv. viceae 3841, whereas the remaining four αr35 genes are extrachromosomal and were identified on the R. etli CFN42 plasmid p42f, R. leguminosarum bv. viceae 3841 plasmid pRL11 and R. leguminosarum bv. trifolii 1325 plasmids pRl132502 and pRl132504. Again, the majority of the αr35 genes appeared to be independent transcription units with recognizable promoters with the exceptions of the chromosomal and plasmidic loci of R. leguminosarum bv. viceae 3841 and R. etli CFN42, respectively, which putatively overlap to annotated ORFs of unpredicted function. S. meliloti 1021 and O. anthropi ATCC49188 αr35 genes occur in complete microsynteny with the flanking genes whereas the genomic regions of the other six αr35 representatives revealed partial or no conservation at all (en.wikipedia.org/wiki/%CE%B1r35_RNA#Genomic_Context).
αr14 and αr15 representatives are differentially regulated in S. meliloti
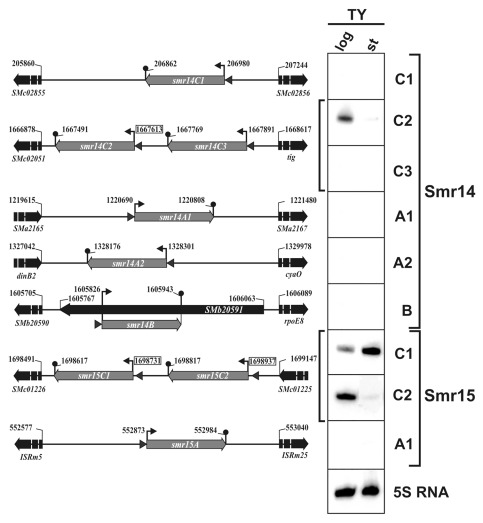
The αr14 and αr15 CMs also identified several related genes in the S. meliloti 1021 genome. A third copy of the Smr15C locus was found in the megaplasmid pSymA (Smr15A) and up to 5 additional copies of the query Smr14C2-encoding gene, were also identified; two of them chromosomally located (Smr14C1 and Smr14C3), two in pSymA (Smr14A1 and Smr14A2) and the remaining one in pSymB (Smr14B) (Fig. 4). Similarly to the situation of Smr15C1/Smr15C2, genes arranged in tandem in the same S. meliloti 1021 IGR encode Smr14C2 and Smr14C3. All the newly predicted Smr14- and Smr15-like sRNAs in the S. meliloti genome are encoded in IGRs, with the exception of Smr14B, which is encoded antisense to the SMb20591 gene (Fig. 4).
Figure 4. Northern analysis of the Smr14 and Smr15 sRNAs in S. meliloti. Maps of the genomic regions (not drawn to scale) of all the genes predicted by the αr14 and αr15 CMs in S. meliloti 1021 are shown to the left of the panels. Numbers denote coordinates of the genes in the genome. Name of the oligonucleotide probes used to hybridize each membrane are indicated to the right and their corresponding nucleotide sequences are listed in Table 2. RNA samples were obtained from logarithmic (log) and stationary phase (st) S. meliloti 1021 cultures in TY broth. 5S RNA was also probed as RNA loading control.
Oligonucleotides specific to all the Smr14 and Smr15 loci were used to probe S. meliloti RNA obtained from log and stationary phase cultures in TY broth (Fig. 4). These experiments confirmed the growth-dependent expression of Smr14C2, Smr15C1 and Smr15C2 transcripts with preferential accumulation of Smr15C1 upon entry of bacteria into stationary phase (Fig. 4). Despite their sequence and structural similarity Smr15C1 and Smr15C2 displayed opposite expression profiles. Strikingly, this set of Northern hybridizations did not reveal sings of expression of any of the other five Smr14 genes whereas the Smr15A transcript was barely detected on gels (Fig. 4). Multiple nucleotide sequence alignments of the promoter regions of all the genes encoding αr15 and αr14 members in species of the Rhizobiales identified diverse conserved motifs that could contribute to the differential expression of these genes in specific biological conditions (en.wikipedia.org/wiki/%CE%B1r14_RNA#Promoter_Analysis; en.wikipedia.org/wiki/%CE%B1r15_RNA#Promoter_Analysis). Supporting this prediction, RNA-Seq of the S. meliloti sRNAs expressed in a number of stress conditions has rendered variable number of reads for the S.meliloti αr14- and αr15-like transcripts, possibly correlating with a diversity of accumulation profiles.9
Discussion
The repertoire of non-coding RNAs expressed by the legume endosymbiont S. meliloti is one of the best characterized among those of its α-proteobacterial counterparts.6-9 However, current information about the function of these transcripts in bacteria is certainly scarce. The first set of sRNAs identified in the reference strain S. meliloti 1021 included eight transcripts with genomic boundaries experimentally determined by independent approaches.6,9 Here, we have performed a comprehensive computational comparative analysis of these eight sRNA sequences to identify conserved structural motifs putatively relevant to their function as well as to assess their conservation patterns in bacterial genomes. CMs derived from alignments of the Smr sRNA homologs first identified Smr22C as the S. meliloti ortholog of the ubiquitous 6S sRNA. This RNA constitutes an example of a well-characterized trans-acting protein-binding sRNA.19 The remaining seven transcripts represent structural and functional novel prokaryotic sRNAs and were collected into six different Infernal models. These CMs were used to accurately identify new members of each family in available sequenced bacterial genomes. This search revealed conservation of the Smr sRNAs in bacterial species belonging to the order of the Rhizobiales within the α-subgroup of proteobacteria and, hence these RNA families were accordingly termed αr. Such a distribution pattern, limited to phylogenetically related bacterial species, is a general feature of the Hfq-dependent base-pairing riboregulators.1 Indeed, the consensus secondary structures deduced from each family model evidenced Hfq-binding and exposed aSD signatures in αr15 and αr14 transcripts as recognizable functional motifs involved in the sRNA-target mRNA interaction. In this regard, it is also noteworthy that previously reported pull-down experiments as well as stability assays on a S. meliloti hfq mutant background independently confirmed the Smr-Hfq interactions predicted by our CMs.17,20
Two particular CMs representing the αr15 and αr45 families rendered partial hits to the Rfam models corresponding to the suhB and speF non-coding RNA elements, respectively. The secondary structure of the αr15 sRNAs is predicted to consist of three hairpin motifs, in good agreement with the mapping of the A. tumefaciens Smr15C1 homolog (AbcR1) by enzymatic probing.21 Furthermore, the aSD-containing 5′ hairpin loop of A. tumefaciens AbcR1 has been shown to be the functional domain of this transcript for targeting the 5′-UTR of the mRNA encoding the GABA-binding protein.21 Confirming these experimental results preliminary predictions of Smr15C1/C2-mRNA interactions in S. meliloti using diverse bioinformatics tools anticipate a major involvement of the 5′ hairpin in target recognition (O. Torres-Quesada and J.I. Jiménez-Zurdo, unpublished results). This 5′ stem loop is a variable or missing domain in suhB-like transcripts. Our comparative analysis revealed a similar situation for the αr35 sRNA family and its variant αr35b. The dispersed occurrence of the αr35 loci in the Rhizobiales points also to the primary hairpin of these molecules as a functional domain, which probably has co-evolved with its target protein or mRNA in these genomes. Some 5′ located sRNA domains have been shown to be critical elements for specific pairing-based mRNA target recognition that can act autonomously when fused to unrelated sRNA molecules.22 Therefore, the structural modules shared by αr15/suhB and αr35/αr35b could be regarded as a kind of α-proteobacteria-specific “structural Legos” which could accommodate autonomous 5′ domains to create functionally diverse sRNAs.23
We have also shown that the S. meliloti Smr45C sRNA and its downstream mRNA containing the cis-regulatory element speF are detected as different RNA species on Northern membranes under several biological conditions. Nonetheless, our comparative analysis also revealed that Smr45C always occurs in a syntenic context with a downstream ornithine decarboxylase-encoding gene in the Rhizobiales (en.wikipedia.org/wiki/%CE%B1r45_RNA). Therefore, it cannot be ruled out that under not yet tested specific biological conditions, probably relevant to polyamine biosynthesis, speF and Smr45C can be transcribed as a single cis-acting RNA element likely controlling translation of the ornithine decarboxylase enzyme. Possible processing of sRNAs from riboswitches was first described in E. coli.24 Furthermore, a dual function of a sRNA as trans- and cis-acting riboregulator has been recently reported for a lysine riboswitch which lies in the 5′-UTR of the lysine transporter gene in Listeria monocytogenes.25
Chromosomal location and conservation of at least one of the flanking protein-coding genes are also dominant features of the intergenic base pairing sRNA loci.1 The αr7, αr9 and αr45 CMs represent new examples of bacterial sRNAs encoded by conserved unique chromosomal genes that occur in extensive microsynteny across phylogenetically related species. However, single-copy genes hardly represent 58% of the total gene content of the S. meliloti genome.4 The genomes of plant-interacting bacteria usually evidence high levels of paralogy suggesting that their expansion through gene duplications has been little constrained during the evolution, facilitating the acquisition of new adaptive functions for life in the soil and within plant cells.2,4 The αr14 and αr15 family members occur in multiple copies in the individual genomes. Multiple sRNA copies are not unusual in bacteria, although the physiological/ecological advantages of these reiterations have been only investigated in a subset of cases.1 Seemingly homologous sRNAs could act either redundantly, serving as backups in critical pathways, additively sensing different stimuli to integrate diverse environmental signals, independently, regulating different set of genes or hierarchically upon each other.26-28 In this work we have investigated the expression in free-living bacteria of the Smr14 and Smr15 genes copies identified by the respective covariance models in S. meliloti 1021. Northern experiments, promoter predictions and reported RNA-Seq data9 provide evidences for the differential regulation of these genes. In particular, the opposite expression patterns of Smr15C1 and Smr15C2 contrast with those of their A. tumefaciens homologs, which encoding genes are similarly arranged in tandem in the circular chromosome of this bacterium but showed identical expression profiles.21 Interestingly, Smr15C1 retained its accumulation pattern in a S. meliloti ΔSmr15C2 derivative and vice versa suggesting that these sRNAs act independently or additively rather than hierarchically as riboregulators in S. meliloti (O. Torres-Quesada and J.I. Jiménez-Zurdo, unpublished). On the other hand, the undetectable expression of some transcripts in our assays, particularly of those grouped within the αr14 sRNA family anticipates that they could be only expressed under not tested specific biological conditions to fulfill different adaptive functions in this bacterium.
In summary, our findings provide a baseline for the forthcoming investigation of the functional plasticity and evolution of the small non-coding RNAs in S. meliloti and related plant-interacting bacteria.
Materials and Methods
Computational tools and methods
In a first step the smr gene sequences were BLASTed with default parameters against all currently available bacterial genomes (1,615 sequences at 20 April 2011; www.ncbi.nlm.nih.gov). The regions exhibiting signiðcant homologies to the query sequence (78–89% similarity) were used to generate automated infernal alignments29 for each family. This initial alignment was hand-curated and manually inspected to deduce a consensus secondary structure for each family. The consensus structure was also independently predicted with the program locARNATE30 in an automatic manner and differences reconciled giving priority to the structural conservation. Given the initial hand-curated structural alignment of close homologs Infernal was used to interrogate the same set of bacterial genomes, searching for new members of the models. The alignment process was repeated during three iterations. The candidates obtained with the Infernal models were selected as members of a given family if their Infernal E-value was e10−03 or lower, or after manual inspection for those with higher Infernal E-values. The hierarchical cluster-tree for each family is derived by WPGMA clustering of the pairwise alignment distances and the optimal number of clusters was calculated from the tree using RNAclust (www.bioinf.uni-leipzig.de/~kristin/Software/RNAclust/). A Stockholm format text file of each family alignment is provided in the links to the family wiki pages at en.wikipedia.org/wiki/Small_noncoding_RNAs_in_the_endosymbotic_diazotroph_%CE%B1-proteobacterium_Sinorhizobium_meliloti.
In order to study the microsinteny of each αr family, we located and extracted the flanking genes of their respective members. Non-annotated ORFs were further annotated using Blast2GO,31,32 and the high-throughput pipelines ProtSweep, and DomainSweep.33 The obtained results were later manually inspected in order to annotate and predict a biological function for these ORFs. In the few cases where the predicted sRNAs overlapped ORFs, the same procedure as with the flanking genes was carried on. ORFs shorter than 30 aa, that neither showed similarity with any database entry, nor motif or signatures when searched against family and motif databases such as Interpro,34 PFAM35 or Smart36 were considered as miss-annotations and thus not registered in the genomic context graph of the corresponding αr family.
Experimental methods
Growth of S. meliloti strain 1021 in TY and MM broths, RNA extraction from free-living and endosymbiotic bacteria and Northern hybridizations were performed as previously described.6 Sequences of the 25-mer oligonuclotides used to probe Northern membranes are detailed in Table 2.
Table 2. Oligonuclotide probes used in Northern hybridizations.
| sRNA | Nucleotide sequence | Target sequencea |
|---|---|---|
| speF |
5′-CTTCAGTAACCCCGGCTGTTGGAGA-3′ |
3,105,282–3,105,258 |
| Smr45C |
5′-CCGCACCGTCGTTGCTTCAAGATGT-3′ |
3,105,328–3,105,304 |
| Smr14C1 |
5′-AACCGACCGAATGCCGGGCGCCGTG-3′ |
206,954–206,930 |
| Smr14C2 |
5′-TGCTTGATCTGATTGGCAACCGGGA-3′ |
1,667,552–1,667,528 |
| Smr14C3 |
5′-ACCGGCGGGCGTCATAAAGGCGATT-3′ |
1,667,818–1,667,794 |
| Smr14A1 |
5′-AACCGATCGGCGTCTTGCGCCGTGG-3′ |
1,220,715–1,220,739 |
| Smr14A2 |
5′-GAGGAAAGGTCGCTCGCATATCGAA-3′ |
1,328,303–1,328,279 |
| Smr14B |
5′-GTGCGCCGGGCTTTCGATCCTGACC-3′ |
1,605,895–1,605,919 |
| Smr15C1 |
5′-GAGGAGAAAGCCGCTAGATGCACCA-3′ |
1,698,728–1698,704 |
| Smr15C2 |
5′-ACTGGGAGGAGAAGCCACCAAAGAT-3′ |
1,698,928–1698,904 |
| Smr15A | 5′-GGAGGAAAACTGCCATGCGCATCAA-3′ | 552,875–552,899 |
a Coordinates of the sequence stretches complementary to each probe in the S. meliloti 1021 genome according to iant.toulouse.inra.fr/bacteria/annotation/cgi/rhime.cgi.
Acknowledgments
C.d.V. and R.R.-Z were supported in part by: the visitor program at Janelia Farm Research Campus (HHMI), the Spanish Ministerio de Ciencia e Innovación (project TIN-2009–13950), the Consejería de Innovación, Investigación y Ciencia de la Junta de Andalucía (project TIC-02788) and the GENIL program (PYR-2010–28). Work at Grupo de Ecología Genética de la Rizosfera was partially funded by ERDF-cofinanced grants CSD2009–00006 (Consolider-Ingenio Program) and AGL2009–07925 from the Spanish Ministerio de Ciencia e Innovación to N.T. and J.I.J.-Z., respectively. O.T.-Q. and A.P. are recipients of Ph.D fellowships from the Spanish Ministerio de Ciencia e Innovación (FPI program) and CSIC (JAE Program), respectively. We thank E. Rivas for helpful discussions, computational support at Janelia Farm Research Campus and careful read of the manuscript. T. Jones for valuable initial discussions.
Glossary
Abbreviations:
- sRNA
bacterial small non-coding RNA
- IGR
intergenic region
- RACE
rapid amplification of cDNA ends
- Smr
S. meliloti sRNA
- TAP
tobacco acid pyrophosphatase
- TSS
transcription start site
- CM
covariance model
- RBS
ribosome binding site
- aSD
anti Shine-Dalgarno
- 5′-UTR
5′ untranslated region
- GABA
γ-amino butyric acid
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/18643
References
- 1.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–28. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batut J, Andersson SG, O'Callaghan D. The evolution of chronic infection strategies in the α-proteobacteria. Nat Rev Microbiol. 2004;2:933–45. doi: 10.1038/nrmicro1044. [DOI] [PubMed] [Google Scholar]
- 3.Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol. 2007;5:619–33. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galibert F, Finan TM, Long SR, Puhler A, Abola P, Ampe F, et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 2001;293:668–72. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- 5.Slater SC, Goldman BS, Goodner B, Setubal JC, Farrand SK, Nester EW, et al. Genome sequences of three Agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J Bacteriol. 2009;191:2501–11. doi: 10.1128/JB.01779-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.del Val C, Rivas E, Torres-Quesada O, Toro N, Jiménez-Zurdo JI. Identification of differentially expressed small non-coding RNAs in the legume endosymbiont Sinorhizobium meliloti by comparative genomics. Mol Microbiol. 2007;66:1080–91. doi: 10.1111/j.1365-2958.2007.05978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulvé VM, Sevin EW, Cheron A, Barloy-Hubler F. Identification of chromosomal alpha-proteobacterial small RNAs by comparative genome analysis and detection in Sinorhizobium meliloti strain 1021. BMC Genomics. 2007;8:467. doi: 10.1186/1471-2164-8-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valverde C, Livny J, Schluter JP, Reinkensmeier J, Becker A, Parisi G. Prediction of Sinorhizobium meliloti sRNA genes and experimental detection in strain 2011. BMC Genomics. 2008;9:416. doi: 10.1186/1471-2164-9-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlüter JP, Reinkensmeier J, Daschkey S, Evguenieva-Hackenberg E, Janssen S, Janicke S, et al. A genome-wide survey of sRNAs in the symbiotic nitrogen-fixing alpha-proteobacterium Sinorhizobium meliloti. BMC Genomics. 2010;11:245. doi: 10.1186/1471-2164-11-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Storz G, Opdyke JA, Zhang AX. Controlling mRNA stability and translation with small, noncoding RNAs. Curr Opin Microbiol. 2004;7:140–4. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 11.Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, et al. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res. 2011;39:D141–5. doi: 10.1093/nar/gkq1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70-4. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corbino KA, Barrick JE, Lim J, Welz R, Tucker BJ, Puskarz I, et al. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome Biol. 2005;6:R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–56. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otaka H, Ishikawa H, Morita T, Aiba H. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc Natl Acad Sci USA. 2011;108:13059–64. doi: 10.1073/pnas.1107050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sauer E, Weichenrieder O. Structural basis for RNA 3′-end recognition by Hfq. Proc Natl Acad Sci USA. 2011;108:13065–70. doi: 10.1073/pnas.1103420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torres-Quesada O, Oruezabal RI, Peregrina A, Jofré E, Lloret J, Rivilla R, et al. The Sinorhizobium meliloti RNA chaperone Hfq influences central carbon metabolism and the symbiotic interaction with alfalfa. BMC Microbiol. 2010;10:71. doi: 10.1186/1471-2180-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacLellan SR, MacLean AM, Finan TM. Promoter prediction in the rhizobia. Microbiology. 2006;152:1751–63. doi: 10.1099/mic.0.28743-0. [DOI] [PubMed] [Google Scholar]
- 19.Wassarman KM. 6S RNA: a regulator of transcription. Mol Microbiol. 2007;65:1425–31. doi: 10.1111/j.1365-2958.2007.05894.x. [DOI] [PubMed] [Google Scholar]
- 20.Voss B, Holscher M, Baumgarth B, Kalbfleisch A, Kaya C, Hess WR, et al. Expression of small RNAs in Rhizobiales and protection of a small RNA and its degradation products by Hfq in Sinorhizobium meliloti. Biochem Biophys Res Commun. 2009;390:331–6. doi: 10.1016/j.bbrc.2009.09.125. [DOI] [PubMed] [Google Scholar]
- 21.Wilms I, Voss B, Hess WR, Leichert LI, Narberhaus F. Small RNA-mediated control of the Agrobacterium tumefaciens GABA binding protein. Mol Microbiol. 2011;80:492–506. doi: 10.1111/j.1365-2958.2011.07589.x. [DOI] [PubMed] [Google Scholar]
- 22.Papenfort K, Bouvier M, Mika F, Sharma CM, Vogel J. Evidence for an autonomous 5′ target recognition domain in an Hfq-associated small RNA. Proc Natl Acad Sci USA. 2010;107:20435–40. doi: 10.1073/pnas.1009784107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunziker A, Tuboly C, Horvath P, Krishna S, Semsey S. Genetic flexibility of regulatory networks. Proc Natl Acad Sci USA. 2010;107:12998–3003. doi: 10.1073/pnas.0915003107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jäger JG, Hüttenhofer A, et al. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–43. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, et al. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–9. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 26.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 27.Tu KC, Bassler BL. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 2007;21:221–33. doi: 10.1101/gad.1502407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–7. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Will S, Reiche K, Hofacker IL, Stadler PF, Backofen R. Inferring Noncoding RNA Families and Classes by Means of Genome-Scale Structure-Based Clustering. PLOS Comput Biol. 2007;3:e65. doi: 10.1371/journal.pcbi.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–6. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 32.Vinayagam A, del Val C, Schubert F, Eils R, Glatting KH, Suhai S, et al. GOPET: a tool for automated predictions of Gene Ontology terms. BMC Bioinformatics. 2006;7:161. doi: 10.1186/1471-2105-7-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.del Val C, Ernst P, Falkenhahn M, Fladerer C, Glatting KH, Suhai S, et al. ProtSweep, 2Dsweep and DomainSweep: protein analysis suite at DKFZ. Nucleic Acids Res. 2007;35:W444-50. doi: 10.1093/nar/gkm364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–5. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–22. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–32. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]