Abstract
Recent studies have revealed extensive genetic variation among isolates of Cryptosporidium parvum, an Apicomplexan parasite that causes gastroenteritis in both humans and animals worldwide. The parasite's population structure is influenced by the intensity of transmission, the host-parasite interaction, and husbandry practices. As a result, C. parvum populations can be panmictic, clonal, or even epidemic on both a local scale and a larger geographical scale. To extend the study of C. parvum populations to an unexplored region, 173 isolates of C. parvum collected in Italy from humans and livestock (calf, sheep, and goat) over a 10-year period were genotyped using a multilocus scheme based on 7 mini- and microsatellite loci. In agreement with other studies, extensive polymorphism was observed, with 102 distinct multilocus genotypes (MLGs) identified among 173 isolates. The presence of linkage disequilibrium, the confinement of MLGs to individual farms, and the relationship of many MLGs inferred using network analysis (eBURST) suggest a predominantly clonal population structure, but there is also evidence that part of the diversity can be explained by genetic exchange. MLGs from goats were found to differ from bovine and sheep MLGs, supporting the existence of C. parvum subpopulations. Finally, MLGs from isolates collected between 1997 and 1999 were also identified as a distinct subgroup in principal-component analysis and eBURST analysis, suggesting a continuous introduction of novel genotypes in the parasite population.
INTRODUCTION
Cryptosporidium parvum and Cryptosporidium hominis are two related species of Apicomplexan protozoa that cause cryptosporidiosis, an enteric infection of humans and animals (30). C. parvum is considered a zoonotic pathogen, as it is often transmitted to humans by environmentally resistant oocysts excreted by ruminants. In contrast, the host range of C. hominis is thought to be restricted to humans. With the exception of a brief diploid phase, C. parvum and C. hominis are haploid. The parasites develop in intestinal epithelial cells of the host, where they undergo consecutive rounds of asexual multiplication. Thereafter, the differentiation and fusion of gametes lead to a transient diploid stage, followed by meiotic division. Meiotic recombination between genetically distinct C. parvum genotypes has been documented in experimental infections (25), but the extent of outcrossing in natural parasite populations appears to vary (12, 13, 26). Mini- and microsatellite markers have provided useful information for studying the population structures of many organisms, including parasites. Studies of Apicomplexan parasites have shown that populations vary from panmictic (random mating among individuals in a population) to clonal (absence of significant gene flow), depending on either transmission intensity (e.g., for Plasmodium falciparum) (2) or host ecology (e.g., for Toxoplasma gondii) (1). The population structure of Cryptosporidium has not been extensively studied, and population genetic studies of this pathogen have been undertaken only in the last decade and in only a limited number of geographic regions (12, 13, 16, 24, 26). Initially it was suggested that C. parvum had a clonal population (3). However, when this model was proposed, the existence of C. hominis was not recognized. Genotypes identified by PCR-restriction fragment length polymorphism (PCR-RFLP) were observed to segregate among two groups, which led to the model of clonality. With the description of C. hominis (15) and the development of microsatellite markers (4, 8, 23, 28), the clonal population model has been questioned. Today a more complex picture is recognized, and the existence of clonal, epidemic, and panmictic populations in various geographic locations has been described (12, 13, 16, 26).
Analyses of Cryptosporidium populations in Europe are, to our knowledge, limited to the British Isles and Spain. Given the availability of DNA samples from a relatively large number of Cryptosporidium isolates from a different location, we undertook an analysis of the genetic structure of C. parvum in Italy using a multilocus typing scheme based on seven polymorphic loci, as used by Mallon and coworkers (12, 13). We assessed the likely impact of genetic exchange in generating genotypic diversity and investigated how the host and the geographical origin and time of collection of the isolates contributed to the parasite population structure.
MATERIALS AND METHODS
Parasite isolates.
Fecal samples from 178 clinical cases (122 from calves, 21 from sheep, 21 from goats, and 14 from humans) were collected between 1997 and 2010 in northern, central, and southern Italy. Details about these isolates are available from the corresponding author upon request. The presence of Cryptosporidium oocysts in these stool specimens was determined using immunofluorescence (Merifluor; Meridian Bioscience, Cincinnati, OH).
Molecular characterization.
Genomic DNA was extracted from positive stools using a commercial kit (QIAamp DNA Stool; Qiagen, Milan, Italy). The DNA was subjected to PCR amplification using the primers for the Cryptosporidium oocyst wall protein (COWP) gene (20). The amplicon was digested with RsaI endonuclease, and on the basis of the RFLP pattern, the source of the DNA identified as C. hominis (formerly type 1) or C. parvum (formerly type 2).
The seven polymorphic loci used in this study were MS1 (11), GP60 (21), and MS9, TP14, MM5, MM18, and MM19 (12, 13, 16). The MS1 marker contains a GGTGGTATGCCA repeat in the heat shock protein 70 gene (cgd2_20) located at positions 3136 to 5184 on chromosome 2. The GP60 marker contains a TCA repeated motif in a 975-bp gene (cgd6_1080) encoding a sporozoite surface protein located at positions 266434 to 267408 on chromosome 6. The MS9 marker contains a TGGACT repeat in a 2,016-bp gene (cgd5_2850) encoding a hypothetical protein located at positions 640137 to 642152 on chromosome 5. The TP14 marker contains a CAA repeat in a 8,421-bp gene (cgd8_1340) encoding a hypothetical protein located at positions 365790 to 374210 on chromosome 8. The MM5 marker contains a TCCTCCTCT repeat located in a 11,418-bp gene (cgd6_4290) located at positions 1002285 to 1013702 on chromosome 6. The MM18 marker contains a GGACCA repeat in the 5,004-bp gene (cgd8_660) located at positions 165295 to 170298 on chromosome 8. The MM19 marker contains a GGAGCT repeat in the 7,230-bp gene (cgd8_4840) located at position 1208520 to 1215749 on chromosome 8.
The sequences of the primers used for amplification were as follows: MS1-1F, 5′-GGAACACCATCCAAGAACCAAAGGT-3′; MS1-1R,5′-TTAGTCGACCTCTTCAACAGTTGG-3′; MS9-9F,5′-GTCTGAGACAGAATCTAGGATCTAC-3′; MS9-9R,5′-GGACTAGAAATAGAGCTTTGGCTGG-3′; TP14-14F,5′-CTAACGTTCACAGCCAACAGTACC-3′; TP14-14R,5′-CAATAAGACCATTATTACCTCC-3′; MM5-5F,5′-CCTGGACTTGGATTTGGACTTACACC-3′; MM5-5R,5′-GGAGAAGATAAGCTAGCCGAATCT-3′; MM18-18F,5′-CTTTCTGGAGGGTTTGTTCCTCC-3′; MM18-18R,5′-CTTCCTGATGATCCAGGCCAAGC-3′; MM19-19F,5′-GATTCTGCAACTTTGAATTCAG-3′; MM19-19R,5′-CCAACCCCGAATTCATTTCCAAC-3′; GP15-15F,5′-GCCGTTCCACTCAGAGGAAC-3′; and GP15-15R,5′-CATTACAAATGAAGTGCCGCA-3′. Reactions were undertaken in a Veriti 96-well thermal cycler (Life Technologies, Carlsbad, CA). Cycling conditions were 94°C for 30 s, 55°C for 30 s, and 72°C for 60 s, for 40 cycles.
Allele and MLG identification.
The size of each PCR product was estimated by electrophoresis on a capillary apparatus (QiaXcel; Qiagen, Milan, Italy) by comparison to size standards, and representative samples of each allele were sequenced to confirm the estimated size. If multiple amplification products were identified in individual samples, only the principal peak was used to define the allele. Each distinct allele was assigned a number, and multilocus genotypes (MLGs) were defined for each isolate by the combination of alleles at the seven loci.
Phylogenetic and population analyses.
Genetic distances between MLGs were calculated with GenAlex version 6.4 (17) using the simple sequence repeat (SSR) distance. This distance metric is based on the difference in length between pairs of alleles. The distance between pairs of MLGs is determined by adding the square of the length difference over all loci. The distance metric was exported to Mega (22) and phylogenetic trees constructed using the neighbor-joining (NJ) method (19).
For principal-component analysis (PCA), the SSR distance was calculated as described above, as was a simple distance metric, the Hamming distance (9). This distance is equal to the number of loci with alleles of different size. A distance matrix based on Hamming distance values between all pair of MLGs was calculated using GenAlex. This matrix and the SSR distance matrix were then analyzed using PCA.
Three variables, i.e., geographic location, year of collection, and host species, were tested for correlation with the clustering of MLGs into related groups (identified by PCA or NJ clustering) by logistic regression using subgroup membership as a dependent variable. This analysis was performed with SPSS Statistics, version 17.0 (IBM, Chicago, IL). To convert the geographic location of each isolate into a continuous variable, the location of each collection site was encoded as the longitude and latitude of the capital of the province where the isolate was collected. This location was plotted on a longitude-versus-latitude scatter plot, and a linear regression was fitted to the locations (r2 = 0.71). For this analysis the 19 isolates from Sardinia were excluded, leaving a total of 159 isolates. The linear regression followed an approximate northwest-to-southeast direction. Each location was then projected perpendicularly on the regression line and the latitude of the projection used in the logistic regression as a continuous variable. Host species were converted into dummy numerical variables and the year of collection entered as a numerical variable.
The program LIAN 3.5 (10) (http://adenine.biz.fh-weihenstephan.de/cgi-bin/lian/lian.cgi.pl) was used to calculate the standardized index of association (IAS). This index, a derivation of the Maynard-Smith index of association (14), measures the strength of the linkage disequilibrium (LD) and is independent of the number of loci analyzed. Linkage analysis between pairs of loci was performed using the web interface of Genepop 4.0 at http://genepop.curtin.edu.au/. This software tests the association of alleles at either of two loci against the null hypothesis that genotypes at one locus are independent from genotypes at the second locus. Nei's genetic distances were calculated using GenAlex. FST values were also calculated using Genepop 4.0. The eBURST software (http://eburst.mlst.net/default.asp) was used to examine the structure of the C. parvum population. Using this method, the clonal nature of related genotypes and putative “founder” genotypes can be identified (7). The most stringent setting was used, and only single-locus variants (SLVs) with 6 out of 7 identical loci were assigned to the same cluster.
RESULTS
Cryptosporidium species identification.
All isolates were initially typed by PCR-RFLP analysis of the COWP gene. The RFLP patterns (data not shown) showed that all 164 animal isolates were C. parvum (122 bovine, 21 ovine, and 21 caprine), whereas the 14 human isolates comprised 9 C. parvum and 5 C. hominis isolates.
Determination of alleles and multilocus genotypes.
A total of 178 isolates were successfully genotyped at all seven micro- and minisatellite loci. For all loci, products representative of each different allele size were sequenced to confirm the size. The number of alleles at each locus ranged from 5 for MS1 to 18 for MM19 (Table 1). The five C. hominis isolates shared the same alleles at five of the seven loci, whereas two alleles were found at the other two loci; these alleles were not observed in C. parvum (Table 1). Because Cryptosporidium parasites are haploid, the presence of more than one amplicon was interpreted as a mixed infection. This interpretation seemed more plausible than the alternative view of nonspecific amplification products, because (i) a BLAST search with each pair of primers always identified a single locus in the C. parvum genome, and (ii) each putative allelic band detected in mixed profiles was also identified as a single allele in other isolates. Based on the genotyping of a total of 173 isolates at 7 loci, 20 mixed profiles were identified, accounting for 1.6% (20 of 1,211) of the investigated loci and for 11.6% of the isolates tested.
Table 1.
Alleles found at each of the seven loci in 164 C. parvum animal isolates and in 14 human isolates (9 C. parvum and 5 C. hominis)
| Locus and allele size, bp (allele no.) | No. (%) of isolates (n = 164) | No. of farms (n = 96) | No. of human isolates (n = 14) |
|
|---|---|---|---|---|
| C. parvum | C. hominis | |||
| MS1 | ||||
| 326 (1) | 2 (1.2) | 2 | ||
| 350 (2) | 51 (31) | 34 | 4 | |
| 362 (3) | 108 (66) | 60 | 5 | |
| 386 (4) | 3 (1.8) | 1 | ||
| 374 (5) | 5 | |||
| MS9 | ||||
| 396 (1) | 1 (0.6) | 1 | ||
| 420 (2) | 3 (1.8) | 1 | ||
| 432 (3) | 15 (9.1) | 9 | ||
| 438 (4) | 22 (13.4) | 11 | 2 | |
| 444 (5) | 13 (7.9) | 6 | ||
| 450 (6) | 94 (57.3) | 68 | 4 | |
| 456 (7) | 12 (7.3) | 11 | 1 | |
| 462 (8) | 4 (2.4) | 3 | 2 | |
| 373 (9) | 5 | |||
| TP14 | ||||
| 297 (1) | 18 (11) | 17 | 1 | |
| 300 (2) | 47 (28.6) | 39 | 3 | |
| 309 (3) | 99 (60.3) | 64 | 4 | |
| 318 (4) | 1 | |||
| 255 (5) | 4 | |||
| 261 (6) | 1 | |||
| MM5 | ||||
| 218 (1) | 1 (0.6) | 1 | 1 | |
| 224 (2) | 7 (4.2) | 7 | ||
| 233 (3) | 64 (39) | 39 | 2 | |
| 242 (4) | 1 (0.6) | 1 | ||
| 260 (5) | 90 (54.9) | 63 | 6 | |
| 275 (6) | 1 (0.6) | 1 | ||
| MM18 | ||||
| 236 (1) | 2 (1.2) | 1 | ||
| 242 (2) | 24 (14.6) | 11 | 2 | |
| 248 (3) | 31 (18.9) | 24 | 2 | |
| 260 (4) | 1 (0.6) | 1 | ||
| 290 (5) | 106 (64.6) | 72 | 5 | |
| 206 (6) | 5 | |||
| MM19 | ||||
| 263 (1) | 2 (1.2) | 1 | ||
| 281 (2) | 11 (6.7) | 6 | 1 | |
| 287 (3) | 62 (37.8) | 47 | 1 | |
| 293 (4) | 10 (6.1) | 8 | 3 | |
| 299 (5) | 1 (0.6) | 1 | 1 | |
| 305 (6) | 1 (0.6) | 1 | ||
| 317 (7) | 11 (6.7) | 7 | ||
| 323 (8) | 19 (11.6) | 13 | 1 | |
| 329 (9) | 1 (0.6) | 1 | ||
| 335 (10) | 22 (13.4) | 15 | 1 | |
| 341 (11) | 2 (1.2) | 2 | ||
| 353 (12) | 7 (4.2) | 3 | 1 | |
| 365 (13) | 6 (3.6) | 4 | ||
| 377 (13) | 2 (1.2) | 1 | ||
| 389 (14) | 3 (1.8) | 3 | ||
| 395 (15) | 2 (1.2) | 1 | ||
| >400 (16) | 2 (1.2) | 2 | ||
| 233 (17) | 4 | |||
| 245 (18) | 1 | |||
| GP15 | ||||
| 306 (1) | 2 (1.2) | 2 | ||
| 315 (2) | 1 | |||
| 318 (3) | 5 (3) | 1 | ||
| 321 (4) | 12 (7.3) | 9 | ||
| 324 (5) | 11 (6.7) | 7 | ||
| 330 (6) | 97 (59.1) | 60 | 6 | |
| 333 (7) | 1 (0.6) | 1 | ||
| 336 (8) | 18 (11) | 17 | 1 | |
| 339 (9) | 13 (7.9) | 11 | 1 | |
| 342 (10) | 2 (1.2) | 2 | ||
| 345 (11) | 1 (0.6) | 1 | ||
| 354 (12) | 2 (1.2) | 1 | ||
| 366 (13) | 5 | |||
A total of 102 distinct MLGs were identified among the 173 C. parvum isolates, and 2 MLGs were observed among the 5 C. hominis isolates (these data are available from the corresponding author upon request). The most frequent C. parvum MLG (MLG 67) was found in 9 isolates (8 from calves and 1 from a lamb), followed by MLG 76, which was found in 8 isolates from calves. Of the 9 MLGs from humans, only 2 (MLG 29 and MLG 64) were found in a lamb and in 2 calves, respectively. It is noteworthy that in a total of 42 isolates from sheep and goats, 34 distinct MLGs were found, and not a single MLG was shared between these two hosts. Moreover, only 4 MLGs (3 from sheep and 1 from a goat) were also found in calves. The frequency of each MLG and the distribution in the four hosts are given in Fig. S1 in the supplemental material.
Overall population analysis.
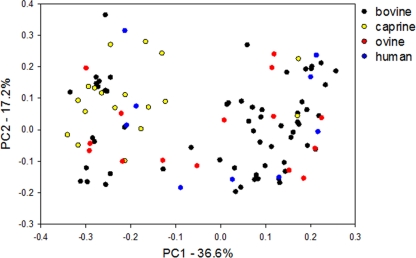
The 7-locus MLGs from 173 C. parvum isolates were analyzed by PCA (based on SSR and Hamming distances) and were found to separate into two subgroups, which did not show direct association with any particular geographic region, a single host species, or year of collection (Fig. 1). Clustering of the MLGs using the NJ algorithm also revealed two main branches of mixed host origin (see Fig. S2 in the supplemental material) which are identical to the subgroups defined by PCA. Thus, the clustering did not show a clear-cut association with host species, year, or geographic origin. However, closer inspection of the phylogenetic tree and PCA showed that a larger number of caprine isolates were within one of the subgroups.
Fig 1.
Principal-component analysis of 102 unique C. parvum MLGs from Italy based on Hamming distance. MLGs are colored according to the host species. The percent variations accounted for by the first and second axes are indicated.
To further investigate the origin of the two subgroups and the possibility that caprine isolates do not cluster randomly, a logistic regression was performed to predict which variables were significantly associated with subgroups. Isolates from Sardinia were excluded because of their unique geographic location outside the main northwest-to-southeast axis. As summarized in Table 2, the odds of an isolate being a member of a subgroup was significantly associated with the year of collection and with the host species but not with the geographical location. The comparison of isolates from each livestock species against those from humans identified goats as a significant predictor of membership of a subgroup.
Table 2.
Logistic regression analysis
| Variable | B | SEa | Wald | df | Significance | eB |
|---|---|---|---|---|---|---|
| Yr | 0.229 | 0.049 | 21.457 | 1 | 0.000 | 1.257 |
| Latitude | 0.096 | 0.160 | 0.358 | 1 | 0.550 | 1.100 |
| Host | 17.914 | 3 | 0.000 | |||
| Host (bovine) | −0.322 | 0.800 | 0.162 | 1 | 0.688 | 0.725 |
| Host (caprine) | −3.060 | 1.021 | 8.989 | 1 | 0.003 | 0.047 |
| Host (ovine) | −1.157 | 0.938 | 1.520 | 1 | 0.218 | 0.314 |
| Constant | −462.083 | 98.314 | 22.091 | 1 | 0.000 | 0.000 |
Standard error of coefficient B.
In order to determine the possible role of genetic exchange, the level of LD was estimated using the standardized index of association IAS, which is a measure of the association between alleles at all pairwise combinations of loci. This index will have a value of zero or be negative in randomly mating populations but have a positive value for nonpanmictic populations. The analysis was carried out at three levels. First, all samples were included (Table 3, all MLG), and second, samples with identical MLGs were represented by a single data point to test whether the population structure is epidemic (Table 3, single MLG). Finally, samples from the same farm with identical MLGs were treated as a single MLG (Table 3, no MLG farm) to test the effect of geographic bias by preventing overrepresentation of MLGs from particular farms. The value of IAS was positive at all three levels, indicating LD (Table 3). Positive values of IAS were also obtained when the C. parvum isolates were separated according to the geographical origin or the host (Table 3). In addition, we tested for LD between pairs of loci using Genepop software. A total of 21 pairwise tests of association between loci were performed, and the corresponding P values were determined for each pair of loci. A total of 15 out of 21 (71%) tests showed a significant (P < 0.05) association (data not shown), further confirming LD in the whole population. From this analysis, one interpretation is that the population (or subpopulations based on host or geography) is not panmictic or epidemic, suggesting that genetic exchange is not occurring at a sufficient frequency to break up allele association between loci. However, two findings suggest that the population is not reproducing exclusively clonally. Firstly the values of the index of association are low (below 0.1) when the populations are subdivided by host or when the whole population is tested for epidemic structure (Table 3). Second, the level of diversity is high, with ∼60% of the genotypes being unique. A working hypothesis to explain these results is that there is both clonal expansion of a set of isolates and some level of genetic exchange.
Table 3.
Analysis of linkage disequilibrium in the Italian C. parvum population
| Population | Analysis | n | IAS | P value | VD >L | LD |
|---|---|---|---|---|---|---|
| Italy | Single MLGa | 102 | 0.0991 | <0.001 | Yes | Yes |
| No MLG farmb | 151 | 0.1393 | <0.001 | Yes | Yes | |
| All MLG | 173 | 0.1385 | <0.001 | Yes | Yes | |
| Northern Italy | All MLG | 58 | 0.1020 | <0.001 | Yes | Yes |
| Single MLG | 33 | 0.0855 | <0.001 | Yes | Yes | |
| Central Italy | All MLG | 97 | 0.1704 | <0.001 | Yes | Yes |
| Single MLG | 63 | 0.1304 | <0.001 | Yes | Yes | |
| Only bovine | All MLG | 131 | 0.1187 | <0.001 | Yes | Yes |
| Single MLG | 73 | 0.0746 | <0.001 | Yes | Yes | |
| Only sheep/goat | All MLG | 42 | 0.1145 | <0.001 | Yes | Yes |
| Single MLG | 34 | 0.0784 | <0.001 | Yes | Yes |
Treating isolates having the same MLG as one individual.
Treating isolates from a single farm having the same MLG as one individual.
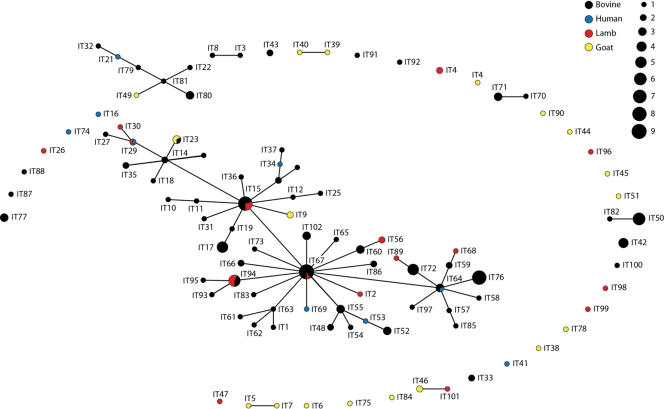
To investigate this hypothesis further, the relationship between MLGs was examined using the eBURST program to explore the occurrence and possible origin of single-locus variants (SLVs). Figure 2 reveals the presence of related clonal clusters with a star-like phylogeny characteristic of clonal expansion. This cluster comprises 55 MLGs mostly of bovine origin, which are closely related to MLG 67, the most abundant MLG in the population, which is present in bovine and ovine samples from three different Italian regions (Piedmont in northern Italy and Umbria and Marche in central Italy). This MLG appears to be the primary founder, with a bootstrap support of 99%. Two other MLGs, MLG 15 and MLG 64, cluster with MLG 67 and, based on the network topology, could be considered subfounders of two further clonal lineages with significant bootstrap values (96% and 88% for MLG 15 and MLG 64, respectively).
Fig 2.
Single-locus variant eBURST network of the Italian C. parvum population. Each MLG is represented by a dot and a unique identifier. The dot diameter is proportional to the number of isolates, as shown in the key. Single-locus variants are connected by lines. MLGs are colored according to host species.
When all isolates were visualized as a population snapshot (by setting the group definition to zero of seven shared alleles), a large number of isolates appeared to lack single-locus variants, a characteristic of populations where genetic exchange is occurring (Fig. 2). Notably, 16 of 18 MLGs from goats appear to have no SLVs in the main cluster, and do not form any clonal complex even when analyzed independently (data not shown). Furthermore, 23 of 27 MLGs from isolates collected between 1997 and 1999 have no SLVs in the main cluster, and only two of these MLGs can be connected with the main cluster even when the less stringent analysis of double-locus variants (DLVs) (5 shared loci of 7) is carried out (data not shown).
Distribution of MLGs in single farms.
The distribution of MLGs in individual farms was examined in two regions, Lombardia (northern Italy) and Marche (central Italy), and a total of 17 MLGs were identified in 37 isolates from 12 farms, corresponding to 1.42 MLGs/farm. The results indicate that a single and unique MLG is present in 7 of 12 farms and that, even if 4 MLGs are found in more than one farm, the MLGs present are distinct between the two regions (data not shown).
Genetic distances and FST values.
Nei's genetic distance (D) was used to quantify the differences between the populations from northern, central, and southern regions of Italy. The genetic distance between northern and central regions was low (D = 0.047), but the distance between northern and southern regions (D = 0.270) and that between central and southern regions (D = 0.278) were significantly higher. The FST values confirmed that the northern and central populations are relatively closely related (FST = 0.023), whereas the differentiation between southern and northern (FST = 0.127) and central (FST = 0.111) populations is again clear.
DISCUSSION
In this study, we genotyped a panel of C. parvum isolates collected in Italy from four host species and over a 10-year period of time using seven polymorphic markers to define MLGs, which were then analyzed with the aim of evaluating the genetic structure of the population.
The first conclusion we draw from this study is the high level of diversity that characterizes Italian isolates of C. parvum, with 102 MLGs identified in 173 isolates typed. Extensive diversity has been reported previously, such as that observed in 23 isolates from Serbia, which were all unique MLGs, and in isolates from Turkey, where 9 different MLGs were found in 11 isolates (26); lower levels appear to characterize isolates from Spain (59 MLGs in 137 isolates) (18) and Scotland (95 MLGs in 297 isolates) (16).
In addition to being genetically diverse, 11.6% of C. parvum isolates from Italy showed evidence of mixed infections (defined as isolate with >1 allele at one or more loci). The level of mixed infection in C. parvum populations varies considerably in published data (16, 18, 24), from no detection in New Zealand (19 human and 7 calf isolates) to 8.2% in Israel (59 calf isolates, 1 goat isolate, and 1 horse isolate), 19% in Uganda (36 human isolates), 20% in Spain (140 calf isolates), 30% in Serbia (23 calf isolates), 53% in Turkey (15 calf isolates), and 10 to 37% in four regions of Scotland (65 human, 211 calf, and 9 sheep isolates). These differences may in part reflect differential sensitivity of either the genotyping method or sets of markers for detecting minority populations or sampling effects (in terms of size and host representation). Differences can also be explained by a reduced frequency of recombination between isolates in countries where modern husbandry practices restrict herd-to-herd transmission. Under such circumstances, specific MLGs may be restricted to individual farms, as observed in Israel and Spain (18, 24) and in this study. However, animal movement can counteract this effect, as reported by Morrison et al. (16), who have clearly shown a strong correlation between parasite genetic diversity in defined areas and animal movement within the country. The balance between these factors may play a role in the dynamics of population structure evolution.
The second conclusion drawn from this study concerns the effect of host and of time of sampling on the overall structure of the parasite population. PCA (Fig. 1) shows the presence of two subgroups, one of which comprises mainly MLGs from goats and from isolates collected between 1997 and 1999. The eBURST diagram of the Italian C. parvum population (Fig. 2) indicates that half of the MLGs (55 of 102) form a network with star-like topologies typical of a clonal population. The remaining 47 MLGs have no single-locus variants within the main cluster of MLGs. Interestingly, 15 of 18 MLGs from goats are found outside the main clonal clusters, independently from their geographical origin or year of collection. Furthermore, MLGs from goats do not form a network even when analyzed independently and instead appear mostly as singletons, indicating a higher degree of genetic divergence. In contrast, only 7 of 16 ovine MLGs are found outside the main clonal cluster, indicating a closer affinity between ovine and bovine MLGs, a finding which is in agreement with the data reported by Mallon et al. (13) from Scotland.
The existence of specific subpopulations of C. parvum in different livestock species was suggested on the basis of unique MLGs observed in isolates from a horse and a goat from Israel (24). A recent study from Spain (18) showed that some alleles commonly found in lambs and goat kids are rarely found in calves from the same region, indicating a similar degree of host association. Therefore, the finding that different subpopulations of the parasite are associated with different livestock species appears to be valid for goats but not for sheep. Given the small number of sheep isolates genotyped so far (21 in this study and 11 in the study by Mallon et al. [13]), this hypothesis needs to be rigorously tested with additional isolates from different livestock species. The identification of C. parvum genotypes which do not appear to infect livestock, often termed “anthroponotic” (12, 13), should also be mentioned in this context. These observations suggest that C. parvum may comprise subpopulations with different host preferences or that husbandry practices for particular host species (e.g., goats) lead to effective isolation of particular C. parvum genotypes and limit opportunities for recombination with the main C. parvum population.
More strikingly, 23 of 27 MLGs from isolates collected between 1997 and 1999 share no SLVs with the main clonal complex (Fig. 2), and only 2 of these 27 MLGs (MLG 16 and MLG 81) are connected with the main complex as double-locus variants (DLVs). These findings suggest that novel genotypes are introduced in the parasite population over time.
A limitation of this study was the insufficient number of humans isolates available for genotyping, which limited the comparison of C. parvum MLGs from humans and animals and an evaluation of zoonotic transmission in Italy. To compare isolates from different hosts, we sequenced a fragment of the GP60 gene from 46 C. parvum isolates from 30 calves, 6 goats, 6 lambs, and 4 humans, and we detected 17 subtypes, 3 of the IId allelic family (in two goats and one human) and 14 of the IIa allelic family (data not shown). Together with previous studies from Italy (5, 6; S. M. Cacciò, unpublished data), these data indicate that GP60 subtypes IIa15G2R1 and IIa18G3R1 circulate in both humans and animals. Analysis of MLGs from a larger number of human C. parvum isolates is needed to assess the importance of zoonotic transmission in Italy.
One important challenge is to understand the relative contributions of recombination and mutation in the evolution of C. parvum populations. In this study, a number of observations are consistent with a predominant role of mutation and asexual reproduction. This model is compatible with the observed linkage disequilibrium, the relative scarcity of mixed genotypes, the confinement of MLGs to individual farms, and the presence of a main clonal complex identified by eBURST. On the other hand, the low IAS values and the fact that a large number of MLGs are not linked (as single-locus variants) to the main cluster can be interpreted as the result of sexual recombination or the presence of divergent populations. The eBURST analysis with computer-simulated populations evolving at a high recombination/mutation ratio shows complex relationships among genotypes and long chaining of single-locus variants (27), features which were observed in areas with a high prevalence of cryptosporidiosis (26) but not in Italy.
One issue that has become apparent during this study is that, at present, data from different studies cannot easily be directly compared, as different typing schemes have been used (29). To advance our understanding of the epidemiology of cryptosporidiosis, a universal multilocus typing method based on polymorphic and unlinked markers is needed. Ideally, an interlaboratory ring trial should be arranged to identify those markers. Results from the present study and previously published studies (see, e.g., reference 16) showed that some markers (e.g., GP60 and MM19) are particularly polymorphic in C. parvum, whereas others (e.g., ML1) have a low level of polymorphism (12, 13, 18) and could be omitted. There are also technical issues to consider, such as the discrimination of alleles having identical size but different nucleotide sequences and the ability of various genotyping methods to detect minority populations in mixed infections (18).
In summary, this study has confirmed that many factors contribute to determine the structure of C. parvum populations, and the results highlight the importance of the host (goat) and time of sampling, at least in Italy. In their extensive study of C. parvum and C. hominis isolates collected from many countries, Tanriverdi et al. (26) reported clear examples of clonal or panmictic populations and, more generally, provided strong evidence for an allopatric model of diversification of these parasites. Our study illustrates how within a single country some populations can undergo clonal expansion whereas others may engage in genetic exchange, resulting in a complex picture that needs to be further investigated by performing longitudinal studies with more extensive sampling.
Supplementary Material
ACKNOWLEDGMENTS
We thank Antonio Scala (Università degli Studi di Sassari, Italy) for supplying isolates from Sardinia.
This work was supported by the European Commission (contract SANCO/2006/FOODSAFETY/032). G.W. acknowledges financial support from the NIAID (grant AI052781). L.J.M. holds a Royal Society University Fellowship.
Footnotes
Published ahead of print 2 March 2012
Supplemental material for this article may be found at http://aem.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Ajzenberg D, et al. 2004. Genetic diversity, clonality and sexuality in Toxoplasma gondii. Int. J. Parasitol. 34:1185–1196 [DOI] [PubMed] [Google Scholar]
- 2. Anderson TJ, et al. 2000. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol. Biol. Evol. 17:1467–1482 [DOI] [PubMed] [Google Scholar]
- 3. Awad-El-Kariem FM. 1999. Does Cryptosporidium parvum have a clonal population structure? Parasitol. Today 15:502–504 [DOI] [PubMed] [Google Scholar]
- 4. Cacciò SM, et al. 2000. A microsatellite marker reveals population heterogeneity within human and animal genotypes of Cryptosporidium parvum. Parasitology 120:237–244 [DOI] [PubMed] [Google Scholar]
- 5. Del Chierico F, et al. 2011. Cases of cryptosporidiosis co-infections in AIDS patients: a correlation between clinical presentation and GP60 subgenotype lineages from aged formalin-fixed stool samples. Ann. Trop. Med. Parasitol. 105:339–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Duranti A, et al. 2009. Risk factors associated with Cryptosporidium parvum infection in cattle. Zoonoses Public Health 56:176–182 [DOI] [PubMed] [Google Scholar]
- 7. Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Feng X, et al. 2000. Extensive polymorphism in Cryptosporidium parvum identified by multilocus microsatellite analysis. Appl. Environ. Microbiol. 66:3344–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hamming RW. 1950. Error detecting and error correcting codes. Bell Syst. Tech. J. 29:147–160 [Google Scholar]
- 10. Haubold B, Hudson RR. 2000. LIAN version 3: a program for detecting linkage disequilibrium in multilocus data. Bioinformatics 16:847–848 [DOI] [PubMed] [Google Scholar]
- 11. Khramtsov NV, Tilley M, Blunt DS, Montelone BA, Upton SJ. 1995. Cloning and analysis of a Cryptosporidium parvum gene encoding a protein with homology to cytoplasmic form Hsp70. J. Eukaryot. Microbiol. 42:416–422 [DOI] [PubMed] [Google Scholar]
- 12. Mallon M, MacLeod A, Wastling J, Smith H, Tait A. 2003. Multilocus of Cryptosporidium parvum type 2: population genetics and sub-structuring. Infect. Genet. Evol. 3:207–218 [DOI] [PubMed] [Google Scholar]
- 13. Mallon M, et al. 2003. Population structures and the role of genetic exchange in the zoonotic pathogen Cryptosporidium parvum. J. Mol. Evol. 56:407–417 [DOI] [PubMed] [Google Scholar]
- 14. Maynard-Smith JM, Smith NH, O'Rourke M, Spratt BG. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. U. S. A. 90:4384–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morgan-Ryan UM, et al. 2002. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J. Eukaryot. Microbiol. 49:433–440 [DOI] [PubMed] [Google Scholar]
- 16. Morrison LJ, et al. 2008. The population structure of the Cryptosporidium parvum population in Scotland: a complex picture. Infect. Genet. Evol. 8:121–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peakall R, Smouse PE. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6:288–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Quílez J, Vergara-Castiblanco C, Monteagudo L, Del Cacho E, Sánchez-Acedo C. 2011. Multilocus fragment typing and genetic structure of Cryptosporidium parvum isolates from diarrheic preweaned calves in Spain. Appl. Environ. Microbiol. 77:7779–7786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 20. Spano F, Putignani L, McLauchlin J, Casemore DP, Crisanti A. 1997. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol. Lett. 150:209–217 [DOI] [PubMed] [Google Scholar]
- 21. Strong WB, Gut J, Nelson RG. 2000. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect. Immun. 68:4117–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 23. Tanriverdi S, Widmer G. 2006. Differential evolution of repetitive sequences in Cryptosporidium parvum and Cryptosporidium hominis. Infect. Genet. Evol. 6:113–122 [DOI] [PubMed] [Google Scholar]
- 24. Tanriverdi S, et al. 2006. Emergence of distinct genotypes of Cryptosporidium parvum in structured host populations. Appl. Environ. Microbiol. 72:2507–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tanriverdi S, Blain JC, Deng B, Ferdig MT, Widmer G. 2007. Genetic crosses in the apicomplexan parasite Cryptosporidium parvum define recombination parameters. Mol. Microbiol. 63:1432–1439 [DOI] [PubMed] [Google Scholar]
- 26. Tanriverdi S, et al. 2008. Inferences about the global population structure of Cryptosporidium parvum and Cryptosporidium hominis. Appl. Environ. Microbiol. 74:7227–7234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Turner KM, Hanage WP, Fraser C, Connor TR, Spratt BG. 2007. Assessing the reliability of eBURST using simulated populations with known ancestry. BMC Microbiol. 7:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Widmer G, Lee Y. 2010. Comparison of single and multilocus genetic diversity in the protozoan parasites Cryptosporidium parvum and C. hominis. Appl. Environ. Microbiol. 76:6639–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Widmer G, Sullivan S. 2012. Genomics and population biology of Cryptosporidium species. Parasite Immunol. 34:61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xiao L, Fayer R, Ryan U, Upton SJ. 2004. Cryptosporidium taxonomy: recent advances and implications for public health. Clin. Microbiol. Rev. 17:72–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.