Abstract
Lytic bacteriophage ATCC 8074-B1 produces large plaques on its host Clostridium sporogenes. Sequencing of the 47,595-bp genome allowed the identification of 82 putative open reading frames, including those encoding proteins for head and tail morphogenesis and lysis. However, sequences commonly associated with lysogeny were absent. ORF 22 encodes an endolysin, CS74L, that shows homology to N-acetylmuramoyl-l-alanine amidases, and when expressed in Escherichia coli, the protein causes effective lysis of C. sporogenes cells when added externally. CS74L was also active on Clostridium tyrobutyricum and Clostridium acetobutylicum. The catalytic domain expressed alone (CS74L1–177) exhibited a similar activity and the same host range as the full-length endolysin. A chimeric endolysin consisting of the CS74L catalytic domain fused to the C-terminal domain of endolysin CD27L, derived from Clostridium difficile bacteriophage ΦCD27, was produced. This chimera (CSCD) lysed C. sporogenes cells with an activity equivalent to that of the catalytic domain alone. In contrast, the CD27L C-terminal domain reduced the efficacy of the CS74L catalytic domain when tested against C. tyrobutyricum. The addition of the CD27L C-terminal domain did not enable the lysin to target C. difficile or other CD27L-sensitive bacteria.
INTRODUCTION
Clostridium sporogenes is environmental bacterium which has been associated with a range of food and silage spoilage problems (13, 30, 39, 46). In cheese spoilage, it can contribute to gas formation and can also compound the damage caused by Clostridium tyrobutyricum, the major cause of cheese blowing (24, 30). It is also physiologically comparable to a nontoxigenic form of proteolytic Clostridium botulinum (29) and as such is often used as a model organism for investigating treatments to reduce C. botulinum contamination. Investigations of methods of control have focused on reducing spore numbers or outgrowth by protocols such as centrifugation and brining (43), polyphosphates and heat treatment (7), acid-blanching and chelation (39), administration of lantibiotics (13, 46), a combination of heat, acidification, and nisin (35), or pulsed pressurization (1). Nitrosyl complexes have also been shown to inhibit the growing cells (20). Bacteriophages and their endolysins represent an additional avenue for control and could be used alone or in combination with current control methods (38).
Bacteriophage endolysins effect the release of new virus particles from an infected cell by attacking the peptidoglycan wall (11, 25). Lysis of Gram-positive bacteria can also be achieved by external application of endolysins, and recent work has demonstrated their potential to reduce infection by a range of bacteria (reviewed in reference 12). Endolysins exhibit a restricted range of activity that is invariably larger than that of the parent bacteriophage but nonetheless relatively specific. Depending on the endolysin, this can encompass all strains within a species, a slightly wider taxonomic grouping, or a broad range of targets. This targeting is commonly thought to be associated with a C-terminal cell wall binding domain, while the hydrolysis of peptidoglycan is achieved by one or more N-terminal catalytic domains. This specificity gives added value to their application as antimicrobials in mixed communities, allowing targeted killing of pathogens or spoilage organisms without disruption of commensal or fermentative microbes.
A number of studies have shown that endolysin domains can be utilized separately; the cell wall binding domain can be isolated and used in conjunction with a reporter or capture system for detection or collection of cells (22, 26, 41), and catalytic domains can be attached to cell wall binding domains from autolysins, other endolysins, or bacteriophage tail-associated peptidoglycan hydrolases to produce new chimeric enzymes with specificities that match the new C-terminal domain (4, 10, 31, 42). In addition, some endolysin catalytic domains retain both their activity and a measure of specificity after removal of their C-terminal domains (9, 28, 32).
In this study, we sequenced the genome of bacteriophage ATCC 8074-B1 (referred to here as Φ8074-B1), which infects C. sporogenes (6), and identified 82 putative open reading frames, many of whose predicted proteins show little homology to existing bacteriophage sequences. The predicted endolysin was expressed in Escherichia coli and demonstrated to exhibit lytic activity against cells of C. sporogenes. A similar level of lytic activity was produced by the N-terminal region of the endolysin expressed either alone or in conjunction with the C-terminal domain of an endolysin active against Clostridium difficile (33).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. sporogenes ATCC 17786, C. difficile NCTC 11204, and C. tyrobutyricum NCIMB 9582 were maintained in Robertson's cooked-meat medium (SGL) at room temperature and grown at 37°C under anaerobic conditions in either RCM broth (Oxoid) or brain heart infusion (BHI) with complements (33). Escherichia coli strains (Invitrogen) were grown in L broth with shaking at 37°C. Other strains were obtained from the NCTC (HPA, London, United Kingdom), the NCIMB (Aberdeen, United Kingdom), or the DSMZ (Braunschweig, Germany) or were kindly donated by Clare Aldus and Sandra Stringer (IFR, Norwich, United Kingdom) and were grown as recommended by DSMZ or in BHI with complements. Strains used to investigate the host range of the endolysin were Anaerococcus hydrogenalis DSMZ 7454, Bacillus amyloliquefaciens 0880, Bacillus cereus NCIMB 11796, Bacillus subtilis ATCC 6633, Clostridium acetobutylicum BL75141, Clostridium bifermentans NCTC 13019, Clostridium butyricum NCIMB 7423 and 8082, Clostridium cellobioparum DSMZ 1351, Clostridium coccoides NCTC 11035, Clostridium innocuum DSMZ 1286, Clostridium leptum DSMZ 753, Clostridium perfringens NCTC 3110, Clostridium ramosum DSMZ 1402, Clostridium sordellii NCTC 13356, C. tyrobutyricum NCIMB 9582, Eubacterium barkeri DSMZ 1223, Listeria innocua NCTC 11288, Listeria ivanovii NCTC 11007, and C. sporogenes ATCC 17886 (also known as 213 [6]), NCIMB 8053, 532, 10696, 9381, 9382, 9383, 12148, 12343, 700933, 701789, 701791, 701792, and 701793, NCDO 1792, and PA 3679.
Propagation and sequencing of Φ8074-B1.
Φ8074-B1 (also known as F1 [6]) was propagated on sensitive strain C. sporogenes ATCC 17786 on RCM supplemented with 5 mM CaCl2 as described previously (34). Genomic DNA was extracted from filtered plate lysates using a λ midikit (Qiagen). Sequencing and assembly of the Φ8074-B1 genome was performed by the DNA Sequencing Facility (University of Cambridge) using 454 sequencing on a Genome Sequencer FLX (Roche) and assembled with the Phred-Phrap program. Open reading frames (ORFs) were determined by Artemis (40) with BLASTP searches (2), and domain searches were performed by InterProScan (49). Amino acid and nucleotide alignments were performed with Vector NTI using the Clustal W algorithm (Invitrogen). Phobius was used to identify transmembrane domains (http://www.ebi.ac.uk/Tools/phobius/), JPred3 (http://www.compbio.dundee.ac.uk/www-jpred/) was used for prediction of secondary structure, and Grease (http://fasta.bioch.virginia.edu/fasta_www2/) was used for Kyte-Doolittle hydropathy plots (23). Electron microscopy to confirm bacteriophage morphology was performed as described previously (33).
Subcloning and expression of Φ8074-B1 endolysin.
The Φ8074-B1 putative endolysin gene, cs74l, was amplified from genomic DNA using Phusion polymerase (Finnzymes) and primers CS74L-F (5′-GGA CTA CAT ATG AAG ATA GGT ATT G [bold letters represent altered nucleotides]; Sigma Genosys) and CS74L-R (5′-TAT TGG GAT CCC TAA ATC CTT), giving a product of 849 bp. The PCR product was restricted, subcloned into the NdeI and BamHI sites of vector pET15b (Novagen), and transformed into E. coli as described previously (33). Positive transformants were selected with ampicillin (100 μg/ml), and after sequence confirmation, construct cs74l-pET15b was transformed into E. coli BL21(DE3) (Invitrogen) for protein expression. The truncated endolysin sequence cs74l1–177 was amplified from cs74l-pET15b using primers T7P from the vector (5′-TAA TAC GAC TCA CTA TAG GG) and CS74L-EAD (5′-TTG ATT CTA GTT TCC AGA TTC T) to insert a stop site after Asn 177. The 687-bp product was subcloned into pCR2.1 (Invitrogen) and then excised with NdeI and XhoI, subcloned into pET15b, and transformed for sequence confirmation and expression as before. The chimeric lysin CSCD nucleotide sequence was produced by splice overlap PCR from cs74l-pET15b and cd27l-pET15b (33). The sequence for the CS74L catalytic domain was amplified using primers T7P and CS_CDC (5′-TTT AAC TCC CTC ATT GTT TCC AGA TTC TCC A). The sequence for the C-terminal domain of CD27L was amplified with CDC_CS (5′-GGA GAA TCT GGA AAC AAT GAG GGA GTT AAA C) and T7T from the vector (5′-GCT AGT TAT TGC TCA GCG G). After amplification and splicing, final products were restricted with NdeI and BamHI and subcloned into pET15b as before.
Measurement of cell lysis.
Endolysin expression was induced in BL21(DE3) cells by isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h as described by the manufacturer (Invitrogen). Crude protein extracts were prepared in either 20 mM sodium phosphate (pH 6), 100 mM HEPES (pH 7), or 20 mM Tris HCl–50 mM NaCl (pH 8) by bead beating as described previously (33) and stored at 4°C. Protein extracts were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting using antibodies to the His tag (Novagen), and His-tagged endolysin was partially purified using Qiagen Fast Start nickel-nitrilotriacetic acid (Ni-NTA) columns, all as described previously (33). Dithiothreitol (DTT; 1 mM) was added to samples before storage. Activity of His-tagged endolysins was assessed by turbidity assays on fresh or frozen cells using 10 μg crude protein extract in 20 mM sodium phosphate (pH 6) or 10 μg partially purified endolysin in elution buffer (50 mM sodium phosphate, 300 mM NaCl, 250 mM imidazole [pH 8.0]). Chicken egg white lysozyme (Sigma) was used as a positive control.
Nucleotide sequence accession number.
The genome of Φ8074-B1 was deposited in GenBank under accession number JQ246028.
RESULTS
Φ8074-B1 genomic analysis.
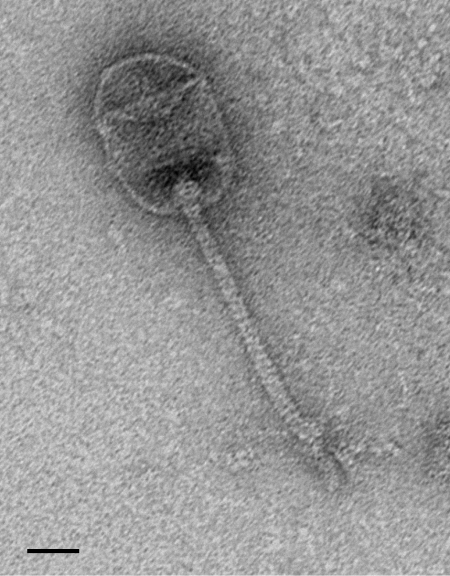
Φ8074-B1 (F1) was previously described as a lytic phage with the ability to form plaques on several strains of C. sporogenes (6). Electron microscopy of phages propagated in strain ATCC 17886 revealed a prolate capsid with icosahedral caps (ca. 70 nm by 46 nm) with a cross-banded and flexible tail (ca. 110 nm long and ca. 7 nm wide) with no visible neck. Microscopy also revealed the presence of short tail fibers. Overall, this indicates that the virus is a member of the family Siphoviridae (Fig. 1) and confirms the morphology described by Betz (5). In addition to the sensitive strains listed previously (6), Φ8074-B1 was able to form plaques on 8 of the 16 C. sporogenes strains tested (Table 1).
Fig 1.
Electron micrograph of Φ8074-B1. The micrograph was taken at a lens setting of 80,000×. Bar, 20 nm.
Table 1.
Lytic activity of CS74L, CS74L1-177, and CSCD on sensitive species
| Bacterium | Lytic activitya |
Φ8074-B1 plaque formationb | |||
|---|---|---|---|---|---|
| CS74L | CS74L1-177 | CSCD | CTP1L | ||
| C. sporogenes ATCC 17886 | 59.9 ± 0.3 | 67.8 ± 0.3 | 69.4 +/-1.3 | − | C |
| C. sporogenes NCIMB 10696 | 84.0 ± 0.3 | 61.6 ± 3.4 | 71.2 ± 0.1 | − | R |
| C. sporogenes NCIMB 532 | 53.6 ± 3.7 | 62.8 ± 0.1 | 67.9 ± 0.4 | − | R |
| C. sporogenes NCIMB 9381 | 77.7 ± 1.8 | 21.5 ± 6.4 | 9.6 ± 3.4 | − | R |
| C. sporogenes NCIMB 9382 | 73.3 ± 4.5 | 33.4 ± 5.1 | 58.0 ± 2.2 | − | C |
| C. sporogenes NCIMB 9383 | 78.9 ± 0.5 | 44.3 ± 2.0 | 75.7 ± 1.1 | − | C |
| C. sporogenes NCIMB 12148 | 40.2 ± 0.7 | 68.1 ± 0.2 | 55.8 ± 0.3 | − | R |
| C. sporogenes NCIMB 12343 | 75.9 ± 5.6 | 70.4 ± 1.6 | 80.2 ± 1.1 | − | T |
| C. sporogenes NCIMB 700933 | 54.9 ± 2.0 | 74.8 ± 0.2 | 74.6 ± 1.9 | − | R |
| C. sporogenes NCIMB 701789 | 35.9 ± 0.3 | 72.2 ± 0.8 | 58.6 ± 3.2 | − | c |
| C. sporogenes NCIMB 701791 | 50.7 ± 4.8 | 75.9 ± 2.5 | 66.9 ± 2.1 | − | R |
| C. sporogenes NCIMB 701792 | 65.9 ± 4.7 | 76.3 ± 1.7 | 68.9 ± 1.2 | − | c |
| C. sporogenes NCIMB 701793 | 58.2 ± 1.9 | 73.6 ± 0.2 | 71.9 ± 0.7 | − | R |
| C. sporogenes NCIMB 8053 | 83.4 ± 0.9 | 52.7 ± 2.5 | 74.9 ± 0.9 | − | R |
| C. sporogenes NCDO 1792 | 51.0 ± 0.5 | 67.5 ± 1.2 | 69.0 ± 0.7 | − | c |
| C. sporogenes PA 3679 | 44.7 ± 0.4 | 68.5 ± 1.8 | 64.5 ± 1.4 | − | c |
| C. acetobutylicum BL75141 | 32.5 ± 0.7 | 31.1 ± 1.1 | 48.5 ± 6.0 | − | R |
| C. tyrobutyricum NCIMB 9582 | 52.7 ± 2.5 | 46.5 ± 7.0 | 16.8 ± 4.2 | 57.6 ± 0.6 | R |
Percent drop in OD at 600 nm in 8 min (mean ± SD), measured over linear lysis of frozen cells incubated with 10 μg partially purified protein. −, not measured.
C, large clear plaques; c, small clear plaques; T, large turbid plaques; R, no plaque formation.
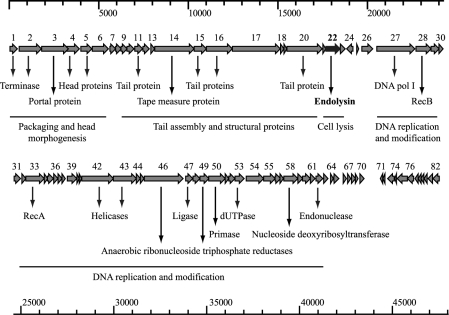
The double-stranded DNA genome of Φ8074-B1 is 47,595 bp, with a 35.4% GC content, and contains 82 putative open reading frames (ORFs). Open reading frames were selected on the basis of proximity to the consensus ribosome binding site, an AG-rich region 5 to 13 nucleotides from the start codon. Most of the ribosome-binding site (RBS) sequences match the most favorable GGAGG sequence (47); AUG is the most common start codon, but some ORFs also have GUG (ORFs 15, 25 and 68), UUG (ORFs 35, 47 and 78), and in one case AUA (ORF 72) as predicted start codons. Most of the proposed ORFs have the same orientation, and several overlap at the predicted starts and ends (Fig. 2). Putative functions were assigned on the basis of BLASTP analysis and/or domain searches (see Table S1 in the supplemental material). In contrast to previously published bacteriophage genomes from members of the Clostridiales, a large number of ORFs did not show high homologies to existing sequences. Those predicted proteins which did show matches to known sequences appeared to be of diverse evolutionary origin, showing similarities to proteins from a wide range of mostly Gram-positive bacteria and phages. No elements associated with lysogeny were identified, and the lack of a prophage state would explain the low level of similar sequences in clostridial genomes. One of the exceptions is the endolysin, ORF 22 (cs74l). A translation of this sequence showed good homology to N-acetylmuramoyl-l-alanine amidases from a variety of sequenced C. botulinum genomes, representing either prophage or autolysin genes. The N-terminal region of CS74L has homology to an N-acetylmuramoyl-l-alanine amidase domain (cd02696, MurNac-LAA; E value, 4e−36), while the C-terminal region showed no homology to the identified cell wall binding domains found in many other endolysins.
Fig 2.
Φ8074-B1 genome map showing predicted ORFs. Arrows indicate the direction of transcription. Proposed functional modules are marked based on BLAST and domain search results (see Table S1 in the supplemental material).
The ORF immediately upstream of the endolysin commonly codes for the holin, forming a two-gene lysis cassette. Unusually, the translation product from the small ORF upstream of ORF 22 does not show any predicted transmembrane domains (TMDs). Computer analysis suggests that its structure is largely helical and slightly hydrophobic. It does share limited homology with part of the predicted holin from ΦCD27, giving amino acid matches of 15.9% (identity) and 26.1% (similarity), but the lack of TMDs argues for either a different function or a different membrane topology. The translation of ORF 21 does have some homology (19.3% identity and 36.8% similarity) to the predicted protein upstream of the sequence with the highest BLASTP homology to the endolysin encoded by ORF 22 (YP_002862711 and YP_002862712, respectively) and to the products of other coding sequences upstream of endolysins with similarity to ORF 22, suggesting that ORF 21 and ORF 22 have been coinherited. However, although holins and lysins would be expected to be cotranscribed, similarity to a consensus promoter sequence was not identified upstream of ORF 21. There were no predicted ORFs overlapping the endolysin sequence.
Both the ORF downstream of cs74l (ORF 23) and a shorter ORF (ORF 25) have one predicted transmembrane domain, but the N termini are predicted to be noncytoplasmic and the C termini cytoplasmic, unlike typical holins of type III (48). BLASTP searches show that the translation product of ORF 23 has similarity to Imm proteins from Clostridium novyi and C. botulinum.
The end of the genome sequence has a section of ORFs reading in the opposite orientation to the majority of the genome. Many of these are short and have limited or no homology to sequences in databases, but most have good matches to the consensus RBS; this section is preceded by a region of ca. 875 bp where none of the short predicted reading frames have recognizable RBS sites. This gap and the change in orientation might suggest some rearrangement of DNA in the past.
CS74L endolysin activity.
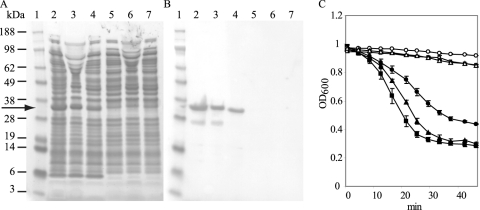
His-tagged CS74L was expressed in E. coli, and crude protein extracts showed clear lytic activity on C. sporogenes 17786 cells (Fig. 3). The most effective extraction buffer in terms of yield and activity was 20 mM sodium phosphate (pH 6); SDS-PAGE and Western analysis of the extracts revealed a ca. 33-kDa band (predicted size, 31.1 kDa) in all extracts, with a second ca. 27-kDa band hybridizing to the His tag antibody in the extracts in 20 mM sodium phosphate (pH 6) and 100 mM HEPES (pH 7) which was absent from extracts in 20 mM Tris HCl, 50 mM NaCl (pH 8). Proteins extracted in HEPES buffer showed a reduced lytic activity. Extracts from cells harboring pET15b alone did not produce appreciable lysis.
Fig 3.
Activity of CS74L in crude protein extracts. (A) SDS-PAGE analysis of crude extracts from E. coli expressing His-tagged endolysin CS74L or empty vector controls. Lane 1, SeeBlue marker (Invitrogen); lanes 2 to 4, E. coli cs74l-pET15b total protein; lanes 5 to 7, E. coli pET15b total protein. Proteins were tested after 4 h induction with IPTG, extracted with 20 mM sodium phosphate (pH 6) (lanes 2 and 5), 100 mM HEPES (pH 7) (lanes 3 and 6), or 20 mM Tris HCl, 50 mM NaCl (pH 8) (lanes 4 and 7). Crude protein samples were loaded at 10 μg per lane. The arrow indicates endolysin CS74L. (B) Western blot analysis of the gel in panel A hybridized to His tag antibody. (C) Turbidity reduction assay of frozen cells of C. sporogenes 17886 incubated with 10 μg crude protein extracts from E. coli expressing cs74l-pET15b (filled symbols) or pET15b (open symbols) extracted in 20 mM sodium phosphate (pH 6) (■), 100 mM HEPES (pH 7) (●), or 20 mM Tris HCl, 50 mM NaCl (pH 8) (▲). Values are means from replicate assays ± standard deviations.
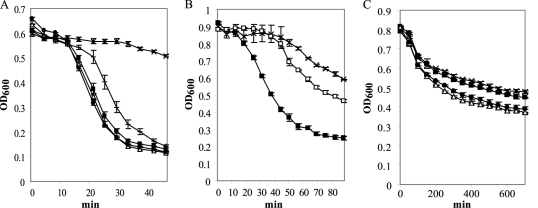
CD27L, which targets C. difficile, is an N-acetylmuramoyl-l-alanine amidase endolysin whose truncated catalytic domain (CD27L1–179) shows an increased activity compared to the full endolysin (32). It shows some similarity to CS74L across the whole length of the proteins (41.7% consensus, 28.4% identity) but is not active against C. sporogenes cells. To investigate the activity and specificity of CS74L further, constructs were produced to express either the CS74L catalytic domain alone (amino acids 1 to 177, covering the region of homology to cd02696 [amino acids 2 to 171]) or a chimera of this domain fused to the region downstream of the catalytic domain of CD27L (amino acids 180 to 270) (Fig. 4). Partially purified extracts of the His-tagged proteins CS74L, truncated CS74L1–177, and chimera CSCD all showed lytic activity on fresh and frozen cells of C. sporogenes 17886 (Fig. 5; Table 1). Lysis of fresh cells commonly occurred after a lag of ca. 15 min. When similar amounts of total protein were added, the truncated catalytic domain CS74L1–177 was slightly more effective than the whole endolysin. This may reflect the higher number of catalytic units in the reaction due to the lower molecular weight. However, unexpectedly CSCD, comprising the CS74L catalytic domain fused to the CD27L C-terminal domain, gave a rate of lysis to similar to that of CS74L1–177. Lysis from 1 μg partially purified CS74L was less than that from 10 μg, and there was a longer lag period before lysis was observed (ca. 30 min) (Fig. 5B). Similar results were obtained with CS74L1–177 and CSCD, while lysis from assays containing 0.1 μg or 0.01 μg lysins was comparable to the gradual autolysis from buffer controls (data not shown).
Fig 4.
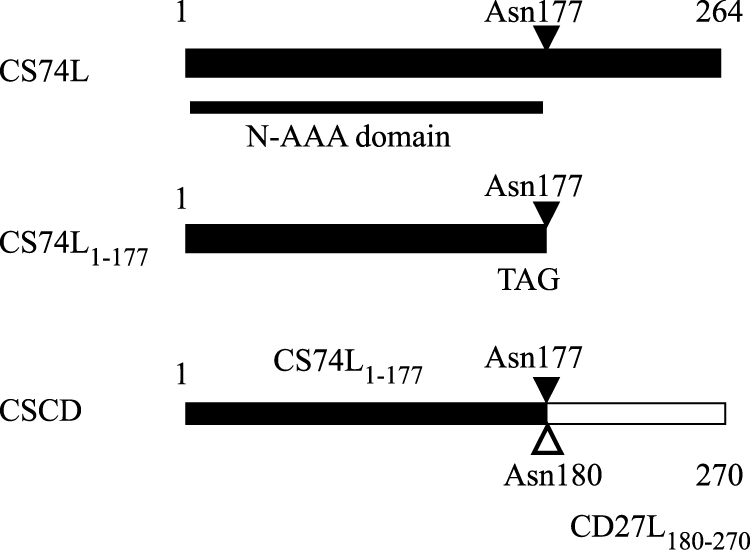
Summary of endolysin truncation and chimera mutants. Portions from endolysin CS74L are in black, and those from CD27L are in white. N-AAA, N-acetylmuramoyl-l-alanine amidase.
Fig 5.
Turbidity reduction assays. Ni-NTA-purified protein extracts were incubated with fresh cells of C. sporogenes ATCC 17886 (A and B) and C. difficile NCTC 11204 (C). (A) ■, 10 μg CS74L; △, 10 μg CS74L1–177; ●, 10 μg CSCD; +, 1,000 U lysozyme, ×, buffer control. (B) ■, 10 μg CS74L; □, 1 μg CS74L; ×, buffer control. (C) ■, 10 μg CS74L; △, 10 μg CS74L1–177; ●, 10 μg CSCD; ×, buffer control. Values are the means for duplicate samples ± standard deviations.
The effect of pH on endolysin activity was investigated using crude protein extracts (Fig. 6). CS74L and CSCD were barely active at pH 4.5 but showed increasing activity to pH 8.5; CS74L1–177 also had little activity at lower pH and a maximum activity at pH 7.5 with a similar activity at pH 8.5.
Fig 6.
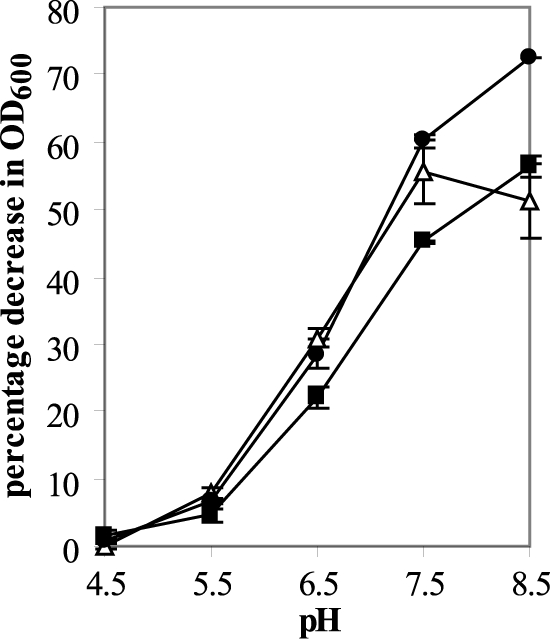
pH profile of endolysin activity. Endolysin activity was measured in a turbidity reduction assay using 10 μg crude protein extract incubated with frozen cells of C. sporogenes ATCC 17886. Results are the means from duplicate assays ± standard deviations and are plotted as the percent decrease in OD at 600 nm over 8 min of linear lysis. ■, CS74L; △, CS74L1–177; ●, CSCD.
All three lysins were active against 15 other strains of C. sporogenes. In more than half of the strains, sensitivity to CS74L1–177 and CSCD was similar and slightly greater than that to CS74L when 10 μg of each partially purified protein was used (Table 1). However, several strains showed a faster lysis with the CS74L, and one strain (NCIMB 9381) showed a much greater resistance to the engineered lysins.
All three lysins were also active on C. tyrobutyricum; CS74L and CS74L1–177 gave rates of lysis similar to that produced by endolysin CTP1L, derived from a bacteriophage of C. tyrobutyricum (34), while CSCD was less effective (Table 1). As CS74L and the glucosyl hydrolase CTP1L have different enzymatic targets, the enzymes were tested both singly and in combination using frozen cells to investigate their potential for synergy. A combination of 5 μg each of CS74L and CTP1L together did indeed give a slightly higher rate of lysis (57.8% ± 1% drop in optical density [OD] over 8 min) than 5 μg of each lysin alone (32.0 ± 10.0% for CS74L and 40.9 ± 0.5% for CTP1L, respectively) but a similar rate of lysis to that from 10 μg CS74L (55.3% ± 0%), indicating a lack of synergy.
CS74L was not effective against C. difficile, and the addition of the C-terminal domain of the C. difficile-targeting endolysin CD27L failed to endow CS74L1–177 with the ability to lyse C. difficile cells. A slight difference between the lysin samples and the buffer controls was noted after an extended incubation, but the effect of CSCD was similar to that of CS74L1–177 (Fig. 5C).
A range of other Gram-positive species were also tested for sensitivity to the lysins. Only C. acetobutylicum showed clear sensitivity to all three lysins, and this was quantitatively similar (Table 1). Other members of the Clostridiales, including C. bifermentans, C. cellobioparum, C. coccoides, C. innocuum, C. leptum, C. perfringens, C. ramosum, C. sordellii, E. barkeri, and the less closely related species B. amyloliquefaciens, B. cereus, B. subtilis, L. innocua, and L. ivanovii, were insensitive to CS74L, CS74L1–177, and CSCD. All three lysins acted on frozen A. hydrogenalis but only after prolonged incubation; 10 μg CS74L took only ca. 8 min to reduce the OD of frozen cells of C. sporogenes by 30%, but the same drop in OD required ca. 370 min with A. hydrogenalis, and although CS74L1–177 and CSCD were more active (ca. 250 and ca. 180 min, respectively), there was still no observable lysis until after 3 h of incubation.
DISCUSSION
C. sporogenes is of interest not only because of its similarity to proteolytic C. botulinum but also in its own right as a spoilage organism. Previous studies identified several bacteriophages capable of infecting C. sporogenes and described Φ8074-B1 (F1) as a tailed bacteriophage with the ability to produce large plaques on several host strains of C. sporogenes (6). Although Betz (5) mentioned preliminary evidence of its ability to lysogenize, F1 was classed as a virulent phage (44). In the present study, no ORFs with homology to sequences associated with lysogeny were identified in the genome sequence of Φ8074-B1; however, the genome contains a large number of sequences for unidentified hypothetical proteins, so the presence of such a module cannot be ruled out completely.
Despite a lack of homology of many of the ORFs to known sequences in databases, the CS74L endolysin sequence was highly conserved with N-acetylmuramoyl-l-alanine amidases from the sequenced genomes of a number of strains of C. botulinum, including some annotated as bacteriophage endolysins. A high conservation of endolysin sequence has been also noted previously in bacteriophages of C. perfringens (36). We failed to identify a typical holin gene in the region of the endolysin gene. Holins usually fall into one of 3 classes based on membrane topology (48). Class I has 3 transmembrane domains (TMD) with the N terminus on the outside and the C terminus inside, class II has two TMDs with both termini inside the cytoplasm, and class III has only a single TMD with the N terminus on the inside and the C terminus outside the membrane. Holin genes are not always located immediately upstream of the endolysin gene: holin coding sequences from one clade of bacteriophages infecting C. perfringens were found downstream of the lysin genes (36), and bacteriophage C2 (ΦC2), which infects C. difficile has a larger lysis cassette, with an ORF with homology to the abiF gene (ORF 37) located between the holin and the lysin sequences (16), while in the T7 phage the holin gene is located in a cluster other than the lysis cassette (48). There are also other examples of bacteriophages where potential classic holins have not been identified (8). Further work to coexpress the lysin with potential holins may help to identify a functional holin gene and demonstrate whether the genome contains a novel type of holin. The abiF gene is thought to confer phage resistance, possibly to prevent superinfection by other phages (16), and so has a functional similarity to the Imm protein. It is possible that ORF 23 is part of the lysis cassette, playing a role similar to that of the ΦC2 ORF 37. It is interesting that the length of the region covering ORFs 21, 22, and 23 and the immediate up- and downstream noncoding regions corresponds to a molecular weight of ca. 0.4 × 106, the same size as a late transcript of Φ8074-B1 identified in a previous work (45).
Endolysin CS74L lysed cells of all 16 strains of C. sporogenes tested but was inactive on a range of other clostridia, with the exception of C. acetobutylicum and C. tyrobutyricum. Truncation of the endolysin to just the proposed catalytic domain produced an active lysin but did not affect the specificity. Other studies reported the maintenance of endolysin activity upon removal of C-terminal domains; in many cases, this truncation led to a notable increase in activity (9, 14, 28, 32), while in others, activity remained similar to or was lower than that of the full-length endolysin (10, 21). In several cases, endolysin catalytic domains retained the same or a similar host range upon removal of the C-terminal domain (9, 18, 28, 32), despite the fact that their peptidoglycan target is usually shared with a number of other species. The difference in lytic activities between CS74L and CS74L1-177 varied between the target strains of C. sporogenes; in the majority of cases, activity was broadly similar or slightly greater, although with some strains the full-length endolysin was clearly more effective. This may reflect strain-specific differences in cell wall decorations that could impact binding to the endolysin, such that in some strains the possession of the correct C-terminal domain is advantageous. In cases where CS74L1-177 was more effective than CS74L, a chimera containing a different C-terminal domain often had a lytic activity similar to that of the truncated endolysin, which implies that the increase is not associated with improved access to the peptidoglycan due to a reduction in size of the protein.
CS74L did not cause significant lysis of C. difficile cells, and the chimera CSCD, consisting of the CS74L catalytic domain cotranslationally fused to the CD27L C-terminal domain, showed only a very limited activity on C. difficile cells which was similar to the activity of CS74L1-177, indicating that the CD27L C-terminal domain did not confer a significant improvement by targeting the endolysin to the cell wall. A number of other studies have demonstrated the ability of endolysin or autolysin C-terminal domains to confer new host specificities on catalytic domains (4, 10, 42). It is possible that the CD27L C-terminal domain does not contain a cell wall binding capability; as with CS74L and CTP1L, there are no recognized cell wall binding motifs such as those found in other lysins (e.g., SH3b and LysM) (4, 19). Alternatively, it could be that the CD27L cell wall binding activity is not fully effective when added externally; since it has evolved to act from inside the cell, it may not be able to access the peptidoglycan or its binding target from the outside due to external structures, such as the extensive array of surface layer proteins. It has been proposed that endolysin C-terminal domains exhibit an inhibitory activity when not bound to the correct target (27, 28). Although the presence of the CD27L C-terminal domain did not reduce activity of the CS74L catalytic domain on C. sporogenes, it did have a negative effect on CSCD lytic activity against C. tyrobutyricum.
Bacteriophages and their endolysins are of increasing interest as possible novel biocontrol agents to reduce usage of antibiotics and to target antibiotic-resistant bacteria. Although bacteriophages infect only a few strains, the use of bacteriophage cocktails has shown promise in the reduction of disease in animals (3) and biocontrol of food-borne pathogens (17). With their broader host range, bacteriophage-derived endolysins have shown potential both as antimicrobials against a range of pathogens and as the basis for novel detection systems (12). These characteristics are also applicable in food protection to reduce spoilage and improve safety. Endolysins have already been demonstrated to reduce pathogens in milk both alone and in combination with lantibiotics (15, 37). Although the use of a protein might be problematic for long-term control, endolysins can be delivered by food-grade organisms such as Lactococcus lactis (14, 33). Coculture of a C. sporogenes strain isolated from silage with a bacteriocin-producing Streptococcus bovis strain can reduce ammonia production, which could be applicable in the prevention of silage spoilage (13). In a similar way, coculture of strains which overproduce and export endolysins might reduce numbers of spoilage organisms or pathogens in such diverse processes as silage fermentation, cheese production, and food preservation. Further analysis of their mode of action, the nature of their specificity, and appropriate delivery systems will aid their use as novel biocontrol agents.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a strategic core grant from the Biotechnology and Biological Science Research Council.
We thank Kathryn Cross (Imaging Platform, IFR) for electron microscopy, Shilo Dickens and John Lester (University of Cambridge) for genome sequencing, and Rob Meijers (EMBL, Hamburg, Germany) for helpful discussions.
Footnotes
Published ahead of print 16 March 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Ahn J, Balasubramaniam VM. 2007. Effects of inoculum level and pressure pulse on the inactivation of Clostridium sporogenes spores by pressure-assisted thermal processing. J. Microbiol. Biotechnol. 17:616–623 [PubMed] [Google Scholar]
- 2. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Atterbury RJ, et al. 2007. Bacteriophage therapy to reduce salmonella colonization of broiler chickens. Appl. Environ. Microbiol. 73:4543–4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Becker SC, Foster-Frey J, Stodola AJ, Anacker D, Donovan DM. 2009. Differentially conserved staphylococcal SH3b_5 cell wall binding domains confer increased staphylolytic and streptolytic activity to a streptococcal prophage endolysin domain. Gene 443:32–41 [DOI] [PubMed] [Google Scholar]
- 5. Betz JV. 1968. Some properties of bacteriophages active on the obligate anaerobe Clostridium sporogenes. Virology 36:9–19 [DOI] [PubMed] [Google Scholar]
- 6. Betz JV, Anderson KE. 1964. Isolation and Characterization of Bacteriophages Active on Clostridium sporogenes. J. Bacteriol. 87:408–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Borch E, Lycken L. 2007. Influence of long-chain polyphosphate and heat treatment on Clostridium cochlearium and Clostridium sporogenes isolated from processed cheese spread. J. Food Prot. 70:744–747 [DOI] [PubMed] [Google Scholar]
- 8. Borysowski J, Weber-Dabrowska B, Gorski A. 2006. Bacteriophage endolysins as a novel class of antibacterial agents. Exp. Biol. Med. (Maywood) 231:366–377 [DOI] [PubMed] [Google Scholar]
- 9. Cheng Q, Fischetti VA. 2007. Mutagenesis of a bacteriophage lytic enzyme PlyGBS significantly increases its antibacterial activity against group B streptococci. Appl. Microbiol. Biotechnol. 74:1284–1291 [DOI] [PubMed] [Google Scholar]
- 10. Donovan DM, et al. 2006. Peptidoglycan hydrolase fusions maintain their parental specificities. Appl. Environ. Microbiol. 72:2988–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Doughty CC, Hayashi JA. 1962. Enzymatic properties of a phage-induced lysin affecting group A streptococci. J. Bacteriol. 83:1058–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fischetti VA. 2010. Bacteriophage endolysins: a novel anti-infective to control Gram-positive pathogens. Int. J. Med. Microbiol. 300:357–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Flythe MD, Russell JB. 2004. The effect of pH and a bacteriocin (bovicin HC5) on Clostridium sporogenes MD1, a bacterium that has the ability to degrade amino acids in ensiled plant materials. FEMS Microbiol. Ecol. 47:215–222 [DOI] [PubMed] [Google Scholar]
- 14. Gaeng S, Scherer S, Neve H, Loessner MJ. 2000. Gene cloning and expression and secretion of Listeria monocytogenes bacteriophage-lytic enzymes in Lactococcus lactis. Appl. Environ. Microbiol. 66:2951–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garcia P, Martinez B, Rodriguez L, Rodriguez A. 2010. Synergy between the phage endolysin LysH5 and nisin to kill Staphylococcus aureus in pasteurized milk. Int. J. Food Microbiol. 141:151–155 [DOI] [PubMed] [Google Scholar]
- 16. Goh S, Ong PF, Song KP, Riley TV, Chang BJ. 2007. The complete genome sequence of Clostridium difficile phage phiC2 and comparisons to phiCD119 and inducible prophages of CD630. Microbiology 153:676–685 [DOI] [PubMed] [Google Scholar]
- 17. Guenther S, Huwyler D, Richard S, Loessner MJ. 2009. Virulent bacteriophage for efficient biocontrol of Listeria monocytogenes in ready-to-eat foods. Appl. Environ. Microbiol. 75:93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Horgan M, et al. 2009. Phage lysin LysK can be truncated to its CHAP domain and retain lytic activity against live antibiotic-resistant staphylococci. Appl. Environ. Microbiol. 75:872–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu S, Kong J, Kong W, Guo T, Ji M. 2010. Characterization of a novel LysM domain from Lactobacillus fermentum bacteriophage endolysin and its use as an anchor to display heterologous proteins on the surfaces of lactic acid bacteria. Appl. Environ. Microbiol. 76:2410–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Joannou CL, et al. 1998. Characterization of the bactericidal effects of sodium nitroprusside and other pentacyanonitrosyl complexes on the food spoilage bacterium Clostridium sporogenes. Appl. Environ. Microbiol. 64:3195–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Korndorfer IP, et al. 2006. The crystal structure of the bacteriophage PSA endolysin reveals a unique fold responsible for specific recognition of Listeria cell walls. J. Mol. Biol. 364:678–689 [DOI] [PubMed] [Google Scholar]
- 22. Kretzer JW, et al. 2007. Use of high-affinity cell wall-binding domains of bacteriophage endolysins for immobilization and separation of bacterial cells. Appl. Environ. Microbiol. 73:1992–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105–132 [DOI] [PubMed] [Google Scholar]
- 24. Le Bourhis AG, et al. 2007. Contribution of C. beijerinckii and C. sporogenes in association with C. tyrobutyricum to the butyric fermentation in Emmental type cheese. Int. J. Food Microbiol. 113:154–163 [DOI] [PubMed] [Google Scholar]
- 25. Loessner MJ. 2005. Bacteriophage endolysins—current state of research and applications. Curr. Opin. Microbiol. 8:480–487 [DOI] [PubMed] [Google Scholar]
- 26. Loessner MJ, Kramer K, Ebel F, Scherer S. 2002. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 44:335–349 [DOI] [PubMed] [Google Scholar]
- 27. Low LY, Yang C, Perego M, Osterman A, Liddington R. 2011. Role of net charge on catalytic domain and influence of cell wall binding domain on bactericidal activity, specificity, and host range of phage lysins. J. Biol. Chem. 286:34391–34403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Low LY, Yang C, Perego M, Osterman A, Liddington RC. 2005. Structure and lytic activity of a Bacillus anthracis prophage endolysin. J. Biol. Chem. 280:35433–35439 [DOI] [PubMed] [Google Scholar]
- 29. Lund BM, Peck MW. 2000. Clostridium botulinum, p 1057–1109 In Lund BM, Baird-Parker TC, Gould GW. (ed), The microbiological safety and quality of food. Aspen Publishers Inc., Gaithersburg, MD [Google Scholar]
- 30. Lycken L, Borch E. 2006. Characterization of Clostridium spp. isolated from spoiled processed cheese products. J. Food Prot. 69:1887–1891 [DOI] [PubMed] [Google Scholar]
- 31. Manoharadas S, Witte A, Blasi U. 2009. Antimicrobial activity of a chimeric enzybiotic towards Staphylococcus aureus. J. Biotechnol. 139:118–123 [DOI] [PubMed] [Google Scholar]
- 32. Mayer MJ, Garefalaki V, Spoerl R, Narbad A, Meijers R. 2011. Structure-based modification of a Clostridium difficile targeting endolysin affects activity and host range. J. Bacteriol. 193:5477–5486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mayer MJ, Narbad A, Gasson MJ. 2008. Molecular characterization of a Clostridium difficile bacteriophage and its cloned biologically active endolysin. J. Bacteriol. 190:6734–6740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mayer MJ, Payne J, Gasson MJ, Narbad A. 2010. Genomic sequence and characterization of the virulent bacteriophage phiCTP1 from Clostridium tyrobutyricum and heterologous expression of its endolysin. Appl. Environ. Microbiol. 76:5415–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Naim F, et al. 2008. Combined effects of heat, nisin and acidification on the inactivation of Clostridium sporogenes spores in carrot-alginate particles: from kinetics to process validation. Food Microbiol. 25:936–941 [DOI] [PubMed] [Google Scholar]
- 36. Oakley BB, et al. 2011. Comparative genomics of four closely related Clostridium perfringens bacteriophages reveals variable evolution among core genes with therapeutic potential. BMC Genomics 12:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Obeso JM, Martinez B, Rodriguez A, Garcia P. 2008. Lytic activity of the recombinant staphylococcal bacteriophage PhiH5 endolysin active against Staphylococcus aureus in milk. Int. J. Food Microbiol. 128:212–218 [DOI] [PubMed] [Google Scholar]
- 38. O'Flaherty S, Ross RP, Coffey A. 2009. Bacteriophage and their lysins for elimination of infectious bacteria. FEMS Microbiol. Rev. 33:801–819 [DOI] [PubMed] [Google Scholar]
- 39. Okereke A, Beelman RB, Doores S. 1990. Control of spoilage of canned mushrooms inoculated with Clostridium sporogenes Pa3679 spores by acid-blanching and EDTA. J. Food Sci. 55:1331–1333 [Google Scholar]
- 40. Rutherford K, et al. 2000. Artemis: sequence visualisation and annotation. Bioinformatics 16:944–945 [DOI] [PubMed] [Google Scholar]
- 41. Sainathrao S, Mohan KV, Atreya C. 2009. Gamma-phage lysin PlyG sequence-based synthetic peptides coupled with Qdot-nanocrystals are useful for developing detection methods for Bacillus anthracis by using its surrogates, B. anthracis-Sterne and B. cereus-4342. BMC Biotechnol. 9:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sheehan MM, Garcia JL, Lopez R, Garcia P. 1996. Analysis of the catalytic domain of the lysin of the lactococcal bacteriophage Tuc2009 by chimeric gene assembling. FEMS Microbiol. Lett. 140:23–28 [DOI] [PubMed] [Google Scholar]
- 43. Su YC, Ingham SC. 2000. Influence of milk centrifugation, brining and ripening conditions in preventing gas formation by Clostridium spp. in Gouda cheese. Int. J. Food Microbiol. 54:147–154 [DOI] [PubMed] [Google Scholar]
- 44. Taylor DE, Guha A. 1974. Asymmetric transcription during development of F1, a bacteriophage specific for Clostridium sporogenes. Virology 59:190–200 [DOI] [PubMed] [Google Scholar]
- 45. Taylor DE, Guha A. 1975. Development of bacteriophage F1 in Clostridium sporogenes: characterization of RNA transcripts. J. Virol. 16:107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wijnker JJ, Weerts EA, Breukink EJ, Houben JH, Lipman LJ. 2011. Reduction of Clostridium sporogenes spore outgrowth in natural sausage casings using nisin. Food Microbiol. 28:974–979 [DOI] [PubMed] [Google Scholar]
- 47. Young M, Minton NP, Staudenbauer WL. 1989. Recent advances in the genetics of the clostridia. FEMS Microbiol. Rev. 5:301–325 [DOI] [PubMed] [Google Scholar]
- 48. Young R. 2002. Bacteriophage holins: deadly diversity. J. Mol. Microbiol. Biotechnol. 4:21–36 [PubMed] [Google Scholar]
- 49. Zdobnov EM, Apweiler R. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.