Abstract
Filter-collected production water samples from a methane-rich gas field in the Cook Inlet basin of Alaska were investigated using whole-cell rRNA-targeted fluorescence in situ hybridization (FISH) and 16S rRNA tag pyrosequencing. Both techniques were consistent in determining the microbial community composition, including the archaeal or bacterial dominance of samples. The archaeal community is dominated by the obligate methylotrophic methanogen genus Methanolobus as well as the nutritional generalist methanogen genus Methanosarcina, which is capable of utilizing acetate, CO2, and methyl-bearing compounds. The most-abundant bacterial groups are Firmicutes, notably of the Acetobacterium genus, and Cytophaga-Flexibacter-Bacteroides species (CFBs) affiliated with the order Bacteroidales. We observed spatial variation among samples in both the percentage of members of Archaea compared to that of members of Bacteria and the dominant members of the bacterial community, differences which could not be explained with the available geochemical data. Based upon the microbial community composition and the isotopic signature of methane associated with the Cook Inlet basin site, we propose a simplified reaction network beginning with the breakdown of coal macromolecules, followed by fermentation and methylotrophic and acetoclastic methane production.
INTRODUCTION
Coal bed methane (CBM) refers to natural gas associated with coal that may be formed by the thermogenic cracking of hydrocarbons or biogenically through the degradation of complex organic matter. This gas may be retained by adsorption to the coal, by storage in associated porous layers and coal fractures, and by dissolution in water (26). Gas composition, reservoir geochemistry, and stable isotopes can be used to discriminate between biogenic and thermogenic sources (11, 36). The stable isotope signature of methane can additionally distinguish between the two main pathways of biogenic methane formation, carbonate reduction and methyl group fermentation (35, 36). CBM represents a significant and growing unconventional source of natural gas (7). Enhancing biogenic conversion of coal to methane by in situ microbial communities requires an understanding of naturally occurring microbial associations and their physiological constraints.
Coal presents a challenge to microbial degradation due to its complex, heterogeneous structure composed of aromatic, polycyclic aromatic, and aliphatic moieties (10). Biological conversion of coal to methane requires a consortium of microorganisms that are capable of depolymerizing coal and fermenting monomers to lower-molecular-weight compounds, such as acetate, H2, CO2, methyl amines, and methyl sulfides, that can be utilized by methanogenic Archaea (4, 10, 14, 27, 32). The specific pathway for coal degradation depends upon the microbial community present. Coal beds show a high diversity of associated members of Bacteria, including significant representation by Firmicutes, Spirochetes, Bacteroidetes, and all subgroups of Proteobacteria (31). Methanogenic Archaea identified in coal beds include species of Methanosarcinales, Methanomicrobiales, and Methanobacteriales, representing all of the known methanogenic pathways (31).
Recent studies have characterized the indigenous coal bed microbial communities using both culturing and 16S rRNA gene cloning (12, 13, 19, 20, 23, 30, 32). These studies provide a glimpse of the natural microbial assemblages associated with coal beds but may not reflect the relative population sizes of the active bacterial and archaeal populations in situ. A better understanding of the geochemical factors that influence microbial community structure and archaeal-bacterial associations is necessary to predict the likely pathways for the breakdown of the complex coal matrix to precursors for methanogenesis. These pathways are relevant to both the industrial production of biogenic methane and the biogeochemistry of archaeal and bacterial associations involved in the degradation of complex organic compounds. Here, we combine 16S rRNA tag pyrosequencing with whole-cell rRNA-targeted fluorescent in situ hybridization (FISH) to generate quantitative microbial diversity estimates and to visualize associations of members of Bacteria and members of Archaea present in coal bed communities in a biogenic gas field in the Cook Inlet basin (CIB), AK.
MATERIALS AND METHODS
Sampling site and geochemical analysis.
Water samples were obtained from production wells of a biogenic gas field in the northern portion of the Cook Inlet basin, AK. The major natural gas reserves consist of 1.8 × 1011 m3 of shallow dry gas (99% methane) occurring in sandstones interbedded with coal from Miocene- to Pliocene-aged rocks in the Kenai group. The δ13Cmethane ranges from −63 to −56‰ and is interpreted to have a biogenic origin (5). Production water samples were analyzed for the concentrations of seven major ions at the Bartlesville Technology Center, ConocoPhillips. Isotopic analysis of gas samples was performed at Isotech, IL. Gas compositional analysis was performed on a Carle AGC400/AGC100. Values for methane δD were measured on an HP6890 coupled with a Delta V Plus GC-P-III interface. Values for methane δ13C were measured on an HP6809 coupled with a Delta Plus Advantage XL via a GC-C-II interface.
Community genomic DNA extraction and 16SrRNA gene tag pyrosequencing.
Production water samples (250 to 500 ml) were filtered through a 47-mm (0.2-μm-pore-size) Durapore membrane filter (Millipore, Billerica, MA). DNA extractions were performed as previously described by Ashby et al. (2). Briefly, filters were sliced into 96 equally sized portions using a sterile scalpel and transferred equally into two 2.0-ml screw-cap centrifuge tubes containing ceramic beads obtained from CeroGlass (Columbia, TN). The bead-beating matrix consisted of one 4-mm glass bead (GSM-40), 0.75 g of 1.4- to 1.6-mm zirconium silicate beads (SLZ-15), and 0.75 g of 1.4- to 1.6-mm zirconium silicate beads (SLZ-15) in 1 ml phosphate buffer (180 mM sodium phosphate, 18 mM EDTA, pH 8.0) and 10% SDS (sodium dodecyl sulfate). Cells were disrupted in a FastPrep FP120 instrument (Bio-101, Vista, CA) as previously described (2). Total genomic DNA was portioned into the supernatant by centrifuging the lysed cells at 13,200 × g for 5 min at 4°C. The supernatants were transferred into new 1.5-ml centrifuge tubes and further purified by adding 500 μl phenol-chloroform-isoamyl alcohol (25:24:1). The aqueous and organic phases were separated by centrifugation at 13,200 × g for 5 min at room temperature. The resulting genomic DNA in the aqueous phase was purified on QIAprep Plasmid Spin columns (Qiagen, Valencia, CA) according to the manufacturer's instructions.
A 600-bp portion of the 16S rRNA gene was amplified by PCR using the bacterial and archaeal primer pair TX-9/1391R (TX-9, 5′-[BioTEG]-GGATTAGAWACCCBGGTAGTC-3′; 1391R, 5′-GACGGGCRGTGWGTRCA-3′) (34) as previously described by Ashby et al. (2). Amplicons were agarose gel purified and quantitated using SYBR green (Invitrogen, Carlsbad, CA). A second round of PCR was performed using fusion primers that incorporated the “A” and “B” 454 pyrosequencing adapters onto the 5′ ends of the TX9 and 1391 primers, respectively. The forward fusion primer also included variable-length barcodes that enabled the multiplexing of multiple samples into a single 454 sequencing run. These amplicons were PAGE purified and quantitated prior to combining into one composite library. The resulting library was sequenced using the standard 454 Life Sciences Lib-L emulsion PCR protocol and Titanium chemistry sequencing (22). Sequences that passed the instrument quality control filters were also subjected to additional filters that required all bases be Q20 or higher and the average of all bases in any read be Q25 or greater. Furthermore, the TX9 primer was trimmed off the 5′ end and the sequences were trimmed on the 3′ end at a conserved site distal to the V6 region (ca. position 1067; Escherichia coli numbering). The rRNA gene sequence fragments were approximately 250 bp in length and included the V5 and V6 regions. The abundances (frequency counts) of all trimmed V5-to-V6 sequences were as follows: CIB-1, 27,410; CIB-2, 25,894; CIB-3, 31,210.
Prior to microbial diversity analysis, the trimmed V5-to-V6 sequences were filtered to include only sequences seen >100 times. This reduced the total numbers of sequences for each sample to 24,039 for CIB-1, 23,780 for CIB-2, and 25,858 for CIB-3. The 100 most abundant sequences per sample were sorted by division (see Table S3 in the supplemental material). The trimmed sequences were considered unique if they differed by one nucleotide and were manually compared to the NCBI database using BLASTn (1). Division assignments were made when the query sequence had >90% identity to sequences with assigned taxonomy.
Whole-cell rRNA-targeted FISH probe design and analysis.
Production water samples (250 to 500 ml) for whole-cell rRNA-targeted FISH were collected using 47-mm (0.2-μm-pore-size) black nucleopore polycarbonate filters (Whatman, United Kingdom) using an in-line filter holder and a luer-lok syringe. Filters were fixed for whole-cell rRNA-targeted FISH in 1% (wt/vol) paraformaldehyde, stored on ice, and processed within 48 h of collection. The filters were washed with 1× phosphate-buffered saline (PBS) and stored in a 1:1 PBS-ethanol solution at −20°C. Whole-cell rRNA-targeted FISH experiments were carried out as described by Macalady et al. (21). Probes were synthesized and labeled at the 5′ end with fluorescent dyes (CY3, CY5, and FLC) from Sigma-Aldrich. After hybridization, cells were stained with 4′,6-diamidino-2-phenylindole (DAPI), mounted with Vectashield (Vectashield Laboratories, United States), and viewed on a Nikon E800 epifluorescence microscope. Images were collected with a Nikon charge-coupled device (CCD) camera using NIS Elements AR 2.30 image analysis software. The real-time deconvolution module in the NIS Elements software was used to aid in cell counts. Large aggregates were not counted because of their three-dimensional (3D) nature and difficulty in resolving individual cells. Microbial diversity estimates were determined from counting a minimum of 1,000 DAPI-stained cells for each sample/probe combination. The standard deviation was determined among separate slides (n ≥ 3 slides).
The oligonucleotide probes and formamide concentrations used in this study are given in Table S1 in the supplemental material. Probes EUBmix, ARCH915, Sarci551, MB311, MG1200b, Chis150, CFX1082, SRB385, and Delta295a have been used previously to analyze environmental samples and were used as described in the references in Table S1 in the supplemental material. Two new probes (Aceto828 and Mlob828) were designed using the ProbeDesign function in the ARB software package. Probe design and testing followed the methods described by Hugenholtz et al. (15), including searches against publicly available sequences with BLAST, ARB, and the online search tool ProbeMatch (http://rdp.cme.msu.edu/probematch/search.jsp) (6). The following pure cultures were obtained for use as positive and negative controls for whole-cell rRNA-targeted FISH: Methanosarcina acetivorans (DSMZ 2834), Methanolobus zinderi SD1 (J.G. Ferry, Pennsylvania State University), Acetobacterium woodii (DSMZ 1030), Desulfosarcina variabilis (DSMZ 2060), Desulfovibrio gigas (DSMZ 1382), Bacteroides coprosuis (NRRL B-41113), Clostridium beijerinckii (NRRL B-597), Clostridium acetobutylicum (NRRL B-527), and Ktedonobacter racemifer (NRRL B-41538).
Principal component analysis of microbial diversity.
Principal component analysis (PCA) was performed on microbial diversity estimates for three Cook Inlet basin samples as well as microbial diversity estimates from four previously described basins (13, 19, 23, 30) by using the default settings of the “princomp” function in the R statistical software package (29). Geochemical data (see Table S2 in the supplemental material) were projected onto the ordination using the “envfit” function (1,000 permutations). The envfit function fits environmental variables onto a previously calculated ordination which identifies gradients related to environmental factors without constraining the ordination. In order to eliminate scaling artifacts, all geochemical data were standardized by the parameter maximum and microbial diversity data were standardized to percent abundance as determined by whole-cell 16S rRNA-targeted FISH counts (see Fig. 4A) or from the microbial diversity of the Cook Inlet basin and four previously described basins (see Fig. 4B) (13, 19, 23, 30).
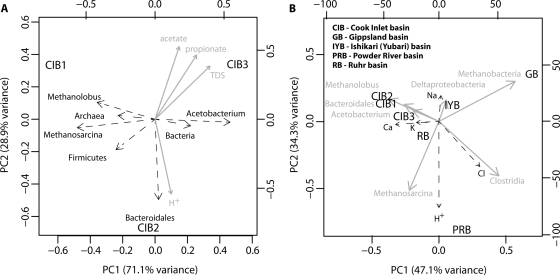
Fig 4.
(A) PCA of Cook Inlet basin microbial diversity estimates (dashed black lines) for three samples (black), with geochemical data (solid gray lines) projected as vectors onto the ordination showing correlation with microbial diversity estimates. (B) PCA of microbial diversity estimates data (solid gray lines) from five sedimentary basins (black), with geochemical data (dashed black lines) projected as vectors onto the ordination showing correlation with microbial diversity estimates data. Geochemical data are normalized by the parameter maximum prior to ordination in order to eliminate scaling artifacts.
RESULTS
Microbial diversity estimates from whole-cell rRNA-targeted FISH and 16S rRNA gene tag pyrosequencing.
A total of 12 whole-cell rRNA-targeted FISH probes (see Table S1 in the supplemental material) were used to analyze the microbial community in the Cook Inlet basin samples CIB-2, CIB-1, and CIB-3. Representative epifluorescence micrographs are shown in Fig. 1. The ratios of members of Archaea to members of Bacteria and the microbial community composition vary between the three samples (Fig. 2). CIB-1 is dominated by members of Archaea, with 56% ± 10.6% of cells hybridizing to ARCH915 and 40.3% ± 8.0% of cells hybridizing to EUBmix. CIB-2 displayed a more even distribution of members of Archaea and members of Bacteria, with 43.4% ± 7.1% of cells hybridizing to ARCH915 and 56.5% ± 9.8% of cells hybridizing to EUBmix. CIB-3 is a Bacteria-dominated site, with 34.3% ± 14.3% hybridizing to ARCH915 and 64.8% ± 4.9% of cells hybridizing to EUBmix.
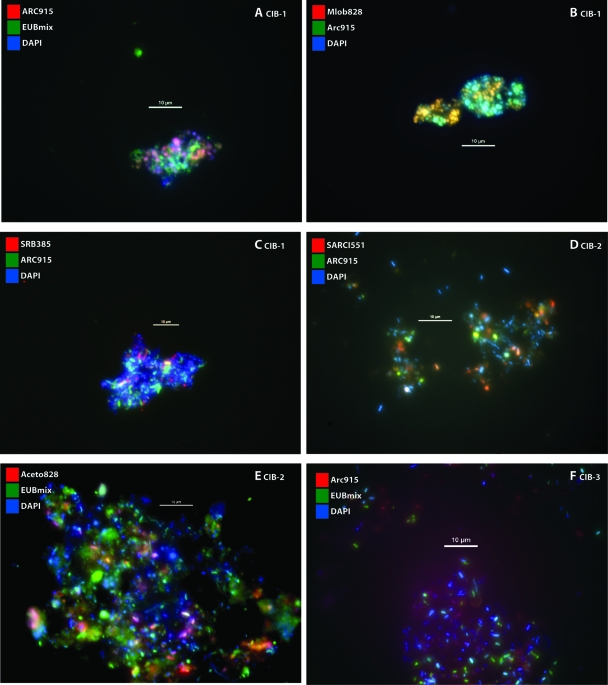
Fig 1.
Representative whole-cell rRNA-targeted FISH micrographs depicting major bacterial and archaeal lineages observed in production water samples from the Cook Inlet basin. (A) Members of Bacteria (green) and members of Archaea (red) in sample CIB-1. (B) Methanolobus spp. (orange) and other members of Archaea (green) in CIB-1. (C) Members of the Firmicutes (yellow-orange) and other members of Bacteria (green) in CIB-1. (D) Methanosarcina spp. (orange) and other members of Archaea (green) in CIB-2. (E) Acetobacterium spp. (red-orange) and other members of Bacteria (green) in CIB-2. (F) Members of Archaea (red) and members of Bacteria (green) in CIB-3. Probe specificities: EUBmix, all Bacteria; ARCH915, most Archaea; DAPI, general DNA stain; Mlob828, genus Methanolobus; SRB385, some Firmicutes; SARCI551, genus Methanosarcina; Aceto828, genus Acetobacterium.
Fig 2.
Microbial communities of the Cook Inlet gas field expressed as percentages of DAPI-stained cells. Bacteria are shown with solid lines, and Archaea are shown with patterned lines. The values are the percentages determined from counting a minimum of 1,000 DAPI-stained cells. Standard deviations of 1.5 to 16% are determined among separate slides (n ≥ 3 slides).
Additional analysis of the archaeal and bacterial populations focused on target groups identified by 16S rRNA tag pyrosequencing. In all three sites, a suite of probes for methanogenic Archaea revealed that the genera Methanolobus and Methanosarcina are the most numerous members of Archaea, with average Methanolobus cell counts greater than average Methanosarcina cell counts in all three samples. CIB-1 consists of 40.8% ± 9.7% Methanolobus and 28.1% ± 9.8% Methanosarcina. CIB-2 consists of 22.3% ± 3.2% Methanolobus and 18.9% ± 8.0% Methanosarcina. CIB-3 consists of 16.0% ± 9.6% Methanolobus and 5.2% ± 16.0% Methanosarcina. No cells are observed to hybridize to probes specific for Methanobacteria or other Methanomicrobiales. Bacterial communities in the three samples are more variable than the archaeal communities. CIB-1 consists of 10.8% ± 1.5% SRB385-hybridizing cells, 8.2% ± 4.2% Bacteroidales, and 6.0% ± 1.9% Acetobacterium. CIB-2 consists of 32.9% ± 5.8% Bacteroidales, 18.5% ± 1.6% Acetobacterium, and 11.7% ± 3.9% SRB385-hybridizing cells. CIB-3 consists of 28.0% ± 3.9% Acetobacterium, 9.6% ± 6.9% Bacteroidales, and 6.1% ± 2.5% SRB385-hybridizing cells. Cells hybridizing to the probe SRB385, which targets some Deltaproteobacteria as well as some Firmicutes, are attributed to Firmicutes rather than Deltaproteobacteria due to the lack of hybridization to Delta495a. Despite evidence for Chloroflexi and Clostridiaceae in the pyrosequencing data, the probes targeting these groups that were utilized in this study, Chis150 and CFX1223, do not hybridize to cells in any of the samples. Although the probes are intended to hybridize to a broad distribution of Chloroflexi or Clostridiaceae, lack of a positive hybridization result may be due to the absence of active cells or to mismatches in the 16S rRNA sequence at the probe site of representative species in these samples. Because we are working with short sequences obtained from V5-to-V6 pyrosequencing, we were not able to confirm that the selected probes targeted sequences in our data set before whole-cell rRNA-targeted FISH experiments were conducted. Additionally, ∼15% of the CIB-1 and ∼20% of the CIB-3 bacterial communities do not hybridize to any of the probes used in this study.
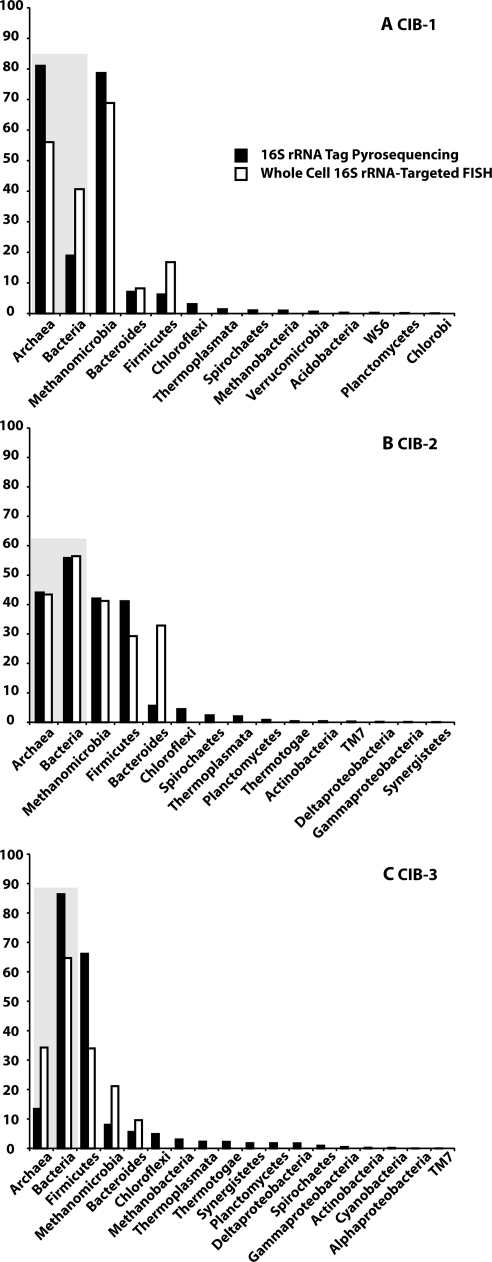
After quality control filtering of 16S rRNA tag pyrosequencing data, the number of sequences per sample was 24,039 for CIB-1, 23,780 for CIB-2, and 25,858 for CIB-3. Whole-cell rRNA-targeted FISH and 16S rRNA tag pyrosequencing, compared in Fig. 3 and Table S3 in the supplemental material, show similar results in the phylum level as well as the domain level community composition. 16S rRNA tag pyrosequencing shows CIB-1 to be dominated by archaeal sequences (81.0%), while sequences from CIB-2 were more evenly distributed between members of Archaea (44.1%) and members of Bacteria (55.9%) and the CIB-3 community is dominated by bacterial sequences (86.4%). Differences in Fig. 3, particularly in the bacterial communities of CIB-2 and CIB-3, may be due to several phyla and additional genera within the Firmicutes that represent less than 2% of 16S rRNA tag pyrosequences and were not detected by whole-cell rRNA-targeted FISH. Additionally, differences in the two techniques may represent PCR primer bias during 16S rRNA gene tag pyrosequencing and biases in 16S rRNA gene copy numbers of groups, such as the Firmicutes, which were detected in lower abundances by whole-cell 16S rRNA-targeted FISH. At the phylum level, the archaeal communities of all three samples are primarily composed of Methanomicrobia, which includes the genera Methanosarcina and Methanolobus. As a percentage of total pyrosequences, Methanomicrobia represent 78.7% of CIB-1, 42.1% of CIB-2, and 8.2% of CIB-3. The most-abundant bacterial phyla are Bacteroides and Firmicutes (7.1% and 6.3%, respectively, in CIB-1; 5.6% and 41.1%, respectively, in CIB-2; and 5.7% and 66.8%, respectively, in CIB-3).
Fig 3.
Comparison of the microbial communities of the Cook Inlet gas field CIB-1 (A), CIB-2 (B), and CIB-3 (C), as determined by 16S rRNA tag pyrosequencing and whole-cell rRNA-targeted FISH. The gray-shaded areas provide a comparison of the percentages of Archaea and the percentages of Bacteria that were determined by each technique.
Despite intense disruption during sampling and sample preparation, a significant fraction of the active microbial cells in epifluorescence photomicrographs of CIB-1 are imaged in close spatial and structural associations that have distinctive archaeal and bacterial phylotypes (Fig. 1A to C). Short filamentous and rod-shaped bacterial cells intertwine with Methanosarcina and Methanolobus cocci, forming large, intact clusters of cells. Rod-shaped cells, identified using whole-cell rRNA-targeted FISH (Fig. 1A to C), are a mixture of Bacteroidales and Acetobacterium, while the filamentous cells are a mixture of SRB385-hybridizing cells and another unidentified lineage. Similar associations of archaeal and bacterial cells are not observed in CIB-2 and CIB-3 (Fig. 1D to F).
Geochemical analysis.
Isotopic data are plotted in the manner of Whiticar (35). The carbon and hydrogen isotope values of methane from these three samples are characteristic of biogenic methanogenesis from acetate fermentation and methyl/methanol utilization (see Fig. S1 in the supplemental material). Water chemistry data (see Table S2 in the supplemental material) showed significant heterogeneity between the three samples, particularly in terms of chloride, iron, and acetate.
Principal component analysis of microbial diversity.
Multivariate statistics were employed using principal component analysis (PCA) to identify trends in the variability of the microbial diversity of the Cook Inlet basin samples and to identify covariance between geochemistry and microbial diversity. A PCA of Cook Inlet basin microbial diversity estimates is shown in Fig. 4A. Principal component 1 (PC1), which explains 71.1% of the variation in the data set, is strongly correlated with the ratio of members of Archaea to members of Bacteria and with the population sizes of Acetobacterium, Methanolobus, and Methanosarcina. PC2 (28.9%) is strongly correlated with Bacteroidales population sizes. Geochemical data (blue vectors) are projected onto the PCA using the envfit function. P values of <0.001 show significant correlations between the ordination and pH and the acetate, propionate, and total dissolved solids (TDS) concentrations, but these parameters are only weakly related to PC1. Although there are positive correlations between calcium and sodium concentrations and bacterial population size and between iron concentrations and archaeal population size, the P values relating these geochemical variables to the PCA are >0.3 and thus are not significant.
A PCA performed using population data from the Cook Inlet, Gippsland, Ishikari (Yubari), Powder River, and Ruhr basins is shown in Fig. 4B. The addition of other basins results in the Cook Inlet samples clustering together. Of the additional sites, the Ishikari (Yubari) and Ruhr basin sites are the nearest in ordination space. This grouping is related to the presence of the Methanolobus and Acetobacterium populations in these sites and their absence in the others. PC1, which explains 46.1% of the observed variance, is most strongly correlated with the population sizes of Methanolobus and Methanobacteria. PC2 (34.3%) is most strongly correlated with the population sizes of Methanosarcina and Clostridia. Geochemical data (blue vectors) are projected onto the PCA using the envfit function. P values for the environmental variables (>0.3) show no significant correlations between the available geochemical data and the ordination of microbial diversity estimates.
DISCUSSION
Isotopic data show that the dominant methanogenic pathway at the Cook Inlet basin study site involves the fermentation of acetate or other methyl group substrates, such as methanol, methyl sulfides, and methyl amines. Carbon and hydrogen isotope values place the methane from the three analyzed wells into the range associated with methyl-type fermentation (see Fig. S1 in the supplemental material). Additionally, cells hybridizing to Methanosarcina and Methanolobus genera-specific probes dominate the observed members of Archaea (Fig. 3). While Methanosarcina species are capable of methanogenesis by acetoclastic, methylotrophic, and CO2 reduction pathways, Methanolobus species are obligate methylotrophs (16). A major component of the bacterial community is the homoacetogen genus Acetobacterium. Homoacetogens produce acetate from the reduction of carbon dioxide with hydrogen. Competition between Acetobacterium spp. and CO2-reducing methanogens (Methanobacteria and Methanomicrobiales) for CO2 and H2 as the substrates may explain the predominance of an acetoclastic or methylotrophic pathway over a carbonate reduction pathway (18).
The close structural association of Methanolobus and Methanosarcina with members of Bacteria in CIB-1 (Fig. 1A to C) is suggestive of syntrophic interactions. The association of methanogens with acetogens (17, 33) or with sulfate-reducing Bacteria (25) has previously been reported in methanogenic communities. A study by Shimizu et al. revealed clones of Methanolobus, Acetobacterium, and Syntrophus spp. in a deep coal seam (30). In CIB-1, a bacterial community composed of Acetobacterium spp., Bacteroidales, and SRB385-hybridizing Firmicutes forms dense aggregates with an archaeal community composed of Methanolobus and Methanosarcina spp. The microbial consortia within these aggregates include constituents capable of macromolecule fermentation (Bacteroidales, Firmicutes), degradation of fermentation intermediates (Bacteroidales, Firmicutes), acetate generation (Acetobacterium spp.), and methanogenesis (Methanosarcina and Methanolobus spp.).
Several other studies of coal bed methane biogeochemistry indicate the presence of acetate-fermenting or methylotrophic methanogen communities on the basis of 16S rRNA gene analysis as well as through carbon and hydrogen isotope analysis of methane (11, 13, 19, 28, 31). Bacterial communities in these studies show high diversity, with considerable contributions from Firmicutes and Bacteroidetes (13, 19, 28, 30, 31). Species of Methanosarcina are frequently identified in both studies showing isotopic evidence of CO2 reduction and those identifying acetate fermentation/methyl group utilization as the dominant methanogenic pathway. While methylotrophic methanogens have been identified in biogenic-methane-producing systems (9, 13, 24, 30), this is the first report of a CBM archaeal community that is dominated by an obligately methylotrophic methanogen genus.
Geochemical factors, including coal maturity, the availability of methyl group substrates, partial pressures of H2 and CO2, and competition for substrates with the bacterial community, may control the archaeal diversity and ultimate methanogenic pathway (8, 18). Multivariate analysis used to visualize trends in complex data of the microbial communities at the three Cook Inlet basin sites and the multiple-basin data set failed to identify significant correlations between microbial community structure and the measured geochemical data. While not statistically significant, sodium, calcium, and iron concentrations correlate with the ratio of members of Bacteria to members of Archaea in the Cook Inlet basin samples. Although not within the original scope of this study, a more complete set of geochemical data, including variables such as redox potential, nutrient and trace metal concentrations, and additional time points, is likely to be a sound investment in future studies in which the goal is to understand geochemical controls on microbial community structure and methane production potential.
Through the combination of the 16S rRNA tag pyrosequencing data and the quantitative whole-cell rRNA-targeted FISH data, we can begin to develop a simplified mechanism for the breakdown of organic matter in coal at the study site. Species of Bacteroidales are capable of the anaerobic degradation of cellulose, polysaccharides, and other complex substrates to simple sugars and other fermentation products (37). Acetobacterium spp. produce acetate from the fermentation of simple sugars as well as through homoacetogenesis with H2 and CO2 (3). Acetoclastic and methylotrophic methanogenesis by Methanosarcina and Methanolobus finally leads to the production of methane. This coal biodegradation scheme retains the general mechanism of macromolecular breakdown followed by fermentation to simple sugars and methanogenesis precursors that was proposed by Strapoc et al. (32) for the Illinois Basin. However, the microbial communities and methanogenic pathway differ in terms of the presence of acetogenic Bacteria and acetoclastic and methylotrophic Archaea.
Although the community compositions and the likely pathways for organic matter degradation for all three Cook Inlet basin sites are similar, we noted significant variation in the proportions of microbial groups present in spatially separated sites within this single production field (Fig. 2 to 4). Microenvironments favoring particular microbes may result from heterogeneity in the coal structure, hydrology, geochemistry, and trace element concentrations. For example, variation in coal structure likely influences the bacterial community. A higher degree of aromatization and condensation may provide a competitive advantage to bacterial hydrocarbon degraders capable of fragmenting and fermenting polycyclic aromatic hydrocarbons over species capable of fermenting cycloalkyl, carboxyl, and methoxy groups. Additionally, more favorable conditions for methanogenic Archaea may be provided by higher concentrations of trace elements such as nickel, which is required for coenzyme F430, a component of the key enzyme involved in methanogenesis. Future efforts to stimulate in situ biogenic CBM production will need to consider differences in community composition as well as geochemistry among fields and at finer scales.
Conclusions.
Quantitative whole-cell rRNA-targeted FISH analyses of a biogenic gas field in the Cook Inlet basin, AK, revealed that the archaeal community is dominated by the obligately methylotrophic methanogen genus Methanolobus as well as the CO2-reducing, methylotrophic and acetoclastic methanogen genus Methanosarcina. The bacterial community is comprised of Acetobacterium, Bacteroidales, and Firmicutes. The microbial community composition, in combination with methane isotope data, indicates that acetate and methyl group fermentation are the dominant methanogenic pathways in the Cook Inlet basin. The substantial variations in the microbial community composition that we observed within this single gas production field suggest that further work will be necessary to understand and predict microbial community dynamics, including measurements of fine-scale heterogeneity in geochemistry, hydrology, and coal structure, in coal bed environments.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by ConocoPhillips, Inc. D.S., B.H., U.L., and M.A. declare their financial interest as a result of their affiliations with ConocoPhillips, Inc., and Taxon Biosciences, Inc., and as a result of being listed in a related patent application. The other authors declare no competing financial interest.
Footnotes
Published ahead of print 16 March 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Ashby MN, Rine J, Mongodin EF, Nelson KE, Dimster-Denk D. 2007. Serial analysis of rRNA genes and the unexpected dominance of rare members of microbial communities. Appl. Environ. Microbiol. 73:4532–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buschhorn H, Durre P, Gottschalk G. 1989. Production and utilization of ethanol by the homoacetogen Acetobacterium woodii. Appl. Environ. Microbiol. 55:1835–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Catcheside DEA, Ralph JP. 1999. Biological processing of coal. Appl. Microbiol. Biotechnol. 52:16–24 [DOI] [PubMed] [Google Scholar]
- 5. Claypool GE, Threlkeld CN, Magoon LB. 1980. Biogenic and thermogenic origins of natural gas in Cook Inlet Basin, Alaska. Am. Assoc. Pet. Geol. Bull. 64:1131–1139 [Google Scholar]
- 6. Cole JR, et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Committee on Management and Effects of Coalbed Methane Development and Produced Water in the Western United States, Committee on Earth Resources, National Research Council 2010. Management and Effects of Coalbed Methane Produced Water in the United States. National Academies Press, Washington, DC [Google Scholar]
- 8. Cord-Ruwisch R, Seitz H-J, Conrad R. 1988. The capacity of hydrogenotrophic anaerobic bacteria to compete for traces of hydrogen depends on the redox potential of the terminal electron acceptor. Arch. Microbiol. 149:350–357 [Google Scholar]
- 9. Doerfert SN, Reichlen M, Iyer P, Wang M, Ferry JG. 2009. Methanolobus zinderi sp. nov., a methylotrophic methanogen isolated from a deep subsurface coal seam. Int. J. Syst. Evol. Microbiol. 59:1064–1069 [DOI] [PubMed] [Google Scholar]
- 10. Fakoussa RM, Hofrichter M. 1999. Biotechnology and microbiology of coal degradation. Appl. Microbiol. Biotechnol. 52:25–40 [DOI] [PubMed] [Google Scholar]
- 11. Flores RM, Rice CA, Stricker GD, Warden A, Ellis MS. 2008. Methanogenic pathways of coal-bed gas in the Powder River Basin, United States: the geologic factor. Int. J. Coal Geol. 76:52–75 [Google Scholar]
- 12. Fry JC, et al. 2009. Prokaryotic populations and activities in an interbedded coal deposit, including a previously deeply buried section (1.6-2.3 km) above ∼150 Ma basement rock. Geomicrobiol. J. 26:163–178 [Google Scholar]
- 13. Green MS, Flanegan KC, Gilcrease PC. 2008. Characterization of a methanogenic consortium enriched from a coalbed methane well in the Powder River Basin, U. S. A. Int. J. Coal Geol. 76:34–45 [Google Scholar]
- 14. Harris SH, Smith RL, Barker CE. 2008. Microbial and chemical factors influencing methane production in laboratory incubations of low-rank subsurface coals. Int. J. Coal Geol. 76:46–51 [Google Scholar]
- 15. Hugenholtz P, Tyson GW, Blackall LL. 2002. Design and evaluation of 16S rRNA-targeted oligonucleotide probes for fluorescence in situ hybridization. Methods Mol. Biol. 179:29–42 [DOI] [PubMed] [Google Scholar]
- 16. Kendall MM, Boone DR. 2006. The order methanosarcinales, p 244–256 In Dworkin M, et al. (ed), The prokaryotes. Springer, New York, NY [Google Scholar]
- 17. Kotelnikova S, Pedersen K. 1998. Distribution and activity of methanogens and homoacetogens in deep granitic aquifers at Äspö Hard Rock Laboratory, Sweden. FEMS Microbiol. Ecol. 26:121–134 [Google Scholar]
- 18. Kotsyurbenko OR, Glagolev MV, Nozhevnikova AN, Conrad R. 2001. Competition between homoacetogenic bacteria and methanogenic archaea for hydrogen at low temperatures. FEMS Microbiol. Ecol. 38:153–159 [Google Scholar]
- 19. Kruger M, et al. 2008. Microbial methane formation from hard coal and timber in an abandoned coal mine. Geomicrobiol. J. 25:315–321 [Google Scholar]
- 20. Li D, Hendry P, Faiz M. 2008. A survey of the microbial populations in some Australian coalbed methane reservoirs. Int. J. Coal Geol. 76:14–24 [Google Scholar]
- 21. Macalady JL, et al. 2006. Dominant microbial populations in limestone-corroding stream biofilms, Frasassi cave system, Italy. Appl. Environ. Microbiol. 72:5596–5609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Margulies M, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Midgley DJ, et al. 2010. Characterisation of a microbial community associated with a deep, coal seam methane reservoir in the Gippsland Basin, Australia. Int. J. Coal Geol. 82:232–239 [Google Scholar]
- 24. Mochimaru H, et al. 2009. Methanolobus profundi sp. nov., a methylotrophic methanogen isolated from deep subsurface sediments in a natural gas field. Int. J. Syst. Evol. Microbiol. 59:714–718 [DOI] [PubMed] [Google Scholar]
- 25. Moser DP, et al. 2005. Desulfotomaculum and Methanobacterium spp. dominate a 4- to 5-kilometer-deep fault. Appl. Environ. Microbiol. 71:8773–8783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nederlof MH. 1988. The scope for natural gas supplies from unconventional sources. Annu. Rev. Energy 13:95–117 [Google Scholar]
- 27. Orem WH, et al. 2010. Organic intermediates in the anaerobic biodegradation of coal to methane under laboratory conditions. Org. Geochem. 41:997–1000 [Google Scholar]
- 28. Penner TJ, Foght JM, Budwill K. 2010. Microbial diversity of western Canadian subsurface coal beds and methanogenic coal enrichment cultures. Int. J. Coal Geol. 82:81–93 [Google Scholar]
- 29. R Development Core Team 2007. R: a language and environment for statistical computing, 2.5.1. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 30. Shimizu S, et al. 2007. Molecular characterization of microbial communities in deep coal seam groundwater of northern Japan. Geobiology 5:423–433 [Google Scholar]
- 31. Strapoc D, et al. 2011. Biogeochemistry of coal-bed methane. Annu. Rev. Earth Planet. Sci. 39:617–656 [Google Scholar]
- 32. Strapoc D, et al. 2008. Methane-producing microbial community in a coal bed of the Illinois Basin. Appl. Environ. Microbiol. 74:2424–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Struchtemeyer CG, Elshahed MS, Duncan KE, McInerney MJ. 2005. Evidence for aceticlastic methanogenesis in the presence of sulfate in a gas condensate-contaminated aquifer. Appl. Environ. Microbiol. 71:5348–5353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tyson GW, et al. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37–43 [DOI] [PubMed] [Google Scholar]
- 35. Whiticar MJ. 1986. Biogenic methane formation in marine and freshwater environments: CO2 reduction vs. acetate fermentation—isotopic evidence. Geochim. Cosmochim. Acta 50:693–709 [Google Scholar]
- 36. Whiticar MJ. 1999. Carbon and hydrogen isotope systematics of bacterial formation and oxidation of methane. Chem. Geol. 161:291–314 [Google Scholar]
- 37. Xu J, et al. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074–2076 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.