Abstract
Many investigations have provided evidence that plant secondary metabolites, especially flavonoids, may serve as signal molecules to trigger the abilities of bacteria to degrade chlorobiphenyls in soil. However, the bases for this interaction are largely unknown. In this work, we found that BphAEB356, the biphenyl/chlorobiphenyl dioxygenase from Pandoraea pnomenusa B356, is significantly better fitted to metabolize flavone, isoflavone, and flavanone than BphAELB400 from Burkholderia xenovorans LB400. Unlike those of BphAELB400, the kinetic parameters of BphAEB356 toward these flavonoids were in the same range as for biphenyl. In addition, remarkably, the biphenyl catabolic pathway of strain B356 was strongly induced by isoflavone, whereas none of the three flavonoids induced the catabolic pathway of strain LB400. Docking experiments that replaced biphenyl in the biphenyl-bound form of the enzymes with flavone, isoflavone, or flavanone showed that the superior ability of BphAEB356 over BphAELB400 is principally attributable to the replacement of Phe336 of BphAELB400 by Ile334 and of Thr335 of BphAELB400 by Gly333 of BphAEB356. However, biochemical and structural comparison of BphAEB356 with BphAEp4, a mutant of BphAELB400 which was obtained in a previous work by the double substitution Phe336Met Thr335Ala of BphAELB400, provided evidence that other residues or structural features of BphAEB356 whose precise identification the docking experiment did not allow are also responsible for the superior catalytic abilities of BphAEB356. Together, these data provide supporting evidence that the biphenyl catabolic pathways have evolved divergently among proteobacteria, where some of them may serve ecological functions related to the metabolism of plant secondary metabolites in soil.
INTRODUCTION
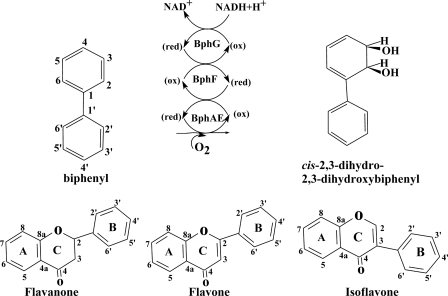
Aryl hydroxylating Rieske-type dioxygenases (ROs) catalyze a cis-dioxygenation of aryl compounds to generate a cis-dihydrodiol metabolite. ROs are promising biocatalysts that metabolize many substituted benzene or diphenyl rings, as well as bicyclic- or tricyclic-fused heterocyclic aromatics, such as quinoline, dibenzofuran, phenanthridine, and flavonoids (3, 4, 15, 22, 29, 30, 33). The biphenyl dioxygenase (BPDO) which catalyzes the first reaction of the bacterial biphenyl catabolic pathway is an RO that has been extensively studied because of its ability to metabolize several polychlorinated biphenyl (PCB) congeners. The BPDO reaction (Fig. 1) requires three components (10, 12, 13). The catalytic component (BphAE) is an RO protein which is a heterohexamer made up of three α (BphA) and three β subunits (BphE). The ferredoxin (BphF) and the ferredoxin reductase (BphG) are involved in electron transfer from NADH to BphAE. The encoding genes for both Burkholderia xenovorans LB400 and Pandoraea pnomenusa B356 are bphA (BphAE α subunit), bphE (BphAE β subunit), bphF (BphF), and bphG (BphG) (6, 36). The α subunit is the one involved in the catalytic activity. It comprises two domains; the Rieske domain containing a 2Fe-2S Rieske cluster receives the electrons from BphF and transfers them to the catalytic mononuclear iron center of the catalytic domain (7).
Fig 1.
Biphenyl dioxygenase reaction (top) and structures of flavone, flavanone, and isoflavone (bottom).
Several investigations have shown that BPDO can metabolize flavonoids (4, 15, 29, 30). These plant secondary metabolites (PSMs) are regarded as very promising for the prevention and treatment of cancers (26) and cardiovascular diseases (35). Plants are currently the major source for these chemicals, but the synthesis of novel derivatives exhibiting improved biological properties is often difficult or impractical (22). Furthermore, in the context of the green chemistry concept, new, more selective and more environmentally friendly approaches to manufacture these biologically specific fine chemicals will be required.
Seeger et al. have shown that B. xenovorans LB400 BPDO (BphAELB400) is able to dihydroxylate several isoflavonoids on ring B (29). BphAELB400 has been extensively investigated because it is regarded as one of the most efficient dioxygenases of natural origin for the degradation of a wide range of chlorobiphenyls. However, in recent years, P. pnomenusa B356 BPDO (BphAEB356) was shown to exhibit superior abilities to degrade several biphenyl analogs, including 2,6-dichlorobiphenyl, 2,4,4′-trichlorobiphenyl, and dichlorodiphenyltrichloroethane (DDT), that BphAELB400 metabolizes poorly (9, 20). Furthermore, preliminary unpublished experiments have suggested that BphAEB356 metabolizes flavonoids more efficiently than BphAELB400. On the other hand, a mutant of BphAELB400, BphAEp4, an evolved BPDO derived from BphAELB400 by the substitutions Thr335Ala Phe336Met, was shown to metabolize a broader range of chlorobiphenyls and dibenzofurans than the parent BphAELB400 (2). The new catalytic properties of this mutant were attributed to the single Thr335Ala substitution. Thr335 exerts constraints on a segment comprised of Val320-Gln322 that lines the catalytic pocket. Replacing Thr335 with Ala releases the constraints on this segment, allowing for more movement during substrate binding (18, 23).
The bacterial metabolism of flavonoids may also have an impact on soil microbiology and on plant-microbe interactions. Many investigations have provided evidence that PSMs may act as signal molecules to trigger the PCB-degrading abilities of soil bacteria (for a review, see reference 34). These signal molecules may have a major impact on the success of rhizoremediation processes aiming at the destruction of PCBs in soil. However, the bases for the PCB-degrading bacterium-plant secondary metabolite interactions are largely unknown. In a recent work (37), we showed that Arabidopsis thaliana root exudates trigger the PCB catabolic abilities of Rhodococcus erythropolis U23A, a rhodococcal rhizobacterium isolated from the rhizosphere of PCB-contaminated-plant roots. Flavanone, one of the major component of these root exudates, was unable to support the growth of strain U23A, but it was metabolized by this strain through its biphenyl catabolic pathway (37). In addition, when used as a cosubstrate with sodium acetate, flavanone was as efficient as biphenyl at inducing the biphenyl catabolic pathway of strain U23A (37). These observations are consistent with the proposed hypothesis whereby flavonoids would act as a signal molecule in soil to modulate the quantity and quality of phenylpropanoids in the rhizosphere (31).
Given the significant impacts the bacterial metabolism of flavonoids may have on green chemistry and on PCB remediation processes and given the preliminary data showing that the two well-characterized P. pnomenusa B356 and B. xenovorans LB400 BPDOs metabolized flavonoids differently, we compared the catalytic properties of BphAELB400 and BphAEB356 toward the simple flavonoids flavone, isoflavone, and flavanone and we assessed the ability of these flavonoids to induce the biphenyl catabolic pathway of these two organisms. In order to get more insights about structural features of BphAE conferring the ability to metabolize these flavonoids, we also docked these chemicals in these protein structures and compared the structure of the docked enzymes with that of BphAEp4.
MATERIALS AND METHODS
Bacterial strains, plasmids, and chemicals.
Wild-type strains P. pnomenusa B356 and B. xenovorans LB400 were described previously (1, 6). All plasmids used in this study were described previously. pET14b[LB400-bphAE] and pET14b[p4-bphAE] carry the genes encoding the wild-type BphAELB400 and its mutant BphAEp4 (Thr335Ala Phe336Met) (2, 17), plasmid pET14b[B356-bphAE] carries the genes encoding BphAEB356 (20), and plasmids pET14b[LB400-bphF] and pET14b[LB400-bphG] carry the genes encoding strain LB400 BphF and BphG (23). Flavone and flavanone were from Sigma-Aldrich, and isoflavone from Indofine Chemical Company, Inc. They were all 99% pure.
Whole-cell assays to determine the ability of P. pnomenusa B356 and B. xenovorans LB400 to metabolize flavanone.
The metabolism of flavanone by resting-cell suspensions of biphenyl-induced cells of P. pnomenusa B356 and B. xenovorans LB400 was examined according to a protocol described previously to investigate the metabolism of flavanone by R. erythropolis U23A (37). Briefly, each strain was grown overnight on medium MM30 (37) containing 3.4 mM biphenyl, and the cells were harvested, washed, and suspended in M9 medium (37) with no carbon source to an optical density at 600 nm (OD600) of 5. This cell suspension was distributed (5-ml amounts) among 50-ml glass tubes, and flavanone was added to a final concentration of 200 μM. The resting-cell suspensions were incubated at either 28°C or 15°C for various periods of between 5 min and 18 h. They were then extracted with ethyl acetate, the organic phase was evaporated, and the residues were treated with n-butylboronate (nBuB) or N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) (Supelco, Sigma-Aldrich) as described previously for gas chromatography-mass spectrometry (GC-MS) analyses (37).
Assays to identify the metabolites produced from flavone, flavanone, and isoflavone by BphAEB356, BphAELB400, and BphAEp4 and to determine their kinetic parameters.
Reconstituted His-tagged BPDO preparations were used in the experiments to identify the metabolites and kinetics of the enzymes and substrates. His-tagged purified enzyme components were produced in recombinant Escherichia coli strains and purified according to published protocols (23). The enzyme assays were performed at 37°C as described previously in a volume of 200 μl in 50 mM morpholinethanesulfonic acid (MES) buffer, pH 6.0, containing 100 nmol substrate (13). The metabolites were extracted at pH 6.0 with ethyl acetate and treated with nBuB or BSTFA for GC-MS analyses.
Catalytic activities were determined by monitoring substrate depletion and metabolite production after 10 min of incubation under the conditions described above. GC-MS peak areas were used to quantify substrate depletion and metabolite production. GC-MS analyses were performed using a Hewlett Packard HP6980 series gas chromatograph interfaced with an HP5973 mass selective detector (Agilent Technologies). The mass selective detector was operated in electron impact ionization (EI) mode and used a quadrupole mass analyzer. The steady-state kinetic parameters of all BphAEs were determined by recording oxygen consumption rates using a Clarke-type Hansatech model DW1 oxygraph (14) for various concentrations of flavonoids between 5 and 150 μM. The kinetic parameters reported in this work were obtained from the analysis of at least two independently produced preparations tested in triplicate.
Assays to assess the ability of flavone, flavanone, and isoflavone to induce the biphenyl catabolic pathway of strains B356 and LB400.
The induction of the biphenyl catabolic pathway of strains B356 and LB400 was assessed on the basis of the amount of 4-chlorobenzoate produced from 4-chlorobiphenyl by resting-cell suspensions previously grown on sodium acetate plus variable concentrations of flavonoids or biphenyl. This assay was performed according to the same method as the previously described assay to assess the ability of flavanone to induce the biphenyl catabolic pathway of R. erythropolis U23A (37). Briefly, cells were grown overnight in medium MM30 amended with 30 mM sodium acetate or with 30 mM sodium acetate plus variable amounts (6 mM, 1 mM, 0.01 mM, or 0.001 mM) of flavone, isoflavone, flavanone, or biphenyl. Cells were harvested and washed in M9 medium without carbon source. The suspensions were adjusted to an OD600 of 1 with M9 medium and distributed in portions of 200 μl into 1.5-ml Eppendorf tubes. 4-Chlorobiphenyl was added to a final concentration of 1.25 mM, and the reaction vials were incubated for 120 min at 28°C in an Eppendorf Thermomixer 5436. The suspensions were then acidified with HCl before the metabolites were extracted with ethyl acetate. The extracts were evaporated, and the residues were derivatized with BSFTA for GC-MS analysis (37).
Docking and structure analysis.
BphAELB400 (RCSB Protein Databank PDB ID 2XRX), BphAEB356 (RCSB Protein Databank PDB ID 3GZX), and BphAEp4 (RCSB Protein Databank PDB ID 2XSH) were used as protein targets, and they were prepared as previously described (20). In the case of BphAELB400 and BphAEp4, we used the structural coordinates of dimer AB for the docking. Ligands were all downloaded as sdf files from PubChem (http://pubchem.ncbi.nlm.nih.gov) and converted into pdb format in Discover Studio Visualizer 2.5. Both proteins and ligands were processed with AutoDockTools to obtain their proper pdbqt format. The searching space for the ligand was centered on the mononuclear iron and contained 20 Å in each x, y, and z direction. Autodock Vina 1.1.2 (24) with the default parameters was used to perform the automatic docking.
RESULTS
Metabolism of flavanone by biphenyl-induced resting cells of strains B356 and LB400.
In a previous report, we showed that although flavanone could not support the growth of R. erythropolis U23A, biphenyl-induced cells of strain U23A metabolized this plant metabolite (37). The induced cells of strain U23A produced small amounts of 2-(2,3-dihydro-2,3-dihydroxyphenyl)chromane-4-one and 2-(3,4-dihydro-3,4-dihydroxyphenyl)chromane-4-one when they were incubated in the presence of flavanone, but the major and ultimate metabolite exhibited mass spectral features corresponding to those of 4-oxo-2-chromanecarboxylic acid (37). Neither strain B356 nor strain LB400 could grow on flavone, isoflavone, or flavanone, but biphenyl-induced cells of both converted flavanone to 4-oxo-2-chromanecarboxylic acid as a dead-end metabolite. 4-Oxo-2-chromanecarboxylic acid was identified from the mass spectral features of its trimethylsilyl (TMS) derivative, which exhibited diagnostically important ions at m/z 264 (M+), 249 (M+ - CH3), 219 (M+ - 3CH3), 205 (M+ - CH3 - O - CO), 174 (M+ - COOTMS), and 131 (M+ - COOTMS - O).
When the resting cell suspensions of strain B356 were incubated at 15°C and for less than 5 min, in addition to 4-oxo-2-chromanecarboxylic acid, small amounts of 2-(2,3-dihydro-2,3-dihydroxyphenyl)chromane-4-one and 2-(3,4-dihydro-3,4-dihydroxyphenyl)chromane-4-one were detected. These two metabolites were identified on the basis of their GC-MS spectral features as described below for the purified enzyme preparation. When the resting cells were incubated at a higher temperature and for longer incubation periods, 4-oxo-2-chromanecarboxylic acid was the only metabolite detected in the culture. This shows that the biphenyl catabolic enzymes of this organism were very efficient in transforming flavanone to 4-oxo-2-chromanecarboxylic acid. In addition, while cells of strain B356 metabolized more than 80% of the added substrate within 5 min at 15°C, cells of strain LB400 metabolized less than 20% of the substrate when they were incubated for 1 h at 28°C. This shows the superiority of strain B356 over strain LB400 in metabolizing flavanone.
Induction of the biphenyl catabolic pathway of strains B356 and LB400 by flavonoids.
Flavanone induction was assessed by monitoring the 4-chlorobenzoate produced from 4-chorobiphenyl which, in both strains B356 and LB400, accumulates as a dead-end metabolite of the biphenyl catabolic pathway. In a recent report, it was shown that the biphenyl catabolic genes are expressed constitutively at low levels during the growth of B. xenovorans LB400 on succinate (28). Furthermore, the level of expression of the pathway enzymes appeared to be influenced by posttranscriptional regulation factors and by the physiological state of the cells, which may significantly influence the chlorobiphenyl degradation abilities of cells (28). In spite of these difficulties, we reasoned that the assay monitoring 4-chlorobenzoate should allow us to determine if strains B356 and LB400 respond similarly to the presence of simple flavonoids during growth on sodium acetate and if the enzymes of the upper biphenyl catabolic pathway are induced by flavonoids.
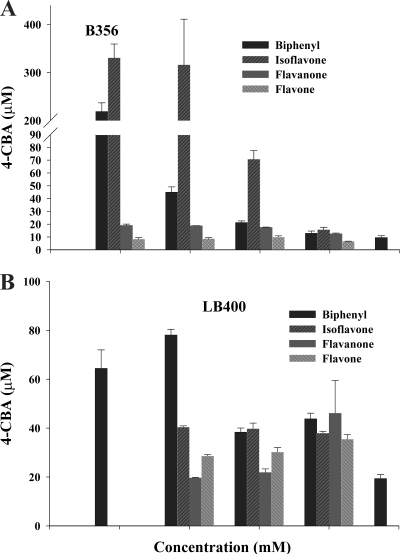
When cells of strain B356 were grown on sodium acetate alone, small amounts of chlorobenzoate were produced in the growth medium (Fig. 2A). However, when cells were grown in the presence of sodium acetate plus variable amounts of biphenyl, the amount of 4-chlorobenzoate varied depending on the amount of biphenyl added to the culture medium (Fig. 2A). This response was similar to that observed for strain U23A grown in the presence of sodium acetate plus biphenyl (37). Cells of strain LB400 grown on sodium acetate alone produced slightly larger amounts of 4-chlorobenzoate than cells of strain B356, and the addition of biphenyl to the growth medium did not induce the biphenyl catabolic enzymes as strongly as for strain B356 (Fig. 2B).
Fig 2.
(A) Amounts (μM) of 4-chlorobenzoic acid (4-CBA) produced when standardized resting cell suspensions of strain B356 were incubated with 1,250 μM 4-chlorobiphenyl for 2 h. (B) Amounts (μM) of 4-chlorobenzoic acid produced when standardized resting cell suspensions of strain LB400 were incubated with 1,250 μM 4-chlorobiphenyl for 2 h. Each strain was previously grown overnight at 28°C in MM30 medium amended with 30 mM sodium acetate or with 30 mM sodium acetate plus the indicated concentration of the indicated flavonoid or of biphenyl. Bars represent means (n = 2) and standard deviations. The protocol for the standardized 4-chlorobiphenyl assay is described in Materials and Methods. Concentrations of 0.0005 and 0.0001 mM were only used for flavanone.
When strain B356 was grown on sodium acetate plus flavanone, for concentrations ranging between 1 mM and 0.01 mM, the amount of 4-chlorobenzoate produced during the assay was not significantly higher than for cells grown on sodium acetate alone (Fig. 2A). Similar results were obtained when cells were grown on sodium acetate plus flavone. However, remarkably, the amounts of 4-chlorobenzoate produced from 4-chlorobiphenyl by resting cells grown on sodium acetate plus isoflavone were significantly higher than those produced for cells grown on sodium acetate plus biphenyl (Fig. 2A). We cannot exclude the possibility that posttranscriptional regulation mechanisms exerted by biphenyl metabolites are responsible for the lower enzyme activity found in cells grown on sodium acetate plus biphenyl. Furthermore, since all three flavonoids are metabolized by whole cells of strain B356, we must exclude the possibility that permeation of flavonoids across the cell membrane/wall had affected the cells' activity for substrate. Therefore, the data show that isoflavone is a good inducer of the biphenyl catabolic enzymes of strain B356. In the case of strain LB400, there was no clear-cut effect for any of the three tested flavonoids that would demonstrate the ability of these flavonoids to induce the biphenyl catabolic pathway. The amounts of 4-chlorobenzoate produced varied slightly in the presence of flavonoids (Fig. 2B), but the amounts produced were not statistically significantly different from those observed in the absence of flavonoids. Therefore, none of the three flavonoids tested significantly influenced the activity of the biphenyl catabolic enzymes of strain LB400, whereas isoflavone was found to act as an inducer of the biphenyl catabolic pathway of strain B356 when it was added as a cosubstrate with sodium acetate.
Metabolites produced from flavonoids by purified BphAEB356, BphAELB400, and BphAEp4.
Since whole cells of strains B356 and LB400 metabolized flavanone differently and since they responded differently to the presence of flavone, flavanone, and isoflavone in the growth medium, we have compared the ability of purified preparations of their biphenyl dioxygenases to metabolize these three plant metabolites. The purified enzymes were prepared from recombinant E. coli cells as described in Materials and Methods. We have also included BphAEp4, a Thr335Ala Phe336Met mutant of BphAELB400 exhibiting an expanded substrate range compared to that of its parent enzyme (2, 18). In a previous report, we showed that replacing Phe336 of BphAELB400, which lines the catalytic pocket, with a residue with a smaller side chain (Met336) increases the space inside the catalytic pocket. In a similar manner, the corresponding Ile334 of BphAEB356 that lines the catalytic pocket is smaller than Phe336 of BphAELB400 and, thus, allows the enzyme to metabolize bulkier substrates, such as DDT (20). Thr335 is at a remove from the substrate; however, changing this residue to the smaller Ala335 relieves intramolecular constraints on Gly321, allowing for significant movement of this residue during substrate binding and thereby increasing the space available to accommodate bulkier substrates (20). In a manner similar to Ala335 of BphAEp4, Gly333 (corresponding to Thr335 of BphAELB400) does not interact with Gly319 (corresponding to Gly321 of BphAELB400). Therefore, although the side chains of Ala335 and Gly333 differ, their effects on the enzyme's structure are likely to be comparable (18).
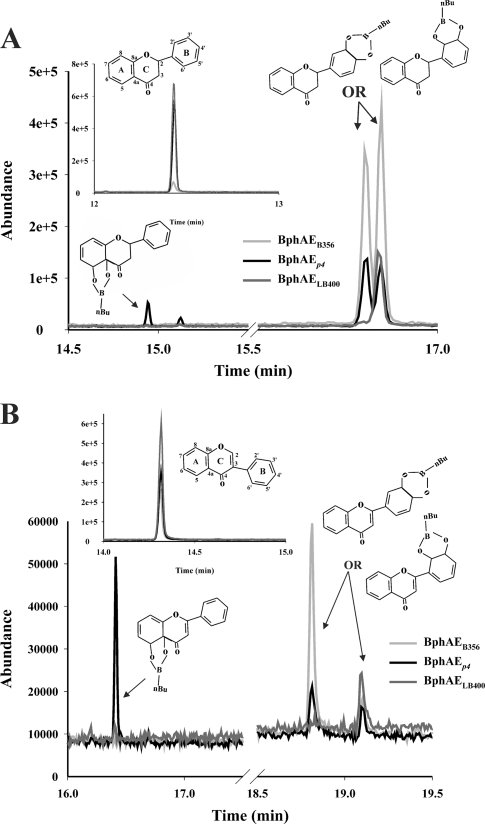
Based on the GC-MS peak area of the remaining substrate, purified preparations of BphAEB356 incubated with 100 nmol flavanone oxidized more than 90 nmol this substrate within 10 min. Under identical conditions, BphAEp4 metabolized approximately 20 nmol flavanone and BphAELB400 metabolized less than 10 nmol of this substrate (Fig. 3A). Consistent with these results, the amount of metabolites generated by BphAEB356 after 10 min of incubation was significantly higher than for the two other enzymes (Fig. 3A). In addition, the pattern of metabolites generated by the three enzymes differed significantly. BphAEB356 produced two metabolites in approximately equal amounts. They both exhibited a very similar mass spectral fragmentation pattern (Table 1). The presence of ions at m/z 147 (M+ - n-BuBO2 - C6H5) and 120 (M+- n-BuBO2 - C6H5 - CH - CH2) was consistent with a dihydroxylation occurring on ring B. BphAELB400 generated only one of the two dihydrodiol metabolites, whereas BphAEp4 produced four dihydrodiol metabolites from flavanone. It produced the two metabolites resulting from the oxidation of ring B, but in addition, it produced two other dihydrodiols that could only result from a hydroxylation of ring A. The mass spectral fragmentation patterns of their butylboronate derivatives are shown in Table 1. The ions at m/z 192 (M+ - C6H5 - C3H3 - O) and 176 (M+ - C6H5 - C3H3 - O2) resulting from the loss of the nonhydroxylated ring B (C6H5) provide evidence that the hydroxylation occurred on ring A. These data were confirmed by the GC-MS analysis of the trimethylsilyl derivatives of the metabolites, which evidenced the formation of two dihydrodiol metabolites from BphAEB356, a single one from BphAELB400, and four dihydrodiol metabolites from BphAEp4 (not shown).
Fig 3.
(A) Total ion chromatogram showing the peaks of metabolites produced from flavanone after 10 min by reconstituted His-tagged BphAEB356 (gray curve), BphAELB400 (dark gray curve), and BphAEp4 (black curve). The inset shows the peak of the substrate remaining in the reaction vial. (B) Total ion chromatogram showing the peaks of metabolites produced from flavone after 10 min by reconstituted His-tagged BphAEB356 (gray curve), BphAELB400 (dark gray curve), and BphAEp4 (black curve). The inset shows the peak of the substrate remaining in the reaction vial.
Table 1.
Mass spectral features of the butylboronate-derived metabolites produced from flavanone and flavone by BphAE-B356, BphAELB400, and BphAEp4
| Substrate | No. of metabolites produced by: |
Metabolite structurea | Oxidized ring | M+ | Other ions | ||
|---|---|---|---|---|---|---|---|
| BphAEB356 | BphAELB400 | BphAEp4 | |||||
| Flavanone | 2 | 1 | 2 | 2-(2,3-Dihydro-2,3-dihydroxyphenyl)chromane-4-one or 2-(3,4-dihydro-3,4-dihydroxyphenyl) chromane-4-one | B | 324 | 308, 267, 240, 224, 147, 120 |
| 2 | 4a,5-Dihydro-4a,5-dihydroxy-2-phenyl chromane-4-one | C | 324 | 308, 267, 240, 224, 192, 176, 131 | |||
| Flavone | 2 | 1 | 2 | 2-(2,3-Dihydro-2,3-dihydroxyphenyl)chromene-4-one or 2-(3,4-dihydro-3,4-dihydroxyphenyl) chromene-4-one | B | 322 | 306, 265, 238, 222, 210, 181, 120 |
| 1 | 4a,5-Dihydro-4a,5-dihydroxy-2-phenyl chromene-4-one | C | 322 | 306, 265, 238, 222, 192, 163, 129 | |||
Structures were tentatively identified on the basis of their mass spectral fragmentation features and on the orientation of the docked substrates in the enzyme catalytic pocket.
BphAEB356 and BphAEp4 metabolized, respectively, 70 nmol and 50 nmol of flavone when they were incubated with 100 nmol of this substrate for 10 min. However, BphAELB400 performed very poorly on flavone, where less than 5% of the added substrate was degraded. As with flavanone, the pattern of metabolites produced from flavone differed significantly among the enzymes (Fig. 3B). Two metabolites were produced when the reaction was catalyzed by BphAEB356, but their proportion differed considerably. Based on the mass spectral fragmentation features of their butylboronate derivatives shown in Table 1, they were identified as 2-(2,3-dihydro-2,3-dihydroxyphenyl)chromene-4-one and 2-(3,4-dihydro-3,4-dihydroxyphenyl)chromene-4-one. On the basis of the docking experiments described below, the major metabolite would result from a hydroxylation of carbons 2′-3′ to generate the 2-(2,3-dihydro-2,3-dihydroxyphenyl)chromene-4-one. BphAEp4 produced three metabolites from flavone (Fig. 3B). The GC-MS features of their butylboronate derivatives are shown in Table 1. Two of the metabolites are identical to those produced by BphAEB356. The mass spectral features of the third one, which is a major metabolite, comprise ions at m/z 192 (M+ - C6H5 - C3H - O) and 163 (M+ - n-BuB - C6H5 - C2H) which are consistent with a hydroxylation on ring A. On the basis of the docking experiments described below, this metabolite would be 4a,5-dihydro-4a,5-dihydroxy-2-phenyl chromene-4-one. BphAELB400 produced trace amounts only of the metabolite corresponding to the peak of 2-(3,4-dihydro-3,4-dihydroxyphenyl)chromene-4-one.
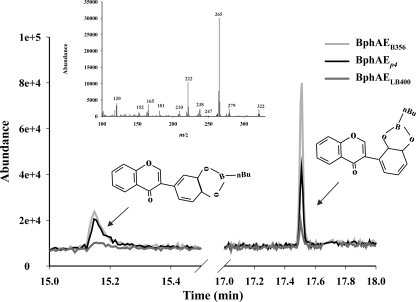
As was the case for the previous two substrates, BphAEB356 performed better than the other two enzymes toward isoflavone. However, in this case, all three enzymes produced the same two metabolites from this flavonoid (Fig. 4). Both of them exhibited a fragmentation pattern comprising ions at m/z 181 (M+ - n-BuBO2 - CO - CH), 165 (M+ - n-BuBO2 - CO - CH - O), and 120 (M+ - n-BuBO2 - C6H5 - C2H) that was consistent with a dihydroxylation of ring B.
Fig 4.
Total ion chromatogram showing the peaks of metabolites produced from isoflavone after 10 min by reconstituted His-tagged BphAEB356 (gray curve), BphAELB400 (dark gray curve), and BphAEp4 (black curve). The inset shows the mass spectrum of the metabolite exhibiting a retention time of 17.5 min. The mass spectrum for the second metabolite is almost identical (not shown).
Kinetic parameters of purified BphAEB356, BphAELB400, and BphAEp4 toward flavone, flavanone, and isoflavone.
The steady-state kinetic parameters of purified preparations of BphAEB356, BphAEp4, and BphAELB400 toward each of the three flavonoids were calculated from the initial oxygen consumption using a Clark-type oxygraph. Notably, for the three substrates, the kcat and kcat/Km values for BphAEB356 were in the range reported (20) for biphenyl (respectively, 4.3 s−1 and 63 × 103 M−1 s−1) when this enzyme was used under the same reaction conditions (Table 2). Consistent with the whole-cell assays described above, flavanone was the best substrate, exhibiting kcat and kcat/Km values that were significantly higher than those previously reported for biphenyl (Table 2). However, flavone and isoflavone were also good substrates for the enzyme since their kinetic parameters were in same range as those reported for biphenyl. Furthermore, for all substrates, the steady-state kinetic parameters of BphAEB356 were significantly higher than those for BphAEp4. The reported kcat and kcat/Km values of BphAEp4 toward biphenyl (1.0 s−1 and 31 × 103 M−1 s−1) (23) were higher than for all three flavonoids. Therefore, although BphAEp4 performed well on these substrates, unlike BphAEB356, biphenyl remains a better substrate than the flavonoids. Consistent with the time point measurement experiments, BphAELB400 was poorly active toward the three flavonoids. The steady-state kinetic parameters obtained with flavone and isoflavone were too low to be determined accurately and therefore are not reported here. Flavanone was the best substrate; the kcat and kcat/Km values obtained with this substrate were significantly lower than the values reported when BphAELB400 was used to metabolize biphenyl under identical conditions (respectively, 0.9 s−1 and 41 × 103 M−1 s−1) (23). Together, these data show that in comparison to the activity of BphAELB400, the double Thr335Ala Phe336Met substitution in BphAEp4 contributed to enhancement of the catalytic activity toward the simple flavonoids. However, these mutations did not allow the enzyme to reach the level of activity of the naturally occurring BphAEB356.
Table 2.
Steady-state kinetic parameters of BphAEB356, BphAELB400, and BphAEp4 toward flavanone, flavone, and isoflavonea
| Substrate, enzyme | Km (μM) | kcat (s−1) | kcat/Km (103 M−1 s−1) |
|---|---|---|---|
| Flavanone | |||
| BphAEB356 | 77.5 ± 4.8 | 9.0 ± 0.4 | 116.1 ± 8.9 |
| BphAEP4 | 27.5 ± 5.7 | 0.60 ± 0.1 | 21.8 ± 4.0 |
| BphAELB400 | 32.1 ± 3.9 | 0.36 ± 0.0 | 11.1 ± 1.3 |
| Flavone | |||
| BphAEB356 | 121.4 ± 7.2 | 4.0 ± 1.3 | 32.9 ± 11.2 |
| BphAEP4 | 21.4 ± 1.4 | 0.08 ± 0.0 | 3.8 ± 0.1 |
| Isoflavone | |||
| BphAEB356 | 15.8 ± 1.0 | 1.2 ± 0.0 | 75.9 ± 4.7 |
| BphAEP4 | 27.6 ± 0.3 | 0.59 ± 0.0 | 21.3 ± 0.0 |
The ± standard deviations of the results for two independently produced enzyme preparations are shown.
Structural analysis of docked flavonoids at active sites of BphAEB356, BphAELB400, and BphAEp4.
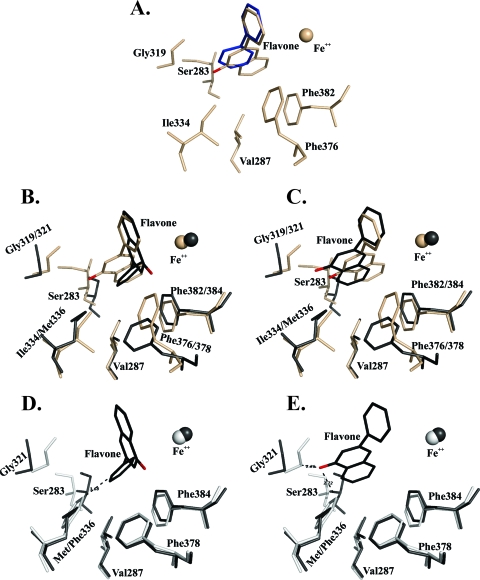
In order to identify the structural features of BphAEB356 and BphAELB400 that explain why the two enzymes catalyze flavone, isoflavone, and flavanone oxidation differently, we docked these flavonoids at their active sites. Since previous reports showed that an induced-fit mechanism was required to bind the substrate productively inside the BPDO catalytic pocket (23), we docked the flavonoids in the substrate-bound form of the enzymes after removing biphenyl (or 2,6-dichlorobiphenyl in the case of BphAEp4). When flavone was docked into BphAEB356, consistent with the biochemical data, the conformation of the top-ranked docked molecule exhibited an orientation that would enable an oxygenation of ring B. Carbons 2′ and 3′ of ring B closely aligned with carbons 2 and 3 of the oxidized ring of biphenyl in the complexed form (Fig. 5A). This suggested that the major metabolite produced from flavone when BphAEB356 catalyzed the reaction would be 2-(2,3-dihydro-2,3-dihydroxyphenyl)chromene-4-one. Therefore, the regiospecificity of BphAEB356 toward flavone would be similar to that of the previously reported BphA1A2(2072) which was obtained by shuffling bphA1 from Pseudomonas alcaligenes with bphA from B. xenovorans LB400 (32). In the case of BphAELB400, none of the docked substrate conformations exhibited a productive orientation toward the catalytic iron. This is consistent with the fact that the catalytic activity of BphAELB400 toward this flavonoid was very low. When BphAEB356 docked with flavone was superposed to the biphenyl-bound form of BphAELB400 (without biphenyl), the chromene oxo group of the docked molecule was at less than 3 Å from both Phe336 and Gly321 of BphAELB400 (not shown). Therefore, the proximity of the chromene oxo group to these two residues probably prevents productive binding to BphAELB400.
Fig 5.
(A) Superposition of catalytic center residues of the flavone-docked (wheat) and biphenyl-bound (blue) forms of BphAEB356. (B and C) Superposition of catalytic center residues of two flavone-docked forms of BphAEp4 (black) and the top-ranked flavone-docked form of BphAEB356 (wheat). (D and E) Superposition of catalytic center residues of two flavone-docked forms of BphAEp4 (black) and the biphenyl-bound form of BphAELB400 (white) after removal of biphenyl. The oxygen of the flavone oxo group is in red.
Unlike the result for BphAEB356, the conformation of the top-ranked docked flavone molecule in BphAEp4 was consistent with the observation that its major metabolite resulted from a dihydroxylation on ring A. On the basis of the orientation of ring A toward the catalytic Fe2+ in the docked form of BphAEp4, the hydroxylation should occur onto carbons 4a and 5 to produce 4a,5-dihydro-4a,5-dihydroxy-2-phenyl chromene-4-one (Fig. 5B). Another of the 10 top-ranked conformations of flavone in BphAEp4's catalytic pocket was similar to but did not superpose exactly with the molecule docked in BphAEB356 (Fig. 5C). In a previous report, the ability of BphAEp4 to oxidize 2,6-dichlorobiphenyl on the meta-para and ortho-meta carbons of biphenyl was attributed to the fact that none of the C-2′–C-3′ or C-3′–C-4′ pairs of carbons were equidistant from the catalytic Fe2+ and they all were within 4.5 Å from it (18). A similar situation was obtained when flavone was docked in BphAEp4. Carbons C-2′ and C-3′ of ring B were not equidistant from the catalytic Fe2+, and they were more distant from it than the corresponding atoms of flavone docked in BphAEB356 (Fig. 5C). This may explain why both 2-(2,3-dihydro-2,3-dihydroxyphenyl)chromene-4-one and 2-(3,4-dihydro-3,4-dihydroxyphenyl)chromene-4-one were produced and why these metabolites were produced in lesser amounts than when BphAEB356 catalyzed the reaction.
As shown in Fig. 5B, when flavone takes an orientation enabling an oxidation of ring A (in black), Phe376 of BphAEB356 is too close (1.5 Å) to the chromene oxo group to allow productive binding of this substrate. Therefore, this residue or others that modulate its conformation exert a strong influence on the regiospecificity of the enzyme toward flavone. In order to confirm that Phe336 and Gly321 prevent the binding of flavone to BphAELB400, we superposed both docked conformations of flavone in BphAEp4 with the biphenyl-bound form of BphAELB400 after biphenyl removal. This is shown in Fig. 5D and E, where it is clear that for both conformations of the docked substrate in BphAEp4, residues Gly321 and Phe336 of BphAELB400 are both too close to the substrate to allow productive binding. Therefore, the Phe336Met and the Thr335Ala substitution are both required to facilitate flavone binding to BphAEp4. However, as shown from the steady-state kinetic parameters of the enzymes, although BphAEp4 can metabolize flavone, its turnover rate of reaction is significantly lower than that of BphAEB356. Therefore, although the two mutations that occurred in BphAEp4 enhanced its catalytic activity toward flavone, other structural features of BphAEB356 that are not present in BphAEp4 are required to facilitate the chemical steps in the catalytic oxygenation reaction. A structural comparison of the catalytic pockets of BphAB356 and BphAEp4 identified Phe376/Phe378 and Ser283/Ile283 as likely candidates to explain the different catalytic properties of the two enzymes (Fig. 5).
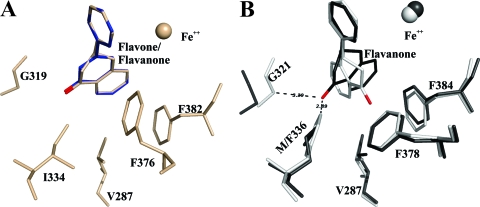
In the flavanone docking experiment, both flavanone and flavone are placed at the same position in BphAEB356 where carbon 2′ and 3′ of ring B superposed almost perfectly with the reactive carbons of biphenyl (Fig. 6A). Biochemical analysis showed that BphAEB356 oxidized both the 2′,3′ and 3′,4′ carbons of ring B (Fig. 3A). Therefore, the binding of BphAEB356 to flavanone can induce other conformations of the substrate that the docking experiment could not reproduce. The docking experiment with BphAEp4 showed that flavanone can take an orientation where ring B superposes exactly with ring B of the docked molecule in BphAEB356 (not shown). However, similar to flavone docking and consistent with the biochemical data, flavanone can also be docked in BphAEp4 in an orientation that would enable a hydroxylation of ring A (not shown). As seen by the observations described above, although BphAELB400 is not as efficient as BphAEp4 in oxidizing flavanone, its activity toward this substrate is more efficient than that toward flavone. Consistent with the biochemical data, automatic docking placed flavanone in an orientation that would allow an oxygenation of ring B by BphAELB400. Structural analysis shows that, unlike the results of the docking experiment done with BphAEp4, in the case of BphAELB400, the chromane moiety of flavanone is oriented such that the oxo group of ring C is distanced from Phe336 and pulled toward Phe378 and Phe384 (Fig. 6B). This shows that the chromane moiety of the molecule reacts differently than the chromene moiety of flavone with the surrounding atoms of the catalytic pocket of BphAELB400. However, the docking experiment has limitations, since it did not allow identification of the protein atoms of BphAELB400 that interact with the chromane moiety of flavanone.
Fig 6.
(A) Superposition of catalytic center residues of the flavanone-docked (wheat) and flavone-docked (blue) forms of BphAEB356. (B) Superposition of catalytic center residues of the top-ranked flavanone-docked form of BphAEp4 (black) enabling the oxidation of ring B and the top-ranked flavanone-docked form of BphAELB400 (white). The oxygen of the flavanone oxo group is in red.
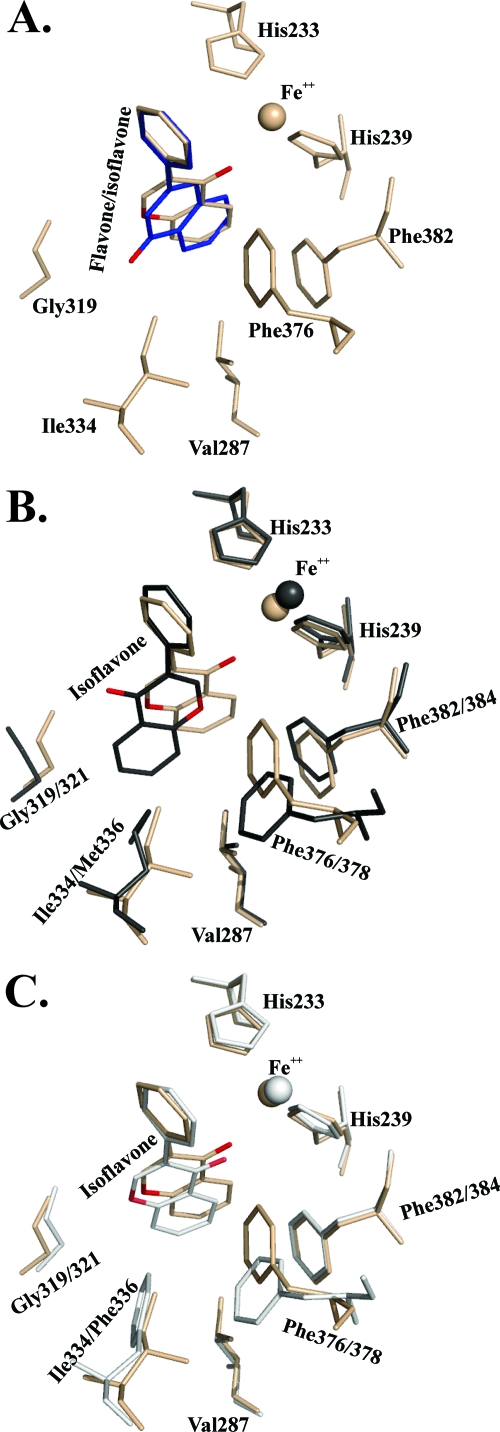
The isoflavone docking experiments are also in agreement with the biochemical data. Ring B of isoflavone superposes well with ring B of flavone in the top-ranked isoflavone-docked form of BphAEB356. The docking experiment suggests that the major metabolite generated by the oxygenation of isoflavone would be 3-(2,3-dihydro-2,3-dihydroxyphenyl)chromene-4-one (Fig. 7A). In the case of BphAEp4, it is not clear why the oxo group is flipped in the opposite orientation for the top-ranked form of isoflavone-docked BphAEp4 (Fig. 7B). There are no apparent constraints that would prevent isoflavone from taking the same conformation as in BphAEB356. This shows that as for flavanone, other structural features that the docking experiment could not identify are likely to be involved in the binding process for this flavonoid. It is also interesting that automatic docking places isoflavone in a productive orientation for BphAELB400 (Fig. 7C). However, in this case, unlike the case for BphAEp4, the oxo group is in an orientation similar to that found in the isoflavone-docked BphAEB356. Since isoflavone was a poor substrate for BphAELB400, it is likely that unidentified structural features that place isoflavone in the opposite orientation in BphAEp4 are required to enable the chemical reactions to proceed. Since in the docked form of BphAEB356, the oxo group is at a distance of approximately 4 Å from Fe2+ and from the two His that coordinate it, the proximity of the oxo group to the iron may hinder the catalytic activity in BphAELB400. The iron is at the interface between two α subunits, and it was shown in a previous work that protein structures surrounding the catalytic iron move during binding and that these structures appeared to be involved in maintaining the integrity of the α3β3 conformation of the enzyme (23). However, more structural analyses of the substrate-bound enzymes will be required to determine more precisely why BphAELB400 has poor activity on isoflavone.
Fig 7.
(A) Superposition of catalytic center residues of the top-ranked isoflavone-docked (wheat) and flavone-docked (blue) forms of BphAEB356. (B) Superposition of catalytic center residues of the top-ranked isoflavone-docked forms of BphAEp4 (black) and of BphAEB356 (wheat). (C) Superposition of catalytic center residues of the top-ranked isoflavone-docked forms of BphAELB400 (white) and of BphAEB356 (wheat). The oxygen of the flavanone oxo group is in red.
DISCUSSION
The perception of flavonoids by plant pathogens and their function as signals in the initiation of legume-rhizobium symbiosis have been well characterized (31). However, the potential impacts of flavonoids on soil and rhizosphere bacteria that do not interact with plants directly in a host-pathogen or symbiotic interaction remain largely unknown. Better insight into this mechanism will help in understanding how plants promote PCB degradation in soil. Many investigations have identified plant secondary metabolites (flavonoids or terpenes) as likely candidates to trigger microbial degradation of PCBs in soil (34). Shaw et al. (31) have hypothesized that PSMs act to shape rhizosphere microbial community structure and, thus, they may have an impact on the rhizosphere function by triggering microbial pathways that can influence the quality and quantity of PSMs in soil. However, this hypothesis remains to be demonstrated.
In a previous report, we showed that the R. erythropolis U23A biphenyl catabolic pathway was induced by flavanone (37). In this work, we showed that isoflavone instead of flavanone was an inducer for the biphenyl catabolic pathway of P. pnomenusa strain B356, whereas none of the three flavonoids induced the biphenyl catabolic pathway of strain LB400. The observation that strains U23A, B356, and LB400 responded differently to simple flavonoids is consistent with divergent regulation mechanisms for their respective biphenyl catabolic operons. Strain LB400's bph operon is controlled by orf0, producing a positive regulator belonging to the GntR family, and by bphR2, producing a LysR-type regulator (5), whereas the biphenyl operons of Rhodococcus globerulus P6 and Rhodococcus jostii RHA1 are regulated through a two-component regulatory system (19, 21). The regulation of strain U23A's bph operon is likely to be very similar to that of other rhodococci. The regulatory system of strain B356's bph operon has not yet been elucidated. However, a gene (orf0B356, GenBank accession number JQ322530) coding for a protein exhibiting 48% homology with BphS of Cupriavidus oxalaticus A5 (previously called Cupriavidus necator A5, Alcaligenes eutrophus, or Ralstonia eutropha A5) (25) and exhibiting homology with other members of the GntR family was found in the genome of strain B356 just upstream of bphG. Sequence alignment of this protein with other members of GntR family proteins showed it clustering with BphS of strain A5 and of Pseudomonas sp. strain KKS102 (not shown) which, unlike Orf0 of strain LB400, were found to be negative regulators (5, 27). The phylogenetic tree obtained when sequences of known BphAs from cultured and uncultured bacteria are aligned (38) shows three branches: two comprise principally Gram-negative proteobacteria, and one comprises exclusively high-GC-content Gram-positive bacteria of the rhodococcal group. It is noteworthy that BphAEB356 clusters with BphA1A2 of strain A5 and of strain KKS102, whereas BphAELB400 belongs to a separate branch. The fact that a similar clustering of the phylogenetic tree is obtained for the deduced amino acid sequences of the regulatory protein and of the first enzyme of the biphenyl catabolic pathway of these strains highlights the possibility that the three biphenyl catabolic pathway clusters may have evolved to serve distinct functions in the environment.
In this work, in order to get more insight about how these pathways interact with simple flavonoids, we compared the metabolism of flavanone, flavone, and isoflavone by BphAEB356 and BphAELB400. Biochemical data showed that, unlike BphAELB400, BphAEB356 is well fitted to metabolize these PSMs. Structural analysis identified two features of BphAELB400 that are responsible for the poor ability of the enzyme to metabolize these flavonoids. As observed in the case of 2,6-dichlorobiphenyl (9, 18) and in the case of DDT (20), Phe336 is too large to enable productive binding with large substrates. In addition, the fact that Gly321 is constrained through a network of hydrogen bonding significantly hinders binding with these substrates (18). In a previous report, we showed that replacing Thr335 with Ala335 in BphAELB400 relieved constraints on the Val320-Gly321-Gln322 segment, allowing for more movement during substrate binding. This feature enables BphAEp4 to accommodate bulkier substrates, such as 2,6-dichlorobiphenyl (18). Similar to Ala335 of BphAEp4, the corresponding residue Gly333 of BphAEB356 is too short to form any contact with this segment (not shown). Therefore, Gly319 of BphAEB356 is more relaxed than Gly321 of BphAELB400, which explains in part why BphAEB356 can metabolize substrates such as 2,6-dichlorobiphenyl (9) or flavonoids that BphAELB400 metabolizes poorly. However, other unidentified structural features influence binding to flavonoids. For example, it is noteworthy that BphAELB400 was found to catalyze the oxidation of flavanone more efficiently than flavone. The superior properties of the enzyme toward flavanone were attributed to the fact that the oxo group of the chromane moiety was placed away from Phe336. This indicates that protein structures involved in the binding process interacted differently with the chromene and chromane moieties of the molecule.
This is also supported by the superior catalytic abilities of BphAEB356 compared to those of BphAEp4 toward flavanone, flavone, and isoflavone in spite of the fact that both BphAEB356 and BphAEp4 contain a smaller amino acid than Phe336 of BphAELB400 at position 336 and, in addition, the fact that the correspondence of Gly321 of BphAEp4 to Gly319 of BphAEB356 is less constrained than that of Gly321 of BphAELB400. The docking experiments did not allow us to identify precisely the BphAEB356 structural features that conferred to the enzyme an ability superior to that of BphAEp4 to catalyze the reaction. In a previous report, it was shown that the helix between residues 282 and 288 moved considerably more toward the substrate during substrate binding with BphAEp4 than with BphAELB400. This movement was attributed to the Thr335Ala substitution that altered the intramolecular hydrogen bonding networks involving residues of this helix (18). It was suggested (9) that the mobile character of this helix may influence binding to substrates larger than biphenyls. Furthermore, residue 283 is at the entranceway of the catalytic pocket and both residue Ile283 of BphAEB356 and residue Ser283 of BphAEp4 are very close (less than 3 Å) to ring A of flavone (Fig. 5) in the flavone-docked enzyme. In the biphenyl-bound form of BphAELB400, Ser283 is far from the substrate. Although its precise role in substrate binding is not clear, residue 283 and the helix to which it belongs appear to be likely candidates for engineering enzymes exhibiting altered substrate specificity and regiospecificity toward flavonoids. However, we cannot exclude other residues that are not in contact with the substrate but that may influence substrate binding by other mechanisms. For example, in a recent report, residues 338 and 409 of BphAELB400 were found to act synergistically to influence the catalytic properties of the enzyme by interacting with residues that are involved in subunit assembly and electron transport (23).
In previous reports, E. coli cells producing P. alcaligenes KF707 BPDO were found to catalyze the hydroxylation of flavone (16) and of flavanone (11). BphA1A2 from strain KF707 is more than 95% homologous to BphAELB400, except that like BphAEB356, residue 335 (corresponding to Phe336 of BphAELB400) is an Ile and residue 334 (corresponding to The335 of BphAELB400) is an Ala. Therefore, with respect to their catalytic properties toward flavonoids, the structural features of BphA1A2KF707 and BphAEp4 are expected to be comparable. However, a BphA1A2KF707 variant was obtained which exhibited enhanced activity toward flavonoids (15). This variant was obtained by the substitutions His255Gln, Val256Ile, Gly266Ala, and Phe277Tyr. It is not clear whether all these residues together or a combination of some of them were required to enhance the activity toward flavonoids. The likely involvement in substrate specificity and selectivity of the mobile loop between residues 240 and 260 that overhang the entranceway of the catalytic pocket has been discussed previously (18). In addition, the likely involvement of residues 266 and 267 in the catalytic properties of BphAEB356 toward DDT has also been discussed (20). Nevertheless, our data with BphAEB356 and BphAELB400 and the data related to BphA1A2KF707 variants show the complexity of the substrate binding process, which involves interaction between the substrate and many protein atoms that either contact the substrate or modulate the conformation of protein structures that are required to enable a productive binding.
Altogether, our investigation identified residues that are involved in substrate binding with simple flavonoids and provided evidence that BphAEB356 has evolved to be better fitted than BphAELB400 to metabolize these PSMs. Moreover, the fact that the biphenyl catabolic pathway of strain B356 was induced by isoflavone provides additional evidence supporting the hypothesis brought forward by Focht (8) and others (31) that the biphenyl catabolic pathways have evolved in bacteria to serve ecological functions, perhaps related to the metabolism of plant secondary metabolites in soil.
In this work, by singling out simple flavonoids and comparing the ability of two well-characterized biphenyl-degrading bacteria to metabolize them, we have shown that both the metabolism of flavonoids and the response to them as signal molecules to trigger the biphenyl catabolic pathway vary considerably among bacteria. This conclusion is significant for the development of more rational approaches for designing efficient rhizoremediation processes. Hence, our data imply that the efficiency of the process will depend on the choice of appropriate bacterial strains responding to the specific PSMs produced by the plants with which they are associated.
ACKNOWLEDGMENTS
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) (grants RGPIN/39579-2007 and STPSC 356996-07)
Footnotes
Published ahead of print 16 March 2012
REFERENCES
- 1. Barriault D, Pelletier C, Hurtubise Y, Sylvestre M. 1997. Substrate selectivity pattern of Commamonas testosteroni strain B-356 towards dichlorobiphenyls. Int. Biodeterior. Biodegradation 39:311–316 [Google Scholar]
- 2. Barriault D, Sylvestre M. 2004. Evolution of the biphenyl dioxygenase BphA from Burkholderia xenovorans LB400 by random mutagenesis of multiple sites in region III. J. Biol. Chem. 279:47480–47488 [DOI] [PubMed] [Google Scholar]
- 3. Boyd DR, Bugg TDH. 2006. Arene cis-dihydrodiol formation: from biology to application. Org. Biomol. Chem. 4:181–192 [DOI] [PubMed] [Google Scholar]
- 4. Chun HK, et al. 2003. Biotransformation of flavone and flavanone by Streptomyces lividans cells carrying shuffled biphenyl dioxygenase genes. J. Mol. Catal. B Enzym. 21:113–121 [Google Scholar]
- 5. Denef VJ, et al. 2004. Biphenyl and benzoate metabolism in a genomic context: outlining genome-wide metabolic networks in Burkholderia xenovorans LB400. Appl. Environ. Microbiol. 70:4961–4970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Erickson BD, Mondello FJ. 1992. Nucleotide sequencing and transcriptional mapping of the genes encoding biphenyl dioxygenase, a multicomponent polychlorinated-biphenyl-degrading enzyme in Pseudomonas strain LB400. J. Bacteriol. 174:2903–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ferraro DJ, Gakhar L, Ramaswamy S. 2005. Rieske business: structure-function of Rieske non-heme oxygenases. Biochem. Biophys. Res. Commun. 338:175–190 [DOI] [PubMed] [Google Scholar]
- 8. Focht DD. 1995. Strategies for the improvement of aerobic metabolism of polychlorinated-biphenyls. Curr. Opin. Biotechnol. 6:341–346 [Google Scholar]
- 9. Gomez-Gil L, et al. 2007. Characterization of biphenyl dioxygenase of Pandoraea pnomenusa B-356 as a potent polychlorinated biphenyl-degrading enzyme. J. Bacteriol. 189:5705–5715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haddock JD, Gibson DT. 1995. Purification and characterization of the oxygenase component of biphenyl 2,3-dioxygenase from Pseudomonas sp. strain LB400. J. Bacteriol. 177:5834–5839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Han J, et al. 2005. Epoxide formation on the aromatic B ring of flavanone by biphenyl dioxygenase of Pseudomonas pseudoalcaligenes KF707. Appl. Environ. Microbiol. 71:5354–5361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hurtubise Y, Barriault D, Powlowski J, Sylvestre M. 1995. Purification and characterization of the Comamonas testosteroni B-356 biphenyl dioxygenase components. J. Bacteriol. 177:6610–6618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hurtubise Y, Barriault D, Sylvestre M. 1996. Characterization of active recombinant his-tagged oxygenase component of Comamonas testosteroni B-356 biphenyl dioxygenase. J. Biol. Chem. 271:8152–8156 [DOI] [PubMed] [Google Scholar]
- 14. Imbeault NY, Powlowski JB, Colbert CL, Bolin JT, Eltis LD. 2000. Steady-state kinetic characterization and crystallization of a polychlorinated biphenyl-transforming dioxygenase. J. Biol. Chem. 275:12430–12437 [DOI] [PubMed] [Google Scholar]
- 15. Kagami O, et al. 2008. Protein engineering on biphenyl dioxygenase for conferring activity to convert 7-hydroxyflavone and 5,7-dihydroxyflavone (chrysin). J. Biosci. Bioeng. 106:121–127 [DOI] [PubMed] [Google Scholar]
- 16. Kim SY, et al. 2003. cis-2′,3′-Dihydrodiol production on flavone B-ring by biphenyl dioxygenase from Pseudomonas pseudoalcaligenes KF707 expressed in Escherichia coli. Antonie Van Leeuwenhoek 84:261–268 [DOI] [PubMed] [Google Scholar]
- 17. Kumar P, et al. 2011. Anaerobic crystallization and initial X-ray diffraction data of biphenyl 2,3-dioxygenase from Burkholderia xenovorans LB400: addition of agarose improved the quality of the crystals. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67:59–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kumar P, et al. 2011. Structural insight into the expanded PCB-degrading abilities of a biphenyl dioxygenase obtained by directed evolution. J. Mol. Biol. 405:531–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Labbe D, Garnon J, Lau PCK. 1997. Characterization of the genes encoding a receptor-like histidine kinase and a cognate response regulator from a biphenyl/polychlorobiphenyl-degrading bacterium, Rhodococcus sp. strain M5. J. Bacteriol. 179:2772–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. L'Abbée JB, Tu YB, Barriault D, Sylvestre M. 2011. Insight into the metabolism of 1,1,1-trichloro-2,2-bis(4-chlorophenyl)ethane (DDT) by biphenyl dioxygenases. Arch. Biochem. Biophys. 516:35–44 [DOI] [PubMed] [Google Scholar]
- 21. Masai E, et al. 1995. Characterization of biphenyl catabolic genes of gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:2079–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Misawa N, et al. 2005. Synthesis of vicinal diols from various arenes with a heterocyclic, amino or carboxyl group by using recombinant Escherichia coli cells expressing evolved biphenyl dioxygenase and dihydrodiol dehydrogenase genes. Tetrahedron 61:195–204 [Google Scholar]
- 23. Mohammadi M, et al. 2011. Retuning Rieske-type oxygenases to expand substrate range. J. Biol. Chem. 286:27612–27621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morris GM, et al. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30:2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mouz S, Merlin C, Springael D, Toussaint A. 1999. A GntR-like negative regulator of the biphenyl degradation genes of the transposon Tn4371. Mol. Gen. Genet. 262:790–797 [DOI] [PubMed] [Google Scholar]
- 26. Nichenametla SN, Taruscio TG, Barney DL, Exon JH. 2006. A review of the effects and mechanisms of polyphenolics in cancer. Crit. Rev. Food Sci. Nutr. 46:161–183 [DOI] [PubMed] [Google Scholar]
- 27. Ohtsubo Y, et al. 2001. BphS, a key transcriptional regulator of bph genes involved in polychlorinated biphenyl/biphenyl degradation in Pseudomonas sp. KKS102. J. Biol. Chem. 276:36146–36154 [DOI] [PubMed] [Google Scholar]
- 28. Parnell JJ, Denef VJ, Park J, Tsoi T, Tiedje JM. 2010. Environmentally relevant parameters affecting PCB degradation: carbon source- and growth phase-mitigated effects of the expression of the biphenyl pathway and associated genes in Burkholderia xenovorans LB400. Biodegradation 21:147–156 [DOI] [PubMed] [Google Scholar]
- 29. Seeger M, et al. 2003. Biotransformation of natural and synthetic isoflavonoids by two recombinant microbial enzymes. Appl. Environ. Microbiol. 69:5045–5050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seo J, et al. 2010. Location of flavone B-ring controls regioselectivity and stereoselectivity of naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4. Appl. Microbiol. Biotechnol. 86:1451–1462 [DOI] [PubMed] [Google Scholar]
- 31. Shaw LJ, Morris P, Hooker JE. 2006. Perception and modification of plant flavonoid signals by rhizosphere microorganisms. Environ. Microbiol. 8:1867–1880 [DOI] [PubMed] [Google Scholar]
- 32. Shindo K, et al. 2003. Enzymatic synthesis of novel antioxidant flavonoids by Escherichia coli cells expressing modified metabolic genes involved in biphenyl catabolism. J. Mol. Catal. B Enzym. 23:9–16 [Google Scholar]
- 33. Shindo K, et al. 2005. Biocatalytic synthesis of monocyclic arene-dihydrodiols and -diols by Escherichia coli cells expressing hybrid toluene/biphenyl dioxygenase and dihydrodiol dehydrogenase genes. J. Mol. Catal. B Enzym. 35:134–141 [Google Scholar]
- 34. Singer A. 2006. The chemical ecology of pollutant biodegradation. Bioremediation and phytoremediation from mechanistic and ecological perspectives, p 5–19 In Mackova M, Dowling DN, Macek T. (ed), Phytoremediation and rhizoremediation. Theoretical background. Springer, Dordrecht, Netherlands [Google Scholar]
- 35. Stangl V, Lorenz M, Stangl K. 2006. The role of tea and tea flavonoids in cardiovascular health. Mol. Nutr. Food Res. 50:218–228 [DOI] [PubMed] [Google Scholar]
- 36. Sylvestre M, et al. 1996. Sequencing of Comamonas testosteroni strain B-356-biphenyl/chlorobiphenyl dioxygenase genes: evolutionary relationships among Gram-negative bacterial biphenyl dioxygenases. Gene 174:195–202 [DOI] [PubMed] [Google Scholar]
- 37. Toussaint JP, Pham TTM, Barriault D, Sylvestre M. 28 December 2011. Plant exudates promote PCB degradation by a rhodococcal rhizobacteria. Appl. Microbiol. Biotechnol. [DOI] [PubMed] [Google Scholar]
- 38. Vézina J, Barriault D, Sylvestre M. 2008. Diversity of the C-terminal portion of the biphenyl dioxygenase large subunit. J. Mol. Microbiol. Biotechnol. 15:139–151 [DOI] [PubMed] [Google Scholar]