Abstract
Paenilide is a novel, heat-stable peptide toxin from Paenibacillus tundrae, which colonizes barley. P. tundrae produced 20 to 50 ng of the toxin mg−1 of cells (wet weight) throughout a range of growth temperatures from +5°C to +28°C. Paenilide consisted of two substances of 1,152 Da and 1,166 Da, with masses and tandem mass spectra identical to those of cereulide and a cereulide homolog, respectively, produced by Bacillus cereus NS-58. The two components of paenilide were separated from those of cereulide by high-performance liquid chromatography (HPLC), showing a structural difference suggesting the replacement of O-Leu (cereulide) by O-Ile (paenilide). The exposure of porcine spermatozoa and kidney tubular epithelial (PK-15) cells to subnanomolar concentrations of paenilide resulted in inhibited motility, the depolarization of mitochondria, excessive glucose consumption, and metabolic acidosis. Paenilide was similar to cereulide in eight different toxicity endpoints with porcine and murine cells. In isolated rat liver mitochondria, nanomolar concentrations of paenilide collapsed respiratory control, zeroed the mitochondrial membrane potential, and induced swelling. The toxic effect of paenilide depended on its high lipophilicity and activity as a high-affinity potassium ion carrier. Similar to cereulide, paenilide formed lipocations, i.e., lipophilic cationic compounds, with K+ ions already at 4 mM [K+], rendering lipid membranes electroconductive. Paenilide-producing P. tundrae was negative in a PCR assay with primers specific for the cesB gene, indicating that paenilide was not a product of plasmid pCER270, encoding the biosynthesis of cereulide in B. cereus. Paenilide represents the first potassium ionophoric compound described for Paenibacillus. The findings in this paper indicate that paenilide from P. tundrae is a potential food-poisoning agent.
INTRODUCTION
The genus Paenibacillus is generally considered a benign colonizer of plant rhizospheres and of no health concern. Paenibacillus spp. have been used and studied as plant growth-promoting agents in agriculture (39, 41, 42, 63). Paenibacilli utilize starch and other plant-related carbohydrates and also proteins such as gelatin (40) and milk (10). They grow at chilled temperatures of <10°C (21, 30), which explains why they appear frequently in harvested vegetables and grains (19), chilled foods (8, 21, 23, 31), natural wood, as well as humus (18, 34). Secondary metabolites of paenibacilli have been investigated for antimicrobial properties with the aim to improve the microbiological safety of foods with extended shelf lives (22, 26, 47, 64, 66, 67). On the other hand, Paenibacillus larvae is known to cause infectious disease in bees (3, 9), and Paenibacillus species in cultures of chilled human blood with potential pathogenicity toward humans have been reported (50, 56). The toxicity of Paenibacillus peptide metabolites toward mammalian cells appears to have been investigated little or not at all. We describe in this paper the mammalian cell toxicity of novel, cereulide-like, heat-stable toxic metabolites, named paenilide and homopaenilide, produced by barley grain isolates of Paenibacillus tundrae over a wide range of temperatures, including +5°C.
MATERIALS AND METHODS
Bacterial strains.
Strains E8a (HAMBI 3232) and E8b were isolated from grains of barley received from the Finnish Food Safety Authority (EVIRA). The barley sample was of interest because it was toxic in a rapid boar sperm bioassay (1), but no toxin had been found to explain this toxicity. Mycotoxins were present in only small amounts (beauvericin and moniliformin at trace amounts and enniatins A, A1, B, and B1 at amounts of 2 to 480 μg kg−1 [36]). Randomly chosen grains from the toxic sample were picked with sterilized tweezers and placed onto tryptic soy agar (TSA) plates. After 30 days of incubation in the dark at 20°C ± 2°C, the bacterial growth around the grains was evaluated for toxicity by a rapid boar sperm bioassay. The isolates were purified and subcultured onto TSA plates (20°C ± 2°C) unless otherwise stated.
Bacillus cereus F4810/72 (HAMBI 2454/SMR-178/CWG52702) (24), B. cereus ATCC 14579T, Bacillus amyloliquefaciens 19b (HAMBI 2660) (4), and B. cereus NS-58 (HAMBI 2450) (5) were from our own collection. Paenibacillus tundrae DSM 21291T was from the DSMZ.
Identification of isolates.
Gram staining, catalase, oxidase, and hydrolysis of starch (TSA with 10 g of soluble starch liter−1) were tested with established methods (32, 44). Growth at +5°C to +45°C (5 to 30 days), tolerance to NaCl (10, 30, or 50 g of NaCl liter−1 for 3 to 12 days at 28°C), and penicillin susceptibility (discs with 5 μg of penicillin) were tested. Motility was observed by microscopy. The hemolytic zone on sheep blood agar (7) was read after 5 days. Growths on Brilliance Bacillus cereus agar and R2A agar were read after 5 to 8 days. For whole-cell fatty acid analysis, biomass grown for 3 days at 28°C on tryptic soy broth agar (tryptic soy broth amended with 15 g liter−1 agar) was processed into fatty acid methyl esters and analyzed by using the MIDI Sherlock microbial identification system with the TSBA 50 library, version 4.5 (MIDI, Inc., Newark, DE), according to the manufacturer's instructions.
For 16S rRNA gene sequencing, whole-cell DNA extracted from biomass was PCR amplified and sequenced by using universal primers pA and pF′ as described previously by Edwards et al. (15). The sequence was assigned to the genus as described previously by Rasimus et al. (54).
The fingerprinting of whole-cell DNA by ribopattern analysis was done as described previously by Apetroaie-Constantin et al. (5), with EcoRI restriction using an automated ribotyper (RiboPrinter microbial characterization system; DuPont Qualicon, Wilmington, DE) and RiboExplorer software, version 2.1.4216.0. The Bacillus cereus-targeted PCR assay using whole-cell DNA and primers S-S-Bc-200-a-S-18 and S-S-Bc-470-a-A-18 was conducted as described previously by Hansen et al. (25). PCR using primers EM1F and EM1R, targeting a 635-bp fragment specific for the cesB gene, encoding a cereulide synthetase of B. cereus, was conducted as described previously by Ehling-Schulz et al. (17). B. cereus F4810/72, B. cereus ATCC 14579T, and B. amyloliquefaciens 19b were used as controls. A Fermentas GeneRuler 100-kb DNA ladder (Thermo Fisher Scientific, Waltham, MA) was used as a molecular size marker.
Isolation and processing of bacterial samples for toxicity assessment and toxin purification.
A rapid boar sperm bioassay (1) was used for toxicity screening of bacterial growth from the barley grains with an exposure time of 0.5 h. Crude cell extracts for the screening were prepared by simply boiling the biomass in methanol as described previously (1). Semipurified extracts for high-performance liquid chromatography (HPLC) fractionation were prepared as follows. A plate-grown biomass (10 to 30 days) of the bacterial isolates harvested into methanol-rinsed glass bottles was subjected to three freeze-thaw cycles, methanol was added (100 mg biomass ml−1), and the bottles were heated in boiling water (10 min), shaken overnight (20°C ± 2°C at 200 rpm), and centrifuged (3,800 rpm for 15 min). The clear supernatant was transferred into a preweighed ampoule, evaporated to dryness, weighed, redissolved in methanol to 10 to 30 mg (dry weight) ml−1, analyzed for toxicity, and/or used for the preparation of purified toxins by HPLC, as described below.
To investigate the cold tolerance of toxin production, biomass grown at +15°C was used to inoculate plates, which were then incubated at +10°C for 26 days. The biomass grown at +10°C was used as the inoculum for subculturing at +5°C ± 2°C for 28 days. Extracts for toxicity testing were then prepared as described above.
Reversed-phase high-performance liquid chromatography (RP-HPLC) and HPLC-mass spectrometry (MS) analyses.
The fractionation of the methanol extracts was carried out with 1100 series liquid chromatography (LC) (Agilent Technology, Wilmington, DE) equipment. The column used was an Atlantis C18 T3 4.6- by 150-mm, 3-μm column (Waters, Milford, MA), isocratically eluted by using 6% 0.1% formic acid (solvent A) and 94% methanol (solvent B) for 12 min, followed by a gradient to 100% solvent B in 1 min and then maintaining 100% solvent B to 35 min at a flow rate of 1 ml min−1. The UV absorption at 205 nm (A205) was used for detection.
HPLC-electrospray ionization ion trap mass spectrometry (ESI-IT-MS) analysis of the HPLC fractions was performed by using an MSD-Trap-XCT_plus ion trap mass spectrometer equipped with an Agilent ESI source and an Agilent 1100 series LC instrument (Agilent Technologies, Wilmington, DE). HPLC-ESI-IT-MS was performed in the positive mode with a mass range of 50 to 2,000 m/z. The HPLC and run conditions are described above. Valinomycin was used as a reference compound for quantification.
Cell toxicity assays.
The boar sperm motility inhibition assay, including all the controls for aging and vehicle-exposed cells and for the repeatability of the assay, was executed as described previously (2). The change in the mitochondrial potential (ΔΨm) and the change in the plasma membrane potential (ΔΨp) in boar spermatozoa were recorded by use of the fluorogenic dye JC-1 (5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide) as described previously by Hoornstra et al. (27). For the estimation of the 100% effective concentration (EC100), 10 microscopic fields at ×10 and ×40 magnifications were inspected. The EC100 for triclosan (the reference toxicant) was 10 μg ml−1 (standard deviation [SD], ±2 μg).
Assays with monolayers of somatic cells.
Cell cultivations and exposures were done in a tissue culture cabinet at 37°C with 5% CO2 in air and a relative humidity (RH) of 100%. Eight-well glass chamber slides (bottom surface, 90 mm2) were seeded with 500 μl of trypsinated monolayers of porcine kidney tubular epithelial (PK-15) cells diluted with RPMI to 1 × 105 cells ml−1. Uniform monolayers consisting of ca. 6 × 105 cells per 9 × 107 μm2 (150 to 250 μm2 per cell) formed in 48 h. The twofold serially diluted test substances or the vehicle only was dispensed into the wells. The glucose content of the wells was measured by using a glucose meter (Precision Xceed; Abbott Diabetes Care Ltd., Berkshire, United Kingdom). In wells that received nothing or the vehicle only, the glucose concentration decreased from an initial concentration (0 h) of 12 mM to 6 mM (24 h), whereas the pH remained stable (ΔpH of <0.2). A glucose concentration after 24 h of ≤3 mM indicated excessive glucose consumption. Prior to the measurement of the pH, the chamber slides were transferred into ambient air for 1 h to allow CO2 to evaporate. A pH drop of ≥0.5 units in the toxin-exposed well compared to the vehicle only was considered proof of acidosis. Acidification was also visible as a change of the indicator color in RPMI medium from pink to yellow. Mitochondrial depolarization was read by a microscope after 26 h of exposure to the toxicants from the monolayers double stained with the membrane-potential-responsive fluorogenic dye JC-1 and with propidium iodide to assess the relaxing of the plasma membrane permeability barrier, similarly to what was reported previously for sperm cells (27).
Assays with proliferating cells for growth inhibition and cytotoxicity.
Forty-eight-hour-old monolayers of PK-15 and murine neuroblastoma (MNA) cells were trypsinated and diluted in RPMI medium to 2 × 105 cells ml−1. The twofold serially diluted test substance (dissolved in methanol) or the vehicle only was dispensed into wells of a 96-well flat-bottomed microplate (Nunc, Roskilde, Denmark) holding 100 μl RPMI medium per well. One hundred microliters of the cell suspension in RPMI was dispensed into each well. After 48 h of exposure, the well contents were inspected by microscopy for cell growth or cytolysis. The EC100 for cytolysis was estimated after inspecting 10 microscopic fields at ×100 and ×400 magnifications. Triclosan was used as the reference toxicant, with an EC100 of 10 μg ml−1 (SD, ±2 μg).
Exposure of isolated rat liver mitochondria.
Rat liver mitochondria (RLM) were isolated from male Wistar rats, as described previously (59), by use of a standard method (37). The mitochondria were washed twice in mannitol buffer (220 mM mannitol, 70 mM sucrose, and 10 mM HEPES-Tris [pH 7.4]), resuspended in the same buffer to 60 to 80 mg protein ml−1, and kept on ice for analysis.
Mitochondrial functions were determined as described previously (59). Mitochondrial swelling was recorded as a decrease in the OD540 (UV-1700 Pharmaspec UV-visible [UV-Vis] spectrophotometer; Shimadzu Corp., Japan). Oxygen uptake was measured with a Clark electrode, and the mitochondrial membrane potential (ΔΨm) was measured with the aid of tetraphenylphosphonium (TPP+) (38) using a TPP+-selective electrode (Niko, Moscow, Russia). Mitochondrial functions were measured with a 1-ml closed chamber at 25°C with magnetic stirring in standard glutamate (5 mM)-malate (5 mM) medium containing 120 mM KCl, 2 mM KH2PO4, and 10 mM HEPES (pH adjusted to 7.3 with a few grains of Trizma base) or in an NaCl medium similar to that described above but with NaCl replacing KCl and NaH2PO4 replacing KH2PO4. The kinetics of the influx of K+ into the mitochondria was measured online as changes in the concentration of [K+] in the external medium with a K+-selective electrode (Nika, Moscow, Russia), as described in detail previously (59, 60).
Measurement of ionophoricity using the black lipid membrane method.
Conductance in a bilayer black lipid membrane (BLM) was measured with a BLM formed in the circular window (0.975 mm2) of a 2-ml Teflon cell, placed into an outer chamber. The Teflon cell and the chamber contained 20 mM Tris-HCl (pH 7.4) and 100 mM KCl or 100 mM NaCl, with an applied voltage of 100 mV. Both the cell and the outer chamber were provided with an electrode, and the conductivity between the two electrodes was measured. For the formation of the BLM, 1.2 mg of total rat liver mitochondrial lipids and 130 μg of cardiolipin were mixed, dried, and dissolved in 80 μl n-decane. A Max 406 operational electrometer amplifier connected to an IBM-compatible computer was used for measuring the membrane current by use of a voltage clamp method described previously (60).
Reagents, media, and test cells.
Valinomycin (≥98% HPLC grade, from Streptomyces griseus; CAS 2001-95-8) was purchased from Sigma-Aldrich (St. Louis, MO). Triclosan (2,4,4′-trichloro-2′-hydroxydiphenyl ether) (CAS 3380-34-5) was obtained from Merck (Darmstadt, Germany). The fluorogenic membrane-potential-responsive dye JC-1 (mg ml−1 in dimethyl sulfoxide [DMSO]) and the DNA dye propidium iodide were obtained from Molecular Probes (Invitrogen, Carlsbad, CA). Tryptic soy agar (TSA) and tryptic soy broth were obtained from Scharlab (Barcelona, Spain), Chromogenic Brilliance Bacillus cereus agar was obtained from Oxoid (Basingstoke, Hampshire, United Kingdom), and R2A agar was obtained from Lab M (Lancashire, United Kingdom). Penicillin discs were obtained from Rosco Diagnostica (Taastrup, Denmark). Blood agar with 5% (vol/vol) sheep blood was obtained from EVIRA (Helsinki, Finland). RPMI 1640 with l-glutamine, heat-inactivated fetal bovine serum albumin, and penicillin-streptomycin (10,000 units penicillin and 10,000 μg ml−1 streptomycin) were obtained from Gibco (Invitrogen, Carlsbad, CA). Trypsin (10×) with Versene for cell detachment was obtained from Lonza (Verviers, Belgium), glucose strips were obtained from Lifescan (Johnson & Johnson, Espoo, Finland), and pH strips (pH 6.5 to 10.0) were obtained from Merck (Darmstadt, Germany). Cereulide (not commercially available) was purified from B. cereus NS-58 as described previously (46, 59). Other chemicals were of analytical quality and were obtained from local suppliers.
Extended commercial boar semen from an artificial insemination station (Faba Sika Ltd., Tuomikylä, Finland) was used as delivered, at 27 × 106 sperm cells ml−1. The epithelial cell line PK-15 from porcine kidney proximal tubules (14) and MNA cells were obtained from EVIRA (Helsinki, Finland). Labtek cultivation chamber slides (catalog number 154534) and a tissue culture cabinet (autosterilizable Heracell 150i) were obtained from Thermo Fisher Scientific (Vantaa, Finland).
Nucleotide sequence accession number.
The 16S rRNA gene sequence (955 nucleotides [nt]) of P. tundrae E8a was deposited in GenBank under accession number JF683621.
RESULTS
Characterization of toxigenic, spore-forming bacteria colonizing barley grains.
Samples from barley grains were inspected for microbial contamination because these samples were found to be toxic in a rapid boar sperm bioassay but contained only low concentrations of mycotoxins or none at all (36). The grains were cultured on TSA plates, and heat-treated (100°C) crude methanol extracts prepared from the obtained colonies were tested for toxicity by the rapid boar sperm bioassay (1). The purified toxic isolates consisted of aerobic Gram-positive rods with polarly located endospores.
Analysis of 16S rRNA gene sequences of two independent isolates (E8a and E8b) with the RDP Classifier tool showed that these isolates belonged to the genus Paenibacillus with 100% confidence. The partial 16S rRNA gene sequence (955 bp) of isolate E8a had the highest level of similarity to type strains of Paenibacillus xylanexedens and P. amylolyticus (similarity score of 1.000 with the RDP SeqMatch tool), and BLAST analysis showed similarity to type strains of P. amylolyticus and P. tundrae (>99.3% similarity). DNA fingerprints of the ribosomal operon area of isolates E8a and E8b were identical, with no matches to any fingerprints available in the DuPont ribopattern library or the ribopattern database of our research facility. Whole-cell fatty acids of isolate E8a contained 12-methyltetradecanoic acid as the main component (61.0%) and smaller amounts of 14-methylpentadecanoic acid (7.8%), tetradecanoic acid (2.5%), 14-methylhexadecanoic acid (4.5%), and cis-5-hexadecenoic acid (2.5%). In the original species description, these four fatty acids distinguish P. tundrae from P. amylolyticus and P. xylanexedens when grown at +28°C (49). Isolate E8a grew on TSA plates at +5°C ± 2°C, +10°C, +15°C, +20°C ± 2°C, +28°C, and +37°C but not at +45°C, and it grew on oligotrophic R2A agar at +20°C ± 2°C. Isolate E8a grew poorly on Bacillus cereus-specific (20) Chromogenic Brilliance Bacillus cereus agar and formed no blue-green colonies. PCR with primers specific for Bacillus cereus group bacteria (S-S-Bc200-aS-18 and S-S-Bc-470-a-18) and for the cereulide synthetase gene cesB (EM1F/EM1R) yielded no amplicons from the DNA of isolate E8a (data not shown). This indicates that isolate E8a did not contain the 288-bp fragment specific for members of the B. cereus group (25) or the 635-bp fragment specific for the cereulide synthetase operon (17). Isolate E8a hydrolyzed starch; grew in the presence of 1%, 3%, and 5% (wt/vol) NaCl; and was nonhemolytic, catalase positive, oxidase negative, and susceptible to penicillin (inhibition zone, 31 mm in diameter).
In summary, the biochemical properties and the DNA-based data from the toxin-producing barley grain isolates E8a and E8b indicate their species identity as Paenibacillus tundrae.
Identification of the toxic substances.
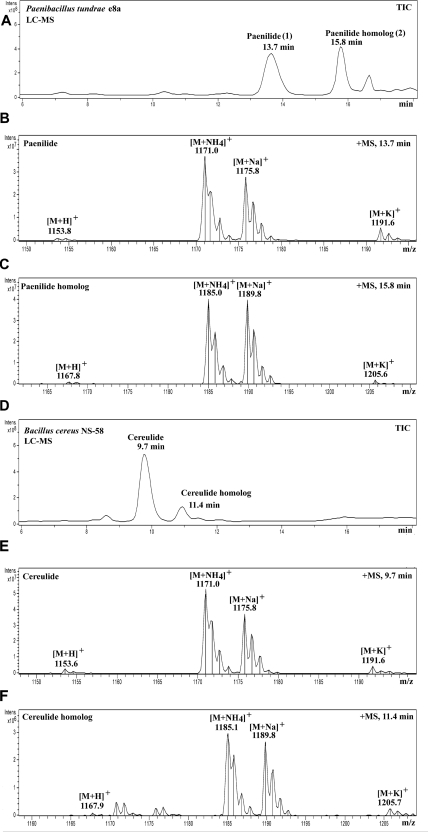
Figure 1A shows the result of the HPLC fractionation of the toxic substances contained in the methanol extracts of P. tundrae E8a. Toxic peak 1 (eluting at 13.7 min) (Fig. 1A) contained the protonated [M + H]+ mass ion at m/z 1,153.8 and the cationized mass ions [M + NH4]+ at m/z 1,171.0, [M + Na]+ at m/z 1,175.8, and [M + K]+ at m/z 1,191.6 (Fig. 1B). Thus, the molecular mass of the toxic substance was 1,152 Da, i.e., identical to that of the known toxin cereulide, isolated from B. cereus NS-58 (Fig. 1E) (46, 52). Toxic peak 2, eluting at 15.8 min, contained the protonated [M + H]+ mass ion at m/z 1,167.8 and the cationized mass ions [M + NH4]+ at m/z 1,185.0, [M + Na]+ at m/z 1,189.8, and [M + K]+ at m/z 1,205.6, matching a molecular mass of 1,166 Da (Fig. 1C), i.e., exactly the same molecular mass as that of the cereulide homolog from B. cereus NS-58 (Fig. 1F) (52, 65). In spite of molecular masses being identical to those of cereulide and its homolog, the retention times of the metabolites produced by P. tundrae E8a upon HPLC analysis under identical conditions were longer. This finding indicates a higher hydrophobicity of the P. tundrae substances (Fig. 1A) than those of cereulide and the cereulide homolog from B. cereus (Fig. 1D). The retention time differences of 4.0 min and 4.4 min, respectively, were confirmed by mixing the P. tundrae E8a substances with cereulide and the cereulide homolog from B. cereus NS-58 before HPLC analysis (not shown). Structural analysis of the P. tundrae E8a substances was continued by tandem mass spectrometry (MS/MS), and cereulide and the cereulide homolog from B. cereus NS-58 were included as references.
Fig 1.
HPLC-MS chromatograms of the toxic methanol extracts of P. tundrae E8a and B. cereus NS-58 (cereulide producing [5]) under the same HPLC conditions. (A) Total ion chromatogram (TIC) of the extract of P. tundrae E8a. Two peaks were found, peaks 1 and 2, that were toxic in the rapid sperm assay, with retention times of 13.7 min (peak 1) and 15.8 min (peak 2). (B) Mass spectrum of peak 1, named paenilide. (C) Mass spectrum of peak 2, named the paenilide homolog (homopaenilide). (D) Total ion chromatogram of the extract of B. cereus NS-58 containing cereulide (9.7 min) and the cereulide homolog (11.4 min). (E) Mass spectrum of cereulide. (F) Mass spectrum of the cereulide homolog.
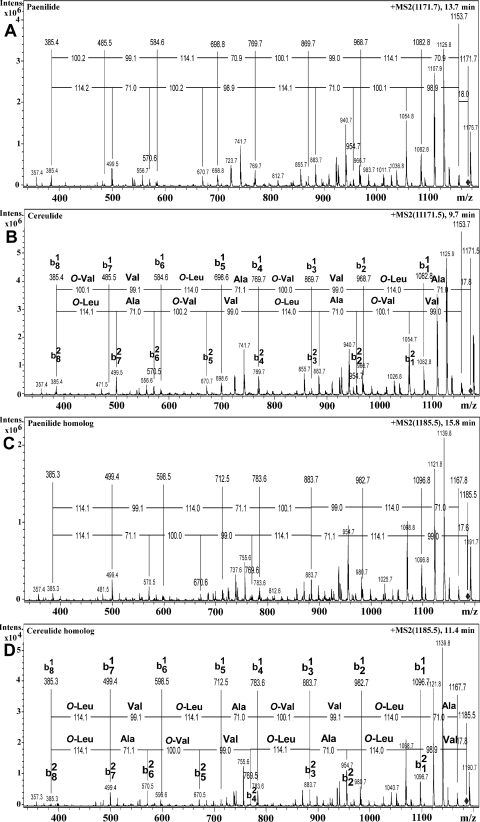
The tandem mass spectrum of the ion [M + NH4]+ at m/z 1,171.7 of toxic peak 1, named paenilide (eluting at 13.7 min), is shown in Fig. 2A, and that of the corresponding mass ion, [M + NH4]+ at m/z 1,171.5 of cereulide (eluting at 9.7 min), is shown in Fig. 2B. Figure 2B also shows that the fragmentation patterns of b1 and b2 series mass ions retrieved from the precursor ion of cereulide correspond to the known sequence of amino and hydroxy acids of cereulide, O-Val–Val–O-Leu–Ala–O-Val–Val–O-Leu–Ala and O-Leu–Ala–O-Val–Val–O-Leu–Ala–O-Val–Val. Identical mass fragments were retrieved from paenilide (Fig. 2A).
Fig 2.
MS/MS analyses of toxic peaks 1 (13.7 min) and 2 (15.8 min) of the semipurified methanol extracts of P. tundrae E8a (shown in Fig. 1A to C) and of similarly prepared extracts from B. cereus NS-58 (shown in Fig. 1D and E). (A) Fragment ions obtained from the precursor ion at m/z 1,171.7 of toxic peak 1 (paenilide; shown in Fig. 1A). (B) Fragmentation patterns of the b1 and b2 series mass ions from the precursor ion at m/z 1,171.5 and corresponding amino and hydroxy acid sequences of cereulide (peak at 9.7 min in Fig. 1D). (C) Fragment ions obtained from the precursor ion at m/z 1,185.5 of toxic peak 2 (paenilide homolog; shown in Fig. 1A). (D) Fragmentation patterns of the b1 and b2 series mass ions from the precursor ion at m/z 1,185.5 and corresponding amino and hydroxy acid sequences of the cereulide homolog (peak at 11.4 min in Fig. 1D). O-Val is 2-hydroxyisovaleric acid, and O-Leu is 2-hydroxyisocaproic acid.
Figure 2D shows b1 and b2 mass ion series obtained from the precursor ion [M + NH4]+ at m/z 1,185.5 of the cereulide homolog from B. cereus NS-58. This cereulide producer thus produces a cereulide homolog with the same masses as those known for B. cereus NC7041 (52). The tandem mass spectrum of toxic peak 2 (eluting at 15.8 min) using [M + NH4]+ at m/z 1,185.5 as a precursor ion (Fig. 2C) produced mass ion series and fragmentation patterns similar to those of the corresponding precursor ion of the cereulide homolog (eluting at 11.4 min). The cereulide homolog consists of two similar tetrapeptides (O-Val–Val–O-Leu–Ala) and one different tetrapeptide (O-Leu–Val–O-Leu–Ala), thus having the cyclic structure of cyclo(O-Val–Val–O-Leu–Ala)2-(O-Leu–Val–O-Leu–Ala). The cereulide homolog has a fragmentation pattern similar to that of cereulide except for the fragment ions b81, b71, b12, and b22, corresponding to the cleavage of O-Leu (114 Da) instead of O-Val in cereulide (Fig. 2D). This explains the 14-Da difference in molecular masses between cereulide (1,152 Da) and its homolog (1,166 Da). A similar difference was seen between P. tundrae E8a toxic metabolites 1 (1,152 Da) and 2 (1,166 Da). In analogy to cereulide and its homolog, the two toxic compounds from P. tundrae, with molecular masses of 1,152 Da and 1,166 Da, were named paenilide and homopaenilide, respectively, and their MS/MS patterns are displayed in Fig. 2A and C. The barley grain isolate P. tundrae E8b was analyzed similarly and found to produce the same toxic substances, paenilide and homopaenilide, but in smaller amounts.
With the mass fragmentation patterns being identical (Fig. 2), how does one explain the higher hydrophobicity of paenilide (change in retention time of +4 min) than that of cereulide? Cereulide contains three 2-hydroxyisocaproic acid residues (O-Leu), whereas the cereulide homolog contains four (52). The mass spectrometry results in Fig. 2A and C show that paenilide may contain three 2-hydroxy-3-methylpentanoic acid residues (O-Ile) and that the paenilide homolog (homopaenilide) may contain four, possibly explaining the longer retention time, when one O-Leu is replaced by one O-Ile.
Mammalian cell toxicity of paenilide compared to cereulide.
Paenilide and homopaenilide (Fig. 1A), purified from P. tundrae strain E8a, were tested for mammalian cell toxicity using three different cell types and a battery of toxicity endpoints. The toxic endpoint concentrations (EC100s) are shown in Table 1. The two toxic peaks (paenilide and homopaenilide) with closely similar structures (Fig. 2A and C) were combined and used as a mixture, indicated as “paenilides” in the results described below. The cereulide preparation, purified from B. cereus NS-58, is indicated similarly as “cereulides,” as it contained the two similar substances cereulide and the cereulide homolog.
Table 1.
Mammalian cell toxicity of paenilides and cereulides, measured with porcine spermatozoa and kidney (PK-15) and murine neuroblastoma cellsa
| Target cell and toxicity parameter | EC100 (ng ml−1) at exposure time of: |
|||||
|---|---|---|---|---|---|---|
| 60 min |
1 day |
2 days |
||||
| Paenilides | Cereulides | Paenilides | Cereulides | Paenilides | Cereulides | |
| Porcine spermatozoa | ||||||
| Loss of motility | 1.4 | 0.4 | 1.1 | 0.2 | 1.0 | 0.1 |
| Loss of ΔΨm | 1.1 | 0.4 | 0.4 | 0.5 | 0.5 | 0.2 |
| Hyperpolarization of ΔΨp | 1.7 | 1.6 | 1.3 | 0.6 | 2.1 | 0.4 |
| Adverse effects on metabolism of: | ||||||
| PK-15 cells | ||||||
| Excessive consumption of glucose | ND | ND | 0.5 | 0.2 | ND | ND |
| Mitochondrial depolarization | ND | ND | 0.7 | 0.4 | ND | ND |
| Metabolic acidification of growth medium (pH decrease from 7.3 to ≤6.8)c | ND | ND | 0.5 | 0.2 | ND | ND |
| Cytolysis | ||||||
| PK-15 cells | ND | ND | 16b | 1,700 | 16b | 6.6 |
| MNA cells | ND | ND | 250b | >1,700 | 1.0b | 1.6 |
Toxicity was assayed by exposing the target cells to purified toxins or heat-treated (100°C) cell-free methanol extracts prepared from a biomass of P. tundrae E8a cells grown on TSA (10 to 30 days at 20°C ± 2°C). The EC100 values are averages based on the results of two to four parallel assays. ND, not determined.
Assay conducted with an extract containing all methanol-soluble substances from the P. tundrae E8a biomass. The amount of paenilides was determined by HPLC analysis. The extract contained 0.1 μg paenilides per mg of methanol-soluble dry substances.
When the cells were exposed to vehicle only or nothing, the change was <0.2 pH units.
The results in Table 1 show that low concentrations (0.5 to 2 ng ml−1) of paenilides caused a loss of motility and depolarized mitochondria of boar spermatozoa and porcine kidney tubular epithelial (PK-15) cells, accelerated glucose consumption, and induced acidosis. These findings indicate that exposure to paenilides from P. tundrae E8a caused mitochondrial dysfunction in germ cells (sperms) as well as somatic cells (PK-15 cells) at below-nanomolar concentrations, similarly to cereulides from B. cereus. In addition, within 1 day, a semipurified methanol extract of P. tundrae E8a killed and inhibited the proliferation of murine neuroblastoma (MNA) and PK-15 cells at EC100s of 250 and 16 ng ml−1, respectively, whereas the mitochondria were depolarized by an EC100 of 0.4 to 1.1 ng ml−1.
The results summarized in Table 1 show that paenilides from P. tundrae E8a equaled cereulides of B. cereus in toxicity toward mammalian cells, displayed by eight different toxicological parameters. Cell extracts of B. cereus ATCC 14579 (cereulide nonproducer) and of P. tundrae DSM 21291T (does not produce paenilide) caused no toxic effects (data not shown). The results indicate that paenilides are the toxic substances emitted by barley grain isolate P. tundrae E8a and that the toxic activity toward boar spermatozoa, porcine kidney epithelial cells, and murine neuroblastoma cells is similarly potent compared to that of cereulides from B. cereus.
Mitochondriotoxic properties of paenilides of P. tundrae studied by use of isolated RLM.
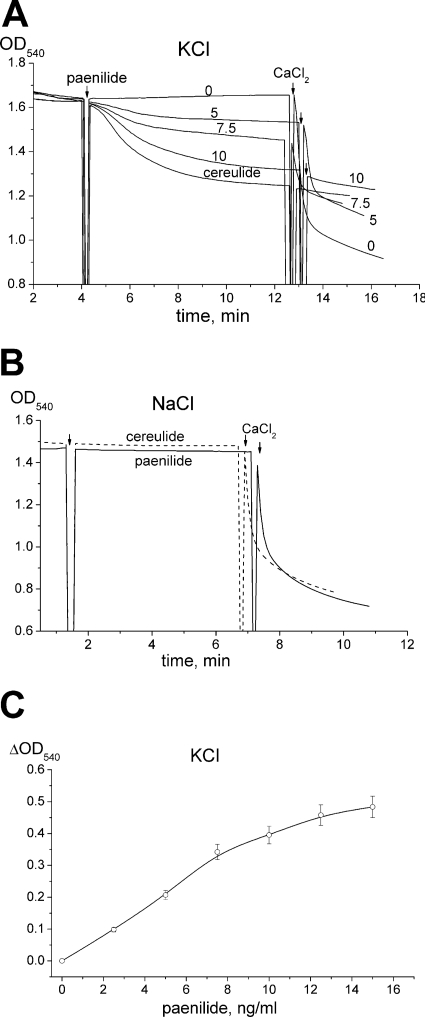
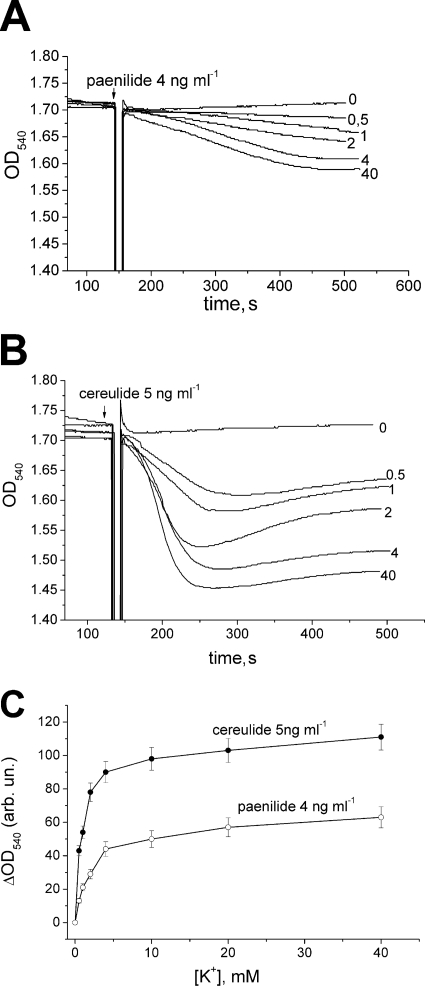
Figure 3 compares paenilide- and cereulide-mediated swelling of RLM, as measured by the decrease in the OD540 in standard glutamate-malate buffer (pH 7.3) prepared with KCl or NaCl. The addition of paenilides (5 to 10 ng ml−1) induced the swelling of energized RLM incubated in KCl medium, similarly to cereulides (7.5 ng ml−1) (Fig. 3A). In NaCl medium, neither the cereulides nor the paenilides induced any swelling of the mitochondria (Fig. 3B). The swelling in KCl medium showed almost a linear dose response to the exposure dose of paenilides (Fig. 3C).
Fig 3.
Paenilide-induced swelling of isolated rat liver mitochondria (RLM). RLM (0.5 mg protein ml−1) were incubated for 3 min at 20°C in glutamate (5 mM)-malate (5 mM) medium with KCl (120 mM) (A and C) or with NaCl (120 mM) (B). Swelling was measured as the decrease of the OD540. (A) Original OD540 traces of the mitochondrial suspension in medium with KCl, spiked with 0, 5, 7.5, or 10 ng ml−1 of paenilides or with 7.5 ng ml−1 of cereulides. The 0 trace is with the vehicle (methanol) only. (B) Original OD540 traces in medium with NaCl, spiked with paenilides (7.5 ng ml−1) or with cereulides (7.5 ng ml−1). At the end of each trace, 300 μM CaCl2 was added to induce the opening of the mitochondrial permeability transition pore (mPTP). (C) Dependence of the maximal ΔOD540 on the dose of paenilides in medium with KCl. Panels A and B represent typical traces from three identical experiments using different mitochondrial preparations. Panel C shows mean values and standard errors (SE) from 3 to 5 experiments.
Calcium was used as a tool to induce the opening of the mitochondrial permeability transition pore (mPTP). As shown in Fig. 3A, the addition of 300 μM calcium in the absence of paenilides (control trace 0) caused a high-amplitude swelling of the mitochondria (visible as a decrease of the OD540), indicating mPTP opening. When paenilides (5 or 7.5 ng ml−1) had been added, the mitochondria responded to the subsequent addition of 300 μM calcium by a decreased amplitude of swelling, indicating that not all added calcium was accumulated by mitochondria due to a reduced potential (Fig. 3A). When paenilides (10 ng ml−1) or cereulides (7.5 ng ml−1) had been used to induce a substantial swelling of the mitochondria, the subsequent addition of 300 μM calcium induced no further changes in the optical density. This suggests that the mitochondrial membrane potential (ΔΨm) was lowered in K+-containing medium by paenilides and cereulides so that the added calcium no longer accumulated.
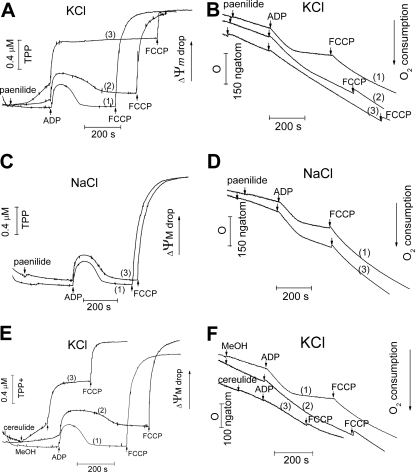
Subsequent experiments with direct measurements of ΔΨm confirmed the above-described conclusion. Figure 4 shows the simultaneous recordings of the mitochondrial membrane potential (ΔΨm) (measured with a TPP+-selective electrode) and the respiration of the mitochondria (on glutamate-malate, measured with an oxygen electrode) from the same cuvette. As shown in Fig. 4, paenilides induced a concentration-dependent decrease of the ΔΨm in KCl (Fig. 4A, rising traces 2 and 3). The dose-response effect was seen from traces 1 (0 ng ml−1 paenilides), 2 (4.5 ng ml−1 paenilides), and 3 (9 ng ml−1 paenilides). After a dose of 9 ng ml−1, the restoration of the ΔΨm, which should occur after the termination of the synthesis of ATP from the added ADP, was no longer observed (Fig. 4A, trace 3). In NaCl-containing medium, paenilides had no effect in excess of that of the vehicle only on the ΔΨm or on the oxidative phosphorylation cycle even at a concentration of 9 ng ml−1 (Fig. 4C, traces 1 and 3).
Fig 4.
Comparison of effects of exposure to paenilides and cereulides on the ΔΨm and oxygen consumption of mitochondria in glutamate-malate medium with KCl or NaCl. Mitochondria (1 mg protein ml−1) were incubated in isotonic glutamate-malate medium with isotonic KCl or NaCl (pH 7.3), as described in the legend of Fig. 3. A TPP+-selective electrode was used to measure the ΔΨm. Oxygen consumption was measured by use of a Clark oxygen electrode. (A and B) Changes in the ΔΨm (A) and oxygen consumption (B) in response to added paenilides, at 4.5 ng ml−1 (trace 2) and 9 ng ml−1 (trace 3), in medium with KCl. Trace 1, vehicle only (methanol). (C and D) ΔΨm and oxygen consumption of mitochondria in medium with NaCl. Trace 1, vehicle only (methanol); trace 2, paenilides at 9 ng ml−1. (E and F) Responses of ΔΨm and mitochondrial oxygen consumption to added cereulides, at 2.5 ng ml−1 (trace 2) or 6.5 ng ml−1 (trace 3), in medium with KCl. Trace 1, vehicle only (methanol). The panels show traces typical of three identical experiments using different mitochondrial preparations. FCCP, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone.
As shown in Fig. 4B, the addition of paenilides to mitochondria incubated in KCl medium, but not in NaCl medium (Fig. 4D), caused a small increase of the respiration rate in state 2, a decrease of the respiration rate in state 3, and an increase of the respiration rate in state 4. This behavior indicates an uncoupling of oxidative phosphorylation and a major loss of respiratory control. The effects of paenilides were almost identical to those of cereulides (Fig. 4A, B, E, and F).
In summary, the results shown in Fig. 3 and 4 indicated that paenilides induced an influx of K+ ions but not of Na+ ions from the external medium into the mitochondrial matrix, resulting in mitochondrial swelling, a decrease of the ΔΨm, an uncoupling of oxidative phosphorylation, and a major loss of respiratory control, explaining the toxic effects described in Table 1.
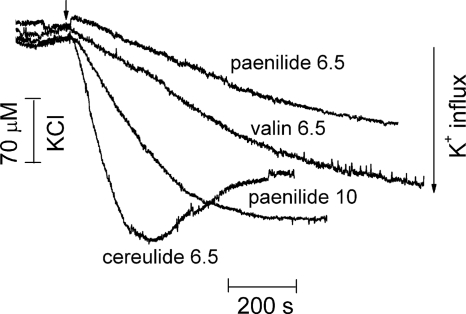
Figure 5 shows the influx of K+ into the mitochondria in real time (one reading per second). The external [K+] concentration was monitored in a suspension of RLM in glutamate-malate buffer with 120 mM NaCl and 200 μM [K+]. The external [K+] concentration remained constant (200 μM) until paenilides, cereulides, or valinomycin was added. Each of these substances initiated an influx of K+, visible as a rapid decrease in the concentration of [K+] in the medium. This observation offers an explanation for all of the observed effects of paenilides on mitochondrial function. Cereulides at a concentration of 6.5 ng ml−1 were slightly more potent than 10 ng ml−1 of paenilides or 6.5 ng ml−1 of valinomycin (Fig. 5). This could be explained by paenilides possessing a lower affinity for K+ than cereulides. This hypothesis was tested by measuring mitochondrial swelling in isotonic medium with a series of rising K+ concentrations (Fig. 6).
Fig 5.
Influx of potassium ions into energized RLM in response to added paenilides, cereulides, or valinomycin. Mitochondria (1 mg protein ml−1) were incubated in glutamate-malate medium (pH 7.3) containing 116 mM NaCl, 4 mM KCl, and 2 mM NaH2PO4. The [K+] concentration was determined with a K+-selective electrode. Paenilides were added at concentrations of 6.5 and 10 ng ml−1. Cereulides and valinomycin (“valin”) were added to 6.5 ng ml−1. The traces shown are representative of three identical experiments using different mitochondrial preparations.
Fig 6.
Dependence of mitochondrial swelling induced by paenilides and cereulides on potassium ion concentrations ranging from 0 to 40 mM in media. RLM (0.5 mg protein ml−1) were incubated in isotonic glutamate-malate medium (pH 7.3) prepared with NaCl plus KCl (120 mM), with various ratios of [K+] to [Na+]. (A and B) Mitochondrial swelling was induced by the addition of paenilides (4 ng ml−1) (A) or cereulides (5 ng ml−1) (B). The panels show the original traces, typical of three identical experiments using different mitochondrial preparations. (C) Concentration dependence of maximal ΔOD540 in response to added paenilides or cereulides with different [K+] concentrations in the external medium (shown on the x axis). Mean values and SE from three experiments are shown.
Figure 6 shows the dependence of paenilide- and cereulide-induced mitochondrial swelling (Fig. 6A and B) on the external concentrations of [K+]. When the amplitude of swelling is plotted against the [K+] concentration (Fig. 6C), it can be seen that [K+] at concentrations as low as 0.5 to 1 mM mediated swelling, and a plateau was reached at 4 mM KCl. The mitochondrial swelling induced by paenilides occurred at a lower rate and at a slightly lower amplitude than that induced by cereulides. This finding is in agreement with the data obtained for K+ influx (Fig. 5). In summary, the results shown in Fig. 5 and 6 confirm the view that the toxicity of paenilides (Table 1) is based on carrier activity with a high affinity for potassium.
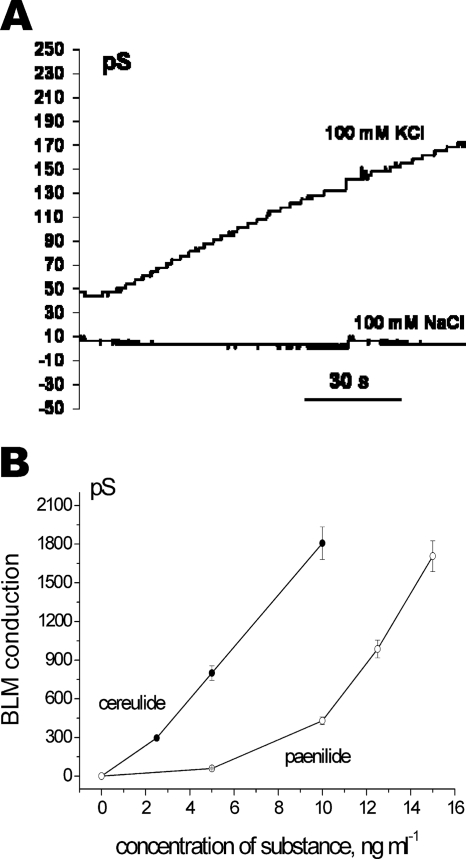
The potassium-specific ionophoricity of paenilides was also shown by experiments with a lipid bilayer (BLM) formed from RLM lipids supplemented with cardiolipin. As shown in Fig. 7A, the addition of paenilides induced an increase in BLM conductivity in KCl buffer but not in NaCl buffer. Paenilides induced electric conduction in the lipid bilayer at concentrations approximately 3-fold higher than those of cereulides (Fig. 7B).
Fig 7.
BLM studies of paenilide-induced electric conductivity of the lipid membrane in K+- and Na+-containing buffers. Changes in the electric conductivity of a lipid membrane, consisting of extracted RLM lipids with cardiolipin added, in response to added paenilides or cereulides was measured by using the BLM technique. (A) Time course of BLM electric conductivity induced by paenilides at 10 ng ml−1. The external chamber medium contained 20 mM Tris-HCl (pH 7.4) with 100 mM KCl or 100 mM NaCl, and the applied voltage was 100 mV. (B) Electric conductivity of the BLM in response to added concentrations of paenilides or cereulides in 100 mM KCl. Mean values and SE from three experiments are shown.
Cold tolerance of paenilide production by Paenibacillus tundrae E8a.
P. tundrae E8a produced 20 to 50 ng of paenilides per mg of biomass (wet weight). The question was asked whether this bacterium would produce significant amounts of toxic paenilides when grown at refrigerated temperatures, such as those prevailing during food storage and transport. Biomass extracts of P. tundrae E8a were similarly toxic (equal EC100 values; SD, 40%) irrespective of the growth temperature (5°C to 28°C). Extracts of P. tundrae DSM 21291T had no toxic effect (EC100 of >250 μg ml−1). The HPLC-MS analysis of the methanol extract of P. tundrae E8a cells grown at +5°C ± 2°C confirmed that paenilide and homopaenilide were present.
DISCUSSION
We showed in this paper that barley grain isolates of P. tundrae produced novel, potassium ionophoric, heat-stable peptide toxins, paenilides, comprising the molecules paenilide (sensu stricto) and paenilide homolog (homopaenilide). These two molecules had MS and MS/MS spectra similar to those of the 36-membered cyclodepsipeptide cereulide and the cereulide homolog (52) but were structurally different from cereulide. Paenilide and homopaenilide were separable from cereulide and its homolog by longer retention times (higher hydrophobicity) by HPLC. The toxic effects and toxic potency of paenilides on porcine sperm cells, porcine kidney tubular epithelial (PK-15) cells, and murine neuroblastoma (MNA) cells were similar to those provoked by cereulide, the most potent heat-stable bacterial food-poisoning toxin described so far. Exposure to subnanomolar concentrations of paenilides depolarized mitochondria, caused excessive glucose consumption, and induced metabolic acidosis in porcine cells. Paenilides are the first reported metabolites from the genus Paenibacillus that have shown a high level of mitochondrial toxicity in mammalian cells.
Metabolic acidosis has been reported to be the major pathological trait in fatal and close-to-fatal cases of human food poisoning caused by cereulide (ΔpH reported to be 0.3 to 0.6) (13, 33, 43, 53, 57). This paper appears to be the first report where metabolic acidosis induced by a bacterial toxin was demonstrated for in vitro-exposed mammalian cells. Interestingly, the results reported in this paper show that exposure to paenilides from P. tundrae E8a provoked mitochondrial dysfunction leading to metabolic acidosis equally as effectively as cereulide. With cereulide, this occurred at exposure concentrations relevant to those in foods associated with severe human food poisonings (35, 48). The findings in this paper indicate that paenilides are also potential food-poisoning agents.
The novel feature of paenilide- and homopaenilide-producing P. tundrae E8a was that the productivity, 20 to 50 ng paenilides per mg of biomass (wet weight), was similar throughout a range of growth temperatures from +5°C to +28°C. Cereulide production by B. cereus is temperature sensitive: no detectable cereulide is produced at temperatures of +10°C or lower (11, 24, 61). The only reported psychrotrophic cereulide producer, Bacillus weihenstephanensis (B. cereus sensu lato), grows but does not produce cereulide at chilled temperatures (61, 62). P. tundrae appears to be the first organism producing a cereulide-like, heat-stable peptide toxin at cold temperatures.
The cyclic peptide cereulide contains three 2-hydroxyisocaproic acid residues (O-Leu), and the cereulide homolog contains four (52). The results in this paper suggest that paenilide contains three 2-hydroxy-3-methylpentanoic acids (O-Ile) and that homopaenilide contains four O-Ile residues. These structural differences could explain the higher hydrophobicity of paenilide and homopaenilide than those of cereulide and the cereulide homolog. Recently, a 0.9-min-longer retention time was observed by RP-HPLC analysis by replacing one O-Leu residue with O-Ile in the otherwise similar 36-membered cyclodepsipeptides bacillistatins 1 and 2, produced by the marine species Bacillus silvestris (51).
Paenilides are the first reported metabolites from the genus Paenibacillus that have shown a high level of mammalian cell toxicity. Paenibacillus was first described as a novel genus with 11 species, distinct from Bacillus, with Paenibacillus polymyxa (formerly Bacillus polymyxa) as the type species (6). The genus has rapidly grown (there are presently >120 validly described species [www.dsmz.de]); novel species have been described mostly from rhizospheres and other plant materials and humus-rich soils (6, 58). Paenibacilli have been shown to produce substances that have antibacterial and/or antifungal properties (12, 41, 42, 63, 66, 67), but mammalian cell toxicity has not been described so far.
Paenilide-producing P. tundrae strain E8a was shown in this paper to be negative in the PCR assay for the cesB gene of the cereulide operon, encoding cereulide synthetase B (16, 55). This finding shows that the paenilides are not products of plasmid pCER270, encoding the biosynthesis of cereulide in B. cereus (16, 28, 55). However, B. cereus isolates that are toxic in the sperm bioassay and that produce a product with a mass fragmentation pattern identical to that of cereulide but with a negative outcome in the assay for the cesB gene have also been reported (29). Since the mass fragmentation patterns of cereulide and paenilide (and their homologs) are identical, it is possible that such strains may have been producers of paenilide.
The results described in this paper show that the toxic action of paenilides is explained by the high affinity of these lipophilic molecules for K+ ions. When bound to K+ ions, paenilides will behave as lipocations, i.e., lipophilic cationic compounds. Lipophilic cationic substances can be expected to migrate from the extracellular fluid across the cytoplasmic membrane into the cell, pulled by the negative charge on the cytoplasmic side. The migration of the K+-charged paenilides was directly demonstrated by the K+-dependent electric conductivity induced by paenilides in a BLM (Fig. 7). Intracellularly, lipocations are known to accumulate inside the mitochondrion, where the negative charges are highest (45). In the present study, this was visible as a depolarization of the mitochondrial membrane potential along with a hyperpolarization of the plasma membrane (Table 1). In the mitochondrion, the toxicity target of paenilides was shown to be the disruption of the mitochondrial respiratory control that led to the uncoupling of oxidative phosphorylation with a concomitant acceleration of glucose consumption, possibly due to the acceleration of glycolysis in the cytoplasm needed for sustained ATP production. Paenilides already became saturated with K+ at [K+] concentrations of 4 mM, i.e., at blood plasma concentrations of K+. These findings indicate that paenilides contained in food may accumulate from the gut into the blood plasma and subsequently become transported to cells and tissues, similarly to cereulide (33, 53, 59).
ACKNOWLEDGMENTS
This work was supported by grants from the Academy of Finland (Center of Excellence Photobiomics grant number 118637), The Finnish Work Environment Fund (grant number 111084), and the Graduate School for Applied Biosciences (ABS).
We acknowledge Marika Jestoi (EVIRA) for collaboration. We thank the Helsinki University Viikki Science Library for excellent information services, the Faculty of Agriculture and Forestry Instrument Centre for technical support, and Leena Steininger, Hannele Tukiainen, and Tuula Suortti for many kinds of help.
Footnotes
Published ahead of print 9 March 2012
REFERENCES
- 1. Andersson MA, et al. 2004. Sperm bioassay for rapid detection of cereulide-producing Bacillus cereus in food and related environments. Int. J. Food Microbiol. 94:175–183 doi:10.1016/j.ijfoodmicro.2004.01.018 [DOI] [PubMed] [Google Scholar]
- 2. Andersson MA, et al. 2010. Boar spermatozoa as a biosensor for detecting toxic substances in indoor dust and aerosols. Toxicol. In Vitro 24:2041–2052 doi:10.1016/j.tiv.2010.08.011 [DOI] [PubMed] [Google Scholar]
- 3. Antunez K, Anido M, Evans JD, Zunino P. 2010. Secreted and immunogenic proteins produced by the honeybee bacterial pathogen, Paenibacillus larvae. Vet. Microbiol. 141:385–389 doi:10.1016/j.vetmic.2009.09.006 [DOI] [PubMed] [Google Scholar]
- 4. Apetroaie-Constantin C, et al. 2009. Bacillus subtilis and B. mojavensis strains connected to food poisoning produce the heat stable toxin amylosin. J. Appl. Microbiol. 106:1976–1985 doi:10.1111/j.1365-2672.2009.04167.x [DOI] [PubMed] [Google Scholar]
- 5. Apetroaie-Constantin C, et al. 2008. Environment driven cereulide production by emetic strains of Bacillus cereus. Int. J. Food Microbiol. 127:60–67 doi:10.1016/j.ijfoodmicro.2008.06.006 [DOI] [PubMed] [Google Scholar]
- 6. Ash C, Priest FG, Collins MD. 1993. Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Proposal for the creation of a new genus Paenibacillus. Antonie Van Leeuwenhoek 64:253–260 [DOI] [PubMed] [Google Scholar]
- 7. Atlas RM. 1993. Handbook of microbiological media. CRC Press, Boca Raton, FL [Google Scholar]
- 8. Carlin F, et al. 2000. Spore-forming bacteria in commercial cooked, pasteurised and chilled vegetable purees. Food Microbiol. 17:153–165 doi:10.1006/fmic.1999.0299 [Google Scholar]
- 9. Chan QW, Melathopoulos AP, Pernal SF, Foster LJ. 2009. The innate immune and systemic response in honey bees to a bacterial pathogen, Paenibacillus larvae. BMC Genomics 10:387 doi:10.1186/1471-2164-10-387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. De Jonghe V, et al. 2010. Toxinogenic and spoilage potential of aerobic spore-formers isolated from raw milk. Int. J. Food Microbiol. 136:318–325 doi:10.1016/j.ijfoodmicro.2009.11.007 [DOI] [PubMed] [Google Scholar]
- 11. Delbrassinne L, et al. 2011. Follow-up of the Bacillus cereus emetic toxin production in penne pasta under household conditions using liquid chromatography coupled with mass spectrometry. Food Microbiol. 28:1105–1109 doi:10.1016/j.fm.2011.02.014 [DOI] [PubMed] [Google Scholar]
- 12. Deng Y, et al. 2011. Isolation and characterization of peptide antibiotics LI-F04 and polymyxin B(6) produced by Paenibacillus polymyxa strain JSa-9. Peptides 32:1917–1923 doi:10.1016/j.peptides.2011.08.004 [DOI] [PubMed] [Google Scholar]
- 13. Dierick K, et al. 2005. Fatal family outbreak of Bacillus cereus-associated food poisoning. J. Clin. Microbiol. 43:4277–4279 doi:10.1128/JCM.43.8.4277-4279.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Echard G. 1974. Chromosomal banding patterns and karyotype evolution in three pig kidney cell strains (PK15, F and RP). Chromosoma 45:133–149 [DOI] [PubMed] [Google Scholar]
- 15. Edwards U, Rogall T, Blocker H, Emde M, Bottger EC. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843–7853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ehling-Schulz M, et al. 2006. Cereulide synthetase gene cluster from emetic Bacillus cereus: structure and location on a mega virulence plasmid related to Bacillus anthracis toxin plasmid pXOI. BMC Microbiol. 6:20 doi:10.1186/1471-2180-6-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ehling-Schulz M, Fricker M, Scherer S. 2004. Identification of emetic toxin producing Bacillus cereus strains by a novel molecular assay. FEMS Microbiol. Lett. 232:189–195 doi:10.1016/S0378-1097(04)00066-7 [DOI] [PubMed] [Google Scholar]
- 18. Elo S, Maunuksela L, Salkinoja-Salonen M, Smolander A, Haahtela K. 2000. Humus bacteria of Norway spruce stands: plant growth promoting properties and birch, red fescue and alder colonizing capacity. FEMS Microbiol. Ecol. 31:143–152 [DOI] [PubMed] [Google Scholar]
- 19. Fangio MF, Roura SI, Fritz R. 2010. Isolation and identification of Bacillus spp. and related genera from different starchy foods. J. Food Sci. 75:M218–M221 doi:10.1111/j.1750-3841.2010.01566.x [DOI] [PubMed] [Google Scholar]
- 20. Fricker M, Reissbrodt R, Ehling-Schulz M. 2008. Evaluation of standard and new chromogenic selective plating media for isolation and identification of Bacillus cereus. Int. J. Food Microbiol. 121:27–34 doi:10.1016/j.ijfoodmicro.2007.10.012 [DOI] [PubMed] [Google Scholar]
- 21. Fromm HI, Boor KJ. 2004. Characterization of pasteurized fluid milk shelf-life attributes. J. Food Sci. 69:M207–M214 [Google Scholar]
- 22. Girardin H, Albagnac C, Dargaignaratz C, Nguyen-The C, Carlin F. 2002. Antimicrobial activity of foodborne Paenibacillus and Bacillus spp. against Clostridium botulinum. J. Food Prot. 65:806–813 [DOI] [PubMed] [Google Scholar]
- 23. Guinebretiere MH, et al. 2001. Identification of bacteria in pasteurized zucchini purees stored at different temperatures and comparison with those found in other pasteurized vegetable purees. Appl. Environ. Microbiol. 67:4520–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Häggblom MM, Apetroaie C, Andersson MA, Salkinoja-Salonen MS. 2002. Quantitative analysis of cereulide, the emetic toxin of Bacillus cereus, produced under various conditions. Appl. Environ. Microbiol. 68:2479–2483 doi:10.1128/AEM.68.5.2479-2483.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hansen BM, Leser TD, Hendriksen NB. 2001. Polymerase chain reaction assay for the detection of Bacillus cereus group cells. FEMS Microbiol. Lett. 202:209–213 [DOI] [PubMed] [Google Scholar]
- 26. He Z, et al. 2007. Isolation and identification of a Paenibacillus polymyxa strain that coproduces a novel lantibiotic and polymyxin. Appl. Environ. Microbiol. 73:168–178 doi:10.1128/AEM.02023-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hoornstra D, Andersson MA, Mikkola R, Salkinoja-Salonen MS. 2003. A new method for in vitro detection of microbially produced mitochondrial toxins. Toxicol. In Vitro 17:745–751 [DOI] [PubMed] [Google Scholar]
- 28. Hoton FM, Andrup L, Swiecicka I, Mahillon J. 2005. The cereulide genetic determinants of emetic Bacillus cereus are plasmid-borne. Microbiology 151:2121–2124 doi:10.1099/mic.0.28069-0 [DOI] [PubMed] [Google Scholar]
- 29. Hoton FM, et al. 2009. Family portrait of Bacillus cereus and Bacillus weihenstephanensis cereulide-producing strains. Environ. Microbiol. Rep. 1:177–183 doi:10.1111/j.1758-2229.2009.00028.x [DOI] [PubMed] [Google Scholar]
- 30. Huck JR, Sonnen M, Boor KJ. 2008. Tracking heat-resistant, cold-thriving fluid milk spoilage bacteria from farm to packaged product. J. Dairy Sci. 91:1218–1228 doi:10.3168/jds.2007-0697 [DOI] [PubMed] [Google Scholar]
- 31. Huck JR, Woodcock NH, Ralyea RD, Boor KJ. 2007. Molecular subtyping and characterization of psychrotolerant endospore-forming bacteria in two New York State fluid milk processing systems. J. Food Prot. 70:2354–2364 [DOI] [PubMed] [Google Scholar]
- 32. Hunt ME, Rice EW. 2005. Microbiological examination, p 9-35–9-52 In Eaton AD, Clesceri LS, Rice EW, Greenberg AE. (ed), Standard methods for the examination of water and wastewater, 21st ed Port City Press, Baltimore, MD [Google Scholar]
- 33. Ichikawa K, et al. 2010. Acute encephalopathy of Bacillus cereus mimicking Reye syndrome. Brain Dev. 32:688–690 doi:10.1016/j.braindev.2009.09.004 [DOI] [PubMed] [Google Scholar]
- 34. Izumi H, Anderson IC, Killham K, Moore ERB. 2008. Diversity of predominant endophytic bacteria in European deciduous and coniferous trees. Can. J. Microbiol. 54:173–179 doi:10.1139/W07-134 [DOI] [PubMed] [Google Scholar]
- 35. Jääskeläinen EL, et al. 2003. In vitro assay for human toxicity of cereulide, the emetic mitochondrial toxin produced by food poisoning Bacillus cereus. Toxicol. In Vitro 17:737–744 [DOI] [PubMed] [Google Scholar]
- 36. Jestoi M, et al. 2004. Presence and concentrations of the Fusarium-related mycotoxins beauvericin, enniatins and moniliformin in Finnish grain samples. Food Addit. Contam. 21:794–802 doi:10.1080/02652030410001713906 [DOI] [PubMed] [Google Scholar]
- 37. Johnson D, Lardy H. 1967. Isolation of liver or kidney mitochondria. Methods Enzymol. 10:94–96 [Google Scholar]
- 38. Kamo N, Muratsugu M, Hongoh R, Kobatake Y. 1979. Membrane potential of mitochondria measured with an electrode sensitive to tetraphenyl phosphonium and relationship between proton electrochemical potential and phosphorylation potential in steady state. J. Membr. Biol. 49:105–121 [DOI] [PubMed] [Google Scholar]
- 39. Lal S, Tabacchioni S. 2009. Ecology and biotechnological potential of Paenibacillus polymyxa: a minireview. Indian J. Microbiol. 49:2–10 doi:10.1007/s12088-009-0008-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Logan NA, et al. 2004. Paenibacillus cineris sp. nov. and Paenibacillus cookii sp. nov., from Antarctic volcanic soils and a gelatin-processing plant. Int. J. Syst. Evol. Microbiol. 54:1071–1076 doi:10.1099/ijs.0.02967-0 [DOI] [PubMed] [Google Scholar]
- 41. Ma M, et al. 2011. Complete genome sequence of Paenibacillus polymyxa SC2, a strain of plant growth-promoting rhizobacterium with broad-spectrum antimicrobial activity. J. Bacteriol. 193:311–312 doi:10.1128/JB.01234-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mageshwaran V, Walia S, Govindasamy V, Annapurna K. 2011. Antibacterial activity of metabolite produced by Paenibacillus polymyxa strain HKA-15 against Xanthomonas campestris pv. phaseoli. Indian J. Exp. Biol. 49:229–233 [PubMed] [Google Scholar]
- 43. Mahler H, et al. 1997. Fulminant liver failure in association with the emetic toxin of Bacillus cereus. N. Engl. J. Med. 336:1142–1148 doi:10.1056/NEJM199704173361604 [DOI] [PubMed] [Google Scholar]
- 44. McFaddin JF. 2000. Biochemical tests for identification of medical bacteria. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 45. Mehta R, Chan K, Lee O, Tafazoli S, O'Brien PJ. 2008. Drug-associated mitochondrial toxicity, p 71–126 In Dykens JA, Will Y. (ed), Drug-induced mitochondrial dysfunction. John Wiley & Sons Inc, Hoboken, NJ [Google Scholar]
- 46. Mikkola R, Saris NE, Grigoriev PA, Andersson MA, Salkinoja-Salonen MS. 1999. Ionophoretic properties and mitochondrial effects of cereulide—the emetic toxin of B. cereus. Eur. J. Biochem. 263:112–117 [DOI] [PubMed] [Google Scholar]
- 47. Naghmouchi K, et al. 2011. Paenibacillus polymyxa JB05-01-1 and its perspectives for food conservation and medical applications. Arch. Microbiol. 193:169–177 doi:10.1007/s00203-010-0654-9 [DOI] [PubMed] [Google Scholar]
- 48. Naranjo M, et al. 2011. Sudden death of a young adult associated with Bacillus cereus food poisoning. J. Clin. Microbiol. 49:4379–4381 doi:10.1128/JCM.05129-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nelson DM, Glawe AJ, Labeda DP, Cann IK, Mackie RI. 2009. Paenibacillus tundrae sp. nov. and Paenibacillus xylanexedens sp. nov., psychrotolerant, xylan-degrading bacteria from Alaskan tundra. Int. J. Syst. Evol. Microbiol. 59:1708–1714 doi:10.1099/ijs.0.004572-0 [DOI] [PubMed] [Google Scholar]
- 50. Ouyang J, et al. 2008. Paenibacillus thiaminolyticus: a new cause of human infection, inducing bacteremia in a patient on hemodialysis. Ann. Clin. Lab. Sci. 38:393–400 [PMC free article] [PubMed] [Google Scholar]
- 51. Pettit GR, et al. 2009. Antineoplastic agents. 570. Isolation and structure elucidation of bacillistatins 1 and 2 from a marine Bacillus silvestris. J. Nat. Prod. 72:366–371 doi:10.1021/np800603u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pitchayawasin S, et al. 2004. Molecular diversity of cereulide detected by means of nano-HPLC-ESI-Q-TOF-MS. Int. J. Mass Spectrom. 235:123–129 doi:10.1016/j.ijms.2004.04.007 [Google Scholar]
- 53. Pósfay-Barbe KM, et al. 2008. Food poisoning as a cause of acute liver failure. Pediatr. Infect. Dis. J. 27:846–847 doi:10.1097/INF.0b013e318170f2ae [DOI] [PubMed] [Google Scholar]
- 54. Rasimus S, Kolari M, Rita H, Hoornstra D, Salkinoja-Salonen M. 2011. Biofilm-forming bacteria with varying tolerance to peracetic acid from a paper machine. J. Ind. Microbiol. Biotechnol. 38:1379–1390 doi:10.1007/s10295-010-0921-4 [DOI] [PubMed] [Google Scholar]
- 55. Rasko DA, et al. 2007. Complete sequence analysis of novel plasmids from emetic and periodontal Bacillus cereus isolates reveals a common evolutionary history among the B. cereus-group plasmids, including Bacillus anthracis pXO1. J. Bacteriol. 189:52–64 doi:10.1128/JB.01313-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Roux V, Raoult D. 2004. Paenibacillus massiliensis sp. nov., Paenibacillus sanguinis sp. nov. and Paenibacillus timonensis sp. nov., isolated from blood cultures. Int. J. Syst. Evol. Microbiol. 54:1049–1054 doi:10.1099/ijs.0.02954-0 [DOI] [PubMed] [Google Scholar]
- 57. Shiota M, et al. 2010. Rapid detoxification of cereulide in Bacillus cereus food poisoning. Pediatrics 125:e951–e955 doi:10.1542/peds.2009-2319 [DOI] [PubMed] [Google Scholar]
- 58. Sirota-Madi A, et al. 2010. Genome sequence of the pattern forming Paenibacillus vortex bacterium reveals potential for thriving in complex environments. BMC Genomics 11:710 doi:10.1186/1471-2164-11-710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Teplova VV, Mikkola R, Tonshin AA, Saris NE, Salkinoja-Salonen MS. 2006. The higher toxicity of cereulide relative to valinomycin is due to its higher affinity for potassium at physiological plasma concentration. Toxicol. Appl. Pharmacol. 210:39–46 doi:10.1016/j.taap.2005.06.012 [DOI] [PubMed] [Google Scholar]
- 60. Teplova VV, Tonshin AA, Grigoriev PA, Saris NE, Salkinoja-Salonen MS. 2007. Bafilomycin A1 is a potassium ionophore that impairs mitochondrial functions. J. Bioenerg. Biomembr. 39:321–329 doi:10.1007/s10863-007-9095-9 [DOI] [PubMed] [Google Scholar]
- 61. Thorsen L, Budde BB, Henrichsen L, Martinussen T, Jakobsen M. 2009. Cereulide formation by Bacillus weihenstephanensis and mesophilic emetic Bacillus cereus at temperature abuse depends on pre-incubation conditions. Int. J. Food Microbiol. 134:133–139 doi:10.1016/j.ijfoodmicro.2009.03.023 [DOI] [PubMed] [Google Scholar]
- 62. Thorsen L, et al. 2006. Characterization of emetic Bacillus weihenstephanensis, a new cereulide-producing bacterium. Appl. Environ. Microbiol. 72:5118–5121 doi:10.1128/AEM.00170-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Trivedi P, Spann T, Wang N. 2011. Isolation and characterization of beneficial bacteria associated with citrus roots in Florida. Microb. Ecol. 62:324–336 doi:10.1007/s00248-011-9822-y [DOI] [PubMed] [Google Scholar]
- 64. Tupinamba G, et al. 2008. Antimicrobial activity of Paenibacillus polymyxa SCE2 against some mycotoxin-producing fungi. J. Appl. Microbiol. 105:1044–1053 doi:10.1111/j.1365-2672.2008.03844.x [DOI] [PubMed] [Google Scholar]
- 65. Wang GYS, Kuramoto M, Yamada K, Yazawa K, Uemura D. 1995. Homocereulide, an extremely potent cytotoxic depsipeptide from the marine bacterium Bacillus cereus. Chem. Lett. 24:791–792 doi:10.1246/cl.1995.791 [Google Scholar]
- 66. Wu XC, et al. 2011. Paenimacrolidin, a novel macrolide antibiotic from Paenibacillus sp. F6-B70 active against methicillin-resistant Staphylococcus aureus. Microb. Biotechnol. 4:491–502 doi:10.1111/j.1751-7915.2010.00201.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wu XC, et al. 2010. Isolation and partial characterization of antibiotics produced by Paenibacillus elgii B69. FEMS Microbiol. Lett. 310:32–38 doi:10.1111/j.1574-6968.2010.02040.x [DOI] [PubMed] [Google Scholar]