Abstract
Screening with hepatitis B surface antigen (HBsAg) is highly recommended for at-risk individuals. Mutations in the HBsAg can result in an inability to detect the virus during routine screening. We describe a hemodialysis patient found to have high levels of hepatitis B virus (HBV) DNA and HBV antibody but negative HBsAg on two routine assays.
CASE REPORT
In May of 2011, a 57-year-old woman with a prior lung transplant and hemodialysis-dependent end-stage renal disease underwent evaluation for renal transplant at an outside facility. She received a left lung transplant in 2000 due to severe pulmonary fibrosis as a consequence of talc toxicity from previous intravenous drug use. Her last reported intravenous drug use was in 1993. As part of her evaluation for kidney transplant, hepatitis serologies were drawn, including hepatitis B surface antibody (anti-HBs), hepatitis B surface antigen (HBsAg), hepatitis B e antigen, and hepatitis B virus (HBV) DNA. One week after evaluation for transplant, the patient presented to a different outside hospital with complaints of fevers, chills, and shortness of breath. She was transferred to this facility for further treatment of pneumonia. As part of routine inpatient dialysis screening, the patient had an HBsAg assay performed in our hospital that returned negative.
However, 1 week following her admission, the results of the HBV workup done as part of her kidney transplant evaluation revealed positive HBsAg, anti-HBs with a titer of 13.18 mIU/ml, positive hepatitis B e antigen, and negative hepatitis B e antibody results and an HBV DNA level of 11,188,000 IU/ml (Table 1). Standard hemodialysis isolation protocols for active HBV infection were instituted. These results prompted review of her records from this institution, which also demonstrated negative HBsAg during previous hospitalizations (7/1999, 1/2006, 2/2010, 8/2010, and 1/2011 [month/year]). The patient's dialysis unit was also contacted regarding previous HBV testing. She had HBsAg, hepatitis B core antibody (anti-HBc), and anti-HBs testing performed at the start of hemodialysis in March of 2010, which revealed a negative HBsAg result and was positive for both anti-HBc and anti-HBs (Table 1). Based on these results, she was classified as immune to HBV, and no further HBV testing was performed by the outpatient dialysis unit. The patient previously had received HBV vaccination with two doses of Recombivax (Merck), the last in February 2006. Because the tests from her kidney transplant evaluation were suggestive of an active HBV infection and testing here failed to reveal the presence of HBsAg, repeat HBV serologies and HBV DNA were sent. HBsAg testing performed as part of the renal transplant workup was with the AxSYM assay from Abbott Diagnostics (Abbott Park, IL), which utilizes microparticle enzyme immunoassay (EIA) technology. HBsAg testing performed in March of 2010 at her previous dialysis unit and at our hospital during this admission was assessed using direct chemiluminescence with the Advia Centaur assay from Bayer Diagnostics (Tarrytown, NY). Due to these discrepant results, HBsAg EIA was repeated using ETI-MAK-2 PLUS (Diasorin, Piscataway, NJ). HBV DNA was assessed using real-time PCR (COBAS AmpliPrep, Roche Diagnostics). Based on the results of these tests, HBV DNA sequencing to evaluate for mutations was performed as follows.
Table 1.
Results of HBV serologic testing
| Test | Dialysis unit | Outside hospital | This hospitalization |
|---|---|---|---|
| HBsAg | Negativea | Positiveb | Negativea/negativec |
| HBeAg | Positive | Positive | |
| HBeAb | Negative | Negative | |
| HBsAb | Reactive | Reactive | Reactive |
| HBsAb Quant (in mIU/ml) | 13.18 | 17 | |
| Hbcore Ab (IgG+IgM) | Reactive | Reactive | Reactive |
| Hbcore IgM | NAd | Nonreactive | |
| HBV DNA (in IU/ml)e | 11,188,000 | 72,800,000 | |
| HBV (log)e | 7.049 | 7.86 |
Test performed using Advia Centaur.
Test performed using Abbott AxSYM.
Performed using ETI-MAK-2PLUS Diasorin kit.
NA, not available.
HBV DNA assessed using real-time PCR.
HBV DNA was extracted from 200 μl of serum using a QIAamp DNA blood minikit (Qiagen, Valencia, CA) according to the manufacturer's instructions. The extracted DNA was eluted in a final volume of 50 μl of the elution buffer supplied. For the first round of a nested PCR, 5 μl of this extraction was amplified with the following primers: 5′-GCCTCATTTTGTGGGTCACCATA-3′ and 5′-AGTTCCGCAGTATGGATCGG-3′. A second round of amplification was performed using 2 μl of the first-round product with the following primers: 5′-TTGGGGTGGAGCCCTCAGGCT-3′ and 5′-GTGGGGGTTGCGTCAGCA-3′.
The amplification conditions have been previously described (1). The amplified product was purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA) according to the manufacturer's instructions and directly sequenced at the Johns Hopkins core sequencing facility using the following primers: 5′-TTGGGGTGGAGCCCTCAGGCT-3′, 5′-GAAGATGAGGCATAGCAGCAGG-3′, 5′-TTGGCCAAAATTCGCAGTC-3′, and 5′-GTGGGGGTTGCGTCAGCA-3′.
The HBV consensus sequence was constructed using SeqScape sequence analysis software (Applied Biosystems, version 2.5). The resulting consensus sequence contained the envelope S gene and the polymerase catalytic units of HBV, overlapping in a frame-shifted manner. The HBV genotype and unique mutations were identified by comparing the consensus sequence to the genotype D reference sequence (GenBank accession number X02496.1) (2) using CodonCode Aligner software (version 3.0.3).
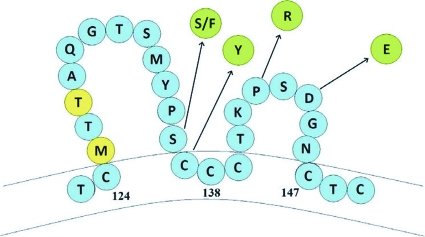
The HBsAg was negative when tested on both the Advia Centaur and ETI-MAK-2PLUS kits. The HBV DNA level was 72,800,000 IU/ml. The other serologies were similar to the previous testing performed at the outside facility (Table 1). At the time these laboratory test samples were drawn, the patient's aspartate aminotransferase (AST), alanine aminotransferase (ALT), and bilirubin levels were within normal limits. The HBV DNA sequencing revealed that the virus was genotype D with envelope mutations as follows: substitution I92I/T (sI92I/T), sY100Y/F, sQ101R, sS136S/F, sC137Y, sP142R, sD144E, and sQ181R. The amino acid substitutions for the “a” determinant of the HBsAg between residues 124 and 147 are included in Fig. 1.
Fig 1.
Hepatitis B surface antigen mutations with amino acid substitutions. The amino acids in blue represent the wild-type for genotype D HBV. The amino acids in yellow are polymorphisms, while those in green are substitutions.
Chronic hepatitis B virus (HBV) infection is common worldwide, with an estimated 350 million people afflicted with the disease and approximately 600,000 deaths annually (9, 20). Screening with hepatitis B surface antigen (HBsAg), hepatitis B core antibody, and hepatitis B surface antibody are highly recommended for at-risk individuals, and early detection is advocated in order to ensure early access to treatment and future monitoring (20).
Mutations in the HBsAg can result in an inability to detect the virus through routine screening methods, leading to occult HBV infections, thereby creating a diagnostic dilemma. The prevalence of occult HBV infections is not well established (18). In one study that included only hemodialysis patients in North America, the prevalence of occult HBV was 3.5% (13). Given the highly infectious nature of the HBV virus, the failure to detect active disease could result in increased exposure risk to personal contacts and to both patients and health care workers in the health care setting.
In this case, the DNA level indicates an active HBV infection despite an undetectable HBsAg on several assays. Importantly, in addition to the HBsAg not being detected, the patient had a protective level of anti-HBs, which was not able to neutralize her virus. If such mutant viruses are more widespread than recognized, this would have significant public health implications for the hemodialysis patient population. These patients could be exposed to a mutant HBV virus that has the potential to infect the immunized population. Though more prominent outside the United States, occult HBV infections in North America (13) and in the United States have been described (17). In occult HBV infections, however, the HBV DNA is typically low (less than 200 IU/ml) and should be distinguished from “false” occult HBV infections (16). In “false” occult HBV infections, HBV variants with mutations occurring in the S gene produce an altered HBsAg that goes undetected by several commercially available FDA-approved assays but maintain viral loads consistent with active infection (16). Though these variants have been reported in European, African, and Asian patient populations (19), these mutants have been described only in limited numbers in the United States, among liver transplant recipients with previously known history of HBV infection or in the setting of donor-positive hepatitis B core antibody and de novo infection despite previous vaccination in the recipient (7, 14). In this patient, the circulating levels of anti-HBs would typically confer immunity. This antibody, derived from either recovery from acute infection or vaccination, is directed against the “a” determinant of the HBsAg, which is located between amino acid residues 124 and 147 (5). Most of the antigenicity of the “a” determinant is located from residues 138 to 147, which is known as the major hydrophilic region (3). Despite the circulating antibody in this case, there is a high level of HBV DNA, indicating that the antibody is not neutralizing the patient's virus. Amino acid substitution within the HBsAg “a” determinant residues is probably the predominant mechanism by which the virus can escape antibody detection, leading to subsequent selection of a mutant strain, subsequent failure of commercially available assays to detect the HBsAg, or both (19). Our patient had multiple envelope mutations within this amino acid range that could be responsible for the failure to detect the HBsAg and for escape of the anti-HBs. In particular, the sD144E substitution that is present in the patient's virus has been implicated as a means of immune escape (10). The inability of certain assays to detect HBsAg mutants has been noted (11, 8, 6, 15). Two studies have found that the Advia Centaur detection assay failed to identify HBsAg variants that were detected by other assays (11, 8). In this patient, the HBsAg was not detected by the Advia Centaur assay or the Diasorin kit but was identified with the Abbott AxSYM assay, which has been noted to better detect these HBV mutants (11). However, this assay does not capture all mutants, and even multivalent HBsAg assays miss some of these mutant strains (12). The accurate detection of HBV is critical since patients with these “false” occult HBV infections could potentially be offered antiviral therapy and should undergo routine surveillance for the development of hepatocellular carcinoma and cirrhosis.
In the dialysis population, such identification is vital, as current recommendations for active HBV infections are to have designated separate rooms for treatment, as well as dedicated machines, instruments, supplies, and medications. Furthermore, staff members caring for patients with active HBV should not care for susceptible patients at the same time (4). Because this patient was classified as being HBV immune based on her initial testing at the start of outpatient hemodialysis, she was not placed in isolation for her treatments. The inability to detect HBsAg by routine screening and lack of clinical evidence suggesting hepatic inflammation could have resulted in a prolonged exposure risk to health care workers and other hemodialysis patients, especially if she harbors a variant capable of vaccine escape.
Furthermore, lack of detection could cause failure to consider necessary therapeutic intervention and future risk management. Of particular concern is that it is unknown how this patient acquired her strain of hepatitis B and the fact that she had multiple negative HBsAg assays over the preceding years. The mechanisms by which she developed active hepatitis B could include a mutation in an already existing but previously undetectable strain (occult hepatitis B infection), with immune escape from the host's either naturally occurring or vaccination-induced immunity, or by acquiring a mutant strain via horizontal transmission that was able to escape the patient's previous hepatitis B vaccination. It is the latter possibility which raises the greatest concern, as it may signify strains of hepatitis B now in the U.S. community capable of escaping both previous hepatitis B immunity and routine HBsAg screening with certain commercial assays. Because of this finding, it is essential that diagnostic assays that can detect these mutants be developed. In populations such as dialysis patients, screening of high-risk individuals should be performed with assays with greater capacity to detect mutant strains of HBsAg. Until assays are developed that can reliably detect these mutant strains, consideration should be given to screening dialysis patients who are anti-HBc positive with HBV DNA.
ACKNOWLEDGMENT
C.L.T. is funded by NIH grants R01AI071820 and R56AI060449.
Footnotes
Published ahead of print 21 March 2012
REFERENCES
- 1. Ayres A, Locarnini S, Bartholomeusz A. 2004. HBV genotyping and analysis for unique mutations. Methods Mol. Med. 95:125–149 [DOI] [PubMed] [Google Scholar]
- 2. Bichko V, Pushko P, Dreilina D, Pumpen P, Gren E. 1985. Subtype ayw variant of hepatitis B virus. DNA primary structure analysis. FEBS Lett. 185:208–212 [DOI] [PubMed] [Google Scholar]
- 3. Carman WF, et al. 1990. Vaccine-induced escape mutant of hepatitis B virus. Lancet 336:325–329 [DOI] [PubMed] [Google Scholar]
- 4. Centers for Disease Control and Prevention 2001. Recommendations for preventing transmission of infections among chronic hemodialysis patients. MMWR Recommend. Rep. 50(RR-5):1–43 [PubMed] [Google Scholar]
- 5. Coleman PF, Chen YC, Mushahwar IK. 1999. Immunoassay detection of hepatitis B surface antigen mutants. J. Med. Virol. 59:19–24 [DOI] [PubMed] [Google Scholar]
- 6. Echevarria JM, Avellon A. 2008. Improved detection of natural hepatitis B virus surface antigen (HBsAg) mutants by a new version of the VITROS HBsAg assay. J. Med. Virol. 80:598–602 [DOI] [PubMed] [Google Scholar]
- 7. Ghany MG, et al. 1998. Hepatitis B virus S mutants in liver transplant recipients who were reinfected despite hepatitis B immune globulin prophylaxis. Hepatology 27:213–222 [DOI] [PubMed] [Google Scholar]
- 8. Gibb R, Nimmo GR, O'Loughlin P, Lowe P, Drummond D. 2007. Detection of HBsAg mutants in a population with a low prevalence of hepatitis B virus infection. J. Med. Virol. 79:351–355 [DOI] [PubMed] [Google Scholar]
- 9. Goldstein ST, et al. 2005. A mathematical model to estimate global hepatitis B disease burden and vaccination impact. Int. J. Epidemiol. 34:1329–1339 [DOI] [PubMed] [Google Scholar]
- 10. Kim KH, et al. 2003. Evolution of hepatitis B virus sequence from a liver transplant recipient with rapid breakthrough despite hepatitis B immune globulin prophylaxis and lamivudine therapy. J. Med. Virol. 71:367–375 [DOI] [PubMed] [Google Scholar]
- 11. La'ulu SL, Roberts WL. 2006. The analytic sensitivity and mutant detection capability of six hepatitis B surface antigen assays. Am. J. Clin. Pathol. 125:748–751 [DOI] [PubMed] [Google Scholar]
- 12. Launay O, et al. 2011. High levels of serum hepatitis B virus DNA in patients with “anti-HBc alone”: role of HBsAg mutants. J. Viral Hepat. 18:721–729 [DOI] [PubMed] [Google Scholar]
- 13. Minuk GY, et al. 2004. Occult hepatitis B virus infection in a North American adult hemodialysis patient population. Hepatology 40:1072–1077 [DOI] [PubMed] [Google Scholar]
- 14. Moraleda G, et al. 2006. De novo HBV infection caused by an anti-HBc positive donor in a vaccinated liver transplant recipient in spite of anti-HBs response. Am. J. Transplant. 6:438–440 [DOI] [PubMed] [Google Scholar]
- 15. Muhlbacher A, et al. 2008. Multicenter study of a new fully automated HBsAg screening assay with enhanced sensitivity for the detection of HBV mutants. Med. Microbiol. Immunol. 197:55–64 [DOI] [PubMed] [Google Scholar]
- 16. Raimondo G, et al. 2008. Statements from the Taormina expert meeting on occult hepatitis B virus infection. J. Hepatol. 49:652–657 [DOI] [PubMed] [Google Scholar]
- 17. Shire NJ, et al. 2007. The prevalence and significance of occult hepatitis B virus in a prospective cohort of HIV-infected patients. J. Acquir. Immune Defic. Syndr. 44:309–314 [DOI] [PubMed] [Google Scholar]
- 18. Tapper EB, Lau DT. 2011. The need for clarity in occult HBV infection. J. Viral Hepat. 18:673–674 [DOI] [PubMed] [Google Scholar]
- 19. Weber B. 2005. Genetic variability of the S gene of hepatitis B virus: clinical and diagnostic impact. J. Clin. Virol. 32:102–112 [DOI] [PubMed] [Google Scholar]
- 20. Weinbaum CM, et al. 2008. Recommendations for identification and public health management of persons with chronic hepatitis B virus infection. MMWR Recommend. Rep. 57:1–20 [PubMed] [Google Scholar]