Abstract
The number of bacterial species estimated to exist on Earth has increased dramatically in recent years. This newly recognized species diversity has raised the possibility that bacterial natural product biosynthetic diversity has also been significantly underestimated by previous culture-based studies. Here, we compare 454-pyrosequenced nonribosomal peptide adenylation domain, type I polyketide ketosynthase domain, and type II polyketide ketosynthase alpha gene fragments amplified from cosmid libraries constructed using DNA isolated from three different arid soils. While 16S rRNA gene sequence analysis indicates these cloned metagenomes contain DNA from similar distributions of major bacterial phyla, we found that they contain almost completely distinct collections of secondary metabolite biosynthetic gene sequences. When grouped at 85% identity, only 1.5% of the adenylation domain, 1.2% of the ketosynthase, and 9.3% of the ketosynthase alpha sequence clusters contained sequences from all three metagenomes. Although there is unlikely to be a simple correlation between biosynthetic gene sequence diversity and the diversity of metabolites encoded by the gene clusters in which these genes reside, our analysis further suggests that sequences in one soil metagenome are so distantly related to sequences in another metagenome that they are, in many cases, likely to arise from functionally distinct gene clusters. The marked differences observed among collections of biosynthetic genes found in even ecologically similar environments suggest that prokaryotic natural product biosynthesis diversity is, like bacterial species diversity, potentially much larger than appreciated from culture-based studies.
INTRODUCTION
Phylogenetic analyses based on 16S rRNA gene sequences show that environmental samples can contain thousands of unique bacterial species, only a small fraction of which are grown using traditional culturing techniques (27). Contrary to the century-old tenant in microbiology that “everything is everywhere; but the environment selects” (4), recent molecular phylogenetic-based biogeography studies have found that ecologically similar yet geographically distant environments can contain distinct consortia of bacterial species (24, 34, 35). Extrapolations from these and other studies have led to predictions that there may be as many as 107 to 109 unique bacterial species on Earth (12, 30). As bacteria are considered one of the world's richest sources of bioactive natural products, such predictions could have profound implications for future drug discovery efforts. However, whether this newly recognized bacterial species diversity corresponds to a modest increase in secondary metabolite biosynthetic diversity or to a radical increase in the number of unexplored biosynthetic systems is currently unknown.
It is possible that there exists a relatively small global collection of secondary metabolite gene clusters that is largely conserved from one location to the next or, conversely, that secondary metabolite gene clusters are to some extent geographically and environmentally constrained, resulting in the presence of largely orthogonal collections of gene clusters in geographically distinct microbiomes. Here we have sought to begin to address this issue by determining whether the collections of biosynthetic genes found in ecologically similar but geographically distinct soil microbiomes differ from sample to sample or whether they are largely conserved across geographic boundaries. The number and diversity of novel natural products that remains to be examined for potentially useful bioactivities is likely to depend heavily on which of these two possibilities is dominant throughout the biosphere.
Secondary metabolite biosynthetic diversity encoded within a soil metagenome is difficult to assess using standard microbiology methods because the majority of environmental bacteria are not readily cultured. Culture-independent or metagenomic methods, which rely on cloning DNA directly from environmental samples, provide a means of exploring secondary metabolism in natural bacterial populations (19). To compare the biosynthetic potentials of different soil microbiomes, environmental DNA (eDNA) extracted directly from three geographically distinct arid soils collected in the American Southwest (the Sonoran Desert of Arizona [AZ], the Anza-Borrego region of the Sonoran Desert of California [AB], and the Great Basin Desert of Utah [UT]) was used to construct three independent eDNA cosmid libraries. Each library contains in excess of 350 gigabases of DNA (∼100,000 bacterial genome equivalents) and is predicted to provide sufficient sequence coverage to capture the major constituents of the respective soil metagenome (2, 3, 6, 16, 17, 21, 22). The enormous size of these cloned soil metagenomes makes it difficult to shotgun sequence to a depth that would provide statistically relevant comparisons of differences in secondary metabolite genes. We have therefore compared secondary metabolism in different soils by pyrosequencing PCR amplified fragments of conserved sequences found in nonribosomal peptide synthetase (NRPS), type I polyketide synthase (PKSI), and type II polyketide synthase (PKSII) gene clusters, three of the most common bacterial natural product biosynthetic systems (Fig. 1A).
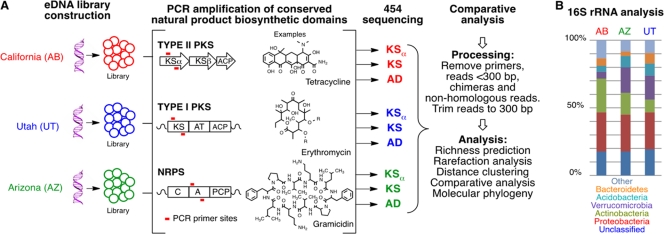
Fig 1.
(A) Overview of the approach used to compare secondary metabolism in different soil microbiomes. Independent environmental DNA libraries were constructed from three different arid soils. DNA from each library was used as the template in PCRs with degenerate primers designed to recognize nonribosomal peptide synthetase AD domains, type I polyketide KS domains, and type II polyketide KSα genes. The resulting amplicons were 454 sequenced, processed, and compared to assess the similarity of the three gene sets derived from different microbiomes. KS, ketosynthase; ACP, acyl carrier protein; AT, acyltransferase; C, condensation domain; A or AD, adenylation domain; PCP, peptide carrier protein. (B) Bar graphs show the frequency at which 16S rRNA genes from different major phyla appeared in each library. As might be expected for metagenomic libraries constructed from ecologically similar soils, 16S rRNA gene analyses indicate that DNA from a very similar distribution of major bacterial phyla was captured in each library.
MATERIALS AND METHODS
Sample collection, DNA isolation, and library construction.
Topsoil, including cryptobiotic crust, was collected from three sites in the Southwestern United States: the Sonoran Desert of Arizona (AZ), the Anza-Borrego section of the Sonoran Desert of California (AB), and the Great Basin Desert of Utah (UT). All samples were collected in the summer months from undisturbed environments that were representative of the most common terrain and vegetation seen in these three deserts. Each sample was sifted through fine mesh to break up large dirt particles and remove rocks and large vegetative material. DNA was then extracted directly from the sifted soils. DNA extraction and library construction were carried out using published protocols (5) (2, 3, 22). Briefly: each dirt sample was incubated at 70°C in lysis buffer (2% sodium dodecyl sulfate [wt/vol], 100 mM Tris-HCl, 100 mM EDTA, 1.5 M NaCl, 1% cetyl trimethyl-ammonium bromide [wt/vol]) for 2 h. Large particulates were then removed by centrifugation (4,000 × g, 30 min), and crude eDNA was precipitated from the resulting supernatant with the addition of 0.6 volumes of isopropyl alcohol. Precipitated DNA was collected by centrifugation (4,000 × g, 30 min), washed with 70% ethanol, and resuspended in a minimum volume of TE (10 mM Tris, 1 mM EDTA [pH 8]). The remaining soil material was separated from the DNA by preparative agarose gel electrophoresis (1% agarose, 0.5× Tris-borate-EDTA, 16 h, 20 V). High-molecular-weight DNA was electroeluted from the agarose, blunt ended (End-It; Epicentre), ligated into the SmaI site of either pWEB or pWEB:TNC, packaged into lambda phage, and transfected into Escherichia coli EC100. Each library was expanded to contain at least 10,000,000 unique eDNA cosmid clones. To facilitate future clone recovery efforts libraries were arrayed as unique 5,000-membered sublibraries consisting of matching minipreps and glycerol stock pairs. Cosmid DNA miniprepped from the pool of clones contained in each library was used for PCR screening as described below. Individual ketosynthase alpha (KSα)-containing clones were recovered from library pools by successive rounds of PCR screening and then sequenced using 454 pyrosequencing.
PCR amplification of domains from each library.
Numerous secondary metabolite biosynthetic gene-specific degenerate primers can be found in the literature. From this pool of degenerate primers, we selected primers designed to recognize conserved regions in nonribosomal peptide synthetase (NRPS) adenylation (AD) domain, type I polyketide synthase (PKSI) ketosynthase (KS) domain, and type II polyketide synthases (PKSII) ketosynthase alpha (KSα) sequences (1, 25, 28). Important features we considered when selecting from the degenerate primers that have appeared in the literature included amplicon length below 1,000 bp, limited homonucleotide stretches in known sequences of the region to be amplified, and robust amplification using standard PCR conditions.
Adenylation domain fragments (∼795 bp) were PCR amplified using primers A3F (5′-GCSTACSYSATSTACACSTCSGG) and A7R (5′-SASGTCVCCSGTSCGGTA) (1). These primers were designed to recognize the conserved regions A3 and A7 in NRPS adenylation domains. Within the bpsA gene from the Amycalopsis balhimycina-derived balhimycin biosynthetic gene cluster, the amplified region corresponds to nucleotides 15217 through 15909 (GenBank accession number Y16952.3). KSα gene fragments (∼672 bp) were amplified using primers KSα-F (5′-TSGCSTGCTTCGAYGCSATC) and KSα-R (5′-TGGAANCCGCCGAABCCGCT) (25). These primers were designed to amplify the most conserved region of KSα genes. In the KSα gene from the Streptomyces coelicolor actinorhodin biosynthesis pathway, the amplified region corresponds to nucleotides 720 through 1332 (GenBank accession number X63449.1). KS domain fragments (∼760 bp) were amplified using primers degKS2F.i (5′-GCIATGGAYCCICARCARMGIVT) and degKS2R.i (5′-GTICCIGTICCRTGISCYTCIAC) (28). These primers were designed to amplify the most conserved regions of PKSI ketosynthase domains, including the active site residues. In the case of the eryAIII gene from erythromycin biosynthesis in Streptomyces erythraea, this amplicon spans nucleotide 11056 through 11735 (GenBank accession number M63677). The 16S rRNA gene V4 hypervariable region (∼207 bp) was amplified using primers 16S-F (5′-AYTGGGYDTAAAGNG) and 16S-R (5′-TACNVGGGTATCTAATCC) (10, 11). Forward and reverse primers incorporated 454-sequencing adaptors (forward primer “A adaptor,” 5′-CGTATCGCCTCCCTCGCGCCATCAG; reverse primer “B adaptor,” 5′-CTATGCGCCTTGCCAGCCCGCTCAG). To allow for sequencing of different genes in the same region of a 454 plate sample, specific tags were added between either the reverse or forward degenerate primer and the 454-sequencing adaptor.
For AD, KSα, and 16S rRNA genes, amplification reactions were carried out using two distinct PCR conditions in an attempt to amplify the most diverse set of eDNA gene sequences. The first reaction mix (20 μl/reaction) contained 100 ng cosmid DNA, 0.5 μM each primer, 200 μM each deoxynucleoside triphosphate (dNTP), 1× Phusion GC buffer (New England BioLabs), 0.2 U Phusion polymerase (New England BioLabs), and 3% dimethyl sulfoxide (DMSO). The second reaction mix contained 1× G buffer (Epicentre), 50 pmol of each primer, 2.5 U Taq polymerase (New England BioLabs), and 100 ng cosmid DNA. For amplifications with the AD and KSα primers, the first reaction mix, used a PCR protocol of 30 cycles consisting of 10 s at 98°C, 30 s at 70°C, and 30 s at 72°C, followed by a final extension at 72°C for 5 min. The second reaction mix, used a PCR protocol of 35 cycles consisting of 1 min at 95°C, 3 min at 72°C, followed by a final extension at 72°C for 5 min. For amplifications with the 16S primers, the first reaction mix used a PCR protocol of 25 cycles consisting of 10 s at 98°C, 30 s at 55°C, and 30 s at 72°C, followed by an extension at 72°C for 5 min. The second reaction mix used a PCR protocol of 35 cycles consisting of 40 s at 95°C, 40 s at 55°C, 40 s at 72°C, followed by extension step at 72°C for 5 min. While AD, KSα, and 16S primers worked successfully under multiple PCR conditions, we could only identify a single condition that worked with the KS domain-specific primers. PCRs (20 μl) using KS domain-specific primers contained 1× G buffer, 50 pmol of each primer, 2.5 U Taq polymerase, and 100 ng cosmid DNA. The amplification reactions performed for 35 cycles consisting of 40 s at 95°C, 40 s at 50°C, 75 s at 72°C, followed by a final extension step at 72°C for 5 min.
454 sequencing and data processing.
PCR products were run on crystal violet-stained gels, and amplicons of correct predicted size were gel purified using Qiagen MinElute columns by following the manufacturer's instructions. The purified PCR products were fluorometrically quantified (PicoGreen Quant-iT; Invitrogen) and analyzed via capillary electrophoresis (DNA 7500; Agilent Technologies). Each purified amplicon was diluted to 109 molecules/μl. Amplicons of the same gene from different libraries were pooled and used as a template for emulsion PCR (emPCR). Parallel pyrosequencing (454 GS-FLX Titanium) of beads from these independent emPCR reactions was performed according to the manufacturer's protocol and processed as described below. Base calls and quality scores were extracted using the 454 GS-FLX Titanium shotgun processing software. Sequences are deposited in the NCBI Sequence Read Archive (SRA) database under accession number SRP008112.
The number of sequences carried forward after each processing step is shown in Table S1 in the supplemental material. Sequences obtained from 454 pyrosequencing (see Table S1, 454 reads) were initially processed and cleaned using the ribosomal database project (RDP) pyrosequencing pipeline (10). This included removal of the primers, removal of reads less than 300 bp, removal of reads containing ambiguous calls, and the trimming of the remaining reads to 300 bp out from the forward primer site (see Table S1, RDPP trim). Potential chimeric sequences were removed using UCHIME (see Table S1, no chimes) (15). Identical sequences were then removed (see Table S1, nonredundant). Each set of sequences was then compared to the appropriate reference sequence database (NRPS-REF, PKSI-REF, and PKSII-REF, see below), and any reads that did not align to a reference over at least 90% of the read with an E value of <10e−10 were removed (see Table S1, ref-homologs). AD sequences were found to be much more divergent than KS or KSα sequences, and therefore an E value cutoff of 10e−5 was used in AD reference database searches. For some analyses, redundant sequences removed early on (see Table S1, nonredundant) were added back to the data set (see Table S1, ref-hom-redun). The ref-hom-redun sequences were clustered at 97% identity using USEARCH to compensate for potential sequencing errors (see Table S1, 97% unique) (14). To search for sequences related to functionally characterized gene clusters, we used unique sequences clustered at 100% identity (ref-homologs).
Reference sequence database preparation for NRPS, PKSI, and PKSII genes (NRPS-REF, PKSI-REF, and PKSII-REF).
To generate a database of known AD, KS, and KSα sequences that could be used to clean our 454 data sets, a primer-based pattern search on NCBI-NT database was performed with forward degenerate primers. From this search, in each case, 300 bp out from the site recognized by the forward primer was cut from the NCBI-NT sequence and this collection of 300-bp fragments was then used in BLASTX queries against all NCBI-NR protein sequences. All BLASTX hits meeting the following criteria were collected and used as reference sequence databases: (i) E value of <10e−10, (ii) alignment of greater than 90% of the length of the translated NCBI-NT derived query sequence, and (iii) greater than 50% identity to the query sequence.
NRPS AD domains, type I polyketide KS domains, and type II polyketide KSα domains are all distantly related to sequences used outside secondary metabolism. In an attempt to avoid populating our reference library with sequences from primary metabolism, annotation data from any sequence that showed an E value of <10e−10 and >90% alignment coverage but lower than 50% identity to a query sequence was manually scrutinized. Among these sequences, only the sequences that were explicitly annotated in the NCBI-NT database as nonribosomal peptide adenylation domains, polyketide type I ketosynthases, or polyketide type II ketosynthases were included in our reference sequence databases (NRPS-REF, PKSI-REF, and PKSII-REF).
16S phylogenetic analysis.
16S rRNA gene hypervariable region reads were processed as described above with a few modifications: (i) reads that did not contain both primers were removed, (ii) chimeric sequences were removed using chimera_bellerophon (MOTHUR) and UCHIME, and (iii) E. coli 16S rRNA gene sequences were explicitly removed from the data set (31). Cleaned 16S reads were classified using RDP classifier. For each 16S data set, a single representative sequence from each of the clusters that formed when grouped at 97% identify (see Table S2 in the supplemental material, 97% unique) was used for RDP-based phylum level classification. The number of sequences carried forward after each processing step is shown in Table S2.
Assignment to similarity-based operational taxonomic units (OTUs) and sequence type richness estimators.
To compute rarefaction curves and sequence type richness estimation with DOTUR we needed to generate a multiple sequence alignment of each 454 data set (29). There were too many sequences to align efficiently using ClustalW. We therefore elected to initially cluster each group of sequences (see Table S1 in the supplemental material, ref-hom-redun) at 85% identity using USEARCH. Representative sequences from each of the resulting clusters were then used to generate multiple sequence alignment. The alignments of KS and KSα sequences were carried out using ClustalW, and for the larger AD data sets, the alignments were carried out using MUSCLE (13). Reference alignments were then used as templates to finally align all of the cleaned reads (see Table S1, ref-hom-redun) using the align_seqs module in MOTHUR (31). Distance matrices were calculated from the resulting alignments using the dist_seqs module in MOTHUR, and then these matrices were used as inputs to compute Shannon-Weaver diversity indices, Chao1 richness estimates, and rarefaction curves with DOTUR (29).
Comparative sequence analyses.
The ref-hom-redun sequence sets for individual genes from different libraries were combined and clustered at 97%, 90%, and 85% sequence identities using USEARCH. The origin of the sequences in each cluster was recorded, and these data are displayed as Venn diagrams (see Fig. 3). BioInfoRx Inc.'s Venn diagram web tool was used for drawing Venn diagrams. The exact number of clades that appears when sequences from each library are clustered at various percent identities is shown in Table S4 in the supplemental material. The number of OTUs generated by DOTUR and USEARCH differ slightly. This is likely due to the different clustering algorithms used by the two programs. DOTUR uses distance matrices based on multiple sequence alignments as a basis for clustering whereas USEARCH uses pairwise alignments and nearest neighbor joining for clustering. The number of clusters predicted by these programs can also vary slightly when starting with different-sized populations of closely related sequences.
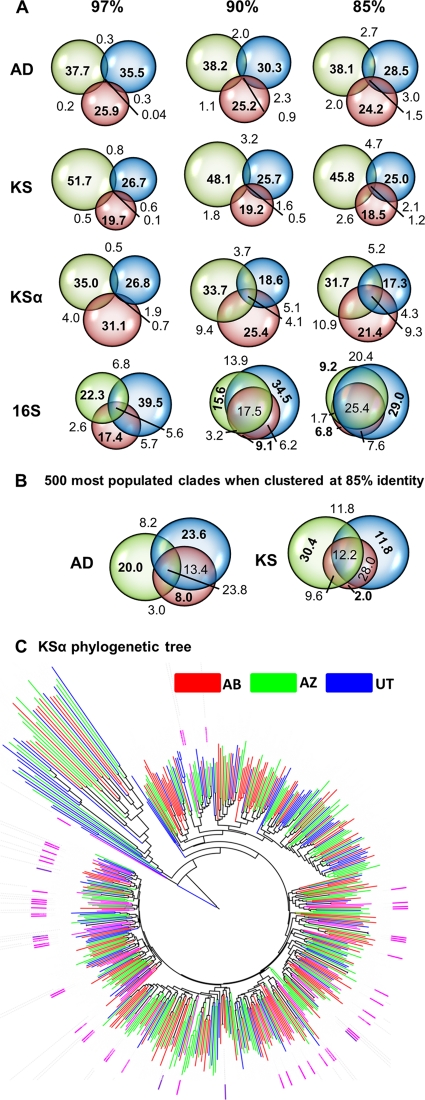
Fig 3.
Comparison of secondary metabolite gene sequences found in three cloned metagenomes. (A) Sequences from all three metagenomes were clustered at various identities, and Venn diagrams were then made to show the percentage of clades containing sequences from each cloned metagenome. Venn diagrams are drawn to scale whenever possible. (B) Even when clustered at 85% identity, a large number of AD and KS clades are sparsely populated. Venn diagrams representing the clustering analysis of only the top 500 most populated AD and KS are shown. Each of these clades contains >20 unique pyrosequencing reads. (C) KSα phylogenetic tree. Functionally characterized KSα sequences (pink) and representative sequences from each OTU that formed when eDNA sequences clustered at 85% identity were aligned using ClustalW and plotted with iTOL. Branches are color coded according to the soil sample that had the most unique sequences in that branch. The KSα gene from resistomycin biosynthesis was used to root the tree. A key advantage of using large libraries rather than crude eDNA to compare microbiomes is that gene clusters associated with novel biosynthetic genes can be recovered and functionally studied. Marked in purple are KSα sequences associated with eDNA-derived gene clusters that have yielded novel secondary metabolites.
Molecular phylogenetic tree construction.
The sets of 97% unique sequences obtained for each gene from all three libraries were pooled and clustered at 85% sequence identity using USEARCH (14). Representative sequences from each clade were aligned, and phylogenetic trees were calculated using the neighbor-joining algorithm of ClustalW (32). Circular phylogenetic trees were plotted using the interactive Tree of Life (iTOL) (23). For KSα, all 492 clusters are drawn on the final tree. Representative sequences from only 500 most-populated clusters were used for the construction of AD and KS trees (see Fig. S1 in the supplemental material).
RESULTS
454 pyrosequencing of cloned AD, KS, and KSα domains.
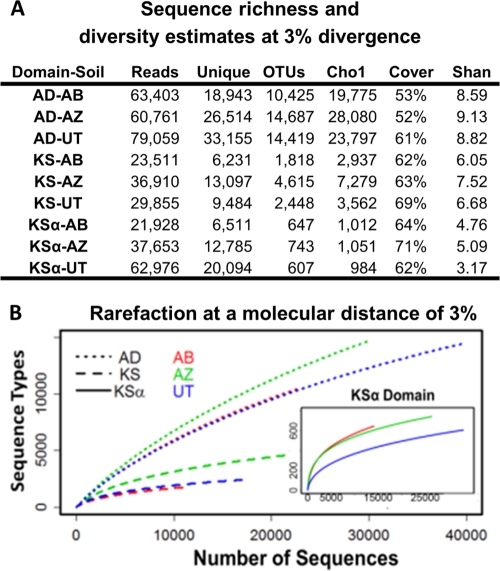
Degenerate primers designed to recognize conserved sequences found in gene clusters encoding nonribosomal peptides, type I polyketides, and type II polyketides were used to PCR amplify secondary metabolite-specific gene sequences from three cloned metagenomes (Fig. 1) (1, 25, 28). The resulting amplicons, corresponding to NRPS adenylation (AD) domain, PKSI ketosynthase (KS) domain, and PKSII ketosynthase alpha (KSα) gene fragments were then pyrosequenced using 454 GS-FLX technology (Fig. 1). In each case, we continued sequencing until Chao1 sequence richness estimates predicted that at least half of the unique genes present within each metagenome had been sequenced (see Discussion below) (Fig. 2A). In total, between 22,000 and 79,000 reads were obtained for each metagenome-specific amplicon (Fig. 2A).
Fig 2.
Sequence richness and diversity estimates. (A) The number of raw reads (reads), unique cleaned reads (unique), and OTUs when grouped at 97% identity are shown for AD, KS, and KSα sequences amplified from each cloned metagenome (AB, AZ, and UT). Chao1 sequence richness estimates are reported at a cutoff of 3%. Shan, Shannon diversity index. Sequences are deposited in the NCBI-SRA database under accession number SRA045798.2. (B) Rarefaction curves (using a 3% cutoff value) for AD, KS, and KSα sequences amplified from each eDNA library.
454-pyrosequencing error rates have been reported to range from less than 1% to as high as 4% (20). Studies looking specifically at 454 GS-FLX sequencing technology report error rates below 3%, even over long read lengths (8). We therefore clustered all processed reads at 97% identity to correct for potential sequencing errors and obtain unique sets of nonredundant gene sequences from each metagenome/amplicon pair. This analysis led to the identification of, on average, 13,177 AD, 2,960 KS, and 666 KSα unique sequences per cloned metagenome (Fig. 2A). If pyrosequencing error rates exceed 3%, the true diversity would be lower than that predicted here. At this depth of sequencing, rarefaction curves for both KS and KSα domains appear to be reaching asymptotes (Fig. 2B). For AD domains, which the Shannon diversity index predicts are the most diverse set of sequences, this does not appear to be the case even though we acquired more than 60,000 AD reads per cloned metagenome (Fig. 2). Chao1 sequence richness estimates calculated at a cutoff of 3% predict that each cloned metagenome contains on average 23,884, 4,493, and 1,016 unique AD, KS, and KSα sequences, respectively (7). As stated above, based on these richness estimates, we have sequenced deep enough to identify over 50% of the unique domains that are accessible from each metagenome using this set of degenerate primers.
454 pyrosequencing and comparison of cloned 16S sequences.
For each library, the percentage of unique 16S rRNA gene fragments corresponding to different major bacterial phyla was calculated (see Table S3 in the supplemental material) and is shown as bar graphs in Fig. 1B. Although these libraries were constructed from soils collected in geographical distinct locations, they contain very similar distributions of major bacterial phyla. Based on 16S rRNA gene sequences, Proteobacteria, Actinobacteria, Verucomicrobia, Acidobacteria, and Bacteroidetes are the most common phyla represented in each library.
Global comparisons of metagenome-derived AD, KS, and KSα amplicons.
For comparison purposes, all of the sequences obtained for a given domain were pooled and then clustered based on sequence identity. The resulting clusters were grouped according to whether they contained sequences from one, two, or all three metagenomes. These relationships are depicted as Venn diagrams in Fig. 3. When sequences from all three soils were clustered at 97% identity, almost all of the resulting clusters were populated with sequences from a single metagenome, indicating that these three soils contain essentially orthogonal sets of secondary metabolite biosynthetic gene sequences. Three percent divergence is commonly used to define unique bacterial species in 16S rRNA gene-based molecular phylogenetic analyses; however, for less conserved sequences like secondary metabolite biosynthetic genes, this metric likely has little functional relevance. It would be more informative to group sequences at similarities corresponding to the point at which two gene sequences have a high likelihood of being derived from gene clusters that encode the production of structurally distinct metabolites. Although for many genes there is unlikely to be a simple linear correlation between sequence divergence and differences in the metabolites encoded by the gene clusters in which these genes are associated, molecular phylogenetic comparisons of microbiome-derived gene sets at different identities should be a useful strategy for comparing secondary metabolite genes derived from different microbiomes (Fig. 3). Even when gene sets from the three metagenomes were clustered at identities as low as 90 and 85%, only a small number of the resulting clusters contained sequences from all three metagenomes (Fig. 3). When clustered at 85% identity, only 1.5% of AD, 1.2% of KS, and 9.3% of KSα clusters contained sequences from all three metagenomes. When grouped at 85% identity, 50% of AD, 37% KS, and 17% KSα clusters are populated with a single pyrosequencing read. While many of these likely represent distinct environmental sequences, it is also possible that some sequences are due to undetected chimera events or higher than predicted sequencing error rates. Venn diagrams constructed using representative sequences from only the 500 most populated AD and KS clades are shown in Fig. 3B. Each of these clades contains more than 20 unique pyrosequencing reads. This subset of clusters shows more extensive metagenome-to-metagenome overlap than the entire population of KS and AD sequences; however, even among these more highly populated clusters we see significant metagenome-specific differences. Although sequence divergence estimates will vary with the length of the sequences being compared as well as the specific region of the gene that is surveyed, the gene fragments examined in this study suggest that these three environments not only contain very few identical sequences but that they actually contain few sequences that are even closely related to each other.
Comparison of eDNA and NCBI-NT database derived sequences.
Forward degenerate primers were used in silico to search fully sequenced cultured bacterial genomes deposited in NCBI-NT for AD, KS, and KSα sequences. In total, 636 AD, 1,455 KS, and 180 KSα sequences were found in this search. These sequences were then computationally rescreened using the appropriate reverse degenerate primers. The remaining 334 AD, 1,303 KS, and 79 KSα were used to assess the phylogenetic specificities of each primer set (see Table S5 in the supplemental material). Sequences recovered with the forward primers were also compared in a clustering analysis with the eDNA derived sequences. NCBI-NT sequences were trimmed to include 300 nucleotides out from the position of the primer, and then they were clustered at different identities together with the eDNA sequences we obtained from all three libraries (Table 1). Even when grouped at identities as low as 85%, only a small fraction (<2%) of clades contained both eDNA and NCBI-NT derived sequences. Secondary metabolite gene sequences found in these three metagenomes not only differ from each other, but they also largely differ from genes previously sequenced from cultured bacteria.
Table 1.
Number of clades that appear when eDNA sequences and NCBI-NT sequences are clustered at different percent identities
| Domain | % Identity | Total | No. of clades (% of total) |
||
|---|---|---|---|---|---|
| NCBI-NT | eDNA | Shared | |||
| AD | 97 | 43,299 | 596 (1.4) | 42,703 (98.6) | 0 (0.00) |
| 90 | 26,454 | 517 (2.0) | 25,937 (98.1) | 11 (0.04) | |
| 85 | 21,534 | 461 (2.1) | 21,073 (97.1) | 14 (0.07) | |
| KS | 97 | 10,083 | 1,278 (12.7) | 8,805 (87.3) | 4 (0.04) |
| 90 | 7,915 | 1,125 (14.2) | 6,790 (85.8) | 4 (0.05) | |
| 85 | 6,818 | 994 (14.6) | 5,824 (85.4) | 8 (0.12) | |
| KSα | 97 | 1,651 | 113 (8.1) | 1,518 (91.9) | 0 (0.00) |
| 90 | 853 | 102 (12.0) | 751 (88.0) | 7 (0.82) | |
| 85 | 578 | 65 (11.2) | 513 (88.8) | 12 (2.08) | |
AD, KS, and KSα phylogenies.
Although the individual soil microbiomes explored in this study contain largely orthogonal sets of biosynthetic gene sequences, genes from these different soil samples do not appear to have radically different evolutionary origins. Clustering KSα sequences from all three metagenomes at 85% identity resulted in 492 unique OTUs. A ClustalW derived phylogenetic tree of a single representative sequence from each OTU is shown in Fig. 3C (23, 32). Environmental DNA derived KSαs fall into clades containing KSα sequences that are known to encode structurally diverse aromatic metabolites, as well as several clades without any functionally characterized KSα sequences (Fig. 3C). Sequences from different metagenomes do not group into metagenome-specific clades but instead distribute throughout the tree, suggesting that KSα sequences from different soils share a set of diverse ancestors. AD and KS sequences were so numerous that even when grouped at a molecular distance of 0.15 it is difficult to display these data as phylogenetic trees. Fig. S1 in the supplemental material contains phylogenetic trees of the 500 most populated AD and KS OTUs. AD and KS sequences show the same general trend as KSα sequences, where sequences from different metagenomes largely distribute uniformly throughout the combined phylogenetic trees.
DISCUSSION
When the degenerate primers used in this study were computationally screened against the NCBI-NT database for AD, KS, and KSα genes, approximately two-thirds of the AD and KS sequences and all of the KSα sequences we identified are from Actinobacteria. The remaining AD and KS sequences are from Proteobacteria, in particular, Pseudomonas and Burkholderia spp. This coincides well with the sources of the deposited sequences encoding these three gene families. Approximately 50% of the AD and KS sequences in the UniProt database are from Actinobacteria, with Proteobacteria being the next most common source (26), and the vast majority of deposited KSα sequences are from Actinobacteria. Even though amplification bias introduced by the degenerate primers has undoubtedly led us to underestimate the true sequence diversity present in each metagenome, the set of eDNA-derived gene sequences amplified by these primers should be representative of the genetic loci encoding secondary metabolism in each soil sample and therefore permit useful comparisons of biosynthetic capacities of these metagenomes.
Although 16S rRNA gene sequence analysis (Fig. 1B; see also Table S3 in the supplemental material) indicates that the three cloned metagenomes analyzed in this study contain DNA from similar distributions of major bacterial phyla, we found that they contain almost completely distinct collections of secondary metabolite biosynthetic gene sequences. Soil-to-soil differences seen in secondary metabolite gene sequences do not appear to be artifacts of the sequencing method, the result of generic variations in DNA sequence makeup such as GC content, or due to very high natural polymorphism rates, as when 16S amplicons from these three metagenomes are clustered in the same manner as the biosynthetic genes, they show significantly higher sample-to-sample overlap at this same range of identities (Fig. 3). In fact, the observed sample-to-sample species overlap of 5 to 10% correlates very well with species overlaps that have been reported in other 16S-based analyses of distantly sampled soils (18).
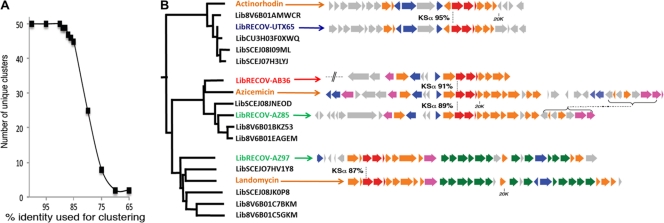
Correlating the observed sequence differences with differences found in the actual secondary metabolites encoded by different metagenomes is complicated by a number of factors, including variations in the rate of evolution of different genes, horizontal gene transfer between bacteria, and the possibility of convergent evolution. In spite of these potential complications, the divergence of functionally characterized KSα gene sequences has been observed to correlate quite well with the production of different structural families by the PKSII gene clusters in which these genes reside, with closely related sequences involved in the biosynthesis of related polyketides and distantly related sequences encoding structurally distinct polyketides (16, 25). While not perfect, the inverse relationship seen between KSα sequence identity and natural product structural diversity is quite robust for functionally characterized KSα gene sequences (Fig. 4A). Although there are outliers to this general trend (37) the functional divergence of characterized PKSII gene clusters correlates well with the divergence of KSα gene sequences by 10 to 15% in identity (Fig. 4A). To see if this observation extended into our metagenomic data set, we recovered and sequenced cosmid clones containing KSα genes that were both closely related (>85% identity) and distantly related (<85% identity) to KSα genes found in functionally characterized gene clusters. In each case where we recovered a cosmid containing a KSα gene that showed high identity (>85%) to a functionally characterized sequence, we found these clones to contain gene clusters that closely resemble the clusters associated with the previously characterized KSα gene in this clade (Fig. 4B). On the other hand, when we examined eDNA clones with KSα genes that diverged by more than 15% from functionally characterized KSα genes, we did not identify any sequences that closely resembled these functionally characterized gene clusters in gene sequence, gene content, or gene organization (see Fig. S2 in the supplemental material).
Fig 4.
Relationship between KSα sequence identity and gene cluster function. (A) The number of distinct groups formed when functionally characterized KSα sequences are clustered at different percent identities is shown. On average, functionally characterized KSα gene sequences (50 in total) that are known to be involved in the biosynthesis of structurally distinct metabolites do not group together when clustered at above 80 to 85% identity. When clustered below 85% identity, this correlation between KSα sequence divergence and small molecule structural divergence is no longer observed. Clustering was carried out using 300-bp KSα gene fragments corresponding to the amplicons that would be produced by the KSα degenerate primers used to access metagenomic sequences. (B) Metagenomic sequences that clustered with functionally characterized KSα genes at 85% identity are shown. A representative eDNA clone containing a KSα gene from each clade was recovered and sequenced. In each case, these clones closely resembled in gene identity, gene complement, and gene organization in the functional characterized gene cluster. The percent identity between KSα genes is shown. Genes are color coded according to the predicted enzymatic function of their products. Red, minimal PKS; blue, regulation and resistance; orange, polyketide biosynthesis; pink, starter biosynthesis; green, sugar biosynthesis; gray, unknown pathway/unrelated enzymes.
Based on the observed correlation between sequence and small molecule diversity, clustering KSα sequences at 85% identity may provide a means of estimating the natural product biosynthesis potential associated with different collections of KSα gene sequences, much in the same way that the clustering 16S sequences at 97% identity is used to gain insights into the species composition of different environmental samples. Based on this metric, the clustering of KSα sequences from a single metagenome at 85% identity could provide a rough estimate of the number of different aromatic polyketides a metagenome might encode, whereas the Venn diagram describing the composition of KSα clades observed when sequences from different metagenomes are clustered together at this same identity (Fig. 3) would represent the relationship between the different collections of type II polyketides encoded by these metagenomes. The 236, 294, and 186 distinct clades (see Table S4 in the supplemental material) observed when AB, AZ, and UT KSα gene sequences were clustered at 85% identity, respectively, suggest that soil metagenomes might encode hundreds of structurally distinct aromatic polyketides each, and the 85% identity Venn diagram further suggests that (i) approximately 10% of the metabolites could be common to all three metagenomes (which is represented by the intersection of the three samples in the Venn diagram), (ii) another 5 to 10% could be shared by any two metagenomes, and (iii) the remainder could be specific to an individual metagenome.
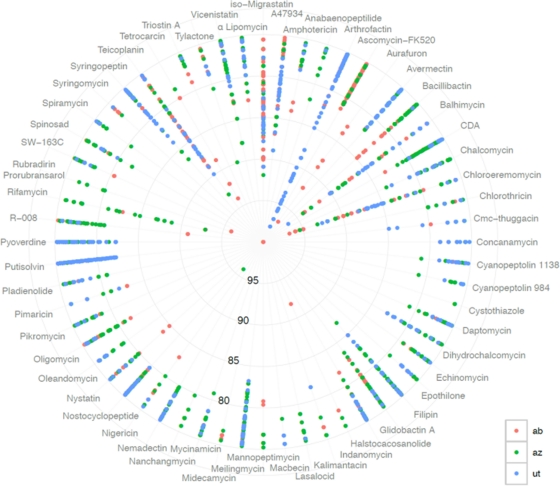
Correlations between sequence divergence and gene cluster function for KS and AD domains are complicated by the fact that multiple KS and AD domains often appear in a single biosynthetic gene cluster. Therefore, even though sequence type richness and diversity estimates (Fig. 2A) indicate environmental KS and AD sequences are more divergent and numerous than KSα sequences, we cannot directly correlate these differences to global differences in the natural product structural diversity a metagenome might encode. Although it is not possible to predict differences in the structures encoded by these metagenomes, a comparison of our eDNA-derived AD and KS sequences to the collection of corresponding domain sequences found in individual functionally characterized PKSI and NRPS gene clusters suggests significant functional differences in the three soil metagenomes (Fig. 5). Domains from some functionally characterized gene clusters only have relatives showing high sequence identity (>75%) in one library (i.e., putsolvin, pyoverdine, mycinamucin, rifamycin, aurafuron, triostin A, etc.), others have relatives in two of the three libraries (i.e., dihydrochalcomycin, bacillibactin, A47934, α-lipomycin, epothilone, etc.), and others have relatives in all three libraries (i.e., balhimycin, chloroeremomycin, tetrocarcin, etc.).
Fig 5.
Comparison of metagenome-derived KS and AD domain sequences with those found in functionally characterized gene clusters. Each metagenomic data set was searched by tBLASTx for KS and AD amplicons that show high sequence identity to similar domains found in functionally characterized PKSI and NRPS gene clusters. Individual spokes of the graph correspond to the collection of identity scores for library-derived amplicons that show greater than 75% identity to any AD or KS domain found in the designated biosynthetic gene cluster. All of the hits shown here have E values of ≤10e−40, and each alignment spans at least 80 amino acid residues. The final image was constructed using R and ggplot2 (36).
Soils contain highly diverse collections of bacteria making them very attractive starting points for both culture-dependent and culture-independent small molecule discovery efforts. We chose to investigate secondary metabolism in geographically distinct yet ecologically similar soils based on the belief that ecologically similar soils would have the highest likelihood of containing related sets of biosynthetic gene sequences. We found, however, that even metagenomes from ecologically similar environments with similar phylum-level 16S rRNA gene distributions (Fig. 1B and 3) can contain almost completely distinct collections of biosynthetic gene sequences. This is likely reflective of the fact that natural product biosynthesis gene content can differ not only between species but also between strains of the same species. In those cases where, based on studies from cultured bacteria, it is possible to speculate about the relationship between gene sequence diversity and secondary metabolite structural diversity, our data suggest that sequences in one soil metagenome are so distantly related to sequences in another metagenome that in most cases they are unlikely to be found in gene clusters that encode the same metabolites. If this holds true for other soil types, the unexplored bacterial biosynthetic diversity present in the Earth's biosphere is, like bacterial species diversity, potentially much larger than predicted previously from fermentation-based analyses (9, 33).
Supplementary Material
ACKNOWLEDGMENT
This work was supported by NIH grant number GM077516. S.F.B. is an HHMI Early Career Scientist.
Footnotes
Published ahead of print 16 March 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Ayuso-Sacido A, Genilloud O. 2005. New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb. Ecol. 49:10–24 [DOI] [PubMed] [Google Scholar]
- 2. Banik JJ, Brady SF. 2008. Cloning and characterization of new glycopeptide gene clusters found in an environmental DNA megalibrary. Proc. Natl. Acad. Sci. U. S. A. 105:17273–17277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Banik JJ, Craig JW, Calle PY, Brady SF. 2010. Tailoring enzyme-rich environmental DNA clones: a source of enzymes for generating libraries of unnatural natural products. J. Am. Chem. Soc. 132:15661–15670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beijerinck MW. 1913. De Infusies en de Ontdekking der Backterien. In van de Koninklijke Jaarboek, van Wetenschappen Akademie Müller, Amsterdam, The Netherlands [Google Scholar]
- 5. Brady SF. 2007. Construction of soil environmental DNA cosmid libraries and screening for clones that produce biologically active small molecules. Nat. Protoc. 2:1297–1305 [DOI] [PubMed] [Google Scholar]
- 6. Chang FY, Brady SF. 2011. Cloning and characterization of an environmental DNA-derived gene cluster that encodes the biosynthesis of the antitumor substance BE-54017. J. Am. Chem. Soc. 133:9996–9999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chao A. 1987. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 43:783–791 [PubMed] [Google Scholar]
- 8. Claesson MJ, et al. 2010. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 38:e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clardy J, Fischbach MA, Walsh CT. 2006. New antibiotics from bacterial natural products. Nat. Biotechnol. 24:1541–1550 [DOI] [PubMed] [Google Scholar]
- 10. Cole JR, et al. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cole JR, et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Curtis TP, Sloan WT, Scannell JW. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. U. S. A. 99:10494–10499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461 [DOI] [PubMed] [Google Scholar]
- 15. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feng Z, Kallifidas D, Brady SF. 2011. Functional analysis of environmental DNA-derived type II polyketide synthases reveals structurally diverse secondary metabolites. Proc. Natl. Acad. Sci. U. S. A. 108:12629–12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Feng Z, Kim JH, Brady SF. 2010. Fluostatins produced by the heterologous expression of a TAR reassembled environmental DNA derived type II PKS gene cluster. J. Am. Chem. Soc. 132:11902–11903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fulthorpe RR, Roesch LF, Riva A, Triplett EW. 2008. Distantly sampled soils carry few species in common. ISME J. 2:901–910 [DOI] [PubMed] [Google Scholar]
- 19. Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. 1998. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5:R245–249 [DOI] [PubMed] [Google Scholar]
- 20. Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8:R143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim JH, et al. 2010. Cloning large natural product gene clusters from the environment: piecing environmental DNA gene clusters back together with TAR. Biopolymers 93:833–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. King RW, Bauer JD, Brady SF. 2009. An environmental DNA-derived type II polyketide biosynthetic pathway encodes the biosynthesis of the pentacyclic polyketide erdacin. Angew. Chem. Int. Ed. Engl. 48:6257–6261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Letunic I, Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128 [DOI] [PubMed] [Google Scholar]
- 24. Martiny JB, et al. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4:102–112 [DOI] [PubMed] [Google Scholar]
- 25. Metsä-Ketelä M, et al. 2002. Molecular evolution of aromatic polyketides and comparative sequence analysis of polyketide ketosynthase and 16S ribosomal DNA genes from various streptomyces species. Appl. Environ. Microbiol. 68:4472–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Minowa Y, Araki M, Kanehisa M. 2007. Comprehensive analysis of distinctive polyketide and nonribosomal peptide structural motifs encoded in microbial genomes. J. Mol. Biol. 368:1500–1517 [DOI] [PubMed] [Google Scholar]
- 27. Rappé MS, Giovannoni SJ. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369–394 [DOI] [PubMed] [Google Scholar]
- 28. Schirmer A, et al. 2005. Metagenomic analysis reveals diverse polyketide synthase gene clusters in microorganisms associated with the marine sponge Discodermia dissoluta. Appl. Environ. Microbiol. 71:4840–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schloss PD, Handelsman J. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Applied and environmental microbiology 71:1501–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schloss PD, Handelsman J. 2004. Status of the microbial census. Microbiol. Mol. Biol. Rev. 68:686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schloss PD, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Watve MG, Tickoo R, Jog MM, Bhole BD. 2001. How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol. 176:386–390 [DOI] [PubMed] [Google Scholar]
- 34. Whitaker RJ, Grogan DW, Taylor JW. 2003. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301:976–978 [DOI] [PubMed] [Google Scholar]
- 35. Whitfield J. 2005. Biogeography. Is everything everywhere? Science 310:960–961 [DOI] [PubMed] [Google Scholar]
- 36. Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer, New York, NY [Google Scholar]
- 37. Zaleta-Rivera K, Charkoudian LK, Ridley CP, Khosla C. 2010. Cloning, sequencing, heterologous expression, and mechanistic analysis of A-74528 biosynthesis. J. Am. Chem. Soc. 132:9122–9128 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.