Abstract
The hepadnavirus encapsidation signal, epsilon (ε), is an RNA structure located at the 5′ end of the viral pregenomic RNA. It is essential for viral replication and functions in polymerase protein binding and priming. This structure could also have potential regulatory roles in controlling the expression of viral replicative proteins. In addition to its structure, the primary sequence of this RNA element has crucial functional roles in the viral lifecycle. Although the ε elements in hepadnaviruses share common critical functions, there are some significant differences in mammalian and avian hepadnaviruses, which include both sequence and structural variations.
Here we present several covariance models for ε elements from the Hepadnaviridae. The model building included experimentally determined data from previous studies using chemical probing and NMR analysis. These models have sufficient similarity to comprise a clan. The clan has in common a highly conserved overall structure consisting of a lower-stem, bulge, upper-stem and apical-loop.
The models differ in functionally critical regions—notably the two types of avian ε elements have a tetra-loop (UGUU) including a non-canonical UU base pair, while the hepatitis B virus (HBV) epsilon has a tri-loop (UGU). The avian epsilon elements have a less stable dynamic structure in the upper stem. Comparisons between these models and all other Rfam models, and searches of genomes, showed these structures are specific to the Hepadnaviridae. Two family models and the clan are available from the Rfam database.
Keywords: encapsidation, epsilon element, hepadnavirus, pgRNA, RNA secondary structure
Hepatitis B Virus
The human hepatitis B virus (HBV) is a major health problem worldwide with an estimated 370 million individuals chronically infected. Chronically infected patients have an increased risk of developing liver cirrhosis and liver cancer resulting in over a million deaths annually.1,2
HBV is a member of the Hepadnaviridae, a family of small hepatotropic DNA viruses. Hepadnaviruses are known to infect certain mammals (orthohepadnavirus) and birds (avihepadnavirus). These viruses have a unique replication lifecycle in that their partially double-stranded DNA genomes are replicated through an RNA intermediate, the pregenomic RNA (pgRNA).3 Hepadnaviruses are related to retroviruses in that they are both retro-transcribing viruses and share some general characteristics.
Current antiviral drugs such as interferon α and nucleoside analogs, while effective in some cases, have problems of limited efficacy and viral resistance after prolonged treatment.4,5 A better understanding between viral and host factors is therefore necessary to facilitate novel antiviral drugs and strategies. A key cis-acting RNA element that acts at several steps in the process is the epsilon (ε) encapsidation signal.
The Structure and Location of ε Elements in Hepadnavirus RNAs
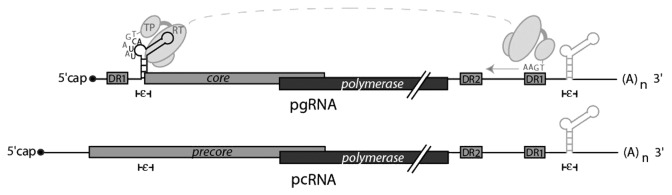
The pgRNA also serves as the mRNA template for the translation of the replicative proteins, the core and polymerase (P) protein.6-10 The pgRNA is one of two greater than genome length mRNAs transcribed from the viral genomes. The other mRNA being the precore RNA (pcRNA), from which the precore protein is translated11,12 (Fig. 1).
Figure 1. A schematic representation of the greater then genome-length HBV pgRNA and pcRNA. Cis RNA elements, namely, epsilon (ε), direct repeat 1 and direct repeat 2 (DR1 and DR2). The ε structure is present at both 5′ and 3‘ termini of the pgRNA, but only the 5′ ε of the pgRNA is selectively recognized for packaging. It facilitates polymerase (P) binding as depicted by the Terminal Domain (TP) and Reverse Transcriptase (RT) domain. The TP domain initiates protein priming at the bulge of the 5′ ε and after initial priming translocates to the 3′ end DR1 acceptor site where complementary base-pairing to the ε donor allows the RT to initiate the minus strand DNA synthesis. The pcRNA is exactly the same as the pgRNA except for a longer 5′ leader, it encodes the precore ORF and also contains the ε element but due to the translation of the precore does not function in encapsidation.
The Functions of the ε Element in Reverse Transcription and Replication
In hepadnaviruses, the processes of reverse transcription and encapsidation of the pgRNA are facilitated by the ε encapsidation signal. The ε element spans a region of approximately 60 nucleotides and is located at both the 5′and 3′ ends of the pgRNA and pcRNA. While both the pgRNA and pcRNA are translated, only the pgRNA is reverse transcribed and encapsidated. Efficient translation of the precore protein across the pcRNA ε element melts the RNA structure and prevents pcRNA encapsidation.13-15 Furthermore, only the 5′ ε of the pgRNA has been shown to be essential for these processes, whereas the 3′ ε which have slightly different conformations is not used.16
During reverse transcription, the 5′ ε element recruits the P protein to the upper stem, then the TP domain of P initiates priming at the conserved bulged UUCA and the synthesis of the minus-strand DNA (Fig. 2). This process involves conformational changes in both the ε structure and bound P protein which open up the base pairing in the upper stem allowing reverse transcription from the bulge.18,19 These conformational changes and recruitment of P protein are also facilitated by cellular chaperones.17,19-21
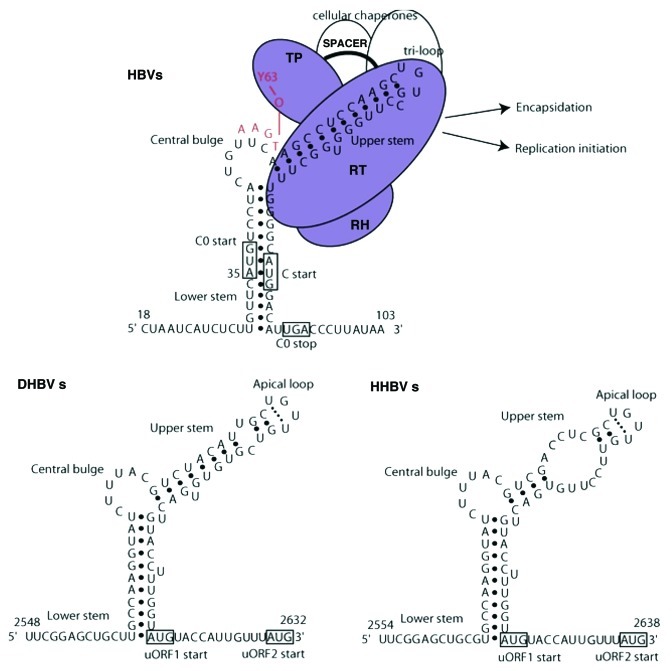
Figure 2. The secondary structure of HBV, DHBV, HHBV ε elements (derived from NMR, structural probing, and functional studies) The ε structure is remarkably conserved throughout the hepadnaviruses. It features two stem-loop structures, a conserved central bulge and an apical loop: tri-loop in human HBV (A) and tetra-loop in duck (B) and heron (C) hepadnaviruses. The core (C) start is included within the ε in HBV and follows it in avian viruses. Additional short upstream ORFS are also found (CO, uORF1, uORF2). The C0 ORF spans the ε structure within the orthohepadnaviruses as represented in HBV. Avian HBV have two similar short conserved uORF (uORF1, uORF2) which start near the end of the ε structure. This figure is adapted with permission from Beck J, et al.17 Also shown are the associated interacting factors involved in the encapsidation process. Domains of the P protein are abbreviated as follows: terminal protein (TP), RNase H domain (RH), reverse transcriptase domain (RT). Open circles represent cellular chaperone also essential in assisting P protein to bind to the ε. The sequence and numbering according to DDBJ accession number AB037684, the number 35 corresponds to nt number 1850 in the ayw subtype. The sequence of the DHBV is from K01834 and the HHBV is from M22056.
Most of the encapsidation process for hepadnaviruses were determined from studies done on the avian Duck hepatitis B virus (DHBV) in vitro.13,20,22,23 In these studies, the sequence and structure at the upper stem, bulge and also sequences at the upper region of the lower stem were shown to be important for both P protein binding and encapsidation.13,24,25 Interestingly, despite its secondary structure the ε region has been targeted effectively by RNAi.26
There are significant variations between the different members of hepadnaviruses. These include notable primary sequence difference within the ε element between the avian and mammalian hepadnaviruses. There are also distinct differences in binding requirements for P protein at the upper stem which is less well based paired in most avian hepadnaviruses (except some DHBV, Fig. 2). In addition, the initiation of DNA synthesis successfully shown in the DHBV system in vitro has so far unable to be shown for HBV, indicating significant differences in the elements.
This study aims to build covariance models of hepadnavirus ε elements that will uniquely identify them. These can be used to investigate the similarities between these models and to other known, or previously undetected, RNA structural elements.
Results
Generation of covariance models for hepadnavirus ε elements
The ε element is well conserved in overall structure between the mammalian and avian hepadnaviruses, despite the viruses having significant genome divergence and differing in the presence or absence of other cis-regulatory elements.27
The hepadnaviruses ε elements share common structural features, namely (1) lower stem, (2) the central bulge, (3) upper stem with the apical loop where the P protein binds (Fig. 2).20 The secondary structure of the HBVε and DHBVε have extensive base–pairing in both stems, while a more open (reduced base-pairing) structure in the upper stem region was observed for the HHBVε (Fig. 2). Despite sharing similarities to the HBVε at the base–paired upper stem, the DHBVε had a less stable (thermally unstable) upper stem than initially believed and could potentially assume an open structure similar to HHBVε under physiological conditions.20 Both the DHBVε and HHBVε have a stable tetra-loop (UGUU) including a non-canonical UU base pair in the apical loop, while the HBVε has a tri-loop (UGU).
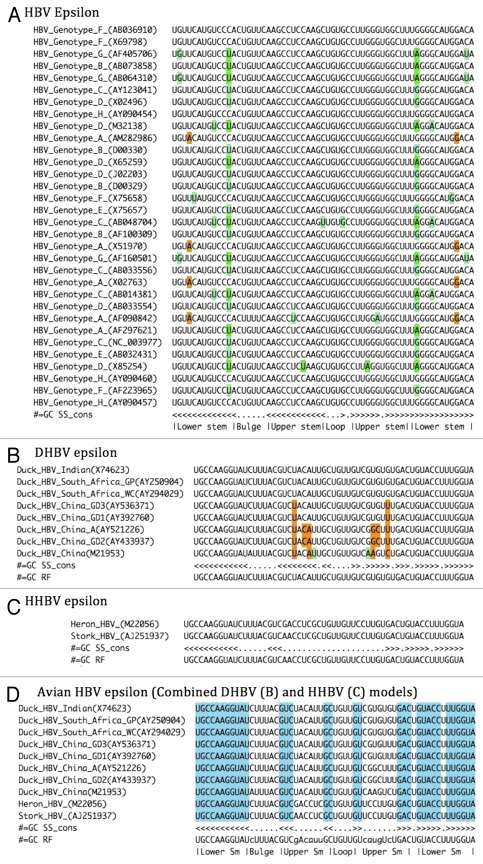
The sequences of the ε element from representative mammalian and avian hepadnaviruses were extracted from public DNA databases (Methods). The sequences were chosen to represent the diversity of HBV genotypes (A-H) in a reference alignment used in Panjaworayan et al.28 Although the genotypes differ by over 8% sequence overall, the ε element is highly conserved due to its multiple functions. The secondary structure is conserved in all 32 members of the reference alignment, except for an A-G mismatch in the middle of the lower stem in all four genotype A viruses (orange in Fig. 3A). This mismatch is unexpected, but non-canonical A-G base pairs can be accommodated with some distortion within an A-helix.29 However, this may indicate structural tolerance at this position. The closely related orthohepadnaviruses (ground squirrel and woodchuck hepatitis virus) ε elements have an inserted C after this point, also indicating tolerance (Methods).
Figure 3. Alignments of families of epsilon elements—HBV (A) DHBV (B) HHBV (C) a combined model AHBV (D). The SS line represents the consensus structure in dot-bracket notation, dots are unpaired, brackets are paired. In A–C, compensating base changes are depicted in green, base pairs incompatible with the consensus structure in orange. In the combined model D, blue shading represents compatibility with the structure line (SS_cons). Stem (Sm) loops and bulges are indicated. These Stockholm format files and models are available in the Supplementary Material.
Some sequences show compensating base changes within the structure (green in Fig. 3A). These changes give independent support for the existence of these base pairs. Orthohepadnaviruses also have two of these compensating base changes, but no additional changes not seen in the HBV alignment. Notably one HBV genotype C (AB048704) has a compensating G-U closing pair adjacent to the apical tri-loop, providing additional covariance support for this pair previously observed in the NMR structure30
A multiple alignment was assembled and manually refined by structure and sequence conservation to form a curated seed alignment (Fig. 3). Alignments of these four elements: HBV ε, DHBV ε, HHBV ε, and a combination of these two—Avian HBV epsilon (AHBV ε), are shown in Figure 3A-D and available in supplementary files. HBV_epsilon (RF01407) and AHBV_epsilon (RF01313) are also available through Rfam with corresponding Wikipedia entries.
Due to the significant sequence difference and function between the ε element of mammalian, heron and duck hepadnaviridae, alignments were initially done for each and separate covariance models built for each family (Fig. 3B, C). For DHBV there are several Chinese isolates for which positions in the upper stem in which the bases are incompatible with a canonical structure (e.g., C-C, C-U, AY433937 China_GD2, orange Fig. 3B). This observation supports the notion that this helix may unstable in DHBV, similar to HHBV (Fig. 3C).25 This contrasts with solution structures of a South African isolate of DHBV (e.g., AY250904, Fig. 3B) that show the upper stem extensively paired in an isolated RNA.31 A combined avian model (Fig. 3D) with less pairing in the upper stem (similar to HHBV, C) permits most pairs to be compatible (blue shading Fig. 3D). These four models were used for further analysis.
Comparison of the ε Models to Each Other to All Other Rfam Families
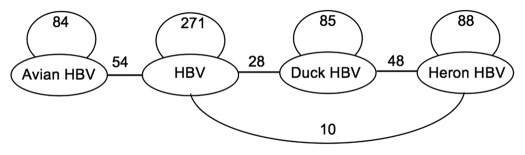
The four covariance models were compared with each other using CMCompare. In a comparison of related and unrelated Rfam models a score of over 20, or E < 1.0 were considered worthy of note. However about 7.4% of pairwise comparisons of Rfam models had scores over 20, and 6.3% over 28.32 The HHBV and DHBV models are most similar (Score 48, Fig. 4), with HBV and DHBV less similar (Score 28). The combined avian model (AHBV, left) showed greater similarity (Score 54) to the HBV model than either alone (Scores 28,10). The maximum possible scores for these models, matches to themselves, are shown in Figure 4 (Scores 84, 271, 85, 88). The elements have sufficient overall functional and structural similarity to form a new clan of Rfam models.
Figure 4. Similarity between the covariance models. The models were compared using CMCompare. Higher scores indicate greater similarity. The Avian model AHBV ε (left) is a combination of the Heron (HHBV epsilon) and Duck (DHBV epsilon) models (right). The maximum similarity score a model could have is that with itself, greater than 20 is likely significant. The next most similar model had score of 26 (see results).
CMCompare was also used to compare the ε models to all other families in Rfam. Weak matches with scores of 20–26 were seen with many other elements, generally a match to stem loop subregion of the alignment. The best matches were between MicC non-coding RNA (RF00121) and HBVε (Score 26), and Equine arteritis virus (EAV) leader TRS hairpin (LTH) (RF00498) and DHBVε (Score 21). Interestingly the EAV hairpin has a role in minus stranded RNA synthesis in that RNA virus.33
However in general, the new ε models are not structurally similar to functionally related replication elements of other viruses (scores <15), for example, Hepatitis C virus or retroviruses (HIV-1 DIS, RF0015). There are nine replication elements in Rfam from human viruses- Entero_CRE (RF00048), Entero_5_CRE, Flavi_CRE (RF00185), HepC_CRE (RF00260), Cardiovirus CRE, Rota_CRE, and plant viruses- Tombus_CRE (RF00510), CTV_rep_sig (RF00193). Although all these replication elements form at least a stem loop structure, they are structurally distinct from the clan proposed here, so are not included as part of the clan.
Searching Sequence Databases for Similar Elements
These four covariance models (Fig. 3 and 4) were calibrated (using cmcalibrate) and used to search on both strands of all the curated RefSeq viral genomes, the viral division of GenBank and RFamSeq10 using cmsearch (Methods). Cmsearch generates a bit score report based on the match of the model to the sequence. It also provides an E value (which corresponds to the expected number of false positives in a database of this size). Hits with E values of <0.1 are considered trustworthy.34
The HBV ε model was built from sequences representing the diversity of the common HBV genotypes. It has significant matches to 6910 sequences in the RFamSeq10 database, all of which are from mammalian HBVs. These matches represent the diversity of HBV genotypes (A-H). The search identified some additional mammalian HBV viruses, e.g., woodchuck HBV. Some apparently diverse matches are due to misclassifications in the EMBL taxonomy: one match is classed by EMBL and RFamSeq10 as being a hepatitis A virus, but is clearly a HBV, one is classed as rock squirrel genome, but is a rock squirrel HBV. There is also a match to a synthetic human HBV containing construct. The next best match in RFamSeq10 is not significant, indicating that good matches to this element are not found in cellular genomes (e.g., the human host genome). Although portions of HBV can be integrated into the human genome, it is not unexpected that the reference human genome does not contain an ε like sequence. Similar results were obtained from the GenBank viral division and RefSeq viruses.
DHBV ε, HHBV ε and the combined AHBV epsilon model all match the same six avian hepadnaviruses in RefSeq viruses (Scores: 53–83, E < 10−9) with 54 hits in RfamSeq10, and 58 hits in the viral division of GenBank. Indeed, the combined avian model identifies the same set with better scores (Score > 40, E < 5 × 10−3). This combined model constitutes Rfam model RF01313. The separate DHBV, HHBV epsilon models presented here are also available (Supplement). Notably the combined model recognizes divergent viruses (e.g., Stork HBV sequences (AJ251937).25
The next best matches in viral genomes are marginal matches to long bacteriophage DNA genomes (DHBV, NC_015289, Score: 24, E = 0.43; HHBV, NC_012697, Score: 22, E = 0.92). The matched regions encode bacteriophage proteins and do not appear to be biologically significant.
There were no significant matches to other retro-transcribing viruses (best Score: 5.0, E = 1.6). There were also no significant matches to HDV, this is not unexpected, as HDV is only dependent on HBV for envelopment and not encapsidation.
Discussion
We have shown that the RNA families of ε replication elements proposed here comprise a clan with both RNA structural and functional similarities. The hepadnavirus ε plays several key roles in the viral lifecycles and has a similar role in the families although they are sufficiently distinct to comprise two or more separate families. Although we tested three separate models it was found that a human and avian model provided the best discrimination between matches and false positives. This is consistent with experimental data that shows that the upper stem in which basepairing may differ between avian viruses is very tolerant to variation25
Therefore this work also suggests that basepairing in the upper stem is not essential.
Although the ε replication element has some similar functions to replication elements from other retro-transcribing and RNA viruses we could detect no significant similarity beyond that expected of a stem-loop structure. This was determined by directly comparing the models to one another. This is consistent with published functional studies where viral or host proteins are specific for the replication elements in the corresponding viruses.35
Searches of known viral and non-viral sequences showed that the models can specifically detect the elements in the context of a viral genome within large databases of sequences. They revealed no significant matches to the mammalian host genomes, although the genomes of duck or other infected birds are not yet available. This type of search with a covariance model is more tolerant of substitutions/covariations within the structure than traditional blastn searches. Therefore this analysis supports the idea that this element is a virus specific target.
The models proposed here are specific for the hepadnaviruses. These models add to the basis for further research into the specific bases and structures required for the ε functions in the viral lifecycle and also potential antiviral strategies targeting these elements. Indeed RNAi strategies against the conserved HBV ε region have been effective despite its secondary structure. This structure was expected to reduce the ability of siRNAs or synthetic miRNAs to target this region.36 In addition, the protein/RNA interaction in replication and the initiation of replication has been a target of anti-HBV drugs.37
Materials and Methods
Sequences were extracted representing the diversity of mammalian and avian hepadnavirus genomes. The principal features of the structures in different functional states were extracted from the literature. For HBV there are over 6000 sequences in public databases but many are identical in this region. The sequences chosen were from a previously published HBV reference set to represent common diversity and are similar to other published genotyping sets for HBV. The sequences of orthohepadnaviruses were also compared (NC_001484, NC_004107). They have an inserted C relative to HBV at position 5 (UGUUCCA) and several compensating changes also found in HBV genomes.
For HHBV and DHBV, fewer sequences are available. There are other members of the avian hepadnaviruses but there is currently too little diversity in the data and insufficient experimental evidence to form an alignment from which to build separate models.
Alignments were done manually using AquaEmacs in Ralee mode guided by structural probing and NMR studies (PDB:2OJ7, 2OJ8, 2K5Z, 2IXY, 2IXZ) and considering the modeling done of the lower stem by other groups.38 These structures were determined by chemical and enzymatic probing and also NMR analyses on HBVε, DHBVε and HHBVε.13,15,23,30,31,39-41 Compatibility with minimum free energy structures was ascertained by folding individual sequences. Covariance models were built from alignments using cmbuild 1.0.2 and calibrated using cmcalibrate. Comparison of models was done with CMCompare.32 The HBV_Epsilon and AHBV_Epsilon have been submitted to Rfam as RF01407 and RF01313, and comprise Rfam clan: CLN00104.
Data sets analyzed. Sequences were searched against each calibrated model using cmsearch (for RefSeq) or Rfam_scan followed by cmsearch for larger databases42 Three data sets were used—(1) RefSeq 47 (12/5/2011) viral genomes—a curated set of viral genomes with limited redundancy (2) the viral division of GenBank (4/7/2011) and (3) the most recent RFAMSEQ, 10, derived by Rfam from EMBL 100 (29/5/2009).
Supplementary Material
HBV_epsilon.stk (S1)
DHBV_epsilon.stk (S2)
HHBV_epsilon.stk (S3)
AHBV_epsilon.stk (S4)
DHBV_epsilon.cm (S5)
HHBV_epsilon.cm (S6)
Supplementary PDF file supplied by authors.
Acknowledgments
Part of this work was supported by a NZ Health Research Council Grant (05/195) to Warren Tate, Elizabeth Poole and CMB and a Human Frontier Science Program grant to Ian Macara, Anne Spang and CMB (RGP0031_2009). Thanks to Jennifer Daub at Rfam for helpful suggestions on the models.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/18649
References
- 1.Chen DS. Toward elimination and eradication of hepatitis B. J Gastroenterol Hepatol. 2010;25:19–25. doi: 10.1111/j.1440-1746.2009.06165.x. [DOI] [PubMed] [Google Scholar]
- 2.World Heath Organisation. Hepatitis B. Available: www.who.int/csr/disease/hepatitis/HepatitisB_whocdscsrlyo2002_2.pdf [Accessed Jun 2011]. 2002.
- 3.Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51–68. doi: 10.1128/MMBR.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tillmann HL. Antiviral therapy and resistance with hepatitis B virus infection. World J Gastroenterol. 2007;13:125–40. doi: 10.3748/wjg.v13.i1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song BC. How to overcome antiviral-resistant hepatitis B virus? Intervirology. 2010;53:29–38. doi: 10.1159/000252781. [DOI] [PubMed] [Google Scholar]
- 6.Fouillot N, Tlouzeau S, Rossignol JM, Jean-Jean O. Translation of the hepatitis B virus P gene by ribosomal scanning as an alternative to internal initiation. J Virol. 1993;67:4886–95. doi: 10.1128/jvi.67.8.4886-4895.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen A, Kao YF, Brown CM. Translation of the first upstream ORF in the hepatitis B virus pregenomic RNA modulates translation at the core and polymerase initiation codons. Nucleic Acids Res. 2005;33:1169–81. doi: 10.1093/nar/gki251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin CG, Lo SJ. Evidence for involvement of a ribosomal leaky scanning mechanism in the translation of the hepatitis B virus pol gene from the viral pregenome RNA. Virology. 1992;188:342–52. doi: 10.1016/0042-6822(92)90763-F. [DOI] [PubMed] [Google Scholar]
- 9.Hwang WL, Su TS. Translational regulation of hepatitis B virus polymerase gene by termination-reinitiation of an upstream minicistron in a length-dependent manner. J Gen Virol. 1998;79:2181–9. doi: 10.1099/0022-1317-79-9-2181. [DOI] [PubMed] [Google Scholar]
- 10.Ryu DK, Ahn BY, Ryu WS. Proximity between the cap and 5′ epsilon stem-loop structure is critical for the suppression of pgRNA translation by the hepatitis B viral polymerase. Virology. 2010;406:56–64. doi: 10.1016/j.virol.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Hwang WL, Su TS. The encapsidation signal of hepatitis B virus facilitates preC AUG recognition resulting in inefficient translation of the downstream genes. J Gen Virol. 1999;80:1769–76. doi: 10.1099/0022-1317-80-7-1769. [DOI] [PubMed] [Google Scholar]
- 12.Cao F, Tavis JE. RNA Elements Directing Translation of the Duck Hepatitis B Virus Polymerase via Ribosomal Shunting. J Virol 2011. [DOI] [PMC free article] [PubMed]
- 13.Beck J, Nassal M. Hepatitis B virus replication. World J Gastroenterol. 2007;13:48–64. doi: 10.3748/wjg.v13.i1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nassal M, Junker-Niepmann M, Schaller H. Translational inactivation of RNA function: discrimination against a subset of genomic transcripts during HBV nucleocapsid assembly. Cell. 1990;63:1357–63. doi: 10.1016/0092-8674(90)90431-D. [DOI] [PubMed] [Google Scholar]
- 15.Beck J, Bartos H, Nassal M. Experimental confirmation of a hepatitis B virus (HBV) epsilon-like bulge-and-loop structure in avian HBV RNA encapsidation signals. Virology. 1997;227:500–4. doi: 10.1006/viro.1996.8329. [DOI] [PubMed] [Google Scholar]
- 16.Rieger A, Nassal M. Specific hepatitis B virus minus-strand DNA synthesis requires only the 5′ encapsidation signal and the 3′-proximal direct repeat DR1. J Virol. 1996;70:585–9. doi: 10.1128/jvi.70.1.585-589.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beck J, Nassal M. Efficient Hsp90-independent in vitro activation by Hsc70 and Hsp40 of duck hepatitis B virus reverse transcriptase, an assumed Hsp90 client protein. J Biol Chem. 2003;278:36128–38. doi: 10.1074/jbc.M301069200. [DOI] [PubMed] [Google Scholar]
- 18.Beck J, Nassal M. Formation of a functional hepatitis B virus replication initiation complex involves a major structural alteration in the RNA template. Mol Cell Biol. 1998;18:6265–72. doi: 10.1128/mcb.18.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tavis JE, Massey B, Gong Y. The duck hepatitis B virus polymerase is activated by its RNA packaging signal, epsilon. J Virol. 1998;72:5789–96. doi: 10.1128/jvi.72.7.5789-5796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nassal M. Hepatitis B viruses: reverse transcription a different way. Virus Res. 2008;134:235–49. doi: 10.1016/j.virusres.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 21.Hu J, Toft DO, Seeger C. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 1997;16:59–68. doi: 10.1093/emboj/16.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu K, Beck J, Nassal M. SELEX-derived aptamers of the duck hepatitis B virus RNA encapsidation signal distinguish critical and non-critical residues for productive initiation of reverse transcription. Nucleic Acids Res. 2004;32:4377–89. doi: 10.1093/nar/gkh772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beck J, Nassal M. Sequence- and structure-specific determinants in the interaction between the rna encapsidation signal and reverse transcriptase of avian hepatitis B viruses. J Virol. 1997;71:4971–80. doi: 10.1128/jvi.71.7.4971-4980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu J, Boyer M. Hepatitis B virus reverse transcriptase and epsilon RNA sequences required for specific interaction in vitro. J Virol. 2006;80:2141–50. doi: 10.1128/JVI.80.5.2141-2150.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmid B, Rosler C, Nassal M. High mutation tolerance in the multifunctional sequence encoding the RNA encapsidation signal of an avian hepatitis B virus and slow evolution revealed by in vivo infection. J Virol. 2011;85:9300–13. doi: 10.1128/JVI.05005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun D, Rosler C, Kidd-Ljunggren K, Nassal M. Quantitative assessment of the antiviral potencies of 21 shRNA vectors targeting conserved, including structured, hepatitis B virus sites. J Hepatol. 2010;52:817–26. doi: 10.1016/j.jhep.2009.10.038. [DOI] [PubMed] [Google Scholar]
- 27.Maguire ML, Loeb DD. cis-Acting sequences that contribute to synthesis of minus-strand DNA are not conserved between hepadnaviruses. J Virol. 2010;84:12824–31. doi: 10.1128/JVI.01487-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panjaworayan N, Roessner S, Firth A, Brown C. HBVRegDB: Annotation, comparison, detection and visualization of regulatory elements in hepatitis B virus sequences. Virol J. 2007;4:136. doi: 10.1186/1743-422X-4-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flodell S, Schleucher J, Cromsigt J, Ippel H, Kidd-Ljunggren K, Wijmenga S. The apical stem-loop of the hepatitis B virus encapsidation signal folds into a stable tri-loop with two underlying pyrimidine bulges. Nucleic Acids Res. 2002;30:4803–11. doi: 10.1093/nar/gkf603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ampt KA, van der Werf RM, Nelissen FH, Tessari M, Wijmenga SS. The unstable part of the apical stem of duck hepatitis B virus epsilon shows enhanced base pair opening but not pico- to nanosecond dynamics and is essential for reverse transcriptase binding. Biochemistry. 2009;48:10499–508. doi: 10.1021/bi9011385. [DOI] [PubMed] [Google Scholar]
- 32.Höner zu Siederdissen C, Hofacker IL. Discriminatory power of RNA family models. Bioinformatics. 2010;26:i453–9. doi: 10.1093/bioinformatics/btq370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Den Born E, Gultyaev AP, Snijder EJ. Secondary structure and function of the 5′-proximal region of the equine arteritis virus RNA genome. RNA. 2004;10:424–37. doi: 10.1261/rna.5174804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–7. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Wimmer E, Paul AV. Cis-acting RNA elements in human and animal plus-strand RNA viruses. Biochem Biophys Acta. 2009;1789:495–517. doi: 10.1016/j.bbagrm.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panjaworayan N, Brown CM. Effects of HBV Genetic Variability on RNAi Strategies. Hepat Res Treat. 2011;2011:367908. doi: 10.1155/2011/367908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin L, Hu J. Inhibition of hepadnavirus reverse transcriptase-epsilon RNA interaction by porphyrin compounds. J Virol. 2008;82:2305–12. doi: 10.1128/JVI.02147-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan HR, Zhu HJ, Song ZR, Zhang XD, Cao H. Molecular modeling of epsilon RNA from hepatitis B virus. Chin Chem Lett. 2009;20:1263–6. doi: 10.1016/j.cclet.2009.05.018. [DOI] [Google Scholar]
- 39.Flodell S, Petersen M, Girard F, Zdunek J, Kidd-Ljunggren K, Schleucher J, et al. Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal. Nucleic Acids Res. 2006;34:4449–57. doi: 10.1093/nar/gkl582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Girard FC, Ottink OM, Ampt KA, Tessari M, Wijmenga SS. Thermodynamics and NMR studies on Duck, Heron and Human HBV encapsidation signals. Nucleic Acids Res. 2007;35:2800–11. doi: 10.1093/nar/gkm131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ampt KA, Ottink OM, Girard FC, Nelissen F, Tessari M, Wijmenga SS. 1H, 13C and 15N NMR assignments of Duck HBV apical stem loop of the epsilon encapsidation signal. Biomol NMR Assign. 2008;2:159–62. doi: 10.1007/s12104-008-9110-z. [DOI] [PubMed] [Google Scholar]
- 42.Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, et al. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res. 2011;39:D141–5. doi: 10.1093/nar/gkq1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HBV_epsilon.stk (S1)
DHBV_epsilon.stk (S2)
HHBV_epsilon.stk (S3)
AHBV_epsilon.stk (S4)
DHBV_epsilon.cm (S5)
HHBV_epsilon.cm (S6)
Supplementary PDF file supplied by authors.