Abstract
Production of the antibiotic tropodithietic acid (TDA) depends on the central phenylacetate catabolic pathway, specifically on the oxygenase PaaABCDE, which catalyzes epoxidation of phenylacetyl-coenzyme A (CoA). Our study was focused on genes of the upper part of this pathway leading to phenylacetyl-CoA as precursor for TDA. Phaeobacter gallaeciensis DSM 17395 encodes two genes with homology to phenylacetyl-CoA ligases (paaK1 and paaK2), which were shown to be essential for phenylacetate catabolism but not for TDA biosynthesis and phenylalanine degradation. Thus, in P. gallaeciensis another enzyme must produce phenylacetyl-CoA from phenylalanine. Using random transposon insertion mutagenesis of a paaK1-paaK2 double mutant we identified a gene (ior1) with similarity to iorA and iorB in archaea, encoding an indolepyruvate:ferredoxin oxidoreductase (IOR). The ior1 mutant was unable to grow on phenylalanine, and production of TDA was significantly reduced compared to the wild-type level (60%). Nuclear magnetic resonance (NMR) spectroscopic investigations using 13C-labeled phenylalanine isotopomers demonstrated that phenylalanine is transformed into phenylacetyl-CoA by Ior1. Using quantitative real-time PCR, we could show that expression of ior1 depends on the adjacent regulator IorR. Growth on phenylalanine promotes production of TDA, induces expression of ior1 (27-fold) and paaK1 (61-fold), and regulates the production of TDA. Phylogenetic analysis showed that the aerobic type of IOR as found in many roseobacters is common within a number of different phylogenetic groups of aerobic bacteria such as Burkholderia, Cupriavidis, and Rhizobia, where it may also contribute to the degradation of phenylalanine.
INTRODUCTION
Due to their metabolic versatility as well as high abundances in marine habitats, the Roseobacter clade represents one of the most important groups of marine bacteria (5, 6, 52). Organisms of the group can utilize a multitude of organic compounds, including carbohydrates, sugar alcohols, organic acids, and amino acids. Many roseobacters are also capable of using aromatic compounds as sole carbon and energy sources (6, 30), which constitute the second most widespread class of organic substrates after carbohydrates. These findings are in accordance with genomic analyses of roseobacters, which revealed a surprisingly high number of pathways for catabolism of structurally diverse aromatic substrates (29, 33, 55). A heterotrophic generalist of this group with a wide substrate spectrum is Phaeobacter gallaeciensis (25). The genus Phaeobacter has received strong interest due to the ability of some species to produce the antibiotic tropodithietic acid (TDA), including our model organism P. gallaeciensis DSM 17395 (3). TDA is a structurally unique sulfur-containing compound with a seven-membered aromatic tropone ring fused to a dithiet moiety, which inhibits growth of marine pathogens such as Vibrio anguillarum (36). We recently showed that TDA biosynthesis in P. gallaeciensis DSM 17395 is regulated by the PgaI-PgaR quorum-sensing system (3).
A substantial part of the aromatic compounds is metabolized by bacteria via the phenylacetyl-coenzyme A (CoA) pathway, such as phenylalanine, phenylacetate, lignin-related aromatic compounds, 2-phenylethylamine, phenylalkanoic acids with an even number of carbon atoms, or even environmental contaminants such as styrene and ethylbenzene (10, 22, 31, 50). Degradation of these molecules is carried out through a large number of peripheral pathways that catalyze the transformation into either phenylacetate or phenylacetyl-CoA, which are catabolized in the central phenylacetate degradation pathway (22). Phenylacetate is activated by the phenylacetyl-CoA ligase (PhAc-CoALs) to phenylacetyl-CoA, the first common intermediate of the phenylacetate pathway (13, 19), and it was recently discovered that all further intermediates are processed as CoA thioesters throughout the phenylacetyl-CoA pathway (50). Phenylacetyl-CoA is the substrate of the multicomponent oxygenase PaaABCDE, which catalyzes the 1,2-epoxidation of the aromatic ring of phenylacetyl-CoA to ring-1,2-epoxyphenylacetyl-CoA (50). These oxygenases form the key multienzyme complex of the central phenylacetyl-CoA pathway (50) that is known to be essential for the catabolism of phenylalanine as well as the synthesis of TDA in Silicibacter sp. strain TM1040 (15). Although the central phenylacetate pathway is well understood (50), nothing is known about genes of the upper pathway leading to phenylacetate or phenylacetyl-CoA, especially in regard to the biosynthesis of TDA in P. gallaeciensis. In this study, we investigated which genes of the upper phenylacetate pathway are involved in the transformation of phenylalanine to phenylacetyl-CoA and therefore in the biosynthesis of TDA. Prior to our investigations, the most likely pathway from phenylalanine to phenylacetyl-CoA seemed to include the initial transamination to phenylpyruvate, its oxidative decarboxylation to phenylacetate (14), and the subsequent activation to phenylacetyl-CoA by a phenylacetyl-CoA ligase (13, 19), which was to our knowledge the only known pathway for bacteria. This assumption was confirmed by the finding that the biosynthesis of the related volatile compound tropone in P. gallaeciensis proceeds from phenylalanine via phenylacetate (51).
In this study, we focused on genes of the upper phenylacetate pathway leading to phenylacetyl-CoA to elucidate the biosynthesis of TDA in P. gallaeciensis DSM 17395. Therefore, we investigated the involvement of the phenylacetyl-CoA ligase in the synthesis of TDA by means of deletion mutants. Furthermore, we identified a gene which is involved in degradation of phenylalanine by P. gallaeciensis and shows similarity to the iorA and iorB genes encoding the indolepyruvate:ferredoxin oxidoreductase (IOR) of archaea.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Strains and plasmids used in this study are listed in Table 1. Phaeobacter gallaeciensis strains were routinely grown on Difco marine broth (MB) 2216 (BD Biosiences, Franklin Lakes, NJ) with shaking at 90 rpm or on a corresponding solid agar medium (17.7 g liter−1 agar) at 28°C (unless indicated otherwise). When required, antibiotics were added to half-strength MB 2216 agar at the following concentrations: 8 μg ml−1 chloramphenicol, 60 μg ml−1 kanamycin sulfate, or 25 μg ml−1 gentamicin sulfate. Escherichia coli strains were grown in Luria-Bertani (LB) medium at 37°C with shaking at 100 rpm or on a corresponding solid agar medium (15 g liter−1 agar). When required, chloramphenicol, kanamycin, or ampicillin was added to a final concentration of 20, 25, or 250 μg ml−1, respectively.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype, phenotype, and/or characteristic(s)a | Source or referenceb |
|---|---|---|
| Strains | ||
| Phaeobacter gallaeciensis strains | ||
| DSM 17395 | Wild-type, antibacterial activity | DSMZ |
| C1 | DSM 17395 ΔpaaK1::Km; Kmr | This study |
| C2 | DSM 17395 ΔpaaK2::Cm; Cmr | This study |
| C3 | DSM 17395 ΔpaaK1::Km ΔpaaK2::Cm; Cmr Kmr | This study |
| WP56 | DSM 17395 ior1::EZTn5; Gmr | This study |
| WP3 | DSM 17395 paaC::EZTn5; Gmr | This study |
| WP69 | DSM 17395 iorR::EZTn5; Gmr | This study |
| WP38 | DSM 17395 pgaI::Gm; Gmr | 3 |
| WP52 | DSM 17395 pgaR:: EZTn5; Gmr | 3 |
| CP12 | C3 ior1::EZTn5; Km Cm Gmr | This study |
| CP51 | C3 paaA::EZTn5; Kmr Cmr Gmr | This study |
| CP65 | C3 paaD::EZTn5; Km Cm Gmr | This study |
| CP33 | C3 paaE::EZTn5; Km Cm Gmr | This study |
| Escherichia coli strains | ||
| DH5α | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | 17 |
| TransforMax EC100D pir-116 | F−mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara-leu)7697 galU galK λ−rpsL (Strr) nupG pir-116(DHFR) | Epicentre, Madison, WI |
| Pseudoalteromonas tunicata | DSM 14096 | DSMZ |
| Plasmids | ||
| pBBR1MCS-2 | Source of kanamycin resistance cassette; Kmr | 20 |
| pBBR1MCS-5 | Source of gentamicin resistance cassette; Gmr | 20 |
| pACYC184 | Source of chloramphenicol resistance cassette; Cmr Tcr | 7 |
| pBluescript KS(+) | Cloning vector; Ampr | MBI Fermentas |
| pBC1a | paaK1 upstream flanking region cloned into pBluescript KS(+); Ampr | This study |
| pBC1b | paaK1 downstream flanking region with the kanamycin cassette cloned into pBC1a; Ampr Kmr | This study |
| pBC2a | paaK2 upstream flanking region cloned into pBluescript KS(+); Ampr | This study |
| pBC2b | paaK2 downstream flanking region with the chloramphenicol cassette cloned into pBC2a; Ampr Kmr | This study |
| EZ-Tn5pMOD3 <R6Kgori/MCS> | Transposon construction vector, Ampr | Epicentre, Madison, WI |
| pMOD3gm | Gentamicin resistance cassette cloned into the SmaI site of EZ-Tn5pMOD3 <R6Kgori/MCS>; Ampr Gmr | 3 |
| pYanni1 | Broad-host-range expression vector; Ampr | 16 |
| pYanni1c | Chloramphenicol resistance cassette cloned into the ScaI site of pYanni1; Cmr Ampr | This study |
| pYanni1IOR | ior1 cloned into pYanni1c; Cmr Ampr | This study |
Amp, ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Tcr, tetracycline resistance; Gmr, gentamicin resistance.
DSMZ, German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany.
Construction of P. gallaeciensis strains C1, C2, and C3.
For deletion of paaK1 (ZP_02144309) and paaK2 (ZP_02146787), primers were designed to amplify chromosomal fragments upstream and downstream of the paaK1 and paaK2 genes. The upstream fragment of the paaK1 gene was amplified from chromosomal DNA of strain DSM 17395 by using the primers 5′-ATCCGCAAGGCCAAGACCGAC-3′ and 5′-CTTCTGACACGGTTTGGCCCGCACCAT-3′, and the upstream fragment of paaK2 was amplified by using the primers 5′-AAGGGCACCATCAGCCCAAC-3′ and 5′-TCCCCTGTCCTGTTGGG-3′. The amplified products were ligated into the SmaI site of pBluescript KS(+) (MBI Fermentas, St. Leon-Rot, Germany), resulting in pBC1a and pBC2a, respectively. The downstream fragment of paaK1 was amplified by using the primers 5′-GGATCCTCTAGAGGCATCCTCCACGCCTTATTC-3′ and 5′-ATTGAGCTCAGATGCACATCCAGCGCATCATTG-3′. The PCR product was subsequently digested with XbaI and XhoI and cloned along with the kanamycin resistance gene (amplified from pBBR1MCS-2 using the primers 5′-TCTAGAAAGCTTCCTTTAGTGAGGGTTAATTGCGC-3′ and 5′-GGATCCTCTAGAGGATGAATGTCAGCTACTGG-3′ and digested with HindIII and XbaI) into the HindIII and XhoI sites of pBC1a, giving rise to plasmid pBC1b. The downstream DNA fragment of paaK2, amplified with the primers 5′-AATGAGGTGCACGTCAGCACCCCGCCTCAGCTC-3′ and 5′-AATGAGCTCGAGGCGTGACTTCCCTGTATTTGC-3′, was digested with ApaLI and XhoI, and the DNA fragment carrying the chloramphenicol resistance gene (amplified from pACYC184 using the primers 5′-ATTGAGGTGCACGAATAAATACCTGTGACGG-3′ and 5′-TATCAAGCTTATTCAGGCGTAGCACCAG-3′ and digested with HindIII and ApaLI) was ligated into pBC2a digested with HindIII and XhoI in a triple ligation mixture, giving rise to plasmid pBC2b. P. gallaeciensis DSM 17395 was transformed by electroporation with the plasmid pBC1b and pBC2b as described by Berger et al. (3), leading to strain C1 (ΔpaaK1::Km) and strain C2 (ΔpaaK2::Cm), respectively. Electroporation of strain C2 with pBC1b and subsequent selection on kanamycin created the double mutant strain C3 (ΔpaaK1::Km ΔpaaK2::Cm) (Table 1).
Transposon mutagenesis and mutant screening.
Random transposon insertions mutants unable to degrade phenylalanine were constructed in P. gallaeciensis strain DSM 17395 and strain C3 by using EZ-Tn5 transposon mutagenesis as described previously (3). Growth of mutants with reduced pigmentation was determined in liquid mineral medium (56) with 5 mM phenylalanine using sterile microtiter plates (Sarstedt, Newton, NC). Two μl of a preculture (MB) were individually inoculated into a 96-well microtiter plate with 200 μl of mineral medium and incubated at 20°C. Selected mutants unable to grow were subsequently screened in liquid mineral medium in triplicates (see below). The locations of the insertions of the transposon were determined by rescue cloning of the candidate mutant as described by Berger et al. (3).
Growth on different carbon sources.
Utilization of sole carbon sources was determined by measuring the optical density at 600 nm (DU520 instrument; Beckman Instruments, Fullerton, CA) in chemically defined mineral medium (56). The carbon compounds tested include phenylacetate (2 mM), phenylpyruvate (5 mM), phenylalanine (5 mM), tyrosine (5 mM), tryptophan (5 mM), and glucose (5 mM). Stock solutions of these substrates (100 mM) were prepared, filter sterilized (0.22-μm-pore-size mixed-cellulose-ester membrane; Carl Roth, Karlsruhe, Germany) and added to the autoclaved mineral medium. P. gallaeciensis DSM 17395 and derived mutants were grown at 28°C on a rotary shaker (90 rpm). Growth on a single carbon source was considered positive after at least two transfers in the same medium.
Antimicrobial activity.
Antimicrobial activity of P. gallaeciensis DSM 17395 and derived mutants was determined by agar diffusion tests as described by Berger et al. (3). Precultures of strain DSM 17395 and derived mutants were grown in 50-ml Erlenmeyer flasks containing 10 ml of mineral medium and transferred twice to fresh medium. Subsequently, cultures were inoculated with 1-ml precultures and grown in 500-ml Erlenmeyer flasks containing 50 ml of mineral medium. All cultures were incubated at 28°C on a rotary shaker (90 rpm) and grown to half-maximal optical density (Table 2) measured at 600 nm (DU520; Beckman Instruments, Fullerton, CA). Activity was expressed as diameter of the inhibitory zone between a paper disk and the bacterial lawn of the target strain. Each sterile-filtered supernatant was analyzed with the agar diffusion test in triplicates.
Table 2.
Half-maximal optical density of cultures of P. gallaeciensis DSM 17395 and derived mutants grown on different sole carbon sources
| Strain | Growth with the indicated sole carbon sourcea |
|||||
|---|---|---|---|---|---|---|
| Glucose |
Phenylalanine |
Phenylacetate |
||||
| Expt. 1 | Expt. 2 | Expt. 1 | Expt. 2 | Expt. 1 | Expt. 2 | |
| DSM 17395 | 0.386 | 0.380 | 0.255 | 0.295 | 0.212 | 0.217 |
| C3 | 0.412 | 0.383 | 0.397 | 0.398 | NG | NG |
| WP56 | 0.41 | 0.41 | NG | NG | 0.23 | 0.245 |
| CP12 | 0.36 | 0.388 | NG | NG | NG | NG |
| CP51 | 0.447 | 0.411 | NG | NG | NG | NG |
Values represent the half-maximal optical density at 600 nm. Experiments were performed in duplicates, and values for both cultures are given. NG, no growth.
Quantification of TDA.
For quantification of TDA, the same sterile-filtered culture fluids as used for determination of the antimicrobial activity (see above) were used, which were kept at −80°C until further analysis. Extraction and high-performance liquid chromatography (HPLC) analysis were performed in duplicates of each 20-ml sample as described previously (3). Extraction, HPLC analysis, and quantification of TDA were carried out by BioViotica Naturstoffe GmbH, Göttingen, Germany.
TDA production in a quorum-sensing-negative background.
P. gallaeciensis DSM 17395 and the derived mutants strains WP38 (pgaI negative) and WP52 (pgaR negative) were grown in chemically defined mineral medium (56). Stock solutions of all carbon sources (alanine, methionine, leucine, aspartate, tryptophan, sodium hexanoate, phenylacetate, phenylalanine, glutamate, pyruvate, Casamino Acids, peptone/yeast extract, glucose, and xylose) were prepared with final concentrations of 100 mM, filter sterilized (0.22-μm-pore-size mixed-cellulose-ester membrane; Carl Roth, Karlsruhe, Germany) and added to the autoclaved mineral medium with a final concentration of 5 mM (for phenylacetate only, a final concentration of 2 mM was used). Precultures and main cultures were grown in 10-ml test tubes at 20°C on a roll mixer (model SP; LTF Labortechnik GmbH & Co. KG, Wasserburg, Germany), each containing 5 ml of mineral medium. Aliquots of 2 ml were collected at the maximal optical density at 600 nm (Table 3) and used for determination of the antimicrobial activity as described above and HSL production as described by Berger et al. (3).
Table 3.
TDA, pigment, and homoserine lactone production of the strains DSM 17395, WP38 (pgaI mutant), and WP52 (pgaR mutant) grown on MB and mineral medium with different sole carbon sources
| Substrate | Strain | OD600a | Activity or growth characteristicb |
||
|---|---|---|---|---|---|
| TDA production | Pigmentation (OD398) | HSL productionc | |||
| MB | DSM 17395 | 0.12 | + | + | + |
| WP38 | 0.12 | − | − | − | |
| WP52 | 0.12 | − | − | + | |
| Peptone + yeast extract | DSM 17395 | 0.18 | + | + | + |
| WP38 | 0.11 | − | − | − | |
| WP52 | 0.10 | − | − | + | |
| Casamino Acids | DSM 17395 | 0.15 | + | + | + |
| WP38 | 0.10 | − | − | − | |
| WP52 | 0.10 | − | − | + | |
| Phenylalanine | DSM 17395 | 0.10 | + | + | + |
| WP38 | 0.11 | + | + | − | |
| WP52 | 0.12 | + | + | + | |
| Phenylacetate | DSM 17395 | 0.05 | + | + | + |
| WP38 | 0.06 | − | − | − | |
| WP52 | 0.05 | − | − | + | |
| Glutamate | DSM 17395 | 0.26 | + | + | + |
| WP38 | 0.23 | − | − | − | |
| WP52 | 0.21 | − | − | + | |
| Pyruvate | DSM 17395 | 0.05 | + | + | + |
| WP38 | 0.05 | − | − | − | |
| WP52 | 0.05 | − | − | + | |
| Glucose | DSM 17395 | 0.22 | + | + | + |
| WP38 | 0.14 | (+) | (+) | - | |
| WP52 | 0.13 | (+) | (+) | + | |
| Xylose | DSM 17395 | 0.13 | + | + | + |
| WP38 | 0.16 | (+) | (+) | - | |
| WP52 | 0.15 | (+) | (+) | + | |
OD600, optical density at 600 nm.
Antimicrobial activity and pigmentation are scored relative to the wild-type strain as follows: +, similar; (+), reduced; −, none.
Homoserine lactone (HSL) production was evidenced as described by Berger et al. (3).
Feeding experiments with 13C-labeled phenylalanine.
A liquid culture of P. gallaeciensis (WP3 or WP56) in half-strength MB 2216 (20 ml), prepared with 100% D2O (99.9% deuterium) instead of H2O and amended with gentamicin sulfate (25 μg ml−1), was grown for 3 days at 28°C and 200 rpm. After the culture was fully grown, [1-13C]phenylalanine, [2-13C]phenylalanine, or [3-13C]phenylalanine was added to a concentration of 1 mm (equivalent to 3.3 mg for a culture volume of 20 ml). The culture was further shaken at 28°C and 200 rpm, and samples (1 ml) were withdrawn after 1 h, 3 h, 6 h, and 24 h. The cells were removed by centrifugation at 13,000 rpm, and the culture supernatant was immediately analyzed by 13C nuclear magnetic resonance (NMR) spectroscopy at 100 MHz.
Complementation of the ior1 mutant.
The ior1 mutant obtained by transposon mutagenesis (see above) was characterized further by reintroducing the wild-type copy of the disrupted gene. The open reading frame (ORF) sequence of ior1 was PCR amplified from chromosomal DNA of P. gallaeciensis DSM 17395 using the primers 5′-GGATCCGAGCTCTTGATGCAAGGGATGTCCCCATG-3′ and 5′-TATCAAGCTTGGCTGATATCAAAGCGTCTCC-3′, digested with EcoICRI and HindIII, and cloned into the SmaI and HindIII sites of pYanni1c, resulting in pYanni1IOR (Table 1). pYanni1c was created prior to that by ligation of the chloramphenicol resistance gene (amplified from pACYC184 [see above]) in the ScaI-site of pYanni1 (16). pYanni1IOR was electroporated into strain WP56. Growth on mineral medium with phenylalanine and antimicrobial activity was determined as described above.
RNA preparation and qRT-PCR.
For isolation of total RNA from cells, the strains DSM 17395 and WP69 were initially grown in 2 ml of mineral medium, from which 200 μl was used as the inoculum of 20 ml of fresh mineral medium in a 100-ml Erlenmeyer flask and incubated at 28°C on a rotary shaker to half-maximal optical density at 600 nm. DSM 17395 was grown on the following carbon sources (the corresponding average and standard deviation of the optical densities of three cultures are given in parenthesis): 5 mM phenylalanine (0.17 ± 0.01), 5 mM glucose (0.32 ± 0.08), 2 mM phenylacetate (0.25 ± 0.01), and both 5 mM glucose supplemented with 1 mM phenylalanine (0.35 ± 0.06). The iorR mutant (WP69) was grown on glucose supplemented with phenylalanine to an optical density of 0.41 ± 0.15. Total RNA was extracted, and quantitative reverse transcription-PCR (qRT-PCR) was performed using a LightCycler 480 as described by Berger et al. (3). For qRT-PCR of paaK1, the primers 5′-CATGGTGACGCCCTCCTA-3′ and 5′-GGGTCCATACCGACCTTGT-3′ (TIB MolBiol, Berlin, Germany) and probe 1 of the Universal ProbeLibrary (Roche Diagnostics GmbH) were used, and for ior1 the primers 5′-GGCTTTGCACAGAAGTTTGG-3′ and 5′-CTCCGGTGTATTGGACAGC-3′ and probe 107 of the Universal ProbeLibrary (Roche Diagnostics GmbH) were used. The values were normalized relative to expression of rpoB (for primers and probe, see reference 3). The reported values show the averages and ranges of three biological replicates, each assayed in triplicate.
Bioinformatic analysis.
Similarity searches were performed using the programs BLAST (1) and FASTA3 (35) against the Swiss-Prot (2), the Refseq (37), and the IMG databases (24). Protein domains were identified using the tools provided by the InterProScan website (38). Phylogenetic analysis was performed with the MEGA4 program suite (49). The evolutionary history was inferred using the minimum-evolution (ME) method (39). The bootstrap consensus tree inferred from 500 replicates is taken to represent the evolutionary history of the taxa analyzed (11). Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The bootstrap values are included in linear versions of the phylogenetic trees in the supplemental material (see Fig. S1 and S2 in the supplemental material). The phylogenetic tree was linearized assuming equal evolutionary rates in all lineages (48). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (57) and are in the units of the number of amino acid substitutions per site. The ME tree was searched using the close-neighbor-interchange (CNI) algorithm (32) at a search level of 1. The neighbor-joining algorithm (40) was used to generate the initial tree. All positions containing gaps and missing data were eliminated from the data set (complete deletion option). There were a total of 922 positions in the final data set.
RESULTS
Relevance of phenylalanine for the production of TDA.
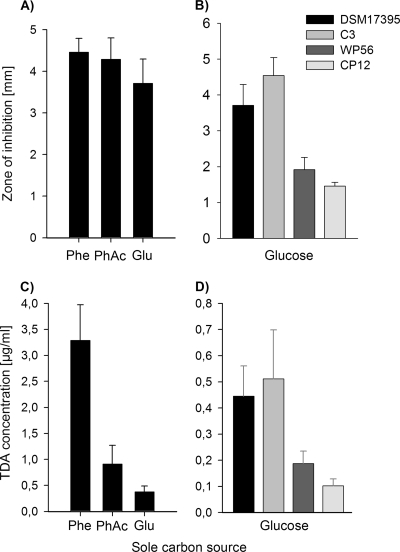
P. gallaeciensis DSM 17395 produces TDA during growth on MB (3). Growth and TDA production also occurred on defined mineral medium with glucose, xylose, glutamate, pyruvate, or different aromatic compounds such as phenylalanine, phenylpyruvate, or phenylacetate as a sole carbon source (Fig. 1A and Tables 3 and 4). TDA quantification by HPLC analysis showed that the level is clearly influenced by the carbon source and that TDA production on aromatic compounds, especially on phenylalanine, is 9-fold enhanced compared to glucose (Fig. 1C). In addition to this carbon source-dependent regulation of TDA, we have previously shown that the biosynthesis of TDA is regulated by N-acyl homoserine lactone (AHL)-mediated quorum sensing when cultures are grown on MB (3). Interestingly, when cultures are grown in mineral medium with phenylalanine as the sole carbon source, the production of TDA is independent from the pgaR-pgaI quorum-sensing system (Table 3).
Fig 1.
Antimicrobial activity and TDA production of the wild-type P. gallaeciensis DSM 17395 and derived mutants C3 (ΔpaaK1::Km ΔpaaK2::Cm), WP56 (ior1::EZTn5), and CP12 (C3 ior1::EZTn5) grown on mineral medium with different sole carbon sources at 28°C. Glu, glucose; Phe, phenylalanine; PhAc, phenylacetate. Antimicrobial activity of cell-free culture fluids was determined in agar diffusion assays in triplicates of the wild type grown in mineral medium with different sole carbon sources (A) and of the wild-type and derived mutants grown on mineral medium with glucose (B). TDA concentration was determined by HPLC analysis of cell-free culture fluids of the wild type grown in mineral medium with different sole carbon sources (C) and of the wild type and mutants grown in mineral medium with glucose (D). The error bars are derived from two parallel cultures determined in each sample in duplicates.
Table 4.
Growth, TDA, and pigment production of strain DSM 17395 and derived mutants on MB and on mineral medium with different substrates
| Strain | Trait | Culture conditiona |
||||
|---|---|---|---|---|---|---|
| MB | Phe | PP | PhAc | Glucose | ||
| DSM 17395 (wild type) | Growth | + | + | + | + | + |
| TDA | + | +* | + | +* | +* | |
| Pigment | + | + | + | + | + | |
| C1 (paaK1 mutant) | Growth | + | + | + | (+) | + |
| TDA | + | + | + | ND | + | |
| Pigment | + | + | + | ND | + | |
| C2 (paaK2 mutant) | Growth | + | + | + | + | + |
| TDA | + | + | + | + | + | |
| Pigment | + | + | + | + | + | |
| C3 (paaK1 paaK2 mutant) | Growth | + | + | + | − | + |
| TDA | + | +* | + | ND | +* | |
| Pigment | + | + | + | ND | + | |
| WP56 (ior1 mutant) | Growth | + | − | − | + | + |
| TDA | (+) | ND | ND | + | (+)* | |
| Pigment | (+) | ND | ND | + | (+) | |
| CP12 (paaK1 paaK2 ior1 mutant) | Growth | + | − | − | − | + |
| TDA | (+) | ND | ND | ND | (+)* | |
| Pigment | (+) | ND | ND | ND | (+) | |
| CP51 (paaK1 paaK2 paaA mutant) | Growth | + | − | − | − | + |
| TDA | − | ND | ND | ND | −* | |
| Pigment | − | ND | ND | ND | − | |
| WP3 (paaC mutant) | Growth | + | − | − | − | + |
| TDA | − | ND | ND | ND | − | |
| Pigment | − | ND | ND | ND | − | |
| WP56(pYanni1IOR) | Growth | + | + | + | ND | ND |
| TDA | + | + | + | ND | ND | |
| Pigment | + | + | + | ND | ND | |
| WP69 (iorR mutant) | Growth | + | − | − | + | + |
| TDA | (+) | ND | ND | ND | (+) | |
| Pigment | (+) | ND | ND | ND | (+) | |
Phe, phenylalanine; PP, phenylpyruvate; PhAc, phenylacetate; ND, not determined or determinable; +, like the wild type; (+), reduced compared to the wild type; −, no growth or production. The asterisk (*) indicates results obtained with cell-free culture fluid tested in agar diffusion assays as well as by HPLC analysis.
Role of phenylacetyl-CoA ligase in P. gallaeciensis.
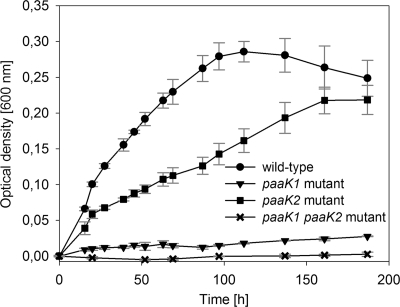
TDA production of the paaA mutant (strain CP51) and the paaC mutant (strain WP3) was completely abolished, as shown by inhibition experiments and/or HPLC analysis (Table 4). Thus, the biosynthesis of TDA is linked to the multicomponent oxygenase PaaABCDE of the central phenylacetate catabolic pathway in P. gallaeciensis, which is in agreement with previous results for Silicibacter sp. TM1040 (20). PaaABCDE uses phenylacetyl-CoA as the substrate (50), which in turn is produced from phenylacetate by the activity of a phenylacetyl-CoA ligase (PhAc-CoAL) (13). Thus, it appeared likely that this enzyme is involved in TDA biosynthesis in P. gallaeciensis. The amino acid sequence of the PhAc-CoAL from E. coli K-12 (accession number P76085) was used for a protein-protein homology search against the genome of DSM 17395. Two proteins were identified in DSM 17395 with 64% identity (PaaK1, ZP_02146787) and 30% identity (PaaK2, ZP_02144309), respectively. To investigate the role of these enzymes in TDA production, we constructed three mutants, i.e., strain C1 (ΔpaaK1::Km), strain C2 (ΔpaaK2::Cm), and the double mutant strain C3 (ΔpaaK1::Km ΔpaaK2::Cm) (Table 1). Subsequently, growth and TDA production of these mutants on different sole carbon sources were compared to levels of the wild type. The paaK1 mutant cultured with phenylacetate showed a strong reduction in growth (Fig. 2). In contrast, growth of the paaK2 mutant was hardly affected, but the double mutant (strain C3) was unable to grow with phenylacetate as the sole carbon source (Fig. 2). This demonstrates that the PhAc-CoAL encoded by the gene paaK1 is the main enzyme of the phenylacetate catabolism and can be only slightly complemented by paaK2. Growth of strains C1, C2, and C3 on phenylalanine or phenylpyruvate was not affected (Table 4), indicating that PhAc-CoAL is not essential for the catabolism of phenylalanine. The double mutant C3 showed no change in antimicrobial activity during growth on MB and mineral medium supplemented with different sole carbon sources other than phenylacetate, i.e., phenylalanine, phenylpyruvate, or glucose, compared to the wild type (Table 4 and Fig. 1B). This is also consistent with the TDA quantification by HPLC analysis, which demonstrated no reduction in TDA synthesis of the double mutant C3 compared to the wild type when grown on glucose as a sole carbon source (Fig. 1D).
Fig 2.
Growth curves of P. gallaeciensis DSM 17395 (wild type) and derived mutants C1 (ΔpaaK1::Km), C2 (ΔpaaK2::Cm), and C3 (ΔpaaK1::Km ΔpaaK2::Cm) with phenylacetate as the sole carbon source. Growth was determined by measuring the optical density at 600 nm in mineral medium at 25°C and 90 rpm. Error bars indicate standard deviations from triplicate trials. No alteration of the optical density was measured in inoculated mineral medium without a carbon source.
Identification of genes of the phenylalanine metabolism involved in TDA production.
The ability of strain C3 to grow on and to produce TDA from phenylalanine or phenylpyruvate suggested the participation of other genes (besides paaK1 and paaK2) coding for enzymes leading to phenylacetyl-CoA. In order to identify these genes, strain DSM 17395 and strain C3 were used for random transposon insertion mutagenesis. On basis of the correlation between TDA production and synthesis of a yellow-brown pigment (4, 15), transformants with no or reduced pigmentation were selected and tested for growth on phenylalanine. This approach yielded several white mutants (WP3, CP33, CP51, and CP65) in which a disruption of one of the multicomponent oxygenase genes paaABCDE was achieved (Table 1). In addition, transposon mutagenesis of the strains DSM 17395 and C3 yielded mutants with reduced pigmentation that were also unable to grow on phenylalanine and phenylpyruvate (WP56 and WP69) or on phenylalanine, phenylpyruvate, and phenylacetate (CP12) (Table 4). In the mutants WP56 and CP12, the transposon interrupted a gene (accession number ZP_02146432) with a length of 3,432 bp located on the chromosome that was subsequently investigated in detail (see below). Analysis of strain WP69 revealed a transposon insertion in the adjacent gene (ZP_02146431) encoding a deduced product with homology to transcriptional regulatory proteins containing a helix-turn-helix DNA-binding domain.
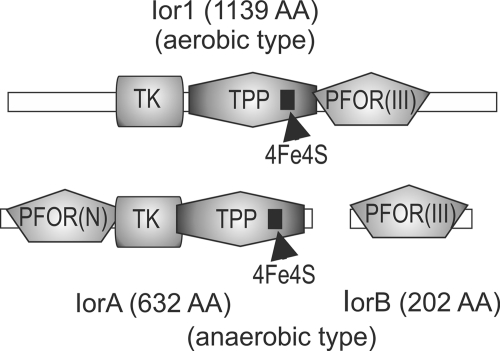
A BLAST search with the deduced protein sequence of ZP_02146432 within the RefSeq database revealed 182 hits which were sorted into two classes, as follows: class 1, hits to proteins (IorA) of strictly and facultatively anaerobic organisms (i.e., clostridia, sulfate-reducing bacteria, and archaea) and, class 2, proteins (Ior1) of aerobic alpha- and betaproteobacteria. The sequences of the proteins were used to construct group-specific hidden Markov models (HMMs) with IorA corresponding to class 1 and Ior1 corresponding to class 2 (the HMMs are available as supporting material). Based on the HMMs, an hmmpfam search on the uniprot database revealed 419 significant hits for the IorA model and 377 hits for the Ior1 model. The results contained five Swiss-Prot entries, including the archaeal indolepyruvate:ferredoxin oxidoreductases (IOR) of Thermococcus kodakaraensis KOD1 (formerly named Pyrococcus sp. strain KOD1), Pyrococcus furiosus, and Methanococcus, which are the only characterized proteins of this class. The archaeal IOR catalyzes the ferredoxin-dependent oxidative decarboxylation of different aryl pyruvates, such as phenylpyruvate, indolepyruvate, and p-hydroxyphenylpyruvate, to their corresponding arylacetyl-CoA derivatives under anaerobic conditions. Analysis of the genomic context revealed that all iorA genes of anaerobic organisms are accompanied by a smaller gene, iorB (Fig. 3). The size of the aerobic Ior protein exceeds the combined sizes of the anaerobic proteins IorA plus IorB. All protein sequences contain the Pfam domains necessary for an oxidative decarboxylation, i.e., a typical ferredoxin-type [4Fe-4S] cluster and a conserved thiamine pyrophosphate (TPP)-binding domain as shown for IOR of archaea (23, 44, 45). An alignment of the two anaerobic proteins with the single aerobic protein based on these Pfam domains showed remarkable similarities. Apparently, the aerobic Ior protein represents a fusion of the anaerobic IorA/IorB proteins (Fig. 3). Based on these similarities of the protein domains, the respective target gene of the transposon mutation of strains WP56 and CP12 (ZP_02146432) has been designated ior1.
Fig 3.
Pfam domain-based alignment of the aerobic and anaerobic type IORs. Corresponding domains are displayed in the same shading; PFOR(N) is described as interaction domain between IorA and IorB. TPP, thiamine pyrophosphate binding domain; TK, transketolase; 4Fe4S, ferredoxin-type cluster; PFOR(III), pyruvate-ferredoxin oxidoreductase domain III superfamily. AA, amino acids.
Phylogenetic analysis of Ior1.
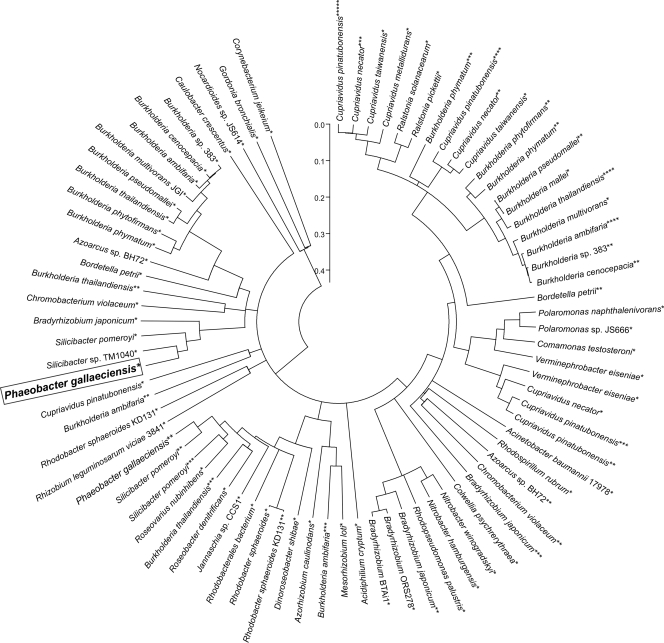
A subset of protein sequences from the database searches was selected from closed genomes and used in a phylogenetic analysis of the two protein families Ior1 and IorA. In total, 79 sequences from 47 strains of the aerobic protein family (Fig. 4; see also Fig. S2 in the supplemental material) and 72 sequences of 46 species of the anaerobic protein family were used in a phylogenetic analysis (see Fig. S1). The closest orthologs of Ior1 were found within the genus Silicibacter (Fig. 4). Many genomes, especially of the Burkholderia and the Cupriavidis genera and some rhizobia, contain paralogous copies of Ior1 from P. gallaeciensis (Fig. 4). Within both protein families the orthologous copies of the proteins from different genomes exhibit more similarities to each other than to the paralogous copies within the same genome (Fig. 4; see also Fig. S1). To investigate the relation of the aerobic and the anaerobic protein families, subsequences of the two Ior paralogs from P. gallaeciensis DSM 17395, which contain the corresponding Pfam domains found in both enzyme classes, have been included in the phylogenetic tree of the anaerobic enzyme type. The two P. gallaeciensis DSM 17395 protein sequences cluster together, indicating that they are more similar to each other than to any of the proteins of the anaerobic organisms (see Fig. S1).
Fig 4.
Evolutionary analysis of 79 proteins of the aerobic Ior1 family from 47 different taxa. The Ior1 from this study is boxed. Paralogous proteins are indicated by a corresponding number of asterisks. The relative evolutionary distance of the nodes is represented by the scale. Accession numbers for the specified protein sequences are provided in the supplemental material.
Effects of ior1 mutation on phenylalanine metabolism and TDA production.
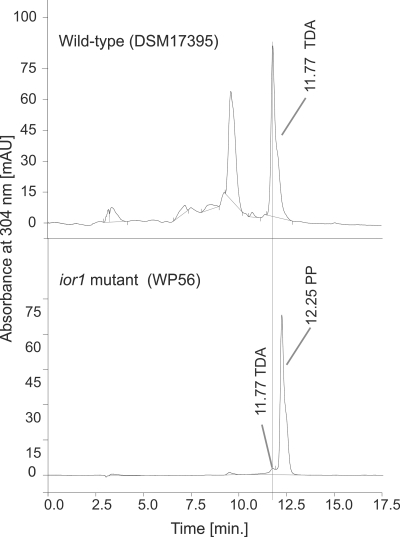
Since the ior1 mutant is deficient in growing on phenylalanine or its transamination product phenylpyruvate (Table 4), the respective gene product, Ior1, must play a crucial role in phenylalanine catabolism. The HPLC chromatogram of culture extracts from the ior1 mutant (WP56) grown in minimal medium with glucose showed an additional peak at a retention time of 12.3 min that was not observed in wild-type extracts (Fig. 5). The respective metabolite was identical to phenylpyruvate in terms of HPLC retention time and UV spectrum, suggesting that the ior1 mutant is unable to catabolize phenylpyruvate. Thus, it is most likely that Ior1 uses phenylpyruvates as substrates, similar to the archaeal IOR (23).
Fig 5.
HPLC chromatograms of ethyl acetate extracts from sterile-filtered culture fluids of the wild type (DSM 17395) and the ior1 mutant (WP56) grown on mineral medium with glucose. The HPLC peak at 11.8 min resulted from TDA, and that at 12.3 min resulted from phenylpyruvate (PP).
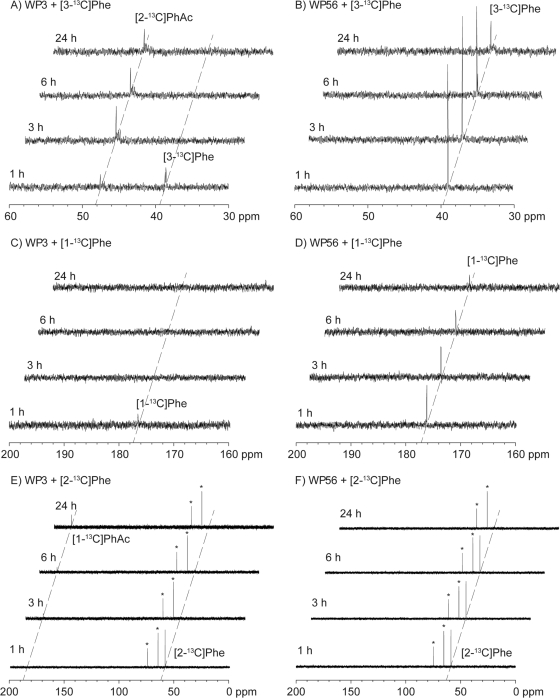
The function of Ior1 was clarified in feeding experiments using isotopically labeled [3-13C]phenylalanine, [1-13C]phenylalanine, and [2-13C]phenylalanine. The paaC mutant (WP3), which is unable to produce TDA or to grow on phenylalanine (Table 4), showed the complete consumption of [3-13C]phenylalanine (chemical shift of 13C-enriched carbon, δ = 39.2 ppm) within the first 3 h after supply (Fig. 6A). The expected initial product [3-13C]phenylpyruvate was not observable, probably due to its rapid conversion into [2-13C]phenylacetyl-CoA by Ior1, which is intact in this mutant. Since the following step of the phenylacetate degradation pathway was blocked by a transposon insertion into paaC, no further catabolism along the phenylacetate degradation pathway was possible, and, consequently, the accumulation of [2-13C]phenylacetate was observed. The enrichment of [2-13C]phenylacetate instead of its CoA derivative is explainable by a limitation of coenzyme A availability, resulting in hydrolysis of the primary product [2-13C]phenylacetyl-CoA to [2-13C]phenylacetate, either by an enzyme-catalyzed reaction or by a spontaneous reaction. In contrast, the ior1 mutant (WP56) did not show any production of [2-13C]phenylacetate, and the consumption of [3-13C]phenylalanine was much slower (Fig. 6B). This is in agreement with a blockage of phenylpyruvate degradation in this mutant (Fig. 5). However, the respective signal for [3-13C]phenylpyruvate was also not visible in this experiment, probably because the amount produced was too low to be detectable under these experimental conditions. Alternative yet unidentified degradation mechanisms may prevent the accumulation of detectable amounts of [3-13C]phenylpyruvate in this mutant, or the accumulating phenylpyruvate may inhibit the further degradation of phenylalanine, giving a reasonable explanation for the much slower degradation of this amino acid in the ior1 mutant (strain WP56) than in the paaC mutant (strain WP3). A feeding experiment with [1-13C]phenylalanine showed that the paaC mutant (strain WP3) degraded [1-13C]phenylalanine within the first 3 h, but since the 13C labeling is lost during the conversion of the transamination product [1-13C]phenylpyruvate into phenylacetate, no product peak was visible (Fig. 6C). In contrast, degradation of [1-13C]phenylpyruvate by the ior1 mutant (strain WP56) was again much slower (Fig. 6D). Feeding of [2-13C]phenylalanine to the WP3 mutant resulted in the rapid depletion of this substrate and the concomitant enrichment of [1-13C]phenylacetate (Fig. 6E), whereas in the same feeding experiment with the WP56 mutant, a much slower degradation of phenylalanine and no significant accumulation of its degradation products became evident (Fig. 6F).
Fig 6.
13C NMR spectra obtained in the feeding experiments with 13C-labeled phenylalanine isotopomers. 13C NMR spectra were recorded from samples withdrawn from liquid cultures 1, 3, 6, and 24 h after addition of 13C-labeled phenylalanine isotopomers to WP3 and WP56, as indicated. Phe, phenylalanine; PhAc, phenylacetate, Asterisks indicate 13C signals of an impurity (glycerol).
All mutants unable to grow on phenylalanine or phenylpyruvate could be grown on glucose, which is a precursor for phenylalanine via the shikimate pathway. Therefore, it was possible to test the mutants indirectly for their ability to make TDA from phenylpyruvate via the shikimate pathway by growing them on its biosynthetic precursor glucose. The ior1 mutant strain WP56 showed reduced antimicrobial activity compared to the wild type when grown on glucose (Fig. 1B). This reduced antibacterial activity was due to a 60% reduced TDA production during growth on glucose compared to growth of the wild type (Fig. 1D). These data confirmed that ior1 is involved in TDA biosynthesis of P. gallaeciensis DSM 17395, but the residual TDA synthesis of the ior1 mutant gave evidence for the activity of an alternative pathway toward TDA.
To investigate whether the gene products of the paaK1 and paaK2 genes were responsible for the residual TDA production of the ior1 mutant (WP56), the triple mutant CP12 (ΔpaaK1::Km ΔpaaK2::Cm ior1::EZTn5) was analyzed. Strain CP12 was unable to grow on phenylalanine, phenylpyruvate, or phenylacetate but grew on MB 2216 or glucose (Table 4). The antimicrobial activity and TDA production of strain CP12 during growth on glucose were significantly reduced compared to levels for the ior1 mutant (ca. 45% reduction) but still not completely abolished (Fig. 1B and D). The deficiency of the ior1 mutant (strain WP56) to catabolize phenylalanine and the reduction in TDA production were fully restored by gene complementation with a plasmid-borne copy of ior1 (pYanni1IOR).
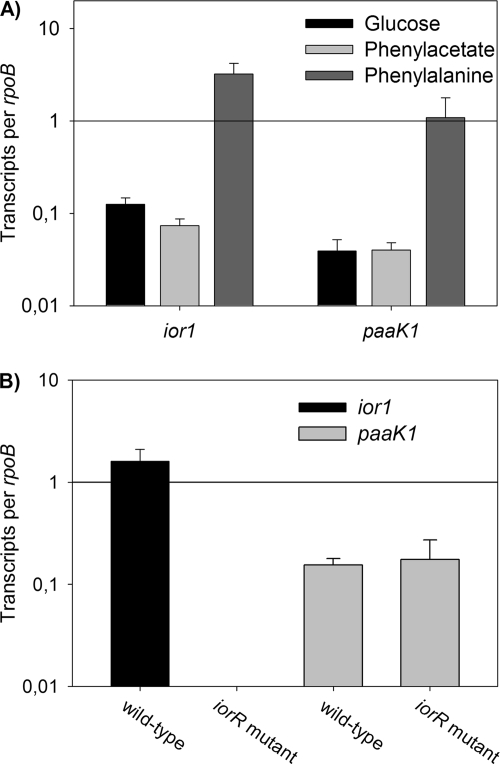
Phenylalanine induces expression of ior1 and paaK1.
In order to study the transcriptional regulation of ior1 and paaK1, qRT-PCR was used. The relative transcription levels of ior1 and paaK1 were analyzed with RNA isolated from DSM 17395 cells grown in mineral medium with phenylalanine, phenylacetate, or glucose. The transcription of ior1 was clearly affected by the carbon source since the transcription level of ior1 was 26-fold higher during growth on phenylalanine than the transcription level of cells grown on glucose, while the transcription levels during growth on phenylacetate and glucose were almost the same (Fig. 7A). Interestingly, nearly the same results were found for the transcription levels of paaK1, indicating that its expression is upregulated not by its native substrate phenylacetate but rather by phenylalanine.
Fig 7.
Transcriptional regulation of ior1 and paaK1. (A) Relative transcription levels of ior1 and paaK1 of the wild-type P. gallaeciensis DSM 17395 grown in mineral medium with glucose, phenylacetate, and phenylalanine as determined using qRT-PCR. (B) Relative transcription levels of ior1 and paaK1 of the wild type (DSM 17395) and the iorR mutant (WP69) grown in mineral medium with 5 mM glucose and 1 mM phenylalanine. The transcription levels are shown as the relative expression level of each target gene compared to that of rpoB. Error bars are derived from three independent cultures, each assayed in triplicates.
Transcription of ior1 depends on iorR.
Strain WP69 has an insertion in the gene ZP_02146431 (iorR). It was obvious that iorR encodes a transcriptional regulator for the adjacent ior1 gene since the iorR mutant showed the same phenotype as the ior1 mutant (Table 4). To confirm this, we compared the transcription level of ior1 of the iorR mutant upon induction with phenylalanine (addition of 1 mM phenylalanine to a culture grown on glucose) to the ior1 transcription level of the wild type by qRT-PCR. In the iorR-negative background, no transcripts of ior1 were detected, suggesting that iorR is essential for the transcription of ior1 (Fig. 7B). In contrast, the transcription level of paaK1 in the iorR mutant was identical to the transcription level of the wild type, indicating that IorR regulates only the expression of ior1.
DISCUSSION
We found that the phenylacetyl-CoA ligase encoded by the gene paaK1 is the main enzyme of the phenylacetate catabolism in P. gallaeciensis strain DSM 17395 and can be only slightly complemented by paaK2 (Fig. 2). This indicates that the PhAc-CoAL encoded by paaK2 acts slowly on phenylacetate and has possibly another, yet unknown, substrate specificity. The phenylacetyl-CoA ligases characterized from other organisms so far are highly specific for phenylacetate and show only low activity with other substrates (10, 26, 28), whereas several phenylacyl-CoA ligases with different substrate specificities for phenylacetic acid, phenylpropanoic acid, or phenylcinnamic acid are present in Pseudomonas putida (53). The phenylacetyl-CoA ligases in DSM 17395 are not essential for the catabolism of phenylalanine and phenylpyruvate since strain C3 (ΔpaaK1::Km ΔpaaK2::Cm) was able to grow on these substrates (Table 4). The multicomponent oxygenase (PaaABCDE), which uses phenylacetyl-CoA as the substrate, however, was shown to be essential for the degradation of phenylalanine, phenylpyruvate, and phenylacetate (Table 4). This demonstrates that degradation of these substrates depends on enzymes of the central phenylacetate pathway (Fig. 8). Production of TDA was also not affected by deletion of paaK1 and paaK2 (Table 4 and Fig. 1D). The transcription of paaK1 was induced when cells were grown on phenylalanine but not during growth on phenylacetate (Fig. 7A). It was shown for E. coli that phenylacetyl-CoA is the inducer for the expression of the phenylacetyl-CoA ligase in its phenylacetate catabolon (13), and this could also explain the induction of paaK1 expression upon phenylalanine addition.
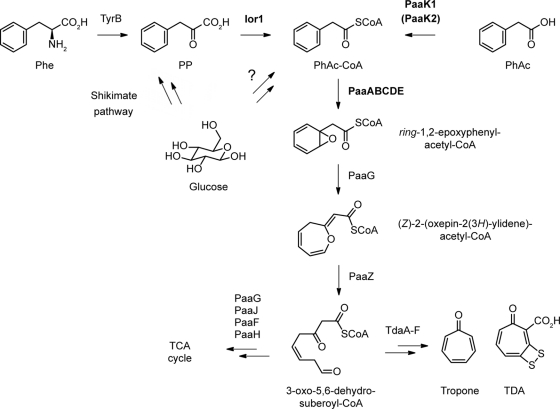
Fig 8.
Degradation of phenylalanine and biosynthesis of TDA in P. gallaeciensis. Phe, phenylalanine; PP, phenylpyruvate; PhAc-CoA, phenylacetyl-CoA; PhAc, phenylacetate; TDA, tropodithietic acid; TCA cycle, tricarboxylic acid cycle. Genes investigated in this study are shown in bold. Unknown reactions with respect to enzyme functions are indicated by question marks.
Due to the inability of the ior1 mutant (WP56) to degrade phenylalanine and phenylpyruvate, it became obvious that no other enzyme is present in strain DSM 17395, which can efficiently catabolize phenylpyruvate under the given conditions. This is also indicated by the accumulation of phenylpyruvate in the culture fluid during growth of the ior1 mutant on phenylalanine (Fig. 5). NMR spectroscopic investigations using 13C-labeled phenylalanine isotopomers demonstrated that this amino acid is transformed into phenylacetyl-CoA by Ior1 (Fig. 6 and 8). The transamination of phenylalanine to phenylpyruvate could be catalyzed by TyrB (ZP_02144572), which shows similarity to the aromatic amino acid aminotransferase of archaea. A catabolic route via free phenylacetate as described for Thauera aromatica (41) or Aromatoleum aromaticum EbN1 (54), however, does not exist in strain DSM 17395. This is also supported by a lack of genes in the genome of strain DSM 17395 encoding a phenylpyruvate decarboxylase, such as pdc of strain EbN1 (54) or ipdC of Azospirillum brasilense (46). Growth of the ior1 mutant on tyrosine and tryptophan indicated that ior1 is not essential for the catabolism of other aromatic amino acids, but this does not exclude the possibility that Ior1 can convert other aryl pyruvates such as indolepyruvate and p-hydroxyphenylpyruvate, like the archaeal IOR (23). The paralogous protein Ior2 in P. gallaeciensis is the most likely candidate involved in the catabolism of the aromatic amino acids tyrosine and/or tryptophan.
In addition to the essential function of Ior1 in the catabolism of phenylalanine, the enzyme plays a considerable role in the production of TDA since the TDA production decreased by 60% in an ior1-negative background (Fig. 1D). In contrast, no reduced TDA production was observed for strain C3 (ΔpaaK1::Km ΔpaaK2::Cm). Thus, it is obvious that the biosynthesis of TDA proceeds via phenylacetyl-CoA, mainly produced by the activity of Ior1 (Fig. 8). However, another thus far unknown enzyme producing phenylacetyl-CoA must exist, which allows the residual TDA synthesis in an ior1 paaK1 paaK2 mutant background. For P. putida production of phenylacetyl-CoA from other substrates, e.g., phenylalkanoic acids with an even number of carbon atoms, was described (34). This is also possible for P. gallaeciensis due to the presence of different AMP-dependent ligases of unknown substrate specificity in the genome (e.g., paaK2, ZP_02146955, or ZP_02146795). However, the next step, the ring oxidation by the multicomponent oxygenase encoded by paaABCDE (12), is critical for TDA production, as was shown by the disruption of paaA and paaC (Table 4). This is in agreement with data obtained for Silicibacter sp. strain TM1040 (15) and shows that the biosynthesis of TDA depends on the central phenylacetate catabolic pathway (paaABCDE). The phenylacetate catabolic pathway is probably also involved in the biosynthesis of other secondary metabolites in P. gallaeciensis, as proposed for production of the recently described roseobacticides, which are likely synthesized from the precursors tropone and phenylacetyl-CoA (43). Thus, the supply of precursors from the phenylacetyl-CoA pathway for the biosynthesis of different secondary metabolites underlines the importance of this pathway for the success of these roseobacters.
Based on the phylogenetic analysis, it is tempting to speculate on the history of the protein class Ior. The data might reflect a two-step evolution beginning within the anaerobic enzyme class, which is, considering the sequence similarities and the taxonomic distance of the harboring genomes, more diverse than the aerobic enzyme family. Starting points are genes encoding the two anaerobic proteins IorA and IorB. In the first step, the corresponding genes might have been distributed via horizontal gene transfer (HGT) among a set of completely diverse amino acid-degrading anaerobic organisms, including the ancestors of the archaea analyzed here, proteobacteria and clostridia. It is known that under anoxic conditions, ferredoxin-reducing decarboxylation reactions might contribute to the energy metabolism of the species hosting the iorAB operons (18). HGT of energy-conserving enzymes between archaea and bacteria has been observed in such archaea as Methanosarcina mazei GOE1 (9) and bacteria as Desulfobacterium autotrophicum HRM2 (47). In the second step, in one of the facultatively anaerobic strains of the alpha- or betaproteobacteria, a fusion event of iorA and iorB occurred, resulting in the ancestor of ior1. Fusions of proteins carrying functional domains have been suggested, for instance, in energy conserving enzymes as heterodisulfide reductase (Hdr) and hydrogenase (MvhD) in D autotrophicum HRM2 (47). The ior gene finally was subject to gene duplications which generated the ancestors of the observed paralogous genes, which afterwards were distributed into the aerobic strains. One of these proteins finally evolved into Ior1 and adapted to the role investigated here in TDA synthesis and phenylalanine metabolism.
Phenylalanine seems to play an important role in the regulation of TDA, which was illustrated by the fact that TDA production was regulated independently from the PgaR-PgaI quorum-sensing system when strain DSM 17395 was grown on phenylalanine as a carbon source (Table 3). The question of how phenylalanine leads to an independency of quorum sensing control of TDA production remains currently unanswered. However, in several bacteria the production of secondary metabolites is regulated hierarchically (21, 27, 42). Thus, it is possible that the phenylalanine-dependent regulation acts at a different level in a regulation cascade than the cell density-dependent regulation to affect the production of TDA. Furthermore, phenylalanine drastically promoted TDA production in P. gallaeciensis (Fig. 1C). The effect of stimulation of secondary metabolite production could be due to the increasing amount of a limiting precursor, the induction of one or more biosynthetic genes of the biosynthesis pathway of a secondary metabolite, or both (8). The increasing amount of phenylacetyl-CoA formed from phenylalanine via ior1 subsequently stimulates TDA biosynthesis. This is only possible because phenylalanine induces the expression of ior1 (Fig. 7A), which leads to an increase in the limiting primary metabolite, phenylacetyl-CoA, for biosynthesis of TDA. Phenylacetyl-CoA is produced from phenylpyruvate by the activity of Ior1, from phenylacetate by the activity of a phenylacetyl-CoA ligase (PaaK1), and from a further unknown pathway (Fig. 8). Redundancy of different upper pathways involved in TDA production may guarantee the supply of phenylacetyl-CoA as a precursor for TDA under different nutritional and physiological conditions and is an expression of the importance of the secondary metabolite TDA for the benefit of P. gallaeciensis in its ecological context.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Volkswagen-Stiftung, VW-Vorab, Lower Saxony 11-7651-13-4/06 (ZN2235), by the Transregional Collaborative Research Centre Roseobacter (Transregio TRR 51) funded by the Deutsche Forschungsgemeinschaft, by an Emmy Noether fellowship of the Deutsche Forschungsgemeinschaft (to J.S.D.), and by a scholarship of the Fonds der Chemischen Industrie (to N.L.B.).
We thank S. Thole and D. Kalhoefer for expert bioinformatics advice and W. Wackernagel for providing plasmids.
Footnotes
Published ahead of print 9 March 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Bairoch A, Boeckmann B, Ferro S, Gasteiger E. 2004. Swiss-Prot: juggling between evolution and stability. Brief. Bioinformatics 5:39–55 [DOI] [PubMed] [Google Scholar]
- 3. Berger M, Neumann A, Schulz S, Simon M, Brinkhoff T. 2011. Tropodithietic acid production in Phaeobacter gallaeciensis is regulated by N-acyl homoserine lactone-mediated quorum sensing. J. Bacteriol. 193:6576–6585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brinkhoff T, et al. 2004. Antibiotic production by a Roseobacter clade-affiliated species from the German Wadden Sea and its antagonistic effects on indigenous isolates. Appl. Environ. Microbiol. 70:2560–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brinkhoff T, Giebel HA, Simon M. 2008. Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch. Microbiol. 189:531–539 [DOI] [PubMed] [Google Scholar]
- 6. Buchan A, Gonzalez JM, Moran MA. 2005. Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71:5665–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chang AC, Cohen SN. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Demain AL. 1998. Induction of microbial secondary metabolism. Int. Microbiol. 1:259–264 [PubMed] [Google Scholar]
- 9. Deppenmeier U, et al. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4:453–461 [PubMed] [Google Scholar]
- 10. Erb TJ, Ismail W, Fuchs G. 2008. Phenylacetate metabolism in thermophiles: characterization of phenylacetate-CoA ligase, the initial enzyme of the hybrid pathway in Thermus thermophilus. Curr. Microbiol. 57:27–32 [DOI] [PubMed] [Google Scholar]
- 11. Felsenstein J. 1985. Confidence-limits on phylogenies—an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed] [Google Scholar]
- 12. Fernandez C, Ferrandez A, Minambres B, Diaz E, Garcia JL. 2006. Genetic characterization of the phenylacetyl-coenzyme A oxygenase from the aerobic phenylacetic acid degradation pathway of Escherichia coli. Appl. Environ. Microbiol. 72:7422–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ferrandez A, et al. 1998. Catabolism of phenylacetic acid in Escherichia coli. Characterization of a new aerobic hybrid pathway. J. Biol. Chem. 273:25974–25986 [DOI] [PubMed] [Google Scholar]
- 14. Fujioka M, Morino Y, Wada H. 1970. Metabolism of phenylalanine (Achromobacter eurydice). Methods Enzymol. 17:585–596 [Google Scholar]
- 15. Geng HF, Bruhn JB, Nielsen KF, Gram L, Belas R. 2008. Genetic dissection of tropodithietic acid biosynthesis by marine roseobacters. Appl. Environ. Microbiol. 74:1535–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Graupner S, Wackernagel W. 2000. A broad-host-range expression vector series including a Ptac test plasmid and its application in the expression of the dod gene of Serratia marcescens (coding for ribulose-5-phosphate 3-epimerase) in Pseudomonas stutzeri. Biomol. Eng. 17:11–16 [DOI] [PubMed] [Google Scholar]
- 17. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 18. Herrmann G, Jayamani E, Mai G, Buckel W. 2008. Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J. Bacteriol. 190:784–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ismail W, et al. 2003. Functional genomics by NMR spectroscopy. Phenylacetate catabolism in Escherichia coli. Eur. J. Biochem. 270:3047–3054 [DOI] [PubMed] [Google Scholar]
- 20. Kovach ME, et al. 1995. 4 new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 21. Lu J, et al. 2009. The distinct quorum sensing hierarchy of las and rhl in Pseudomonas sp. M18. Curr. Microbiol. 59:621–627 [DOI] [PubMed] [Google Scholar]
- 22. Luengo JM, Garcia JL, Olivera ER. 2001. The phenylacetyl-CoA catabolon: a complex catabolic unit with broad biotechnological applications. Mol. Microbiol. 39:1434–1442 [DOI] [PubMed] [Google Scholar]
- 23. Mai X, Adams MW. 1994. Indolepyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. A new enzyme involved in peptide fermentation. J. Biol. Chem. 269:16726–16732 [PubMed] [Google Scholar]
- 24. Markowitz VM, et al. 2010. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 38:D382–D390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martens T, et al. 2006. Reclassification of Roseobacter gallaeciensis Ruiz-Ponte et al. 1998 as Phaeobacter gallaeciensis gen. nov., comb. nov., description of Phaeobacter inhibens sp. nov., reclassification of Ruegeria algicola (Lafay et al. 1995) Uchino et al. 1999 as Marinovum algicola gen. nov., comb. nov., and emended descriptions of the genera Roseobacter, Ruegeria and Leisingera. Int. J. Syst. Evol. Microbiol. 56:1293–1304 [DOI] [PubMed] [Google Scholar]
- 26. Martinez-Blanco H, Reglero A, Rodriguez-Aparicio LB, Luengo JM. 1990. Purification and biochemical characterization of phenylacetyl-CoA ligase from Pseudomonas putida. A specific enzyme for the catabolism of phenylacetic acid. J. Biol. Chem. 265:7084–7090 [PubMed] [Google Scholar]
- 27. McGowan SJ, et al. 2005. Carbapenem antibiotic biosynthesis in Erwinia carotovora is regulated by physiological and genetic factors modulating the quorum sensing-dependent control pathway. Mol. Microbiol. 55:526–545 [DOI] [PubMed] [Google Scholar]
- 28. Mohamed ME. 2000. Biochemical and molecular characterization of phenylacetate-coenzyme A ligase, an enzyme catalyzing the first step in aerobic metabolism of phenylacetic acid in Azoarcus evansii. J. Bacteriol. 182:286–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moran MA, et al. 2007. Ecological genomics of marine roseobacters. Appl. Environ. Microbiol. 73:4559–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moran MA, Miller WL. 2007. Resourceful heterotrophs make the most of light in the coastal ocean. Nat. Rev. Microbiol. 5:792–800 [DOI] [PubMed] [Google Scholar]
- 31. Navarro-Llorens JM, et al. 2005. Phenylacetate catabolism in Rhodococcus sp strain RHA1: a central pathway for degradation of aromatic compounds. J. Bacteriol. 187:4497–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, NY [Google Scholar]
- 33. Newton RJ, et al. 2010. Genome characteristics of a generalist marine bacterial lineage. ISME J. 4:784–798 [DOI] [PubMed] [Google Scholar]
- 34. Olivera ER, et al. 1998. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc. Natl. Acad. Sci. U. S. A. 95:6419–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pearson WR. 2000. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol. Biol. 132:185–219 [DOI] [PubMed] [Google Scholar]
- 36. Porsby CH, Nielsen KF, Gram L. 2008. Phaeobacter and Ruegeria species of the Roseobacter clade colonize separate niches in a Danish turbot (Scophthalmus maximus)-rearing farm and antagonize Vibrio anguillarum under different growth conditions. Appl. Environ. Microbiol. 74:7356–7364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pruitt KD, Tatusova T, Maglott DR. 2007. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35:D61–D65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Quevillon E, et al. 2005. InterProScan: protein domains identifier. Nucleic Acids Res. 33:W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rzhetsky A, Nei M. 1992. A simple method for estimating and testing minimum-evolution trees. Mol. Biol. Evol. 9:945–967 [Google Scholar]
- 40. Saitou N, Nei M. 1987. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 41. Schneider S, Fuchs G. 1998. Phenylacetyl-CoA:acceptor oxidoreductase, a new alpha-oxidizing enzyme that produces phenylglyoxylate. Assay, membrane localization, and differential production in Thauera aromatica. Arch. Microbiol. 169:509–516 [DOI] [PubMed] [Google Scholar]
- 42. Schuster M, Greenberg EP. 2006. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296:73–81 [DOI] [PubMed] [Google Scholar]
- 43. Seyedsayamdost MR, Case RJ, Kolter R, Clardy J. 2011. The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat. Chem. 3:331–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Siddiqui MA, Fujiwara S, Imanaka T. 1997. Indolepyruvate ferredoxin oxidoreductase from Pyrococcus sp. KOD1 possesses a mosaic: structure showing features of various oxidoreductases. Mol. Gen. Genet. 254:433–439 [DOI] [PubMed] [Google Scholar]
- 45. Siddiqui MA, Fujiwara S, Takagi M, Imanaka T. 1998. In vitro heat effect on heterooligomeric subunit assembly of thermostable indolepyruvate ferredoxin oxidoreductase. FEBS Lett. 434:372–376 [DOI] [PubMed] [Google Scholar]
- 46. Spaepen S, et al. 2007. Characterization of phenylpyruvate decarboxylase, involved in auxin production of Azospirillum brasilense. J. Bacteriol. 189:7626–7633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Strittmatter AW, et al. 2009. Genome sequence of Desulfobacterium autotrophicum HRM2, a marine sulfate reducer oxidizing organic carbon completely to carbon dioxide. Environ. Microbiol. 11:1038–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Takezaki N, Rzhetsky A, Nei M. 1995. Phylogenetic test of the molecular clock and linearized trees. Mol. Biol. Evol. 12:823–833 [DOI] [PubMed] [Google Scholar]
- 49. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 50. Teufel R, et al. 2010. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc. Natl. Acad. Sci. U. S. A. 107:14390–14395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thiel V, et al. 2010. Identification and biosynthesis of tropone derivatives and sulfur volatiles produced by bacteria of the marine Roseobacter clade. Org. Biomol. Chem. 8:234–246 [DOI] [PubMed] [Google Scholar]
- 52. Wagner-Döbler I, Biebl H. 2006. Environmental biology of the marine Roseobacter lineage. Annu. Rev. Microbiol. 60:255–280 [DOI] [PubMed] [Google Scholar]
- 53. Ward PG, O'Connor KE. 2005. Induction and quantification of phenylacyl-CoA ligase enzyme activities in Pseudomonas putida CA-3 grown on aromatic carboxylic acids. FEMS Microbiol. Lett. 251:227–232 [DOI] [PubMed] [Google Scholar]
- 54. Wöhlbrand L, et al. 2007. Functional proteomic view of metabolic regulation in “Aromatoleum aromaticum” strain EbN1. Proteomics 7:2222–2239 [DOI] [PubMed] [Google Scholar]
- 55. Yan D, Kang J, Liu D-Q. 2009. Genomic analysis of the aromatic catabolic pathways from Silicibacter pomeroyi DSS-3. Ann. Microbiol. 59:789–800 [Google Scholar]
- 56. Zech H, et al. 2009. Growth phase-dependent global protein and metabolite profiles of Phaeobacter gallaeciensis strain DSM 17395, a member of the marine Roseobacter-clade. Proteomics 9:3677–3697 [DOI] [PubMed] [Google Scholar]
- 57. Zuckerkandl E, Pauling L. 1965. Evolutionary divergence and convergence in proteins, p 97–166 In Bryson V, Vogel HJ. (ed), Evolving genes and proteins. Academic Press, New York, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.